The large-scale automation of neuroscience has enabled the construction of genome-wide atlases, of which the Allen Brain Atlas (ABA), which allows the 3D visualization of the expression profile of 21,500 genes in the male mouse brain down to single-cell level resolution, is the most comprehensive (1). In PNAS, Mahfouz et al. (2) use the ability of the ABA to pinpoint the anatomical locations of expressed genes to uncover transcripts whose expression profiles correlate with those of steroid receptors, to begin to understand their function and specificity of action in different brain regions. This study shows that mapping combinatorial interactions among specific sets of genes represents a significant leap forward in our understanding of how tissue specificity for a given signaling pathway is determined, and in identifying the potential relationship between otherwise unrelated brain areas in terms of the adaptive response to specific biological and environmental challenges.
Steroid receptors are pleiotropic transcription factors belonging to the superfamily of nuclear receptors, whose activity is induced by steroid hormones: lipophilic signaling molecules derived from cholesterol and primarily produced by the gonads and the adrenal cortex. In the mammalian brain, steroid hormones mediate the feedback from these steroid-generating organs on the neuroendocrine hypothalamus to control bodily functions (reproduction, metabolism, stress, inflammation, osmoregulation), but also play a fundamental organizational role during brain development, trigger adult brain plasticity, and are involved in cognitive and emotional regulation (3–10).
The idea of having an anatomical map combined with a quantitative expression map of nuclear receptor genes dates back to 2007, when Gofflot et al. created an interactive database of 49 nuclear receptor genes spanning more than 100 different regions of the mouse brain (11). These researchers used two complementary approaches to meet the challenge of obtaining both cell-level resolution and an unbiased expression profile of large anatomical regions: real-time PCR provided a broad estimate of nuclear receptor expression levels in selected brain regions, and spatial expression patterns were more closely studied using high-resolution in situ hybridization (ISH). The study by Mahfouz et al. (2) significantly extends this approach to identify novel aspects of steroid hormone action via the spatial correlation of the expression patterns of steroid receptors with those of genes that could potentially be steroid hormone targets or even downstream receptor coregulators.
Six well-studied steroid receptors were chosen by Mahfouz et al. (2) to costar in the “guilt by association” play; their expression profiles, although already reported in the literature, were validated using 3D spatial gene-expression data from the ABA (1). Based on the “guilt by association” principle, genes with similar spatial expression profiles are assumed to share similar biological functions, forming a neighborhood network of potential partners (12). Hence, for each receptor, Mahfouz et al. (2) ranked potential partner genes based on their spatial coexpression in various brain structures in different parts of the brain, and tested the assumption that genes strongly coexpressed within a given brain region are related to a localized functional role of the steroid receptor. Genome-wide coexpression analyses indeed showed the strong coexpression of known glucocorticoid receptor (GR) transcriptional targets in the hippocampus and known estrogen receptor-α (ESR1) transcriptional targets in the hypothalamus. Interestingly, these analyses revealed an unexpected coexpression of Esr1-related genes with Esr1 outside known sites of action of estrogens, calling into question our understanding of the coordinated response of the brain to this gonadal steroid. Equally intriguing is the finding that among the top 10 genes coexpressed with Gr, as well as known glucocorticoid-responsive genes across the whole brain, none were strongly coexpressed in the hypothalamus, indicating that GR signaling in the hypothalamus is distinct from GR signaling in the rest of the brain. These results show that spatial coexpression analysis has great potential for the identification of novel steroid receptor targets and putative region-specific pathways or gene networks. One should, however, be very careful when interpreting these unanticipated coexpression patterns; these representations do not necessarily reflect a causal relationship, but rather a likelihood of the association of certain genes that could indeed identify an actual functional relationship after further validation via expression measurement techniques.
Alternatively, the coexpression patterns described by Mahfouz et al. (2) could pinpoint potential steroid receptor coregulators rather than target genes, because each correlation merely indicates a possible associative, rather than causative, link between expressed genes. To explore this possibility, the authors analyzed the coexpression patterns of each and every steroid receptor as well as a set of published nuclear receptor coregulators. The results reveal a great heterogeneity of coexpression patterns across distinct brain regions, pointing toward selective, region-specific coregulation. For example, Pias2 and Ncoa4, two coactivators of GR and of the androgen receptor (AR), were found to be highly coexpressed with Gr in midbrain and hypothalamic regions, respectively, but not with Ar, although the relative abundance of its transcript is higher than that of Gr in these regions. Conversely, the authors uncovered the high coexpression of the mineralocorticoid receptor (Mr) with several nuclear receptor coregulators not thought to regulate MR function in the hippocampus, thus identifying putative novel coregulators of the MR pathway in this brain region. These results highlight the region-specific actions of coregulators and support Mahfouz et al.’s notion that a brain-wide qualitative approach measuring mRNA levels using ISH could indeed be used to identify the spatially restricted regulation of steroid receptor function.
An important limitation of this approach, however, lies in the fact that it relies on the quality of the ISH, which is insufficient for some genes and datasets. Consequently, there is a risk of false-negative results that goes hand in hand with the use of a genome-wide approach to identify region-specific targets and coregulators. This is something that Mahfouz et al. (2) do not neglect to underline. However, their findings constitute a rich resource for the further prediction and validation of upstream or downstream genes using quantitative approaches, like quantitative PCR (qPCR) or ChIP analysis.
The strength and utility of the approach proposed by Mahfouz et al. (2) is further demonstrated by the prediction of Magel2 as a transcriptional target for ESR1. Brain estrogen receptors α were selectively activated using the synthetic estrogen diethylstilbestrol
Mapping combinatorial interactions among specific sets of genes represents a significant leap forward in our understanding of how tissue specificity for a given signaling pathway is determined.
(DES) in castrated male mice; this resulted in an increase in mRNA expression for estrogen-responsive genes. Among the 10 genes most significantly coexpressed with Esr1, they identified Magel2, a previously unidentified ESR1 target that shows strong coexpression with Esr1 in the hypothalamus according to the ABA database. The measurement of Magel2 expression using qPCR and quantitative double ISH in the hypothalamus after DES-induced activation revealed a significant increase in Magel2 mRNA levels after DES treatment, predicting that Magel2 could be a target of ESR1. The loss of Magel2 expression has been shown to contribute to several aspects of Prader-Willi syndrome (13), including hypogonadotropic hypogonadism (13, 14), providing support for a link with estrogen regulation. As noted above, though, coexpression analyses remain a rather indirect measurement of interaction, and ChIP assays followed by next-generation sequencing to identify ESR1 binding sites in the Magel2 promoter region will be required to fully validate this hypothesis.
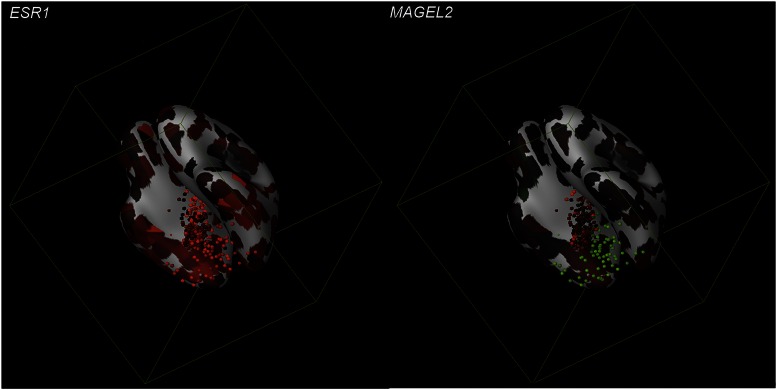
The findings of Mahfouz et al. (2) convincingly show that the spatial correlation of steroid receptors with genome-wide mRNA expression across different regions of the mouse brain using web-based repositories provides a novel in silico assay with which to explore novel aspects of steroid hormone action and obtain a glimpse of how the brain, by integrating ever-fluctuating combinatorial levels of circulating steroid hormones, orchestrates the adaptive response of the organism. Could this study be the stepping-stone to deciphering even more unknown steroid receptor pathways and networks not only in the mouse brain but also in the human brain (Fig. 1)? Certainly one could envision that this method will act as a catalyst for the elucidation of the molecular mechanisms underlying steroid actions, but also for more efficient drug production against neuroendocrine disorders. Clearly, as exciting as this possibility is, there is a great deal of work yet to be done to overcome its limitations in terms of the sensitivity of the high-resolution ISH, and improve its predictive power. Still, as neuroscientists we cannot but acknowledge the unparalleled opportunity for prediction that this technique represents, and that too of using data already available in the literature and online databases, to explore uncharted territories in brain function.
Fig. 1.
Three-dimensional gene expression in the human brain. The model shows the expression of ESR1 and MAGEL2 mapped onto a ventral view of a 3D-reference atlas reconstruction. The expression of these genes is indicated by the dots. The color of the dots indicates the expression level: green (low expression) through red (high expression). Images were produced by the Brain explorer 2 software application, an interactive version of the Allen Human Brain Atlas.
Footnotes
The authors declare no conflict of interest.
See companion article on page 2738.
References
- 1.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 2.Mahfouz A, et al. Genome-wide coexpression of steroid receptors in the mouse brain: Identifying signaling pathways and functionally coordinated regions. Proc Natl Acad Sci USA. 2016;113:2738–2743. doi: 10.1073/pnas.1520376113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simerly RB. Wired for reproduction: Organization and development of sexually dimorphic circuits in the mammalian forebrain. Annu Rev Neurosci. 2002;25:507–536. doi: 10.1146/annurev.neuro.25.112701.142745. [DOI] [PubMed] [Google Scholar]
- 4.Bakker J, Brock O. Early oestrogens in shaping reproductive networks: Evidence for a potential organisational role of oestradiol in female brain development. J Neuroendocrinol. 2010;22(7):728–735. doi: 10.1111/j.1365-2826.2010.02016.x. [DOI] [PubMed] [Google Scholar]
- 5.Israel JM, Cabelguen JM, Le Masson G, Oliet SH, Ciofi P. Neonatal testosterone suppresses a neuroendocrine pulse generator required for reproduction. Nat Commun. 2014;5:3285. doi: 10.1038/ncomms4285. [DOI] [PubMed] [Google Scholar]
- 6.Nugent BM, et al. Brain feminization requires active repression of masculinization via DNA methylation. Nat Neurosci. 2015;18(5):690–697. doi: 10.1038/nn.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEwen BS, et al. Mechanisms of stress in the brain. Nat Neurosci. 2015;18(10):1353–1363. doi: 10.1038/nn.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handa RJ, Weiser MJ. Gonadal steroid hormones and the hypothalamo-pituitary-adrenal axis. Front Neuroendocrinol. 2014;35(2):197–220. doi: 10.1016/j.yfrne.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gore AC, Martien KM, Gagnidze K, Pfaff D. Implications of prenatal steroid perturbations for neurodevelopment, behavior, and autism. Endocr Rev. 2014;35(6):961–991. doi: 10.1210/er.2013-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McEwen BS, Gray JD, Nasca C. 60 Years of neuroendocrinology: Redefining neuroendocrinology: Stress, sex and cognitive and emotional regulation. J Endocrinol. 2015;226(2):T67–T83. doi: 10.1530/JOE-15-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gofflot F, et al. Systematic gene expression mapping clusters nuclear receptors according to their function in the brain. Cell. 2007;131(2):405–418. doi: 10.1016/j.cell.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, et al. Study of gene function based on spatial co-expression in a high-resolution mouse brain atlas. BMC Syst Biol. 2007;1:19. doi: 10.1186/1752-0509-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaaf CP, et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet. 2013;45(11):1405–1408. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eiholzer U, et al. Hypothalamic and gonadal components of hypogonadism in boys with Prader-Labhart-Willi syndrome. J Clin Endocrinol Metab. 2006;91(3):892–898. doi: 10.1210/jc.2005-0902. [DOI] [PubMed] [Google Scholar]