Significance
Balancer chromosomes are highly rearranged chromosomes that suppress recombination and are an important tool in Drosophila genetics, yet their precise molecular structure is unknown. Here we characterize the inversion breakpoints of the X chromosome balancer FM7, and provide evidence that rare double-crossover events with balanced homologs can occur. These rare exchange events do not undermine the use of balancers, but lead to diversity among balancers. We also provide genomic evidence that unequal exchange between duplicated regions underlies reversion at the Bar locus. Our work demonstrates the power of genome sequencing to understand the molecular nature of classical genetic resources, and cautions that mutations maintained over balancers in regions susceptible to exchange should be checked regularly to prevent their loss.
Keywords: balancer chromosome, inversion, recombination, duplication, unequal exchange
Abstract
Multiply inverted balancer chromosomes that suppress exchange with their homologs are an essential part of the Drosophila melanogaster genetic toolkit. Despite their widespread use, the organization of balancer chromosomes has not been characterized at the molecular level, and the degree of sequence variation among copies of balancer chromosomes is unknown. To map inversion breakpoints and study potential diversity in descendants of a structurally identical balancer chromosome, we sequenced a panel of laboratory stocks containing the most widely used X chromosome balancer, First Multiple 7 (FM7). We mapped the locations of FM7 breakpoints to precise euchromatic coordinates and identified the flanking sequence of breakpoints in heterochromatic regions. Analysis of SNP variation revealed megabase-scale blocks of sequence divergence among currently used FM7 stocks. We present evidence that this divergence arose through rare double-crossover events that replaced a female-sterile allele of the singed gene (snX2) on FM7c with a sequence from balanced chromosomes. We propose that although double-crossover events are rare in individual crosses, many FM7c chromosomes in the Bloomington Drosophila Stock Center have lost snX2 by this mechanism on a historical timescale. Finally, we characterize the original allele of the Bar gene (B1) that is carried on FM7, and validate the hypothesis that the origin and subsequent reversion of the B1 duplication are mediated by unequal exchange. Our results reject a simple nonrecombining, clonal mode for the laboratory evolution of balancer chromosomes and have implications for how balancer chromosomes should be used in the design and interpretation of genetic experiments in Drosophila.
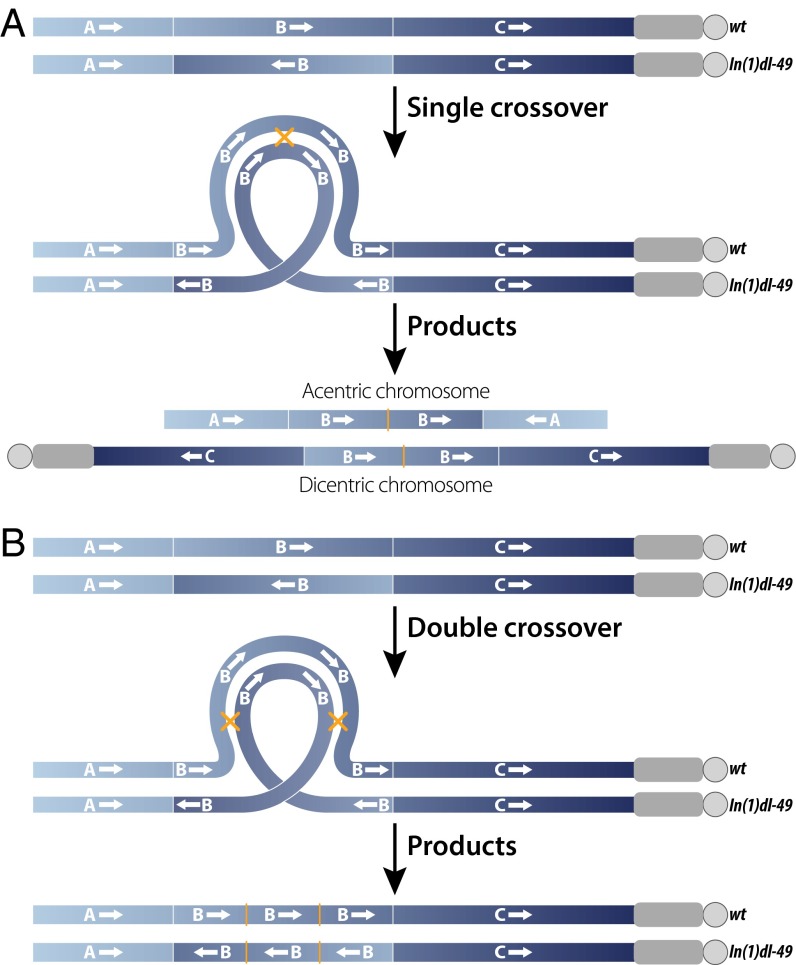
Balancer chromosomes are genetically engineered chromosomes that suppress crossing over with their homologs and are used for many purposes in genetics, including construction of complex genotypes, maintenance of stocks, and estimation of mutation rates. Balancers typically carry multiple inversions that suppress genetic exchange or result in the formation of abnormal meiotic products if crossing over does occur (Fig. 1A); for example, single crossovers inside the inverted segment create acentric or dicentric chromosomes that will fail to segregate properly during meiosis or large deletions or duplications that will likely result in inviable gametes (1, 2). Balancers also often carry recessive lethal or sterile mutations to prevent their propagation as homozygotes as well as dominant markers for easy identification. First developed for use in Drosophila melanogaster, balancer chromosomes remain some of the most powerful tools for genetic analysis in this species (3).
Fig. 1.
Consequences of a single or double crossover between a WT X chromosome and an X chromosome carrying a single inversion, In(1)dl-49. Euchromatin is shown in blue, heterochromatin is shown in gray, and centromeres are depicted as circles. Thin white lines mark locations of inversion breakpoints, and yellow crosses/thin lines mark locations of crossover events. (A) A single crossover event within the inverted segment results in the formation of chromosomes with deletions and zero (acentric) centromeres or duplications and two (dicentric) centromeres, neither of which will segregate properly during meiosis. (B) A double crossover within an inverted segment results in intact chromosomes with one centromere that will segregate properly during meiosis.
Despite their widespread use, very little is known about the organization of Drosophila balancer chromosomes at the molecular level. Since their original syntheses decades ago, balancers have undergone many manipulations, including the addition or removal of genetic markers. Moreover, rare recombination events can cause spontaneous loss of deleterious alleles on chromosomes kept over balancers in stock, as well as loss of marker alleles on balancer chromosomes themselves (3). Likewise, recent evidence has shown that sequence variants can be exchanged between balancer chromosomes and their wild type (WT) homologs via gene conversion during stock construction or maintenance (4, 5). Thus, substantial variation may exist among structurally identical balancer chromosomes owing to various types of sequence exchange.
To gain insight into the structure and evolution of balancer chromosomes, we have undertaken a genomic analysis of the most commonly used X chromosome balancer in D. melanogaster, First Multiple 7 (FM7). We have focused on FM7 because this X chromosome balancer series lacks lethal mutations and thus can be easily sequenced in a hemizygous or homozygous state. In addition, the FM7 chromosome has been shown to pair normally along most of its axis with a standard X chromosome, providing a structural basis for possible exchange events (6). Moreover, although details of how early balancers in D. melanogaster were created are not fully recorded, the synthesis and cytology of the FM7 series is reasonably well documented (3).
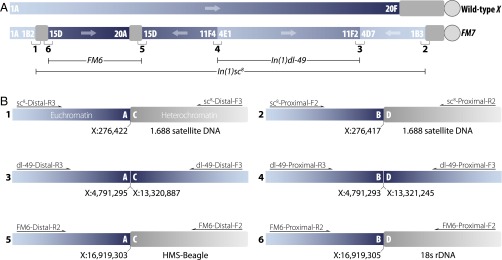
The earliest chromosome in the FM7 series, FM7a, was constructed using two progenitor X chromosome balancers, FM1 and FM6, to create a chromosome carrying three inversions—In(1)sc8, In(1)dl-49, and In(1)FM6—relative to the WT configuration (7, 8) (Fig. 2A). Subsequently, a female-sterile allele of singed (snX2) was introduced onto FM7a to create FM7c, which prevents the loss of balanced chromosomes carrying recessive lethal or female-sterile mutations (9). More recently, versions of FM7a and FM7c have been generated that carry transgene insertions that allow the determination of balancer genotypes in embryonic or pupal stages (10–14).
Fig. 2.
Structure of the FM7 balancer chromosome. Euchromatin is shown in blue, and heterochromatin is shown in gray. (A) Schematic view of the organization of WT and FM7 X chromosomes. FM7 contains three inversions—In(1)sc8, In(1)dl-49, and In(1)FM6—relative to WT. The six breakpoint junctions for the three inversions are numbered 1–6 and are shown in detail in B. (B) Location and organization of inversion breakpoints in FM7. Each inversion has two breakpoints that can be represented as A/B and C/D in the standard WT arrangement and as A/C and B/D in the inverted FM7 arrangement, where A, B, C, and D represent the sequences on either side of the breakpoints. Locations of euchromatic breakpoints are on Release 5 genome coordinates, and the identity of the best BLAST match in FlyBase is shown for heterochromatic sequences. Primers used for PCR amplification are shown above each breakpoint; details are provided in Methods and Datasets S2 and S3. Forward and reverse primers are named with respect to the orientation of the assembled breakpoint contigs, not the orientation of the WT or FM7 X chromosome.
To identify the inversion breakpoints in FM7 balancers and to study patterns of sequence variation that may have arisen since the origin of the FM7 series, we sequenced genomes of eight D. melanogaster stocks carrying the FM7 chromosome (four FM7a and four FM7c). We discovered several megabase-scale regions in which FM7c chromosomes differ from one another, which presumably arose via double-crossover (DCO) events from balanced chromosomes (Fig. 1B). These DCOs eliminate the female-sterile snX2 allele in the centrally located In(1)dl-49 inversion and are expected to confer a fitness advantage to sn+ chromosomes, either by allowing propagation of sn+ FM7 as homozygotes in females or by sn+ FM7 males outcompeting snX2 FM7 males in culture. We found that loss of the snX2 allele is common in FM7c chromosomes by screening other FM7c-carrying stocks at the Bloomington Drosophila Stock Center. We also identified the breakpoints of the B1 duplication carried on FM7, and found direct molecular evidence for the role of unequal exchange in the origin and reversion of the B1 allele (15–19). Our results provide clear evidence that the common assumption that balancers are fully nonrecombining chromosomes is incorrect on a historical timescale, and that substantial sequence variation exists among balancer chromosomes in circulation today.
Results
Identification of FM7 Inversion Breakpoints.
The inversions carried by FM7 that confer the ability to suppress recombination were generated by X-ray mutagenesis and characterized using genetic and cytogenetic data in the pregenomic era, and thus the precise locations and molecular nature of their breakpoints remain unknown. To better understand the genomic organization of FM7 chromosomes, we used whole-genome sequencing to identify breakpoints for the three inversions present on FM7: In(1)sc8, In(1)dl-49, and In(1)FM6 (Fig. 2A). Based on cytological data, it is known that both breakpoints of In(1)dl-49 lie in euchromatic regions (20–22); however, for both In(1)sc8 and In(1)FM6, one breakpoint is euchromatic, and the other lies in centric heterochromatin (22–27).
Our general strategy to identify breakpoint regions was as follows. We sequenced eight FM7-carrying stocks to ∼50-fold coverage with paired-end Illumina data and mapped reads to the D. melanogaster reference genome; summary statistics are provided in Dataset S1. We identified clusters of split or discordantly mapped reads from all stocks in the vicinity of expected breakpoint locations based on previous cytological data, then performed de novo assembly of split/discordant reads and their mate pairs (i.e., reads from the other end of the same paired-end sequenced fragments). We then used breakpoint contigs identified by sequence analysis to design PCR amplicons that span breakpoints, and Sanger-sequenced the resulting PCR amplicons to verify breakpoint assemblies. Using this approach, we were able to map euchromatic breakpoints of all three inversions on the FM7 chromosome to reference genome coordinates, and also characterize the sequence composition of the heterochromatic breakpoints for both In(1)sc8 and In(1)FM6 (Fig. 2B).
The distal breakpoint of the X-ray–induced In(1)sc8 inversion has been localized near bands 1B2-3 between the achaete (ac) and scute (sc) genes (22–26, 28). We identified a cluster of split/discordant reads in FM7 stocks around X:276,500 (predicted band 1A7) of the type expected in the vicinity of an inversion breakpoint. Split/discordant reads from ±1.5 kb around the putative In(1)sc8 inversion breakpoint (which map to the A and B regions) and their mate pairs (which map to the C and D regions) were extracted from all FM7 strains, pooled together, and assembled to identify candidate A/C and B/D breakpoint sequences. BLAST analysis of the resulting assembly revealed two contigs of 506 bp and 551 bp. The euchromatic components of these contigs mapped to nucleotides X:276,417–276,422 in the Release 5 genome sequence between ac and sc, within an intron of CG32816. The heterochromatic components of these contigs contained copies of the 1.688 satellite DNA repeat (29) that covers approximately one-half of the X chromosome centric heterochromatin (30).
We used the locations and sequences of candidate breakpoints for In(1)sc8 to design PCR primers that yielded amplicons in all stocks carrying In(1)sc8, but not in stocks lacking this inversion (Dataset S2). Sanger sequencing of PCR amplicons spanning breakpoint regions confirmed the sequence of A/C and B/D de novo assemblies. Comparison of A/C and B/D fragments revealed a 6-bp sequence (TTTCGT) from the ac–sc region that is present at both breakpoint junctions, suggesting that the X-ray–induced inversion event created a small, staggered break at the euchromatic end. Our candidate A/C and B/D breakpoint regions also had strong BLAST hits to an In(1)sc8 A/C junction from the Dp(1;f)1187 minichromosome and the corresponding WT A/B junction identified in a previous study (31). Both our A/C fragment and that obtained by Glaser and Spradling (31) map the euchromatic part of the distal In(1)sc8 breakpoint to the same location in the D. melanogaster euchromatin and contain 1.688 satellite DNA in their heterochromatic part (Dataset S3).
In(1)dl-49 is an X-ray–induced inversion (32) with both distal and proximal breakpoints in euchromatic regions at bands 4D7–E1 and 11F2–4, respectively (20–22). We identified clusters of split/discordant reads for the distal breakpoint near X:4,791,300 (predicted band 4D5) and for the proximal breakpoint from approximately X:13,321,200–13,321,900 (predicted band 11F6). These candidate breakpoint intervals were also identified using Breakdancer (33), an independent method that is able to predict inversions with two euchromatic breaks. We extracted split/discordant reads within ±1.5 kb of each of the putative In(1)dl-49 breakpoint intervals plus their mate pairs, pooled reads from both breakpoints, then performed de novo assembly, followed by PCR and Sanger sequencing (Dataset S3). As expected, PCR amplification was successful in stocks carrying In(1)dl-49, but failed in stocks lacking In(1)dl-49 (Dataset S2).
Sanger sequencing verified the sequence of the A/C and B/D breakpoint assemblies for In(1)dl-49. Both the proximal and distal breakpoints were found in unique genomic regions, with the distal break occurring between X:4,791,293 and X:4,791,295 in an intron of CG42594 and the proximal break occurring from X:13,320,887 to X:13,321,245 in an intergenic region between SET domain-containing 2 (Set2) and Neuropilin and tolloid-like (Neto) (Fig. 2B). The breakpoint in the A/C fragment contained a small (3 bp) duplication not present in the reference genome, suggesting repair of a small staggered break during the inversion process. A 358-bp deletion was found in the B/D fragment, possibly due to resection during the repair event, which explains why the split/discordant reads for the proximal breakpoint mapped to an interval in the reference genome rather than to a single point.
The distal euchromatic breakpoint of the X-ray–induced In(1)FM6 was reported to be near band 15D-E (22, 27). We identified a cluster of split/discordant reads near X:16,919,300 (predicted band 15D3) in all FM7 stocks, and used these reads and the corresponding reads from the other end of the same paired-end sequenced fragments for de novo assembly. PCR using primers based on the two resulting putative A/C and B/D contigs validated that this breakpoint was present in all FM7 stocks, but not in stocks lacking the In(1)FM6 inversion (Dataset S2). Sanger sequencing of amplicons verified the predicted breakpoint sequences (Dataset S3). Euchromatic components of the A/C and B/D fragments mapped to the same location within an intron of CG45002 and revealed that the inversion introduced a 1-bp deletion (X:16,919,304) (Fig. 2B).
The heterochromatic part of the In(1)FM6 A/C fragment contains sequence from the transposable element HMS-Beagle (34), and the heterochromatic part of the B/D fragment contains 18S rDNA sequence, consistent with the proximal breakpoint being in X chromosome centric heterochromatin (35). The fact that the heterochromatic regions in the A/C and B/D fragments are not the same sequence suggests either a complex breakage/repair event following irradiation or postinversion rearrangement of sequences at either the A/C or B/D breakpoint. Nevertheless, the structure of the euchromatic junctions for the In(1)sc8, In(1)dl-49, and In(1)FM6 inversions carried on FM7 together show that X-ray–induced mutagenesis can often generate rearrangements with relatively precise breakpoints.
Recombination Generates Sequence Variation Among FM7 Chromosomes.
It is widely believed that balancers seldom undergo recombination (36, 37), giving rise to the idea that they should diverge from each other clonally and thus accumulate deleterious mutations under Muller’s ratchet (38). However, previous studies have shown that sequence exchange can occur, albeit rarely, both into and out of balancer chromosomes (4, 5), although the frequency and genomic scale of such events is unknown. To test whether ongoing sequence exchange between balancers and homologous chromosomes has occurred since the original synthesis of the first FM7 chromosome, we identified variants present on only one of the eight FM7 chromosomes in our sample. Unique variants that differentiate one FM7 from all others in our sample can arise either by de novo mutation or by recombination events that donate sequences from homologous chromosomes to balancers (by either gene conversion or crossing over); however, crossing over is the only mechanism that can explain the large contiguous tracts of sequence variation unique to individual FM7 chromosomes.
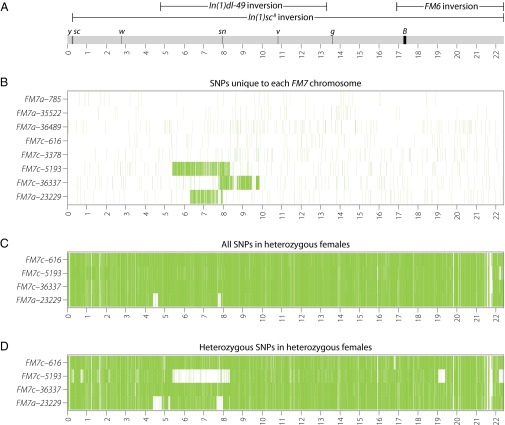
As shown in Fig. 3B, we observed megabase-scale tracts of unique variation on three of the eight FM7 chromosomes (FM7c-5193, FM7c-36337, and FM7a-23229), superimposed on a relatively even distribution of unique variants along the remainder of the chromosome. Notably, all of these tracts of unique variation were contained within the In(1)dl-49 inversion, spanned the sn locus, and were found only in sn+ stocks. These tracts of variants were not caused by placement of the snX2 allele onto FM7a to create FM7c (9), since FM7cs marked with snX2 (FM7c-616 and FM7c-3378) do not differ substantially in their SNP profile from FM7as in the sn region (Fig. S1B). In fact, similarity between FM7a and the original FM7c is expected in the sn region, because an In(1)dl-49 chromosome was a progenitor of FM7a (7, 8), the snX2 allele arose on an In(1)dl-49 chromosome (39), and an snX2-marked In(1)dl-49 served as the donor to move snX2 onto FM7a to create FM7c (9).
Fig. 3.
Recombination generates sequence diversity among FM7 balancer chromosomes. (A) Schematic of the WT X chromosome showing the locations of inversions (oriented with respect to the reference genome, not FM7), visible genetic markers, and Release 5 genome coordinates (in Mb). (B) Heatmap of unique SNPs found in only one FM7 chromosome in our sample. The density of unique SNPs is plotted in 5-kb windows with a 5-kb offset. The three large tracts of unique SNPs on FM7c-5193, FM7c-36337, and FM7a-23229 are contained fully within In(1)dl-49 and replace the snX2 allele with the WT sequence. The FM7a-23229 chromosome is a mislabeled FM7c (Fig. S1B). (C) Heatmap of all SNPs found in heterozygous female samples carrying FM7 balancers over different balanced X chromosomes. Genotypes of balanced X chromosomes are listed in Dataset S1. Small tracts in which few SNPs are present in FM7a-23229 arise because of common ancestry among the X chromosomes in FM7, the balanced chromosome, and the ISO-1 reference genome (Fig. S1C). (D) Heatmap of heterozygous SNPs found in heterozygous female samples carrying FM7 balancers over different balanced X chromosomes. LOH is observed for a large tract in FM7c-5193 that corresponds to the large tract of unique variants for this chromosome shown in B. LOH is also observed in FM7c-5193 for two deletions in the balanced chromosome [Df(1)JA27 and an uncharacterized deletion on the Df(1)JA27 chromosome], and for tracts in FM7a-23229 that share ancestry with y1-ncdD and ISO-1 (Fig. S1C).
Fig. S1.
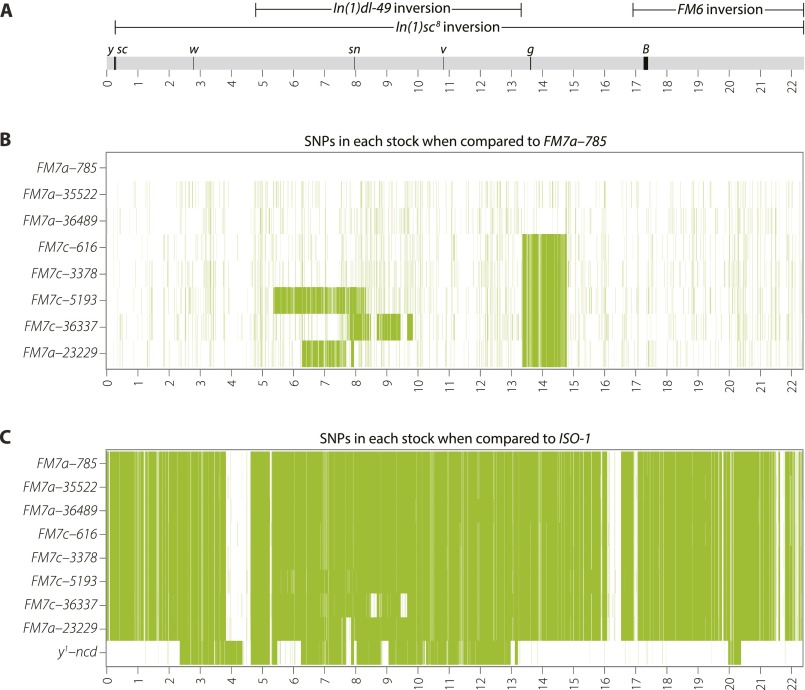
Polymorphisms are evident both among FM7 stocks and relative to the ISO-1 reference genome. (A) Schematic of the WT X chromosome, showing the locations of inversions (oriented with respect to the reference genome, not FM7), marker genes, and Release 5 genome coordinates (in Mb). (B) Heatmap of SNPs detected in the eight FM7 stocks used in this study when using FM7a-785 as a genome reference. Increased SNP density covering the sn region in stocks FM7c-5193, FM7c-36337, and FM7a-23229 indicates the region replaced by a DCO event. Note the increased SNP density between 13,369,185 Mb and 14,812,237 Mb present in all FM7c stocks (and the mislabeled FM7a-23229) that defines the haplotype containing g4 present on FM7c. (C) Heatmap of SNPs detected among all FM7 stocks and y1-ncdD compared with the ISO-1 reference genome. Sequence diversity among the eight FM7 stocks is apparent at this scale as differing levels of SNP density surrounding the sn locus. Blocks of similarity between all FM7’s and ISO-1 suggest a common ancestor for these regions. Blocks of diminished SNP density (in white) due to shared ancestry between FM7a-23229 and y1-ncdD are shown in Fig. 3D as an apparent absence of SNPs.
The nature of the snX2 allele was not determined in earlier studies (40); however, we identified a cluster of split/discordant reads at X:7,878,402–7,878,413 arising from the insertion of an F-element in the second coding exon of sn that is present only in the sn− stocks FM7c-616 and FM7c-3378. We propose that this F-element insertion is the lesion that causes the snX2 allele. In addition, if the tracts of variants in FM7c-5193, FM7c-36337, and FM7a-23229 arose from movement of snX2 onto FM7c, then they would not be unique. Rather, they would form a haplotype shared by all other FM7c chromosomes, as is observed in the region surrounding the g locus (Fig. S1B). The FM7c g haplotype on FM7a-23229 is unexpected, and suggests that this balancer is actually an FM7c that has been mislabeled as FM7a because of its sn+ phenotype. Taken together, these results indicate that all chromosomes with large tracts of unique SNPs are FM7cs that lack the snX2 allele.
The number of unique single nucleotide variants expected on each FM7 chromosome if they evolved clonally and independently under de novo mutation alone since their origin in 1968 (7, 8) to the time that our lines were sequenced is ∼150 (45 y * 26 generations/year * 22 × 106 bp * 5.8 × 10−9 mutations/bp/generation) (41). Shared ancestry among chromosomes in our sample, such as for the FM7c chromosomes that were generated several years later (9), would decrease the number of unique variants observed from this expectation. The number of unique variants observed for five out of eight FM7 chromosomes (56–152 unique SNPs) was less than or nearly equal to the expected value under independent clonal evolution with de novo mutation alone. However, the number of unique variants observed for FM7c-5193, FM7c-36337, and FM7a-23229 (between 541 and 3,564 unique SNPs) was more than threefold higher than expected under clonal evolution with mutation alone, suggesting that the action of additional processes, such as gene conversion or crossing over, is required to explain these observations.
The large tracts of unique variation on FM7c-5193, FM7c-36337, and FM7a-23229 range between 1.7 and 3.0 Mb in length and encompass 195–356 genes. Given that the average tract length of gene conversion in D. melanogaster is ∼350–450 bp (42, 43), we propose that the large tracts of unique variants on FM7c-5193, FM7c-36337, and FM7a-23229 arose by independent DCOs from unrelated chromosomes onto different FM7 balancer chromosome lineages that replaced snX2 with sn+.
The most obvious donor for sequence exchange onto a balancer chromosome is the chromosome with which it is kept in stock. To test whether the large tracts of unique sequence variation seen on FM7 chromosomes are the result of recombination with their homolog in stock, we sequenced heterozygous females from the three stocks with putative DCO events (FM7c-5193, FM7c-36337, and FM7a-23229) and from one negative control with no putative DCO event (FM7c-616). If a recent exchange event occurred between the balanced chromosome and its homolog, we would expect to see a loss of heterozygosity (LOH) in the region where the two chromosomes underwent recombination. As shown in Fig. 3C, the distribution of all SNPs (both homozygous and heterozygous variants) in heterozygous samples was high and relatively constant across the entire X chromosome for three of the four stocks, with two small regions in FM7a-23229 yielding a paucity of SNPs because of shared ancestry between all FM7s and the y1 chromosomes in both ISO-1 and the balanced chromosome (Fig. S1C). Analysis of heterozygous SNPs in heterozygous females (Fig. 3D) showed a relatively uniform distribution of heterozygous SNPs across the X chromosome, with clear LOH in the exact region of the predicted exchange event for FM7c-5193, but not for FM7c-36337 or FM7a-23229.
These results indicate that recent exchange between FM7c-5193 and its balanced homolog can explain the large tract of unique variants on this chromosome. However, the predicted exchange events for FM7c-36337 or FM7a-23229 must have occurred sometime in the past with different chromosomes other than those with which they are currently kept in stock.
Intriguingly, all three putative DCOs are contained within the central In(1)dl-49 inversion, occur on FM7c chromosomes, and replace the female-sterile snX2 allele present on the original FM7c (9) with a WT allele. Although DCOs fully within the In(1)dl-49 regions are rare (2, 44), such events would lead to viable FM7-bearing gametes (Fig. 1B). Furthermore, replacement of the female-sterile snX2 allele with sn+ would be expected to generate FM7 chromosomes with a fitness advantage relative to the ancestral FM7c, and thus these rare recombinant chromosomes could quickly increase in frequency in stock. Loss of snX2 could lead to a fitness advantage by allowing propagation of sn+ FM7 as homozygotes in females, although this would lead to a loss of balanced mutations in culture, which occurs only rarely. Alternatively, sn+ FM7c males may have a fitness advantage in crowded cultures relative to snX2 FM7c males, who have bristle and mechanosensory defects (22, 45). We favor the advantage of sn+ FM7c males in culture as the predominant mechanism by which sn+ FM7c chromosomes replace snX2 FM7c chromosomes, because FM7cs likely have accumulated other female-sterile mutations over time, which would reduce the fitness of homozygous sn+ FM7c females even in the absence of snX2.
To address how often loss of snX2 occurs in FM7c chromosomes, we screened and classified the sn phenotype in males from 630 stocks carrying an FM7c chromosome in the Bloomington Drosophila Stock Center (Dataset S4). Of 630 stocks labeled as carrying FM7c, 82 (13%) had the revertant sn+ phenotype in B-eyed males, consistent with loss of the female-sterile snX2 allele on FM7c chromosomes by DCO with a balanced homolog inside the In(1)dl-49 inversion while maintained in stock. Of these 82 stocks, only 16 (20%) had any previous evidence of snX2 reversion in their genotype or description, underscoring how commonly the snX2 reversion may occur without notice. The genotypes of these sn+ stocks have now been updated in the Bloomington Drosophila Stock Center database.
Because at least one of the FM7a stocks that we sequenced (FM7a-23229) was in reality an FM7c stock mislabeled as an FM7a stock, the lack of snX2 on FM7 chromosomes could simply reflect the fact that these chromosomes are actually FM7as mislabeled as FM7cs, rather than a true loss of snX2 by a DCO inside In(1)dl-49 on FM7c. To resolve these alternatives, we took advantage of the fact that all bona fide FM7cs are expected to carry the same allele at the garnet locus (g4), whereas all FM7as should lack this marker. Within the mutant g gene on all FM7c (and FM7a-23229) chromosomes (Fig. S1B), we found a diagnostic 24-bp deletion that spans an intron–exon junction and results in a frame shift in the RB and RD transcripts (FBtr0331709 and FBtr0073842), and also ablates the ATG start codon of the RF transcript (FBtr0331710). We tested 76 of the 82 revertant sn+ stocks labeled as FM7c in the Bloomington Drosophila Stock Center database for the presence or absence of this putative g4-causing deletion by PCR and Sanger sequencing. We found that 71 of 76 (93%) of the sn+ stocks screened by PCR and Sanger sequencing carried the g4 allele present on all FM7c chromosomes (Dataset S4), indicating that the majority of these are bona fide FM7cs and thus are truly revertants.
Because g lies outside the In(1)dl-49 inversion and sn resides inside it, it is highly unlikely that one DCO event could have replaced both snX2 and g4 in any of the five putative FM7c sn+ stocks lacking the g4 deletion. Therefore, we conclude that these five stocks have been mislabeled as FM7cs when in fact they are actually FM7as. Thus, the vast majority of sn+ stocks labeled as FM7cs in the Bloomington Drosophila Stock Center are indeed FM7cs, but mislabeling of FM7 subtypes (a vs. c) occurs in approximately 7% of stocks. Overall, these results support the conclusion that the DCOs within the In(1)dl-49 interval occur at an appreciable frequency, endangering mutations in homologous chromosomes kept in stock over balancer chromosomes and leading to sequence diversity among FM7c balancers in circulation today.
Origin and Reversion of the B1 Allele.
X chromosome balancers, including FM7, carry the B1 allele, a dominant mutation affecting eye morphology, discovered more than 100 years ago (46). B1 is an unusual allele that reverts to WT at a high frequency in females (47, 48) through either interchromosomal or intrachromosomal unequal exchange (15, 16, 18, 19). B1 is known to revert on FM7 (3), and previous work suggests that B1 reversion rates may be higher in inverted X chromosomes (19, 44). B1 has been shown to be associated with a tandem duplication of a large segment containing cytological bands 16A1–7, and B1 revertants lack this duplicated segment (49, 50). Muller (17) argued that B1 arose by an unequal exchange between two sister chromatids or homologous chromosomes, rather than through a duplicative insertion event, as suggested by Bridges (49).
Muller’s model for the origin of B1 is supported by the work of Tsubota et al. (51), who used a P-element–induced revertant of B1 to clone the putative breakpoint of the B1 duplication. Those authors found a roo transposable element located exactly at the breakpoint between the two duplicated segments, and proposed that the B1 allele originated by unequal exchange between roo elements located at 16A1 and 16A7 on two different homologous chromosomes (51) (Fig. 4A). The exact nature of the B1 rearrangement remains to be clarified, however, given that the 16A7 breakpoint of B1 identified by Tsubota et al. (51) contains a short segment of DNA not found in WT flies. Moreover, neither the genomic extent nor the gene content of the B1 duplication has been investigated in the context of modern genomic data.
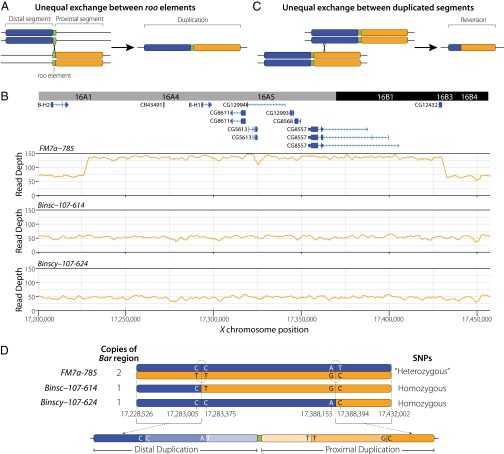
Fig. 4.
Genomic evidence for the role of unequal exchange at the Bar locus. (A) Model for the origin of the B1 allele by unequal exchange (17) between two different roo transposable elements (51). The distal and proximal segments of the B1 duplication are shown in blue and orange, respectively, and roo elements are shown in green. (B) Genome annotation and depth of coverage for X chromosome balancers carrying B1 (FM7a-785) and WT revertants (Binsc-107–614 and Binscy-107–624). Note the twofold increase in depth that starts downstream of B-H2 and ends upstream of CG12432 in the FM7a-785 chromosome carrying B1 that is lacking in Binsc-107–614 and Binscy-107–624 revertants. (C) Model for the reversion of the B1 allele to WT by unequal exchange between the two duplicated regions. The model shows an interchromosomal exchange event (15, 16); however, intrachromosomal exchange events are also possible (18, 19). (D) Schematic of sequence variants in B1 chromosomes (FM7a-785) and WT revertants (Binsc-107–614 and Binscy-107–624). Sequences from the distal and proximal duplicated regions in B1 chromosomes map to the same coordinates in the reference genome, resulting in apparent heterozygosity. The two revertant chromosomes are characterized by different haplotypes of homozygous SNPs. Sequences shared by both revertants at their 5′ and 3′ ends can be used to define the boundaries of unequal exchange events and partially phase the distal and proximal haplotypes, respectively. Diagnostic SNPs from fragments that span the junctions of putative unequal exchange events then can be used to phase haplotypes on both sides of both exchange junctions in the B1 chromosomes (dotted arcs), which together with the sequence of the revertants can be used to assign the location of each exchange event to the appropriate revertant stock.
We identified the precise genomic limits of the B1 duplication on the basis of a contiguous 203,476-bp region between X:17,228,526 and X:17,432,002 with twofold greater sequencing depth in all eight FM7 stocks sequenced (Fig. 4B). Sequences flanking the duplicated interval correspond exactly to the B1 breakpoints identified by Tsubota et al. (51). We found that previous uncertainty in the WT configuration of the 16A7 B1 breakpoint region reported by Tsubota et al. (51) is due to inclusion of phage DNA in their sequence. The B1 duplicated interval contains the BarH1 (B-H1) homeodomain gene, which has been shown to be involved in the Bar eye phenotype (52, 53), plus seven other predicted protein-coding genes and a putative ncRNA gene (CR43491) that likely corresponds to the T1/T2 or BarA transcript identified previously (52, 54). As predicted by Higashijima et al. (55), the B1 breakpoint lies in an intergenic region upstream of B-H1 and downstream of BarH2 (B-H2) (Fig. 4B), a related homeodomain gene also involved in eye morphogenesis (52). Thus, the B1 duplication on FM7 chromosomes carries an intact B-H2–B-H1 locus, plus an additional copy of B-H1 fused downstream of CG12432 (Fig. 4B).
Tsubota et al. (51) proposed that unequal exchange between two roo insertions at different positions on homologous chromosomes caused the B1 duplication (Fig. 4A). To provide an independent assessment of this hypothesis, we extracted split/discordant reads and their mate pairs in the ±1.5-kb intervals at either end of the duplicated segment, then performed de novo assembly as above for the FM7 inversions, and recovered two contigs spanning the 16A1 and 16A7 sides of the B1 breakpoint. Both of these contigs contained roo sequences that began after the exact point at which alignment to the reference genome ended. We used long-range PCR to amplify an ∼8-kb fragment spanning the breakpoint from the end of 16A7 to the beginning of 16A1 in FM7-carrying stocks, but not in WT stocks (Dataset S2). Sanger sequencing of the 5′ and 3′ ends of this breakpoint-spanning fragment revealed a roo element in the expected location and orientation (Dataset S3). Taken together, these results confirm the work of Tsubota et al. (51), showing that the B1 breakpoint contains a roo element in the 5′ to 3′ orientation located precisely at the junction between the duplicated segments.
Our genomic data also allow us to investigate sequence variation directly within the B1 duplication, which provides new insights into the origin and reversion of the B1 allele. Analysis of sequence variation in the region duplicated in B1 revealed a large number of “heterozygous” SNPs in each hemizygous or homozygous FM7 stock (minimum, 1,242; maximum, 1,250). Heterozygous SNPs in hemizygous or homozygous stocks can arise from calling variants in duplicated regions that are mapped to the same single-copy interval of the reference genome (56). This apparent heterozygosity in the B1 interval implies the existence of substantial sequence divergence between the two ancestral haplotypes that underwent unequal exchange to form the original B1 allele, providing independent support for the origin of B1 by unequal exchange between two homologous chromosomes rather than two sister chromatids (51). In addition, the heterozygous SNP profile was nearly identical among all eight FM7 stocks, supporting a single origin for the B1 allele, consistent with the historical record (46).
These heterozygous variants also give us a rich set of molecular markers that, together with depth of coverage in the B region, can be used to investigate the mechanism of B1 reversion. If reversion of the B1 allele is due to either interchromosomal or intrachromosomal unequal exchange (15, 16, 18, 19), then we would expect a twofold reduction in the depth of coverage to be associated with loss of heterozygosity across the entire B1 duplicated region in revertant chromosomes (Fig. 4C). To test this hypothesis, we identified two X chromosome balancer stocks carrying reversions of B1 (Binsc-107–614 and Binscy-107–624) and sequenced their genomes. As expected, the depth of coverage in both B1 revertants was at WT levels across the B1 interval X:17,228,526–17,432,002 (Fig. 4B). In addition, no high-quality heterozygous SNPs or split/discordant reads were observed in the B1 interval in either revertant. These results demonstrate that the duplicated segment is strictly associated with the B phenotype, as shown previously at the cytological level (49, 50).
Comparison of the single-copy haplotypes in the two revertants revealed likely sites of unequal exchange (Fig. 4D). Binsc-107–614 and Binscy-107–624 haplotypes in the B1 interval contained the same SNPs from X:17,228,526–17,283,005 and again from X:17,388,394–17,432,002, but differed from each other in the central X:17,283,375–17,388,155 interval. This result indicates that unequal exchange must have occurred in a 370-bp window between X:17,283,005 and X:17,283,375 in one stock, and in a 239-bp window between positions X:17,388,155 and X:17,388,394 in the other stock. This result also implies that the haplotype from the beginning of B1 to 17,283,005 is from the 5′ duplicated segment, and that the haplotype from X:17,388,394 to the end of B1 is from the 3′ duplicated segment.
Because the SNPs defining the sites of unequal exchange were close to one another, we were able to phase haplotypes from the distal and proximal duplicates using read-pair data in nonrecombinant FM7 “heterozygotes.” Knowing the phase and location of both nonrecombinant haplotypes in the B1 duplication allowed us to infer that unequal exchange occurred between X:17,283,005 and X:17,283,375 in Binsc-107–614, and independently between X:17,388,155 and X:17,388,394 in Binscy-107–624. Taken together, these data provide definitive genomic evidence that B1 reversion is associated with unequal exchange among duplicated segments directly within the B1 interval.
Discussion
Our work provides detailed insight into the structure and diversity of the most commonly used X chromosome balancer in D. melanogaster, FM7. We mapped and characterized breakpoints of the three large inversions present on FM7 and identified major sequence differences in the vicinity of g between the two subtypes of FM7 (FM7a and FM7c). Surprisingly, we identified megabase-scale tracts of unique sequences in different FM7cs that likely arose from DCOs removing the snX2 allele within the In(1)dl-49 inversion. We also found that loss of the snX2 allele affected a substantial proportion of FM7c chromosomes at the Bloomington Drosophila Stock Center. Finally, we clarified the molecular organization of the B1 allele carried on FM7, and found definitive genomic evidence for the origin and reversion of B1 by unequal exchange. In contrast to the prevailing notion of balancers as clonal nonrecombining chromosomes, our results provide evidence that rare recombination events have led to large-scale sequence differences among balancers currently used by Drosophila researchers.
Our work has a number of implications for the design and interpretation of experiments that use X chromosome balancers in D. melanogaster. Knowing the precise molecular location of inversion breakpoints on FM7 reveals regions of the X chromosome that are more or less susceptible to exchange events. Furthermore, the fact that many FM7cs carry megabase-scale tracts of unique variation, and that a substantial proportion of FM7 chromosomes are mislabeled, should motivate researchers to characterize which FM7 subtype their stocks actually carry. Characterization of an FM7 subtype may be carried out by PCR and Sanger sequencing of g, or by simply crossing the FM7 chromosome in question to a stock carrying a loss-of-function allele of g and scoring the eye phenotype of heterozygous females. The genomic scale of sequence differences among FM7 subtypes is sufficiently large such that, without controlling properly for FM7 subtype, the effects attributed to balanced chromosomes in heterozygotes could arise from differences in the FM7 genetic background.
Our finding that reversion of the female-sterile snX2 allele by DCO in the In(1)dl-49 interval is common suggests that researchers should be cautious when using FM7c for long-term stock maintenance of mutations in this region. We advise that replicate copies of such stocks be maintained and periodically checked for sn+, B1 males that could indicate breakdown of the balanced chromosome by a DCO event. Alternatively, such mutations could be maintained using attached-X stocks instead of balancer chromosomes (3). Unavoidable DCOs within the In(1)dl-49 region that remove the snX2 allele on FM7c may motivate the synthesis of a new generation of female-sterile X chromosome balancers, perhaps by introducing additional inversions inside the In(1)dl-49 interval on FM7c. Although our work documents that DCOs do occur within FM7 on a historical timescale, we emphasize that such events are not sufficiently common to impair the utility of FM7s as balancer chromosomes in routine genetic analysis.
The present study also demonstrates the value of sequencing classical stocks of D. melanogaster to uncover the molecular basis of uncharacterized mutations and better understand the genetic background of mutant stocks. Despite the availability of a nearly complete, richly annotated genome sequence, more than 1,000 existing classical mutations in D. melanogaster have not been associated with gene models or linked to genomic sequences. Here we have identified the causal molecular basis of three classical inversions, In(1)sc8, In(1)dl-49, and In(1)FM6; mapped the locations of the B1 duplication and Df(1)JA27 deletion (356-kbp deletion from X:19,043,642 to X:19,399,862); proposed candidates for the lesions that cause the g4 and snX2 alleles; and identified an uncharacterized deletion in the Df(1)JA27 chromosome (X:22,164,372–heterochromatin).
Further analysis of our genomic data should provide insight into the molecular basis of additional mutations carried by these strains, including the sites of transgene insertions that mark some FM7 balancer chromosomes (10–14). Sequencing classical laboratory stocks also can lead to the identification of mislabeled strains (e.g., that FM7a-23229 is in fact a FM7c chromosome) and unreported genotypes (e.g., sn+ in FM7a-23229), and thereby reduce sources of unwanted experimental variation. Thus, systematic sequencing of stocks in the Bloomington Drosophila Stock Center could improve the precision of Drosophila genetics and, in conjunction with extensive phenotypic information in FlyBase, provide a powerful model for developing workflows to identify rare disease variants in humans.
Future work on second and third chromosome balancers is needed to generalize the findings reported here, although such studies will be more challenging because genomic analysis will need to be performed in heterozygotes. Sequencing larger samples of FM7 chromosomes also could provide deeper insight into the mechanisms of exchange in highly inverted chromosomes (2, 44). Here we identified 71 FM7c sn+ stocks that are bona fide FM7cs likely to have undergone DCO with a balanced stock, which should provide a rich sample for studying how DCOs are distributed relative to the locations of breakpoints in inversion heterozygotes. Likewise, sequencing of additional B1 revertants can now be used as a model system to study unequal exchange at the molecular level, especially given our finding that the two duplicated regions in B1 differ by numerous variants. By generating a large sample of B1 revertants in heterozygotes that differ from FM7 outside the B1 interval, it will be possible to precisely measure the relative contribution of interchromosomal and intrachromosomal unequal exchange events, and to understand how unequal exchange events are distributed across the duplicated region. More in-depth analysis of sequence variation among FM7 chromosomes also could lead to insights into gene conversion between balancers and balanced chromosomes (4, 5), as well as into whether the predicted accumulation of deleterious mutations on balancers is observed at the molecular level (38). Finally, sequencing a larger panel of FM7 chromosomes and more primitive X chromosome balancers could shed light on the ancestral state of FM7 at the time of its origin, as well as how inversions were integrated within inversions to create the founders of the FM series (57).
Methods
Fly Stocks Used.
The X chromosome balancer stocks used in this experiment were obtained from either the Bloomington Drosophila Stock Center or the Drosophila Genetic Resource Center. Dataset S1 lists stock identifiers. The y1-ncdD stock used as a parental X chromosome in the construction of the ISO-1 reference genome strain (58) was obtained from Jim Kennison. Full genotypes of stocks as labeled at the outset of this project are listed in Dataset S1 and are referred to in the text by their abbreviated genotype followed by their stock number (where available). All flies were kept on standard cornmeal-molasses and maintained at 25 °C.
DNA Preparation and Whole-Genome Sequencing.
DNA was prepared from 10 adult hemizygous FM7-carrying Bar eyed males for stocks FM7a-785, FM7a-23229, FM7a-35522, FM7a-36489, FM7c-616, FM7c-3378, Binsc-107–614, and Binscy-107–624. Because of the poor viability of FM7-carrying hemizygous males in FM7c-5193 and FM7c-36337, DNA was prepared from a mixture of 10 adult hemizygous FM7 males and homozygous FM7 females for these two samples. Ten heterozygous adult females were used for the FM7c-616, FM7c-5193, FM7c-36337, and FM7a-23229 heterozygous samples, and 10 adult hemizygous yellow males were used for the y1-ncdD sample. All DNA samples were extracted using the Qiagen DNeasy Blood & Tissue Kit (catalog no. 69504). Flies were starved for 4 h before freezing at –80 °C for at least 1 h before DNA extraction. Then 600- to 800-bp fragments of DNA were selected after shearing, and libraries were prepared using a Nextera DNA Sample Prep Kit (Illumina; catalog no. FC-121-1031) following the manufacturer’s directions. Hemizygous males and homozygous females from stocks FM7a-785, FM7a-23229, FM7a-35522, FM7a-36489, FM7c-616, FM7c-5193, FM7c-3378, and FM7c-36337 were sequenced as 100-bp paired-end samples on an Illumina HiSeq 2500 system. Heterozygous females from stocks FM7c-616, FM7c-5193, FM7a-23229, and FM7c-36337 were sequenced as 150-bp paired-end samples on an Illumina HiSeq 2500 system. Hemizygous males from stocks y1-ncdD, Binsc-107–614, and Binscy-107–624 were sequenced as 150-bp paired-end samples on an Illumina NextSeq.
Genome Alignment and SNP Calling.
Alignment to the UCSC Genome Bioinformatics dm3 version of the Release 5 D. melanogaster reference genome sequence was performed using bwa (version 0.7.7-r441) (59). Variants were called using SAMtools and BCFtools version 0.1.19–44428cd (60). Indels and low-quality SNPs (qual <200) were filtered out of single-sample variant call format (VCF) files. Unique SNPs were identified by also filtering out heterozygous SNPs from single-sample VCF files and merging samples to identify SNPs present in only one sample using VCFtools version 0.1.12b and visualized as heatmaps using R version 3.1.3.
Identification, Assembly, and Validation of Rearrangement Breakpoints.
Rearrangement breakpoints were identified using three strategies. For the In(1)sc8, In(1)dl-49, In(1)FM6, and B1 breakpoints, split/discordant X chromosome read pairs were identified using samblaster (61) and visualized using the UCSC Genome Browser (62). Clusters of split/discordant reads corresponding to putative breakpoints were identified in the approximate locations where rearrangements were expected based on classical work. Original fastq sequences of split/discordant reads and their mate pairs from ±1.5 kb around putative breakpoints from the same rearrangement were then merged from all eight FM7 stocks into a single per-rearrangement file. SOAPdenovo2 version 2.04 was then used to perform de novo assemblies for both breakpoints of each rearrangement at the same time using a kmer size of 41 or 51 for the In(1)sc8, In(1)dl-49, and In(1)FM6 inversions and a kmer size of 73 for the B1 duplication breakpoint (63).
To identify the In(1)dl-49 inversion, we also ran Breakdancer version 1.4.4 (33) using default options, with the exception that only the X chromosome was analyzed (-o X), and any event with fewer than 10 supporting reads was ignored (−r 10). For the B1 duplication, we also identified an interval with the expected twofold higher read-depth coverage in the location where the duplication was expected to be found (Fig. 4B) (22).
Contigs spanning candidate breakpoints were used to design PCR primers on either side of each candidate breakpoint region using Primer3 (64). PCR was performed using Phusion DNA polymerase (New England BioLabs; catalog no. M0530L) with a 62 °C annealing temperature and 45-s extension time. PCR products were purified from a gel using the QIAquick PCR Purification Kit (Qiagen; catalog no. 28106) and Sanger sequenced. Long-range PCR of the junction between the two duplicated B1 regions was performed using the Qiagen LongRange PCR Kit (catalog no. 206402) using 250 ng of genomic DNA, with a 59 °C annealing temperature, and 9-min extension time.
Screen for sn Reversion in FM7 Stocks at the Bloomington Drosophila Stock Center.
We visually screened 630 stocks from the Bloomington Drosophila Stock Center that were labeled as carrying FM7c for the presence or absence of the sn phenotype in B males. Eighty-two stocks yielded B, sn+ males and were classified as putative FM7c revertants. To determine whether putative FM7c revertants were in fact mislabeled FM7as, we screened 76 of these putative FM7c revertants for the presence of a diagnostic 24-bp deletion associated with the g4 allele present on all bona fide FM7cs. The primers used to amplify a fragment spanning the g4 deletion were garnet_F2 (5′-ACACCCGCATCGTATTGATT-3′) and garnet_R2 (5′-CCAGTTGGCTGAAACTGAAA-3′). DNA was prepared by placing single B, sn+ males in a standard fly squish buffer (50 μL of 1 M Tris pH 8.0, 0.5 M EDTA, 5 M NaCl) plus 1 μL of 10 mg/mL proteinase K. Extracts were then placed in a thermocycler at 37 °C for 30 min and 95 °C for 2 min, followed by a 4 °C hold. PCR was performed using 4 μL of fly squish product in a total volume of 50 μL. Fragments were amplified using Phusion polymerase (New England BioLabs; catalog no. M0530L). Reaction conditions were in accordance with the manufacturer’s instructions except for a 64 °C annealing temperature and a 45-s extension time. PCR amplicons were Sanger sequenced, and the resulting sequences were aligned to the reference genome to determine the presence or absence of the 24-bp deletion.
Supplementary Material
Acknowledgments
We thank Jim Kennison for the y1-ncdD stock; Kate Malanowski, Kendra Walton, and Anoja Perera for expert assistance with DNA sequencing; Angela Miller for assistance with editing and figure preparation; John Merriam, Dan Lindsley, Jim Kennison, Alexander Konev, and Andreas Prokop for helpful discussions; members of the R.S.H. and C.M.B. laboratories for constructive comments on the manuscript; and GitHub for providing free private repositories that enabled this collaboration. R.S.H. is supported by the Stowers Institute for Medical Research. K.R.C. was supported by National Institutes of Health Grant P40 OD018537. C.M.B. was supported by Human Frontier Science Program Young Investigator Grant RGY0093/2012.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the European Bioinformatics Institute Sequence Read Archive, www.ebi.ac.uk/ena/ (project no. PRJEB11499).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601232113/-/DCSupplemental.
References
- 1.Beadle GW, Sturtevant AH. X chromosome inversions and meiosis in Drosophila melanogaster. Proc Natl Acad Sci USA. 1935;21(6):384–390. doi: 10.1073/pnas.21.6.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novitski E, Braver G. An analysis of crossing over within a heterozygous inversion in Drosophila melanogaster. Genetics. 1954;39(2):197–209. doi: 10.1093/genetics/39.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M, Golic K, Hawley RS. Drosophila—A Laboratory Handbook. 2nd Ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. [Google Scholar]
- 4.Blumenstiel JP, et al. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics. 2009;182(1):25–32. doi: 10.1534/genetics.109.101998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper JL, et al. Retention of induced mutations in a Drosophila reverse-genetic resource. Genetics. 2008;180(1):661–667. doi: 10.1534/genetics.108.092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong WJ, McKim KS, Hawley RS. All paired up with no place to go: Pairing, synapsis, and DSB formation in a balancer heterozygote. PLoS Genet. 2005;1(5):e67. doi: 10.1371/journal.pgen.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merriam JR. FM7: First multiple seven. Dros Inf Serv. 1968;43:64. [Google Scholar]
- 8.Merriam JR. FM7: A “new” first chromosome balancer. Dros Inf Serv. 1969;44:101. [Google Scholar]
- 9.Merriam JR, Duffy C. First multiple seven now contains sn[x2] for better balancing. Dros Inf Serv. 1972;48:43–44. [Google Scholar]
- 10.Le T, et al. A new family of Drosophila balancer chromosomes with a w−dfd-GMR yellow fluorescent protein marker. Genetics. 2006;174(4):2255–2257. doi: 10.1534/genetics.106.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casso D, Ramírez-Weber F, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev. 2000;91(1-2):451–454. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- 12.Abreu-Blanco MT, Verboon JM, Parkhurst SM. Cell wound repair in Drosophila occurs through three distinct phases of membrane and cytoskeletal remodeling. J Cell Biol. 2011;193(3):455–464. doi: 10.1083/jcb.201011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lattao R, Bonaccorsi S, Guan X, Wasserman SA, Gatti M. Tubby-tagged balancers for the Drosophila X and second chromosomes. Fly (Austin) 2011;5(4):369–370. doi: 10.4161/fly.5.4.17283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pina C, Pignoni F. Tubby-RFP balancers for developmental analysis: FM7c 2xTb-RFP, CyO 2xTb-RFP, and TM3 2xTb-RFP. Genesis. 2012;50(2):119–123. doi: 10.1002/dvg.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturtevant AH, Morgan TH. Reverse mutation of the Bar gene correlated with crossing over. Science. 1923;57(1487):746–747. doi: 10.1126/science.57.1487.746. [DOI] [PubMed] [Google Scholar]
- 16.Sturtevant AH. The effects of unequal crossing over at the Bar locus in Drosophila. Genetics. 1925;10(2):117–147. doi: 10.1093/genetics/10.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller HJ. Bar duplication. Science. 1936;83(2161):528–530. doi: 10.1126/science.83.2161.528-a. [DOI] [PubMed] [Google Scholar]
- 18.Peterson HM, Laughnan JR. Intrachromosomal exchange at the Bar locus in Drosophila. Proc Natl Acad Sci USA. 1963;50(1):126–133. doi: 10.1073/pnas.50.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabay SJ, Laughnan JR. Recombination at the bar locus in an inverted attached-X system in Drosophila melanogaster. Genetics. 1973;75(3):485–495. doi: 10.1093/genetics/75.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Painter TS. The morphology of the X chromosome in salivary glands of Drosophila melanogaster and a new type of chromosome map for this element. Genetics. 1934;19(5):448–469. doi: 10.1093/genetics/19.5.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoover ME. Cytogenetic analysis of nine inversions in Drosophila melanogaster. Z Vererbungsl. 1938;74(1):420–434. [Google Scholar]
- 22.Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. Academic Press; San Diego, CA: 1992. [Google Scholar]
- 23.Sidorov BN. A study of step-allelomorphism in Drosophila melanogaster: A case of origination of an allelomorph of scute producing simultaneously characters of “hairy wing” (translation). Zhurnal eksperimental noi biologii i meditsiny. 1931;7:28–40. [Google Scholar]
- 24.Muller HJ, Prokofyeva AA. Continuity and discontinuity of the hereditary material. Dokl Akad Nauk SSSR NS. 1934;4:74–83. [Google Scholar]
- 25.Patterson JT, Stone WS. Some observations on the structure of the scute-8 chromosome of Drosophila melanogaster. Genetics. 1935;20(2):172–178. doi: 10.1093/genetics/20.2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson JT. The mechanism of mosaic formation in Drosophila. Genetics. 1933;18(1):32–52. doi: 10.1093/genetics/18.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grell RF, Lewis EB. New mutants report. Dros Inf Serv. 1956;30:71. [Google Scholar]
- 28.Campuzano S, et al. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985;40(2):327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh T, Brutlag D. Sequence and sequence variation within the 1.688 g/cm3 satellite DNA of Drosophila melanogaster. J Mol Biol. 1979;135(2):465–481. doi: 10.1016/0022-2836(79)90447-9. [DOI] [PubMed] [Google Scholar]
- 30.Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134(4):1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glaser RL, Spradling AC. Unusual properties of genomic DNA molecules spanning the euchromatic-heterochromatic junction of a Drosophila minichromosome. Nucleic Acids Res. 1994;22(23):5068–5075. doi: 10.1093/nar/22.23.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller HJ. 1926. Laboratory notebook. Muller mss., 1910–1967, The Lilly Library Manuscript Collections (Series VI, Box 2, 1926–1927. X-ray mutation experiment starting Fall of 1926), Indiana University Libraries, Bloomington, IN.
- 33.Chen K, et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6(9):677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder MP, et al. A transposable element that splits the promoter region inactivates a Drosophila cuticle protein gene. Proc Natl Acad Sci USA. 1982;79(23):7430–7434. doi: 10.1073/pnas.79.23.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tartof KD, Dawid IG. Similarities and differences in the structure of X and Y chromosome rRNA genes of Drosophila. Nature. 1976;263(5572):27–30. doi: 10.1038/263027a0. [DOI] [PubMed] [Google Scholar]
- 36.Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: Behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116(5):1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes SE, et al. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 2009;5(1):e1000348. doi: 10.1371/journal.pgen.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Araye Q, Sawamura K. Genetic decay of balancer chromosomes in Drosophila melanogaster. Fly (Austin) 2013;7(3):184–186. doi: 10.4161/fly.24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bender HA. Studies on the expression of various singed alleles in Drosophila melanogaster. Genetics. 1960;45(7):867–883. doi: 10.1093/genetics/45.7.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paterson J, O’Hare K. Structure and transcription of the singed locus of Drosophila melanogaster. Genetics. 1991;129(4):1073–1084. doi: 10.1093/genetics/129.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445(7123):82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- 42.Miller DE, et al. A whole-chromosome analysis of meiotic recombination in Drosophila melanogaster. G3 (Bethesda) 2012;2(2):249–260. doi: 10.1534/g3.111.001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilliker AJ, et al. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics. 1994;137(4):1019–1026. doi: 10.1093/genetics/137.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sturtevant AH, Beadle GW. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics. 1936;21(5):554–604. doi: 10.1093/genetics/21.5.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cant K, Knowles BA, Mooseker MS, Cooley L. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J Cell Biol. 1994;125(2):369–380. doi: 10.1083/jcb.125.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tice SC. A new sex-linked character in Drosophila. Biol Bull. 1914;26(4):221–230. [Google Scholar]
- 47.May HG. Selection for higher and lower facet numbers in the bar-eyed race of Drosophila and the appearance of reverse mutations. Biol Bull. 1917;33(6):361–395. [Google Scholar]
- 48.Zeleny C. The direction and frequency of mutation in the bar-eye series of multiple allelomorphs of Drosophila. J Exp Zool. 1921;34(2):202–233. [Google Scholar]
- 49.Bridges CB. The Bar “gene” A duplication. Science. 1936;83(2148):210–211. doi: 10.1126/science.83.2148.210. [DOI] [PubMed] [Google Scholar]
- 50.Muller HJ, Prokofyeva AA, Kossikov KV. Unequal crossing-over in the bar mutant as a result of duplication of a minute chromosome of Drosophila. Comptes Rendus (Doklady) de l'Acad Sci URSS. 1936;1(10):83–88. [Google Scholar]
- 51.Tsubota SI, Rosenberg D, Szostak H, Rubin D, Schedl P. The cloning of the Bar region and the B breakpoint in Drosophila melanogaster: Evidence for a transposon-induced rearrangement. Genetics. 1989;122(4):881–890. doi: 10.1093/genetics/122.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higashijima S, et al. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 1992;6(1):50–60. doi: 10.1101/gad.6.1.50. [DOI] [PubMed] [Google Scholar]
- 53.Kojima T, et al. Identification of a different-type homeobox gene, BarH1, possibly causing Bar (B) and Om(1D) mutations in Drosophila. Proc Natl Acad Sci USA. 1991;88(10):4343–4347. doi: 10.1073/pnas.88.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norris E, Sanders M, Crumety V, Tsubota SI. The identification of the Bs breakpoint and of two possible Bar genes. Mol Gen Genet. 1992;233(1-2):106–112. doi: 10.1007/BF00587567. [DOI] [PubMed] [Google Scholar]
- 55.Higashijima S, Michiue T, Emori Y, Saigo K. Subtype determination of Drosophila embryonic external sensory organs by redundant homeo box genes BarH1 and BarH2. Genes Dev. 1992;6(6):1005–1018. doi: 10.1101/gad.6.6.1005. [DOI] [PubMed] [Google Scholar]
- 56.Remnant EJ, et al. Gene duplication in the major insecticide target site, Rdl, in Drosophila melanogaster. Proc Natl Acad Sci USA. 2013;110(36):14705–14710. doi: 10.1073/pnas.1311341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis EB, Mislove RF. New mutants report. Dros Inf Serv. 1953;27:57–58. [Google Scholar]
- 58.Brizuela BJ, Elfring L, Ballard J, Tamkun JW, Kennison JA. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics. 1994;137(3):803–813. doi: 10.1093/genetics/137.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faust GG, Hall IM. SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30(17):2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenbloom KR, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43(Database issue):D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li R, et al. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 64.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.