Abstract
In winegrapes (Vitis spp.), fruit quality traits such as berry color, total soluble solids content (SS), malic acid content (MA), and yeast assimilable nitrogen (YAN) affect fermentation or wine quality, and are important traits in selecting new hybrid winegrape cultivars. Given the high genetic diversity and heterozygosity of Vitis species and their tendency to exhibit inbreeding depression, linkage map construction and quantitative trait locus (QTL) mapping has relied on F1 families with the use of simple sequence repeat (SSR) and other markers. This study presents the construction of a genetic map by single nucleotide polymorphisms identified through genotyping-by-sequencing (GBS) technology in an F2 mapping family of 424 progeny derived from a cross between the wild species V. riparia Michx. and the interspecific hybrid winegrape cultivar, ‘Seyval’. The resulting map has 1449 markers spanning 2424 cM in genetic length across 19 linkage groups, covering 95% of the genome with an average distance between markers of 1.67 cM. Compared to an SSR map previously developed for this F2 family, these results represent an improved map covering a greater portion of the genome with higher marker density. The accuracy of the map was validated using the well-studied trait berry color. QTL affecting YAN, MA and SS related traits were detected. A joint MA and SS QTL spans a region with candidate genes involved in the malate metabolism pathway. We present an analytical pipeline for calling intercross GBS markers and a high-density linkage map for a large F2 family of the highly heterozygous Vitis genus. This study serves as a model for further genetic investigations of the molecular basis of additional unique characters of North American hybrid wine cultivars and to enhance the breeding process by marker-assisted selection. The GBS protocols for identifying intercross markers developed in this study can be adapted for other heterozygous species.
Introduction
The inherent role of the enological properties of a winegrape cultivar is undeniable in the final taste of wine. While both growing conditions and choice of vinification procedures influence eventual wine properties [1–4], the grapevine cultivar has arguably the largest effect on wine quality [5, 6]. Understanding the genetic controls of enologically important traits will assist breeders, viticulturists, and enologists to delineate strategies for improving wine quality.
Grapevine is a cross pollinating, highly heterozygous crop that exhibits inbreeding depression in the form of diminished seed viability and reduced vine vigor. Because of this, grapevine breeders typically employ breeding schemes based on background selection for V. vinifera fruit quality and foreground selection for introgressed traits [7, 8], which are applied under a framework of modified-backcrossing [2, 8].
Winegrape breeding began in earnest in the 19th C., as breeders attempted to develop grapevines with improved pest and disease resistance by cross-hybridizing popular V. vinifera winegrape cultivars with resistant North American wild species such as V. aestivalis Michx, V. cinerea (Engelm. ex A. Gray) Engelm. ex Millard, V. labrusca L., V. riparia Michx and V. rupestris Scheele [9].While these introgressions have positive adaptive traits including disease resistance, pest resistance, and cold hardiness [5, 10, 11], they often have inferior fruit composition for winemaking [9, 12].
Common quality parameters for wine grapes include titratable acidity (TA), soluble solids (SS) content as a proxy for sugar concentration, and yeast assimilable nitrogen (YAN) [13]. Sugars increase during berry ripening, and an SS of 20–24% w/w (Brix) is typically targeted for production of a dry table wine with 11–14% v/v alcohol. Conversely, malic acid (MA)–one of the two major grape organic acids that contribute to TA–is respired during berry ripening [14]. While SS contents of wild Vitis and V. vinifera are comparable, MA concentrations in wild Vitis are generally much higher–often in excess of 10 g/L at harvest [15]–and wines produced from Vitis interspecific hybrids are frequently reported to have excessive acidity and sourness [5] if not ameliorated before bottling. This situation is exacerbated in cooler climates where hybrid grapes are more widely used, such as the northern United States, because of slower MA respiration [16]. Thus, a key goal following trait introgression is combining lower MA content with appropriate sugar content. YAN is also widely measured in winegrapes prior to fermentation, since insufficient concentrations (< 150 mg/L) can lead to stuck fermentations and off-odor formation [17], while excessive concentrations can lead to formation of ethyl carbamate, a known carcinogen [1]. YAN concentrations at both extremes have been reported in interspecific hybrids [18], and breeding cultivars with appropriate YAN content is desirable.
Genetic controls of these key compositional factors have only recently been investigated in grapevine [19]. The formation of MA in the grape berry involves the glycolysis and citric acid (TCA) cycle and is regulated by the malate metabolism pathway. The diverse pathways in metabolic processes and environmental factors, such as temperature during ripening, all contribute to the complexity for dissecting the molecular basis of MA content. Two recent studies reported several QTLs controlling total SS and acid content [20, 21]. However, the identified QTLs could only explain a small percentage of the total phenotypic variance (< 17%), and candidate genes could not be identified. QTL analysis of YAN in grape berries has not been attempted, even though YAN is known to vary considerably among both V. vinifera [22] and Vitis spp. hybrids [18].
Given the heterozygosity and inbreeding depression in Vitis, QTL mapping studies generally use F1 full-sib progenies and the pseudo-testcross method [23], with 70 to 300 progeny and from 100 up to 1,826 molecular markers [20, 24–27]. Previously, a family of 119 F2 progeny was developed from selfing an F1 individual derived from the cross V. riparia × Vitis hybrid ‘Seyval’ [28], and a linkage map based on 120 simple sequence repeats (SSRs) was developed [29]. ‘Seyval’ is a complex interspecific hybrid of V. vinifera (55% by pedigree) with wild species native to the south-central United States and is a relatively cold hardy white wine cultivar, resistant to disease and the phylloxera aphid Daktulosphaira vitifoliae (Fitch 1855). V. riparia has been used to breed for early acclimation and freezing tolerance in hybrid winegrapes [28]. The acclimation and dormancy characteristics of V. riparia and ‘Seyval’ have been studied at the physiological, transcriptomic and metabolomic levels providing additional gene level characterization [28–32].
Genotyping by sequencing (GBS) is a marker platform that uses restriction enzymes to reduce genome complexity prior to next generation sequencing. GBS has become widely used due to several benefits: simultaneous marker discovery and genotyping, high sample throughput and scalability, and high resolution at a low per-marker cost [33]. However, due to arbitrary sampling of sites and the high level of multiplexing typical in GBS, sequencing depth is usually reduced, leading to missing data and heterozygote undercalling, in which heterozygous sites are undersampled and wrongly interpreted as homozygous [34]. While heterozygote undercalling has been irrelevant to GBS applications in inbred species, it is a critical problem in grapevine and other highly heterozygous and diverse species, requiring the development of new methodologies.
The present study demonstrates the use of novel GBS analysis methods to develop an improved linkage map for the F2 mapping family derived from V. riparia × ‘Seyval’ [35]. This GBS map was validated by analysis of the berry skin color locus, previously characterized [36–38]. Our results indicate this is an effective approach for mapping QTL and identifying candidate genes in Vitis, including preliminary results for SS, MA and YAN.
Materials and Methods
Plant material
The F2 mapping family was generated as described in Fennell et al. [28]. The F2 progeny were derived from self-pollination of a single hermaphrodite F1 individual (16-9-2) generated from a cross of V. riparia (USDA PI 588259) × ‘Seyval’ (Seyve-Villard 5–276). One hundred nineteen F2 seedlings were germinated from seed, grown in the greenhouse, cycled into dormancy and cuttings taken for vegetative propagation. After chilling fulfillment, cuttings were rooted and vines grown for field establishment as well as maintenance in a controlled environment. Ecodormant one-year-old vines were planted in the field in spring 2005, trained and maintained for fruiting. A first genetic map for this family was published in 2009 using SSR markers [29]. Fruit from the F1 parent, the V. riparia grandparent, and 65 fruiting progeny with greater than 20 clusters were harvested 30 days post veraison, and 150 g of randomly collected berries were flash frozen and sent to Cornell University for berry quality phenotyping in 2013. Fruit ripening parameters particularly SS and acid content are impacted by changes in temperature. This population had ripening times spanning early to late season; therefore, harvesting at 30 days post veraison avoided low night temperature exposure that would have impacted the late season genotypes. Additional F2 seeds were generated in 2011 and 2012. Three hundred fourteen additional seedlings were germinated in 2013 and genotyped for increased map resolution.
Phenotyping
Frozen berry samples (150 g) from each individual were destemmed, allowed to partially thaw, and homogenized in a 250-mL stainless steel Waring blender (Waring laboratory science, Stamford, CT, USA) at medium speed for 30 s followed by high speed for 30 s. A 10 g portion of berry slurry was transferred to a 15-mL plastic centrifuge tube and then frozen at -20°C. Prior to analysis, samples were thawed. Juice soluble solids (Brix) were analyzed by refractometer (Misco Model #PA203X; Misco, Cleveland, OH USA). The MA was measured on an Agilent 1260 infinity series HPLC using a previously reported method [39]. The YAN was measured as the sum of ammonium and primary amino nitrogen using commercial enzymatic assays (Unitech Scientific, LLC., Hawaiian Gardens, CA, USA) on a ChemWell model 2900 analyzer (Awareness Technologies, Palm City, FL, USA). A subset of MA analyses (one out of every ten individuals) were subjected to either injection replication or extraction replication. The relative standard deviation (%RSD) for MA analyses was 0.4% and 0.7% for injection alone or extraction/injection, respectively. A set of standards and blanks was run every ten samples to verify the retention time and response factor of analytes being measured. An analogous approach was used for replication of YAN analyses. Technical replicates of SS measurements were not conducted due to the high reproducibility of the method.
Genotyping
For each vine, a single small leaf (less than 1 cm diameter) was placed in a tube of a Costar 96-well cluster tube collection plate (Corning Life Sciences, Tewksbury, MA, USA). Each 96-well plate consisted of 91 genotype-specific samples, two sets of duplicates and one blank well. The location of the blank well in each plate was unique as a plate identity control. Leaf tissues were maintained at 4°C during and after harvest. In the laboratory, two stainless steel genogrinder beads were placed in each tube and plates were frozen at -80°C. Tissue grinding took place in a Geno/Grinder 2000 (OPS Diagnostics LLC, Lebanon NJ, USA) with 96-well plates agitated in pairs at 400X speed for 1 minute. Plates were then stored at -80°C until processing with DNeasy 96-well DNA extraction kits (Qiagen, Valencia CA, USA). Modifications were made to the manufacturer’s protocol to improve DNA quality and quantity as follows: 1) PVP-40 (2% w/v) was added to the AP1 lysis buffer prior to heating; and 2) visual inspection for complete re-suspension of the sample pellet of each 8-tube strip was added to the agitation step following AP1 addition.
Genotyping-by-sequencing (GBS) was performed as described by Elshire et al. (2011) [35], integrating four 96-well plates across 384 barcodes for library preparation and sequencing. For SNP calling, the raw sequence data for the 424 F2 progeny plus the F1 progenitor (16-9-2) was processed through the TASSEL 3.0 GBS pipeline [33] using the 12X.v2 V. vinifera ‘PN40024’ reference genome [40] from The French-Italian Public Consortium (https://urgi.versailles.inra.fr/Species/Vitis/Data-Sequences/Genome-sequences) for alignment and the Burrows–Wheeler Aligner (BWA) mem [41] with default parameters. The output consisted of variant call format (VCF) file version 4.1 [42] including SNPs present in at least 40% of the progeny and with a minor allele frequency (MAF) ≥ 0.1. Subsequently, the VCF was filtered using vcftools ver. 1.12a [42] and TASSEL [43] versions 3.0 and 4.0. A total of 291,453 SNPs were identified in 424 F2 progeny by TASSEL 3.0, then a custom filtering process was applied for alignment. The filtering was based on keeping sites with a minimum read depth of 6 and 75% completeness by site across progeny and by progeny across sites. Results were output as a TASSEL hapmap file. Finally, using a custom perl script (S1 File) markers heterozygous in the F1 progenitor and with a co-dominant 1:2:1 segregation among the F2 progeny were identified by a chi-squared (χ2) goodness-of-fit test at α≤0.01. These were reformatted to be imported in JoinMap® 4.1.
Map construction
For map construction, the intercross markers described above were coded as hk×hk (assumption of no dominance because the gametic phases were unknown) to be imported in JoinMap4® 4.1 Build: 31jul13.4feb11. Cross-pollination (CP) cross type was used to estimate linkages [44]. This type of marker segregation is particularly useful when performing linkage mapping in outbreeding species because the markers are symmetrical with respect to parental segregation and because they allow construction of integrated linkage maps even when the identification of tri or tetra-allelic markers is insufficient [45]. For the present study, these markers, in conjunction with the multipoint maximum likelihood algorithm implemented in JoinMap® [46], allowed linkage calculation and phasing determination, which enabled us to recode marker genotypes based on predicted phase, and then perform QTL analyses as in an F2 experimental cross.
Prior to grouping and ordering, markers with highly significant (p≤0.00001) deviation from Mendelian expectations and loci duplicating genetic information were filtered in JoinMap®. For grouping, a linkage-independence LOD ≥10 was used. For ordering and genetic location determination, the Maximum Likelihood mapping algorithm and Kosambi mapping function were applied, in addition to three rounds of map optimization. Given the amount of missing data allowed, genetic distance inflation occurred among markers. Thus, to keep the most reliable markers and their locations while limiting distance inflation, a threshold of 2.5 cM for the nearest neighbor stress (N.N. stress) was considered. This is an empirical threshold determined through the linkage mapping of 16 full-sib families analyzed in the USDA-NIFA Specialty Crops Research Initiative VitisGen project (www.vitisgen.org).
Sixty-five F2 progeny were included for evaluation of berry skin color, for which black (pigmented) and white (nonpigmented) were recoded as 1 and 0, respectively. QTL analysis was performed using the standard interval mapping procedure (EM algorithm) with binary model in the package R/qtl [47] version 1.36.5 for R ver. 3.1.2 [48] and using F2 as the cross type. One thousand permutations at α = 0.05 were executed to calculate the LOD threshold. The additive and dominance QTL effects were estimated taking missing genotype information into account. Missing genotypes were simulated given observed marker data by the hidden Markov model (sim.geno function in R/qtl). Reports were generated for maximum LOD score, 1.5-LOD support interval in cM, and the physical location in the reference genome in Mbp, as well as the percentage of variation explained (R2).
The QTL mapping of the quantitative traits SS, MA and YAN was performed in two stages: 1) for each trait independently using composite interval mapping through the package R/qtl [47] version 1.36.5 for R ver. 3.1.2 [48]; and 2) joint QTL mapping for multi-trait trials in a single environment, using the routines QMTQTLSCAN, QMTESTIMATE and QMTBACKSELECT [49] integrated in the statistical package GenStat for Windows 17th [50].
For the three traits, phenotypic information was used from fruit of 63 progeny harvested in 2013. The normality of the distributions was tested by the Shapiro-Wilks tests. Logarithm (Log) transformation was applied for results validation. The QTL detected were similar using either transformed or original data. The markers were coded according to the recommendations of the software used (A, B and H for R/qtl, and as 1|1, 2|2 and 1|2 in GenStat). QTL mapping was executed as described for the berry skin color in R/qtl. The 1.8-LOD support intervals were reported as suggested by “A Guide to QTL Mapping” [51] for mapping traits for an intercross family.
In the case of the joint QTL mapping in GenStat, the joint QTL mapping was performed using the phenotypic data for each combination of two traits (MA-SS, MA-YAN and SS-YAN). Cofactors were placed every 1.51 cM along the genome, resulting in placement of 861 cofactors along the genetic map. The model used for the first QTL scan was , where yij is the value of trait j for genotype i, Tj is the trait main effect, F is the set of cofactors, xifadd and xiadd are the additive genetic predictors of genotype i at the cofactor positions and at the tested position, respectively. The associated effects are denoted by cjfadd and αjadd for cofactors and tested positions, respectively. xifdom and xidom are dominance genetic predictors of genotype i at the cofactor positions and at the tested position, respectively, with associated effects cjfdom, and αjdom. TEij refers to the residual error of the trait j for genotype i. The fit of the unstructured model was iteratively performed through Restricted Maximum Likelihood (REML), using 1000 iterations. The QTL threshold was set at a significance level of 0.05 using the Li and Ji method [52] with minimum cofactor proximity of 50 cM and minimum separation of selected QTLs of 30 cM.
Subsequently for the interval mapping and estimation of QTL effects, the model was simplified to: , where yij is the value of trait j for genotype i, Tj is the trait main effect, xiladd are the additive genetic predictors of genotype i for locus l, and αjladd are the associated effects. xildom are the dominance genetic predictors, and αjldom are the associated effects. The method to fit these models follow the ideas of Malosetti et al. [53] and Boer et al. [54]. Both mixed models consider the genotypes as random terms, while the genetic predictors as fixed effects. The genetic predictors can be considered as genotypic covariables informing the genotypic composition of a genotype at a specific chromosome locus [55]. GTij is assumed to follow a multi-Normal distribution with mean vector 0, and a variance covariance matrix Σ, which was modeled during the QTL mapping procedure.
Finally, to approach the causality between traits MA and SS (causal, reactive, independent, full), a causal model selection test [56] was implemented using the function cmst in the R package qtlhot version 0.9.0 [57].
Results
Marker development
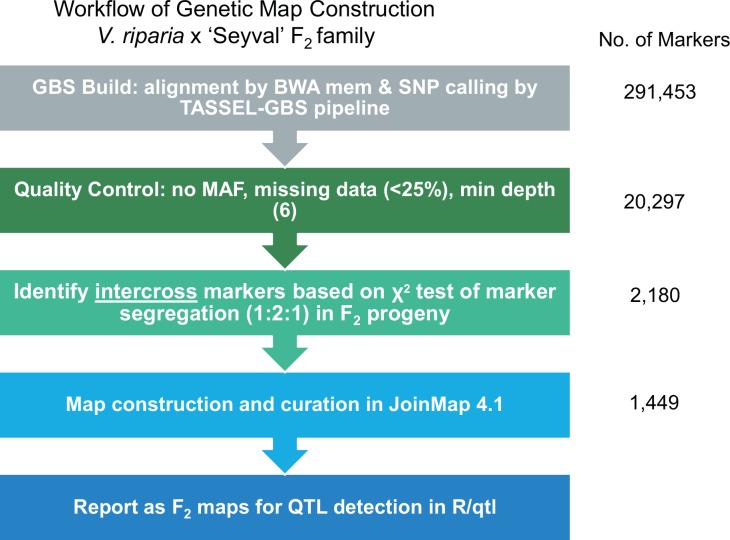
A total of 291,453 quality single nucleotide polymorphisms (SNPs) were identified in 424 F2 progeny relative to the V. vinifera ‘PN40024’ reference genome, version 12X.v2. Subsequently SNP filtering by minimum read depth and missing data rate yielded 2180 intercross markers with 1:2:1 segregation in the progeny, as determined by a χ2 test of marker segregation. The markers were discovered and coded as hk×hk using a custom perl script (S1 File). Further marker curation and genetic map construction were performed in JoinMap® 4.1, resulting in a final set of 1,449 markers. The workflow for marker development and linkage map construction in the F2 mapping family are presented in Fig 1.
Fig 1. Workflow of marker development and genetic map construction in the V. riparia × ‘Seyval’ F2 family.
The left panel shows the five main steps with filtering parameters, and the right panel shows the numbers of markers resulting from each step.
Linkage map construction
Linkage analysis using LOD 10.0 grouped the 1449 markers into 19 linkage groups (LG) with a total map size of 2423.9 cM and a map density (average distance between markers) of 1.67 cM (Fig 2). Given the grapevine genome size of about 458.8 Mb [40], the current genetic map is about 0.189 Mb/cM. Based on a comparison of the genetic map to the reference genome (physical map), all linkage groups (except Chr15) had an estimated genome coverage greater than 90%, and the average genome coverage was 95.1% (S2 File). To check marker order, the genetic positions of markers within each LG were compared with physical coordinates on the reference genome (S3 File). Marker order was conserved between the genetic and physical map.
Fig 2. Linkage map of F2 mapping family derived from V. riparia × ‘Seyval’.
The linkage map consists of 1,449 markers with a total map size of 2423.9 cM. The 19 chromosomes are labeled at the top of each linkage group. Genetic positions (cM) and physical locations (‘PN40024’ reference version 12X.v2) are listed to the left and right of each chromosome, respectively.
Compared to an SSR map previously developed for the same F2 family [29], this GBS map represented a nine-fold improvement in resolution from 14.9 cM to 1.67 cM between markers and an 11% increase in the average genome coverage, from 84% to 95% (Table 1). In addition, there were six gaps greater than 20 cM in the previous SSR map, while the largest gap in the GBS map was 13.5 cM.
Table 1. Comparison between the previously published SSR-based map and the GBS-based map.
| SSR map | GBS map | |
|---|---|---|
| No. of progeny | 119 | 424 |
| No. of markers | 120 | 1449 |
| No. of linkage groups | 21 | 19 |
| Map size (cM) | 1784 | 2424 |
| Average distance between markers (cM) | 14.87 | 1.67 |
| Average genome size per cM (Mb/cM) | 0.257 | 0.189 |
| Genome coverage (%) | 83.7 | 95.1 |
Berry color mapping for map validation
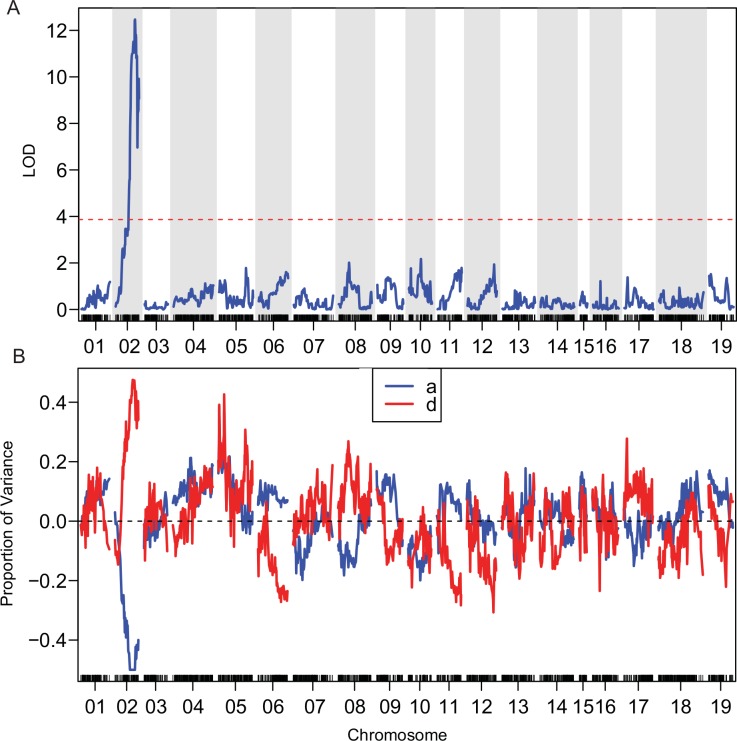
Berry skin color is controlled by the VvmybA1 gene (Chr02: 14,179,266–14,180,746) [58]. This well-studied trait was used to validate the soundness of the F2 map construction strategy. Berry skin color was recorded for 65 F2 progeny and showed a 51:14 ratio of black:white (pigmented:nonpigmented), which was not significantly different from 3:1 (Chi-squared test, p = 0.80, S4 File), confirming the trait was controlled by one completely-dominant gene. Only one significant QTL, with a peak LOD score of 12.47 (threshold = 3.9), was identified on LG02, and explained up to 90% of the phenotypic variance for berry skin color (Fig 3A). The 1.5-LOD support interval spanned the genetic map at 71.06–90.85cM, corresponding to Chr02 6.3–14.3Mbp and encompassed the VvmybA1 locus (Table 2). The estimated additive effect was 0.5 (a = 0.50), equal to the estimated dominant deviation (d = 0.46), which perfectly matched the one-locus, complete-dominance model and thereby validated the F2 map construction strategy (Fig 3B).
Fig 3. Berry color segregation used for genetic map validation.
(A) QTL for berry skin color is confirmed on Chr02 of the genetic map. Permutation tests were carried out to identify the 95% confidence threshold, and the significance threshold for the LOD score is presented as a horizontal red dashed line. (B) The additive effect (a, blue line) and dominant deviation (d, red line) are estimated across the whole genome.
Table 2. Summary of QTL analyses in V. riparia × ‘Seyval’ F2 family.
| Trait | No. of individuals phenotyped | Linkage Group (LG) | Peak position (cM) a | Physical position of the nearest marker | LOD score | LOD interval (cM) b | LOD interval (physical location) | Additive effect c | Dominant effect | R2 d |
|---|---|---|---|---|---|---|---|---|---|---|
| Berry skin color | 65 | 2 | 85.40 | 9,126,568 | 12.47 | 71.06–90.85 | 6,360,259–14,300,736 | -0.50 | 0.47 | 89.92 |
| Yeast assimilable nitrogen (YAN, mg/L) | 63 | 7 | 100.44 | 18,819,915 | 6.48 | 94.87–106.85 | 16,987,038–20,565,550 | -52.55 | -38.91 | 22.81 |
| Malic acid concentration (MA, g/L) | 63 | 6 | 70.24 | 7,985,435 | 6.24 | 62.95–75.23 | 7,343,698–8,358,059 | 2.50 | 1.26 | 26.19 |
| Total soluble solids content (SS, %w/w) | 63 | 6 | 46.18 | 5,498,696 | 4.76 | 35.85–54.71 | 4,258,443–6,497,323 | -2.16 | 0.0811 | 19.14 |
| MA/SS ratio | 63 | 6 | 32.10 | 3,857,140 | 5.93 | 27.07–35.85 | 3,524,817–4,258,443 | 0.17 | 0.00262 | 26.04 |
| Joint analysis of MA and SS | 63 | 1 | 49.94 | 5,061,702 | 6.55 | 49.10–51.12 | 5,026,255–5,123,842 | MA -0.40 | 0.34 | 7.93 |
| SS -0.14 | 0.20 | 1.02 | ||||||||
| Joint analysis of MA and SS | 63 | 6 | 75.23 | 8,358,059 | 4.23 | 70.21–80.83 | 7,985,435–11,876,418 | MA 0.60 | 0.28 | 18.37 |
| SS 0.62 | -0.24 | 19.43 |
a Position of likelihood peak (highest LOD score).
b LOD interval refers to 1.8-LOD support interval, except for Berry skin color, for which a 1.5-LOD support interval was used.
c Additive effect: A positive value means the higher value of the trait due to allele from grandparent2, ‘Seyval’. A negative value means the higher value of the traits due to allele from grandparent1, V. riparia. The units depend on the traits.
d R2 (coefficient of determination): percentage of phenotypic variance explained by the QTL.
QTL mapping for enological traits
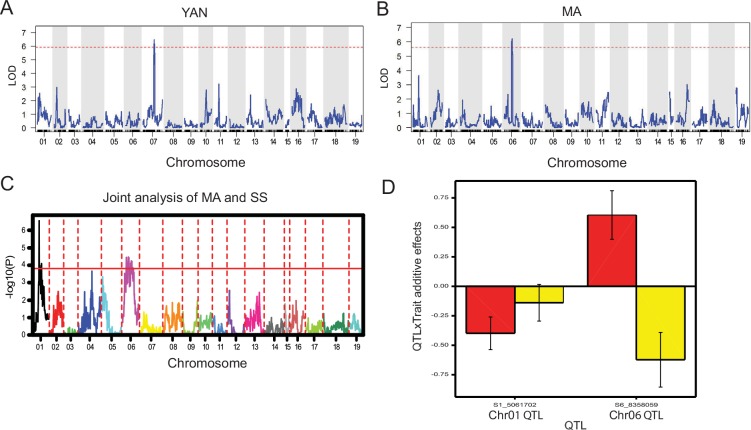
QTL mapping of the enological traits YAN, MA and SS was conducted using composite interval mapping (CIM) and the Kosambi mapping function. Each phenotype followed a normal distribution (S4 File). A QTL corresponding to YAN was identified on LG07 with a peak at 100.44 cM (LOD = 6.48; Fig 4A). This YAN QTL spanned a physical interval from 17.0–20.6 Mbp (1.8-LOD support interval) and explained 22.8% of phenotypic variance (Table 2). The additive effect of the QTL was negative, indicating that higher concentrations of YAN came from the grandparent 1, V. riparia. For MA, a single QTL was observed on LG06 with a peak at 70.24 cM (LOD = 6.24). This MA QTL contained a relatively narrow 1.8-LOD support interval from 7.3–8.4 Mbp and explained 26.2% of the phenotypic variance (Fig 4B, Table 2). In contrast to YAN, the positive additive effect of MA refers to higher concentration in the grandparent 2, ‘Seyval’. For the ratio of MA/SS, a single QTL was identified on LG06, peaking at 32.10 cM (LOD = 5.93). This MA/SS QTL spanned a relatively narrow interval between 3.5 and 4.3 Mbp and explained 26.0% of the phenotypic variance (Table 2). The additive effect was positive, indicating that higher MA/SS ratios came from ‘Seyval’, and the dominant effect was negligible.
Fig 4. QTL mapping for quantitative enological traits.
(A) QTL mapping of yeast assimilable nitrogen (YAN, mg/L). (B) QTL mapping of malic acid concentration (MA, g/L). (C) The joint QTL mapping of MA (g/L) and total soluble solids content (SS, %w/w). Permutation tests were carried out to identify the 95% confidence threshold, and the significance thresholds of LOD score is presented as a horizontal red line in A-C. (D) The interaction between MA (g/L) and SS (%w/w) QTLs and trait additive effects. Red bars represent additive effects of MA and yellow bars represent additive effects of SS. A positive value means the higher value of the trait is due to an allele from grandparent 2, ‘Seyval’. A negative value means the higher value of the trait is due to an allele from grandparent 1, V. riparia.
Since the QTL analysis was performed on data for a single year and single location, data transformation and multiple QTL detection methods and software programs were applied to confirm the results. The joint analysis of SS and MA showed evidence of two QTLs affecting both traits (Fig 4C). One QTL was detected at LG01 at the 49.94cM (5.0 Mbp), for which the additive effects indicated that the traits came from the grandparent 1, V. riparia (Table 2). This QTL explained around 7.9% and 1.0% of the phenotypic variance for MA and SS, respectively. Another QTL on LG06 at 75.23cM (8.3 Mbp) also showed high evidence for influencing both traits simultaneously; however, the additive positive effect on MA came from grandparent 2, ‘Seyval’, and the additive effect on SS came from V. riparia. This particular QTL was noteworthy since it explained around 18.3 and 19.4% of the phenotypic variance of MA and SS, respectively, and co-localized with the MA QTL identified in the CIM analysis. No significant dominance effect was observed for either trait. The interaction between QTL and trait additive effects was significant (Fig 4D). Thus for SS, both QTLs showed negative additive effects (-0.14 and -0.62, for LG01 and LG06, respectively), while for MA, the QTL on LG01 showed a negative effect and the QTL on LG06 positive (-0.40 and 0.60, respectively).
The results from the causal model selection hypothesis tests showed that the values for Bayesian Information Criterion (BIC) and Akaike Information Criterion (AIC) did not agree in their most adequate model (S5 File). The AIC value suggested that the full model with nine parameters was best, and that the traits were affected by more than genetic factors. The BIC value suggested that considering SS as phenotype 1, the best model was a causal effect of SS on MA. When MA was considered as phenotype 1, the BIC value indicated the reactive model was best, confirming that MA was reacting to a causal effect of SS.
Discussion
Over the last ten years, linkage map construction has been widely used in grapevine genetic research, with markers primarily being amplified fragment length polymorphisms (AFLPs), SSRs and SNPs [27]. Although SSRs have been widely used in grapevine linkage map construction, the method is relatively time consuming and costly, typically resulting in low marker density. For example, 119 individuals from the mapping family used here were previously genotyped using 115 SSRs [29], which was much less than that required to capture all the informative recombination. That SSR map was unable to cover the whole genome and resulted in six gaps larger than 20 cM due to uneven distribution, which would result in information loss if causal alleles were located in the missing region.
In contrast, GBS-based SNPs can provide high-density genetic maps and are conducive to high-throughput genotyping. The genetic map for this F2 population (Fig 2, Table 1) consisted of 1,449 markers 12.6-fold increase in marker density compared to the earlier SSR map. The maximum spacing between markers was reduced to 13.5 cM, and 15 out of 19 chromosomes have maximum gaps less than 10 cM, resulting in an average distance of 1.67 cM/marker. The GBS markers are almost evenly distributed across the genome, and cover 95% of the ‘PN40024’ reference genome. This high-resolution genetic map should better detect recombination events. Since linkage disequilibrium (LD) is low in Vitis due to its heterozygosity and diversity [11, 59], greater marker density increases the odds of finding markers for specific traits, or even to fine map candidate causal genes. Further, the GBS protocol is well established, publicly available, and can be scaled efficiently up to the current 384-plex barcoding system [35], at which the total genotyping cost is currently less than $15 per sample. Thus, GBS-SNP markers are a time- and cost-efficient method for high resolution genetic map construction and potentially for marker-assisted selection. However, current GBS pipelines have been designed for inbred germplasm with very low residual heterozygosity and do not provide methodology for obtaining intercross markers, for which heterozygous materials are enriched and for which the marker phase is not known.
The co-dominant intercross markers of an F2 mapping family enabled the capture of additional meiotic events relative to the F1 pseudo-testcross mapping approach. Furthermore, in an F1 family, only QTLs in heterozygous allelic states in the parents with large allele-effect differentials can be detected [60], which complicates the estimation of additive effects contributed by each parent. Thus, F2 families with at least 200 progeny genotyped with intercross markers have superior linkage map accuracy [61, 62] and allow the estimation of additive, and to some extent dominant, genetic effects contributed per parent [63]. There are two types of markers in an F2 mapping family: 1:2:1 segregation of co-dominant markers, which are more informative than 3:1 segregation of dominant markers.
Here, we addressed several specific challenges associated with GBS analysis, particularly of a heterozygous family. First, retrieving useful GBS markers for map generation poses a challenge, since there is still a gap between the standard GBS variant calling pipeline TASSEL and commercial map construction software JoinMap. Also, heterozygous genotypes may be called due to sequencing errors or genome structure variance such as paralogs or tandem duplication. When imputation is difficult to apply on highly heterozygous species, pre-filtering and marker type identification and selection need to be carefully performed. The custom perl script (S1 File) provided in this paper identified markers with a co-dominant segregation (i.e. 1:2:1) in the progeny and verified each marker against the allelic state of the progenitor. To our knowledge, this is the first publication using intercross SNP markers for map construction in an F2 progeny in grapevine. The perl script can be used for marker development in other highly heterozygous species, such as apple, poplar and willow.
To validate the marker development and map construction strategy, the trait of berry color was subjected to QTL analysis (Fig 3). Berry color was selected for map validation for two reasons: (1) it is a highly heritable binary trait that is easy to phenotype; and (2) the genetic basis is well studied. Specifically, the VvmybA1 gene (Chr02: 14,179,266–14,180,746) regulates anthocyanin pigment biosynthesis in grapes [64], and a retrotransposon-induced mutation in VvmybA1 has been identified as the molecular basis of white cultivars of V. vinifera [58]. The QTL detected at 14.2 Mbp on Chr02 aligned perfectly with VvmybA1, indicating our approach was effective, even with a small sample size of 65 phenotyped progeny. The GBS genetic map was also compared to a physical map based on the reference genome V. vinifera ‘PN40024’ (S3 File). The majority of co-linearity indicated that marker order is well conserved even across the genome segments of diverse Vitis species represented in the parents, including V. vinifera, V. rupestris, V. aestivalis var lincecumii and V. riparia. These markers can be used as anchors to improve the ‘PN40024’ reference genome assembly by filling existing gaps or incorporating random contigs. However, there are some chromosomal regions that show weaker correlation between genetic and physical maps, e.g. the upper end of Chr16. These regions might indicate species-specific genome structure variation, such as chromosome rearrangement, transposable elements and tandem duplication. V. vinifera is the only species in the Vitis complex with a whole genome sequence already published, and the de novo genome assembly for other Vitis species can provide insight into unique characteristics arising from adaption to local conditions. Thus, this interspecific genetic map can be viewed as an early step toward a grapevine pan-genome, as pursued in maize [64].
Once the genetic map was generated and validated, three berry traits related to winegrape quality were chosen for QTL analysis. YAN–or the sum of nitrogen contained in primary amino acids and ammonium–is the nitrogenous component of grapes that can be utilized by yeast during alcoholic fermentation. Proper nitrogen concentrations are of interest to winemakers since nitrogen deficiency is associated with sluggish or stuck fermentations, increased production of off-aroma compounds like H2S, and decreased formation of desirable odorants like esters [1, 65]. Conversely, excess YAN can cause several problems, most notably formation of carcinogenic ethyl carbamate. Typically, ammonium is accumulated first, and decreases during ripening with corresponding increases in primary amino nitrogen [1]. Total YAN changes relatively little during berry ripening–across a wide range of cultivars and sites, YAN concentrations at commercial harvest averaged only 25% higher and were well correlated with values measured up to 5 weeks prior [66]. However, genotype can have a profound effect on YAN–up to a three-fold variation among vinifera cultivars (75 mg/L vs. 220 mg/L) was observed in a survey of cultivars in one growing region [66], and much larger variations were observed among interspecific hybrids [18]. We observed a YAN range in our mapping family (100–600 mg/L) greater than this previous survey, further confirming the importance of genetics in YAN.
Currently there is little information on the genetics of YAN and this, to our knowledge, is the first YAN associated QTL in grapevine. The QTL results in this study revealed one locus associated with YAN on LG07 (LOD = 6.48) from 16,987,038 to 20,565,550 bp, within the 1.8-LOD support interval (Table 2). Thirteen genes relevant to nitrogen metabolism were identified within the interval and can be used to further explore the trait. Because berries appear to accumulate ammonium, which is subsequently metabolized to primary amino acids, the presence of ammonium transporter genes in this list (AMT2) is of particular interest. The altered function of these genes could limit either N uptake by the vines or YAN accumulation in the berries. The estimated additive effect of the QTL indicated the V. riparia grandparent contained a higher concentration of YAN than ‘Seyval’ and may provide a novel tool for research into the nature of nitrogen content in grape berries.
Both MA and SS are critical parameters for evaluating fruit maturity, where the primary component of SS in mature fruit is hexose sugars (fructose, glucose). The key biochemical pathways associated with MA and hexose sugar metabolism are well-established [67]. Pre-veraison, sucrose imported into grape berries via the phloem is used as the primary metabolic substrate. Concurrently, a portion of malate from the citric acid cycle is transported and stored in vacuoles, where it serves as a key contributor (along with tartaric acid) to the high TA of unripe grapes [68]. During veraison and ripening, grapes accumulate sugars, and shift to using MA as an energy source [14]. Thus, a key change associated with veraison is an increase in SS (caused primarily by inversion of imported sucrose and accumulation of resulting hexose sugars) coinciding with a reduction in TA (caused primarily because of MA respiration). Grape crosses with non-domesticated Vitis in their background, e.g. V. riparia, can have unacceptably high TA values and MA concentrations (>10 g/L) even at normal harvest SS values (>20 Brix) [5]. The family used in this study shows this problem with over 50% of individuals having MA > 10 g/L (S4 File).
Because MA and SS are co-regulated, QTL analyses of individual traits were complemented by two other approaches 1) QTL analysis of the ratio, MA/SS, and 2) joint (simultaneous multi-trait) analysis of MA and SS. Both techniques have advantages over the trait-by-trait analysis, the most relevant being the increase in power to identify and accurately locate QTLs [69–72]. However, both approaches also provide additional information that is not easily obtained when traits are analyzed separately, such as the power to identify loci involved in the regulation at specific metabolic branch-points and infer interaction among genetic components for the exhibition of the traits.
The analysis of two traits as a ratio creates some difficulties, because when two traits are identically normally distributed, their ratio follows a Cauchy distribution as opposed to the normal distribution expected by most algorithms [71]. However, the mapping of these ratio-traits is relevant because it allows researchers to test and infer the interaction between traits, exemplified here by the interaction of compounds that have relevance for harvest decision-making in the field and fermentation must manipulations in the winery. From a genetics perspective, the analysis of ratio-traits allows the potential opportunity to integrate and discover independent, closely-linked, and pleiotropic loci [73] when the separated traits exhibit a significant positive or negative correlation. For example, MA/SS ratio would be less sensitive than the individual traits to changes in berry size.
In our study, the analysis of the MA/SS ratio showed a QTL on LG06 distal from the QTLs identified for SS and MA when analyzed separately (Table 2, S6 File). The LOD-based evidence (4.77–6.24) and amount of variance explained (19–26%) was comparable for the traits analyzed individually or as a ratio. The interpretation of the additive and dominance effects suggested that the higher value for the ratio could be due to the genetic contribution from ‘Seyval’; however, since this ratio can be interpreted as a biochemical interaction, the interpretation may require additional inference to suggest what is being genetically inherited.
The joint analysis of multiple traits includes all traits of interest simultaneously in a single model. This approach can provide insights into fundamental genetic mechanisms underlying trait relationships such as pleiotropy and close linkage, genotype-by-environment (G × E) interactions, and the possible trade-off given for negatively correlated traits [70]. This joint analysis approach generated a list of 14 candidate genes within the QTL region. The joint analysis showed a QTL detected on Chr06 with 1.8-LOD support interval from 70.2–80.8 cM (Table 2). The corresponding physical interval, Chr06:7,985,435–11,876,418, covered the region of grapevine orthologs of the aluminum activated malate transporter, ALMT1. This gene corresponds to the Ma1 and Ma2 loci in apple, which are malic acid transporters controlling the acidity level in apple fruits [74]. Our observation aligns with a QTL for MA at 74.1 cM on Chr06, reported by another group using a Vitis spp. mapping family [20]. The QTL explained 17% of the variance for MA in one year of a three year study. This group did not implicate ALMT1 as a candidate gene in Chr06, although they did observe a more minor QTL on Chr18, which they suggested could be related to a different ALMT. Interestingly, significant QTL were not detected for other genes expected to control MA concentrations. For example, MA respiration rate, the rate at which grape berries metabolize MA after veraison, is dependent on NAD dependent malate enzyme [68], but no QTL was detected at its genetic location. QTL analyses of MA are complicated because high MA at harvest could arise from either high MA accumulation pre-veraison or slow MA respiration, and future studies could likely benefit from having multiple time points, i.e. pre- and post-veraison.
The analysis of multiple traits simultaneously implies advantages when causal relationships between the studied traits and genes or among traits exist; however, when the trait is affected by a gene through a transitional trait, the advantages almost disappear [72]. For such a reason the testing of the causal relationships (correlation, pleiotropy, or close linkage) between or among traits must be addressed to yield outputs that can be interpreted biologically. Also, the addition of more traits to the joint QTL analysis does not result in more detection power, but may give rise to spurious signals since additional parameters will be fitted in the model [75]. However, to break unfavorable linkages of genes involved in the exhibition of economically important traits, the separation of closely linked loci is needed to eliminate pleiotropic negative effects [75].
In this study, the test for causal relationships between SS and MA was pursued through causal model selection hypothesis tests [56]. The results from CMST showed that a causal relationship clearly exists between MA and SS with a common genetic component (QTL), as the model for independence was not significant in any of the tests. AIC values were minimized for the full model, indicating SS and MA share more than one common genetic component. While, BIC values were minimized for models showing a casual effect of a QTL controlling SS also affecting MA. The discrepancy between AIC and BIC is not a concern for the interpretation of the results in the light of the empirical experience for these traits. Therefore, the results are compatible with the empirical experience, in which the balance between MA and SS is intertwined and influenced by several other factors, such as environment (particularly temperature), field management (irrigation, fertilization), and biology (additional biochemical interactions). Hence, genetic research, breeding, and probably biochemistry approaches, should consider MA and SS simultaneously, as well as the possible interactions with non-genetic factors.
Conclusions
The results presented here confirmed the GBS mapping strategy for an expanded F2 mapping family in grapevine. The protocol presented here filters heterozygous markers and retrieves high quality intercross markers which are phased during linkage map development and allows QTL analysis using an F2 genetic design, which is not common for grapevine genetic research. In addition, the protocols for calling intercross GBS markers could be used for other heterozygous species. The linkage map provided qualitative and quantitative improvements over a previous SSR map for marker density and genome coverage while maintaining co-linearity with the reference genome. The availability of the linkage map facilitated the genetic study of winegrape quality traits, including a well-characterized binary trait (berry color), more complicated traits (malic acid, soluble solids) and a neglected trait (yeast assimilable nitrogen). Enhanced marker resolution provided the opportunity to conduct joint analysis of malic acid and soluble solids and results indicated these compounds are intertwined and should be considered jointly in future genetic studies. The resources generated here can further guide the study and manipulation of fruit harvest, must processing and wine fermentation, and may also contribute to decision making for breeding and cultivar development of grapevines. The novel GBS analysis methods presented here could have broad impact for F2 mapping in diverse heterozygous species.
Supporting Information
Perl code to identify markers from TASSEL hapmap file that are (1) heterozygous in the F1 progenitor and (2) show co-dominant 1:2:1 segregation among the F2 progeny by a chi-squared (χ2) goodness-of-fit test at α≤0.01.
(PL)
Summary statistics of the V. riparia × ‘Seyval’ F2 GBS-based linkage map.
(DOCX)
X-axes represent physical coordinates in the reference genome V. vinifera ‘PN40024’ version 12X.v2 (Mb). Y-axes represent genetic coordinates (cM).
(PDF)
(A) Sixty five F2 progeny were measured for berry skin color. White (nonpigmented) is coded as 0 and black (pigmented) is coded as 1. (B-D) Distribution frequency for quantitative enological traits in 63 F2 progeny.
(PDF)
Log-likelihood (LogLik), Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) evaluated for each of the causal models considered in CMST. In the column “Test”, a simple directed acyclic diagram is shown as a representation of the test performed. In this particular case, a genetic component-QTL (Q) influences either of the traits (MA or SS). The arrows represent the direction of the influence, which can go from Q to either trait or from trait to trait. Influence of trait upon Q is not considered in this approach. The values associated with the most likely models are highlighted in bold.
(DOCX)
QTL mapping for (A) total soluble solids content (SS, %w/w) and (B) the ratio of malic acid concentration (MA, g/L) to total soluble solids content (SS, %w/w). Permutation tests were carried out to identify 95% confidence thresholds, and the significance threshold of LOD score is presented as a horizontal red dashed line.
(PDF)
The sequence of markers in the final map.
(FASTA)
Acknowledgments
The authors thank Kalley Besler and Mani Awale for fruit collection and preprocessing and the South Dakota State University Agricultural Experiment Station NE Hansen Research Farm where the grapevine population is maintained.
Data Availability
All relevant data are within the paper and its Supporting Information files. All sequence information for the sequence markers are published in S7 File.
Funding Statement
This work was supported by the United States Department of Agriculture, National Institute of Food and Agriculture Specialty Crops Research Initiative Award No. 2011-51181-30635, BIR, LCD, AYF, GLS, JJL, AKM. This fund supported JFR and DCM. This work was also supported by the National Science Foundation Award No. IIA1355423, South Dakota Research Innovation Center and a gift from J. Lohr Vineyards & Wines to AYF. SY was supported by a grant from the National Grape and Wine Initiative to BIR. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the NSF, USDA or other funding entities. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bell S- J, Henschke PA. Implications of nitrogen nutrition for grapes, fermentation and wine. Aust J Grape Wine Res. 2005; 11(3): 242–95. 10.1111/j.1755-0238.2005.tb00028.x [DOI] [Google Scholar]
- 2.Mahanil S, Ramming D, Cadle-Davidson M, Owens C, Garris A, Myles S, et al. Development of marker sets useful in the early selection of Ren4 powdery mildew resistance and seedlessness for table and raisin grape breeding. Theoret Appl Genet. 2012; 124(1): 23–33. 10.1007/s00122-011-1684-7 [DOI] [PubMed] [Google Scholar]
- 3.Sacks GL, Acree TE, M.T. K, Vanden Heuvel JE, Wilcox WF, editors. Tell me about your childhood”–the role of the vineyard in determining wine flavour chemistry. 15th Australian wine industry technical conference; 2013; Sydney, New South Wales, Australia.
- 4.Ugliano M, Henschke P. Yeasts and wine flavour In: Moreno-Arribas MV, Polo MC, editors. Wine chemistry and biochemistry: Springer New York; 2009. p. 313–92. [Google Scholar]
- 5.Reisch B, Owens C, Cousins P. Grape In: Badenes ML, Byrne DH, editors. Fruit breeding. Handbook of plant breeding. Springer US; 2012. p. 225–262. [Google Scholar]
- 6.Young PR, Vivier MA. Genetics and genomic approaches to improve grape quality for winemaking In: Reynolds AG, editor. Managing wine quality: Woodhead Publishing; 2010. p. 316–364. [Google Scholar]
- 7.Di Gaspero G, Cattonaro F. Application of genomics to grapevine improvement. Aust J Grape Wine Res. 2010; 16: 122–130. 10.1111/j.1755-0238.2009.00072.x [DOI] [Google Scholar]
- 8.Sawler J, Reisch B, Aradhya MK, Prins B, Zhong GY, Schwaninger H, et al. Genomics assisted ancestry deconvolution in grape. PLoS One. 2013; 8(11): e80791 10.1371/journal.pone.0080791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson RS. Wine science: Principles and applications. 4th ed. San Diego: Academic Press; 2014. [Google Scholar]
- 10.Myles S. Improving fruit and wine: what does genomics have to offer? Trends Genet. 2013; 29(4): 190–196. 10.1016/j.tig.2013.01.006 [DOI] [PubMed] [Google Scholar]
- 11.Myles S, Boyko AR, Owens CL, Brown PJ, Grassi F, Aradhya MK, et al. Genetic structure and domestication history of the grape. Proc Natl Acad Sci USA. 2011; 108(9): 3530–3535. 10.1073/pnas.1009363108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Q, Gates MJ, Lavin EH, Acree TE, Sacks GL. Comparison of odor-active compounds in grapes and wines from Vitis vinifera and non-foxy American grape species. J Agric Food Chem. 2011; 59(19): 10657–10664. 10.1021/jf2026204 [DOI] [PubMed] [Google Scholar]
- 13.Boulton R, Singleton V, Bisson L, Kunkee R. Preparation of musts and juice Principles and practices of winemaking: Springer US; 1999. p. 65–101. [Google Scholar]
- 14.Volschenk1a H, Van Vuuren H, Viljoen-Bloom M. Malic acid in wine: origin, function and metabolism during vinification. S Afr J Enol Vitic. 2006; 27(2): 123–136. [Google Scholar]
- 15.Kliewer WM. Concentration of Tartrates, Malates, Glucose and Fructose in the Fruits of the Genus Vitis. Am J Enol Vitic. 1967; 18(2): 87–96. [Google Scholar]
- 16.Jackson D. Climate, monographs in cool climate viticulture-2. Daphne Brasell NZ. 2001. [Google Scholar]
- 17.Brice C, Sanchez I, Bigey F, Legras JL, Blondin B. A genetic approach of wine yeast fermentation capacity in nitrogen-starvation reveals the key role of nitrogen signaling. BMC Genomics. 2014; 15:495 10.1186/1471-2164-15-495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart ACH. Nitrogen composition of interspecific hybrid and Vitis vinifera wine grapes from the Eastern United States. Ph.D. Dissertation. Purdue University; 2013. Available: http://docs.lib.purdue.edu/dissertations/AAI3592130/.
- 19.Song S, del Mar Hernández M, Provedo I, Menéndez C. Segregation and associations of enological and agronomic traits in Graciano × Tempranillo wine grape progeny (Vitis vinifera L.). Euphytica. 2014; 195(2): 259–277. 10.1007/s10681-013-0994-z [DOI] [Google Scholar]
- 20.Chen J, Wang N, Fang L-C, Liang Z-C, Li S-H, Wu B-H. Construction of a high-density genetic map and QTLs mapping for sugars and acids in grape berries. BMC Plant Biol. 2015; 15: 28 10.1186/s12870-015-0428-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viana AP, Riaz S, Walker MA. Genetic dissection of agronomic traits within a segregating population of breeding table grapes. Genet Mol Res. 2013; 12(2): 951–964. 10.4238/2013.April.2.11 [DOI] [PubMed] [Google Scholar]
- 22.Huang Z, Ough CS. Effect of Vineyard Locations, Varieties, and Rootstocks on the Juice Amino Acid Composition of Several Cultivars Am J Enol ViticAm J Enol Vitic. 1989; 40(2): 135–139. [Google Scholar]
- 23.Grattapaglia D, Bertolucci FL, Sederoff RR. Genetic mapping of QTLs controlling vegetative propagation in Eucalyptus grandis and E. urophylla using a pseudo-testcross strategy and RAPD markers. Theor Appl Genet. 1995; 90(7–8): 933–947. 10.1007/BF00222906 [DOI] [PubMed] [Google Scholar]
- 24.Adam-Blondon AF, Roux C, Claux D, Butterlin G, Merdinoglu D, This P. Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor Appl Genet. 2004; 109(5): 1017–1027. 10.1007/s00122-004-1704-y [DOI] [PubMed] [Google Scholar]
- 25.Barba P, Cadle-Davidson L, Harriman J, Glaubitz JC, Brooks S, Hyma K, et al. Grapevine powdery mildew resistance and susceptibility loci identified on a high-resolution SNP map. Theor Appl Genet. 2014; 127(1): 73–84. 10.1007/s00122-013-2202-x [DOI] [PubMed] [Google Scholar]
- 26.Doligez A, Adam-Blondon AF, Cipriani G, Di Gaspero G, Laucou V, Merdinoglu D, et al. An integrated SSR map of grapevine based on five mapping populations. Theor Appl Genet. 2006; 113(3): 369–82. 10.1007/s00122-006-0295-1 [DOI] [PubMed] [Google Scholar]
- 27.Wang N, Fang LC, Xin HP, Wang LJ, Li SH. Construction of a high-density genetic map for grape using next generation restriction-site associated DNA sequencing. BMC Plant Biol. 2012; 12:148 10.1186/1471-2229-12-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fennell A, Mathiason K, Luby J. Genetic segregation for indicators of photoperiod control of dormancy induction in Vitis species. Acta Hortic. 689 2005; p. 533–540. 10.17660/ActaHortic.2005.689.66 [DOI] [Google Scholar]
- 29.Garris A, Clark L, Owens C, McKay S, Luby J, Mathiason K, et al. Mapping of photoperiod-induced growth cessation in the wild grape Vitis riparia. J Am Soc Hortic Sci. 2009; 134(2): 261–272. [Google Scholar]
- 30.Fennell AY, Schlauch KA, Gouthu S, Deluc LG, Khadka V, Sreekantan L, et al. Short day transcriptomic programming during induction of dormancy in grapevine. Front. Plant Sci. 2015; 6:834 10.3389/fpls.2015.00834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathiason K, He D, Grimplet J, Venkateswari J, Galbraith DW, Or E, et al. Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Funct Integr Genomics. 2009; 9(1): 81–96. 10.1007/s10142-008-0090-y [DOI] [PubMed] [Google Scholar]
- 32.Sreekantan L, Mathiason K, Grimplet J, Schlauch K, Dickerson JA, Fennell AY. Differential floral development and gene expression in grapevines during long and short photoperiods suggests a role for floral genes in dormancy transitioning. Plant Mol Biol. 2010; 73(1–2): 191–205. 10.1007/s11103-010-9611-x [DOI] [PubMed] [Google Scholar]
- 33.Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, et al. TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. PLoS One. 2014; 9(2): e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swarts K, Li HH, Navarro JAR, An D, Romay MC, Hearne S, et al. Novel Methods to Optimize Genotypic Imputation for Low-Coverage, Next-Generation Sequence Data in Crop Plants. Plant Genome. 2014; 7(3). 10.3835/plantgenome2014.05.0023 [DOI] [Google Scholar]
- 35.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS One. 2011; 6(5): e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azuma A, Kobayashi S, Mitani N, Shiraishi M, Yamada M, Ueno T, et al. Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor Appl Genet. 2008; 117(6): 1009–1019. 10.1007/s00122-008-0840-1 [DOI] [PubMed] [Google Scholar]
- 37.Cardoso S, Lau W, Dias JE, Fevereiro P, Maniatis N. A Candidate-Gene Association Study for Berry Colour and Anthocyanin Content in Vitis vinifera L. PLoS One September 28 2012;7(9): e46021 10.1371/journal.pone.0046021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salmaso M, Malacarne G, Troggio M, Faes G, Stefanini M, Grando MS, et al. A grapevine (Vitis vinifera L.) genetic map integrating the position of 139 expressed genes. Theor Appl Genet. 2008; 116(8): 1129–1143. 10.1007/s00122-008-0741-3 [DOI] [PubMed] [Google Scholar]
- 39.Castellari M, Versari A, Spinabelli U, Galassi S, Amati A. An improved HPLC method for the analysis of organic acids, carbohydrates, and alcohols in grape musts and wines. J Liq Chromatogr Relat Technol. 2000; 23(13): 2047–2056. 10.1081/Jlc-100100472 [DOI] [Google Scholar]
- 40.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007; 449(7161): 463–467. 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25(14):1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011; 27(15): 2156–8. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007; 23(19): 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 44.Van Ooijen J. JoinMap® 4. Software for the calculation of genetic linkage maps in experimental populations. Version 4.1. Wageningen, The Netherlands: Kyazma; 2006.
- 45.Wu R, Ma C-X, Painter I, Zeng Z-B. Simultaneous maximum likelihood estimation of linkage and linkage phases in outcrossing species. Theoretical Popul Biol. 2002; 61(3): 349–363. 10.1006/tpbi.2002.1577 [DOI] [PubMed] [Google Scholar]
- 46.Van Ooijen J. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res. 2011; 93(5): 343–349. 10.1017/S0016672311000279 [DOI] [PubMed] [Google Scholar]
- 47.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003; 19(7): 889–890. 10.1093/Bioinformatics/Btg112 [DOI] [PubMed] [Google Scholar]
- 48.R Core Team. R: A language and environment for statistical computing. Version 3.1.2. Vienna, Austria: R Foundation for statistical computing; 2014.
- 49.Boer MP, Malosetti M, Welham SJ, Thissen JTNM. Part 2: Statistical genetics and QTL estimation. In: Payne RW, Harding SA, Murray DA, Soutar DM, Baird DB, Glaser AI, et al., editors. The guide to GenStat release 17. Hemel Hempstead, UK: VSN International; 2014.
- 50.VSN International. GenStat for Windows 17th edition Hemel Hempstead, UK: VSN International; 2014. [Google Scholar]
- 51.Broman KW, Sen S. A guide to QTL mapping with R/qtl: Springer; 2009. [Google Scholar]
- 52.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005; 95(3): 221–7. 10.1038/Sj.Hdy.6800717 [DOI] [PubMed] [Google Scholar]
- 53.Malosetti M, Voltas J, Romagosa I, Ullrich SE, van Eeuwijk FA. Mixed models including environmental covariables for studying QTL by environment interaction. Euphytica. 2004;137(1):139–145. 10.1023/B:Euph.0000040511.46388.Ef [DOI] [Google Scholar]
- 54.Boer MP, Wright D, Feng L, Podlich DW, Luo L, Cooper M, et al. A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics. 2007; 177(3): 1801–1813. 10.1534/genetics.107.071068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynch M, Walsh B. Genetics and analysis of quantitative traits Sunderland, Mass: Sinauer; 1998. [Google Scholar]
- 56.Neto EC, Broman AT, Keller MP, Attie AD, Zhang B, Zhu J, et al. Modeling causality for pairs of phenotypes in system genetics. Genetics. 2013; 193(3): 1003–1013. 10.1534/Genetics.112.147124/-/Dc1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neto EC, Yandell BS. qtlhot: Inference for QTL Hotspots. 2013. R package version 0.9.0. Available: http://CRAN.R-project.org/package=qtlhot.
- 58.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004; 304(5673): 982 10.1126/science.1095011 [DOI] [PubMed] [Google Scholar]
- 59.Myles S, Chia J-M, Hurwitz B, Simon C, Zhong GY, Buckler E, et al. Rapid genomic characterization of the genus Vitis. PLoS One. 2010. January 13, 2010; 5(1): e8219 10.1371/journal.pone.0008219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plomion C, Durel C. Estimation of the average effects of specific alleles detected by the pseudo-testcross QTL mapping strategy. Genet Sel Evol. 1996; 28(3): 1–13. 10.1186/1297-9686-28-3-223 [DOI] [Google Scholar]
- 61.Lorieux M, Perrier X, Goffinet B, Lanaud C, de León DG. Maximum-likelihood models for mapping genetic markers showing segregation distortion. 2. F2 populations. Theor Appl Genet. 1995; 90(1): 81–89. 10.1007/BF00220999 [DOI] [PubMed] [Google Scholar]
- 62.Staub JE, Serquen FC, Gupta M. Genetic markers, map construction, and their application in plant breeding. HortScience. 1996; 31(5): 729–741. [Google Scholar]
- 63.Van Ooijen J, Jansen J. Genetic mapping in experimental populations. Cambridge: Cambridge University Press; 2013. [Google Scholar]
- 64.Morgante M, De Paoli E, Radovic S. Transposable elements and the plant pan-genomes. Curr Opin Plant Biol. 2007; 10(2):149–155. 10.1016/j.pbi.2007.02.001 [DOI] [PubMed] [Google Scholar]
- 65.Ugliano M, Kolouchova R, Henschke PA. Occurrence of hydrogen sulfide in wine and in fermentation: influence of yeast strain and supplementation of yeast available nitrogen. J Ind Microbiol Biotechnol. 2011; 38(3): 423–429. 10.1007/s10295-010-0786-6 [DOI] [PubMed] [Google Scholar]
- 66.Nisbet MA, Martinson TE, Mansfield AK. Accumulation and Prediction of Yeast Assimilable Nitrogen in New York Winegrape Cultivars. Am J Enol Vitic. 2014; 65(3): 325–332. 10.5344/ajev.2014.13130 [DOI] [Google Scholar]
- 67.Kanellis A, Roubelakis-Angelakis K. Grape Biochemistry of fruit ripening: Springer; 1993. p. 189–234. [Google Scholar]
- 68.Sweetman C, Deluc LG, Cramer GR, Ford CM, Soole KL. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry. 2009; 70(11–12): 1329–1344. 10.1016/j.phytochem.2009.08.006 [DOI] [PubMed] [Google Scholar]
- 69.Banerjee S, Yandell BS, Yi NJ. Bayesian quantitative trait loci mapping for multiple traits. Genetics. 2008; 179(4): 2275–2289. 10.1534/Genetics.108.088427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu CW, Li ZK, Xu SZ. Joint mapping of quantitative trait loci for multiple binary characters. Genetics. 2005; 169(2): 1045–1059. 10.1534/Genetics.103.019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang RQ, Li JH, Xu SZ. Mapping quantitative trait loci for traits defined as ratios. Genetica. 2008; 132(3): 323–329. 10.1007/S10709-007-9175-0 [DOI] [PubMed] [Google Scholar]
- 72.Zhu W, Zhang H. Why do we test multiple traits in genetic association studies? J Korean Stat Soc. 2009; 38(1): 1–10. 10.1016/j.jkss.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang RQ, Jin TB, Li WB. Mapping genome-wide QTL of ratio traits with Bayesian shrinkage analysis for its component traits. Genetica. 2010; 138(8): 853–860. 10.1007/S10709-010-9468-6 [DOI] [PubMed] [Google Scholar]
- 74.Khan SA, Beekwilder J, Schaart JG, Mumm R, Soriano JM, Jacobsen E, et al. Differences in acidity of apples are probably mainly caused by a malic acid transporter gene on LG16. Tree Genet Genomes. 2013; 9(2): 475–487. 10.1007/S11295-012-0571-Y [DOI] [Google Scholar]
- 75.Jiang C, Zeng ZB. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics. 1995; 140(3): 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Perl code to identify markers from TASSEL hapmap file that are (1) heterozygous in the F1 progenitor and (2) show co-dominant 1:2:1 segregation among the F2 progeny by a chi-squared (χ2) goodness-of-fit test at α≤0.01.
(PL)
Summary statistics of the V. riparia × ‘Seyval’ F2 GBS-based linkage map.
(DOCX)
X-axes represent physical coordinates in the reference genome V. vinifera ‘PN40024’ version 12X.v2 (Mb). Y-axes represent genetic coordinates (cM).
(PDF)
(A) Sixty five F2 progeny were measured for berry skin color. White (nonpigmented) is coded as 0 and black (pigmented) is coded as 1. (B-D) Distribution frequency for quantitative enological traits in 63 F2 progeny.
(PDF)
Log-likelihood (LogLik), Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) evaluated for each of the causal models considered in CMST. In the column “Test”, a simple directed acyclic diagram is shown as a representation of the test performed. In this particular case, a genetic component-QTL (Q) influences either of the traits (MA or SS). The arrows represent the direction of the influence, which can go from Q to either trait or from trait to trait. Influence of trait upon Q is not considered in this approach. The values associated with the most likely models are highlighted in bold.
(DOCX)
QTL mapping for (A) total soluble solids content (SS, %w/w) and (B) the ratio of malic acid concentration (MA, g/L) to total soluble solids content (SS, %w/w). Permutation tests were carried out to identify 95% confidence thresholds, and the significance threshold of LOD score is presented as a horizontal red dashed line.
(PDF)
The sequence of markers in the final map.
(FASTA)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All sequence information for the sequence markers are published in S7 File.