Significance
Here we provide what is, to our knowledge, the first gene map of the type I IFN region of any bat species with the sequence of the type I IFN locus of the Australian black flying fox, Pteropus alecto. The bat IFN locus contains fewer IFN genes compared with any other mammal sequenced to date, including only three IFN-α genes. We also demonstrate that bat IFN-α genes are constitutively expressed in unstimulated bat tissues and cells and that their expression is unaffected by viral infection. This unusual pattern of IFN-α expression has not been described in any other species to our knowledge and has important implications for the role of innate immunity in the ability of bats to coexist with viruses in the absence of disease.
Keywords: interferon, innate immunity, bat immunology
Abstract
Bats harbor many emerging and reemerging viruses, several of which are highly pathogenic in other mammals but cause no clinical signs of disease in bats. To determine the role of interferons (IFNs) in the ability of bats to coexist with viruses, we sequenced the type I IFN locus of the Australian black flying fox, Pteropus alecto, providing what is, to our knowledge, the first gene map of the IFN region of any bat species. Our results reveal a highly contracted type I IFN family consisting of only 10 IFNs, including three functional IFN-α loci. Furthermore, the three IFN-α genes are constitutively expressed in unstimulated bat tissues and cells and their expression is unaffected by viral infection. Constitutively expressed IFN-α results in the induction of a subset of IFN-stimulated genes associated with antiviral activity and resistance to DNA damage, providing evidence for a unique IFN system that may be linked to the ability of bats to coexist with viruses.
Bats harbor a number of emerging and reemerging viruses, many of which are highly pathogenic in humans and other species, including henipaviruses (Hendra and Nipah), coronaviruses (SARS-CoV), rhabdoviruses (rabies and lyssaviruses), and filoviruses (Ebola and Marburg), but cause no clinical signs of disease in bats (1). In addition, bats are capable of clearing experimental infections in vivo with henipaviruses and lyssaviruses at doses of infection that are lethal in other mammals (2, 3). The mechanisms responsible for the ability of bats to coexist with viruses remain poorly understood (4).
The interferon (IFN) system provides the first line of defense against viral infection in vertebrates. There are three types of IFNs in mammals, designated types I, II, and III, which differ in their amino acid sequences and the receptor complex they signal through. Type I and type III IFNs are induced directly in response to viral infection and are key cytokines capable of inducing an “antiviral state” in infected and neighboring cells. Type I IFNs include IFN-α, IFN-β, IFN-ω, IFN-ε, IFN-ζ, IFN-κ, IFN-τ, and IFN-δ, that signal through the IFN-α receptor (IFN-αR) that consists of IFN-αR1 and IFN-αR2 chains (5).
All type I IFN genes (with the exception of IFN-κ) are located within the boundaries of IFN-β and IFN-ε, which spans ∼400 kb in humans and 360 kb in mice (6–9). Among type I IFNs, IFN-α and IFN-β proteins account for the majority of the antiviral response generated following viral infection (10). IFN-α and IFN-β expression are normally undetectable in the absence of infection but are rapidly induced following viral infection or treatment with synthetic ligands, including dsRNA (11). A low level of constitutively expressed IFN-α mRNA has been detected in humans and germ-free mice (12–14). In humans, IFN-α1 and IFN-α2 transcripts are present in normal spleen, liver, and kidney (13). However, as the spontaneous IFN-α and IFN-β proteins are expressed at very low levels, even the most sensitive assays fail to detect them (15). In addition to direct antiviral activity (although at a very low level), constitutively expressed IFN-α is believed to play a role in priming the IFN response, rendering cells “ready to go” by stimulating amplified IFN-α/β production in response to viral infection and enhanced responses to other cytokines (16, 17). In the promoter regions of human IFN-α genes, three modules that are responsible for binding to IFN regulatory factors (IRFs) 3 and 7 determine the induction profile of different IFN-αs. For constitutively expressed human IFN-α1, it is believed that binding of IRF3 to the unique module II (also called module C) in the promoter region leads to weak endogenous expression. The promoter regions of all other human IFN-α genes (except IFN-α13) use modules I and III for binding to IRF3 or IRF7, respectively (18, 19).
IFN-α and IFN-β proteins bind to the IFN-αR and trigger the phosphorylation of STAT1 and STAT2, which then forms a ternary complex with IRF9 to form the tripartite transcription factor IFN-stimulated gene (ISG) factor 3 (ISGF3) and drives the expression of ISGs (5). However, continuous exposure of cells to a low level of IFN-β, which often occurs in cancers, leads to steady-state increased expression of an unphosphorylated form of ISGF3 (U-ISGF3), which in turn leads to the expression of a subset of ISGs also induced by ISGF3. This response can extend resistance to virus infection and render cells resistant to DNA damage (20). The U-ISGF3–associated ISGs are driven by distinct IFN-stimulated response elements and include Mx1, ISG15, and STAT1 (20).
Few studies have been performed to understand the mechanisms responsible for the ability of bats to coexist with viruses. The sequencing of two bat genomes (Pteropus alecto and Myotis davidii) has revealed several genes involved in the DNA repair and innate immunity pathways that have undergone positive selection in bats compared with other mammals, providing evidence that the evolution of flight could have had inadvertent consequences for the innate immune system of bats (21). Studies have also inferred the existence of seven IFN-α genes in Pteropus vampyrus (22), eight IFN-α subtypes (or alleles) in Dobsonia viridis, and one IFN-ω and IFN-κ in Eptesicus serotinus (23, 24). However, as only low-coverage bat genomes have been used to identify IFNs for these studies, the exact genome structure of type I IFN family members is yet to be confirmed. Current knowledge on bat type I IFN responses is also very preliminary, with descriptions of type I and III IFN induction following polyinosinic:polycytidylic acid (polyI:C) stimulation of bat cells (25). Evidence for unique expression patterns of IFN-related genes have also been described in P. alecto, including the constitutive expression of IRF7 and a wider distribution of the type III IFN receptor consistent with the constitutive activation of some aspects of the innate immune system (26, 27). In this study, we report what is, to our knowledge, the first systematic characterization of the bat type I IFN locus and comparison with other species. We also describe the unique constitutive expression of IFN-α and ISGs in unstimulated bat tissues and cells, a finding that may have implications for the ability of bats to coexist with viruses in the absence of disease.
Results
Sequencing and Annotation of P. alecto Type I IFN Genomic Locus.
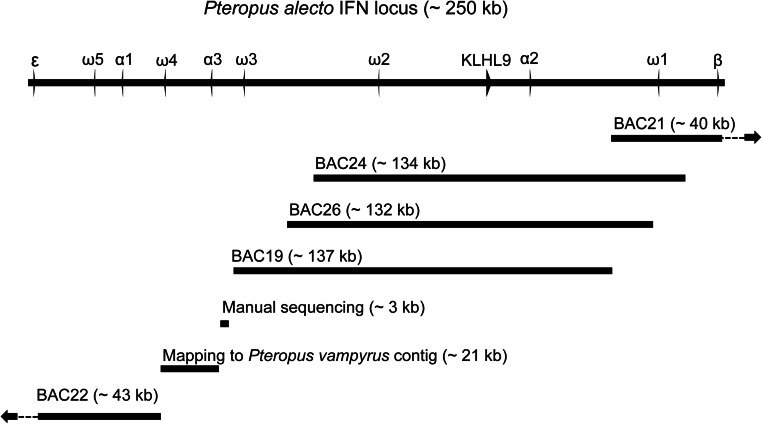
Two scaffolds (scaffold95 and scaffold222) corresponding to the partial type I IFN locus were identified in the P. alecto whole-genome sequence (21). Scaffolds 95 and 222 span 25.14 Mb and 4.344 Mb, respectively, and each contains type I IFN genes. However, these scaffolds did not overlap and therefore did not cover the entire type I IFN locus. To obtain the complete sequence of the type I IFN locus, a P. alecto BAC library was used to identify the remaining type I IFN region. The BAC library was screened with overgo probes corresponding to IFN-β, IFN-ε and kelch-like 9 (KLHL9), yielding a total of seven BACs corresponding to the IFN region. BAC end sequences were determined for the positive BAC clones using Sanger sequencing to determine whether any of the BAC clones overlapped with each other or with the genomic scaffolds. A total of five BAC clones were chosen for further long-read pyrosequencing and analysis. The five positive BAC clones were assembled into a single scaffold 433 kb in length with a gap of 21 kb, which was filled by cloning (3 kb) and using data from the closely related bat, P. vampyrus (scaffold 12130) and raw reads from the P. alecto genome (Fig. S1). No IFN genes were identified in the region 89 kb upstream of IFN-β (proximal 5′ end) or 94 kb downstream of IFN-ε (most 3′ end).
Fig. S1.
Assembly and composition of sequences used to construct the P. alecto IFN locus. ORFs within the IFN locus are shown as arrows. The image is drawn to scale.
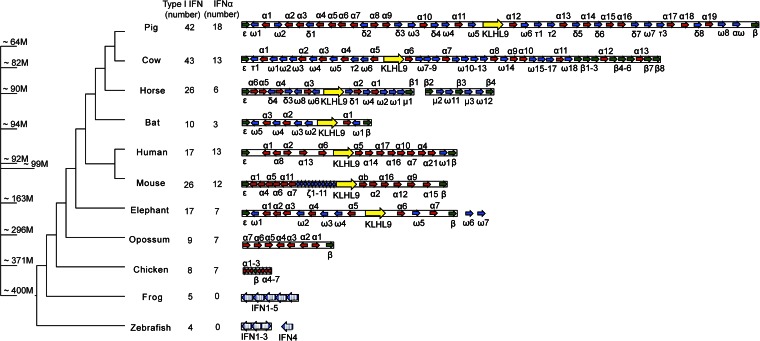
The P. alecto type I IFN locus was compared with the corresponding region from the genomes of 10 other vertebrates. The size of the IFN locus ranged from ∼25 kb in fish to 1 Mb in pig, with a trend toward increasing size through evolution as shown in Fig. 1. The only two exceptions were chicken and bat, both of which have shorter IFN loci of 30 kb and 250 kb, respectively.
Fig. 1.
Vertebrate type I IFN gene family among species. Type I IFN loci in selected vertebrate species (loci drawn to scale). IFN genes are annotated and labeled (not drawn to scale). The blocked arrows represent IFN ORFs, and directions indicate strand of the genes. IFN-α (red), IFN-ε, and IFN-β (green), other intron-less type I IFNs (blue), and the non-IFN gene, KLH9 (yellow), are shown. The intron-containing fish and frog IFNs are shown in large blue blocked arrows, with white columns to indicate the exon/intron boundaries. The unplaced IFN containing fragments outside the major IFN locus for some species are also shown. The numbers of type I IFNs (including IFN-κ) and IFN-α counts for each species are shown on the right. The phylogenetic tree on the left was drawn according to TimeTree, and the approximate divergence times are labeled (M, million years) (38).
A total of 10 genes with intact ORFs, including three IFN-α genes, one IFN-β, one IFN-ε gene, and five IFN-ω genes, were identified in the assembled bat IFN scaffold. In addition, a single copy of IFN-κ was found on a separate scaffold in the P. alecto genome (scaffold 14). The pattern of a single copy of IFN-κ and IFN-ε is conserved across all species (8). Consistent with the expansion in the genomic size of the IFN locus, gene duplication has occurred in the vertebrate type I IFN family in a step-wise manner, from only four type I IFNs at the basal branch such as in fish to 42 in pig. However, bats do not follow this trend and have only 10 type I IFN loci, three of which are IFN-α genes. Of the species that contain IFN-α genes, bats have the fewest IFN-α family members compared with any other mammalian genome studied (Fig. 1).
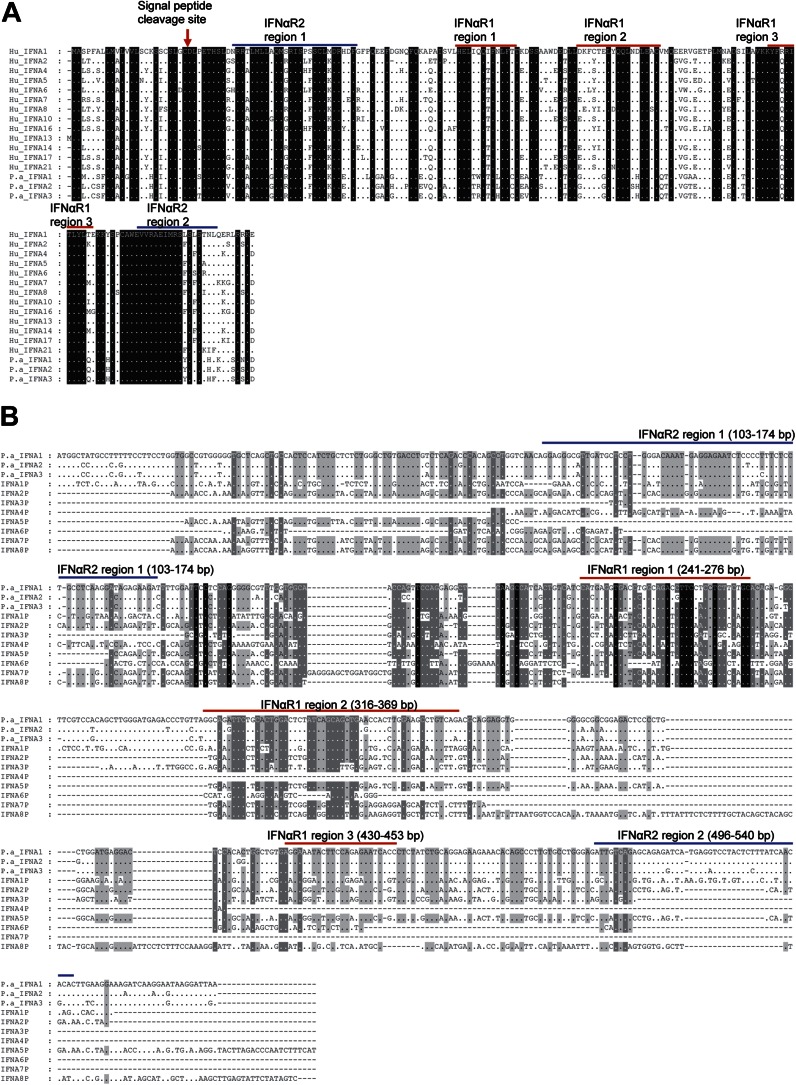
The deduced amino acid sequences of the three bat IFN-α genes share a number of features with IFN-α genes from humans and other mammals, including predicted signal peptides and conserved binding domains for IFN-αR1 and IFN-αR2 for activation of downstream signaling (Fig. S2A). They share 93–96% similarity to each other and 79–85% similarity to human IFN-α genes at the amino acid level.
Fig. S2.
(A) Alignment of P. alecto IFN-α putative protein sequences with human IFN-α proteins. The putative signal peptide cleavage site is indicated with a red arrow (prediction from CBS prediction server; www.cbs.dtu.dk/services/). (B) Alignment of P. alecto pseudotyped and functional IFN-α genes. Alignment was performed by using ClustalX and visualized by using Genedoc. Dots indicate similarity; dashes indicate gaps. The conserved regions that are predicted to be responsible for IFN-αR binding are also shown. The prediction was performed as published by Thomas et al. (53).
The bat IFN locus contains an additional eight IFN-α loci that appear to be pseudogenes (IFN-αP). This number is larger compared with humans or mice, which each have five IFN-αPs. Nucleotide alignment of the bat IFN-αP sequences with the three presumably functional IFN-α genes show that many of the IFN-αPs contain conserved partial IFN-αR binding domains, consistent with the likelihood that they once encoded functional IFN-α proteins (Fig. S2B).
Evolution of Bat IFN-α Families.
IFN-α and IFN-ω shared a common ancestor ∼130 Mya and are interspersed with each other on the mammalian IFN locus (28) (Fig. 1). To determine the evolutionary pressures responsible for the diversification of type I IFN genes, we performed an evolutionary analysis of IFN-α and IFN-ω families across eight mammalian species. The ratio of nonsynonymous (dN) to synonymous (dS) changes (dN/dS ratio) was measured to examine the selection pressures on the bat IFN-α and IFN-ω genes. For the bat ancestor, the dN/dS ratio was 0.54, which is similar to the selection pressures on ancestral IFN-α genes from other species including pigs (0.58), horses (0.34), humans (0.24), and mice (0.85). The purifying selection of bat IFN-α genes indicates its functional conservation and importance. Interestingly, positive selection for IFN-ω was observed at the ancestor of bats (dN/dS ratio = 1.07), and its selection pressure was higher than any nonbat mammalian type I IFN genes.
IFN-α Maintains a Constitutive and Ubiquitous Expression Pattern in Bat Tissues and Cells.
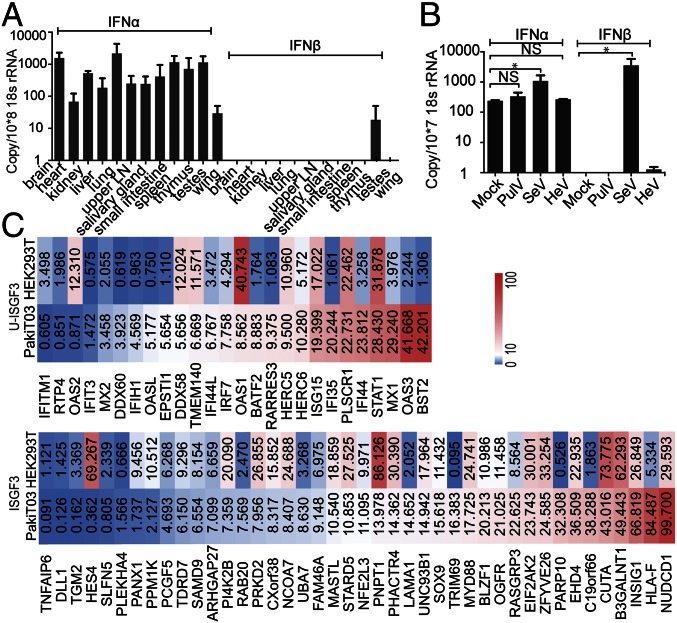
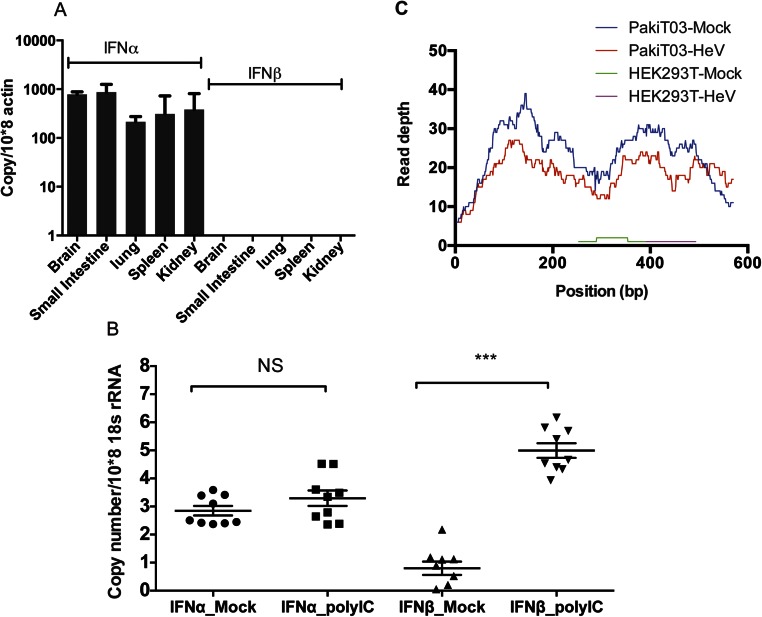
We then examined the IFN-α mRNA expression in comparison with IFN-β in tissues from three apparently healthy wild-caught P. alecto bats. As shown in Fig. 2A, P. alecto IFN-β was undetectable across all tissues tested with the exception of testes. In contrast, primers capable of detecting all three bat IFN-α genes demonstrated significant expression of IFN-α in all bat organs tested, with lung and brain the highest and wing the lowest (Fig. 2A). To determine whether the constitutive expression of IFN-α is P. alecto-specific, a second bat species, the lesser short nosed fruit bat (Cynopterus brachyotis) was also tested. Similar to the P. alecto tissues, IFN-α was expressed constitutively in tissues from C. brachyotis in contrast to undetectable levels of IFN-β across all tissues tested (Fig. S3A).
Fig. 2.
Bat IFN-α has a constitutive and ubiquitous expression pattern. qRT-PCR detection of IFN-α and IFN-β mRNA expression in 12 P. alecto tissues (n = 3) (A). LN, lymph node. The error bars represent SD. (B) Bat PulV, HeV, and SeV were used to infect PaKiT03 cells. IFN-α and IFN-β mRNA expression was detected 6 h post infection. Two-sample t tests assuming unequal variance were used to compare IFN expression in response to viral infection. Data represent the mean and SE from three experiments (*P < 0.05). (C) Transcription profile of selected ISGs in uninfected PaKiT03 and HEK293T cells. Data illustrate average normalized fragments per kilobase of transcript per million mapped reads (FPKM) across four RNAseq replicates in PaKiT03 cells compared with HEK293T cells. ISGF3, ISG genes that are induced only by ISFG3; U-ISGF3, ISG genes that are induced by unphosphorylated ISGF3 (20).
Fig. S3.
Bat IFN-α has a constitutive and ubiquitous expression pattern. (A) qRT-PCR detection of IFN-α and IFN-β mRNA expression in five C. brachyotis tissues (n = 2). The expression level was normalized to housekeeping gene actin. The error bars represent SD. (B) Expression of IFN-α and IFN-β mRNA following polyI:C stimulation of bat primary cell lines. The primary cells include lung, liver, heart, kidney, small intestine, brain, fetus, salivary gland, and muscle. Data represent the mean and SE of duplicates from each cell line. Two-sample t tests for unequal variances were used to compare the treated and mock-treated samples. NS, nonsignificant (***P < 0.001). (C) The transcription profile of IFN-α in PaKiT03 and HEK293T cells in mock samples or 8 h following HeV infection. Data show read counts per nucleotide for IFN-α.
The inducibility of bat IFN-α and IFN-β was then compared in primary cell lines derived from nine different P. alecto tissues before and after transfection with polyI:C for 3 h. The responses of primary cells confirmed our finding from the bat tissues demonstrating that IFN-α maintains a constitutive expression pattern in unstimulated bat primary cells. However, upon polyI:C treatment, IFN-α was not significantly induced. This is in clear contrast to IFN-β, which was highly induced in polyI:C-treated bat cells (Fig. S3B).
To examine the production patterns of bat IFN-α and IFN-β in response to viral challenge, we used two bat viruses and a mouse paramyxovirus to infect P. alecto kidney cell line PaKiT03 cells. Both Hendra virus (HeV) and Pulau virus (PulV) are bat-borne viruses carried by Pteropus bats. Sendai virus (SeV; Cantell strain) is a mouse paramyxovirus and is used in IFN research because of its ability to induce type I IFN through the production of defective interfering particles (29). Only SeV infection resulted in significant induction of IFN-β (P < 0.05). The absence of IFN-β induction is likely the result of antagonism of the IFN-β response by bat-borne viruses as reported previously for HeV (30). In contrast, IFN-α was significantly induced by SeV (P < 0.05) but to a lesser extent compared with the induction of IFN-β. Infection of bat cells with the two bat-borne viruses, HeV and PulV, caused no change in the constitutive IFN-α expression pattern (Fig. 2B). RNA sequencing (RNAseq) data available from HeV-infected human (HEK293T) and bat (PaKiT03) cells was used to confirm our findings (31). In bat cells, the constitutive IFN-α expression pattern was confirmed by using read depth counts of IFN-α transcripts in uninfected cells and showed little change following HeV infection. In contrast, few IFN-α transcripts were detected in infected or uninfected human cells (Fig. S3C). As a comparison, read mapping of RNAseq data from uninfected human and bat cells failed to detect IFN-β in either cell line. To confirm that the bat cells were not harboring an unrecognized infection, we used BLASTX to query the RNAseq data for the presence of sequences corresponding to known pathogens. Among the 64 million paired end reads in our dataset, no transcripts showed significant homology to known viruses or microbes. Even unknown viruses would be expected to show some sequence similarity to known virus families, as described previously for RNAseq data from bat tissues (32). This further supports our conclusion that the constitutive expression of IFN-α in bats is not associated with active viral infection.
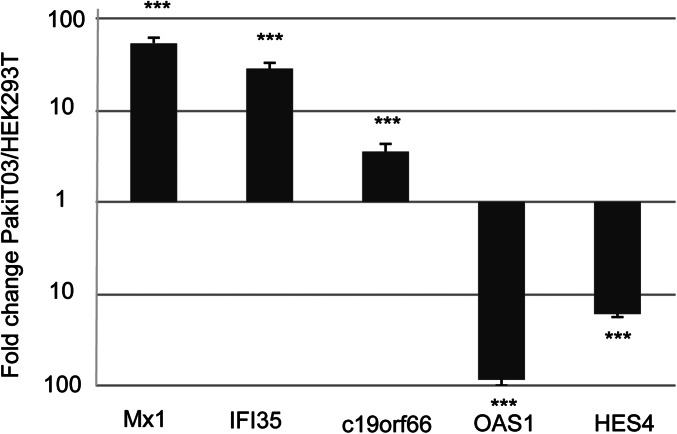
Although the constitutive expression of bat IFN-α at the protein level has not been confirmed as a result of the absence of a bat-specific antibody, a high level of IFN-α protein expression would be expected to lead to the induction of ISGs. In human cells, continuous IFN-β exposure has been shown to lead to steady-state induction of the U-ISGF3–dependent proteins, with no sustained increase in other IFN-β–induced proteins (20). To determine whether the constitutive expression of IFN-α in bat cells resulted in induction of U-ISGF3–associated genes, we compared the expression of ISGF3-dependent and U-ISGF3–dependent transcripts in RNAseq data from uninfected human (HEK293T) and bat (PaKiT03) cells. Previous analyses describing U-ISGF3 and ISGF3-induced ISGs in human cells were used as the basis for distinguishing bat ISGs in the present study (20). Expression was calculated using normalized read counts based on four replicates of RNAseq data from each cell line. Using a cutoff of 1.5-fold up-regulation between cell lines, 61.5% (16 of 26 genes) of U-ISGF3–dependent ISGs were expressed at a higher level in bat compared with human cell lines, compared with only 23.0% (6 of 26) that had higher expression in human cells. Conversely, 40.5% (17 of 42) of ISGF3-dependent ISGs displayed higher expression in human compared with bat cells, and only 33.3% (14 of 42) were higher expression in bat cells (Fig. 2C). The U-ISGF3–associated ISGs with the highest expression in bats included well-known antiviral proteins including bone marrow stromal cell antigen 2 (BST2; also known as tetherin) and Mx1. The expression of a subset of genes that were up-regulated in either bat or human cells was validated by using quantitative RT-PCR (qRT-PCR), confirming the pattern obtained from the RNAseq dataset (Fig. S4).
Fig. S4.
qRT-PCR detection of Mx1, IFI35, c19orf66, OAS1, and HES4 mRNA expression in uninfected PaKiT03 and HEK293T cells. Gene expression was calculated and compared by using the standard curve methods. The expression level was normalized to the housekeeping gene 18s rRNA. All values are the mean of at least three independent experiments, and error bars indicate SDs. Data represents the ratio of fold change in mRNA expression in PaKiT03 cells relative to HEK293T cells (***P < 0.001, one-tailed Student t test).
IFNα2 and IFNα3 Are the Main Constitutively Expressed Bat IFNs.
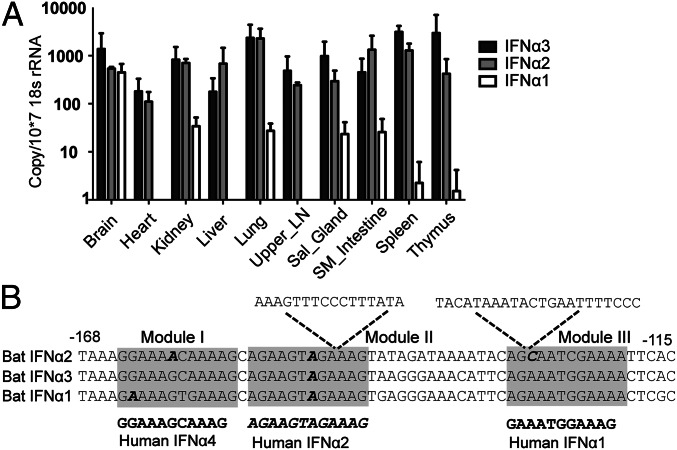
To test which bat IFN-α gene is constitutively expressed, TaqMan quantitative PCR (qPCR) assays were used to distinguish the three bat IFN-α genes in P. alecto tissues. IFN-α distribution among bat organs from three individual bats demonstrates that IFN-α2 and IFN-α3 are constitutively expressed in all organs tested, whereas IFN-α1 is expressed to a lesser extent and only in a subset of tissues. IFN-α2 and IFN-α3 displayed a similar expression pattern across most organs with the exception of the thymus, in which IFN-α3 was higher (Fig. 3A). These data confirm that P. alecto has three expressed IFN-α genes, of which IFN-α2 and IFN-α3 contribute to the majority of the constitutive expression of IFN-α.
Fig. 3.
IFN-α2 and IFN-α3 maintain a constitutive expression pattern. (A) IFN-α subtype mRNA expression in 10 P. alecto bat organs (n = 3) in sequence-specific TaqMan qPCR. The expression was normalized to the housekeeping gene 18S rRNA. The error bars represent SD. Sal_gland, salivary gland; SM_intestine, small intestine. (B) Sequence comparison of putative bat IFN-α gene promoters. The two sequence insertions in bat IFN-α2 promoter are indicated. Three IRF binding modules that are important for human IFN-α induction were predicted in the bat promoter regions (shadowed) (18). The human reference modules are shown in bold and the reported nonfunctional module is in bold and italic (19).
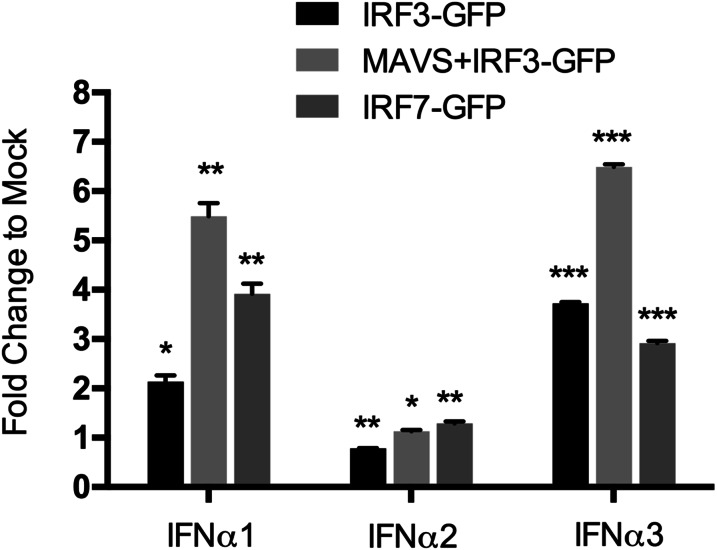
For human IFN-α1, the unique promoter structure, and the simultaneous recruitment of IRF3 with the transcriptional coactivators CBP and p300, leads to a weak expression of endogenous IFN-α1 (18). To explore whether constitutively expressed bat IFN-α genes also have unique promoters, we analyzed the proximal promoters of bat IFN-α genes. A region 200 bp upstream of the putative translation start which contains the three IRF binding modules in human and mouse IFN-α genes was chosen for this analysis (19, 33). The three modules were identified in bat IFN-α1 and IFN-α3, and modules I and III were conserved with those of functional human IFN-α genes whereas module II was identical to that of human IFN-α2, which is nonfunctional (19). In contrast, the bat IFN-α2 promoter contains mutations within module I and nucleotide insertions within modules II and III, which would render it unable to bind to IRFs (Fig. 3B). Promoter assays demonstrated that only IFN-α1 and IFN-α3 responded to IRF3 and IRF7, whereas IFN-α2 failed to respond even in the presence of mitochondrial antiviral-signaling protein (MAVS), which is known to stimulate IRF activation (19) (Fig. S5). These findings are consistent with bat IFN-α2 being regulated by factors other than IRF3 and IRF7 to maintain its constitutive expression pattern.
Fig. S5.
P. alecto IFN-α promoter luciferase assays. The putative bat IFN-α gene promoters (as indicated in Fig. 3B) were cloned into the pGL4.1 luciferase reporter plasmid. PaKiT03 cells were transiently transfected with 200 ng of expression plasmids for bat IRF3-GFP (with or without 10 ng bat MAVS-GFP) or IRF7-GFP with 100 ng bat IFN-α promoter plasmids and 50 ng pRL-Tk plasmid. GFP vector plasmid was used at 200 ng per well as a negative control. After 30 h, cells were analyzed for promoter activity by reporter gene assay. All values are the mean of at least three independent experiments, and error bars indicate SEs (*P < 0.05, **P < 0.01, and ***P < 0.001, two-tailed unpaired Student t test).
P. alecto IFN-α Proteins Are Functional.
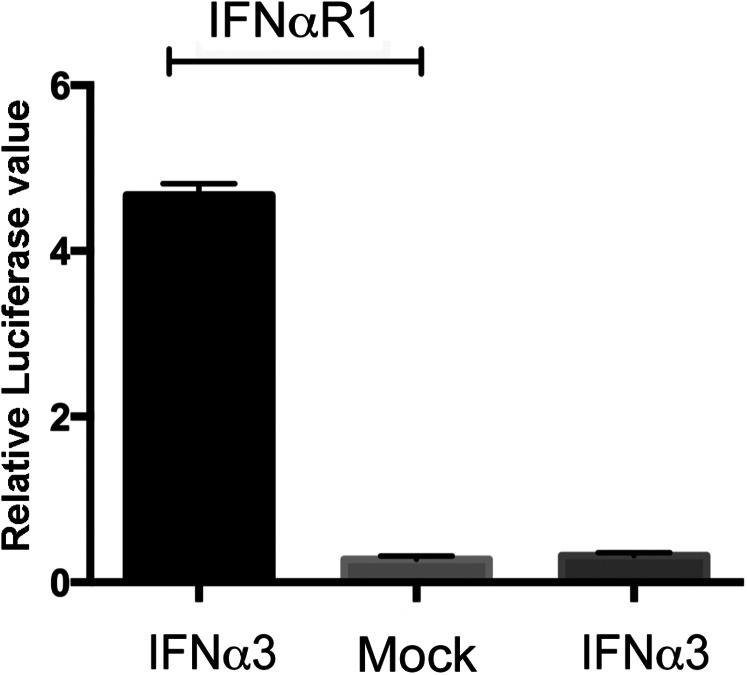
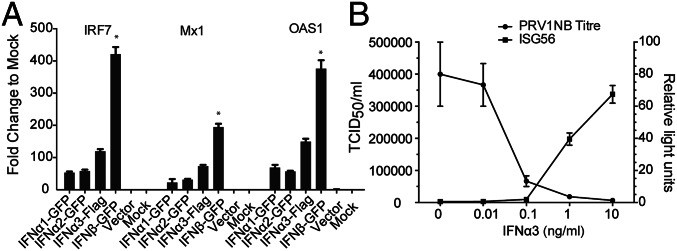
To assess the functionality of P. alecto IFN-α proteins, plasmids encoding the three individual IFN-α ORFs were transiently transfected into human HEK293T cells. We chose HEK293T cells because of their high transfection efficiency, and also because the human IFN-αR cannot respond to bat IFN and trigger downstream signaling (Fig. S6). Cell supernatant was collected as IFN-α conditioned medium after confirming the successful expression of each protein (Fig. S7). Bat IRF7, Mx1, and OAS1 were used as indicators of IFN-α functionality. Compared with untreated or vector mock-treated PaKiT03 cells, all three IFN-α proteins successfully induced ISGs, demonstrating that all three bat IFN-α proteins are potentially functional. As a positive control, recombinant bat IFN-β also induced ISG production in PaKiT03 cells (Fig. 4A). Although approximately similar quantities of each IFN protein were used, IFN-β resulted in higher ISG induction compared with the three IFN-α proteins (P < 0.05). This result may reflect a higher binding capacity to the IFN-αR as reported for human IFN-β (34).
Fig. S6.
Bat IFN-α signals through bat but not human IFN-αR. HEK293T cells were cotransfected with HISG54 promoter plasmid, RL-TK with or without bat IFN-αR1 expression plasmids. Twenty-four hours posttransfection, cells were mock-treated or treated with bat IFN-α containing HEK293T supernatant derived from bat IFN-α3 ectopically expressed in HEK293T cells. Another 6 h later, luciferase activity was measured. Data shows the mean of two experiments. Error bars indicate SDs.
Fig. S7.
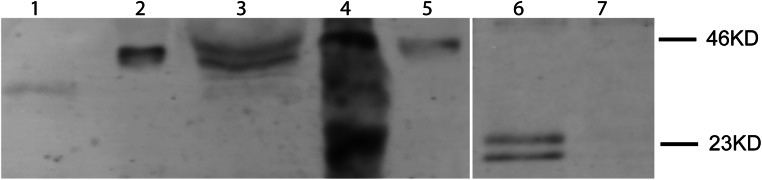
P. alecto recombinant IFN expression. Conditioned media from HEK293T cells transfected with plasmids encoding pCAGGS-GFP (GFP control, lane 1), pcDNA6.2/C-emGFP–IFN-α1 (lane 2), -IFN-α2 (lane 3), or -IFN-β (lane 5) were evaluated by immunoblotting with anti-GFP antibody. Conditioned media from HEK293T cells transfected with pCAGGS/FLAG-IFN-α3 plasmid (lane 6) or mock-transfected (lane 7) were evaluated by using anti-FLAG antibody. The molecular weight marker (lane 4) is shown with additional labeling in kilodaltons on the right. The approximate molecular weights of the proteins are as follows: GFP, 27 kDa; IFN-α1–GFP, 46 kDa; IFN-α2–GFP, 46 kDa; IFN-α3–FLAG, 23 kDa (migrating as a doublet as a result of posttranslational modification); and IFN-β-GFP, 46 kDa.
Fig. 4.
Bat IFN-α proteins are functional. (A) Bat IFN-α proteins induce an ISG response. Bat IFN-α and IFN-β protein containing supernatant produced in HEK293T cells were used to treat PaKiT03 cells. Six hours later, cells were collected for qRT-PCR detection of mRNA expression of IRF7, Mx1, and OAS1. Supernatant from empty vector-transfected or mock-transfected HEK293T cells were used as controls. Data show fold changes compared with mock and represent the mean and SD from two experiments. The difference in ISG induction in response to IFN-β was calculated in comparison with ISG induction by each of the three individual IFN-α proteins (*P < 0.05). (B) Bat IFN-α3 blocked PRV1NB replication in a dose-dependent manner. IFN-α3 protein was added at varying doses to PaKiT03 cells before PRV1NB infection, and 50% tissue culture infective dose (TCID50) assays were performed. In a parallel experiment, PaKiT03 cells were transfected with bat ISG56 promoter reporter plasmid, and luciferase activity was analyzed after treatment with varying doses of IFN-α3. Fold activation was determined by dividing the relative light units of each experimental sample by the relative light units of media alone. Data represent the mean and SE of triplicate experiments.
As IFN-α3 was the most abundant IFN-α in bat tissues, it was chosen to examine the antiviral activity of bat IFN-α. The antiviral activity of bat IFN-α3 was assessed on Pteropine orthoreovirus NB (PRV1NB)-infected PaKiT03 cells. PRV1NB is a biosafety level 2 reovirus carried by Pteropus bats that is easily cultured and tested for viral titer (35). IFN-α3 protected PaKiT03 cells from viral-induced cytopathogenic effect when applied 24 h before adding PRV1NB. IFN-α3 showed antiviral activity and ISG56 inducibility in a dose-dependent manner, and the activity disappeared at 0.01 ng/mL (Fig. 4B). These results demonstrate that bat IFN-α3 could protect bat cells from PRV1NB viral infection.
Discussion
Type I IFNs provide the first line of defense against viral infection and are typically expressed only at low levels in unstimulated cells but are rapidly induced following infection. An increase in the size of the IFN locus has been accompanied by the evolution of a family of IFN-α genes that each display distinct roles in the antiviral immune response of most mammals. Paradoxically, bats, which are important reservoirs for a variety of viruses, have a contracted IFN locus and have only three functional IFN-α loci that are expressed constitutively in the absence of viral infection. The constitutive expression of bat IFN-α results in the up-regulation of a distinct subset of ISGs that may have implications for the ability of bats to coexist with viruses and resist DNA damage associated with flight.
The bat type I IFN locus was remarkably contracted in the bat genome at ∼250 kb compared with other eutherian mammals that range from 350 kb (mouse) to 1,000 kb (pig). The smaller genome size of flying mammals has been speculated to be related to the evolution of flight (36), with bats and birds having smaller genomes compared with other species (37). However, the contraction of the IFN locus is striking, with only three functional IFN-α genes in the bat genome compared with 7–18 IFN-α loci in other mammals. Contraction of the bat IFN locus appears to have occurred after the divergence of bats from ungulates ∼80 Mya (38). The presence of eight IFN-α pseudogenes provides further evidence for the contraction of the bat IFN locus from a large IFN-α family in the ancestral bat genome. The dN/dS ratio, indicative of purifying selection pressures shaping ancestral bat IFN-α emphasizes that the three functional bat IFN-α genes are conserved and functionally important to the host. In comparison, bat IFN-ω genes experienced positive selection at the ancestral branch, suggesting host–pathogen antagonism has continued through the long coevolutionary history of bats and viruses.
Type I IFN mRNA and proteins have been detected in tissues of healthy mice maintained in pathogen-free environments, but only in extremely low quantities (17). In humans, IFN-α1 mRNA is detectible in healthy spleen, liver, and kidney, but not in other organs (13). Bats are unusual in that IFN-α mRNA is detectible across all organs from apparently healthy individuals of at least two bat species. In contrast, IFN-β is barely detectable. Furthermore, stimulation of bat cells with the dsRNA ligand polyI:C results in the up-regulation of IFN-β while the expression of IFN-α mRNA remained similar to that of unstimulated cells 3 h following stimulation. Our previous work also showed extremely low induction of IFN-α in response to polyI:C, up to a maximum of only approximately threefold at 9 h post transfection of bat lung cells (25). These findings suggest that IFN-α is not significantly up-regulated in response to cytoplasmic dsRNA sensing in bats. Nevertheless, the high baseline levels of IFN-α mean that substantial quantities of IFN-α mRNA can be detected even in the absence of immune stimulation. Similarly, Rousettus aegyptiacus lung cells had an apparent low level of constitutive IFN-α expression even before stimulation, and significant levels of IFN-α mRNA were not present until 8 h after polyI:C treatment (39). Thus, IFN-α in bats forms two layers of protection: the constitutive and the induced IFN-α. The human IFN-α response peaks at 2, 8, and 12 h following SeV infection (18), whereas, in bats, the IFN-α response is constitutively activated and further induced 8–9 h post infection (25). The two layers of the response are anticipated to provide bats with immediate protection but also allow them to react with a higher response when stimulated. Whether the two layers of the bat IFN-α response are dependent on each other or have different antiviral functions remains to be determined.
Very low levels of constitutively expressed IFN-α/IFN-β in humans and mice are believed to play a role in priming downstream responses, rather than having a direct role in antiviral immunity (16, 17). However, the ability of recombinant bat IFN-α3 to inhibit viral replication is consistent with constitutive IFN-α having a direct role in antiviral immunity in vivo. HeV has been demonstrated to block IFN production and signaling in bat cells (30), but does not affect the basal expression of IFN-α. RNAseq analysis supported our conclusion that bat IFN-α is constitutively expressed at a much higher level than human IFN-α, and is almost unaffected by HeV infection. Thus, the basal expression of bat IFN-α appears to be capable of avoiding the consequences of viral antagonism (at least by HeV and PulV) that leads to inhibition of the IFN-β response (30).
Constitutive expression of IFN-α would be expected to result in the corresponding induction of ISGs. To test this hypothesis, we used the available RNAseq dataset to compare expression of ISGs in human and bat cells (31). Human cells continually exposed to IFN-β express a distinct subset of ISGs that are driven by U-ISGF3, which leads to extended resistance to virus infection and DNA damage (20). The ISG response of bat cells appears to be enriched in ISGs associated with U-ISGF3. These ISGs were previously annotated, thus confirming they are counterparts to the corresponding human genes (21). Among the ISGs are well-characterized intracellular antiviral factors BST-2 (tetherin), which has been reported to restrict replication of HIV, Ebola, and Marburg viruses, and Mx1, which is recognized as having broad-spectrum antiviral activity against many RNA viruses (including influenza virus) and some DNA viruses (40, 41). Such ISGs may provide a “switched-on” defense mechanism to blunt virus replication and potentially viral pathogenesis in bats. The ISGs driven by U-ISGF3 do not appear to mediate the acute inflammatory responses often associated with IFN responses and may therefore contribute to the ability of bats to tolerate high levels of IFN-α with no pathological consequences. Furthermore, this subset of ISGs has been linked to resistance to DNA damage in human cells (20). The evolution of a prolonged ISG response in bats may be yet another adaptation caused by the evolution of flight that has had inadvertent consequences for antiviral immunity (20).
Of the three functional IFN-α loci in the bat genome, all three show some level of expression in bat tissues and cells, but IFN-α2 and IFN-α3 account for the majority of the constitutive expression pattern. IRF3 and IRF7 drive IFN expression in humans and other species by binding to unique IRF modules in the promoter regions (18). Curiously, despite the high expression of IFN-α2, no intact IRF binding elements were present in the promoter region of this gene. Furthermore, only module I and III appear to be potentially functional in the promoter regions of IFN-α3 and IFN-α1. Similar observations have been made in mouse IFN-α13 and IFN-ε, in which the IFN promoters have lost modules for IRF binding but maintain a constitutive expression pattern in certain organs (thymus, spleen, and spinal cord for IFN-α13; female reproductive tract for IFN-ε) (42, 43). In both cases, IFN gene expression is independent of viral infection. It is possible that the constitutively expressed IRF7 of bats contributes to the constitutive expression of bat IFN-α3 and IFN-α1 (27). However, the absence of IRF binding modules in IFN-α2 and the failure of IRF3 or IRF7 to induce IFN-α2 is consistent with the possibility that other transcription factors drive expression of bat IFN-α genes. Differences in the induction of IFNs may provide the opportunity to avoid antagonism by viruses that target IFN production pathways (44).
IFN genes evolve by gene duplication and deletion, and the loss of genes indicates their functions have become redundant, as observed in birds, which have a highly contracted IFN family (45, 46). Natural selection can result in mutations that favor less than the complete repertoire of functional genes, often with favorable consequences. This has been termed the “less-is-more” hypothesis (47). The contraction of the type I IFN family in bats with corresponding changes in their expression patterns is consistent with this hypothesis. Bats use fewer IFN-α genes to efficiently perform the functions of as many as 13 IFN-αs in other species and have a system that is constitutively ready to respond to infection.
In summary, we present an evolutionarily unique bat type I IFN locus with the discovery of only three functional bat IFN-α genes. Although bats have fewer IFN-α family members, the constitutive and ubiquitous expression pattern of IFN-α in bats may provide bats with a highly effective system for controlling viral replication.
Methods
Bat Tissues and Cells.
Tissues were collected from P. alecto and C. brachyotis bats as described in SI Methods. Details of the P. alecto primary and immortalized cell lines and culture conditions are described in SI Methods. All animal experiments were conducted following guidelines approved by the Australian Animal Health Laboratory (AAHL) ethics committee (AEC1389 and AEC1557) or Singapore animal ethics committee [B01/12 (A4) 12].
Viral Infection.
PaKiT03 cells were mock-infected or infected with HeV, Sendai virus, or PulV as described in SI Methods. IFN-α viral protection assays were performed in PaKiT03 cells as described in SI Methods.
IFN Locus Sequencing and Annotation.
Detailed descriptions of the sequencing, annotation, and comparative analysis of the bat type I IFN locus with other species are provided in SI Methods.
Comparative and Evolutionary Analysis of the Mammalian Type I IFN Locus and IFN Genes.
Comparative analysis of the bat IFN locus was performed with the corresponding genomic region from other vertebrates and is described in SI Methods and Tables S1–S9. Evolutionary analyses were performed by using sequence alignments of IFN-ω and IFN-α genes across a variety of vertebrates and is described in SI Methods.
Table S1.
Comparative genomics of the type I IFN locus across species
| Summary species | Location | Genome released | Annotated by |
| Pig | Chr 1 | Aug.2011 (SGSC Sscrofa10.2/susScr3) | Local tBlastn |
| Cow | Chr 8 | Jun.2014 (Bos_taurus_UMD_3.1.1/bosTau8) | Publication data, Map Viewer |
| Horse | Chr 23, unplaced scaffold | Sep.2007 (Broad/equCab2) | Publication data |
| Bat | Bat IFN locus | BAC sequencing | Local tBlastn |
| Human | Chr 9 | Dec.2013 (GRCH38/hg38) | Publication data, Map Viewer |
| Mouse | Chr 4 | Dec.2011 (GRCm38/mm10) | Publication data, Map Viewer |
| Elephant | Chr 6 | Jul.2009 (Broad/loxAfr3) | Local tBlastn |
| Opossum | Chr 6 | Oct.2006 (Broad/monDom5) | NCBI Map Viewer |
| Chicken | Chr Z | Nov.2011 (ICGSC Gallus_gallus-4.0/galGal4) | NCBI Map Viewer |
| Frog | Scaffold GL172684 | Nov.2009 (JGI 4.2/xenTro3) | NCBI Map Viewer |
| Zebrafish | Chr 3, Chr 12 | Jul.2010 (Zv9/danRer7) | NCBI Map Viewer |
Table S9.
Coordinates of the type I IFN locus in the bat genome
| Gene name | P.a IFN locus, start..end | Strand |
| IFN-β | 1.0.2358 | N |
| IFN-ω1 | 21678.0.22238 | P |
| IFN-α2 | 67866.0.68435 | N |
| KLHL9 | 82273.0.84126 | N |
| IFN-ω2 | 122948.0.123250 | P |
| IFN-ω3 | 171573.0.172130 | P |
| IFN-α3 | 183390.0.183959 | P |
| IFN-ω4 | 200209.0.200766 | P |
| IFN-α1 | 215571.0.216140 | P |
| IFN-ω5 | 225670.0.226128 | P |
| IFN-ε | 247308.0.247889 | N |
| IFN-κ | Outside locus | — |
Table S2.
Coordinates of the type I IFN locus in the zebrafish genome
| Gene name | Chr 3, start..end | NCBI ID | Strand |
| IFN-1 | 22232222.0.22235514 | NM_207640.1 | N |
| IFN-2 | 22222004.0.22226020 | NM_001111082.1 | N |
| IFN-3 | 22215034.0.22217388 | NM_001111083.1 | P |
| Chr 12 (start..end) | — | — | |
| IFN4 | 4876950.0.4882012 | NM_001161740 | N |
Table S3.
Coordinates of the type I IFN locus in the frog genome
| Gene name | GL172684, start..end | NCBI ID | Strand |
| IFN-1 | 2515253.0.2518416 | BN001167.1 | N |
| IFN-2 | 2494902.0.2498823 | BN001168.1 | N |
| IFN-3 | 2483442.0.2485943 | BN001169.1 | N |
| IFN-4 | 2471564.0.2473408 | BN001170.1 | N |
| IFN-5 | 2455797.0.2459041 | BN001171.1 | N |
Table S4.
Coordinates of the type I IFN locus in the chicken genome
| Gene name | ChrZ, start..end | Ensembl ID | Strand |
| IFN-α1 | 7258168.0.7258879 | ENSGALT00000043889 | P |
| IFN-α2 | 7246275.0.7246856 | ENSGALT00000046096 | P |
| IFN-α3 | 7240494.0.7241075 | ENSGALT00000043767 | P |
| IFN-β | 7230966.0.7231577 | ENSGALT00000039477 | P |
| IFN-α4 | 7236522.0.7237103 | ENSGALT00000043672 | P |
| IFN-α5 | 7244444.0.7245782 | ENSGALT00000043280 | P |
| IFN-α6 | 7254170.0.7254932 | ENSGALT00000021627 | P |
| IFN-α7 | 7260251.0.7260863 | ENSGALT00000044893 | P |
Table S5.
Coordinates of the type I IFN locus in the opossum genome
| Gene name | Chr 6, start..end | NCBI ID | Strand |
| IFN-β | 35511639.0.35511085 | XP_001373462 | N |
| IFN-α1 | 35538139.0.35537588 | CAM33514 | N |
| IFN-α2 | 35563135.0.35563686 | CAM33518 | P |
| IFN-α3 | 35588681.0.35589232 | CAM33515 | P |
| IFN-α4 | 35599779.0.35600330 | XP_001373547 | P |
| IFN-α5 | 35626780.0.35626229 | XP_001373586 | N |
| IFN-α6 | 35662828.0.35662277 | XP_001373622 | N |
| IFN-α7 | 35687402.0.35686851 | CAM33513 | N |
| IFN-κ | — | XP_007498406 | — |
Table S6.
Coordinates of the type I IFN locus in the elephant genome
| Gene name | Chr 6, start..end | NCBI ID | Strand |
| IFN-ε | 120359.0.120850 | XP_010586201.1 | P |
| IFN-ω1 | 151025.0.151570 | XP_003407380.1 | N |
| IFN-α1 | 168323.0.168955 | XP_003407699.1 | N |
| IFN-α2 | 189358.0.189954 | XP_003407700.1 | N |
| IFN-α3 | 234118.0.234696 | XP_003407380.1 | N |
| IFN-ω2 | 297337.0.297870 | XP_003407701.1 | N |
| IFN-α4 | 313969.0.314544 | XP_003407702.1 | N |
| IFN-ω3 | 329412.0.329858 | XP_010586351.1 | N |
| IFN-ω4 | 344728.0.345273 | XP_003407382.1 | N |
| IFN-α5 | 377328.0.377927 | XP_003407703.1 | N |
| IFN-α6 | 431420.0.432004 | XP_003407706.1 | P |
| IFN-ω5 | 466084.0.466635 | XP_003407383.1 | P |
| IFN-α7 | 517076.0.517549 | XP_010586204.1 | P |
| IFN-β | 536069.0.536455 | XP_003407384.1 | P |
| IFN-ω6 | S509 (3713.0.4243) | XP_003423618.1 | — |
| IFN-ω7 | S599 (21322.0.21864) | XP_003423647.1 | — |
| IFN-κ | — | XP_003407372.1 | — |
Table S7.
Coordinates of the type I IFN locus in the mouse genome
| Gene name | Chr 4, start..end | NCBI ID | Strand |
| IFN-β1 | 88522025.0.88522794 | NM_010510 | N |
| IFN-α15 | 88557673.0.88558245 | NM_206870 | N |
| IFN-α9 | 88591813.0.88592385 | NM_010507 | N |
| IFN-α12 | 88602580.0.88603376 | NM_177361 | N |
| IFN-α16 | 88676287.0.88676856 | NM_206867 | N |
| IFN-α2 | 88683207.0.88683779 | NM_010503 | N |
| IFN-αb | 88690599.0.88691268 | NM_008336 | N |
| KLHL9 | 88718292.0.88722508 | NM_172871 | N |
| IFN-ζ11 | 88760774.0.88761584 | NM_001085531 | P |
| IFN-ζ10 | 88773834.0.88774801 | NM_001243155 | P |
| IFN-ζ9 | 88776762.0.88777729 | NM_001243165 | P |
| IFN-ζ8 | 88779850.0.88780660 | NM_001161608 | P |
| IFN-ζ7 | 88782131.0.88783592 | NM_197889 | P |
| IFN-ζ6 | 88785054.0.88786515 | NM_001085532 | P |
| IFN-ζ5 | 88787977.0.88789438 | NM_001098840 | P |
| IFN-ζ4 | 88790895.0.88792356 | NM_001098841 | P |
| IFN-ζ3 | 88793742.0.88795297 | NM_001085533 | P |
| IFN-ζ2 | 88797237.0.88798204 | NM_001243166 | P |
| IFN-ζ1 | 88805531.0.88808380 | NM_001243167 | P |
| IFN-α7 | 88816225.0.88816800 | NM_008334 | P |
| IFN-α11 | 88819919.0.88820565 | NM_008333 | P |
| IFN-α6 | 88827416.0.88827985 | NM_206871 | P |
| IFN-α5 | 88835525.0.88836094 | NM_010505 | P |
| IFN-α4 | 88841816.0.88842459 | NM_010504 | P |
| IFN-α1 | 88850087.0.88850656 | NM_010502 | P |
| IFN-ε | 88879538.0.88880201 | NM_177348 | N |
| IFN-κ | — | NP_954608.1 | — |
Table S8.
Coordinates of the type I IFN locus in the human genome
| Gene name | Chr 9, start..end | NCBI ID | Strand |
| IFN-β1 | 21077105.0.21077944 | NM_002176 | N |
| IFN-ω1 | 21140632.0.21142145 | NM_002177 | N |
| IFN-α21 | 21165637.0.21166660 | NM_002175 | N |
| IFN-α4 | 21186618.0.21187599 | NM_021068 | N |
| IFN-α7 | 21201469.0.21202205 | NM_021057 | N |
| IFN-α10 | 21206181.0.21207143 | NM_002171 | N |
| IFN-α16 | 21216373.0.21217311 | NM_002173 | N |
| IFN-α17 | 21227243.0.21228222 | NM_021268 | N |
| IFN-α14 | 21239202.0.21239979 | NM_002172 | N |
| IFN-α5 | 21304614.0.21305313 | NM_002169 | N |
| KLHL9 | 21331019.0.21335430 | NM_018847 | N |
| IFN-α6 | 21350318.0.21350887 | NM_021002 | N |
| IFN-α13 | 21367372.0.21368076 | NM_006900 | N |
| IFN-α2 | 21384255.0.21385397 | NM_000605 | N |
| IFN-α8 | 21409147.0.21410185 | NM_002170 | P |
| IFN-α1 | 21440454.0.21441316 | NM_024013 | P |
| IFN-ε | 21480839.0.21482313 | NM_176891 | N |
| IFN-κ | — | NP_064509 | — |
Analysis of IFN and ISG Transcript Abundance.
RNAseq datasets from P. alecto PaKiT03 and human HEK293T cells obtained from mock and HeV infection are described in SI Methods (31). Analyses to determine changes in transcript abundance of IFNs and ISGs are described in SI Methods. qRT-PCR validation of gene expression was performed on total RNA from tissues or cells as described previously (25) and described in SI Methods. Primers are listed in Table S10.
Table S10.
Primer and overgo sequences
| Gene | Primer | Sequence (5′-3′) | Application | PrimerBank ID |
| P.a IFN-α1 | IFN-α1–1F | ATGGCCCTGCCCTGTTC | Cloning to pcDNA6.2/EmGFP | — |
| IFN-α1–1R | ATCCTTAYTCCTTGATCTTTCC | Cloning to pcDNA6.2/EmGFP | — | |
| IFN-α1–2F | ACCCGGCAGACCTTCCA | TaqMan qPCR | — | |
| IFN-α1–2R | CGTTGATAAGGAGTAGGATCTCATGAC | TaqMan qPCR | — | |
| IFN-α1p | AGAGGTCTCGTCCGCAGCTTGGGAT | TaqMan qPCR probe | — | |
| P.a IFN-α2 | IFN-α2–1F | ATGGCTATGCCTTTTTCC | Cloning topcDNA6.2/EmGFP | — |
| IFN-α2–1R | ATCCTTAYTCCTTGATCTTTCC | Cloning to pcDNA6.2/EmGFP | — | |
| IFN-α2–2F | CAAGGATAGAGAAGATTTTGGATTCCT | TaqMan qPCR | — | |
| IFN-α2–2R | AGTCCAGTGCAGAATCTGCCTAA | TaqMan qPCR | — | |
| IFN-α2p | CATCCATGAGGTGACCTGGCAGACC | TaqMan qPCR probe | — | |
| P.a IFN-α3 | IFN-α3–1F | GCAGAATTCCCACCATGGCCCTGCCCTGT | Cloning to pCAGGS | — |
| IFN-α3–1R | GCACTCGAGTTACTTGTCGTCATCGTCTTTGTAGTCATCCTTACTCTTTGA | Cloning to pCAGGS | — | |
| IFN-α3–2F | CTGGGACAAATGAGGAGAATCTCT | TaqMan qPCR | — | |
| IFN-α3–2R | ACGACAGCGATGGCTTGAA | TaqMan qPCR | — | |
| IFN-α3p | CATTTCATGGCAACCCCTTCCAGGA | TaqMan qPCR probe | — | |
| C.b IFN-α | C.b_IFN-α-1F | ATGCTCCTGGGACAAATGAG | SYBR qPCR | — |
| C.b_IFN-α-1R | GAAGAGGTGGAAGGTCTGC | SYBR qPCR | — | |
| P.a IFN-β | IFN-β-1F | ATGACCAACAGGTGCATCCTC | Cloning to pcDNA6.2/EmGFP | — |
| IFN-β-1R | GTTTTGGAGGTATTCTGTAAG | Cloning to pcDNA6.2/EmGFP | — | |
| IFN-β-ova | ACAAGGACAGGATGGACTTCAAGC | BAC library probing | — | |
| IFN-b-ovb | TTAATCTCCGCAGGGAGCTTGAAG | BAC library probing | — | |
| C.b IFN-β | C.β_IFN-β-2F | AACTTGATAGGCAGATGGACC | SYBR qPCR | — |
| C.β_IFN-β-2R | AGTGTCGTGCTTTCCCAG | SYBR qPCR | — | |
| P.a 18s rRNA | 18s-1F | CGGCTACCACATCCAAGGAA | TaqMan qPCR | — |
| 18s-1R | GCTGGAATTACCGCGGCT | TaqMan qPCR | — | |
| 18sp | TGCTGGCACCAGACTTGCCCTC | TaqMan qPCR probe | — | |
| P.a IFN-ε | IFN-εpa | TGATTAGCAAGTACTTCTTTGAAG | BAC library probing | — |
| IFN-εpb | AGCAGCACCAACACAACTTCAAAG | BAC library probing | — | |
| P.a KLHL9 | KLHL9pa | GGGTGATGGAGATGAAATCTTCCC | BAC library probing | — |
| KLHL9pb | CATAGCTCTGTGAACGGGGAAGAT | BAC library probing | — | |
| pCC1F | GGATGTGCTGCAAGGCGATTAAGTTGG | BAC end sequencing | — | |
| pCC1R | CTCGTATGTTGTGTGGAATTGTGAGC | BAC end sequencing | — | |
| gDNA | gDNA-1F | CATGGACTTGCCTGGAAGCCCAATCTC | Genome DNA PCR | — |
| gDNA-1R | CCACAGAGTAAATGGATCCACTT | Genome DNA PCR | — | |
| Pa Mx1 | Pa Mx1F | GTTGCAGATGTATGGCATGG | SYBR qPCR | — |
| Pa Mx1F | AACAGCCGAGTGTTGCTCTT | SYBR qPCR | — | |
| Hu Mx1 | Hu Mx1F | GTTTCCGAAGTGGACATCGCA | SYBR qPCR | 222136618c1 |
| Hu Mx1R | CTGCACAGGTTGTTCTCAGC | SYBR qPCR | — | |
| Pa IFI35 | Pa IFI35F | TTTGGGAAGACGAAGAATGG | SYBR qPCR | — |
| Pa IFI35R | GATCCGATCTCAGCCTTCTG | SYBR qPCR | — | |
| Hu IFI35 | Hu IFI35F | AACAAAAGGAGCACACGATCA | SYBR qPCR | 306922357c1 |
| HuIFI35R | CTCCGTTCCTAGTCTTGCCAA | SYBR qPCR | — | |
| Pa OAS1 | Pa OAS1F | AGCTGGAAGCCTGTCAAAAA | SYBR qPCR | — |
| Pa OAS1R | GTAGCTTCTCGGCACCTGAC | SYBR qPCR | — | |
| Hu OAS1 | Hu OAS1F | AGTTGACTGGCGGCTATAAAC | SYBR qPCR | 74229012c3 |
| Hu OAS1R | GTGCTTGACTAGGCGGATGAG | SYBR qPCR | — | |
| HES4 | HES4F | TCAAGACCCTCATCCTGGAC | SYBR qPCR (specific for human and bat) | — |
| HES4R | ACCTCTGCCAGACACTCGTT | SYBR qPCR (specific for human and bat) | — | |
| c19orf66 | Hu_ c19orf66_F | CTCCATCGTGTACGGGGTAAA | SYBR qPCR (specific for human and bat) | 154350197c1 |
| Hu_ c19orf66_R | GGTCCTGCTTCATATCCTCTGGT | SYBR qPCR (specific for human and bat) | — |
ISG Induction and Antiviral Activity of Bat IFN-α.
Details of the cloning and expression of recombinant P. alecto IFN-α (IFN-α1–3) and IFN-β are described in SI Methods. The activity of the recombinant IFN-α proteins was determined by their ability to induce the production of ISGs and inhibit virus-mediated cytolysis as described in SI Methods.
Luciferase Promoter Assays.
Details of the luciferase promoter assays used to test the ability of the three bat IFN-α genes to respond to IRF3 and IRF7 are described in SI Methods.
SI Methods
IFN Locus Sequencing and Annotation.
The Pteropus alecto whole-genome sequence was interrogated for type I IFNs and conserved flanking genes by using the BLAST algorithm. Scaffolds containing IFN genes were reannotated, and the identification of ORFs were confirmed by using BLAST. The P. alecto scaffolds described in this study are scaffold14 (NW_006436810), scaffold95 (NW_006441071), and scaffold222 (NW_006433716).
The P. alecto BAC library was constructed by Amplicon Express using genomic DNA extracted from the liver of a wild-caught adult male bat. The IFN-positive BACs were screened using overgoes designed based on P. alecto IFN-β and IFN-ε which mark the boundaries of the type I IFN locus in other species, and KLHL9, which is located within the IFN locus and is highly conserved across species (7, 9). Overgoes were designed by using overgo maker (bioinf.wehi.edu.au/cgi-bin/overgomaker) using sequences identified in the P. alecto whole genome. All primer and overgo sequences are provided in Table S10.
Hybridization of high-density BAC filters was performed as follows: briefly, overgo probes were labeled with 32P dCTP using the Prime It II labeling kit (Stratagene) following the manufacturer’s instructions. High-density BAC library filters were hybridized overnight with pools of radioactively labeled overgos in Church buffer at 65 °C. Positive clones were further screened by PCR using gene-specific primers (Table S10), and single end libraries were constructed using the GS FLX Titanium Rapid Library Preparation Kit (GS FLX+ Series-XL+; Roche) on selected clones, which were subsequently sequenced using the Roche 454 platform with the FLX+ long read chemistry (Roche). BAC end sequencing was also performed on all clones by Sanger sequencing by using sequencing primers found in the CopyControl pCC1BAC vector (Table S10).
Raw reads were filtered, trimmed, and assembled using a combination of CLC genomics 6.5.2 (CLC Bio) and SeqMan Pro-11.2.1 (DNASTAR). The raw reads were trimmed (trim function in CLC) using the following parameters: ambiguous limit, 2; ambiguous trim, yes; quality limit, 0.1; quality trim, yes; remove 3′ nucleotide, no; remove 5′ nucleotide, no. quality control (QC) report showed 75% of the reads have the Phil’s Read Editor (PHRED) score higher than 20. Reads were then mapped to the reference vector (BAC vector with HindIII cut), and the unmapped reads (IFN reads) were collected for downstream analysis. The mapping function was used according to the following parameters: similarity score, 0.9; length fraction, 0.5. The IFN reads (unmapped from previous step) were de novo assembled with word size of 64 and bubble size of 600, which generated multiple large contigs. These contigs were then assembled using SeqMan Pro to generate larger contigs. Finally, all reads were mapped back to the contigs using the mapping function in CLC Genomics. To fill the gaps generated after assembly, a 3-kb contig was amplified by using genomic DNA from the P. alecto kidney cell line (PaKiT03) using primers listed in Table S10 and sequenced using Sanger sequencing. Sequences from the boundary of the BAC contigs were used to search against the Pteropus vampyrus genome (version 79; Ensembl database) using BLAT. A 21-kb region within scaffold 12130 that shared more than 97% similarity to these end sequences was identified. P. alecto raw reads were mapped against this 21-kb region to generate a consensus before assembly with the two P. alecto contigs to obtain a final scaffold of ∼250 kb.
The 250-kb scaffold was used to create a local BLAST database in CLC (BLAST function). P. alecto IFN-ω and IFN-α sequences obtained from the bat genome annotation were used as baits for TBLASTN search against this local database (21). For the confirmed IFN sequences, domain prediction was further performed by using the National Center for Biotechnology Information (NCBI) Web conserved domain search tool (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). IFN-α genes with incomplete IFN-αR1 or IFN-αR2 binding domains were classified as IFN pseudogenes. Four bat IFN-αPs have full-length IFN-α reading frames but were disrupted by frameshift mutations (IFN-αP1, 2, 7, and 8), whereas the other four have incomplete reading frames and are likely nonfunctional genes (IFN-αP3, 4, 5, and 6).
Nucleotide sequence data from this manuscript have been deposited in the GenBank database with the following accession numbers: bat BAC clones 19–21 (KT384435–KT384439) and cloned 3-kb bat IFN region (KT384440). RNAseq data from uninfected HEK293T and PaKiT03 cells have been deposited in the Sequence Read Archive and have been assigned the accession numbers SRP067312 and SRP067371, respectively.
Comparative Analysis of Mammalian Type I IFN Locus.
For comparative purposes, the IFN locus was obtained from the genomes of 10 species by using the Ensembl database: zebrafish (Danio rerio, version GRCz10), western clawed frog (Xenopus tropicalis, version JGI 4.2), chicken (Gallus gallus, version Galgal4), South American opossum (Monodelphis domestica, version monDom5), elephant (Loxodonta africana, version Loxafr3.0), mouse (Mus musculus, version GRCm38.p3), human (Homo sapiens, version GRCh38.p2), horse (Equus caballus, version Equ Cab 2), cow (Bos taurus, version UMD3.1), and pig (Sus scrofa, version Sscrofa10.2). Annotations were already available for the human, mouse, and horse IFN locus (6, 7). For cow, opossum, chicken, frog, and zebrafish, NCBI Map Viewer was used to obtain annotations of the IFN regions. For pig and elephant, for which no reliable annotation was available, a local annotation was performed in a similar manner to the bat by using IFN-β and IFN-ε to mark the boundaries of the IFN locus. A phylogenetic tree was drawn by using FigTree v1.4.2 (tree.bio.ed.ac.uk/software/figtree/), and the approximate divergence times were shown according to TimeTree (38).
Evolutionary Analyses.
Detection of recombinants was performed by using recombination detection program 4 (RDP4) (48), implementing the RDP and GENECONV methods with a significance cutoff of P = 0.05 and Bonferroni correction. Because of the large number of sequences recovered from different taxa, only representative sequences with potential coding ability (i.e., not interrupted by indels or premature stop codons) were selected. As we were interested in the functional evolution of mammalian IFNs, sequences commonly shared (perfectly aligned) by different taxon were selected with stop codons removed. IFN-ω and IFN-α genes across species were collected and aligned by using multiple sequence comparison by log expectation (MUSCLE). Recombination tests were carried out on the aligned sequences of IFN-ω and IFN-α. Sequences were considered to be potential recombinants if one of the aforementioned methods returned a significant value, and thus were not considered for the following evolutionary analyses. All sequences were aligned and a conserved region was selected across species, consisting of 102 species and 531 bp in length. A nonbootstrapped maximum likelihood phylogenetic reconstruction was performed by using PhyML 3.0 (49), under GTR+I+Γ model. Tests for selection on ancestral IFNs of different taxa were inferred by using PAML 4.8 (50), under a free-ratio model, which allows each branch having different dN/dS values. The ratio of nonsynonymous to synonymous changes (i.e., dN/dS) was measured to examine the selection pressures on the bat IFN-α and IFN-ω genes. A dN/dS ratio of <1 is indicative of purifying selection, whereas a ratio of >1 indicates positive selection.
P. alecto Cell Lines.
The establishment and culture of P. alecto bat cell lines and culture conditions have been described previously (51). Cell lines used in the present study had been passaged multiple times with no evidence of cytopathic effect. The bat primary cells were stimulated with polyI:C (Invivogen) as described previously (51). Three viruses, HeV (Hendra virus/Australia/Horse/1994/Hendra), Sendai virus (Cantell strain), or PulV were used for infection of PakiT03 cells. HeV and PulV are bat-borne viruses, and culture conditions in bat cell lines have been described previously (25, 30).
Analysis of IFN, ISG, and Viral Transcript Abundance in RNAseq Data.
To understand changes in transcript abundance, IFN-α RNAseq reads from PaKiT03 or HEK293T (mock- or HeV-infected for 8 h) cells were mapped separately against bat IFN-α3 or human IFN-α6. The number of IFN-α reads mapped were compiled and counted by using SAMtools, version 0.1.18 (52). Data were illustrated in read counts per nucleotide.
A similar strategy was used to compare ISG expression between uninfected PaKiT03 and HEK293T cells using four replicates of RNAseq data from each cell line. One replicate from each cell line has been described previously (31). Total RNA was isolated from a further three replicates of uninfected PaKiT03 and HEK293T cells using the RNeasy kit (Qiagen) with DNase I treatment per the manufacturer’s instructions. mRNA was sequenced as 100-bp paired-end reads on a HiSEq.2000 system (Illumina). Resulting reads were trimmed for adapters and quality-assessed using FastQC. Pair-end reads were reference-mapped against previously assembled de novo transcriptomes for human and P. alecto by using Bowtie2 (31). The number of reads mapping to each transcript was obtained by using SAMtools. Normalized FPKM values were calculated for each transcript, averaged across the four replicates, and used for comparison between the two cell lines. When multiple transcripts were observed for a single gene, the sum of FPKM values across the isoforms were used. The gene expression profile was illustrated by using a heat map generated using Cluster 3.0 and TreeView software (taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Analysis of RNAseq Data for Viral Transcripts.
A previously annotated RNAseq dataset consisting of 64 million paired end sequences (31) was used to search for evidence of viral (including microbial) transcripts in uninfected (mock) PaKiT03 cells. We excluded identical viral transcripts, including those from endogenous retroviruses, present in both bat and human cell lines. This left a total of 378 transcripts previously annotated as unconfirmed viral transcripts in PaKiT03 cells that were not present in the corresponding dataset from HEK293T cells. These transcripts were mapped back to the P. alecto genome (GenBank assembly accession no. GCA_000325575.1), resulting in 316 matches, using CLC Genomics Workbench 8.5.1 (https://www.qiagenbioinformatics.com/; “map reads to reference” algorithm; default settings except similarity fraction set at 0.8 and nonspecific matches ignored). The remaining unmapped 62 transcripts were subjected to BLASTX and BLASTN against the NCBI nonredundant database, resulting in all transcripts matching bat genomic sequences at high identity (expect value E < 0.001).
qRT-PCR.
SYBR Green qRT-PCR primers for P. alecto IFN-α, IFN-β, 18s rRNA, actin, IRF7, Mx1, and OAS1 have been described previously (35). The P. alecto IFN-α primers used for SYBR Green qRT-PCR detected all three IFN-α genes identified in the genomic sequence. These primers demonstrated similar amplification efficiency for each of the three IFN-α genes based on standard curves produced using IFN-α plasmids as the template. Primers corresponding to OAS1, HES4, IFI35, Mx1, and C19orf66 were used to validate gene expression in bat (PaKiT03) and human (HEK293T) cells. P. alecto primers were designed by using Primer3 software based on sequences identified in the RNAseq dataset, and human primers were obtained from the human primer bank database (https://pga.mgh.harvard.edu/primerbank/). Cynopterus brachyotis IFN-β primers for SYBR Green qRT-PCR were designed by using Primer Express 3.0 based on IFN-β ORF conserved regions among multiple species. TaqMan qRT-PCR primers (and probes) for P. alecto IFN-α1, 2, and 3 were designed to distinguish the three bat IFN-α genes. TaqMan primers for the three IFN-α genes and 18s rRNA were designed using Primer Express 3.0 (Applied Biosystems) with default parameter settings. All data were normalized relative to the housekeeping genes (18S rRNA, GAPDH, or actin) as indicated. The expression level of the target genes was calculated by using the standard cures method or fold induction compared with mock (OAS1, Mx, IRF7) or copy number relative to the housekeeping gene (IFN-α, IFN-β). All primers are listed in Table S10.
ISG Induction and Antiviral Activity of Bat IFN-α.
Conditioned media from HEK293T cells transfected with plasmids encoding pcDNA6.2/C-emGFP–IFN-α1, -IFN-α2 and -IFN-β, and pCAGGS/FLAG–IFN-α3 were evaluated by immunoblotting with rabbit polyclonal anti-GFP or mouse monoclonal anti-FLAG antibody (Sigma). Approximately equivalent amounts of each protein were used based quantification on a Western blot by using ImageJ densitometry software (Fig. S7) from IFN-conditioned medium was diluted at 1:2 and used to treat the PaKiT03 cells. Supernatant from HEK293T cells cultured under normal conditions or transfected with vector alone without insertion were used as negative controls. Cells were incubated at 37 °C for 6 h and collected into buffer RLT (Qiagen) for extraction of total RNA. The expression of known bat ISGs including IRF7, Mx1, and OAS1 were determined by qRT-PCR as described previously (27, 35).
IFN-α3–Flag proteins were also purified by affinity chromatography from the supernatant of HEK293T cells transiently transfected with pCAGGS/FLAG–IFN-α3 expression plasmid. The protein was tested for antiviral activity in PaKiT03 cells. Cells were seeded in 96-well plates at 1 × 105 cells per well and incubated with serial dilutions of IFN-α from 0 to 10 ng/mL at 37 °C for 24 h. Medium was then replaced with the bat orthoreovirus PRV1NB-containing supernatant at a multiplicity of infection of 0.1 and incubated for 1 h at 37 °C. Virus-containing supernatant was then replaced with culture medium and incubated for another 24 h in a humidified atmosphere of 5% CO2 in air at 37 °C. Culture supernatant was collected for TCID50 testing in PaKiT03 cells.
A P. alecto ISG56 promoter plasmid was constructed by inserting 600 bp upstream of ISG56 translation start into the pGL4.1 vector (Promega). Serial dilutions of IFN-α proteins as described earlier were used to treat PaKiT03 cells transfected for 24 h with the bat ISG56 luciferase promoter. Six hours post treatment, cells were collected in passive lysis buffer and tested for luciferase activity by using the dual luciferase reporter assay system (Promega) using a Thermo Fluoroskan Ascent FL machine.
IFN Promoter Assays.
PaKiT03 cells were transfected by using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions. Approximately 1 × 105 PaKiT03 cells per well were transiently cotransfected with 200 ng of expression plasmid for bat IRF3-GFP (with or without 10 ng bat MAVS-GFP) or IRF7-GFP with 100 ng bat IFN-α promoter plasmid and 50 ng pRL-Tk plasmid (Promega) as an internal control in a 24-well plate. GFP vector plasmid was used at 200 ng per well as a negative control. Cells were harvested 30 h posttransfection and lysed using passive lysis buffer. Luciferase activities were determined using the dual-luciferase assay system (Promega) using a Thermo Fluoroskan Ascent FL machine.
Acknowledgments
We thank Susanne Wilson for P. alecto tissue collection and Yok Teng Chionh and Dolyce Low Hong Wen for C. brachyotis tissue collection and RNA extraction. This work was supported in part by National Institutes of Health Institutional Development Award Programme of the National Centre for Research Resources Grant P20RR018754 (to M.L.B.), Australian Research Council Future Fellowship FT110100234 (to M.L.B.), a Commonwealth Scientific and Industrial Research Organization Chief Executive Officer Science Leaders Award (to L-.F.W.), and Singaporean National Research Foundation Competitive Research Programme Grant NRF-CRP10-2012-05 (to L-.F.W.), and by the National Collaborative Research Infrastructure Strategy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Nucleotide sequence data have been deposited in the GenBank database [accession nos. KT384435–KT384439 (bat BAC clones 19–21) and KT384440 (cloned 3-kb bat IFN region)]. RNA sequence data have been deposited in the Sequence Read Archive [accession nos. SRP067312 (uninfected HEK293T cells) and SRP067371 (PaKiT03 cells)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518240113/-/DCSupplemental.
References
- 1.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: Important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19(3):531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Middleton DJ, et al. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus) J Comp Pathol. 2007;136(4):266–272. doi: 10.1016/j.jcpa.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Sétien AA, et al. Experimental rabies infection and oral vaccination in vampire bats (Desmodus rotundus) Vaccine. 1998;16(11-12):1122–1126. doi: 10.1016/s0264-410x(98)80108-4. [DOI] [PubMed] [Google Scholar]
- 4.Baker ML, Schountz T, Wang LF. Antiviral immune responses of bats: A review. Zoonoses Public Health. 2013;60(1):104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Weerd NA, Nguyen T. The interferons and their receptors--distribution and regulation. Immunol Cell Biol. 2012;90(5):483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Detournay O, Morrison DA, Wagner B, Zarnegar B, Wattrang E. Genomic analysis and mRNA expression of equine type I interferon genes. J Interferon Cytokine Res. 2013;33(12):746–759. doi: 10.1089/jir.2012.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy MP, Owczarek CM, Jermiin LS, Ejdebäck M, Hertzog PJ. Characterization of the type I interferon locus and identification of novel genes. Genomics. 2004;84(2):331–345. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 9.Walker AM, Roberts RM. Characterization of the bovine type I IFN locus: Rearrangements, expansions, and novel subfamilies. BMC Genomics. 2009;10:187. doi: 10.1186/1471-2164-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borden EC, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6(12):975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ragg H, Weissmann C. Not more than 117 base pairs of 5′-flanking sequence are required for inducible expression of a human IFN-alpha gene. Nature. 1983;303(5916):439–442. doi: 10.1038/303439a0. [DOI] [PubMed] [Google Scholar]
- 12.Vogel SN, Fertsch D. Endogenous interferon production by endotoxin-responsive macrophages provides an autostimulatory differentiation signal. Infect Immun. 1984;45(2):417–423. doi: 10.1128/iai.45.2.417-423.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tovey MG, et al. Interferon messenger RNA is produced constitutively in the organs of normal individuals. Proc Natl Acad Sci USA. 1987;84(14):5038–5042. doi: 10.1073/pnas.84.14.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton JA, Whitty GA, Kola I, Hertzog PJ. Endogenous IFN-alpha beta suppresses colony-stimulating factor (CSF)-1-stimulated macrophage DNA synthesis and mediates inhibitory effects of lipopolysaccharide and TNF-alpha. J Immunol. 1996;156(7):2553–2557. [PubMed] [Google Scholar]
- 16.Taniguchi T, Takaoka A. A weak signal for strong responses: Interferon-alpha/beta revisited. Nat Rev Mol Cell Biol. 2001;2(5):378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 17.Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36(2):166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Génin P, Lin R, Hiscott J, Civas A. Differential regulation of human interferon A gene expression by interferon regulatory factors 3 and 7. Mol Cell Biol. 2009;29(12):3435–3450. doi: 10.1128/MCB.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Génin P, Vaccaro A, Civas A. The role of differential expression of human interferon--a genes in antiviral immunity. Cytokine Growth Factor Rev. 2009;20(4):283–295. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Cheon H, et al. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32(20):2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339(6118):456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kepler TB, et al. Chiropteran types I and II interferon genes inferred from genome sequencing traces by a statistical gene-family assembler. BMC Genomics. 2010;11:444. doi: 10.1186/1471-2164-11-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X, et al. Anti-lyssaviral activity of interferons κ and ω from the serotine bat, Eptesicus serotinus. J Virol. 2014;88(10):5444–5454. doi: 10.1128/JVI.03403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He G, He B, Racey PA, Cui J. Positive selection of the bat interferon alpha gene family. Biochem Genet. 2010;48(9-10):840–846. doi: 10.1007/s10528-010-9365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou P, et al. Type III IFNs in pteropid bats: Differential expression patterns provide evidence for distinct roles in antiviral immunity. J Immunol. 2011;186(5):3138–3147. doi: 10.4049/jimmunol.1003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou P, et al. Type III IFN receptor expression and functional characterisation in the pteropid bat, Pteropus alecto. PLoS One. 2011;6(9):e25385. doi: 10.1371/journal.pone.0025385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou P, et al. IRF7 in the Australian black flying fox, Pteropus alecto: Evidence for a unique expression pattern and functional conservation. PLoS One. 2014;9(8):e103875. doi: 10.1371/journal.pone.0103875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts RM, Liu L, Guo Q, Leaman D, Bixby J. The evolution of the type I interferons. J Interferon Cytokine Res. 1998;18(10):805–816. doi: 10.1089/jir.1998.18.805. [DOI] [PubMed] [Google Scholar]
- 29.Ito Y, Hosaka Y. Component(s) of Sendai virus that can induce interferon in mouse spleen cells. Infect Immun. 1983;39(3):1019–1023. doi: 10.1128/iai.39.3.1019-1023.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virtue ER, Marsh GA, Baker ML, Wang LF. Interferon production and signaling pathways are antagonized during henipavirus infection of fruit bat cell lines. PLoS One. 2011;6(7):e22488. doi: 10.1371/journal.pone.0022488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wynne JW, et al. Proteomics informed by transcriptomics reveals Hendra virus sensitizes bat cells to TRAIL-mediated apoptosis. Genome Biol. 2014;15(11):532. doi: 10.1186/s13059-014-0532-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dacheux L, et al. A preliminary study of viral metagenomics of French bat species in contact with humans: Identification of new mammalian viruses. PLoS One. 2014;9(1):e87194. doi: 10.1371/journal.pone.0087194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Civas A, Dion M, Vodjdani G, Doly J. Repression of the murine interferon alpha 11 gene: Identification of negatively acting sequences. Nucleic Acids Res. 1991;19(16):4497–4502. doi: 10.1093/nar/19.16.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaks E, Gavutis M, Uzé G, Martal J, Piehler J. Differential receptor subunit affinities of type I interferons govern differential signal activation. J Mol Biol. 2007;366(2):525–539. doi: 10.1016/j.jmb.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 35.Zhou P, Cowled C, Wang LF, Baker ML. Bat Mx1 and Oas1, but not Pkr are highly induced by bat interferon and viral infection. Dev Comp Immunol. 2013;40(3-4):240–247. doi: 10.1016/j.dci.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Hughes AL, Hughes MK. Small genomes for better flyers. Nature. 1995;377(6548):391. doi: 10.1038/377391a0. [DOI] [PubMed] [Google Scholar]
- 37.Smith JD, Gregory TR. The genome sizes of megabats (Chiroptera: Pteropodidae) are remarkably constrained. Biol Lett. 2009;5(3):347–351. doi: 10.1098/rsbl.2009.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Hedges SB. TimeTree2: Species divergence times on the iPhone. Bioinformatics. 2011;27(14):2023–2024. doi: 10.1093/bioinformatics/btr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omatsu T, et al. Induction and sequencing of Rousette bat interferon alpha and beta genes. Vet Immunol Immunopathol. 2008;124(1-2):169–176. doi: 10.1016/j.vetimm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haller O, Kochs G. Human MxA protein: An interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res. 2011;31(1):79–87. doi: 10.1089/jir.2010.0076. [DOI] [PubMed] [Google Scholar]
- 41.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 42.van Pesch V, Michiels T. Characterization of interferon-alpha 13, a novel constitutive murine interferon-alpha subtype. J BiolChem. 2003;278(47):46321–46328. doi: 10.1074/jbc.M302554200. [DOI] [PubMed] [Google Scholar]
- 43.Fung KY, et al. Interferon-ε protects the female reproductive tract from viral and bacterial infection. Science. 2013;339(6123):1088–1092. doi: 10.1126/science.1233321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Randall RE, Goodbourn S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 45.Kaiser P, et al. A genomic analysis of chicken cytokines and chemokines. J Interferon Cytokine Res. 2005;25(8):467–484. doi: 10.1089/jir.2005.25.467. [DOI] [PubMed] [Google Scholar]
- 46.Lovell PV, et al. Conserved syntenic clusters of protein coding genes are missing in birds. Genome Biol. 2014;15(12):565. doi: 10.1186/s13059-014-0565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olson MV. When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64(1):18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin D, Rybicki E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics. 2000;16(6):562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 49.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 50.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 51.Crameri G, et al. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One. 2009;4(12):e8266. doi: 10.1371/journal.pone.0008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas C, et al. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 2011;146:621–632. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]