Abstract
Previously, we and others mapped an increased homologous recombination activity within the subgenomic promoter (sgp) region in brome mosaic virus (BMV) RNA3 (A. Bruyere et al., J. Virol. 74:4214-4219, 2000; R. Wierzchoslawski et al., J. Virol. 77:6769-6776, 2003). In order to correlate sgp-mediated recombination and transcription, in the present work we used BMV RNA3 constructs that carried altered sgp repeats. We observed that the removal or extension of the poly(U) tract reduced or increased recombination, respectively. Deletion of the sgp core hairpin or its replacement by a different stem-loop structure inhibited recombination activity. Nucleotide substitutions at the +1 or +2 transcription initiation position reduced recombination. The sgp core alone supported only basal recombination activity. The sites of crossovers mapped to the poly(U) region and to the core hairpin. The observed effects on recombination did not parallel those observed for transcription. To explain how both activities operate within the sgp sequence, we propose a dual mechanism whereby recombination is primed at the poly(U) tract by the predetached nascent plus strand, whereas transcription is initiated de novo at the sgp core.
Viral RNA recombination plays an important role during rearrangements of viral RNAs and provides an efficient tool for the repair of their genomic sequences (12, 12A, 36, 40). In a variety of RNA viruses, including the bromovirus brome mosaic virus (BMV), coronaviruses, poliovirus, carmoviruses, tombusviruses, and flaviviruses (7, 8), RNA recombination events seem to occur via a copy choice mechanism, where the replicase enzyme changes templates during RNA synthesis. The evidence is based on the effects of RNA sequence modifications and the participation of replicase proteins (49) in recombination, both in vivo and in vitro (14, 16, 18, 53). In retroviruses, there seem to be three, non-mutually exclusive copy choice mechanisms: forced (strong-stop) strand transfer, pause-driven strand transfer, and pause-independent (RNA structure-driven) strand transfer (26). Other proposed mechanisms are cleavage-religation (20, 37, 40) and transesterification (12, 12A, 20).
BMV is a tripartite RNA virus, where RNA components 1 and 2 (RNA1 and RNA2) encode, respectively, replicase proteins 1a and 2a, while RNA3 encodes a movement protein (3a) and a coat protein (CP) (4). The CP is expressed from subgenomic (sg) RNA4.
In BMV, the frequency of homologous intersegmental recombination within the 3′ noncoding region is approximately 10 times higher than that of nonhomologous crossovers (17, 38). Most of the 3′ crossovers are precise (38, 39) and are concentrated within GC-rich sequences followed by downstream AU-rich regions (38-40). The imprecise crossovers pinpoint the actual crossover sites. A proposed model suggests that the BMV RNA-dependent RNA polymerase (RdRp) pauses (stalls) at AU-rich sequences and then switches onto the acceptor template, with the upstream GC-rich domain facilitating the rehybridization of the detached nascent strand (39, 40).
Besides recombination among different RNA segments, homologous recombination activity between RNA3 molecules has been mapped to the intercistronic region (11). This consists of the subgenomic promoter (sgp) on the minus strand of RNA3 (32) and the 1a-RNA3 binding site on the plus strand (50). Detection of crossovers was possible by coinfection with pairs of RNA3 variants carrying marker mutations at different positions. Further work has pinpointed the crossovers to the sgp sequence, and it was also observed that RNA recombination and transcription events overlapped within the sgp (11, 15, 19, 54). The sgp is recognized by the replicase enzyme that binds internally to minus strands (46) and synthesizes the sg RNA4. Structurally, the sgp has a modular composition which includes GC-rich and AU-rich “enhancing” regions, a poly(U) stretch, a core region with the RdRp-binding stem-loop structure (41), and a downstream portion (31, 50). Such a multidomain nature of sgp's has been confirmed for other RNA viruses (34).
In this work we expand the analysis of the sgp region in BMV RNA3 in order to determine in vivo the functional correlation between transcription and recombination. By using specially designed RNA3 vectors, we observed a variety of effects on recombination and transcription activities. The removal or the extension of the poly(U) tract reduced or increased the frequency of crossovers, respectively. Both the deletion of the sgp core hairpin and its replacement by a different stem-loop structure inhibited recombination activity. Nucleotide substitutions at +1 or +2 transcription initiation positions reduced recombination. The sgp core alone supported lower recombination activity. The sites of crossovers mapped to the poly(U) region and to the core hairpin. The observed effects on recombination did not parallel those on transcription, demonstrating that transcription and recombination utilized different sgp sequences (and thus mechanisms), although partial overlapping of the active sequences is possible.
MATERIALS AND METHODS
Materials.
Plasmids pB1TP3, pB2TP5, and pB3TP7 (28) were used as templates to synthesize in vitro the infectious transcripts of wild-type (wt) BMV RNA1, RNA2, and RNA3, respectively, by using the MEGAscript T7 kit (Ambion, Austin, Tex.). Plasmids ID2 to ID13 (see below) were used to synthesize mutant RNA3 transcripts. Moloney murine leukemia virus reverse transcriptase and restriction enzymes were obtained from Promega Corp. (Madison, Wis.).
Generation of ID RNA3 constructs and in vitro transcription.
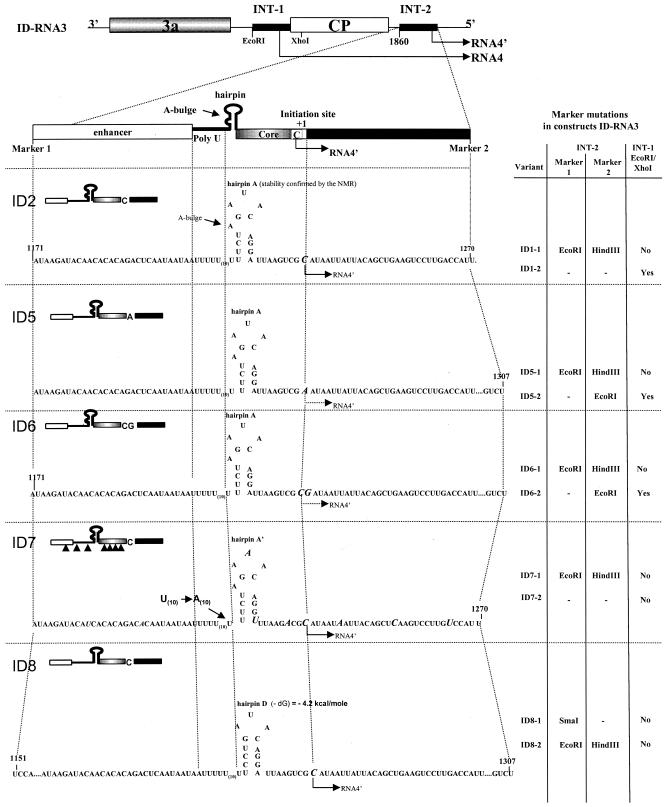
Figure 1 shows the sequences of ID RNA3 constructs that carry insert repeats of the sgp sequence at the INT-2 locus. The repeats were introduced downstream of the CP open reading frame (ORF) (at the INT-2 locus) by ligation of PCR-amplified (from the original wt pB3TP7 plasmid [28]) cDNA products at the BamHI site of plasmid SF-25 (11, 54). In several cases the constructs were obtained by sequential PCR, using the product of the first PCR as a primer for the second PCR. Plasmid SF-25 was used for PCR amplification of INT-2 insertions in constructs ID2 through ID6, ID9, and ID12, while in constructs ID7, ID8, ID10, ID11, and ID13, the INT-2 inserts were amplified from pairs of overlapping primers. Each ID RNA3 was generated in two versions that differed by flanking marker restriction sites at the INT-2 locus, as shown in Fig. 1 and 2. The oligonucleotide primers used for each PCR amplification are not specified, since it should not be a problem for other laboratories to recreate any ID sequence.
FIG. 1.
BMV RNA3 constructs (ID RNA3s) used in these studies. A schematic of the genetic organization of BMV RNA3 is shown at the top. Thin lines, noncoding regions; solid rectangles show the locations of the sgp sequences. Shaded and open rectangles, ORFs for the movement protein (3a) and CP, respectively. Arrows mark the regions of the sg RNA4 and RNA4′ transcriptions. EcoRI and XhoI marker restriction sites are shown at INT-1. Below the RNA3 schematic, the elements of the sgp insert of INT-2 [the enhancer, the poly(U) tract, the hairpin, the core region, the initiation site, and a downstream sequence] are shown. Below the INT-2 schematic, the nucleotide sequences of constructs ID2 to ID13 are given, with structures represented on the left and locations of marker restriction sites in the two variants of each construct tabulated on the right. ID2 carries the wt sequence; ID5 and ID6 carry mutations at positions +1 and +2, respectively; ID7 carries nine single marker mutations (indicated by italicized capital letters); ID8 carries hairpin D; ID9 has no poly(U) tract; ID10 has no enhancer and no poly(U) elements, so the core sequence forms a new hairpin B structure by pairing with the adjacent upstream region; ID11 carries a replacement of hairpin A with the 3′ hairpin C; ID12 carries an extended poly(U) tract; and ID13 has no hairpin structure. Nucleotide positions of junction sites are given according to the numbering of the wt sequence, as counted from the 5′ end. Thin V-shaped lines represent deleted elements. For the ID7 RNA3, only one variant was created (ID7-1), and it was tested for recombination with ID1-1 (described previously by Wierzchoslawski et al. [54]).
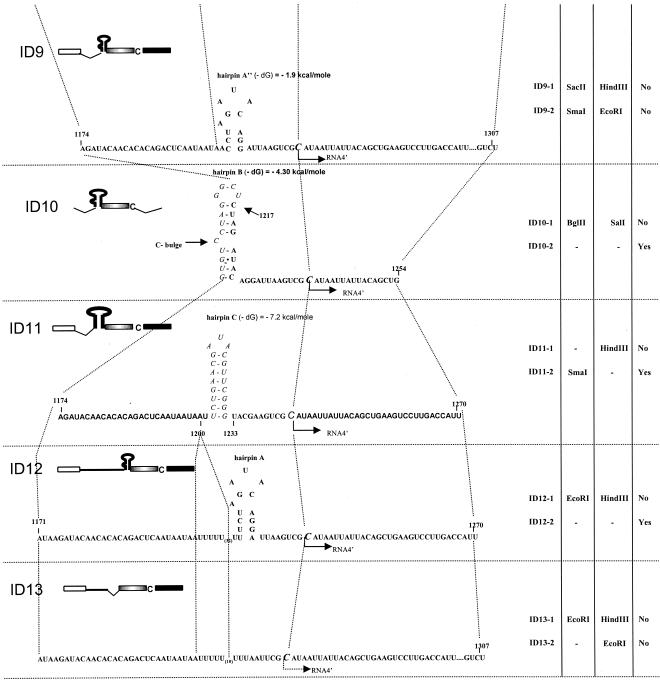
FIG. 2.
General illustration of homologous crossovers occurring between two coinoculated variants of ID RNA3s. At the top, the regions of sgp-mediated crossovers at the INT-2 locus, the INT-1 locus, and the control CP ORF are shown schematically (not to scale). The elements of the RNA3 molecule are represented as in Fig. 1; arrows indicate homologous crossovers. The types of ID RNA3s carrying specific restriction sites at four restriction marker positions (indicated by circles, triangles, diamonds, and inverted triangles) are given in the balloons (see also the restriction marker table in Fig. 1, right side). Below the crossover schematic, the molecules of two parental ID RNA3 variants [designated generally as ID RNA3(1) and ID RNA3(2)] are shown on the left, whereas the predicted types of progeny RNA3-RNA3 recombinants that arise due to crossovers within the recombination regions (see also Table 1) are shown on the right. Solid rectangles, INT-1 and INT-2 regions; shaded and open rectangles, ORFs.
The ID RNA3 constructs carried various modifications within the sgp elements. The INT-2 insert of ID2 RNA3 (54) comprised the wt sgp sequence, including the core, the poly(U) tract, and part of the enhancer domain (Fig. 1). Construct ID5 carried a C-to-A mutation at the +1 start site, whereas construct ID6 carried an A-to-G mutation at the +2 site; both mutations were constructed in order to determine the role of transcription initiation site nucleotides. Construct ID7 carried multiple nucleotide substitutions at 5- to 7-nucleotide (nt) intervals, at positions that had been determined in vitro not to interfere with transcription (46), to map the positions of crossovers. Construct ID8 carried a deletion of the A-1220 residue, which removed the bulge from the core stem-loop structure, as well as the U-1217-to-C substitution, which further stabilized the structure (Fig. 1, hairpin D); both these mutations have been reported previously to increase transcription activity (22, 23). Construct ID9 carried a deletion of the poly(U) tract, whereas ID10 carried a larger deletion of both the enhancer region and the poly(U) tract, leaving only the core region. Both constructs were used to determine the functionality of these elements in both transcription and recombination. Construct ID11 carried a 24-nt stem-loop structure derived from the 3′-tRNA-like terminus of BMV RNA3, replacing the wt core stem-loop (Fig. 1, hairpin C), a modification that has been reported to increase transcription (22, 23). Construct ID12 carried a prolonged poly(U) tract of 40 nt to test if the poly(U) tract was responsible for crossovers. Finally, construct ID13 carried a deletion of nt 1218 to 1232, which participate in the formation of the core stem-loop structure, found to bind to the RdRp complex (22, 23). In addition, constructs ID2, ID5, ID6, ID10, ID11, and ID12 carried the EcoRI and XhoI marker mutations at flanking positions of the wt sgp locus (INT-1) in order to determine the recombination frequency at INT-1 (Fig. 1 and 2).
The resulting plasmids were linearized at the 3′ end of the RNA3 sequence, and the capped RNAs were synthesized in vitro by T7 RNA transcription with the MEGAscript kit (36), followed by phenol-chloroform extraction and ethanol precipitation. After centrifugation, the RNA pellet was air dried and dissolved in RNase-free water. The integrity of the RNA was determined electrophoretically in denaturing (formamide-formaldehyde) agarose gels, and the amounts were measured with a UV spectrophotometer.
Determination of recombination frequency in vivo.
To determine recombination frequency during infection, pairs of ID RNA3 variants were coinoculated together with wt RNA1 and wt RNA2 on Chenopodium quinoa leaves, as previously described (11). Briefly, a mixture of each transcript in 15 μl of the inoculation buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.1% Celite) was inoculated mechanically by rubbing fully expanded leaves. In each experiment, four separate leaves per plant (two to three plants) were inoculated. Each inoculation experiment was repeated two or three times. The infected plants were maintained in greenhouse conditions, as described previously (38-40). Local lesions were counted, and the lesion tissue was cut out 10 days postinoculation, quickly frozen in liquid nitrogen, and stored at −80°C.
The recombination frequency was determined as the fraction of recombinants in a total number of cDNA clones that were amplified by reverse transcription-PCR (RT-PCR) from total-RNA extracts of the combined local lesion tissue (36). The oligonucleotides used for amplification were RW-Pst (5′-AAAACTGCAGCCAATGGTCTCTTTTAGAGATTTACAGTG-3′) and RW-Pst1 (5′-AAAACTGCAGCCAACTTACCTTACAACGGCGTGTTGAG-3′) for constructs with mutations at INT-1; for constructs without mutations at INT-1, RW-Pst and PN1 (5′-CTGAAGCAGTGCCTGCTAAGGCGGTC-3′) were used (PstI sites are boldfaced; sequences corresponding to BMV are underlined). PCR products were purified on QIAquick PCR purification spin columns (QIAGEN Inc.) and ligated into the 3′-U overhangs of the pDrive cloning vector (in the QIAGEN PCR cloning kit, catalog no. 231122). The cDNA clones were then digested with marker restriction enzymes, and those carrying the recombinant sequences were counted over those carrying the parental sequences (Fig. 2; Table 1). Some of the clones were sequenced to elucidate the precision of crossovers and to confirm the insert size.
TABLE 1.
Recombination frequencies for coinoculations with pairs of ID RNA3s
| Coinfected ID RNA3 variants | Total no. of clones tested (C)a | No. of parental clonesb | No. of homologous recombinant clones (% of C)
|
||
|---|---|---|---|---|---|
| INT-1c | INT-2d | Control region (CP)e | |||
| ID2-1 × ID2-2 (wt sgp) | 71 | 20 | 17 (24) | 18 (25) | 16 (22) |
| ID5-1 × ID5-2 (+1 C to A) | 28 | 15 | 8 (28) | 3 (10) | 2 (7) |
| ID6-1 × ID6-2 (+2 A to G) | 36 | 32 | 8 (22) | 4 (11) | 2 (5) |
| ID7-1 × ID1-1 (multiple mutations) | 46 | 37 | NA | 9 (20) | NA |
| ID8-1 × ID8-2 (deletion of A-bulge) | 36 | 14f | NA | 22f (60) | NA |
| ID9-1 × ID9-2 [Δ poly(A)] | 56 | 54 | NA | 2 (4) | NA |
| ID10-1 × ID10-2 (core) | 36 | 14 | 9 (26) | 6 (18) | 7 (19) |
| ID11-1 × ID11-2 (3′-hairpin) | 27 | 27g | 0 (0) | 0 (0) | 0 (0) |
| ID12-1 × ID12-2 [long poly(A)] | 30 | 11 | 5 (17) | 12 (40) | 2 (7) |
| ID13-1 × ID13-2 (Δ core hairpin) | 27 | 27h | NA | 0 (0) | NA |
Total number of insert-bearing clones analyzed per coinfection experiment. The cDNA clones from total progeny RNA were generated by RT-PCR using BMV RNA3- specific primers as described in Materials and Methods.
Determined by digestion with marker restriction enzymes.
Recombinant clones at INT-1 were first selected by digestion with marker restriction enzymes and then sequenced at the INT-1 region. NA, not applicable.
Recombinant clones at INT-2 were first selected by digestion with marker restriction enzymes and then sequenced at the INT-2 region.
Recombinant clones between the INT-1 and INT-2 sites (CP ORF) were first selected by digestion with marker restriction enzymes and then sequenced through the CP ORF region. See Fig. 1 and 2 and legends for further explanations.
With deletions.
Of the 27 parental clones, 66% were ID7-2 and 34% were ID7-1.
Of the 27 parental clones, 55% were ID13-1 and 45% were ID13-2.
Control RT-PCRs.
Two control RT-PCRs were performed. First, each counterpart of the particular pair of RNA3 mutants was inoculated separately on C. quinoa plants at the same time as the coinoculation experiment; the progeny RNA was isolated from separate infections, combined together, and subjected to RT-PCR, followed by cloning and restriction analysis and/or sequencing. Second, the in vitro-transcribed BMV RNA3 preparations were mixed pairwise, 100 ng each, and used directly as templates for RT-PCR amplification. The resulting PCR fragments were cloned and analyzed.
RNA analyses.
Extraction and Northern hybridization analyses of total RNAs from infected plants were carried out as described previously (18). To prepare a BMV-specific probe for hybridization experiments, an 83-nt fragment common to the 3′ ends of all four BMV genomic components (positioned between nucleotides 1870 and 1953 on BMV RNA3) was PCR amplified from plasmid pB3TP7 with oligonucleotides RP5 (5′-AACTGCAGGTAGAGACCCTGTCCAGG-3′) and RP3 (5′-CGGAATTCAACCACGACTATCAGTTATCAG-3′) carrying PstI and EcoRI restriction sites, respectively (restriction sites are boldfaced; sequences corresponding to BMV are underlined). The PCR product was first cut with the PstI and EcoRI restriction enzymes and then ligated into the pGEM-3Z cloning vector from Promega. The resulting RW-INT(−) plasmid was linearized with PstI and used as a template for synthesis of an RNA probe by using the MAXIscript T7 kit (Ambion) and [α-32P]CTP from Amersham Biosciences. Hybridization signals were quantified in a STORM-3 PhosphorImager from Molecular Dynamics by using ImageQuant software (version 5.0) from Amersham-Biosciences.
RESULTS
Effect of mutations at INT-2 on BMV RNA accumulation.
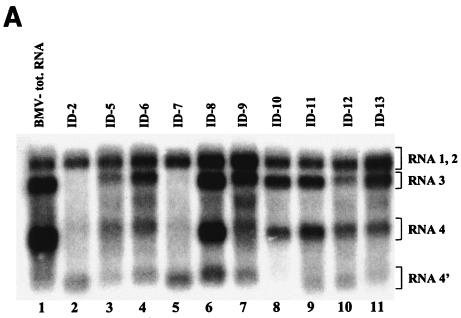
All the RNA3 ID constructs of Fig. 1 were infectious to C. quinoa plants (data not shown), and the INT-2 sgp inserts were maintained during infection in all but ID8 progeny RNAs (Table 1; Fig. 4). To determine how sgp sequence modifications affected BMV RNA accumulation and INT-2 transcription activity, the progeny RNAs were harvested at the time that secured an equal accumulation of the virus for all of the constructs, i.e., 10 days postinfection, as established previously (54). The leaf tissues were quickly frozen in liquid nitrogen immediately after harvesting to prevent RNA degradation, and then the total RNAs were extracted from equal weights (100 mg) of the combined local lesion tissue by using a standard RNA isolation protocol. The RNA was analyzed by Northern blotting with an RNA probe that was complementary to an internal 3′ proximal region common among four BMV RNAs, to avoid detection of putative 3′ BMV RNA degradation fragments.
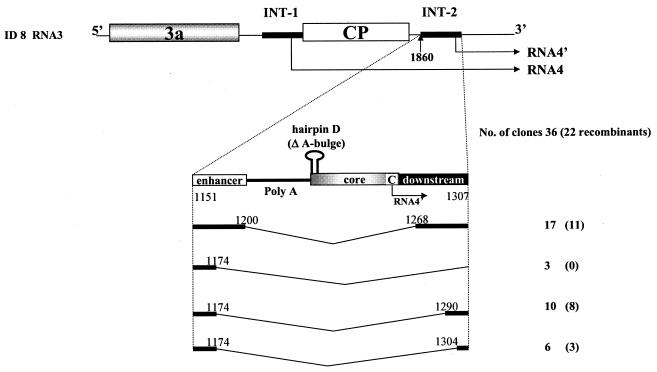
FIG. 4.
Results of sequencing of RT-PCR-generated cDNA clones of progeny BMV RNA3 extracted from ID8-1 × ID8-2-coinfected leaves. Schematic representations of ID8 RNA3, and of the sgp INT-2 insert with the modified hairpin D (see Fig. 1), are shown at the top. Elements are represented as in Fig. 1. Below the schematics are representations of deletion variants. Thin bent lines, deleted sequences. Nucleotide positions (according to wt RNA3 numbering) are shown on either side. The number of clones (recombinant clones) representing each particular sequence is given on the right.
Figure 3A shows that, depending on the length of the 3′-end insert at INT-2, some constructs produced longer RNA4′ than others, and that the relative concentrations of RNA3, RNA4, and RNA4′ differed among the ID mutants. Specifically, for the wt INT-2 sgp (in ID2), the relative amounts of RNA3 and RNA4, but also RNA4′, were lower than those shown previously for ID2 (54). This could be due to the use of a probe that was more selective than the 3′ probe used before. The remaining constructs formed two groups (Fig. 3). In one group, the concentrations of RNA3 and RNA4 were higher than that of RNA4′, reflecting reduced (or abolished) transcription at INT-2. This group included ID5 (C-to-A mutation at +1) (Fig. 3, lane 3), ID6 (A-to-G mutation at +2) (lane 4), ID8 (deleted A-bulge) (lane 6), ID9 [deleted poly(U) tract] (lane 7), ID10 (core region only) (lane 8), ID11 (stem-loop structure derived from the 3′-tRNA-like terminus) (lane 9), ID12 [carrying a prolonged poly(U) tract] (lane 10), and ID13 (deleted core hairpin) (lane 11). A similar trend was observed for the control wt BMV RNA (Fig. 3A, lane 1). The second group consisted of ID2, which carried the unmodified insert at INT-2, and ID7, which carried multiple point mutations at neutral positions. For both constructs, the concentration of RNA4′ reached higher levels than those of RNA3 or RNA4 (Fig. 3, lanes 2 and 5, respectively), possibly because of functional competition between INT-1 and INT-2 (see Discussion).
FIG. 3.
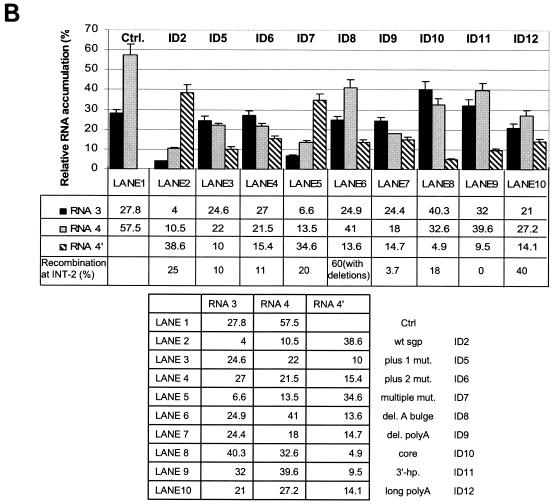
(A) Northern blot analysis of the accumulation of BMV RNA3 ID constructs. Leaves were coinoculated mechanically with in vitro-transcribed wt RNA1 and RNA2 and with RNA3 ID mutants (as indicated above the gel). Total (tot.) RNA was extracted from the infected tissue, separated electrophoretically in a 1% agarose-formamide-formaldehyde gel, and blotted to a nylon membrane (HybondN+; Amersham), and BMV RNA sequences were detected by using a 32P-labeled RNA probe that was complementary to an 83-nt fragment common to the 3′ ends of all four BMV RNAs (nt 1870 to 1953 on the BMV RNA3 positive strand). Lanes 2 to 11, RNAs extracted from the local lesion tissues of C. quinoa. Lane 1 (control), RNA from systemically infected barley seedlings. The positions of BMV RNA components are indicated on the right. (B) Histogram showing the quantification of the accumulation of RNA3 and sg RNA4 and RNA4′. The Northern blot autoradiogram shown in panel A was scanned in the PhosphorImager and subjected to computer-based densitometry by using the ImageQuant (version 5.0) program from Amersham Biosciences. Bars represent the percentages which the band intensities for RNA3, RNA4, and RNA4′ contributed to the total intensity of all BMV RNA components. The numbers below show the actual results of quantification and the observed recombination frequencies at INT-2 (as a reference; repeated from Table 1).
Mutations at the transcription initiation site.
First we tested the role of nucleotides at the transcription initiation site by using RNA3 constructs ID5 and ID6 (Fig. 1). The constructs were inoculated pairwise (ID5-1 × ID5-2 and ID6-1 × ID6-2) on C. quinoa, and the general schematics of expected crossovers are shown in Fig. 2. Total RNA was extracted 10 days postinoculation, the cDNA products were amplified by RT-PCR and ligated into the PUC-19 cloning vector, and the inserts were sized by electrophoresis in an agarose gel (data not shown), as described in Materials and Methods. Subsequently, the clones were digested with restriction enzymes to reveal double-marker or no-marker recombinants (54) (Fig. 2). The recombinant nature of these clones was further confirmed by sequencing, revealing no imprecise sequences. Among 28 clones derived from ID5-1 × ID5-2 coinfection (Table 1), 3 were recombinants at INT-2 (10% frequency of recombination), 8 were recombinants at INT-1 (28%), 2 recombined within the CP control region (7%; this recombination frequency was determined based on the cosegregation of the 5′ marker of the INT-1 region and the 3′ marker of the INT-2 region), and 15 carried parental sequences. ID6-1 × ID6-2 coinfection yielded 36 clones, with 32 parental sequences, 4 recombinants at INT-2 (11%), 8 recombinants at INT-1 (22%), and 2 recombinants within the CP region (5%). These results revealed the role in recombination of nucleotides downstream of the core hairpin (see Discussion). Scanning with the PhosphorImager (Fig. 3) demonstrated that the level of sg RNA4′ (that is, the level of transcription) was reduced for both ID5 and ID6, to 10 and 15.4% of the level of all BMV RNAs in the sample, respectively. These results suggested that wt cytidylate at +1 and wt adenylate at +2 participated in transcription initiation, which confirmed earlier observations (46).
Mapping crossover sites within the sgp sequence.
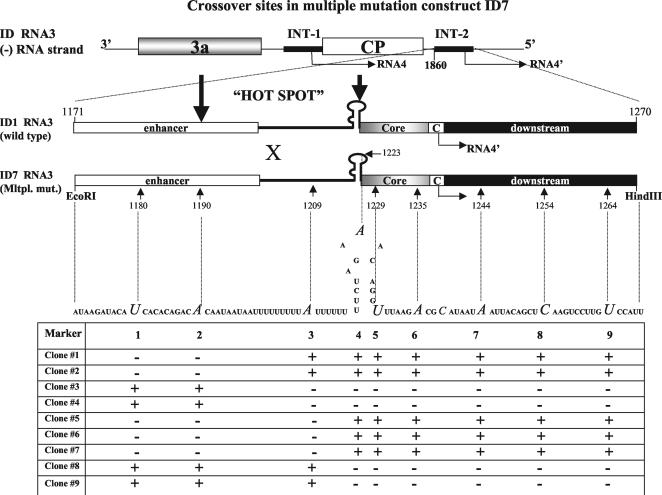
Next, we attempted to localize more precisely the sites of crossovers within the sgp sequence. Since the crossovers were precise and homologous, single-nucleotide mutations were introduced as markers at INT-2 in the parental construct ID7, at the following positions: 1180 (substitution of U for A), 1190 (A for T), 1209 (A for U), 1223 (A for U), 1229 (U for A), 1235 (A for U), 1244 (A for U), 1254 (C for G), and 1264 (U for A) (see Fig. 5). This covered the core region, the core hairpin, and the poly(U) tract. The substitutions were at transcriptionally neutral locations, as determined by Siegel et al. (46). The substitution of A for U (position 1223) changed the AUA triloop into an AAA triloop within the sg core hairpin (hairpin A′ [Fig. 1]). Northern blot analysis (Fig. 3, lane 5) confirmed that ID7-1 RNA3 supported 34.6% transcription at INT-2. Thus, the transcriptional activity remained unchanged compared to that for the wt INT-2 sgp sequence of ID2. For the latter construct, we have previously reported higher concentrations of RNA4′ (45.8 to 52.3% of all BMV RNAs in the sample [54]). However, the probe used for Northern blotting was complementary to the terminal 3′ region, resulting in the detection of undesired comigrating 3′-degradation products. In this study we used a more internal probe (described in Materials and Methods) to eliminate such background. Interestingly, transcription at INT-1 was dramatically reduced, suggesting some interplay between the INT-1 and INT-2 loci (see Discussion).
FIG. 5.
Mapping of the crossover sites to the poly(U) tract based on ID1 × ID7 coinfection. (Top) Diagram showing the structures of the two ID RNA3 constructs that carry either the nonmutated sgp insert (ID1 RNA3) or the sgp insert with multiple single-nucleotide mutations (ID7 RNA3) at INT-2. All elements are as described in the legend to Fig. 1. The construction of ID1 RNA3 has been described previously (54). Two thick arrows flank the hot spot region. (Bottom) Table shows the presence (+) or absence (−) of marker mutations within the INT-2 repeats of nine RT-PCR-generated cDNA clones (numbered from 1 through 9) derived from the recombinant RNA3 progeny. Marker nucleotides are italicized, and their positions are given below the ID7 RNA3 sequence.
The recombination activity of ID7-1 was tested by coinoculation with construct ID1-1 (described in reference 54), which carried wt sgp at INT-2. Of 40 cDNA clones, 7 represented homologous recombinants and 33 carried parental RNAs (17 ID1-1 and 16 ID7-1 clones). Sequencing of nine clones revealed that the parental distribution of marker mutations was retained up to position 1190 and above position 1223. We concluded that the crossovers occurred between positions 1190 and 1223, and therefore the recombination was mapped to the AU-rich sequence that preceded the core hairpin and to the hairpin itself.
Stabilization of the sgp core hairpin.
Next, we analyzed the sequence elements responsible for directing the crossovers to the region adjacent to the core hairpin. Plants were coinoculated with ID8-1 and ID8-2, carrying a stabilized stem (hairpin D [Fig. 1]), which has been shown previously to support increased transcription (23). Surprisingly, massive deletions were detected at the INT-2 insert: of 36 progeny clones (Table 1), 22 were deletion recombinants while 14 were nonrecombined variants with partial deletions (Fig. 4). The deletions differed in size (between 68 and 136 nt), with the upstream cutoff site at either nt 1174 (19 clones) or nt 1200 (17 clones). The downstream cutoff sites were located at positions between nt 1268 and nt 1310. All the clones lacked the core sequence, so the remaining transcription activity (13.6% [Fig. 3, lane 6]) must have been derived from the unchanged parental ID8 RNA3.
Deletion of the poly(U) tract and of the enhancer region.
To confirm that the poly(U) tract participated in recombination, the poly(U) sequence was deleted in construct ID9. ID9-1 × ID9-2 coinfection produced 56 clones (Table 1), of which only 2 were recombinant clones (4% frequency) while 54 were parental clones (29 ID9-1 and 25 ID9-2 clones). This minimal frequency correlated with low transcription activity at INT-2 (14.7%), showing that the poly(U) tract participated in both events.
To test if the enhancer, poly(U) tract, and downstream regions participated in recombination and/or transcription, constructs ID10-1 and ID10-2 were used; essentially, ID10 carried only the sgp core at INT-2. Analysis of 36 progeny cDNA clones (Table 1) confirmed the uniform size of the INT-2 insert. Restriction analysis of clones demonstrated a reduced recombination frequency at INT-2 (18%; 6 clones) compared to that at wt INT-1 (26%; 9 clones) or at the wt INT-2 sequence of ID2 (25%). Seven clones had recombined within the CP (19%). Among the remaining 14 clones, 8 represented ID10-1 and 6 represented ID10-2 RNA3s, demonstrating equal accumulation or availability for recombination of parental sequences.
Scanning with the PhosphorImager (Fig. 3) demonstrated that the level of ID10 sg RNA4′ (that is, the level of transcription) constituted only 4.9% of all BMV RNAs in the sample, in sharp contrast to the 38.6% share of RNA4′ for ID2 infection.
The fact that the sgp core alone (in ID10) maintained recombination activity, though at a reduced rate, was surprising. To check if this was due to structural factors, the sequence of the INT-2 insert (plus the upstream wt region) was folded with the program M-FOLD (version 3.0). The predicted structure (hairpin B [Fig. 1]) represented a novel tetraloop stable element (−4.30 kcal/mol) carrying a bulged 8-bp stem. In contrast, the wt INT-2 insert (in ID2) formed a less-stable pseudo-triloop hairpin with a bulged 5-bp stem (hairpin A [Fig. 1]) (23). We speculated that the tetraloop structure interfered with transcription to a greater extent than with recombination (see Discussion).
Substitution with 3′ hairpin C.
To test whether different core hairpins could support recombination and/or transcription, the wt hairpin was replaced with a highly stable triloop stem-loop structure (8 bp and −7.20 kcal/mol [Fig. 1, ID11, hairpin C]) that was derived from the 3′ noncoding region of BMV RNA3 (hairpin C, nt 2035 to 2057). Previous in vitro results have shown an increase to 432% in the level of transcription from the sgp carrying the same modification (23). Coinfection with ID11-1 and ID11-2 yielded no recombinant sequences at INT-2 among the 27 cDNA clones analyzed (Table 1): 34 and 66% of the clones had ID11-1 and ID11-2 sequences, respectively. Thus, hairpin C did not support recombination. Also, transcription activity was reduced to 9.5% at INT-2 (Fig. 3, lane 9). Similarly, no recombinants were observed for the control CP region or for the unmodified INT-1 locus of ID11 (Table 1). The lack of recombination at INT-1 despite the increased transcription activity at this locus (39.6%) is interesting; it suggests long-distance INT-1-INT-2 interactions that affect recombination (see Discussion).
Extension of the poly(U) tract.
To further verify the role of the poly(U) tract, we tested if an extended poly(U) region would increase the recombination frequency. Construct ID12 carried 40 U residues at INT-2, compared to 18 U residues in the wt sequence. Indeed, among 30 progeny clones (Table 1) (ID12-1 × ID12-2 coinfection), there were 12 recombinants (40%) at INT-2, 5 recombinants (17%) at INT-1, 2 recombinants (7%) within the control CP region, and 11 parental sequences (5 ID12-1 and 6 ID12-2 clones). Surprisingly, the doubled (compared to wt ID2) recombination frequency did not parallel the transcription activity (Fig. 3), which was decreased to 14% (see Discussion).
Deletion of the core hairpin.
The role of the core hairpin was further studied by deleting the corresponding sequence in construct ID13. ID13-1 × ID13-2 coinoculation yielded 27 clones that carried only the parental sequences at INT-2, almost equally distributed (15 ID13-1 clones [55%] and 12 ID13-2 clones [45%]). This confirmed that the core hairpin was essential for recombination. Similarly, the relative amount of the RNA4′ transcript was reduced to 4.6%, verifying the importance of the core hairpin for transcription (Fig. 3, lane 11).
DISCUSSION
Previously, our laboratory reported homologous recombination activity of the sgp region of BMV RNA3 (11, 54). We have used an RNA3 ID construct that carried a duplication of the sgp sequence so that it could be modified without inactivating the virus. When its transcriptional activity was inhibited, recombination was abolished, suggesting a link between the two functions (54). To study the relationship between transcription and recombination in vivo, in this study we used nine different RNA3 ID constructs. In general, we observed major disproportions between recombination and transcription, and the two functions utilized different sgp portions, which suggested different mechanisms. Moreover, we were able to finely map the crossovers occurring within the sgp and to verify the participation of the sgp regions in transcription. Below we discuss specific points of this research.
The prototype for the RNA3 ID constructs was ID2. It accumulated low levels of RNA3 and RNA4 but high levels of RNA4′ (Fig. 3). This was likely not due to selective RNA degradation, because the samples were quick-frozen prior to RNA extraction, and a selective probe was used for Northern blotting (see Materials and Methods). Rather, this ratio was observed because ID2 carried the wt sgp, which was functional in transcription and might have been preferentially functional at INT-2 (19), generating higher levels of RNA4′. Similar results were obtained with ID7, which carried multiple mutations at transcriptionally neutral positions, confirming previous in vitro data (46). Interestingly, the concentrations of the ID2 and ID7 viruses in infected C. quinoa leaves were similar to each other as well as to those of other ID variants (data not shown). Perhaps, in spite of its low levels, RNA4 secured efficient translation of the viral CP, and thus the final virus concentration was controlled by a different mechanism.
By use of ID7, the crossover sites were mapped to the upper portion of the poly(U) tract, near the core hairpin (Fig. 5). This finding implied that poly(U) and the core hairpin were important during recombination. Indeed, in ID9, in which the poly(U) tract was deleted, recombination activity was abolished (although the lack of recombination could also be due to the presence of a hairpin that was less stable than the wt hairpin). In addition, in ID12, the poly(U) sequence was extended and recombination increased, while in ID13, in which the core hairpin was deleted, recombination was abolished. We then created other ID RNA3s to analyze how poly(U) and the core hairpin interacted during recombination. Surprisingly, ID10, which carried only the core region, supported recombination. We argue that a novel stable tetraloop, hairpin B (Fig. 1), can form in ID10 and induce recombination. Apparently, however, the sequence of the hairpin is more important for recombination than its stability, because construct ID11, which carried a hairpin even more stable than that of ID10, did not support crossovers. Even if this finding does not support the roadblock effect, the pausing of RdRp in ID10 might be achieved through interactions of hairpin B with other viral and/or host proteins (35, 55).
Interestingly, ID8 mediated the highest recombination activity, but the crossovers were nonhomologous, leading to deletions (Fig. 4). We assume that this reflects a selection pressure for variants that are inactive in transcription (at INT-2). Increased transcriptional activity at INT-2 (see reference 22) could be detrimental for viral infection.
For some ID RNA3s, transcriptional activity paralleled recombination activity. For instance, ID5 and ID6 carried mutations at +1 and +2 sites and supported low levels of both activities. Previous in vitro data have revealed that +1 and +2 sites participated in BMV transcription (46). Interestingly, different viruses have different preferences for transcription initiation (3, 44). With regard to recombination, we have assumed that altered RNA folding in ID5 and ID6 affected the crossovers, but the M-FOLD program predicted unchanged core hairpins (data not shown). Perhaps, then, +1 and +2 nucleotides do mediate some distinct, yet to be characterized interactions. It was shown recently by Hema and Kao that the template sequence near the initiation nucleotide can modulate BMV RNA replication (25).
ID9 supported low recombination and transcription activities. The latter confirms the previous observation that the poly(U) tract is required for transcription (2, 48), although the less-stable hairpin could also contribute to the effect. In contrast, ID10 supported minimal transcription (but moderate recombination), which could be due to ineffective binding of RdRp to hairpin B or to the missing poly(U) tract.
The low transcriptional activity found for ID11 contradicts in vitro data reported by Haasnoot et al. (23), although the spacing between the hairpin and the initiation cytidylate was different in their constructs. In addition, some protein factors or cellular structures might be absent from the RdRp preparations but present in infected whole plants. It should also be noted that the experiments were conducted on different hosts (barley versus C. quinoa), which might affect transcription (54).
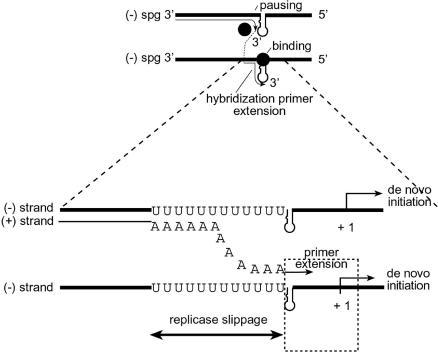
We propose a novel mechanism to explain these data, and especially the mapping of crossovers to the poly(U)-hairpin region (Fig. 6). The model predicts two separate processes, both occurring during plus-strand synthesis: transcription initiates de novo by internal binding of the RdRp complex to the sgp, whereas recombination is facilitated by priming with the reattached nascent plus strand. Both de novo initiation and primer extension activities have been reported in vitro for the RdRp's of BMV (45), flaviviruses (44), carmoviruses (41), and nidoviruses (42, 52). In retroviruses, crossovers occur between the dimeric “kissing loops” (57). Both the length heterogeneity of the detached nascent strand and rehybridization at multiple sites may increase the odds of primer extension at the poly(U) tract. On the other hand, the core region might be required for reattachment of the RdRp complex during strand transfer. With regard to transcription, the basal function of the sgp core is consistent with the results of Adkins et al. (2), but normal levels of transcription require upstream enhancers (19, 24). The proposed model requires biochemical confirmation.
FIG. 6.
Diagrammatic representation of the model of crossovers at the sgp region. Thick lines, minus strands of BMV RNA3; dashed lines, progeny plus strands. Arrows indicate the direction of migration of the BMV replicase enzyme (solid circle). (Top) The RdRp (solid circle) approaches the sgp region (thick line), detaches at the sgp region [e.g., hairpin or poly(U) tract], hybridizes to another RNA molecule, and reinitiates by primer extension with the reannealed nascent plus strand (thin line). (Bottom) The poly(U) area in the minus strand of the sgp. The detached poly(U) region reanneals near the sgp core and extends RNA synthesis. Dotted box, replicase binding region (as determined in vitro).
The poly(U) tract may function as a nonpaired spacer that facilitates the access of the replicase to the template. The unstructured nature of the poly(U) tract [and the poly(A) tract] has been suggested before (27, 31). Nagy and Bujarski have mapped the hot spots for 3′ recombination at AU-rich regions (39), likely due to destabilization (pausing) of the RdRp complex (29, 51) or to RdRp slippage (5, 9), which promotes strand detachment (39). However, the case of ID10 suggests that sometimes poly(U) can be replaced by special RNA folding. Mapping of crossover sites on ID10 is necessary to confirm the structural requirements for recombination.
An alternative mechanism may involve direct reinitiation at the poly(U) tract (in minus strands) in a primer-independent manner. Such a property has been reported for the hepatitis C virus RdRp enzyme from an in vitro study (51). Apart from template sequences, RNA-protein interactions could make the poly(U) tract [the poly(A) tract in plus strands] prone to recombination. For example, certain poly(U)-binding proteins participate in posttranscriptional regulation of gene expression (43). In Escherichia coli, host factors of the Hfq family bind preferentially to single-stranded AU-rich sequences followed by stem-loops, and the binding enhances RNA-RNA pairing (56). In fact, protein-RNA interactions have been found to facilitate RNA-RNA crossovers in human immunodeficiency virus (6, 30).
The recombination frequency at INT-1 remained high for all ID RNA3s tested (ID2, -5, -6, -10, and -12), except ID11, although there were variations in the accumulation (transcription) of RNA4. This additionally demonstrates that different domains are utilized by both functions. The lack of recombination for ID11 might be due to distant interactions between INT-1 and INT-2 sgp's; long-distance interactions have been observed to mediate various functions in other viruses (10, 31, 47). Also, the presence of two copies of hairpin C, a key element for minus-strand synthesis (23), could be detrimental to transcription.
In addition to the sgp, the occurrence of crossovers was studied within a larger control region of the CP ORF (see Table 1), between INT-1 and INT-2 (540 nt) (see also reference 11). Four constructs (ID5, ID6, ID11, and ID12) supported low or no activity, but two constructs (ID2 and ID10) supported higher activity. These results suggest the existence of a recombination hot spot within the CP ORF. Regardless of the mechanism, the discrepancy in length (100 nt for INT-2 versus 540 nt for the CP ORF) demonstrates that the CP ORF was more than five times less efficient at recombination than the sgp hot spot.
Sequencing identified only precise homologous RNA3-RNA3 recombinants at INT-2, but aberrant recombinants at the 3′ noncoding region have been observed previously (38). The difference might be due to the different natures of the 3′ promoter and the sgp (1), e.g., because of more efficient polymerase binding to the sgp than to the 3′ region.
To our knowledge, the BMV sgp represents the first well-characterized recombinationally active sgp. Further experiments will address the nature (competitive or synergistic) of interregulation of the recombination and transcription functions. Besides that of BMV, a recombination activity was observed at the intercistronic region of cowpea chlorotic mottle bromovirus RNA3 (15, 21). Also, sgp-mediated recombination has likely led to the formation of subgroups of luteoviruses (33) and to modular rearrangements in closteroviruses (8). By anticipating the discontinuous process of synthesis of BMV RNA3 plus strands, our model is analogous to the discontinuous mode of sg RNA synthesis in nidoviruses. The difference is that the latter relies on translocation (reminiscent of recombination) of partially synthesized nascent plus strands to the 3′ leader sequence on minus strands (42), while in BMV RNA3 the translocation occurs between corresponding sequences of two essentially identical RNAs. Also, some viruses support crossovers via promoter-like elements, e.g., RNA replication enhancers in turnip crinkle carmovirus (13, 41), but more studies are needed to investigate the possible links between transcription and recombination.
Acknowledgments
We thank Barbara Ball for excellent assistance with the figures.
This work was supported by a grant from the National Science Foundation (MCB-0317039), by the Polish Government through a grant (3 P04A 039 25) from the State Committee for Scientific Studies, awarded to J.B., and by the Plant Molecular Biology Center at Northern Illinois University.
REFERENCES
- 1.Adkins, S., and C. C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, S., R. W. Siegel, J. H. Sun, and C. C. Kao. 1997. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA 3:634-647. [PMC free article] [PubMed] [Google Scholar]
- 3.Adkins, S., S. S. Stawicki, G. Faurote, R. W. Siegel, and C. C. Kao. 1998. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 4.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 5.Ahlquist, P., V. Luckow, and P. Kaesberg. 1981. Complete nucleotide sequence of brome mosaic virus RNA3. J. Mol. Biol. 153:23-38. [DOI] [PubMed] [Google Scholar]
- 6.Balakrishnan, M., P. J. Fay, and R. A. Bambara. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482-36492. [DOI] [PubMed] [Google Scholar]
- 7.Banner, L. R., and M. M. C. Lai. 1991. Random nature of coronavirus RNA recombination in the absence of selection pressure. Virology 185:441-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bar-Joseph, M., G. Yang, R. Gafny, and M. Mawassi. 1997. Subgenomic RNAs: the possible building blocks for modular recombination of Closteroviridae genomes. Semin. Virol. 8:113-119. [Google Scholar]
- 9.Barr, J. N., and G. W. Wertz. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 75:6901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barry, J. K., and W. A. Miller. 2002. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. USA 99:11133-11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruyere, A., M. Wantroba, S. Flasinski, A. Dzianott, and J. J. Bujarski. 2000. Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J. Virol. 74:4214-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bujarski, J. J. (ed.). 1996. Experimental systems of genetic recombination and defective RNA formation in RNA viruses, part I. In Seminars in virology, vol. 7, no. 6. Academic Press, New York, N.Y.
- 12a.Bujarski, J. J. (ed.). 1997. Experimental systems of genetic recombination and defective RNA formation in RNA viruses, part II. In Seminars in virology, vol. 8, no. 2. Academic Press, New York, N.Y.
- 13.Cascone, P. J., T. F. Haydar, and A. E. Simon. 1993. Sequences and structures required for recombination between virus-associated RNAs. Science 260:801-805. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, C. P., and P. D. Nagy. 2003. Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro. J. Virol. 77:12033-12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzianott, A., S. Flasinski, S. Pratt, and J. J. Bujarski. 1995. Foreign complementary sequences facilitate genetic recombination in brome mosaic virus. Virology 208:370-375. [DOI] [PubMed] [Google Scholar]
- 16.Dzianott, A., N. Rauffer-Bruyere, and J. J. Bujarski. 2001. Studies on functional interaction between brome mosaic virus replicase proteins during RNA recombination, using combined mutants in vivo and in vitro. Virology 289:137-149. [DOI] [PubMed] [Google Scholar]
- 17.Figlerowicz, M., and J. J. Bujarski. 1998. RNA recombination in brome mosaic virus, a model plus strand RNA virus. Acta Biochim. Pol. 45:847-868. [PubMed] [Google Scholar]
- 18.Figlerowicz, M., P. D. Nagy, and J. J. Bujarski. 1997. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc. Natl. Acad. Sci. USA 94:2073-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 62:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gmyl, A. P., S. A. Korshenko, E. V. Belousov, E. V. Khitrina, and V. I. Agol. 2003. Nonreplicative homologous RNA recombination: promiscuous joining of RNA pieces? RNA 9:1221-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene, A. E., and R. F. Allison. 1994. Recombination between viral RNA and transgenic plant transcripts. Science 263:1423-1425. [DOI] [PubMed] [Google Scholar]
- 22.Haasnoot, P. C. J., F. T. Brederode, R. C. Olsthoorn, and J. F. Bol. 2000. A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6:708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haasnoot, P. C. J., R. C. Olsthoorn, and J. F. Bol. 2002. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA 8:110-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haasnoot, P. C. J., J. F. Bol, and R. C. Olsthoorn. 2003. A plant virus replication system to assay the formation of RNA pseudotriloop motifs in RNA-protein interactions. Proc. Natl. Acad. Sci. USA 100:12596-12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hema, M., and C. C. Kao. 2004. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 78:1169-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, W. S., T. Rhodes, Q. Dang, and V. Pathak. 2003. Retroviral recombination: review of genetic analyses. Front. Biosci. 8:D143-D155. [DOI] [PubMed] [Google Scholar]
- 27.Huez, G., Y. Cleuter, C. Bruck, L. Van Vloten-Doting, R. Goldbach, and B. Verduin. 1983. Translational stability of plant viral RNAs microinjected into living cells. Influence of a 3′-poly(A) segment. Eur. J. Biochem. 130:205-209. [DOI] [PubMed] [Google Scholar]
- 28.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions of transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 29.Kim, M. J., and C. Kao. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc. Natl. Acad. Sci. USA 98:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klasens, B. I., H. T. Huthoff, A. T. Das, R. E. Jeeninga, and B. Berkhout. 1999. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim. Biophys. Acta 1444:355-370. [DOI] [PubMed] [Google Scholar]
- 31.Marsh, L. E., T. W. Dreher, and T. C. Hall. 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16:981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 33.Miller, W. A., S. P. Dinesh-Kumar, and C. P. Paul. 1995. Luteovirus gene expression. Crit. Rev. Plant Sci. 13:179-211. [Google Scholar]
- 34.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Nagel, R., and M. Ares. 2000. Substrate recognition by eukaryotic RNase III: the double-stranded RNA-binding domain of Rnt1p selectively binds RNA containing a 5′-AGNN-3′ tetraloop. RNA 6:1142-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy, P. D., and J. J. Bujarski. 1992. Genetic recombination in brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J. Virol. 66:6824-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy, P. D., and J. J. Bujarski. 1993. Targeting the site of RNA-RNA recombination in brome mosaic virus with antisense sequences. Proc. Natl. Acad. Sci. USA 90:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagy, P. D., and J. J. Bujarski. 1995. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J. Virol. 69:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy, P. D., and J. J. Bujarski. 1997. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J. Virol. 71:3799-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagy, P. D., C. Ogiela, and J. J. Bujarski. 1999. Mapping sequences active in homologous RNA recombination in brome mosaic virus: prediction of recombination hot spots. Virology 254:92-104. [DOI] [PubMed] [Google Scholar]
- 41.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasternak, A. O., E. Van Den Born, W. J. Spaan, and E. J. Snijder. 2003. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 77:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paynton, B. V. 1998. RNA-binding proteins in mouse oocytes and embryos: expression of genes encoding Y box, DEAD box RNA helicase, and poly(U) binding proteins. Dev. Genet. 23:285-298. [DOI] [PubMed] [Google Scholar]
- 44.Ranjith-Kumar, C. T., L. Gutshall, M. J. Kim, R. T. Sarisky, and C. C. Kao. 2002. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 76:12526-12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranjith-Kumar, C. T., X. Zhang, and C. C. Kao. 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 77:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siegel, R. W., S. Adkins, and C. C. Kao. 1997. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 48.Smirnyagina, E., Y. H. Hsu, N. Chua, and P. Ahlquist. 1994. Second-site mutations in the brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology 198:427-436. [DOI] [PubMed] [Google Scholar]
- 49.Steitz, T. 1998. A mechanism for all polymerases. Nature 391:231-232. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, X. L., R. B. Johnson, M. A. Hockman, and Q. M. Wang. 2000. De novo RNA synthesis catalyzed by HCV RNA-dependent RNA polymerase. Biochem. Biophys. Res. Commun. 268:798-803. [DOI] [PubMed] [Google Scholar]
- 52.Van Vliet, A. L., S. L. Smits, P. J. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White, K. A., and T. J. Morris. 1995. RNA determinants of junction site selection in RNA virus recombinants and defective interfering RNAs. RNA 1:1029-1040. [PMC free article] [PubMed] [Google Scholar]
- 54.Wierzchoslawski, R., A. Dzianott, S. Kunimalayan, and J. J. Bujarski. 2003. A transcriptionally active subgenomic promoter supports homologous cross-overs in a plus-strand RNA virus. J. Virol. 77:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeffman, A., S. Hassard, G. Varani, and A. Lever. 2000. The major HIV-1 packaging signal is an extended bulged stem-loop whose structure is altered upon interaction with the Gag polyprotein. J. Mol. Biol. 297:877-893. (Erratum, 301:1315.) [DOI] [PubMed]
- 56.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, J., and Y. Ma. 2001. Evidence for retroviral intramolecular recombinations. J. Virol. 75:6348-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]