Abstract
Processing of the GagPol polyprotein precursor of human immunodeficiency virus type 1 (HIV-1) is a critical step in viral assembly and replication. The HIV-1 protease (PR) is translated as part of GagPol and is both necessary and sufficient for precursor processing. The PR is active only as a dimer; enzyme activation is initiated when the PR domains in two GagPol precursors dimerize. The precise mechanism by which the PR becomes activated and the subsequent initial steps in precursor processing are not well understood. However, it is clear that processing is initiated by the PR domain that is embedded within the precursor itself. We have examined the earliest events in precursor processing using an in vitro assay in which full-length GagPol is cleaved by its embedded PR. We demonstrate that the embedded, immature PR is as much as 10,000-fold less sensitive to inhibition by an active-site PR inhibitor than is the mature, free enzyme. Further, we find that different concentrations of the active-site inhibitor are required to inhibit the processing of different cleavage sites within GagPol. Finally, our results indicate that the first cleavages carried out by the activated PR within GagPol are intramolecular. Overall, our data support a model of virus assembly in which the first cleavages occur in GagPol upstream of the PR. These intramolecular cleavages produce an extended form of PR that completes the final processing steps accompanying the final stages of particle assembly by an intermolecular mechanism.
The human immunodeficiency virus type 1 (HIV-1) Gag precursor contains the structural proteins of the viral core: matrix (MA), capsid (CA), and nucleocapsid (NC), as well as the p6 protein (12, 32). The coding domains of the viral protease (PR), reverse transcriptase (RT), and integrase (IN) reside within the Pol domain of GagPol (32). GagPol itself is a fusion between the Gag and Pol polyproteins and is translated as the result of a −1 translational frameshift (14, 41). Processing of these precursors is accomplished by the viral protease encoded within GagPol without assistance from a cellular protease. Both the order and kinetics of cleavage as well as the extent of precursor processing appear to be critical steps in the generation of fully infectious, appropriately assembled viral particles (21, 28, 36, 54).
The protease of HIV-1 is an aspartic protease and is functional only as a dimer; dimerization results in the formation of a binding cleft in which each of the two catalytic aspartic acids is contributed by one of the monomers. Because the protease is active only as a dimer, two of the GagPol precursors must themselves dimerize during virus assembly so that their protease domains can dimerize, become active, and process the precursors. The functional consequence of this arrangement is that precursor processing is initiated by precursor dimerization and subsequent activation of the protease embedded within the precursors (31).
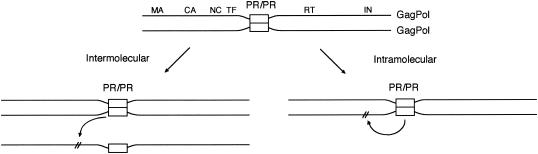
Despite the critical role that the initiation of precursor processing plays in viral replication, the events surrounding these first cleavages by the precursor-embedded, immature viral PR are poorly understood. For example, it is uncertain whether the initial cleavages mediated by the PR are inter- or intramolecular (Fig. 1). To characterize the initiation of precursor processing and protease activation, we examined the in vitro processing of a full-length GagPol precursor that is cleaved by its endogenous protease. Our studies indicate that the initial cleavages are intramolecular. These findings are consistent with previously published biochemical and in vitro processing data and suggest a mechanism for maintaining the order of precursor processing during virus assembly.
FIG. 1.
Schematic models of intermolecular (cis) versus intramolecular (trans) processing for the initial processing of the GagPol precursor. Previous results indicated that initial cleavage of the GagPol precursor by the activated GagPol PR occurs at two sites: p2/NC site (M377/M378) and TF F440/L441 (34). Cleavage of these GagPol sites could conceivably occur by either an intermolecular mechanism (left) or an intramolecular mechanism (right). Note that processing of the Gag precursor can occur only by an intermolecular mechanism since the Gag precursor lacks an embedded protease.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
The construction of pGPfs and pGPfs-PR was previously described (34). HIV-1 sequences were derived from an HXB isolate of HIV-1 (accession no. NC 001802) (40). Briefly, pGPfs contains a single GagPol open reading frame downstream of the bacteriophage T7 promoter in vector pIBI20 (International Biotechnologies). pGPfs-PR contains an additional catalytic mutation (D25A) of the protease domain that renders PR inactive. In both plasmids, a continuous GagPol open reading frame was created by site-directed mutagenesis to reproduce exactly in amino acid sequence the major GagPol product found in virions (10, 14). pGag1 contains the Gag and Pol open reading frames and produces full-length pr55 Gag and pr160 GagPol in an approximately 20:1 ratio by translational frameshift during translation in vitro (34). pGagS was previously described and produces only the pr55 Gag product upon translation in vitro (35, 36). Site-directed mutagenesis was performed as described previously (1, 22). All mutations were confirmed by direct sequencing by the dideoxy termination method prior to use.
In vitro assays for the proteolytic processing of Gag.
Transcription and translation of pGPfs, pGPfs-PR, pGag1, or pGag were performed in rabbit reticulocyte lysate (RRL) with the TNT system (Promega) in 50-μl reaction mixtures with 20 μCi of [35S]cysteine (>1,000ci/mM; Amersham Pharmacia Biotech). For coexpression of Gag-GagPol or GagPol-GagPol containing various mutations, plasmids were premixed prior to transcription-translation to express the respective protein products in ratios according to the molecular mass of the products. The concentration of all products expressed in RRL is estimated at approximately 1 nM (data not shown).
For cis-protease processing reactions, transcription and translation of pGPfs-PR-based constructs proceeded for 2 h at 30°C. Two-microliter aliquots were removed at various times, and the reaction was stopped by the addition of an equal volume of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (Invitrogen). Products of the processing reaction were heated to 70°C prior to separation on NuPage 4 to 12% Bis-Tris gradient gels as recommended by the manufacturer (Invitrogen). Gels were fixed in 10% acetic acid, dried before autoradiography, and captured for densitometric analysis on a Molecular Dynamics Storm model 800 phosphoimager.
Competitive inhibition of cis- and trans-protease processing reactions by ritonavir.
Ritonavir (ABT-538; Abbott) was serially diluted in dimethyl sulfoxide to provide a 100× stock of inhibitor (1% final concentration of dimethyl sulfoxide). For cis-protease processing reactions, transcription and translation were performed as described above in a 50-μl total volume with 0.5 μl of ritonavir stock added prior to transcription and translation. The trans-protease processing reactions were performed in 25 mM Na2HPO4-25 mM NaCl-1 mM dithiothreitol (pH 7.0) buffer. Reaction mixtures in a 50-μl volume contained 0.5 μl of ritonavir stock and 8 μl of RRL containing full-length PR(−) D25A GagPol (GPfs-PR) were preincubated for 10 min on ice prior to the addition of purified PR (325 nM final concentration) to initiate the reaction. Reaction mixtures were incubated for 10 min at 30°C. Fifty percent inhibitory concentrations (IC50s) for the inhibition of the p2/NC (M377/M378) and TF F440/L441 sites were calculated as follows: first, densitometric analysis was performed on the respective gels, and relative protein ratios for precursors and products were calculated based on their known compositions and the number of labeled cysteine residues of each. An additional normalization was used to set the percentage of uncleaved substrate to 0% in the control reaction mixture with no added inhibitor. Inhibition curves were fitted with GraphPad Prism by the use of triplicate independent measurements and fitted to sigmoidal curves with a variable Hill slope. Error was calculated as standard error of the mean. The concentration of inhibitor necessary for 50% inhibition of each site (IC50) was extrapolated from plots of the percent uncleaved substrate versus inhibitor concentration. This procedure for the estimation of IC50s is a modification of an earlier procedure previously used to estimate the differences in the rates of cleavage of HIV-1 Gag processing sites (35, 36).
Expression and purification of HIV PR.
Recombinant wild-type HIV PR was expressed in Escherichia coli, purified from inclusion bodies, and refolded as described previously (11, 34). Total yield of purified protease was 5 to 10 mg/liter of E. coli culture in a buffer consisting of 25 mM sodium phosphate (pH 7.0), 25 mM NaCl, 0.2% β-mercaptoethanol, and 10% glycerol. The final concentration of PR was 0.1 mg/ml before storage at −70°C. The percentage of active sites in the prepared protease was determined by active-site inhibition (50) with the tight-binding inhibitor ABT-538 [11; J. Levin, 1996, Ritonavir (Norvir). Proceedings to the Community Symposium on Protease Inhibitors: Current and Future Use, http://www.hivpositive.com/f-Treatment/5-Treatments/f-protease/f-JL-ProteaseReport/c-1D.html] as described previously (34) Briefly, ABT-538 inhibited cleavage of PR-negative GagPol by 400 nM trans-protease (by monomer mass) at an IC50 of <100 nM, indicating that >50% of the protease was present in the active form.
RESULTS
Expression of the full-length GagPol precursor results in processing by the endogenous PR.
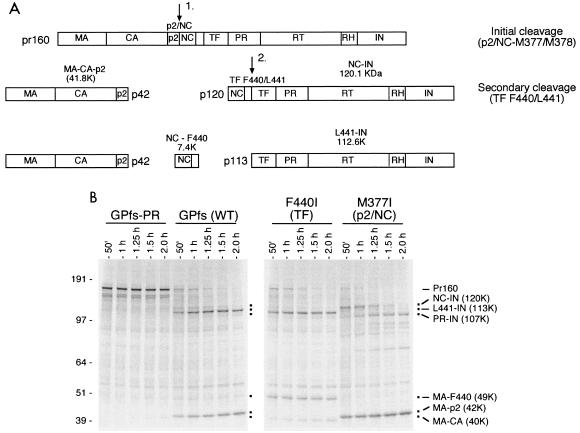
We examined the processing of a full-length GagPol precursor by its endogenous PR in an RRL system in order to evaluate the earliest steps in PR activation and GagPol cleavage. Expression of the full-length GagPol precursor in an RRL system resulted in ordered processing of the first two processing sites (p2/NC M377/378 and transframe F440/L441) by the embedded viral PR (34). Three fragments were produced: a 42-kDa fragment containing the MA, CA, and p2 proteins; a small (7.4-kDa) protein consisting of the viral NC with an 8-amino-acid C-terminal extension; and a 113-kDa protein containing the transframe region, the PR itself, the RT, and IN (schematic in Fig. 2A). To identify the cleavage sites, we took advantage of the observation that the substitution of a β-branched amino acid at the P1 position of a PR cleavage site blocks cleavage at that site (35-37, 42, 52) by HIV-1 protease (the P1 position is the first residue upstream of the scissile bond) (45). GagPol precursors containing these substitutions produce a readily distinguishable cleavage pattern (34), indicating that the substituted site was blocked for cleavage. We also noted alternative site selection when the preferred site was blocked. The alternative site chosen was close to the preferred site and was a site not typically cleaved by activated GagPol protease in vitro. For example, a Phe-to-Ile alteration at position 440 blocked cleavage at F440/L441 within the transframe region. As expected, the 113-kDa cleavage intermediate comprised of (from the amino terminus) TF L441-PR-RT-RH-IN coding domains was not observed. In its place was a 107-kDa species which extended from the N terminus of PR through the end of IN (Fig. 2B, F440I). Similarly, the introduction of a Met-to-Ile alteration at amino acid 377 of the p2/NC (M377/M388) cleavage site resulted in the disappearance of the expected band at 42 kDa (MA-CA-p2) (Fig. 2B, M377I). In its place, a smaller, 40-kDa band representing enhanced cleavage at an alternate site upstream (CA/p2 L363/A364) was seen (Fig. 2B). These results are consistent with cleavage patterns that we have identified previously (34).
FIG. 2.
Initial cleavages of the GagPol precursor in vitro after activation of the protease. Full-length GagPol was expressed in vitro as described in Materials and Methods. (A) Schematic for the ordered processing of the GagPol precursor after protease activation. Initial cleavage occurred at the p2/NC site (M377/M378) followed by rapid cleavage of the TF F440/L441 site. Significant cleavage of the other GagPol sites was not observed with wild-type precursor in vitro. The observed protein products with their calculated molecular masses based on sequence are shown. (B) Identification of the location of the initial cleavages and the effect of site-specific blocking mutation on ordered processing of GagPol by the activated protease. The full-length GagPol precursor was generated in vitro. Aliquots were removed at the indicated time and separated by SDS-PAGE. Processing of the wild-type precursor (GPfs) by the embedded protease is shown. Inactivation of the protease in GagPol with D25A (GPfs-PR) prevents processing of the Gag precursor. The effect of inhibiting cleavage at the p2/NC or the TF F440/L441 site with site-specific P1 Ile substitutions is shown. Major products of the GagPol precursor are denoted by square dots. The composition and calculated molecular masses (in kilodaltons) of the products based on published sequences are on the right. Products are presented in abbreviated form by their N- and C-terminal domains only according to accepted nomenclature (24). Numbers at left are molecular masses in kilodaltons. WT, wild type.
The PR embedded within GagPol is relatively insensitive to inhibition by an active-site inhibitor.
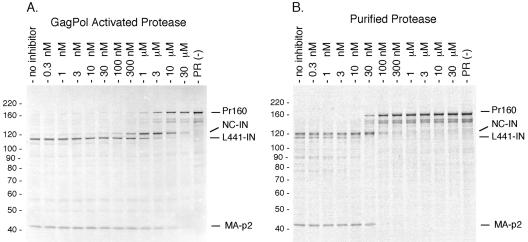
We used this full-length GagPol processing system to characterize the sensitivity of the GagPol PR to a competitive active-site inhibitor (ritonavir). Specifically, we examined processing of GagPol by the embedded PR in the presence of increasing concentrations of ritonavir. We found that each of the two processing sites was inhibited to different degrees by ritonavir (Fig. 3A). We observed a 50% inhibition of processing at the slower TF F440/L441 site at approximately 189 nM ritonavir; in contrast, a higher concentration of ritonavir (5.17 μM) was required to achieve the same level of inhibition of processing for the faster p2/NC site (Fig. 3A). These values are significantly higher than the subpicomolar inhibitor constant (Ki) values for ritonavir derived from purified, mature protease (20, 29).
FIG. 3.
Comparison of the effects of the competitive inhibitor ritonavir on activated GagPol protease and trans protease. (A) Wild-type GagPol was translated in vitro for 2 h in duplicate reactions containing increasing concentrations of ritonavir (ABT-538) to monitor the effects of the drug on activity of GagPol protease. The protease within GagPol activates and cleaves the precursor at the primary p2/NC and secondary TF F440/L441 sites. The concentration of GagPol in the reaction mixture is approximately 1 nM. The concentration of ritonavir is given above the lanes. (B) Effect of ritonavir on trans-cleavage of the GagPol precursor in vitro. A 325 nM concentration of mature recombinant protease monomers was added in trans to PR D25A mutated GagPol (160 pM) with various concentrations of ritonavir (above). Reactions shown were stopped at 10 min of incubation. The gels shown are representative of triplicate experiments. Products are presented in abbreviated form by their N- and C-terminal domains only. Numbers at left of each panel are molecular masses in kilodaltons.
We sought to compare the sensitivity of the mature PR to inhibition by an active-site inhibitor with the results obtained with the embedded PR utilizing GagPol as a substrate. This trans-processing reaction was accomplished by adding purified PR in trans to a GagPol construct encoding a PR inactivated by an Asp-to-Ala alteration at the enzyme active site (GagPol PR−). In contrast to the results obtained above with an active embedded PR (Fig. 3A), exogenously added PR was much more sensitive to inhibition by ritonavir (Fig. 3B). For the purified PR processing the full-length GagPol PR− in trans, cleavage at the slower TF F440/L441 site was 50% inhibited at a ritonavir concentration of 18 nM and that at the faster p2/NC site was inhibited at 64 nM (Fig. 3B). Of note, for these studies, although the concentration of wild-type GagPol (and, therefore, the endogenous PR) was approximately 1 nM, 325 nM dimeric PR was added to the GagPol PR− construct in order to visualize processing. Therefore, processing of the precursor by the embedded protease was as much as 10,000-fold less sensitive to inhibition by this active-site inhibitor than was processing of the precursor in trans by purified, mature protease.
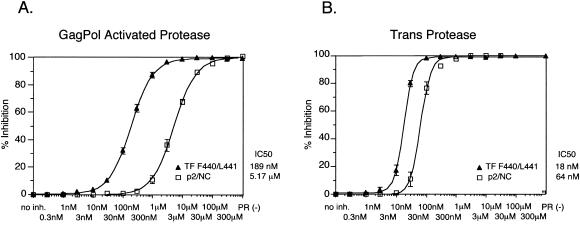
We also compared the concentrations of ritonavir required to inhibit cleavage at two different sites in GagPol (M377/M378 and F440/L441). A graphic representation of the data from Fig. 3 demonstrates that the concentration of ritonavir required to inhibit cleavage by 50% differed by more than 27-fold for the two sites (5.17 μM versus 189 nM) for the endogenous protease (Fig. 4A). In contrast to the results obtained with the endogenous PR, ritonavir inhibition of purified PR added in trans differed by only 3.5-fold across the two cleavage sites (64 versus 18 nM; Fig. 4B).
FIG. 4.
Comparison of the susceptibilities of cleavage at the p2/NC and TF sites to inhibition for cis versus trans cleavage. Plots show the percent inhibition for the individual p2/NC and TF F440/L441 sites for GagPol protease (A) or trans-protease (B) with increasing concentrations of ritonavir. Plots were derived from densitometric analysis of SDS-polyacrylamide gels as described in Materials and Methods. The estimated IC50s for the inhibition of the individual sites for the GagPol protease are 189 nM for TF F440/L441 and 5.17 μM for p2/NC. The estimated IC50s for the trans protease are 18 nM for TF F440/L441 and 64 nM for p2/NC. Curves were determined as explained in Materials and Methods with a minimum of three independent replicates. Error bars indicate standard errors of the means.
The initial cleavages within the GagPol precursor are intramolecular.
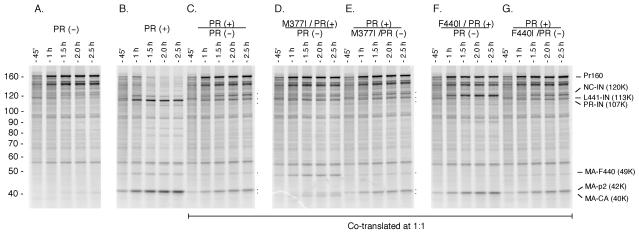
We characterized the extent of intra- versus intermolecular processing of the full-length GagPol further through mixing experiments in which equivalent amounts of substituted GagPol constructs were coexpressed. In these experiments, we cotranslated GagPol constructs encoding various combinations of PR active-site substitutions (GagPol PR−) and/or processing-site mutations (M377I and F440I) (Fig. 5). For these experiments, we took advantage of our observation that the introduction of these processing-site substitutions produces distinct cleavage patterns.
FIG. 5.
Cleavage of the p2/NC and TF F440/L441 sites by the activated GagPol protease occurs by an intramolecular mechanism rather than an intermolecular mechanism. GagPol or equal amounts of two GagPol species were expressed in vitro, and aliquots were taken at the indicated time prior to SDS-PAGE. (A) Wild-type GagPol-containing protease inactivated at the catalytic aspartate (D25A). (B) Wild-type GagPol. (C to G) trans-complementation test in which equal amounts of two GagPol species were cotranslated as shown above the panel. (C) Expression of wild-type and PR D25A protease. Efficient trans-complementation would be expected to result in 75% inhibition of GagPol precursor processing as shown by the persistence of the full-length precursor (75%) and reduced amounts of the 42-kDa MA-p2 product (25%). (D and E) A mutation that blocks cleavage of the p2/NC site (M377I) was placed on either the GagPol with an active PR monomer (D) or the GagPol with an inactive PR monomer (E). Processing of the p2/NC site, as shown by the 42-kDa product, is observed only when the unblocked site is located on the same precursor as the active protease, indicating an intramolecular cleavage mechanism (E). (F and G) A mutation that blocks cleavage of the TF F440/L441 site (F440I) was placed on either the GagPol with an active PR monomer (F) or the GagPol with an inactive PR monomer (G). Generation of the 113-kDa L441-IN product is seen only when the unblocked site is located on the same precursor as the activated protease, indicating that cleavage occurs by an intramolecular mechanism. Numbers at left are molecular masses in kilodaltons.
In summary, we observed that the pattern of cleavage was dictated by the construct containing the active PR domain (Fig. 5). Overall, we found that coexpression of GagPol constructs that contained an active PR and wild-type processing sites with a construct that contained an inactivated PR and a blocked processing site showed a wild-type pattern of cleavage. Alternatively, if the active PR domain was present on the construct with a mutated cleavage site, a cleavage pattern characteristic of the mutated cleavage site was observed (Fig. 5).
For example, we expressed a GagPol construct containing an active PR and a Met-to-Ile blocking alteration at position 377 (M377I/PR+) together with a GagPol construct containing an inactive PR and the wild-type methionine at position 377 (PR−) (Fig. 5D). The pattern that we observed was similar to the pattern seen when the GagPol with the M337I substitution was expressed alone (Fig. 2B). Consistent with intramolecular cleavage of the precursor containing both the active PR and the M377I substitution, there was no evidence that the p2/NC site was cleaved. Rather, the 42-kDa band (MA-CA-p2) was not seen and was replaced by the 40-kDa band representing the MA-CA product (Fig. 2B, M377I).
In contrast, coexpression of the wild-type GagPol construct (PR+) with a construct containing the M377I substitution and the D25A PR substitution (M377I/PR−) resulted in a wild-type pattern of cleavage (Fig. 5E). Similar results were obtained when constructs containing combinations of the Phe-to-Ile alteration at position 440 with either active or inactive PR were coexpressed (Fig. 5F and G). In all cases, a wild-type cleavage pattern was observed for the constructs with wild-type cleavage sites in which the PR was intact and an altered pattern of processing was observed when the intact PR active site was paired with a mutated cleavage site on the same construct (Fig. 5).
Of note, overall processing was significantly diminished by the coexpression of a construct which contained a wild-type PR with a construct containing an active-site-substituted PR (Fig. 5C, PR+/PR−). If the mutated protease was exerting a true trans-dominant inhibitory effect, absence of protease activity would be expected in 75% of the GagPol expressed due to the formation of PR−/PR− and PR−/PR dimers. Thus, the observed reduction in PR activity was similar to what would be expected if true transdominant complementation were achieved during coexpression.
Coexpressed Gag precursor is not processed by the GagPol PR in trans.
During viral assembly, two precursors are produced, Gag and GagPol. Gag is translated at a level approximately 20-fold greater than that of GagPol and encodes the structural proteins of the viral core. Because the Gag precursor terminates before the PR coding domain, during virus assembly, the Gag precursor must be cleaved by the PR in trans, either by the PR dimer embedded within the GagPol precursor or by the mature, fully processed PR dimer. To characterize the activity of the embedded PR further, we extended our observations to the processing of the Gag precursor (Fig. 6).
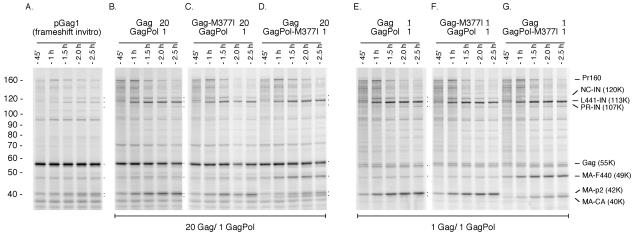
FIG. 6.
Cotranslation of the Gag and GagPol precursors. pGag1 (A) or Gag and GagPol (B to G) were cotranslated, and aliquots were taken at the indicated times prior to SDS-PAGE. (A) Translation of the Gag and Pol open reading frames (pGag1) in RRL resulted in the expression of pr55 Gag and pr160 GagPol by a translational frameshift mechanism (14). The Gag/GagPol ratio is approximately 20:1. Cleavage of the p2/NC site is evident by the generation of the 42-kDa MA-CA-p2 product. (B) Cotranslation of Gag and GagPol in a 20:1 ratio via separate plasmids. (C and D) A mutation that blocks cleavage of the p2/NC site (M377I) was placed on either Gag (C) or GagPol (D) and expressed at a 20:1 ratio. Processing of the p2/NC site, as shown by the 42-kDa product, is observed only when the unblocked site is located on the GagPol precursor (E). (F and G) Gag and GagPol were expressed in a 1:1 ratio in vitro. A mutation that blocks cleavage of the p2/NC site (M377I) was placed on either Gag (F) or GagPol (G). Cleavage of the p2/NC site, as shown by the 42-kDa MA-CA-p2 product, was observed only when the unblocked site was located on the GagPol precursor (G). Numbers at left are molecular masses in kilodaltons.
The two precursors were coexpressed at a 20:1 Gag-to-GagPol ratio in the RRL. Overall, the 55-kDa Gag precursor did not appear to undergo processing to any significant extent in these experiments. Coexpression of the wild-type Gag construct together with GagPol did not influence GagPol processing (Fig. 6B). Similarly, there was no evidence that a Gag precursor containing an M377I substitution was processed in trans (Fig. 6C). Further, processing of a GagPol precursor containing an M377I alteration was unaffected by the presence of Gag (Fig. 6D). Therefore, despite the 20-fold excess of Gag precursor, no intermolecular cleavage was apparent in this system. As expected, similar results were obtained when the Gag and GagPol precursors were expressed at a ratio of 1:1 (Fig. 6E to G). We have previously shown that the Gag precursor is efficiently cleaved when PR is added in trans (35, 36, 47).
DISCUSSION
As is the case for all retroviruses, each of the cleavages within the HIV-1 GagPol precursor is accomplished by the viral PR itself, without assistance from a cellular protease. Therefore, at least for the initial cleavages, the PR embedded within GagPol must dimerize and become active as part of the precursor. Because activation of the viral protease and initiation of precursor processing are absolute requirements for the production of infectious particles, we sought to determine whether these critical initial cleavages are inter- or intramolecular.
Our studies using the spontaneous in vitro processing of full-length GagPol yielded several new insights into the initiation of precursor processing. First, at least for the initial cleavages, the PR embedded within GagPol is considerably less sensitive to the effects of an active-site PR inhibitor than is the purified enzyme added exogenously. Specifically, we examined processing of GagPol by the embedded PR (wild-type GagPol) in the presence of increasing concentrations of ritonavir. We compared these results with the effects of ritonavir on a PR− GagPol construct processed in trans by purified wild-type PR added exogenously. In these experiments, the exogenously added PR which processes the precursor in trans was approximately 10,000-fold more sensitive to inhibition than the immature PR was. This is about the difference one would expect between an intermolecular (exogenous PR cleaving a GagPol substrate) and an intramolecular (PR cleaving sites within its own GagPol precursor) cleavage (2, 15, 16).
Second, we determined the amount of ritonavir necessary to inhibit cleavage at two sites within GagPol (M377/M378 and F440/L441) by 50%. These IC50 calculations demonstrate that, for the wild-type GagPol precursor, sensitivity to ritonavir differs by 27.3-fold between the two sites. This is much greater than differences reported for the in vitro cleavage by purified PR of peptides representing a range of native PR cleavage sites (51). Of note, we found a much smaller difference (3.6-fold) in the sensitivity of those same sites to inhibition when cleavage was accomplished by purified PR added exogenously. The extent to which cleavage at these sites exhibited a differential sensitivity to inhibition indicates that the two sites are not equally accessible to the GagPol-embedded PR or reflects a difference in cleavage-site affinity. These differences in accessibility appear to be much less prominent for trans processing by the exogenously added PR since the sensitivity of inhibition was more similar for the two cleavage sites. These results indicate that the context of the cleavage site in GagPol plays an important role in determining the efficiency of cleavage and further support the idea that the initial cleavages by the PR within the precursor are intramolecular.
Finally, coexpression experiments were used to characterize the initial steps in precursor processing further. For these studies, we combined full-length GagPol with Gag constructs containing mutations that either inactivated the PR with an active-site substitution or blocked one of the cleavage sites. Because two active-site aspartic acids are required to produce an active enzyme, GagPol dimers composed of at least one active-site mutant-containing construct are inactive. This design enabled us to determine whether the early GagPol cleavages are inter- or intramolecular. Under all conditions examined, cleavage at each of the two sites required that an active PR domain be present on the same GagPol molecule as the cleavage site. Of note, the GagPol-associated PR was unable to cleave a coexpressed Gag precursor. Therefore, for these initial processing events, we found no evidence for intermolecular cleavage of either precursor.
The validity of these studies is limited by the extent to which this in vitro processing system mimics the precursor cleavage that accompanies virus assembly in vivo. There are several lines of evidence that suggest that the spontaneous processing of in vitro-translated precursors is an appropriate model for processing in vivo. First, expression of GagPol in RRL avoids possible complications of a truncated product, nonphysiological conditions, or overexpression of products. Several groups (23, 30, 48) have observed that Gag or Pol precursors produced in RRL multimerize into higher-order components similar to those observed in virions. These studies indicate that the concentration of GagPol precursors in this system is high enough to support multimerization. Other lines of evidence suggest that the spontaneous processing of in vitro-translated precursors is an appropriate model for processing in vivo. Carter and coworkers (6, 33, 57) also observed spontaneous activation and cleavage of a Gag-PR construct. Others (13, 38) have noted efficient membrane association of translated Gag in RRL. Campbell et al. (4, 5) found that components within the reticulocyte lysate are required for proper multimerization of purified HIV Gag into 110-nm particles in vitro and later linked that requirement to specific phospholipids. Finally, we report here that all the expressed GagPol produced in RRL is eventually cleaved at the two processing sites, suggesting efficient dimerization and multimerization leading to activation of the viral PR.
The data described here extend studies reported by several other groups. In an extensive analysis of the activation of the PR from another retrovirus, Rous sarcoma virus, Vogt and coworkers have demonstrated that the N-terminal amino acids of the PR are critical for enzyme activation (43, 44, 46). Additional studies conducted in the Rous sarcoma virus system by Leis and coworkers also suggest that the cleavage specificity of the embedded PR differs from that seen with the mature viral enzyme (56). Biochemical studies by Louis et al. and others examined processing at the N terminus of a PR-containing construct with short stretches of native sequence at its N and/or C termini (7, 26, 27, 55). Their data suggest that this cleavage site is processed through an intramolecular interaction. We have expanded on these earlier studies by examining processing in the context of the full-length Gag and GagPol precursors. The use of the full-length precursors, rather than truncated constructs, for the study of PR activation is particularly appropriate in light of data suggesting that sequences within GagPol outside of the PR play a role in supporting enzyme activation (8, 34). We also amplify these observations by comparing GagPol processing by the endogenous PR with that by the exogenous PR and by evaluating processing at more than one cleavage site. This experimental design allows us to identify differences in the efficiency with which different sites are processed by the endogenous PR and reveals that the context of the cleavage site sequence, or perhaps the PR domain, appears to be important for cleavage by the endogenous but not the exogenously added PR.
Our conclusion that the initial precursor cleavages are intramolecular is also consistent with an evaluation of Gag and GagPol processing reported by Lever and coworkers. In these studies, Kaye and Lever found that expression of a Gag construct that was extended to include the PR produced virus-like particles comprised of appropriately processed viral proteins (19). In contrast, full-length, frameshifted GagPol with an inactive PR (HVPGPpr−) expressed alone in COS-1 cells was neither exported into the supernatant nor processed. They further found that coexpression of the PR-containing Gag construct together with HVPGPpr− rescued release of the full-length GagPol. Of particular relevance for the studies presented here, the HVPGPpr− rescued by the Gag construct remained unprocessed. In these virus-like particles, only the PR-containing Gag precursor was processed; there was no evidence of intermolecular processing of the GagPol precursor by the PR fused to the Gag precursor.
The reason that processing fails to proceed beyond the initial GagPol cleavages in our RRL system is unclear. This may reflect merely a concentration of precursors that is too low to support precursor dimerization. This seems unlikely, since the concentration of GagPol is adequate for the dimerization of the PR domains on the two precursors. Alternatively, it is possible that intermolecular cleavages can be accomplished only by a GagPol processing intermediate in which the PR is more active. For example, using a construct in which the PR was extended at the N and C termini, Wondrak and Louis (55) demonstrated that N-terminal cleavage of the protease produces an enzyme whose activity is similar to that of the mature enzyme. Such a more active intermediate may be required for intermolecular processing and may appear only during the structural rearrangements that accompany the late stages of virus assembly. Similarly, the absence of Gag processing may reflect either a concentration of PR and Gag substrate that is too low to support processing or an inadequately processed PR-containing GagPol precursor.
Taken together with studies of PR activation and virus assembly (3, 9, 17, 18, 25, 39, 44, 46, 49, 53, 57), our findings suggest a model of PR initiation whereby the initial cleavages performed by the immature PR dimer are intramolecular and occur at the membrane of the infected cell at the time of virus budding. These intramolecular cleavages are coordinated with budding of the particle from the surface of the cell and release a more active GagPol processing intermediate that contains an active PR. Once the particle buds from the cell, this extended protease intermediate is released so that Gag and GagPol cleavages may occur in trans. Studies are currently under way to examine the significance of these findings for virus replication and assembly.
Acknowledgments
We gratefully acknowledge helpful discussions with G. Schatz, V. M. Vogt, and J. Clemente. Ritonavir was obtained from the NIH AIDS Research and Reference Reagent Program. Purified HIV-1 protease was obtained from S. Gulnik.
This work was supported by NIH grant RO1 GM GM66681-01 and the UNC Center for AIDS Research.
REFERENCES
- 1.Bebenek, K., and T. A. Kunkel. 1989. The use of native T7 DNA polymerase for site-directed mutagenesis. Nucleic Acids Res. 17:5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruice, T. C., and S. Benkovic. 1966. Bioorganic mechanisms, p. 119-125. W. A. Benjamin, New York, N.Y.
- 3.Burstein, H., D. Bizub, M. Kotler, G. Schatz, V. M. Vogt, and A. M. Skalka. 1992. Processing of avian retroviral gag polyprotein precursors is blocked by a mutation at the NC-PR cleavage site. J. Virol. 66:1781-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, S., R. J. Fisher, E. M. Towler, S. Fox, H. J. Issaq, T. Wolfe, L. R. Phillips, and A. Rein. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. USA 98:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, C., and G. Zybarth. 1994. Processing of retroviral Gag polyproteins: an in vitro approach. Methods Enzymol. 241:227-253. [DOI] [PubMed] [Google Scholar]
- 7.Co, E., G. Koelsch, Y. Lin, E. Ido, J. A. Hartsuck, and J. Tang. 1994. Proteolytic processing mechanisms of a miniprecursor of the aspartic protease of human immunodeficiency virus type 1. Biochemistry 33:1248-1254. [DOI] [PubMed] [Google Scholar]
- 8.Dautin, N., G. Karimova, and D. Ladant. 2003. Human immunodeficiency virus (HIV) type 1 transframe protein can restore activity to a dimerization-deficient HIV protease variant. J. Virol. 77:8216-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatlin, J., S. J. Arrigo, and M. G. Schmidt. 1998. HIV-1 protease regulation: the role of the major homology region and adjacent C-terminal capsid sequences. J. Biomed. Sci. 5:305-308. [DOI] [PubMed] [Google Scholar]
- 10.Gorelick, R. J., and L. E. Henderson. 1994. Part III: analyses, p. 2-5. In G. Myers, B. Korber, S. Wain-Hobson, K. T. Jeang, L. Henderson, and G. Pavlakis (ed.), Human retroviruses and AIDS. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 11.Gulnik, S. V., L. I. Suvorov, B. Liu, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 34:9282-9287. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, L. E., R. C. Sowder, T. D. Copeland, S. Oroszlan, and R. E. Benveniste. 1990. Gag precursors of HIV and SIV are cleaved into six proteins found in the mature virions. J. Med. Primatol. 19:411-419. [PubMed] [Google Scholar]
- 13.Hermida-Matsumoto, L., and M. D. Resh. 1999. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55gag and p17MA. J. Virol. 73:1902-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacks, T., M. D. Power, F. R. Maslarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frame-shifting in HIV-1 gag/pol expression. Nature (London) 331:280-283. [DOI] [PubMed] [Google Scholar]
- 15.Jencks, W. P. 1975. Binding energy, specificity, and enzymic catalysis: the circe effect. Adv. Enzymol. Relat. Areas Mol. Biol. 43:219-410. [DOI] [PubMed] [Google Scholar]
- 16.Jencks, W. P. 1969. Catalysis in chemistry and enzymology, p. 7-41. McGraw-Hill, New York, N.Y.
- 17.Kaplan, A. H., J. A. Zack, M. Knigge, D. A. Paul, D. J. Kempf, D. W. Norbeck, and R. Swanstrom. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J. Virol. 67:4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193:661-671. [DOI] [PubMed] [Google Scholar]
- 19.Kaye, J., and A. Lever. 1996. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J. Virol. 70:880-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klabe, R. M., L. T. Bacheler, P. J. Ala, S. Erickson-Viitanen, and J. L. Meek. 1998. Resistance to HIV protease inhibitors: a comparison of enzyme inhibition and antiviral potency. Biochemistry 37:8735-8742. [DOI] [PubMed] [Google Scholar]
- 21.Krausslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125-139. [DOI] [PubMed] [Google Scholar]
- 23.Lee, Y. M., B. Liu, and X. F. Yu. 1999. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J. Virol. 73:5654-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leis, J., D. Baltimore, J. M. Bishop, J. Coffin, E. Fleissner, S. P. Goff, S. Oroszlan, H. Robinson, A. M. Skalka, H. M. Temin, and V. Vogt. 1988. Standardized and simplified nomenclature for proteins common to all retroviruses. J. Virol. 62:1808-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindhofer, H., K. von der Helm, and H. Nitschko. 1995. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology 214:624-627. [DOI] [PubMed] [Google Scholar]
- 26.Louis, J. M., G. M. Clore, and A. M. Gronenborn. 1999. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 6:868-875. [DOI] [PubMed] [Google Scholar]
- 27.Louis, J. M., E. M. Wondrak, A. R. Kimmel, P. T. Wingfield, and N. T. Nashed. 1999. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 274:23437-23442. [DOI] [PubMed] [Google Scholar]
- 28.Mervis, R. J., N. Ahmad, E. P. Lillehoj, M. G. Raum, F. H. Salazar, H. W. Chan, and S. Venkatesan. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J. Virol. 62:3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 30.Moody, M. D., S. C. Pettit, W. Shao, L. Everitt, D. D. Loeb, C. A. Hutchison III, and R. Swanstrom. 1995. A side chain at position 48 of the human immunodeficiency virus type-1 protease flap provides an additional specificity determinant. Virology 207:475-485. [DOI] [PubMed] [Google Scholar]
- 31.Navia, M. A., and B. M. McKeever. 1990. A role for the aspartyl protease from the human immunodeficiency virus type 1 (HIV-1) in the orchestration of virus assembly. Ann. N. Y. Acad. Sci. 616:73-85. [DOI] [PubMed] [Google Scholar]
- 32.Oroszlan, S., and T. B. Luftig. 1990. Retroviral proteinases. Curr. Top. Microbiol. Immunol. 157:153-185. [DOI] [PubMed] [Google Scholar]
- 33.Partin, K., G. Zybarth, L. Ehrlich, M. DeCrombrugghe, E. Wimmer, and C. Carter. 1991. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 88:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettit, S. C., S. Gulnik, L. Everitt, and A. H. Kaplan. 2003. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J. Virol. 77:366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettit, S. C., G. J. Henderson, C. A. Schiffer, and R. Swanstrom. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J. Virol. 76:10226-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettit, S. C., J. Simsic, D. D. Loeb, L. Everitt, C. A. Hutchison III, and R. Swanstrom. 1991. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 266:14539-14547. [PubMed] [Google Scholar]
- 38.Platt, E. J., and O. K. Haffar. 1994. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc. Natl. Acad. Sci. USA 91:4594-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quillent, C., A. M. Borman, S. Paulous, C. Dauguet, and F. Clavel. 1996. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology 219:29-36. [DOI] [PubMed] [Google Scholar]
- 40.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R.-S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 3:57-69. [DOI] [PubMed] [Google Scholar]
- 41.Reil, H., H. Kollmus, U. H. Weidle, and H. Hauser. 1993. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J. Virol. 67:5579-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richards, A. D., L. H. Phylip, W. G. Farmerie, P. E. Scarborough, A. Alvarez, B. M. Dunn, P. H. Hirel, J. Konvalinka, P. Strop, L. Pavlickova, V. Kostka, and J. Kay. 1990. Sensitive, soluble chromogenic substrates for HIV-1 proteinase. J. Biol. Chem. 265:7733-7736. [PubMed] [Google Scholar]
- 43.Schatz, G., I. Pichova, and V. M. Vogt. 1997. Analysis of cleavage site mutations between the NC and PR Gag domains of Rous sarcoma virus. J. Virol. 71:444-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schatz, G. W., J. Reinking, J. Zippin, L. K. Nicholson, and V. M. Vogt. 2001. Importance of the N terminus of Rous sarcoma virus protease for structure and enzymatic function. J. Virol. 75:4761-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schechter, I., and A. Berger. 1967. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27:157-162. [DOI] [PubMed] [Google Scholar]
- 46.Sellos-Moura, M., and V. M. Vogt. 1996. Proteolytic activity of purified avian sarcoma and leukemia virus NC-PR protein expressed in Escherichia coli. Virology 221:335-345. [DOI] [PubMed] [Google Scholar]
- 47.Sheng, N., S. C. Pettit, R. J. Tritch, D. H. Ozturk, M. M. Rayner, R. Swanstrom, and S. Erickson-Viitanen. 1997. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 71:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spearman, P., and L. Ratner. 1996. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J. Virol. 70:8187-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart, L., and V. M. Vogt. 1994. Proteolytic cleavage at the Gag-Pol junction in avian leukosis virus: differences in vitro and in vivo. Virology 204:45-59. [DOI] [PubMed] [Google Scholar]
- 50.Tomasselli, A. G., M. K. Olsen, J. O. Hui, D. J. Staples, T. K. Sawyer, R. L. Heinrikson, and C. C. Tomich. 1990. Substrate analogue inhibition and active site titration of purified recombinant HIV-1 protease. Biochemistry 29:264-269. [DOI] [PubMed] [Google Scholar]
- 51.Tozser, J., I. Blaha, T. D. Copeland, E. M. Wondrak, and S. Oroszlan. 1991. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 281:77-80. [DOI] [PubMed] [Google Scholar]
- 52.Tozser, J., I. T. Weber, A. Gustchina, I. Blaha, T. D. Copeland, J. M. Louis, and S. Oroszlan. 1992. Kinetic and modeling studies of S3-S3′ subsites of HIV proteinases. Biochemistry 31:4793-4800. [DOI] [PubMed] [Google Scholar]
- 53.Wan, M., M. Takagi, B. N. Loh, X. Z. Xu, and T. Imanaka. 1996. Autoprocessing: an essential step for the activation of HIV-1 protease. Biochem. J. 316:569-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wondrak, E. M., and J. M. Louis. 1996. Influence of flanking sequences on the dimer stability of human immunodeficiency virus type 1 protease. Biochemistry 35:12957-12962. [DOI] [PubMed] [Google Scholar]
- 56.Xiang, Y., T. W. Ridky, N. K. Krishna, and J. Leis. 1997. Altered Rous sarcoma virus Gag polyprotein processing and its effects on particle formation. J. Virol. 71:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zybarth, G., and C. Carter. 1995. Domains upstream of the protease (PR) in human immunodeficiency virus type 1 Gag-Pol influence PR autoprocessing. J. Virol. 69:3878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]