Significance
Electronic computers are extremely powerful at performing a high number of operations at very high speeds, sequentially. However, they struggle with combinatorial tasks that can be solved faster if many operations are performed in parallel. Here, we present proof-of-concept of a parallel computer by solving the specific instance {2, 5, 9} of a classical nondeterministic-polynomial-time complete (“NP-complete”) problem, the subset sum problem. The computer consists of a specifically designed, nanostructured network explored by a large number of molecular-motor-driven, protein filaments. This system is highly energy efficient, thus avoiding the heating issues limiting electronic computers. We discuss the technical advances necessary to solve larger combinatorial problems than existing computation devices, potentially leading to a new way to tackle difficult mathematical problems.
Keywords: parallel computing, molecular motors, NP complete, biocomputation, nanotechnology
Abstract
The combinatorial nature of many important mathematical problems, including nondeterministic-polynomial-time (NP)-complete problems, places a severe limitation on the problem size that can be solved with conventional, sequentially operating electronic computers. There have been significant efforts in conceiving parallel-computation approaches in the past, for example: DNA computation, quantum computation, and microfluidics-based computation. However, these approaches have not proven, so far, to be scalable and practical from a fabrication and operational perspective. Here, we report the foundations of an alternative parallel-computation system in which a given combinatorial problem is encoded into a graphical, modular network that is embedded in a nanofabricated planar device. Exploring the network in a parallel fashion using a large number of independent, molecular-motor-propelled agents then solves the mathematical problem. This approach uses orders of magnitude less energy than conventional computers, thus addressing issues related to power consumption and heat dissipation. We provide a proof-of-concept demonstration of such a device by solving, in a parallel fashion, the small instance {2, 5, 9} of the subset sum problem, which is a benchmark NP-complete problem. Finally, we discuss the technical advances necessary to make our system scalable with presently available technology.
Many combinatorial problems of practical importance, such as the design and verification of circuits (1), the folding (2) and design (3) of proteins, and optimal network routing (4), require that a large number of possible candidate solutions are explored in a brute-force manner to discover the actual solution. Because the time required for solving these problems grows exponentially with their size, they are intractable for conventional electronic computers, which operate sequentially, leading to impractical computing times even for medium-sized problems. Solving such problems therefore requires efficient parallel-computation approaches (5). However, the approaches proposed so far suffer from drawbacks that have prevented their implementation. For example, DNA computation, which generates mathematical solutions by recombining DNA strands (6, 7), or DNA static (8) or dynamic (9) nanostructures, is limited by the need for impractically large amounts of DNA (10–13). Quantum computation is limited in scale by decoherence and by the small number of qubits that can be integrated (14). Microfluidics-based parallel computation (15) is difficult to scale up in practice due to rapidly diverging physical size and complexity of the computation devices with the size of the problem, as well as the need for impractically large external pressure.
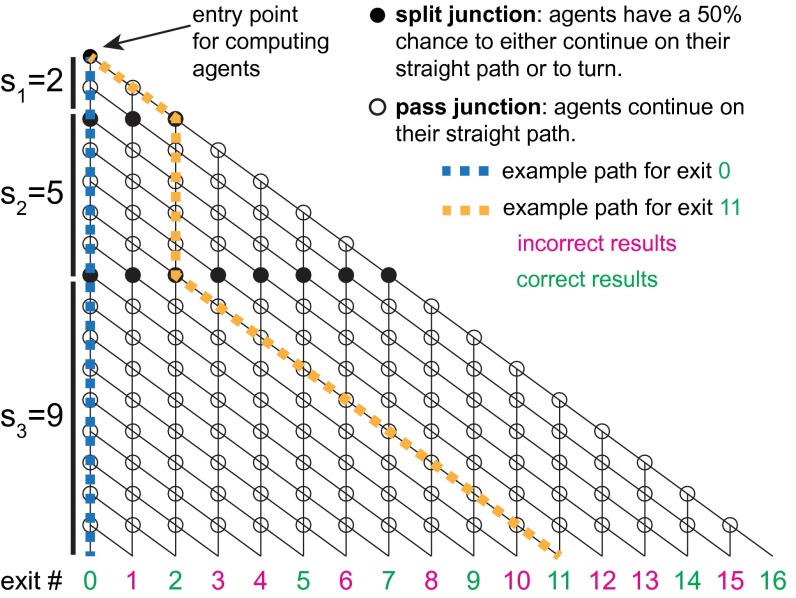
Here, we propose a parallel-computation approach, which is based on encoding combinatorial problems into the geometry of a physical network of lithographically defined channels, followed by exploration of the network in a parallel fashion using a large number of independent agents, with very high energy efficiency. To demonstrate operational functionality, we applied it to a small instance of a benchmark classical nondeterministic-polynomial-time complete (NP-complete) problem (16), the subset sum problem (SSP) (Fig. 1). This problem asks whether, given a set S = {s1, s2, ..., sN} of N integers, there exists a subset of S whose elements sum to a target sum, T. More formally, the question is whether there is a solution where wi {0, 1}, for any given T from 0 to . To find all possible subset sums by exploring all possible subsets requires the testing of 2N different combinations, which––even for modest values of N––is impractical on electronic computers because of exponentially increasing time requirements (SI Appendix, section S1). Although more sophisticated algorithms exist (17–19), none of these avoids the exponentially growing exploration time, a property that is harnessed in some cryptography systems to generate encoded messages (20).
Fig. 1.
Computation network for the SSP {2, 5, 9}. The agents enter the network from the top-left corner. Filled circles represent split junctions where it is equally probable that agents continue straight ahead or turn. Empty circles represent pass junctions where agents continue straight ahead. Moving diagonally down at a split junction corresponds to adding that integer (numbers 2 and 9 for the yellow example path). The actual value of the integer potentially added at a split junction is determined by the number of rows of junctions until the next split junction. The exit numbers correspond to the target sums T (potential solutions) represented by each exit; correct results for this particular set {2, 5, 9} are labeled in green, and incorrect results (where no agents will arrive) are labeled in magenta. The working principle is also detailed in Movie S1.
Our approach replaces the requirement for exponentially growing time needed by traditional, electronic computers to solve NP-complete problems, with the requirement for an exponentially growing number of independent computing agents. We use a proof-of-concept device to successfully solve the specific three-variable instance {2, 5, 9} of the SSP. Key technical advancements necessary to scale up our approach to be of practical relevance include the need to reduce error rates and to supply sufficiently many computing agents. We identify several possible approaches to address these requirements.
Results
In our network encoding of the SSP, the channel-guided unidirectional motions of agents are equivalent to elementary operations of addition, and their spatial positions in the network are equivalent to “running sums.” Starting from an entrance point at one corner of the network (Fig. 1, Top Left), agents are guided unidirectionally downward by the channels in vertical or diagonal directions. Two types of junctions were designed to regulate the motion of agents in the network: (i) “split junctions,” where agents are randomly distributed between two forward paths, and (ii) “pass junctions,” where agents are guided onward to the next junction along the initial direction. The vertical distance (measured as the number of junctions) between two subsequent rows of split junctions represents an integer from the set S. The process of an agent moving straight downward from a given split junction is equivalent to excluding the corresponding si from the summation, whereas traveling diagonally downward is equivalent to including that si in the subset sum. A solution Σwi si = TJ to the SSP is represented by an agent choosing a path to one of the exit nodes in the network (bottom row in Fig. 1). If a sufficiently large number of agents is used, all possible paths are explored, and therefore all possible subset sums of S are generated, simultaneously.
We implemented the proposed computational approach with biological agents that satisfy the following requirements: The agents (i) are available in large numbers at negligible cost; (ii) are self-propelled and thus do not require a global, external driving force; (iii) operate independently of each other to ensure parallel exploration; (iv) have small dimensions to enable use in high-density networks with high computing power per unit area; (v) move rapidly to maximize computational speed; and (vi) move in a predominantly forward direction (to ensure low error rates). In particular, we used cytoskeletal filaments (actin filaments and microtubules), which are propelled by molecular motors (myosin II and kinesin-1, respectively) along a surface in gliding motility assays (21, 22). Both kinds of cytoskeletal filaments have small diameters (∼10 nm for actin filaments and ∼25 nm for microtubules) and move at high speeds (5–10 µm s−1 for actin filaments driven by fast myosin II from skeletal muscle, and ∼0.5–1 µm s−1 for microtubules driven by kinesin-1). The filaments are guided unidirectionally (23, 24) along lithographically defined channels, which are functionalized with molecular motors, and whose roofs are open to supply the motors with biochemical fuel (adenosine 5′-triphosphate, ATP) by diffusion from the surrounding buffer fluid (25, 26), allowing for a distributed energy supply. The width of the channels was set to below 200 nm and 250 nm, for actin filaments and microtubules, respectively, which were observed to reliably guide cytoskeletal filaments (27–29).
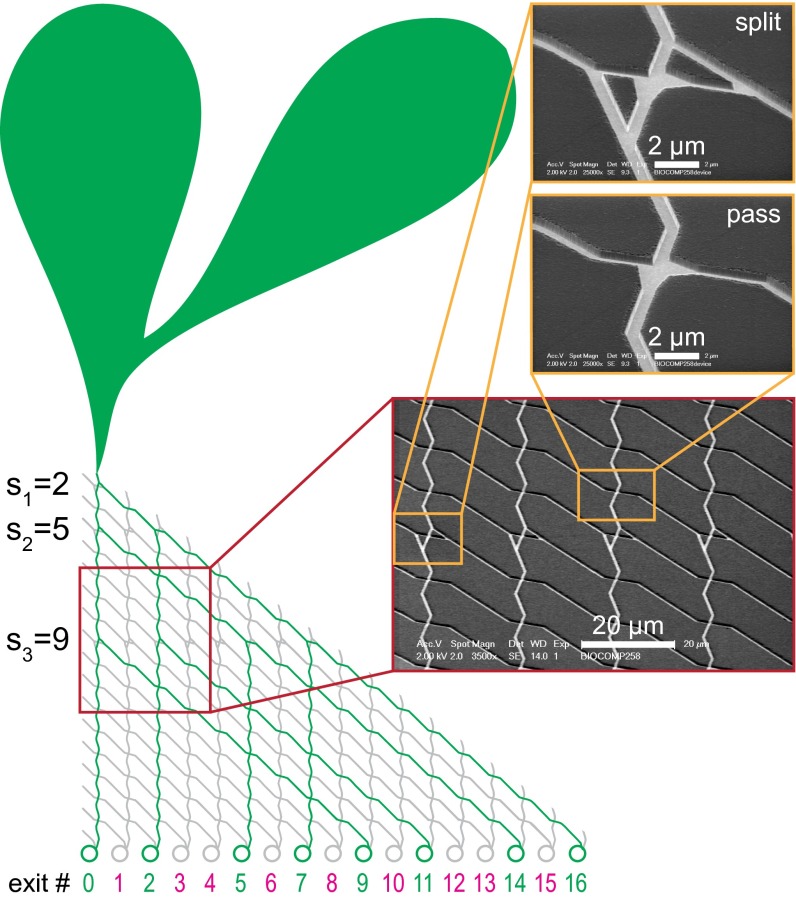
The general layouts of the networks used in our devices for microtubule–kinesin- and actin–myosin–based computations are shown in Fig. 2 and SI Appendix, Fig. S2.1, respectively. To start the computation, the filaments are collected from the bulk solution and guided to enter the network through loading zones (large tear-shaped areas in Fig. 2 and SI Appendix, Fig. S2.1). The computational networks comprise a set of standardized rectangular lattices, each containing two isomorphic unit cells representing the split junctions and pass junctions (Fig. 2, Inset and SI Appendix, Fig. S2.1). This standardized structure facilitates the future encoding of problems of any size and the scalability of fabrication, e.g., through step-and-repeat deep-UV lithography, for which the fabrication of channels with widths of 200 nm (as reported here), or below, is readily achievable.
Fig. 2.
Device layout of a computation device for the SSP {2, 5, 9}. Schematic of the actual device layout used for microtubules, including the loading zones for the microtubules (green balloon-like areas), the channels traversed by the microtubules during calculation (green lines), and the channels that should not be traversed (gray lines). Exit numbers corresponding to correct results are shown in green; numbers corresponding to incorrect results are shown in magenta. The circles at each exit are designed to store filaments for easy readout. (Insets) Scanning electron micrographs of parts of the network used for microtubules, showing a split- and a pass junction. See SI Appendix, section S2 for a corresponding schematic layout of the device that was used with actin filaments, including details of the rectifiers used for that device.
To account for different stiffness and size of actin filaments compared with microtubules, we optimized the device design and the junction geometries individually for each filament system with the assistance of numerical modeling and simulation (SI Appendix, section S4). After traversing the network, the filaments emerge at exits corresponding to the target sums and are either recycled back to the entrance point (actin–myosin device; SI Appendix, section S2) or collected (microtubule–kinesin device; Fig. 2). The networks were fabricated by electron-beam lithography on SiO2 substrates to obtain the required resolution and fidelity (see Materials and Methods Summary for fabrication details).
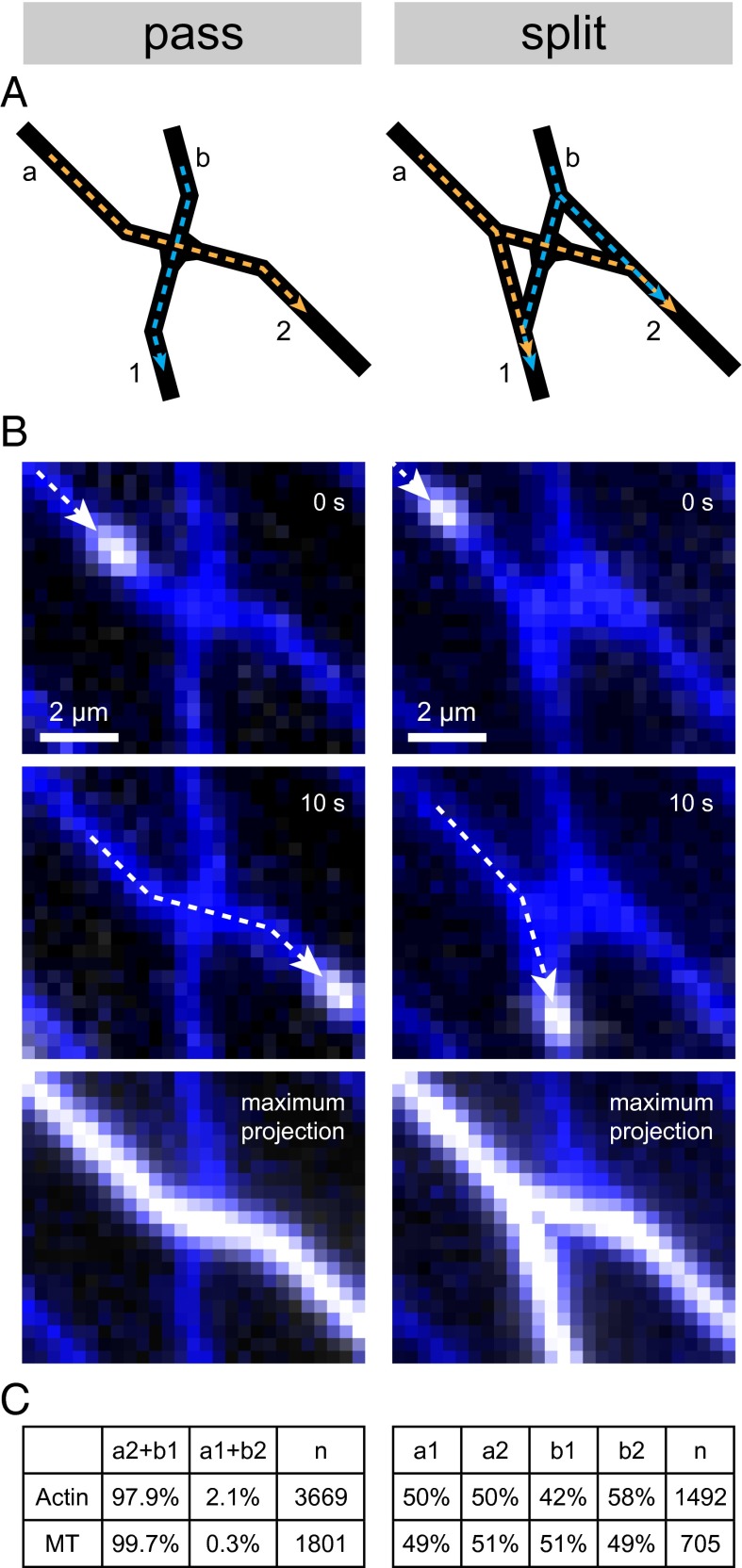
The minimization of computation errors requires that the error rates of pass junctions are as low as possible, i.e., that filaments do not progress along erroneous paths and emerge at exit nodes not corresponding to target sums (SI Appendix, section S1). In contrast, the error rates of split junctions (designed to yield a 50:50 split) are less critical to computational performance because these junctions mainly serve to distribute agents across the network, thus ensuring that no solution is missed. Experiments (see Materials and Methods Summary for experimental procedures and imaging details) confirmed that the junction designs in our devices fulfill these performance requirements (Fig. 3). Fluorescently marked cytoskeletal filaments traversing individual split junctions and pass junctions (Fig. 3A) were tracked using fluorescence microscopy (Fig. 3B). Statistical analysis of the motion of actin filaments and the microtubules showed that 97.9% and 99.7%, respectively, took the correct (straight) paths through pass junctions, whereas split junctions distributed filaments approximately evenly (Fig. 3C and SI Appendix, section S5).
Fig. 3.
Performance of pass junctions (Left) and split junctions (Right) with actin filaments and microtubules as agents. (A) Schematic drawing of a pass junction (Left) and a split junction (Right). Entrance channels for agents moving diagonally in the network are labeled a, whereas entrance channels for agents moving straight downward are labeled b. Exit channels are labeled 1 for straight downward and 2 for diagonally moving agents, respectively. Intended paths through the junctions are indicated by yellow (agents entering diagonally) and blue (agents entering straight downward) dotted lines. (B) Fluorescence micrographs of microtubules moving in a pass- (Left) or a split junction (Right). Paths of the microtubules in previous frames and direction of movement are indicated by white dotted lines and arrows, respectively. The bottom row of images shows maximum projections of several agents moving through the respective junctions. (C) Analysis of the error rates. For pass junctions, agents moving from entrance a to exit 2 and agents moving from b to 1 were behaving correctly (column a2 + b1). For split junctions, agents from each entrance were intended to be split evenly between exits (ideally, both would be 50%). Columns headed n denote the number of filaments analyzed for each junction type. MT, microtubules. See SI Appendix, section S6 for details.
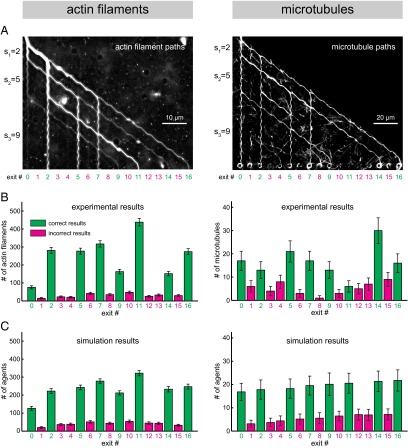
Our proof-of-concept experiments on the set {2, 5, 9} show that both the actin–myosin system and the microtubule–kinesin system can be used to solve a combinatorial problem by parallel computation (Fig. 4). Superimposed fluorescence micrographs show time-integrated paths of the fluorescently labeled filaments (Fig. 4A). These images demonstrate that the filaments traversed the network from the entrance point to the exit nodes that represent correct results (Fig. 4A, green exit numbers). Statistical analysis of the number of filaments leaving each exit (obtained by counting the filaments in image sequences tracking each filament) confirms that both types of agents found all of the correct results and that significantly more agents (P < 0.002; unpaired two-tailed t test) of both types exited nodes corresponding to correct results than incorrect results (Fig. 4B). The experimental data are in good agreement with those obtained by Monte Carlo simulations (Fig. 4C and SI Appendix, section S6), which are based on the experimentally measured error rates at each junction type (obtained separately for actin filaments and microtubules; Fig. 3C and SI Appendix, section S5).
Fig. 4.
Solving the SSP {2, 5, 9} by actin filaments (Left) and microtubules (Right). (A) Average (Left) and maximum (Right) projections of several hundred typical fluorescence micrographs of actin filaments (Left) and microtubules (Right) moving through a {2, 5, 9} device. An example of the computation (for a device using actin filaments) is presented in Movie S2. (B) Experimental results obtained from 2,251 actin filaments (Left; total experiment time: 26 min) and 179 microtubules (Right; total experiment time: 180 min). Error bars represent the counting error (√n). Total experiment time refers to the time required for the given number of agents to enter and traverse the network (see also SI Appendix, section S1). We note that the overall performance of the microtubule device was to some extent inferior to the actin device due to an accidental obstruction in a channel leading to exit 11 (causing a lower number of filaments reaching this exit), and due to a number of filaments landing at random points of the network in the channels where they were transported with high probability by the processive kinesin motors (increasing the number of filaments reaching the wrong exits). Both issues will be remedied in a next generation of devices by avoidance of fabrication errors, working in a cleanroom environment, and microfluidic focusing of the filaments in solution to the landing zones, respectively (see also SI Appendix, section S6 for more details on these sources of error). (C) Monte Carlo simulation results (mean ± SD of 100 simulations; see SI Appendix, section S6 for simulation details) for actin filaments (Left) and microtubules (Right) using the actually measured error rates of the pass junctions and measured splitting ratios of the split junctions (Fig. 3C). In A–C, green numbers and bars represent correct results, and magenta numbers and bars represent incorrect results.
Discussion
We developed a parallel-computation approach based on encoding combinatorial problems into the geometry of physical networks. We showed that these networks can be manufactured lithographically and explored using independent agents. Using such a device, we demonstrated the solution of one particular three-variable instance of the SSP.
Notably, once the device is loaded with the required number of agents, the effective computational time for NP-complete problems grows only polynomially, e.g., as N2 if the elements of S are approximately equidistantly spaced. This is in contrast to traditional, sequentially operating, electronic computers, where the time required to explore every possible solution sequentially would scale exponentially as 2N. However, it is inherent to combinatorial and NP-complete problems (assuming P! = NP) that the exploration of the entire solution space requires the use of exponentially increasing amounts of some resource, such as time, space, or material. In the present case this fundamental requirement manifests itself in the number of agents needed, which grows exponentially with 2N. Effectively we are trading the need of time for the need of molecular mass.
The error rates of this first device are too large for scaling up to problems containing more than ∼10 variables (see SI Appendix, section S1 for a more detailed scaling analysis).
Nevertheless, we argue that our approach has the potential to be more scalable in practice than other approaches because it offers several advantages: (i) Myosin II and kinesin-1 molecular motors use a distributed energy supply (ATP in the surrounding solution), thus eliminating the need for external forces (such as pressure or an electric potential) to drive the computation. This need inherently prevents, for example, microfluidic approaches from scaling up, because in these devices the pressures needed to pump fluid through the network become prohibitively large for large N (30). (ii) The molecular motors operate in a highly energy-efficient manner. As a result, the approach demonstrated here consumes orders of magnitude less energy per operation compared with both electronic and microfluidic computers, eliminating issues related to the dissipation of heat. Specifically, we estimate an energy cost of 2–5 × 10−14 J per operation for a molecular-motor-based device compared with about 3–6 × 10−10 J per operation for the most advanced electronic computers, or an estimated minimum of 10−12 J per operation for microfluidics-based computers (SI Appendix, section S7). (iii) The networks in which the problems are encoded in our approach are planar and comprise standardized modules, therefore being fully scalable with existing technology (see SI Appendix, section S1 for a more detailed discussion). Thus, they avoid potential engineering challenges associated with building large-scale 3D microfluidics devices.
Most importantly, however, we foresee several practical solutions to managing the need for exponentially increasing numbers of agents that we highlighted above. (i) The number of agents can self-adjust to the problem size. Specifically, cytoskeletal filaments can self-replicate as they traverse the network by enzymatic splitting and simultaneous elongation (31, 32). Alternatively, self-propelled, dividing microorganisms can be used as agents (33–35). Thus, the larger the network, the more the agents will multiply, in an exponential fashion. Any kind of agent multiplication scheme will also solve potential problems with sequential feeding of the agents into the network through a single entrance, which represents a bottleneck for large N. Moreover, for cytoskeletal filaments, the splitting and elongation rates will be limited by the global concentrations of enzymes and filament subunits, respectively. Thus, multiplication will be negatively regulated in parts of the network where the agent density is high (i.e., above the density needed for successful computation), consequently counteracting the risk of channel clogging. (ii) The NP-complete problem is encoded into a planar, physical network and not into the agents themselves. This simplifies fabrication and encoding because the network that encodes the problem grows polynomially, whereas the exponentially scaling number of cytoskeletal filaments can be fabricated in bulk. This is in contrast to DNA computing, where large amounts of DNA need to be specifically synthesized for each problem. Furthermore, because of their generic nature, agents can continue to explore the network as long as ATP is available, which means they can be by recirculated and used more than once (SI Appendix, Fig. S2.1). (iii) Because our device implements basic addition operations, it can benefit from existing optimized algorithms and can be easily combined with a conventional computer to form a hybrid device. For example, an electronic computer could be used to, first, solve a subset of the largest numbers in the SSP. Then, the solutions calculated by the computer would be passed on to a set of biological computers described here, drastically reducing the number of agents needed because the biological computer solves only that part of the problem that overwhelms the electronic computer. Furthermore, the time needed to feed the filaments into the network would be further reduced because many entrances can be used in parallel. An extended analysis of scaling and energy considerations is presented in SI Appendix, sections S1 and S7, respectively.
Summarizing, the technical advances that would be necessary for a future device able to challenge an electronic computer are (i) scaling up of the physical network size from currently ∼100 × 100 µm2 to wafer size, which is achievable by current patterning technology. (ii) Reduction of the filament feeding time, which can be achieved by using networks with multiple entrances, or by self-replicating filaments, see above. (iii) Reduction of pass-junction error rates. We expect that this can be realized by simulation-driven design (such as described in SI Appendix, section S4), by evolutionary algorithms for designing the junction geometries (36, 37), or by using 3D geometries such as bridges or tunnels (38) which would offer zero error rates at pass junctions. (iv) To circumvent the inherent difficulties of tracking large numbers of individual filaments, automatic readout schemes at exits of interest can be used (39). (v) Programmable devices which can flexibly encode different problems could be achieved by using heat-controlled (40) or electrostatic (41) gates in only one programmable type of junction instead of the two (isomorphic) static junctions. (vi) Finally, filaments can be prevented from attaching to or detaching from the network by using closed channels with porous openings for allowing the supply of ATP (42).
The potential practical relevance of our approach goes beyond solving SSP, because all NP-complete problems can be converted to one another using a polynomial-time conversion (7, 43) and due to the general nature of our SSP computation network (namely a computer that can perform addition, and, by using right-to-left diagonals, also subtraction). Therefore, our approach has the potential to be general and to be developed further to enable the efficient encoding and solving of a wide range of large-scale problems. Accomplishing this would move forward (but not remove) the limit of the size of combinatorial problems that can be solved.
Materials and Methods Summary
Please see the SI Appendix for a detailed description of the materials and methods.
Fabrication of Computational Networks for Use with the Actin–Myosin System.
Electron-beam lithography (EBL) was used for pattern formation in a poly(methyl methacrylate) (PMMA) resist on a SiO2-coated Si substrate. After development and O2-plasma-ashing [to ensure that the PMMA was hydrophilic and therefore unable to support motility (27)], the sample was silanized with trimethylchlorosilane to promote motility on the floor of the exposed SiO2 substrate (44). Wetting of the surface was performed to reduce the possibility of air bubbles forming in the channels (45).
Fabrication of Computational Networks for Use with the Microtubule–Kinesin System.
A silicon wafer was sputter-deposited with Au, sandwiched between two Ti adhesion layers. Next, a quartz layer was deposited, followed by a TiW layer and a ZEP520 positive-tone electron-beam resist layer. After exposure in an EBL system and development, the TiW, the quartz, and the upper Ti layers were etched by reactive ion etching down to the Au layer. Finally, the resist residue and the TiW were removed.
Actin–Myosin in Vitro Motility Assays.
The in vitro motility assays were performed at 26–29 °C, as described previously (46). Briefly, the flow cell was preincubated with (i) heavy meromyosin (47) (120 µg mL−1) for 4 min; (ii) 1 mg mL−1 bovine serum albumin for 1 min; (iii) rhodamine-phalloidin–labeled actin (48) filaments (10-nM monomeric concentration) for 1 min. The flow cell was washed both before and after actin filament incubation. Next, the flow cell was incubated with rigor solution (without ATP) for initial observations. Motility was initiated by introducing a MgAdenosine-5′-triphosphate (MgATP)-containing assay solution.
Microtubule–Kinesin in Vitro Motility Assays.
Microtubule–kinesin gliding assays were performed using full-length kinesin-1 (kinesin) from Drosophila (49) and rhodamine-labeled tubulin (50) by following a procedure (51) that was upgraded for motility in nanochannels (52). The SiO2 surface of the computational chip was passivated with 2-[Methoxy(poly-ethyleneoxy) propyl] trimethoxysilane] to prevent protein binding anywhere except on the gold bottom of the channels. Flow cells were perfused with (i) casein-containing solution (0.5 mg ml−1, 5 min); (ii) kinesin solution (2 nM, 5 min); and (iii) motility solution containing 1 mM ATP and rhodamine-labeled, taxol-stabilized.
Imaging Methods.
Rhodamine-labeled cytoskeletal filaments were observed using inverted fluorescence microscopes. Images were recorded with electron-multiplying charge-coupled device cameras and analyzed with ImageJ (imagej.nih.gov/ij/). Microtubule paths were tracked with software developed in-house (53).
Supplementary Material
Acknowledgments
Dan V. Nicolau Jr. thanks Grant Shoffner and Yue Shark Yu from University of California, Berkeley, for help in running stochastic simulations. Financially supported by the European Union Seventh Framework Programme (FP7/2007-2011) under Grant Agreements 228971 [Molecular Nano Devices (MONAD)] and 613044 [Parallel computing based on designed networks explored by self-propelled, biological agents (ABACUS)]; Defense Advanced Research Projects Agency under Grant Agreement N66001-03-1-8913; by NanoLund; by the Miller Foundation; by the Swedish Research Council (Projects 621-2010-5146 and 2010-4527); The Carl Trygger Foundation, German Research Foundation within the Cluster of Excellence Center for Advancing Electronics Dresden and the Heisenberg Program; and by Linnaeus University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510825113/-/DCSupplemental.
References
- 1.Nam GJ, Sakallah KA, Rutenbar RA. A new FPGA detailed routing approach via search-based Boolean satisfiability. Ieee T Comput Aid D. 2002;21(6):674–684. [Google Scholar]
- 2.Fraenkel AS. Complexity of protein folding. Bull Math Biol. 1993;55(6):1199–1210. doi: 10.1007/BF02460704. [DOI] [PubMed] [Google Scholar]
- 3.Pierce NA, Winfree E. Protein design is NP-hard. Protein Eng. 2002;15(10):779–782. doi: 10.1093/protein/15.10.779. [DOI] [PubMed] [Google Scholar]
- 4.Hopfield JJ, Tank DW. “Neural” computation of decisions in optimization problems. Biol Cybern. 1985;52(3):141–152. doi: 10.1007/BF00339943. [DOI] [PubMed] [Google Scholar]
- 5.Nicolau DV, et al. 2006. Molecular motors-based micro- and nano-biocomputation devices. Microelectron Eng 83(4-9 Spec. Iss.):1582-1588.
- 6.Adleman LM. Molecular computation of solutions to combinatorial problems. Science. 1994;266(5187):1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 7.Lipton RJ. DNA solution of hard computational problems. Science. 1995;268(5210):542–545. doi: 10.1126/science.7725098. [DOI] [PubMed] [Google Scholar]
- 8.Mao C, LaBean TH, Relf JH, Seeman NC. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature. 2000;407(6803):493–496. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 9.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332(6034):1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 10.Beaver D. Computing with DNA. J Comput Biol. 1995;2(1):1–7. doi: 10.1089/cmb.1995.2.1. [DOI] [PubMed] [Google Scholar]
- 11.Braich RS, Chelyapov N, Johnson C, Rothemund PWK, Adleman L. Solution of a 20-variable 3-SAT problem on a DNA computer. Science. 2002;296(5567):499–502. doi: 10.1126/science.1069528. [DOI] [PubMed] [Google Scholar]
- 12.Ouyang Q, Kaplan PD, Liu S, Libchaber A. DNA solution of the maximal clique problem. Science. 1997;278(5337):446–449. doi: 10.1126/science.278.5337.446. [DOI] [PubMed] [Google Scholar]
- 13.Reif J, LaBean T, Sahu S, Yan H, Yin P. 2005. Design, simulation, and experimental demonstration of self-assembled DNA nanostructures and motors. Unconventional Programming Paradigms, eds Banâtre J-P, Fradet P, Giavitto J-L, Michel O, Lecture Notes in Computer Science (Springer, Berlin), Vol 3566, pp 173–187.
- 14.Ladd TD, et al. Quantum computers. Nature. 2010;464(7285):45–53. doi: 10.1038/nature08812. [DOI] [PubMed] [Google Scholar]
- 15.Chiu DT, Pezzoli E, Wu H, Stroock AD, Whitesides GM. Using three-dimensional microfluidic networks for solving computationally hard problems. Proc Natl Acad Sci USA. 2001;98(6):2961–2966. doi: 10.1073/pnas.061014198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garey MR, Johnson DS. Computers and Intractability: A Guide to the Theory of NP-Completeness. W. H. Freeman & Co.; New York: 1979. p. 338. [Google Scholar]
- 17.Caprara A, Kellerer H, Pferschy U. The multiple subset sum problem. SIAM J Optim. 2001;11(2):308–319. [Google Scholar]
- 18.Kellerer H, Mansini R, Pferschy U, Speranza MG. An efficient fully polynomial approximation scheme for the Subset-Sum Problem. J Comput Syst Sci. 2003;66(2):349–370. [Google Scholar]
- 19.Schnorr CP, Euchner M. Lattice basis reduction: Improved practical algorithms and solving subset sum problems. Math Program. 1994;66(2):181–199. [Google Scholar]
- 20.Kate A, Goldberg I. Generalizing cryptosystems based on the subset sum problem. Int J Inf Secur. 2011;10(3):189–199. [Google Scholar]
- 21.Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342(6246):154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 22.Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci USA. 1986;83(17):6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiratsuka Y, Tada T, Oiwa K, Kanayama T, Uyeda TQ. Controlling the direction of kinesin-driven microtubule movements along microlithographic tracks. Biophys J. 2001;81(3):1555–1561. doi: 10.1016/S0006-3495(01)75809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vikhorev PG, et al. Diffusion dynamics of motor-driven transport: Gradient production and self-organization of surfaces. Langmuir. 2008;24(23):13509–13517. doi: 10.1021/la8016112. [DOI] [PubMed] [Google Scholar]
- 25.Hess H, Clemmens J, Qin D, Howard J, Vogel V. Light-controlled molecular shuttles made from motor proteins carrying cargo on engineered surfaces. Nano Lett. 2001;1(5):235–239. [Google Scholar]
- 26.Nicolau DV, Suzuki H, Mashiko S, Taguchi T, Yoshikawa S. Actin motion on microlithographically functionalized myosin surfaces and tracks. Biophys J. 1999;77(2):1126–1134. doi: 10.1016/S0006-3495(99)76963-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundberg M, et al. Actin filament guidance on a chip: Toward high-throughput assays and lab-on-a-chip applications. Langmuir. 2006;22(17):7286–7295. doi: 10.1021/la060854i. [DOI] [PubMed] [Google Scholar]
- 28.Clemmens J, Hess H, Howard J, Vogel V. Analysis of microtubule guidance in open microfabricated channels coated with the motor protein kinesin. Langmuir. 2003;19(5):1738–1744. [Google Scholar]
- 29.Reuther C, Hajdo L, Tucker R, Kasprzak AA, Diez S. Biotemplated nanopatterning of planar surfaces with molecular motors. Nano Lett. 2006;6(10):2177–2183. doi: 10.1021/nl060922l. [DOI] [PubMed] [Google Scholar]
- 30.Fuerstman MJ, et al. Solving mazes using microfluidic networks. Langmuir. 2003;19(11):4714–4722. [Google Scholar]
- 31.Orlova A, Prochniewicz E, Egelman EH. Structural dynamics of F-actin: II. Cooperativity in structural transitions. J Mol Biol. 1995;245(5):598–607. doi: 10.1006/jmbi.1994.0049. [DOI] [PubMed] [Google Scholar]
- 32.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274(47):33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 33.Cho H, et al. Self-organization in high-density bacterial colonies: Efficient crowd control. PLoS Biol. 2007;5(11):e302. doi: 10.1371/journal.pbio.0050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson KL, et al. Fungi use efficient algorithms for the exploration of microfluidic networks. Small. 2006;2(10):1212–1220. doi: 10.1002/smll.200600105. [DOI] [PubMed] [Google Scholar]
- 35.Tero A, et al. Rules for biologically inspired adaptive network design. Science. 2010;327(5964):439–442. doi: 10.1126/science.1177894. [DOI] [PubMed] [Google Scholar]
- 36.Rupp B, Nédélec F. Patterns of molecular motors that guide and sort filaments. Lab Chip. 2012;12(22):4903–4910. doi: 10.1039/c2lc40250e. [DOI] [PubMed] [Google Scholar]
- 37.Sunagawa T, Tanahashi A, Downs ME, Hess H, Nitta T. In silico evolution of guiding track designs for molecular shuttles powered by kinesin motors. Lab Chip. 2013;13(14):2827–2833. doi: 10.1039/c3lc41103f. [DOI] [PubMed] [Google Scholar]
- 38.Lard M, ten Siethoff L, Generosi J, Månsson A, Linke H. Molecular motor transport through hollow nanowires. Nano Lett. 2014;14(6):3041–3046. doi: 10.1021/nl404714b. [DOI] [PubMed] [Google Scholar]
- 39.Lard M, ten Siethoff L, Månsson A, Linke H. Tracking actomyosin at fluorescence check points. Sci Rep. 2013;3:1092. doi: 10.1038/srep01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroeder V, Korten T, Linke H, Diez S, Maximov I. Dynamic guiding of motor-driven microtubules on electrically heated, smart polymer tracks. Nano Lett. 2013;13(7):3434–3438. doi: 10.1021/nl402004s. [DOI] [PubMed] [Google Scholar]
- 41.van den Heuvel MG, de Graaff MP, Dekker C. Molecular sorting by electrical steering of microtubules in kinesin-coated channels. Science. 2006;312(5775):910–914. doi: 10.1126/science.1124258. [DOI] [PubMed] [Google Scholar]
- 42.Graczyk M, Balaz M, Kvennefors A, Linke H, Maximov I. 2012. Optimization of a self-closing effect to produce nanochannels with top slits in fused silica. J Vac Sci Technol, B 30(6):06FF09-1–06FF09-4.
- 43.Karp RM. Reducibility among combinatorial problems. In: Miller RE, Thatcher JW, editors. Complexity of Computer Computations. Plenum; New York: 1972. pp. 85–103. [Google Scholar]
- 44.Sundberg M, et al. Silanized surfaces for in vitro studies of actomyosin function and nanotechnology applications. Anal Biochem. 2003;323(1):127–138. doi: 10.1016/j.ab.2003.07.022. [DOI] [PubMed] [Google Scholar]
- 45.Bunk R, et al. Guiding motor-propelled molecules with nanoscale precision through silanized bi-channel structures. Nanotechnology. 2005;16(6):710–717. [Google Scholar]
- 46.Sundberg M, et al. Selective spatial localization of actomyosin motor function by chemical surface patterning. Langmuir. 2006;22(17):7302–7312. doi: 10.1021/la060365i. [DOI] [PubMed] [Google Scholar]
- 47.Kron SJ, Toyoshima YY, Uyeda TQ, Spudich JA. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 1991;196:399–416. doi: 10.1016/0076-6879(91)96035-p. [DOI] [PubMed] [Google Scholar]
- 48.Pardee JD, Spudich JA. Purification of muscle actin. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- 49.Coy DL, Wagenbach M, Howard J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem. 1999;274(6):3667–3671. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- 50.Hyman A, et al. Preparation of modified tubulins. In: Vallee RB, editor. Methods in Enzymology. Vol 196. Academic; Cambridge, MA: 1991. pp. 478–485. [DOI] [PubMed] [Google Scholar]
- 51.Nitzsche B, et al. Studying kinesin motors by optical 3D-nanometry in gliding motility assays. Methods Cell Biol. 2010;95:247–271. doi: 10.1016/S0091-679X(10)95014-0. [DOI] [PubMed] [Google Scholar]
- 52.van den Heuvel MGL, Butcher CT, Smeets RMM, Diez S, Dekker C. High rectifying efficiencies of microtubule motility on kinesin-coated gold nanostructures. Nano Lett. 2005;5(6):1117–1122. doi: 10.1021/nl0506554. [DOI] [PubMed] [Google Scholar]
- 53.Ruhnow F, Zwicker D, Diez S. Tracking single particles and elongated filaments with nanometer precision. Biophys J. 2011;100(11):2820–2828. doi: 10.1016/j.bpj.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.