Significance
Programmed cell death plays a central role in host defense in plants, animals, and fungi, but the extent to which cell death modalities are evolutionarily related and mechanistically similar in different kingdoms is unclear. The involvement of prion-like mechanisms in host defense and cell death cascades has been reported in animals and fungi. In fungi, a cell death-inducing pore-forming protein termed HET-S is activated by amyloid templating. Here we characterize a protein termed HELLP, which behaves analogously to HET-S as a membrane-targeting cell death-inducing protein regulated by amyloid templating. We find that HELLP is homologous to MLKL, the protein involved in execution of necroptotic cell death in mammals, thus revealing transkingdom conservation of amyloid-regulated programmed cell death.
Keywords: amyloid, prion, programmed cell death, incompatibility, necroptosis
Abstract
Recent findings have revealed the role of prion-like mechanisms in the control of host defense and programmed cell death cascades. In fungi, HET-S, a cell death-inducing protein containing a HeLo pore-forming domain, is activated through amyloid templating by a Nod-like receptor (NLR). Here we characterize the HELLP protein behaving analogously to HET-S and bearing a new type of N-terminal cell death-inducing domain termed HeLo-like (HELL) and a C-terminal regulatory amyloid motif known as PP. The gene encoding HELLP is part of a three-gene cluster also encoding a lipase (SBP) and a Nod-like receptor, both of which display the PP motif. The PP motif is similar to the RHIM amyloid motif directing formation of the RIP1/RIP3 necrosome in humans. The C-terminal region of HELLP, HELLP(215-278), encompassing the motif, allows prion propagation and assembles into amyloid fibrils, as demonstrated by X-ray diffraction and FTIR analyses. Solid-state NMR studies reveal a well-ordered local structure of the amyloid core residues and a primary sequence that is almost entirely arranged in a rigid conformation, and confirm a β-sheet structure in an assigned stretch of three amino acids. HELLP is activated by amyloid templating and displays membrane-targeting and cell death-inducing activity. HELLP targets the SBP lipase to the membrane, suggesting a synergy between HELLP and SBP in membrane dismantling. Remarkably, the HeLo-like domain of HELLP is homologous to the pore-forming domain of MLKL, the cell death-execution protein in necroptosis, revealing a transkingdom evolutionary relationship between amyloid-controlled fungal programmed cell death and mammalian necroptosis.
Programmed cell death (PCD) plays a central role in response to nonself and host defenses in animals, plants, fungi, and bacteria (1–4). Altruistic cell suicide hinders pathogen replication and promotes survival in multicellular organisms, and provides benefits at the population level in unicellular microbes. Questions surrounding the classification, evolutionary origin, and level of transkingdom conservation of PCD modalities remain open and often polemic (5). Classification systems of PCD types are based on cytological or molecular markers that can differ between phyla, making it difficult to merge cell death processes into unifying categories (5).
In filamentous fungi, a form of PCD termed heterokaryon incompatibility occurs in response to conspecific nonself (3). Cell death by incompatibility takes place following fusion of cells from genetically distinct individuals and is controlled by het genes. One of the best-characterized incompatibility systems involves a cell death-inducing protein termed HET-S, which is controlled by amyloid templating. HET-S of Podospora anserina exhibit an N-terminal cell death execution HeLo domain and a C-terminal regulatory prion forming domain (PFD) that adopts a specific β-solenoid amyloid fold, characterized by the stacking of two pseudorepeats of an elementary amyloid motif (6–8). When the HET-S PFD is converted to the β-solenoid fold, the HeLo domain undergoes refolding, and an N-terminal transmembrane helix (TMH) is exposed, causing HET-S to behave as a pore-forming toxin (9). On transconformation, HET-S relocates from the cytoplasm to the cell membrane, where it exerts toxicity (10).
Conversion of the HET-S PFD region can be achieved by two means. The first described mechanism occurs in the context of HET-S/[Het-s] incompatibility (9). [Het-s] is a variant form of HET-S that has lost the pore-forming activity and can stably propagate as a prion. [Het-s] and HET-S strains are incompatible, because [Het-s] acts as a seed converting the PFD of HET-S, thereby leading to activation of the HeLo cell death-inducing domain. A second, recently described mode of activation involves a Nod-like receptor protein (NLR), termed NWD2, encoded by the gene adjacent to het-S in the genome (11, 12). The NWD2 NLR displays a 21-aa N-terminal region homologous to the HET-s amyloid motif. On binding of a specific ligand to NWD2, these N-terminal extensions adopt an amyloid fold and convert HET-S to the toxic pore-forming state.
The NWD2/HET-S system is one of several examples in which activation of cell death and host defense cascades relies on prion-like and/or amyloid polymerization mechanisms (13, 14). Activation of ASC (apoptosis-associated speck-like protein with a caspase recruitment domain) by the NLRP3 human NLR depends on a prion-like polymerization of the PYRIN domain and the HET-s-prion motif is able to substitute for the PYRIN signaling domain in NLRP3-dependent ASC activation (13). Another example is the mammalian RIP1/RIP3 kinase complex controlling necroptosis, assembled via short amyloid motifs termed RHIM (15). One downstream effector of the RIP1/RIP3 complex in necroptosis is the mixed lineage kinase domain-like protein (MLKL), which is a target of RIP3 (16). In fungi, we have identified several other gene clusters analogous to Nwd2/het-S in which an NLR and a putative cell death effector protein are encoded by adjacent genes and in which the N-terminal extension of the NLR protein is homologous to the C-terminal end of the effector (12, 17). One such proposed prion motif, termed PP, was found to be associated with HeLo domain proteins, lipases, and regulatory NLRs (12). Herein we characterize a protein identified in Chaetomium globosum bearing this motif, termed HELLP.
Results
Identification of a PP Motif Gene Cluster.
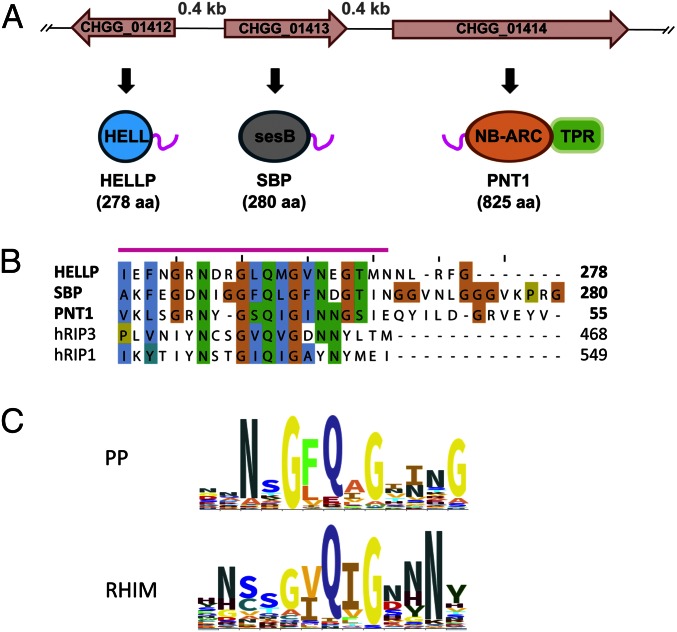
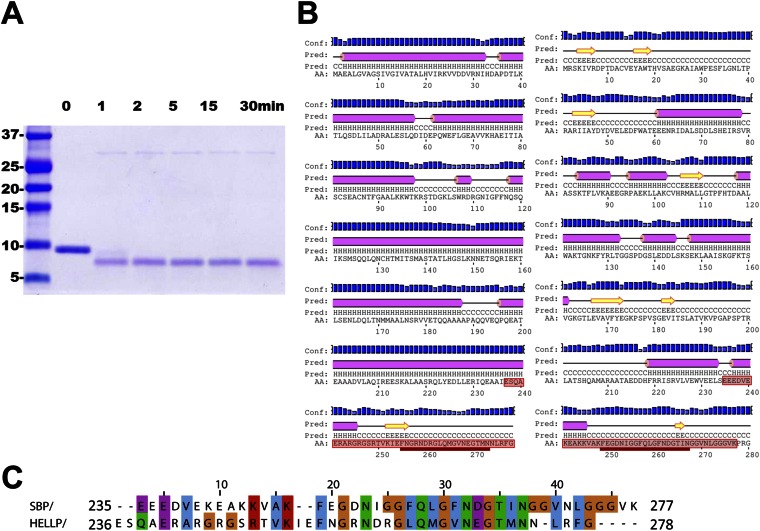
We identified in C. globosum a gene cluster with three adjacent genes, CHGG_01412, CHGG_01413, and CHGG_01414, encoding proteins with PP motifs, corresponding to a HeLo-like domain protein (HELLP), a putative lipase (SBP) and a NLR (PNT1), respectively (Fig. 1A). An N-terminal transmembrane helix (TMH) is predicted in HELLP that shows homology to the HET-S TMH (Fig. S1). The SBP lipase is homologous to SesB, a protein involved in propagation of the σ infectious element of Nectria hematococca (18, 19). HELLP, SBP, and PNT1 share a region of homology of 20–21 residues corresponding to the PP motif and located C-terminally in HELLP and SBP and N-terminally in PNT1. The similarity in genome and domain architecture between the het-S/Nwd2 and Hellp/Sbp/Pnt1 gene clusters suggests that HELLP may represent an HET-S analog activated by amyloid templating via the PP motif. It has been proposed that the HET-s prion motif is evolutionarily related to the RHIM amyloid motif controlling necroptotic signaling (20). The PP motif of HELLP shows similarity to human RIP1 and RIP3 RHIM motifs (Fig. 1B). The RHIM and PP motif signatures share a central common G-φ-Q-φ-G core centered on a conserved Q residue and flanked by N residues (Fig. 1C).
Fig. 1.
The PP motif gene cluster. (A) Representation of the CHGG_1412/13/14 locus with three adjacent genes, one encoding a HELL domain protein termed HELLP, one encoding a predicted lipase related to SesB (SBP), and one encoding a NB-ARC/TPR STAND protein (PNT1). The PP motif is represented as a pink line. (B) Alignment of the PP motif region of HELLP, SBP, and PNT1 together with a portion of the RHIM motif of human RIP1 and RIP3. (C) Sequence signatures for the central region of PP and the RHIM motif generated with Skylign. Letter size corresponds to level of conservation.
Fig. S1.
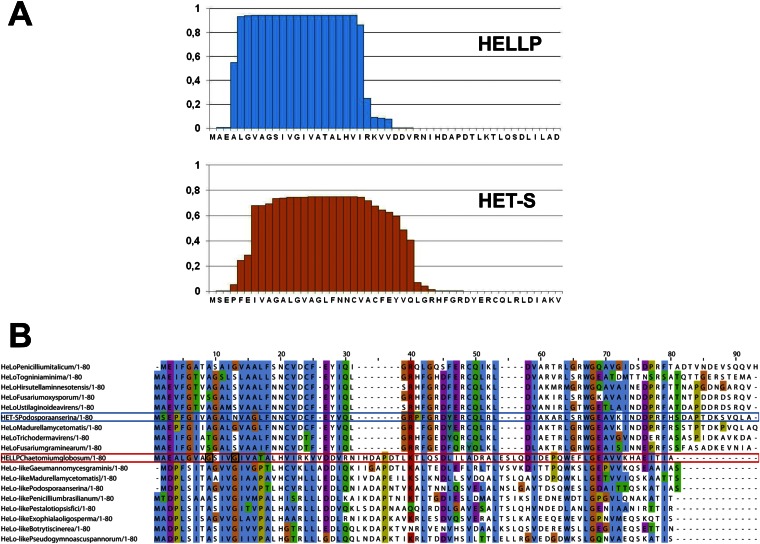
Conservation of a predicted N-terminal transmembrane helix in HELLP. (A) Prediction of a transmembrane helix for HELLP and HET-S using TMHMM. The probability of transmembrane helix formation for N-terminal residues is given. (B) Alignment of the N-terminal regions of HeLo and HeLo-like domain proteins. HET-S and HELLP and the eight strongest homologs of each protein identified in the National Center for Biotechnology Information’s nr database were aligned. HELLP and HET-S are boxed in red and blue, respectively. The G9 and G13 residues of HELLP are boxed in black.
HELLP Behaves as an HET-S Analog.
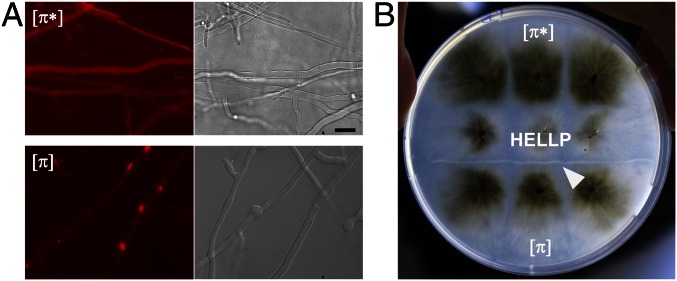
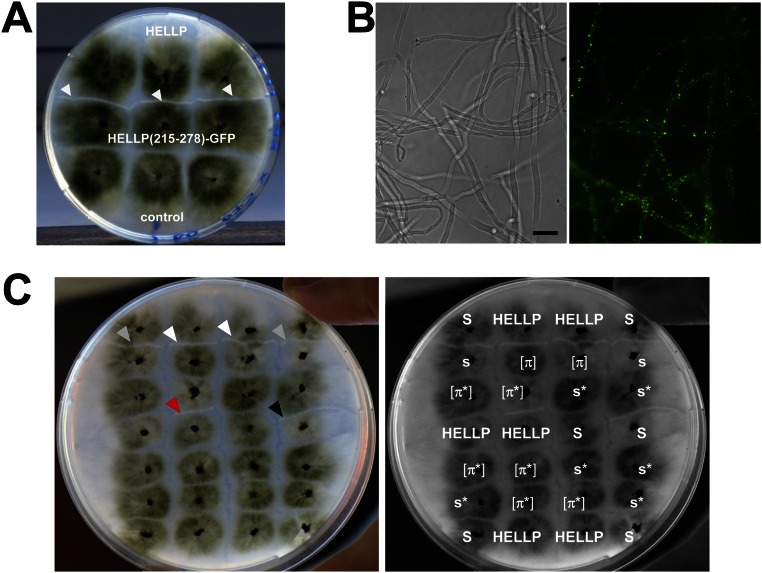
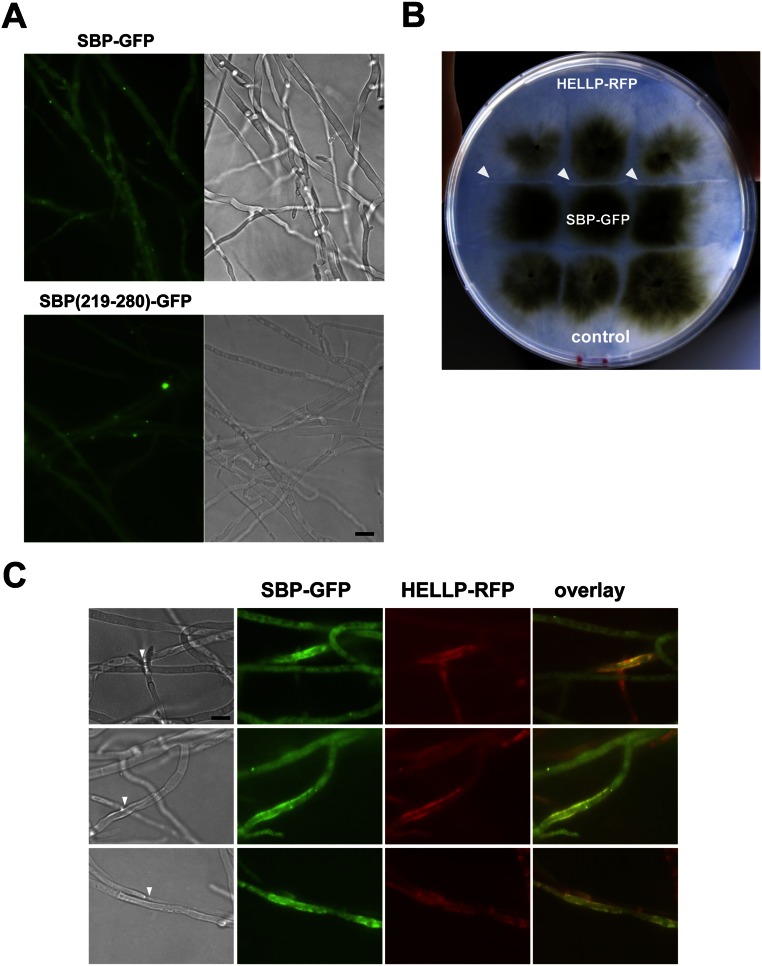
We hypothesized that HELLP might behave as HET-S, and thus attempted to recreate an incompatibility system, analogous to [Het-s]/HET-S, based on HELLP. HET-S cannot form a prion, because amyloid conversion of HET-S leads to membrane targeting and toxicity, preventing prion propagation. The HeLo domain must be mutated or truncated to allow for prion formation (7, 8). We reasoned that if HELLP is a HET-S analog, then truncation of the HELL domain should allow formation of a PP-based prion, and that strains bearing this prion should be incompatible with strains expressing full-length HELLP. A truncated HELLP construct, HELLP(172-278)-RFP, was expressed in P. anserina. HELLP(172-278)-RFP fluorescence was initially diffuse, but dot-like and elongated aggregates formed on further subculturing (Fig. 2A). Strains expressing HELLP(172-278)-RFP were then confronted to strains expressing full-length HELLP-RFP (Fig. 2B). Barrage formation, indicative of an incompatibility reaction, occurred specifically in confrontations with strains displaying HELLP(172-278)-RFP aggregates. A shorter construct, HELLP(215-278)-GFP, also led to dot formation and to incompatibility with full-length HELLP (Fig. S2).
Fig. 2.
HELLP(172-278)-RFP forms a prion, incompatible with full-length HELLP. (A) Micrograph of a P. anserina strain expressing a HELLP(172-278)-RFP fusion protein in either the soluble [π*] state (Upper) or the aggregated [π] state (Lower). (Scale bar: 5 μ.) (B) Confrontation of a strain expressing full-length HELLP and expressing HELLP(172-278)-RFP in the soluble state [π*] or aggregated state [π] as noted. Note the formation of a barrage (marked by a white arrow) specifically in the confrontation with the strain expressing HELLP(172-278)-RFP in the aggregated state [π].
Fig. S2.
Expression of HELLP(215-278)-GFP and prion propagation of HELLP(172-278)-RFP. (A) Strains expressing HELLP(215-278)-GFP from a barrage reaction (marked by white arrows) when confronted to strains expressing full-length HELLP (HELLP-RFP). The control was an untransformed ΔPa_5_8070 strain. (B) HELLP(215-278)-GFP forms aggregates when expressed in P. anserina. (Scale bar: 5 μ.) (C) Lack of cross-infection between [π] and [Het-s] prions. The same picture is shown with and without labeling for clarity. (Right) Strains were confronted on solid medium. [π] strains produce a barrage to strains expressing HELLP (white arrowheads), and [Het-s] strains (s) produce a barrage to [Het-S] strains (S) (gray arrowheads). On confrontation with a [π] strain, a [π*] strain acquires the [π] phenotype and produces a barrage to HELLP (red arrowhead). A [Het-s*] strain (s*) confronted to a [Het-s] acquires the [Het-s] phenotype and produces a barrage to [Het-S] (black arrowhead); however, [Het-s] strains do not convert [π*], and [π] strains do not convert [Het-s*].
Strains expressing truncated HELLP display two phenotypic states that we term [π*] and [π]. [π*] strains show diffuse fluorescence and are compatible with HELLP, whereas [π] strains show aggregates and are incompatible with HELLP (Fig. 2 A and B). [π*] strains were systematically converted to the [π] state when confronted with [π] strains (Fig. S2). The rate of spontaneous [π] formation was dependent on the growth medium (Table S1). There was no cross-infection between [π] and [Het-s] prions. [π] strains did not convert [Het-s*] to the [Het-s] state, and [Het-s] strains did not convert [π*] to the [π] state (Fig. S2C).
Table S1.
Rate of spontaneous [π] formation in HELLP(172-278) strains cultivated on different media
| Medium | Total tested, n | [π], n (%) |
| DO | 228 | 7 (3) |
| DO sorbitol | 100 | 60 (60) |
| SUHMEA | 96 | 96 (100) |
HELLP Relocates to the Membrane, and Mutations in the TMH Abolish Incompatibility.
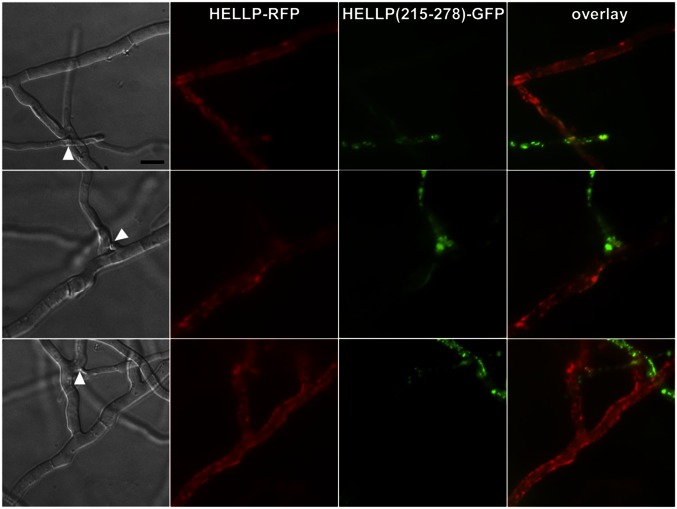
HET-S relocates from the cytoplasm to the cell membrane on conversion of the PFD region to the β-solenoid prion fold (9–11). We tested whether HELLP also relocates to the cell membrane on interaction with the prion form of the PP motif. A strain expressing HELLP-RFP was confronted to a strain expressing HELLP(215-278)-GFP in the [π] prion state. HELLP-RFP had a diffuse cytoplasmic localization in homokaryotic mycelium and relocated to the cell membrane region in fusion cells on interaction with HELLP(215-278)-RFP prion aggregates (Fig. 3 and Fig. S3A).
Fig. 3.
HELLP-RFP localizes at the cell periphery on interaction with aggregated HELLP(215-278)-GFP. On fusion of cells coexpressing HELLP-RFP and HELLP(215-278)-GFP under the aggregated state [π], HELLP-RFP relocalizes to the membrane region. The white arrowhead marks the position of the cell fusion point between the HELLP-RFP and HELLP(215-278)-GFP strains. (Scale bar: 5 μ.)
Fig. S3.
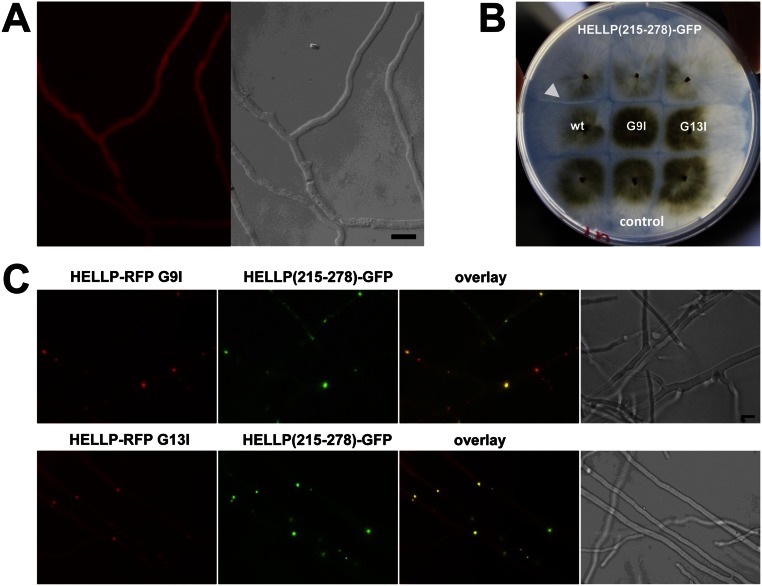
Localization of HELLP-RFP in homokaryotic mycelium and localization of HELLP-RFP point mutants. (A) HELLP-RFP is expressed in a wild-type strain and shows a diffuse cytoplasmic localization. (Scale bar: 5 μ.) (B) HELLP-RFP G9I and G13I mutants do not produce a barrage reaction to HELLP(215-278)-GFP, whereas wild-type HELLP-RFP does (white arrowhead). (C) Localization of HELLP-RFP G9I and HELLP-RFP G13I in fusion cells with strains expressing HELLP(215-278)-GFP. (Scale bar: 5 μ.)
The N-terminal TMH of HET-S plays a critical role in membrane targeting and toxicity (9). To investigate the role of the predicted HELLP TMH, we mutated two glycines, part of a putative GxxxG glycine-zipper motif, (a motif involved in TMH multimerization (21) into large hydrophobic residues (G9I and G13I) predicted to disrupt the glycine zipper (Fig. S1). Both mutants no longer produced a incompatibility reaction to [π] (Fig. S3B), and instead of being targeted to the membrane, HELLP-RFP G9I and G13I mutants formed cytoplasmic aggregates that partially colocalized with HELLP(215-278)-GFP (Fig. S3C).
The Prion-Forming Domain of HELLP Forms Amyloid Fibrils in Vitro.
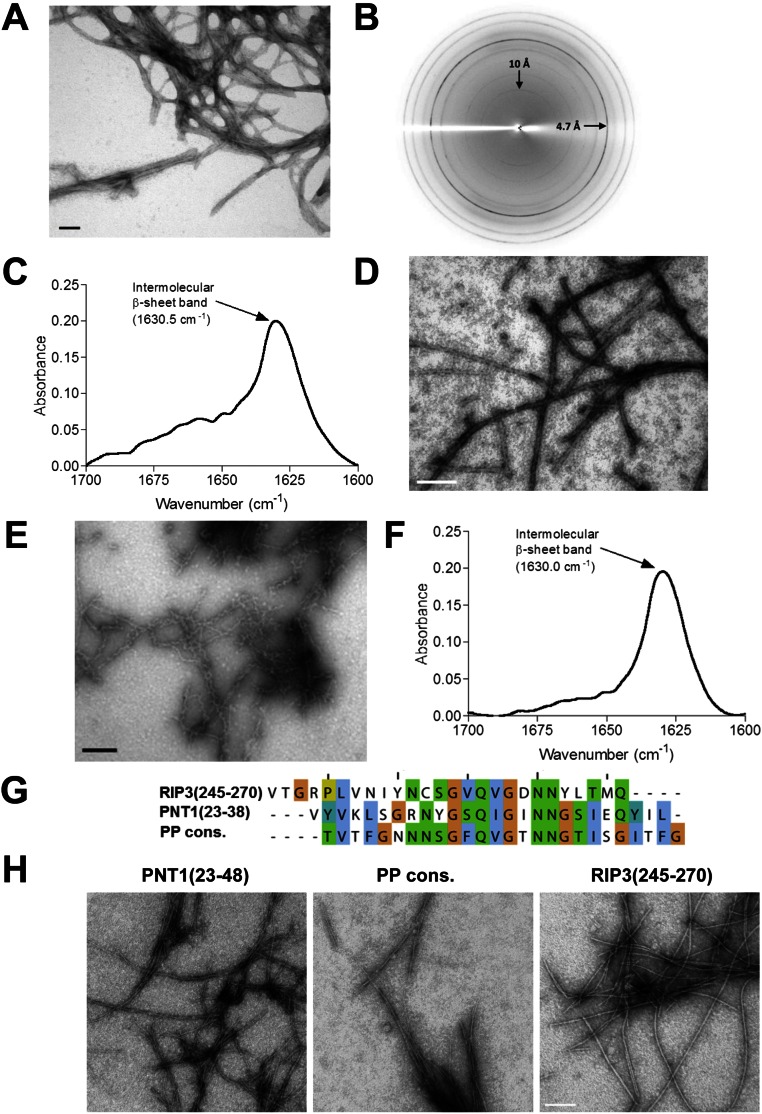
In vitro, purified HELLP(215-278) spontaneously assembled into ∼5-nm-wide fibrils (Fig. S4A). In X-ray diffraction experiments, the fibrils presented a reflection at 4.7 Å, typical of a cross-β structure (Fig. S4B). When analyzed by spectroscopy, fibrils showed a single strong amide I band (around 1,630 cm−1), usually associated with parallel β-sheet structures in amyloid aggregates (Fig. S4C). As described previously for HET-s(218-289) (22), HELLP(215-278) assembled into fibrils in denaturing conditions (8 M urea) as well (Fig. S4D). The HELLP(215-278) fibrils showed a proteinase K-resistant core (Fig. S5). Two main products corresponding to regions 238–278 and 236–278 were identified. The proteinase K-resistant core encompassed the PP motif per se and extended N-terminally into a highly charged region not conserved among HELLP, SBP, and PNT1. We transfected [π*] strains with HELLP(215-278) fibrils to assay their prion infectivity (Table S2). On transfection of [π*] strains with HELLP(215-278) fibrils, [π] formation rate was strongly increased, indicating that HELLP(215-278) fibrils assembled in vitro display prion infectivity.
Fig. S4.
HELLP(215-278), SBP(219-280), and PNT1(23-38) form amyloid fibrils. (A) Electron micrograph of negatively stained HELLP(215-278) fibrils. (Scale bar: 30 nm.) (B) X-ray diffraction of HELLP(215-278) fibrils. Note the major diffraction at 4.7 Å. (C) FTIR analysis of HELLP(215-278) fibrils showing a major band at 1,630 cm−1 indicative of the presence of parallel stacked β-sheets. (D) Electron micrograph of HELLP(215-278) fibrils formed in 8 M urea. (Scale bar: 100 nm.) (E) Electron micrograph of negatively stained SBP(219-280) fibrils. (Scale bar: 30 nm.) (F) FTIR analysis of SBP(219-280) fibrils showing a major band at 1,630 cm−1. (G) Sequence and alignment of three 26-aa peptides related to the PP motif and analyzed by EM. (H) Electron micrograph of the assemblies formed by the synthetic peptide as given in E. (Scale bar: 100 nm.)
Fig. S5.
Proteinase K-resistant core of HELLP(210-278) and SBP(219-280) fibrils. (A) SDS/PAGE analysis of HELLP(215-278) fibrils after proteinase K digestion. Fibrils were digested for the indicated times. (B) Secondary structure prediction for HELLP and SBP using PSIPRED. The proteinase K-protected region as identified by mass spectrometry is boxed in red. The position of the PP motif is underlined in red. The beginning of the proteinase K-resistant region extends into the last predicted α-helix of the HELL domain. Note that the electrophoretic mobility of the peptides on SDS/PAGE (in A) is slightly aberrant (with an apparent mass of ∼7 kDa), possibly owing to the high content in charged residues in the N-terminal part of the protected region. (C) Alignment of the proteinase K-resistant regions of HELLP and SBP fibrils.
Table S2.
HELLP(215-278) fibrils induce the [π] state
| HELLP(215-278) fibrils | No protein control | |||||
| Experiment | Total tested, n | [π], n | Conversion, % | Total tested, n | [π], n | Conversion, % |
| 1 | 30 | 14 | 46.6 | 30 | 0 | 0 |
| 2 | 30 | 24 | 80 | 30 | 1 | 3.3 |
| 3 | 40 | 29 | 72.5 | 40 | 3 | 7.5 |
| 4 | 40 | 37 | 92.5 | 40 | 4 | 10 |
| 5 | 30 | 13 | 43.3 | 30 | 3 | 10 |
| All, mean ± SD | 66.8 ± 21.6 | 6.1 ± 4.5 | ||||
Solid-State NMR Analysis of HELLP Fibrils.
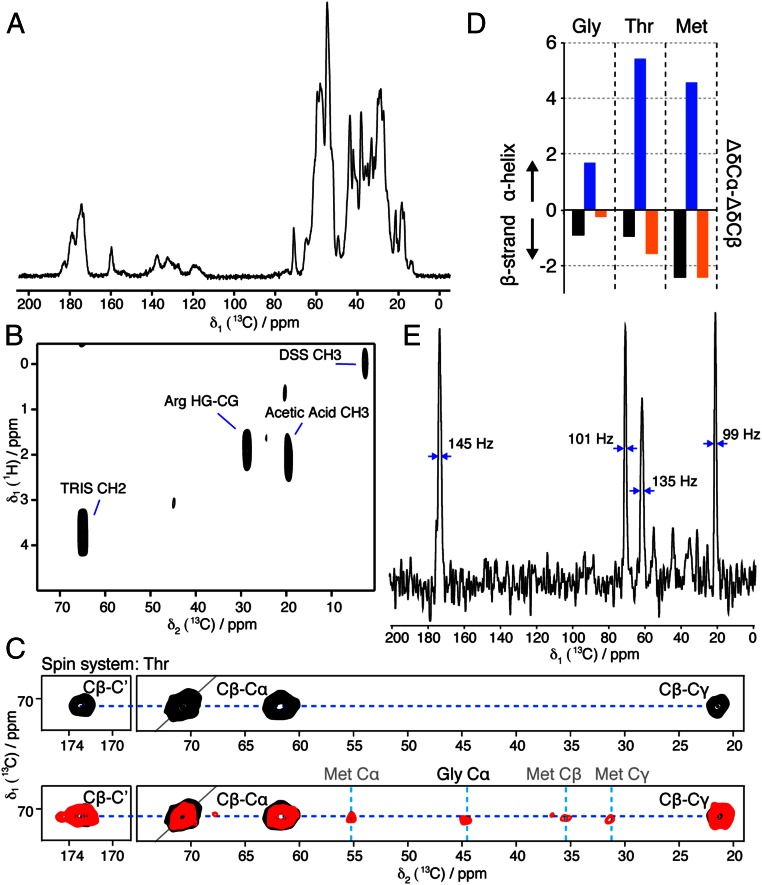
We carried out solid-state NMR (SSNMR) experiments to characterize the structural order in HELLP(215-278) fibrils. A 1D 13C cross-polarization (CP) experiment (Fig. 4A) showed strong signals, indicating that a significant part of the protein was immobilized in the fibrils. Multidimensional MAS SSNMR was conducted on 15N and 13C isotope- labeled HELLP fibrils. A 2D SSNMR insensitive nuclei enhanced by polarization transfer (INEPT) experiment was performed to detect mobile residues. The 2D 1H-13C INEPT (Fig. 4B) revealed the buffer carbon signals and a side-chain arginine (Arg) carbon, indicating the absence of residues that execute nearly isotropic motion on time scales much shorter than 1 μs.
Fig. 4.
Conformational analysis of HELLP(215-278) fibrils by solid-state NMR. (A) 1D 13C-detected CP spectrum. (B) 2D 1H-13C INEPT spectrum. (C) Excerpt of 2D PDSD ssNMR 13C-13C spectra centered around the Thr resonance frequency (Upper, 30 ms mixing time; Lower, 150 ms mixing time), optimized to detect intraresidual (30 ms) and sequential (150 ms) 13C-13C correlations. All peak assignments of the Thr271 spin system except for the diagonal Cα-Cα peaks are annotated. The additional peaks visible in the red spectrum reveal interresidual contacts of the Thr spin system. Assignments of Gly270 and Met272 are annotated above the spectral excerpt. The complete aliphatic spectral region of the 2D PDSD (30 ms mixing time) is shown in Fig. S5D. (D) Secondary ssNMR chemical shifts ΔδCα-ΔδCβ of the Gly270-Thr271-Met272 amino acid stretch in HELLP(215-278) (black), revealing their β-strand secondary structure. The secondary chemical shifts of a Gly-Thr-Met stretch in typical α-helical and β-strand conformations (blue and orange, respectively) are plotted for eye guidance. (E) 1D trace of the 2D PDSD (150 ms mixing time) at the position of the dotted blue line indicated in C, Lower. The line widths of intense signals are denoted.
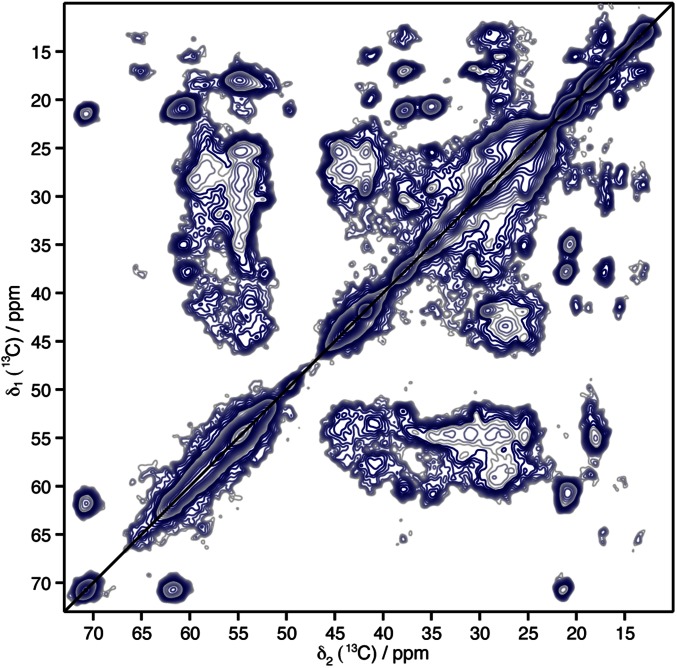
Two-dimensional 13C-13C experiments (Fig. 4C) were recorded using proton driven spin-diffusion (PDSD) mixing times of 30 and 150 ms, leading to isolated cross-peaks encoding intraresidue and interresidue correlations, respectively. Several residue spin systems were observable based on their typical chemical shifts, including threonine (Thr), isoleucine, alanine, and valine. A detailed analysis was possible for one apparent Thr residue, because the SSNMR Cα-Cβ and Cβ-Cγ resonance peaks of Thr residues lay in an isolated part of the 2D 13C-13C correlation spectra. We could assign the Cα, Cβ, and Cγ resonance frequencies of only one Thr (Fig. 4C, Upper). The sequential contacts connecting the Thr residue to its amino acid neighbors were detected in a 2D 13C-13C PDSD spectrum designed to detect interresidual 13C-13C correlations (Fig. 4C, Lower) with a PDSD mixing time of 150 ms (full 2D spectrum shown in Fig. S6). We could tentatively assign the glycine (Gly270)-Thr271 amino acid pair located in the proteinase K core region 238–278, because SSNMR Gly Cα resonance peaks are very specific and only one Gly-Thr pair is present in the HELLP(215-278) primary sequence. The neighboring methionine (Met) 272 Cα, Cβ, and Cγ resonance frequencies were assigned owing to their sequential contacts with the isolated Thr Cβ frequency (Fig. 4B). The three-residue stretch, Gly270-Thr271-Met272, of the fibrillar core region located in the PP motif appeared to adopt a β-strand conformation in HELLP(215-278) fibrils, as demonstrated by the secondary chemical shift values (Fig. 4C). The 1D trace of the 150-ms PDSD spectrum is shown in Fig. 4E to indicate the spectral resolution and sensitivity, representative of the decent structural order and rigidity of the amyloid core.
Fig. S6.
Solid-state NMR PDSD spectrum of HELLP(215-278) fibrils, recorded at 300-MHz 1H frequency with an MAS rate of 11 kHz.
The PP Motif Regions of SBP and PNT1 Form Amyloids.
The PP motif was also found at the C terminus of SBP encoded by the gene adjacent to hellp (Fig. 1). Recombinant SBP(219-280) also formed fibrils in vitro with a parallel β-sheet FTIR signature and a proteinase K-resistant core encompassing residues 235–277 (Fig. S4 E and F and Fig. S5). A 26-aa peptide corresponding to the PP motif of PNT1 [PNT1(23-48)] also formed fibrils, as did two other PP-related peptides, corresponding to a PP motif consensus sequence and to the C-terminal part of the RHIM motif of human RIP3 [RIP3(245-270)] (Fig. S4 G and H).
HELLP Targets SBP to the Plasma Membrane.
We also analyzed in vivo aggregation of SBP and the SBP/HELLP interaction. Full-length SBP and the region encompassing SBP(219-280) were expressed in vivo as GFP fusion proteins. SBP(219-280)-GFP led to the formation of dot-like aggregates (Fig. S7A). For full-length SBP-GFP, diffuse cytoplasmic localization and dot formation was detected, suggesting that full-length SBP also can be converted to an aggregated state (Fig. S7A). To explore a possible interaction between HELLP and SBP, a strain expressing HELLP-RFP was confronted to a strain expressing SBP-GFP and displaying dots. A barrage formation was observed in the confrontation zone, indicative of a cell death reaction (Fig. S7B). In HELLP/SBP fusion cells, relocalization of both SBP and HELLP from the cytoplasm to the membrane was observed (Fig. S7C), suggesting that SBP aggregates are able to activate HELLP, and that HELLP allows targeting of SBP at the membrane. The combined and synchronized activation of HELLP and SBP lipase, together with the possibility of colocalizing SBP with the membrane-bound HELLP, might synergize membrane disruptive activity.
Fig. S7.
Expression of SBP-GFP in P. anserine. (A) P. anserina strains expressing SBP-GFP or SBP(219-280)-GFP and showing dot formation. (Scale bar: 5 μ.) (B) Strains expressing SBP-GFP produce a barrage reaction to strains expressing HELLP-RFP (white arrowheads). The control strain is an untransformed ΔPa_5_8070 strain. (C) In fusion cells between strains expressing HELLP-RFP and SBP-GFP and displaying SBP-GFP aggregates, SBP and HELLP colocalize in the plasma membrane region. The recipient strain background is ΔPa_5_8070. The arrows point to the presumed fusion points between the two strains. (Scale bar: 5 μ.)
The HELL Domain Is Homologous to the Pore-Forming Domain of MLKL.
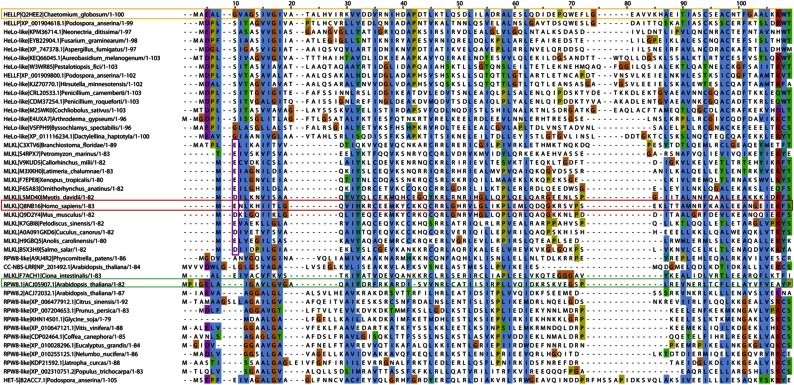
The HELL domain occurs in fungi as an N-terminal domain of NLRs or associated with amyloid-forming motifs (17, 19). To identify distant homologs of HELL outside of the fungal kingdom, we performed profile hidden Markov model searches and found hits in chordate MLKL, the cell death execution protein in mammalian necroptosis, the target of a phosphorylation by the RIP1/RIP3 complex (16, 23–26). The homology between HELLP and MLKL lies in the N-terminal 4HB domain responsible for cell death execution by membrane permeation. In HHPred searches, human MLKL was the first hit to HELLP, and HELLP scored significantly better to MLKL than to HET-S (P = 6.8 ×10−7 to MLKL and 9.2 × 10−4 to HET-S). Fig. 5 shows the alignment of fungal HELL domain proteins and chordate MLKL homologs.
Fig. 5.
Homology between the HELL domain and the pore-forming domain of MLKL. Shown is alignment of the HELL domain of HELLP (and various fungal homologs) with the 4HB domain region of various MLKL homologs from different phylogenetically diverse chordate species. The C. globosum HELLP sequence is boxed in orange, and the human MLKL sequence is boxed in red. The secondary structure of human MLKL (after PDB ID code Q8NB16) is given below the alignment. Alignment was generated with MAFFT with default settings.
Sequence homology began in the N terminus in the region predicted as TMH in HELLP, and then roughly correlated with the secondary structure elements of the 4HB region of MLKL. Conservation of a negatively charged residue in position 2 or 3 was common to MLKL, HeLo, and HELL domains. Homology between HELL and N-terminal domains of plant NLRs was detected as well. These N-terminal domains are of the non-TIR, CCR-type and are related to the RPW8 domain mediating cell death and membrane targeting (27). HELL, HeLo, and CCR (RPW8) domains occur in similar domain architectures particularly as N-terminal domains of NLRs, but also associated with kinase domains, with the latter architecture representing the MLKL domain architecture. As observed in the MLKL and HELL sequences, a negatively charged residue was frequently found close to the N termini in RPW8 sequences (Fig. S8). In HELL- and RPW8-related sequences, the region corresponding to the predicted TMH helix was similar in length, and both types of sequences contained GXXXG-type motifs. This region was shorter in the MLKL sequences. These analyses revealed homology among the chordate MLKL 4HB domain, the fungal HELL domain, and plant RPW8-related domains, three types of domains that have membrane-targeting activity in host-defense related processes.
Fig. S8.
Alignment of chordate MLKL homologs, plant RPW8 domain proteins, and fungal HeLo-like domain proteins. Phylogenetically diverse chordate MLKL homologs, plant RPW8 proteins, and fungal HeLo-like domain protein sequences were in alignment with MAFFT with default settings. The HELLP, mouse MLKL, and Arabidopsis RPW8.1 sequences are boxed in orange, red, and green, respectively. Conserved negatively charged residues at position +2 in MLKL homologs are boxed in purple.
Discussion
HET-S is a cell death-execution protein regulated by amyloid templating (11). We have proposed the existence in fungi of other cell death-effector proteins activated by amyloid signaling (12). Here we have characterized one such protein, HELLP, displaying an N-terminal HELL domain and a C-terminal PP motif. Like het-S, the gene encoding HELLP lies adjacent to a gene encoding an NLR with an N-terminal amyloid-forming motif. We found that the HELL domain is also an amyloid-controlled membrane-targeting cell death-inducing domain. Based on the similarity between HELLP and HET-S, it is reasonable to propose that the HELL domain also might function as a pore-forming domain, and that this activity involves the predicted TMH region. Like the HET-s amyloid motif, the PP motif should be viewed as a functional amyloid under evolutionary constraints to optimize and maintain function.
Of note is the resemblance of the PP motif to the RHIM motif (15). The PP and RHIM motifs share a pseudopalindromic organization centered on a Q residue, and both obey the general expression N-(x)1–2-G-φ-Q-φ-G-(x)1–2-N, raising the possibility of long-term evolutionary conservation of functional amyloid motifs. However, owing to the low sequence complexity and short size of these motifs, an alternate scenario of convergent evolution toward an amyloid-forming ability cannot be ruled out. The RHIM motif has been proposed to be related to the HET-s motif (20). Compared with the HET-s motif, the PP motif has greater sequence similarity to RHIM. Moreover, like RHIM and unlike HET-s, the PP motif occurs as a single motif rather than as two pseudorepeats.
The HELL domain exhibits homology to the 4HB domain of MLKL, responsible for membrane targeting in necroptotic cell death. Thus, mammalian necroptosis and this form of fungal PCD appear to be evolutionarily related and to use conserved protein domains for cell death execution. In both mechanisms, a terminal cell death execution domain functions by altering plasma membrane integrity (and the PCD signaling cascade relies on formation of amyloid complexes). Necroptosis and fungal PCD involving HeLo or HELL proteins appear to be related processes, indicating that this form of cell dismantle is evolutionarily ancient and plays a central role in the control of cell fate and defense from fungi to humans and plants. The current mechanistic models for MLKL and HeLo domain membrane targeting are incomplete but contrasting. In the case of the HeLo domain, the N-terminal TMH is proposed to be a central element in membrane binding and disruption, and in the case of the MLKL domain, it is reported that most of the 4HB domain inserts into the membrane (9, 26). Future studies are needed to determine the mechanistic similarities and differences in membrane alteration by MLKL, HeLo, and HELL domain proteins.
Materials and Methods
Prion Propagation and Incompatibility Assays.
Incompatibility phenotypes were determined by confronting strains of solid corn meal agar medium. Prion propagation was assayed as the ability to transmit the [π] prion from a [π]-donor strain to a [π*] prion-free tester strain after confrontation on solid medium. Protein transfection experiments with amyloid fibrils of recombinant HELLP(215-278) protein were performed by spotting 100 μL of a 1 mg mL−1 HELLP(215-278) fibril suspension onto a [π*] mycelium. The mycelium was then cut with a scalpel blade, and the regenerating mycelium was tested for the ability to produce a barrage reaction to a strain expressing full-length HELLP.
Microscopy.
P. anserina hyphae were inoculated on solid medium and cultivated for 24–72 h at 26 °C. The medium was then cut out, placed on a glass slide, and examined with a Leica DMRXA microscope equipped with a Micromax CCD (Princeton Instruments) controlled by Metamorph 5.06 software (Roper Scientific). The microscope was fitted with a Leica PL APO 100× immersion lens.
SSNMR Spectroscopy.
SSNMR experiments were performed with 300- and 800-MHz 1H Larmor frequency spectrometers (Bruker Biospin) using 3.2- and 4-mm MAS probes, respectively, filled with ∼7 mg and ∼14 mg of fibril samples. The MAS frequency was set between 7 and 20 kHz. Sample temperature was set to 6 °C with the internal reference DSS (28). A ramped CP with a 1-ms contact time was used for the 1H-13C polarization transfer. The 1D 1H-13C CP was recorded at 800 MHz 1H Larmor frequency with an MAS frequency of 20 kHz. An acquisition time of 20 ms was used for 512 scans, with an interscan delay of 3 s. The 2D 13C-13C PDSD spectra were recorded at 300 MHz 1H Lamor frequency with an MAS rate of 11 kHz . 13C-13C polarization transfer was achieved with PDSD with mixing times of 30 ms to correlate intraresidue carbon atoms and 150 ms to connect sequential atoms. Proton decoupling with a frequency of 85 kHz was applied during acquisition times, using the SPINAL-64 decoupling sequence (29). Acquisition times of 20 ms (t1) × 24 ms (t2) (for the 30-ms mixing time) and 17 ms (t1) × 24 ms (t2) (for the 150-ms mixing time) were chosen for the indirect and direct dimensions, respectively, resulting in a total experiment time of ∼6.5 d. A 1D trace was extracted at the positions of the Thr resonances, indicated by a dashed blue line in Fig. 4C, from the 2D PDSD (for the 150-ms mixing time). Mobile residues were probed using a 2D 1H-13C INEPT experiment recorded at 300 MHz 1H Lamor frequency for a total experiment time of ∼2 d. Random coil chemical shifts for the secondary chemical shift calculation were as reported previously (30). Spectra were analyzed and figures prepared using the CCPNMR analysis software (31).
SI Materials and Methods
Strains, Plasmids, and Media.
The P. anserina strains used in this study were wild-type het-s, het-S, and het-s° strains and ΔPa_5_8070. Pa_5_8070 encodes a HELLP ortholog in P. anserina, and this background was used to avoid potential interference with expression of C. globosum HELLP. The ΔPa_5_8070 strain was obtained after disruption of the gene Pa_5_8070 and replacement of its ORF with the nourseothricin-resistance gene nat. The growth medium for barrage assays and prion transmission assays was standard corn meal agar DO medium. The ΔPa_5_8070 or het-s° strain was used as a recipient strain for the expression of our molecular constructs. Strains were transformed by a pGEM-T (Promega)-derived vector, and expression of the molecular fusion was under the constitutive gpd promoter. The hellp gene was amplified with oligonucleotides 5′-GCTTAATTAAATGGCCGAAGCTCTTGGTGTTGC-3′ and 5′-ATCGTTAACTCCAAAGCGAAGGTTGTTC-3′. The hellp(172-278) gene fragment was amplified with oligonucleotides 5′-GCTTAATTAAATGGCCGCTTTGAATAGCCGTGTTG-3′ and 5′-ATCGTTAACTCCAAAGCGAAGGTTGTTC-3′. The PCR products were cloned first in a TOPO-XL-PCR plasmid (Invitrogen) by a TA cloning and RFP PCR product amplified with oligonucleotides 5′-ATGGTTAACAGATCTATGGCCTCCTCCGAGGACG-3′ and 5′-TTGCGGCCGCTTAGGCGCCGGTGGAGTGGCG-3′ was cloned downstream to them using HpaI/NotI restriction sites. The fusion genes hellp-RFP and hellp(172-278)-RFP were the digested and ligated to the pOP plasmid using the PacI/NotI restriction sites, creating the plasmids pOP-HELLP-RFP and pOP-HELLP(172-278)-RFP. The fragment hellp(215-278) was cloned downstream of the GFP encoding sequence using the NotI/BamHI restriction sites to generate the plasmid pGB6-HELLP(215-278)-GFP. The same types of molecular fusions were created for the sbp gene and sbp(219-280). The sbp sequence was amplified with oligonucleotides 5′-TGCAAAGCGCGGCCGCATGCGCTCCAAGATTGTCCGTG-3′ and 5′-GAAAATATGGATCCTCACCCCCGAGGTTTGACCC-3′ and cloned downstream of the GFP to generate the plasmid pGB6-SBP-GFP. The plasmid pGB6-SBP(219-280)-GFP was generated by an inverse PCR reaction.
The plasmids pET24-HELLP(215-278)6His and pET24-SESBpp(219-280)6His were used for protein expression in E.coli. The gene fragment hellp(215-278) was amplified with oligonucleotides 5′-AGCCATATGAAAGCCCTGGCGGCTTCG-3′ and 5′-ATCTCGAGTCCAAAGCGAAGGTTGTTC-3′, and the PCR product was cloned into the pET24a expression vector (Novagen) using the NdeI/XhoI restriction sites. The gene fragment sesbpp(219-280) was amplified with oligonucleotides 5′-CATATGCGACGCATCTCGCGGGTTTTGG-3′ and 5′-CTCGAGCCCCCGAGGTTTGACCC-3′, and the PCR product was cloned into the pET24a expression vector (Novagen) using the NdeI/XhoI restriction sites.
Protein Preparation.
HELLP(215-278) and SBP(219-280) proteins were expressed in Escherichia coli BL21-CodonPlus-RP competent cells and purified using an ÄKTA purifier core system (GE Healthcare). Both proteins had a C-terminal 6-histidine tag and were expressed as insoluble proteins and purified under denaturing conditions using Qiagen columns. Yields were in the range of 10 mg L−1 of culture. Proteins were eluted in 8 M urea, 50 mM Tris⋅HCl pH 8, 150 mM NaCl, and 200 mM imidazole. Elution buffer was replaced by overnight dialysis at 4 °C in Milli-Q water. Synthetic peptides were synthesized at >99% purity by Genosphere Biotechnologies. Fibril formation was carried out at room temperature and occurred spontaneously after the synthetic peptide was solubilized in Milli-Q water at a concentration of 3 mg mL−1.
Proteinase K Digestion Assays.
Sample preparation for proteinase K digestion was as described above. The reactions were performed in 100 µL of Milli-Q water with 100 µg of protein incubated with 2.5 µg of proteinase K at 37 °C for various times ranging from 1 min to 30 min. The digestion was stopped by adding urea to a final concentration of 8 M. Protein samples were analyzed by SDS/PAGE using the Mini-PROTEAN Tetra System (Bio-Rad). Proteins were migrated onto 16% Tris-Tricine gels, followed by Coomassie blue staining.
Mass Spectrometry of HELLP Proteinase K Digestion Products.
A 30 μM solution of aggregated HELLP(215-278) was digested for 40 min at 37 °C with 2.5 μg mL–1 proteinase K in 50 mM Tris⋅HCl pH 8.0 and 150 mM NaCl. The reaction mixture was centrifuged for 10 min at 10,000 × g, after which the pellet fraction was resuspended in H2O. To dissociate the remaining aggregated material, powder urea was added to 80 µL of each sample to reach a final concentration of 8 M (approximate final volume, 110 µL). Then 30 µL of each sample was diluted in 30 µL of water, desalted, and concentrated onto a C18-reverse phase microchromatography column (ZipTip C18; Millipore). The elution was obtained with 3 µL of 70% acetonitrile in water in 0.1% aqueous trifluoroacetic acid. The uncleaved HELLP aggregated form and its proteinase K digestion products were analyzed separately on a MALDI-TOFTOF mass spectrometer (Ultraflex III; Bruker) operated in MS mode, with α-cyano-4-hydroxy-cinnamic acid (Sigma-Aldrich) as a matrix (5 mg/mL solution in 50% acetonitrinile in 0.1% aqueous trifluoroacetic acid). The dried droplet method was used for sample loading on stainless steel targets.
Monoisotopic and average masses of proteinase K digestion products were measured in the reflectron mode and the linear mode, respectively, using an internal calibration with a mixture of oxidized insulin B chain alkylated by iodoacetamide (theoretical monoisotopic mass, 3,511.75 Da) and insulin AB (theoretical monoisotopic mass, 5,729.6 Da) (Sigma-Aldrich). The MS/MS mode was used to confirm the sequence of N-terminal and C-terminal peptides produced by Endolysin C and trypsin proteolysis, respectively. The protein-containing slices were destained in a solution of 25 mM NH4HCO3/50% ACN and then rinsed twice in ultrapure water. They were then shrunk in 100% ACN for 10 min. ACN was removed, and the gel pieces were dried at room temperature, covered with the trypsin or endolysine C solution (10 ng/mL, in 40 mM NH4HCO3), rehydrated at room temperature for 10 min, and incubated overnight at 37 °C, with rotary shaking. The supernatants were collected, and an extraction solution of ACN/HCOOH/H2O (60:1:40, vol:vol:vol) was poured onto the gel slices, which were agitated for 15 min. This extraction step was repeated twice. The supernatants were pooled, concentrated in a vacuum centrifuge to a final volume of 25 µL, acidified by the addition of 1.5 mL of 5% HCOOH, and stored at −20 °C. These samples were analyzed by MS or MS/MS on the spectrometer described above.
Attenuated Total Reflectance-FTIR Assays.
The amyloid-like samples were previously lyophilized using a LyoQuest (Telstar). Then secondary structure analyses of the lyophilized amyloid fibers were performed by attenuated total reflectance (ATR) in conjunction with FTIR using a Bruker Tensor 27 FTIR spectrometer (Bruker Optics) with a Golden Gate MKII ATR accessory. Each spectrum consists of 20 independent scans, measured at a spectral resolution of 2 cm−1 within the range of 1,500–1,800 cm−1. All spectral data were acquired and normalized using Bruker OPUS MIR Tensor 27 software. Finally, PeakFit (Systat Software) peak separation analysis software was used to identify peak centers, followed by Gaussian fitting to quantify the deconvoluted peaks of lyophilized samples.
Diffraction Assays.
The amyloid fibers were lyophilized using a LyoQuest freeze dryer (Telstar). Then lyophilized powders were mounted on a 200-µm nylon loop and kept at room temperature (19 °C). The samples were mounted at 150 mm from a Mar 345dtb detector in a Rigaku 007 X-ray generator working at a wavelength of 1.541 Å. The samples were exposed for 600 s. The rotation angle during collection was 5°.
Bioinformatic Methods.
Sequence alignments were performed with Clustal Omega or MAFFT (www.ebi.ac.uk). To generate the HMM profile signatures for PP and RHIM, PSI-BLAST searches were performed (blast.ncbi.nlm.nih.gov/Blast.cgi) using the consensus PP sequence (15). The RHIM motif of human RIP3 and matching sequences were recover aligned with Clustal Omega, and the alignment was analyzed with Skylign (skylign.org). Secondary structure predictions were performed with PSIPRED (bioinf.cs.ucl.ac.uk/psipred/). Hidden Markov model searches were performed using HHPred (toolkit.tuebingen.mpg.de) and Jackhhmer (www.ebi.ac.uk/Tools/hmmer/search/phmmer), both with default settings.
Acknowledgments
We thank Dr. David Gajan for technical assistance with the NMR measurements. We acknowledge the support provided by the Crystallography Platform of the Institute of Molecular Biology of Barcelona, and particularly Dr. Joan Pous, for help with diffraction assays and spectra analyses. Financial support for this research from the TGIR-RMN-THC Fr3050 CNRS for conducting the research is gratefully acknowledged. This work was supported, in whole or in part, by L’Agence Nationale de la Recherche (Grants STANDPRION ANR-11-BSV8-0001, to S.J.S; ANR-13-PDOC-0017-01, to B.H.; and ANR-14-CE09-0020-01, to A.L.), IdEx Bordeaux (PEPS 2014, to A.L. and S.J.S.), Fondation pour la Recherche Médicale (Grant FRM-AJE20140630090, to A.L.), the FP7 Program (Grant FP7-PEOPLE-2013-CIG, to A.L.) and the European Research Council under the European Union's Horizon 2020 Program (Starting Grant Agreement 105945, to A.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522361113/-/DCSupplemental.
References
- 1.Dickman MB, Fluhr R. Centrality of host cell death in plant–microbe interactions. Annu Rev Phytopathol. 2013;51:543–570. doi: 10.1146/annurev-phyto-081211-173027. [DOI] [PubMed] [Google Scholar]
- 2.Blander JM. A long-awaited merger of the pathways mediating host defence and programmed cell death. Nat Rev Immunol. 2014;14(9):601–618. doi: 10.1038/nri3720. [DOI] [PubMed] [Google Scholar]
- 3.Pinan-Lucarré B, Paoletti M, Clavé C. Cell death by incompatibility in the fungus Podospora. Semin Cancer Biol. 2007;17(2):101–111. doi: 10.1016/j.semcancer.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Makarova KS, Anantharaman V, Aravind L, Koonin EV. Live virus-free or die: Coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Doorn WG. Classes of programmed cell death in plants, compared to those in animals. J Exp Bot. 2011;62(14):4749–4761. doi: 10.1093/jxb/err196. [DOI] [PubMed] [Google Scholar]
- 6.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319(5869):1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 7.Balguerie A, et al. Domain organization and structure–function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22(9):2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenwald J, et al. The mechanism of prion inhibition by HET-S. Mol Cell. 2010;38(6):889–899. doi: 10.1016/j.molcel.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seuring C, et al. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol. 2012;10(12):e1001451. doi: 10.1371/journal.pbio.1001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathur V, Seuring C, Riek R, Saupe SJ, Liebman SW. Localization of HET-S to the cell periphery, not to [Het-s] aggregates, is associated with [Het-s]-HET-S toxicity. Mol Cell Biol. 2012;32(1):139–153. doi: 10.1128/MCB.06125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daskalov A, et al. Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold. PLoS Biol. 2015;13(2):e1002059. doi: 10.1371/journal.pbio.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daskalov A, Paoletti M, Ness F, Saupe SJ. Genomic clustering and homology between HET-S and the NWD2 STAND protein in various fungal genomes. PLoS One. 2012;7(4):e34854. doi: 10.1371/journal.pone.0034854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156(6):1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153(2):287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150(2):339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148(1-2):213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 17.Daskalov A, Dyrka W, Saupe SJ. Theme and variations: Evolutionary diversification of the HET-s functional amyloid motif. Sci Rep. 2015;5:12494. doi: 10.1038/srep12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graziani S, Silar P, Daboussi MJ. Bistability and hysteresis of the “Secteur” differentiation are controlled by a two-gene locus in Nectria haematococca. BMC Biol. 2004;2:18. doi: 10.1186/1741-7007-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyrka W, et al. Diversity and variability of NOD-like receptors in fungi. Genome Biol Evol. 2014;6(12):3137–3158. doi: 10.1093/gbe/evu251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajava AV, Klopffleisch K, Chen S, Hofmann K. Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci Rep. 2014;4:7436. doi: 10.1038/srep07436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller BK, Subramaniam S, Senes A. A frequent, GxxxG-mediated, transmembrane association motif is optimized for the formation of interhelical Cα-H hydrogen bonds. Proc Natl Acad Sci USA. 2014;111(10):E888–E895. doi: 10.1073/pnas.1319944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabaté R, et al. Prion and non-prion amyloids of the HET-s prion forming domain. J Mol Biol. 2007;370(4):768–783. doi: 10.1016/j.jmb.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Murphy JM, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39(3):443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 24.Cai Z, et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol. 2014;16(1):55–65. doi: 10.1038/ncb2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24(1):105–121. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su L, et al. A plug release mechanism for membrane permeation by MLKL. Structure. 2014;22(10):1489–1500. doi: 10.1016/j.str.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collier SM, Hamel LP, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact. 2011;24(8):918–931. doi: 10.1094/MPMI-03-11-0050. [DOI] [PubMed] [Google Scholar]
- 28.Böckmann A, et al. Characterization of different water pools in solid-state NMR protein samples. J Biomol NMR. 2009;45(3):319–327. doi: 10.1007/s10858-009-9374-3. [DOI] [PubMed] [Google Scholar]
- 29.Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson. 2000;142(1):97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci. 2002;11(4):852–861. doi: 10.1110/ps.3180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]