Significance
Among all regular nucleotides, GTP is commonly regarded as the sole signaling ligand associated with G proteins. However, the ability of the YchF subfamily (an unconventional G protein) to bind and hydrolyze both GTP and ATP poses a major question on the role of ATP binding. Through X-ray crystallography, we showed the different specific conformational changes caused by the binding of OsYchF1 to ATP versus GTP. A mutation that precludes the binding/hydrolysis of ATP also prevents OsYchF1 from functioning as a negative regulator of plant defense responses, demonstrating the specific role of ATP-binding/hydrolysis in disease resistance. This discovery will have a significant impact on our understanding of the structure–function relationships of the YchF subfamily of G proteins in all kingdoms of life.
Keywords: P-loop NTPase, ATP-binding protein, YchF, G protein, X-ray crystallography
Abstract
G proteins are involved in almost all aspects of the cellular regulatory pathways through their ability to bind and hydrolyze GTP. The YchF subfamily, interestingly, possesses the unique ability to bind both ATP and GTP, and is possibly an ancestral form of G proteins based on phylogenetic studies and is present in all kingdoms of life. However, the biological significance of such a relaxed ligand specificity has long eluded researchers. Here, we have elucidated the different conformational changes caused by the binding of a YchF homolog in rice (OsYchF1) to ATP versus GTP by X-ray crystallography. Furthermore, by comparing the 3D relationships of the ligand position and the various amino acid residues at the binding sites in the crystal structures of the apo-bound and ligand-bound versions, a mechanism for the protein’s ability to bind both ligands is revealed. Mutation of the noncanonical G4 motif of the OsYchF1 to the canonical sequence for GTP specificity precludes the binding/hydrolysis of ATP and prevents OsYchF1 from functioning as a negative regulator of plant-defense responses, while retaining its ability to bind/hydrolyze GTP and its function as a negative regulator of abiotic stress responses, demonstrating the specific role of ATP-binding/hydrolysis in disease resistance. This discovery will have a significant impact on our understanding of the structure–function relationships of the YchF subfamily of G proteins in all kingdoms of life.
GTP-binding proteins (G proteins) play important roles in diverse fundamental biological processes in living organisms, including signal transduction, cell division, development, intracellular transport, translation, and others (1, 2). The functions and working mechanisms of some ancestral G proteins, which are believed to play essential roles in cellular processes, are still unknown. One such group is the Obg family, which features an N-terminal glycine-rich sequence, followed by a G domain for nucleotide binding and a C-terminal TGS (ThrRS, GTPase, and SpoT) domain that has nucleic acid binding affinity (3).
A unique subgroup within the Obg family, the YchF proteins, exhibit relaxed nucleotide-binding specificities (4–6). All G proteins contain a G domain (composed of the G1–G5 motifs) for GTP binding and hydrolysis (4). The G4 motif (canonical sequence: NKxD) confers the specificity for binding GTP (4). In YchFs, the noncanonical G4 sequence (NxxE) is correlated with the loss of nucleotide-binding specificities (4, 5). In some cases, ATP is the preferred ligand (4, 5). However, the physiological significance of the ancient and evolutionarily conserved YchF proteins having the relaxed binding specificities for both ATP and GTP is still unknown.
Only a few reports touch on the biological functions of YchF proteins and most focus on human and microorganisms (7–9). The YchF proteins may play a role in iron utilization and regulation of the Ton system in Brucella melitensis (7), and in the growth of the procyclic forms of Trypanosoma cruzi (5). The human YchF homolog hOLA1 suppresses cellular oxidative responses (9) but enhances heat shock responses (10). In plants, we have shown that rice (OsYchF1) and Arabidopsis (AtYchF1) YchF proteins function as negative regulators in both abiotic stress responses (11) and plant-defense responses (12).
The structures of bacterial, yeast, and human YchF proteins were reported (4, 6). In this study, we have successfully resolved the first crystal structure of a plant YchF homolog (OsYchF1) and, in addition, its cocrystal structures with AMPPNP (adenylyl-imidodiphosphate) and GMPPNP (guanylyl-imidodiphosphate). Our structures explain the dual specificity of OsYchF1 for both ATP and GTP. Using site-directed mutagenesis, we converted the noncanonical G4 motif of OsYchF1 to the canonical sequence of most regular G proteins to test the effect of losing the ATP-binding ability, while retaining GTP binding, on its physiological functions in both plant-defense responses and abiotic-stress responses.
Results
Structural Basis of Nucleotide Recognition by OsYchF1.
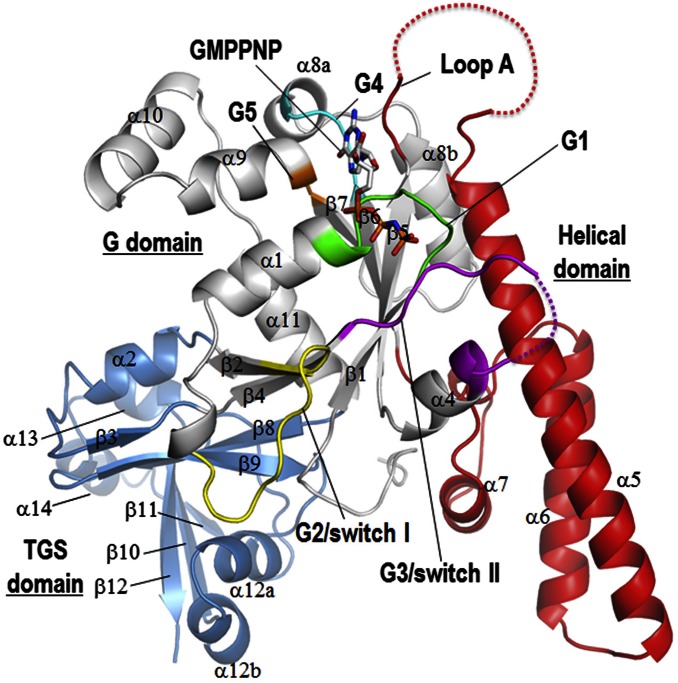
We resolved the structure of the apo forms of OsYchF1 and its complex with AMPPNP and GMPPNP by using X-ray crystallography. The statistics for diffraction data collection and structure refinement are summarized in Table S1. There are three distinct domains in OsYchF1 (Fig. 1 and Fig. S1): (i) the G domain, which consists of a six-stranded β-sheet with strand order 2-4-1-5-6-7 and has a structural signature for a TRAFAC class G protein, with strand β2 being antiparallel to strand β4; (ii) a helical domain, inserted between strand β5 and strand β6 of the G domain; and (iii) the TGS (ThrRS, GTPase, SpoT) domain, located at the C terminus (Fig. 1). Because this result is the first resolved structure to our knowledge of any plant YchF homologs, we compared it to other YchF homologs from Homo sapiens, Haemophilus influenzae, Thermus thermophilus, and Schizosaccharomyces pombe with Cα rmsd values of 1.995, 2.312, 3.394, and 2.850 Å, respectively (PDB ID codes: 2OHF, 1JAL, 2DBY, and 1NI3), and found that all these YchF homologs have similar overall structures.
Table S1.
Data collection and refinement statistics of X-ray crystallography
| apo-OsYchF1 at pH 6.5 (PDB ID code 5EE0) | apo-OsYchF1 at pH 7.85 (PDB ID code 5EE1) | AMPPNP-OsYchF1 (PDB ID code 5EE3) | GMPPNP-OsYchF1 (PDB ID code 5EE9) | |
| Data collection | ||||
| Wavelength, Å | 0.9795 | 0.9793 | 0.9790 | 0.9786 |
| Space group | P 1211 | P 22121 | P 212121 | P 212121 |
| Cell dimensions | ||||
| a, b, c, Å | 58.6, 66.9, 66.9 | 56.0, 70.4, 117.4 | 70.1, 112.4, 115.0 | 70.5, 112.1, 115.7 |
| α, β, γ, ° | 90.0, 112.7, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution, Å | 50–2.21 (2.25–2.21) | 50–2.55 (2.59–2.55) | 50–2.90 (2.95–2.90) | 50–2.75 (2.80–2.75) |
| Rmerge, % | 7.7 (59.0) | 6.7 (80.0) | 8.0 (55.3) | 10.0 (48.4) |
| I/σ | 19.0 (2.0) | 17.2 (1.9) | 19.2 (1.9) | 12.6 (2.4) |
| Completeness, % | 97.4 (99.2) | 99.7 (99.9) | 99.9 (98.3) | 98.5(99.1) |
| Redundancy | 4.8 (4.1) | 5.4 (5.6) | 7.6 (5.5) | 4.5 (3.9) |
| Refinement | ||||
| Resolution, Å | 32.84–2.20 (2.26–2.20) | 50.01–2.53 (2.59–2.53) | 50.01–2.90 (2.97–2.90) | 50.01–2.75 (2.82–2.75) |
| No. of reflections | 17,505 (569) | 14,816 (754) | 19,709 (1346) | 22,711 (1384) |
| Rwork/Rfree, % | 23.5/25.8 | 22.9/26.3 | 22.7/26.1 | 22.9/26.2 |
| No. of atoms | ||||
| Protein | 2,832 | 2,791 | 5,496 | 5,433 |
| Ligands | — | — | 62 | 32 |
| Water | 112 | 23 | 18 | 256 |
| B factors | ||||
| Protein | 23.0 | 38.6 | 65.7 | 43.4 |
| Ligands | — | — | 87.1 | 54.5 |
| Water | 33.2 | 27.0 | 45.4 | 35.6 |
| rms deviations | ||||
| Bond lengths, Å | 0.007 | 0.01 | 0.009 | 0.005 |
| Bond angles, ° | 1.13 | 1.34 | 1.34 | 0.89 |
| Ramachandran plot,* % | 93.5/5.8/0.6/0.0 | 92.2/7.8/0/0 | 92.1/7.9/0/0 | 91.5/8.5/0/0 |
Values in parentheses are for highest-resolution shells.
Residues in: most favored, additional allowed, generously allowed, and disallowed regions of the Ramachandran plot, respectively.
Fig. 1.
The general structure of OsYchF1. A cartoon representation of GMPPNP-OsYchF1 structure showing its three domains: a G domain (white), a helical domain (red), and a C-terminal TGS domain (blue). The G1–G5 motifs of the G domain are color-coded in green, yellow, magenta, cyan, and orange, respectively. Secondary structure elements are assigned by using DSSP and are numbered according to the convention used in the structures of human OLA1 (4) and Haemophilus influenzae YchF (6). Note that helix α12b is not observed in these YchF homologs. A helical domain comprising helixes α5, α6, and α7 is inserted between strands β5 and β6 and is a structural signature of YchF proteins.
Fig. S1.
Sequence alignments of YchFs. Sequences of OsYchF1, human OLA1 (hOLA1), and YchFs from T. thermophilus (ThYchF), S. pombe (SpYchF), and H. influenzae (HiYchF) were aligned by using the program MUSCLE (36). The G1–G5 motifs and domain boundaries are indicated below the alignment. The secondary structural elements are numbered according to the convention used in HiYchF (6) and hOLA1 (4). Note: Helix α3 is unfolded in GMPPNP-OsYchF1 and in apo-OsYchF1 at pH 6.5.
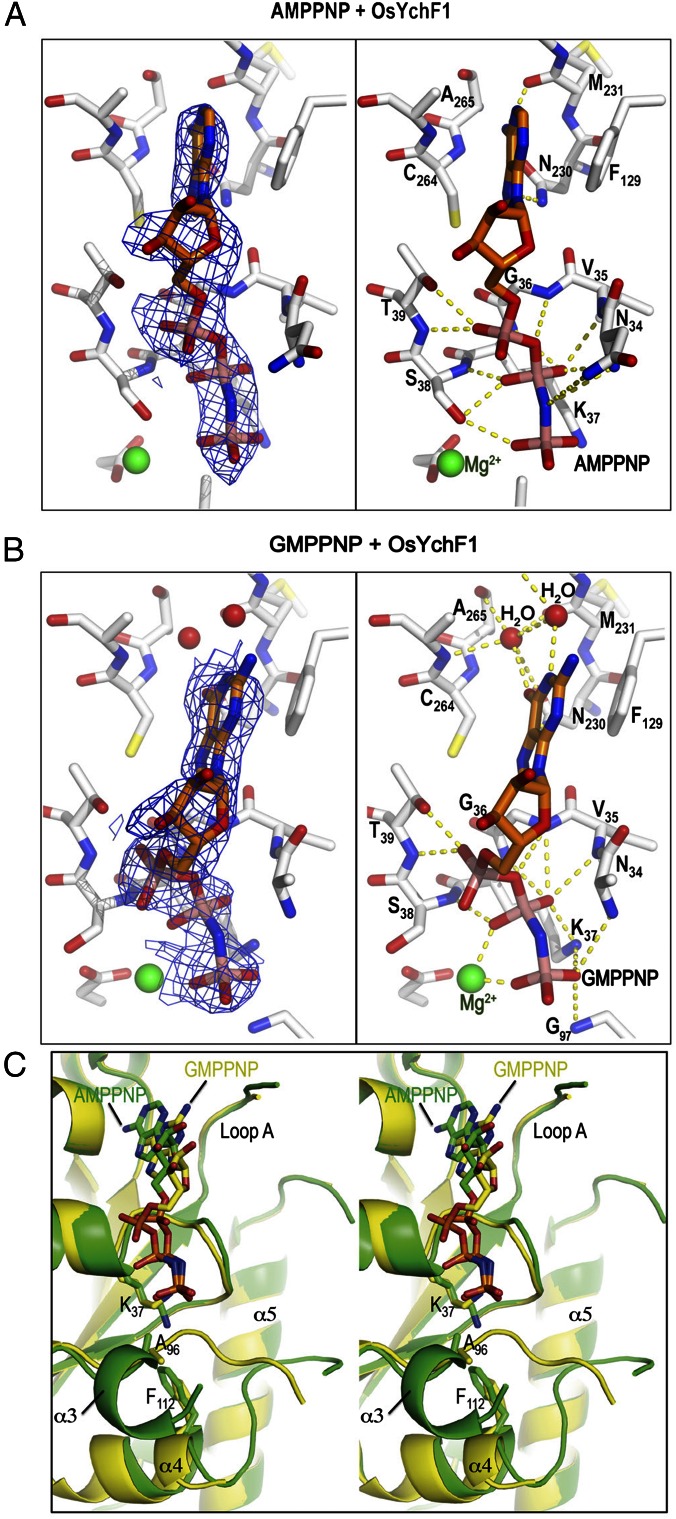
We previously established that OsYchF1 could hydrolyze both ATP and GTP (11, 12). To understand the structural basis of nucleotide recognition, we compared the structures of OsYchF1 cocrystallized with AMPPNP and GMPPNP at pH 7.5 and at pH 8.0, respectively (Table S1 and Fig. 2). In both cocrystal structures, the electron density of ligands is clearly visible at the nucleotide-binding site (Fig. 2 A and B). Like other NTPases, the phosphate groups are recognized by the G1 motif. The conserved residues 34NVGKST39 create a cradle-like structure that forms a number of hydrogen bonds with the oxygen atoms of the α-, β-, and γ-phosphates of AMPPNP and GMPPNP. The conserved interaction of the P-loop Lys-37 (via NHζ) with the γ-phosphate is observed. However, the binding of NHζ of Lys-37 to the β-phosphate is only observed in the GMPPNP-OsYchF1 structure. The bases are sandwiched between the G5 motif (Cys-264, Ala-265) and an invariant phenylalanine residue Phe-129, which is located at the beginning of loop A connected to the helical domain. The aromatic ring of Phe-129 packs nicely onto the purine ring, which stabilizes the binding of the nucleotide. In GMPPNP-OsYchF1, a Mg2+ is coordinated by the β- and γ-phosphates, whereas in AMPPNP-OsYchF1, the Mg2+ is coordinated by the γ-phosphates and the side chain of Ser-38 in the G1 motif (Fig. 2).
Fig. 2.
Structures of OsYchF1 cocrystallized with AMPPNP and GMPPNP. (A and B) Structural diagrams of OsYchF1 cocrystallized with AMPPNP (A) and GMPPNP (B). The “omit” electronic density of nucleotides are contoured at 1σ at Left, whereas hydrogen bonds between the protein and the nucleotides are indicated as yellow dotted lines at Right. (C) A stereo diagram showing the structural comparison of OsYchF1 in complex with AMPPNP (PDB ID code: 5EE3, green) and GMPPNP (PDB ID code: 5EE9, yellow). The guanine base is bound to the binding pocket ∼2 Å shallower than the adenine base. Binding of GMPPNP induces the unfolding of the helix α3 so that the backbone amide of Gly-97 of the G3/switch II region can form a hydrogen bond with the γ-phosphate of GMPPNP.
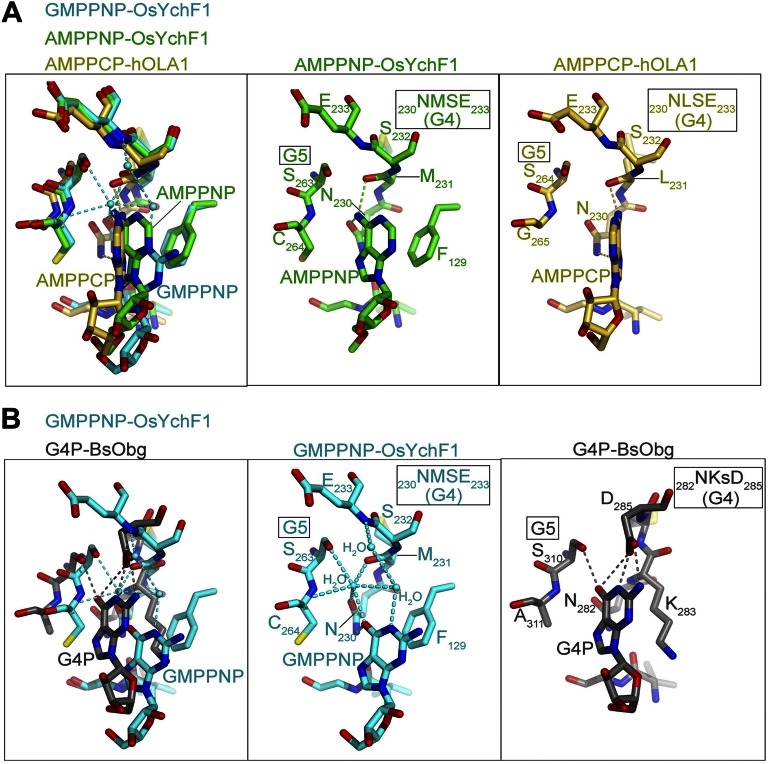
Our structures of OsYchF1 in complex with AMPPNP and GMPPNP explain the structural basis of the dual specificity of YchF proteins, which lack the canonical G4 motif (NKxD) of GTPases. In the OsYchF1 structures, the Glu-233 of the G4 motif (230NMSE233) points away from the binding pocket to provide room to accommodate binding of both ATP and GTP (Fig. S2). In the AMPPNP-OsYchF1 structure, the amine group at N6 of AMPPNP forms two hydrogen bonds with Asn-230 and Met-231 in the G4 motif, supporting the recognition for the adenine base (Fig. S2A). In the GMPPNP-OsYchF1 structure, the guanine base was bound in the binding pocket ∼2 Å shallower than the adenine base so that the purine ring of GMPPNP was more exposed to the solvent (Fig. 2B and Fig. S2). The interactions between the guanine base and the G4 and G5 motifs are mainly water-mediated hydrogen bonds, with the exception of a direct hydrogen bond between O6 of GMPPNP and the side chain of Asn-230 (Fig. 2B and Fig. S2). This mode of recognition is in sharp contrast to that found in canonical GTPases like BsObg, where the conserved Asp-285 and Ser-310 of the G4 and G5 motif form direct interactions with the guanine base (Fig. S2B). Although the overall structures of GMPPNP-OsYchF1 and AMPPNP-OsYchF1 are similar (with pairwise Cα rmsd values in the range of 0.4–1.1 Å), we could discern a notable structural difference in the conformations of Lys-37, P-loop and G3/switch II (Fig. 2C). In particular, the G3/switch II residues (95IAGLV99) form an additional helix, α3, in the AMPPNP-OsYchF1 structure. Binding of GMPPNP induces the unfolding of α3 so that the amide of Gly-97 is repositioned to form a hydrogen bond with the γ-phosphate of GMPPNP (Fig. 2B).
Fig. S2.
Structural comparisons of OsYchF1 with its homologs. (A) Structural comparison of OsYchF1 and hOLA1. The crystal structure of human OLA1 (hOLA1; yellow) in complex with AMPPCP (PDB ID code: 2OHF) is superimposed on the structure of AMPPNP-OsYchF1 (green) and GMPPNP-OsYchF1 (cyan). (B) The noncanonical G4 motif (NMSE) of OsYchF1 is structurally distinct from the canonical G4 motif (NKxD). The crystal structure of Bacillus subtilis Obg (BsObg; gray) in complex with the guanine nucleotide (PDB ID code: 1LNZ) is superimposed on the structure of GMPPNP-OsYchF1 (cyan).
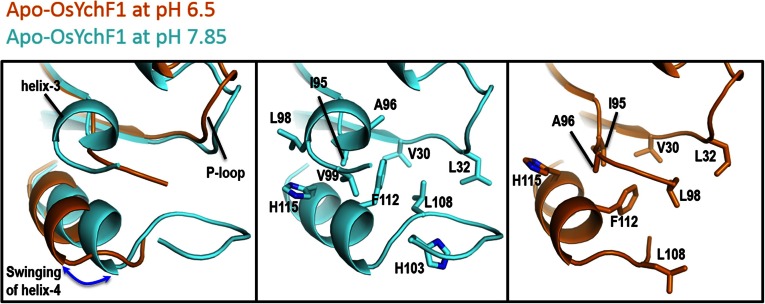
We have also determined the structures of the apo form of OsYchF1 at pH 6.5 and pH 7.85 (Table S1 and Fig. S3). Whereas the structure of apo-OsYchF1 at pH 7.85 is virtually identical to that of AMPPNP-OsYchF1 at pH 7.5, we found that there are significant conformational changes in the G3/switch II region of apo-OsYchF1 at pH 6.5. In particular, a higher pH induces the formation of helix α3 and the swinging of helix α4 toward the nucleotide-binding site, so that Phe-112 and His-115 make a number of hydrophobic contacts with Ile-95, Ala-96, Leu-98, and Val-99 of helix α3. At pH 6.5, it is anticipated that the imidazole ring of His-115 is protonated, thereby disrupting these hydrophobic interactions that stabilize α3. As a result, α3 is unfolded and α4 swings away from the nucleotide-binding site (Fig. S3). In addition, His-103, which is partially buried in the switch II loop, may also be involved in the pH-dependent conformational changes.
Fig. S3.
Structural changes in Apo-OsYchF1 at pH 6.5 and pH 7.85. A lower pH induces the unfolding of helix-3 and the swinging of helix-4 away from the nucleotide-binding sites.
The Noncanonical G4 Motif of OsYchF1 Is Essential for Its ATP Binding and ATPase Activity.
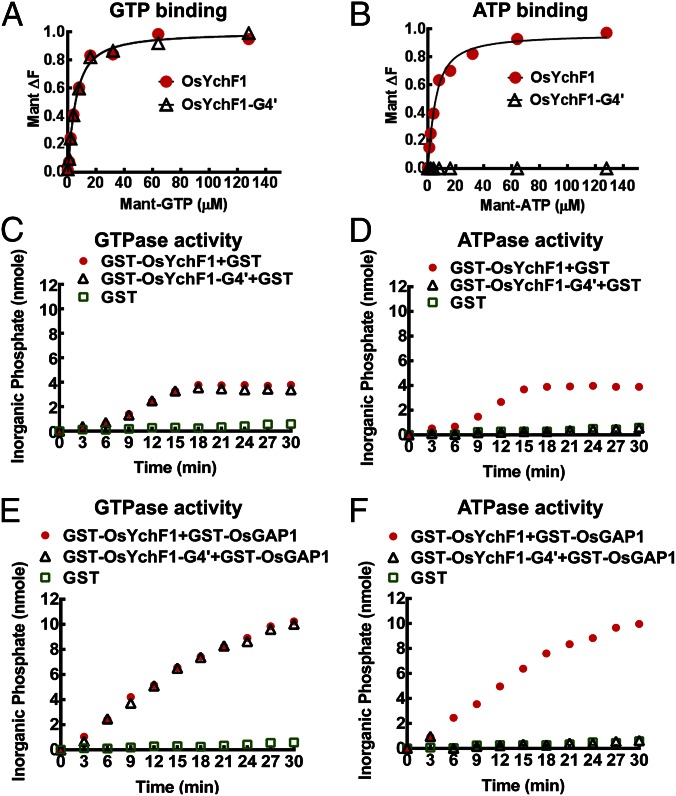
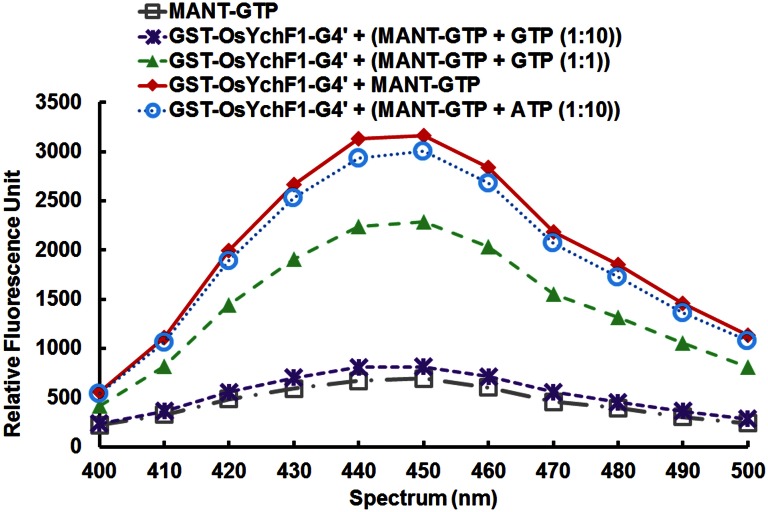
To show that the noncanonical G4 motif of OsYchF1 (230NMSE233) is essential for ATP binding, site-directed mutagenesis was performed to convert this motif to the canonical G4 motif of other P-loop GTPases (i.e., NKSD), and the mutant was designated as OsYchF1-G4′. We performed titration experiments with 2′,(3′)-O-[N-methylanthraniloyl] derivatives of ATP (Mant-ATP) and GTP (Mant-GTP), fluorescent analogs of ATP and GTP, respectively, which are commonly used in nucleotide-binding measurements (13). In this assay, OsYchF1 can bind to both GTP and ATP with similar affinities (Fig. 3 A and B) and the Kd values of OsYchF1 for binding Mant-GTP and Mant-ATP were 3.7 ± 0.6 and 3.6 ± 0.5 μM, respectively. However, OsYchF1-G4′ did not show any binding activity toward Mant-ATP (Fig. 3B) but exhibited a similar titration curve (Fig. 3A) and a similar Kd (3.760 ± 0.39 μM) for binding Mant-GTP to the wild-type OsYchF1 protein. Competition assays were also performed to confirm ATP/GTP binding by incubating Mant-GTP–charged OsYchF1-G4′ with either GTP or ATP (Fig. S4). The GST-OsYchF1-G4′ protein was first charged with Mant-GTP to give a characteristic fluorescent signal, whereas free Mant-GTP did not give any fluorescence. Successful competition was indicated by elimination of the increase in Mant-GTP fluorescent signals. Only GTP, but not ATP, could compete with Mant-GTP to bind to GST-OsYchF1-G4′. In contrast, similar experiments using the wild-type OsYchF1 showed competition by both GTP and ATP (12). Together, our results indicate that the noncanonical G4 motif in the native OsYchF1 is essential for ATP binding.
Fig. 3.
GTP/ATP binding and GTPase/ATPase activities of OsYchF1 and OsYchF1-G4′. (A and B) OsYchF1 and OsYchF1-G4′ were titrated with Mant-GTP (A) or Mant-ATP (B). GST-OsYchF1 and GST-OsYchF1-G4′ fusion proteins were expressed in E. coli, purified, and the GST tag was removed before the titration experiments. The final concentration of the target protein in the binding assays was approximately 6.5 µM. Emission signals were collected at a wavelength of 445 nm. For both A and B, circles, OsYchF1 protein; triangles, OsYchF1-G4. Data fitting was performed by using Prism 6.01v (GraphPad Software). OsYchF1 was able to bind both ATP and GTP with similar affinities, whereas OsYchF1-G4′ was only able to bind GTP. (C–F) The hydrolytic powers of GST-OsYchF1-G4′ and GST-OsYchF1 toward GTP were comparable (C), but only GST-OsYchF1 could hydrolyze ATP (D). GST-OsGAP1 could further enhance the GTPase activity of both GST-OsYchF1-G4′ and GST-OsYchF1 (E), but could only enhance the ATPase activity of GST-OsYchF1 but not that of GST-OsYchF1-G4′ (F). GST controls did not exhibit either GTPase or ATPase activities. For C–F, circles, GST-OsYchF1 protein; triangles, GST-OsYchF1-G4; squares, GST only.
Fig. S4.
OsYchF1-G4′ could not bind to ATP. Mant-GTP binding affinities of the GST-OsYchF1-G4′ fusion protein was examined by monitoring the emission of fluorescent signals (diamonds). The increases in fluorescence signals were eliminated when challenged with 1-fold unlabeled GTP (triangles) and 10-fold unlabeled GTP (crosses), but not 10-fold unlabeled ATP (circles). Unbound Mant-GTP gave minimal signals (squares).
Next, we tested whether the loss of ATP binding capacity in OsYchF1-G4′ would result in the elimination of the ATPase activity by using GST-fusion proteins. When GTP was used as the substrate, there was no apparent difference between the hydrolytic activities of GST-OsYchF1 and GST-OsYchF1-G4′ (Fig. 3 C and E). This finding showed that the GST-OsYchF1-G4′ is a functional protein. Moreover, the GTPase activities of both fusion proteins could be enhanced in the presence of OsGAP1 (a positive regulator of OsYchF1; refs. 12 and 14). However, when ATP was used, only GST-OsYchF1 exhibited hydrolytic activity, which could be further stimulated by OsGAP1. No ATPase activity could be observed when GST-OsYchF1-G4′ was used, even in the presence of OsGAP1 (Fig. 3 D and F). As a negative control, GST itself did not exhibit any GTP/ATPase activity or GTP/ATPase-activating activity (Fig. 3 C–F).
The ATP-Binding Activity of OsYchF1 Contributes to Its Negative Role in Plant-Defense Responses.
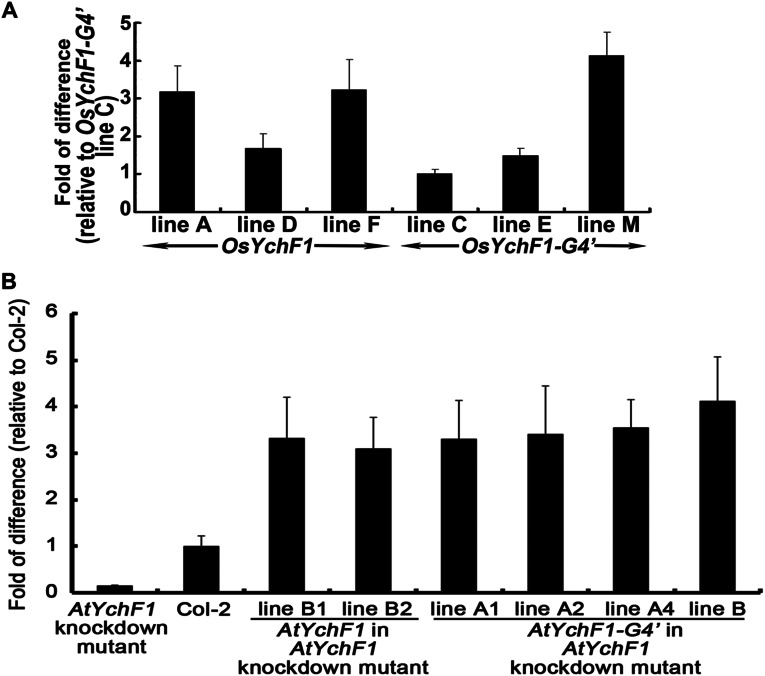
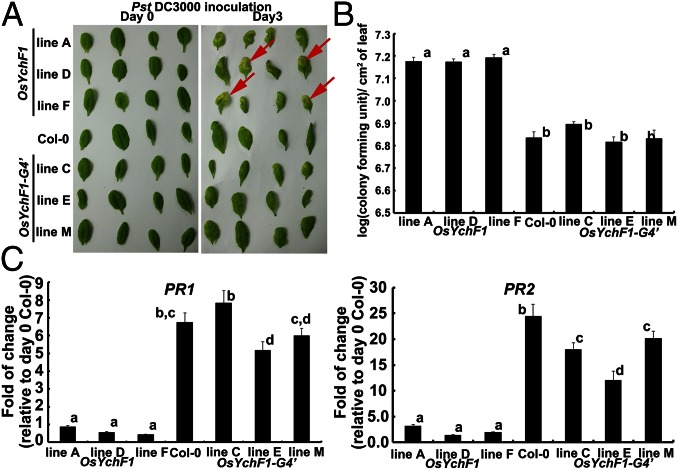
So far, one question still remains: What is the biological significance of ATP binding by OsYchF1? Now that we have discovered the relationship between the amino acid sequence in the G4 motif of OsYchF1 and its ability to bind ATP, we wanted to understand the physiological significance of its unique ATP-binding ability. We previously showed that the ectopic expression of OsYchF1 in Arabidopsis thaliana increases the sensitivity of the transgenic plants toward the bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000 (Pst DC3000) (12). Could the ability to bind and hydrolyze ATP have to do with its negative role in plant disease resistance? Using transgenic plants, we compared the effects of expressing OsYchF1 and OsYchF1-G4′ on the plant-defense responses in A. thaliana after inoculation with Pst DC3000. Three independent transgenic A. thaliana lines each of the native OsYchF1 or the OsYchF1-G4′ variant were used for the functional assays. The expression of the target genes in transgenic lines expressing the native OsYchF1 or OsYchF1-G4′ was confirmed (Fig. S5A). Consistent with our previous data, transgenic A. thaliana-expressing OsYchF1 exhibited a higher sensitivity toward Pst DC3000 than the wild type (Col-0; Fig. 4A). The area of pathogen-induced necrosis in infected leaves was much more extensive in OsYchF1-expressing lines (Fig. 4A). This more severe disease symptom was accompanied by a higher pathogen titer (Fig. 4B) and the repression of defense marker genes, PR1 and PR2 (Fig. 4C). However, OsYchF1-G4′ transgenic plants did not show any significant difference in disease symptoms compared with the untransformed wild type (Col-0) in terms of the degree of necrosis, pathogen titer, and the expression of defense marker genes (Fig. 4). This result demonstrates that the ATP-binding activity of OsYchF1 contributes to its negative role in plant-defense responses.
Fig. S5.
Expression of transgenes in different transgenic Arabidopsis lines. (A) Expressions of the transgenes in three independent lines, each of OsYchF1- and OsYchF1-G4′-transformed A. thaliana as measured by quantitative RT-PCR (qRT-PCR). The relative fold of difference was determined by setting the lowest gene expression level (line C of OsYchF1-G4′) to 1. (B) Expressions of the transgene in two and four independent lines of AtYchF1- and AtYchF1-G4′-transformed A. thaliana AtYchF1 knockdown mutant as measured by qRT-PCR. The relative fold of difference was determined by setting wild-type, Col-2, to 1. Error bar = SD. n = 3 or 4.
Fig. 4.
The ectopic expression of OsYchF1 but not OsYchF1-G4′ elevated disease symptoms caused by a bacterial pathogen (Pst DC3000). (A) Disease symptoms were observed 3 d after the inoculation of Pst DC3000 via syringe infiltration. More lesions (indicated by red arrows) were observed on the leaves of transgenic Arabidopsis lines expressing OsYchF1, whereas the extents of lesions observed in OsYchF1-G4′ transgenic lines were similar to the untransformed wild-type Col-0. (B) Pathogen titers were determined by counting colony forming units per cm2 of leaf surface area 3 d after inoculation. Error bars: SDs. n = 3 plants. All three transgenic lines expressing OsYchF1 had significantly higher pathogen titers than those expressing OsYchF1-G4′ or Col-0 (wild type). Values associated with the same letters (a or b) were not significantly different from one another (one-way ANOVA, P < 0.05). (C) The expressions of defense marker genes (PR1 and PR2) were estimated by real-time PCR. Error bar: SD. n = at least 3 samples. The transgenic lines expressing OsYchF1 did not show significant levels of induction of PR1 and PR2 after Pst DC3000 inoculation, compared with the wild-type Col-0 at day 0, whereas those expressing OsYchF1-G4′ showed high levels of PR1 and PR2 induction similar to Col-0. All experiments were performed twice, and similar results were obtained.
For further confirmation, we also overexpressed the Arabidopsis homolog of OsYchF1 (AtYchF1) and the G4-mutated form of AtYchF1 in an AtYchF1-knockdown mutant (Fig. S5B). The AtYchF1-knockdown mutant exhibited a higher resistance toward Pst DC3000, with less severe disease symptoms, lower pathogen titers, and higher expressions of defense marker genes, compared with the untransformed parent Col-2. Whereas the overexpression of AtYchF1 in this mutant led to significantly reduced plant defense responses, the effects of overexpressing AtYchF1-G4′ in the same mutant were significantly lower (Fig. S6). Because the expression levels of AtYchF1 and AtYchF1-G4′ in the respective transgenic lines were similar, this result supports our notion that the G4 motif (and its binding to ATP) in plant YchF1 proteins is important for their negative functions in plant-defense responses.
Fig. S6.
Transgenic lines overexpressing AtYchF1 exhibited significantly higher disease symptoms caused by a bacterial pathogen (Pst DC3000) compared with transgenic lines overexpressing AtYchF1-G4′. A full-length clone of AtYchF1 (AT1G30580) was amplified from Col-0 cDNA with gene-specific primers (Table S2) and mutations at the G4 motif of AtYchF1 (AtYchF1-G4′; from NMSE to NKSD) were accomplished via PCR-mediated mutagenesis by using two mutagenesis primers (Table S2). The two clones were each inserted into a binary vector and transformed into the AtYchF1-knockdown mutant (TAIR: CS855214) by vacuum infiltration (Materials and Methods). Two and four independent lines of AtYchF1 and AtYchF1-G4′ overexpressors, respectively, were used in this study. Two independent lines of OsYchF1 overexpressors were used as controls. (A) Disease symptoms were observed 3 d after the inoculation of Pst DC3000 via syringe infiltration on 4-wk-old seedlings. More lesions were observed on the leaves of transgenic lines overexpressing AtYchF1 than on transgenic lines overexpressing AtYchF1-G4′, the AtYchF1-knockdown mutant, or the untransformed wild types, Col-0 and Col-2. (B) Pathogen titers were determined by counting colony-forming units per cm2 of leaf surface area 3 d after inoculation. Error bars: SDs. n = 4 pots, each pot with 1–2 plants. Transgenic lines overexpressing AtYchF1 exhibited significantly higher pathogen titers than transgenic lines overexpressing AtYchF1-G4′ and all other control lines. Values associated with the same letters (a or b) are not significantly different from one another (one-way ANOVA, P < 0.05). (C) The expressions of defense marker genes (PR1 and PR2) were estimated by real-time PCR. The transgenic lines overexpressing AtYchF1 showed lower levels of PR1 and PR2 expression after the inoculation of Pst DC3000, compared with transgenic lines overexpressing AtYchF1-G4′ and all other control lines. Error bar: SD. n = at least 3 samples. All experiments were performed twice and similar results were obtained.
The ATP-Binding Activity of OsYchF1 Is Not Required for Its Negative Role in Abiotic Stress Responses.
Our previous works also showed that OsYchF1 played a negative role in abiotic stress responses in plants (11), and the ectopic expression of OsYchF1 in transgenic cells and transgenic plants reduced salinity tolerance. We therefore examined whether this function is also related to the ability of OsYchF1 to bind ATP by using the OsYchF1-G4′ mutant. To test this hypothesis, we used the same transgenic OsYchF1 and OsYchF1-G4′ Arabidopsis lines used in the pathogen inoculation experiments (see above) for stress treatments.
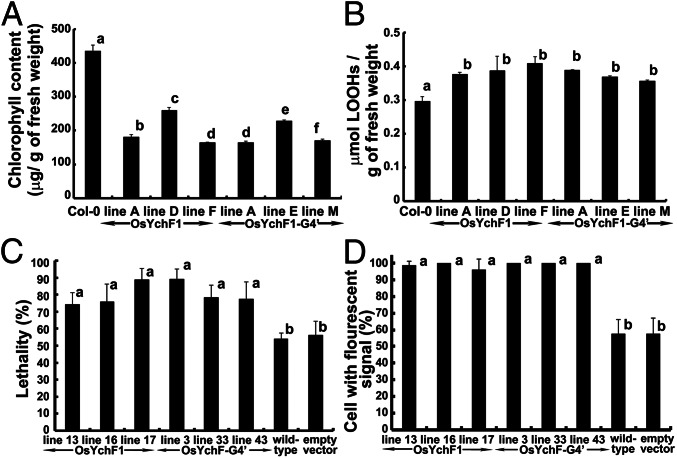
Upon treatment with 150 mM NaCl, the degree of chlorosis (quantified by the decrease in chlorophyll content) was more severe in both the OsYchF1- and OsYchF1-G4′-expressing lines compared with the untransformed control (Col-0) (Fig. 5A). The level of lipid peroxidation (a result of reactive oxygen species (ROS) accumulation under salinity stress) was also significantly higher in both the OsYchF1 and OsYchF1-G4′ transgenic lines than Col-0 (Fig. 5B). This result indicated that the OsYchF1-G4′ mutant protein was expressed properly in the transgenic plants and functioned the same way as the unaltered OsYchF1.
Fig. 5.
The ectopic expression of both OsYchF1 and OsYchF1-G4′ reduced the tolerance toward salinity stress. (A and B) The chlorophyll content (A) and the degree of lipid peroxidation (B) were examined by using 10-d-old OsYchF1- or OsYchF1-G4′-transgenic Arabidopsis seedlings further grown on MS agar medium supplemented with 150 mM NaCl for 14 d. All of the transgenic lines showed significantly different results from Col-0 in both sets of experiments, but the difference between the OsYchF1 and the OsYchF1-G4′ transgenic lines was either not significant (B) or inconclusive (A) (one-way ANOVA, P < 0.05). (C and D) Using transgenic BY-2 cells, cell viability (C), indicated by Trypan blue staining, and ROS contents (D), monitored by the florescent dye H2DCFDA, were used as stress parameters. Independent OsYchF1 (lines 13, 16, and 17) and OsYchF1-G4′ (lines 3, 33, and 43) transgenic BY-2 cell lines were compared with the wild-type cell line and the transgenic cell line containing the empty vector. The percentage of dead cells and florescence signals were minimal without NaCl treatment. Upon NaCl treatment, the percentages of dead cells and of cells with fluorescent signals from all six transgenic lines expressing either the OsYchF1 or the OsYchF1-G4′ recombinant constructs were significantly higher than the wild-type cell line and the transgenic line with the empty vector (one-way ANOVA, P < 0.05). Values with the same letter (a or b) are not significantly different. All experiments were performed twice, and similar results were obtained. For A and B, at least three sample sets (at least two seedlings each) were collected. For C and D, the number of cells counted were >80 and >40, respectively (each data point representing counts from at least six photos).
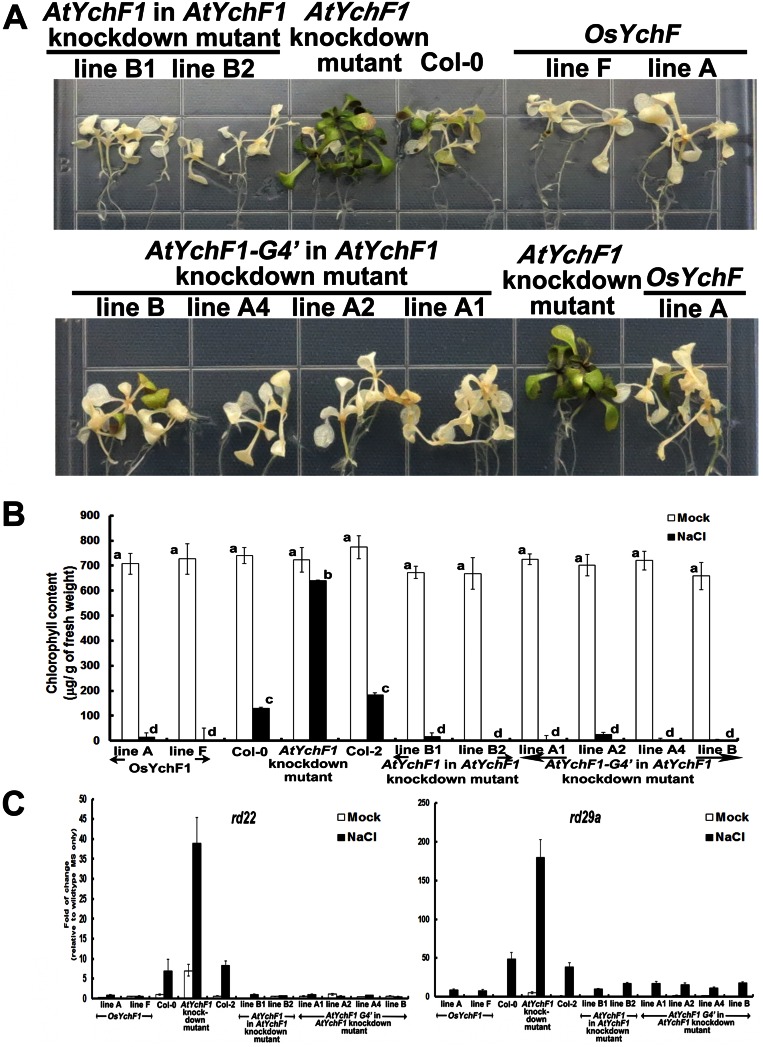
We also performed similar experiments by using transgenic A. thaliana-expressing AtYchF1 and AtYchF1-G4′ in an AtYchF1-knockdown mutant (Fig. S7). Under salt stress, the AtYchF1-knockdown mutant exhibited a higher tolerance, with less severe chlorosis symptoms, and higher expression of stress responsive genes. However, expression of either AtYchF1 or AtYchF1-G4′ in this mutant both led to a severe reduction in salt tolerance, supporting the hypothesis that binding to ATP is not essential for the negative functions of plant YchF1 proteins in salt tolerance.
Fig. S7.
The ectopic expression of both AtYchF1 and AtYchF1-G4′ reduced the tolerance toward salinity stress. The same transgenic lines as described in Fig. S6 were used in this study. Phenotypes of leaf chlorosis (A) and chlorophyll contents (B) were examined by using 10-d-old AtYchF1- or AtYchF1-G4′-transgenic Arabidopsis seedlings further grown on MS agar medium supplemented with 150 mM NaCl for 7 d. Transgenic lines overexpressing AtYchF1 or AtYchF1-G4′ showed significantly reduced salt tolerance. There were no significant difference (one-way ANOVA, P < 0.05) between AtYchF1 and AtYchF1-G4′ transgenic lines. (C) Expressions of abiotic stress responsive genes rd22 and rd29a were observed 1 d after salt treatment by using real-time PCR (Table S2). All experiments were performed twice, and similar results were obtained.
To get a more detailed picture, we examined the abiotic stress response at the cellular level. Transgenic tobacco BY-2 cells stably expressing either OsYchF1 or OsYchF1-G4′ were generated. When subjected to salinity stress (150 mM NaCl), the amounts of dead cells were much higher in both OsYchF1- and OsYchF1-G4′-expressing cell lines compared with the untransformed wild-type cells (Fig. 4C). The level of ROS accumulation under salinity stress inside the BY-2 cells was detected by prestaining with a chemical probe (H2DCFDA) and subsequent microscopic observations after stress treatments. Fluorescent signals observed inside the cells indicated the presence of ROS. Consistent with the results of dead cell counts (Fig. 4C), the accumulation of cellular ROS was also significantly higher in the OsYchF1 and OsYchF1-G4′ transgenic cells than the untransformed wild type or cells transformed with an empty vector (Fig. 4D).
In both the transgenic plant and transgenic cell systems, OsYchF1 and OsYchF1-G4′ exhibited similar effects under salinity stress, showing that the ability to bind ATP is not essential for OsYchF1 to function as a negative regulator of salt stress responses.
Discussion
In animals (human and protozoa), YchF homologs are primarily regarded as ATPases (4, 5, 15). However, a crystallographic study and subsequent binding assays suggested the presence of a GTP binding site in the H. influenza YchF homolog (6). However, there have never been any structural and functional analyses of the same YchF protein binding to both ATP and GTP until now. Compared with other YchF homologs, one unique feature of the rice OsYchF1 protein is that it exhibits similar binding affinities and hydrolytic activities toward both ATP and GTP (Fig. 3). This result enables the structural and functional comparison of OsYchF1 binding to different nucleotide ligands. The Kd of OsYchF1 for ATP is similar to that of the Escherichia coli YchF homolog, but the Kd for GTP is much lower (15). However, the GTP binding affinity of OsYchF1 is low compared with other GTPases (16–18), indicating that the mode of GTP binding to OsYchF1 may not be the same as other GTPases because of the absence of the canonical G4 motif.
Here, we proposed a model for the mechanism of ATP/GTP binding by OsYchF1 through analyzing the crystal structures of apo-bound and ligand-bound versions of the protein (Figs. 1 and 2) and elucidated the critical elements within the nucleotide binding site that allow for the dual specificity. It is interesting to point out that the human homolog of OsYchF1, hOLA1, can only bind ATP. Structural comparisons reveal that hOLA1 has a bulkier Trp-243 residue (compared with a Phe-243 in OsYchF1) that makes the base-binding pocket more crowded than OsYchF1, which may explain the difference in nucleotide specificity between the two proteins.
In YchF, the conserved lysine and aspartate in the canonical G4 motif (NKxD) are replaced by a hydrophobic residue and glutamate, respectively. More importantly, the carboxylate group of Glu-233 of the G4 motif is more than 8.2 Å away from the guanine base and does not have any direct interaction with the bound nucleotide. Therefore, the G4 motif of OsYchF1 is unlikely to distinguish guanine from adenine bases (Fig. S2). However, the structure of the G5 motif of OsYchF1 remains similar to that of the typical Obg protein (Fig. S2B). Actually, the G5 motif is involved in guanine nucleotide recognition, through water-mediated hydrogen bond formation with the O6 atom of GTP. The shallower binding of the guanine base may explain why the binding of GTP could still be accommodated by the mutation in the G4 motif (Fig. 2B and Fig. S2).
The structural data also showed that binding to ATP versus GTP caused notable rearrangements in the G3/switch II motif (Fig. 2). It is likely that the observed structural differences can be exploited by the effector proteins of OsYchF1 so that differential binding of ATP and GTP can lead to distinct biological consequences. The presence of the noncanonical G4 motif in all YchF proteins allows us to design targeted mutations, with the aim to construct a mutant form of OsYchF1 that loses the ATP binding but retains the GTP binding ability. Here, we showed that by mutating its G4 motif from the noncanonical sequence (NMSE) to the canonical sequence specific for GTP binding (NKSD), the ability of OsYchF1 to bind/hydrolyze ATP is abolished.
Both animal and plant YchF homologs are involved in abiotic stress responses (9, 11). Our data showed that the loss of ATP binding in OsYchF1 did not abolish such functions (Fig. 5). However, we showed that ATP binding contributes to (Fig. 4) the negative role of OsYchF1 in plant-specific defense responses (12).
Our data therefore gave the first report to our knowledge demonstrating the importance of ATP binding in determining the different functions of YchF homologs. ATP is traditionally believed to play a role in signal transduction mainly as the substrate of kinases (19). Subsequent research has shown that ATP itself is also an extracellular signaling molecule, a neurotransmitter that is detected by neuroreceptors (20). In this report, we provided evidence to reveal that, similar to the interaction between GTP and G proteins, ATP can also act as a ligand to control the functions of P-loop NTPases. The signaling roles of ATP may be more complicated than what we have previously thought.
Materials and Methods
Protein Expression and Purification of OsYchF1.
The OsYchF1 cDNA fragment was subcloned into an in-house constructed pRSETA-HisSUMO vector for high-level expression in E. coli BL21 (DE3). Target protein expression, extraction, and purification in the bacterial system were performed as described (21) except that isopropyl β-d-thiogalactopyranoside (IPTG) with a final concentration of 1.0 mM was added when OD600 reached 0.6, and 350 mM imidazole in lysis buffer was used to elute the target protein. The HisSUMO tag was removed with SUMO protease SenP1C (22).
X-Ray Crystallography of OsYchF1.
Crystallizations were performed at 4 °C by hanging-drop vapor diffusion method. Apo-OsYchF1 at pH 6.5 and pH 7.85 was concentrated to 15 mg/mL, and crystals were obtained in a buffer containing 16% (g/100 mL) PEG 3350, 0.1 M Bis-Tris pH 6.5, 0.2 M MgCl2, and 16.5% (g/100 mL) PEG 3350, 0.1 M Hepes pH 7.85, 0.2 M MgCl2, respectively. Crystals for the OsYchF1–AMPPNP and OsYchF1–GMPPNP complexes were obtained by first mixing purified OsYchF1 (17.5 and 20.0 mg/mL, respectively) with 3 mM AMPPNP or 5 mM GMPPNP. OsYchF1-AMPPNP crystals were grown in a buffer containing 18% (g/100 mL) PEG 3350, 0.1 M Hepes pH 7.5, and 0.2 M MgCl2. OsYchF1-GMPPNP crystals were grown in a buffer containing 0.1 M Tris pH 8.0, 17% (g/100 mL) PEG 3350, and 0.2 M MgCl2.
Crystals were cryoprotected in mother liquor containing 20% (vol/vol) ethylene glycol. Diffraction data were collected at the beamline BL17B and BL17U at the Shanghai Synchrotron Radiation Facility, beamline NE3A at the Photon Factory (KEK), and beamline 1W2B at the Beijing Synchrotron Radiation Facility. Data were indexed, integrated, and scaled by using HKL2000 (23). The initial phases of native OsYchF1 were found by molecular replacement with PHASER (24) using the structure of human OLA1 (PDB ID code 2OHF) as the search template. The model was then further refined by iterative cycles of automated and manual model building using REFMAC5 (25) and COOT (26). Using the native OsYchF1 structure as the search model, molecular replacement solutions of the complex structures were found by using PHASER. The model was built manually in the program COOT (26), and refinement was carried out with REFMAC5 (25). Data collection and processing statistics are shown in Table S1.
DNA and RNA Manipulation, Site-Directed Mutagenesis.
DNA sequencing, RNA extraction, reverse transcription, and PCR (regular and real-time) were performed according to a previous report (27). The full-length AtYchF1 cDNA clone was amplified from total cDNA of Col-0 by gene-specific primers. The relative gene expression was calculated by using the 2-ΔΔCT method (28) and normalized with the A. thaliana UBQ10 gene (29). The G4 motif of OsYchF1 or AtYchF1 was converted from the noncanonical sequence (NMSE) to the canonical sequence (NKSD) by using PCR site-directed mutagenesis with specially designed primers. Detailed primer information for the cloning and production of recombinant constructs is given in Table S2.
Table S2.
Primers and corresponding cycle profiles used in the experiments
| Primer name | Sequence |
| Primers for real-time PCR of A. thaliana PR1* | 5′- AACTACAACTACGCTGCGAACAC -3′; |
| 5′- CTTCTCGTTCACATAATTCCCAC -3′ | |
| Primers for real-time PCR of A. thaliana PR2* | 5′- CGCCCAGTCCACTGTTGATA -3′; |
| 5′- ACCACGATTTCCAACGATCC -3′ | |
| Primers for real-time PCR of A. thaliana UBQ10* | 5′- GGCCTTGTATAATCCCTGATGAATAAG -3′; |
| 5′- AAAGAGATAACAGGAACGGAAACATAGT -3′ | |
| Primers for amplifying the amino-terminal part with a EcoRI site in front of the start codon up to the G4-mutated region of OsYchF1† | 5′- AGTGAATTCATGCCGCCCAAGGCGTCCAAG -3′ |
| 5′- CCTTATCACTCTTGTTCACC -3′ | |
| Primers for amplifying the carboxyl-terminal part from the G4-mutated region of OsYchF1 with a SalI site added after the stop codon† | 5′- GGTGAACAAGAGTGATAAGG -3′ |
| 5′- CCGTCGACTCACTTCTTTCCACCTCCAGAC -3′ | |
| Primers for cloning AtYchF1 (AT1G30580) | 5′- ATCTAGAATGCCTCCGAAAGCCAAAGC -3′ |
| 5′- ACTCGAGTCATTTCTTCCCACCACC -3′ | |
| Mutagenesis primers for constructing AtYchF1-G4’ | 5′- CTCTGTCATTCTTGTTAATC -3′ |
| 5′- GATTAACAAGAATGACAGAG -3′ | |
| Primers for real-time PCR of Atrd22 (At5g25610) | 5′- CCCTTTCGTGTATAACTATGC -3′ |
| 5′- ACCATCCTCAGCGTTAAACC -3′ | |
| Primers for real-time PCR of Atrd29a (At5g52310) | 5′- GACTTTGACTCTGTTCTCGGTA -3′ |
| 5′- TTTTCCAGCTCAGCTCCT -3′ |
Real-time PCR cycling: 95 °C for 5 min; 40 cycles of 95 °C for 10 s, 60 °C for 30 s.
The amino and carboxyl terminals of G4-mutated OsYchF1 were amplified by PCR separately: 94 °C for 5 min; 30 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min 30 s; an additional polymerization step at 72 °C for 10 min. PCR products were then pooled and further amplified by adding forward and reverse primers with EcoRI and SalI sites with the same cycle profile. PCR products were finally purified and cloned into the EcoRV site of pBluescript KSII (+) via the TA cloning technique. Subsequently, an EcoRI/SalI-digested fragment was released and subcloned into pGex-4T-1 to form an in-frame fusion between OsYchF1-G4′ and the GST tag. For transformation into A. thaliana or BY2 cells, the OsYchF1-G4′ fragment in pBluescript KSII (+) was released by cutting with XhoI and XbaI and subcloned into the binary vector.
Nucleotide Binding Assays and GTPase/ATPase Activity.
GST fusion proteins were expressed by subcloning target cDNAs (encoding native OsYchF1 or OsYchF1-G4′) into the pGEX-4T-1 vector (GE Healthcare) to form in-frame fusion proteins with GST. Protein expression in the E. coli host (BL21) was induced by adding 0.5 mM IPTG to the growth medium and cultivating at 25 °C overnight. The protein was purified by using the MagneGST Protein Purification System (V8603; Promega).
Nucleotide-binding assays were performed as described (30). One hundred nanomolar 2′/3′-O-(N-methyl-anthraniloyl) guanosine 5′ triphosphate, trisodium salt (Mant-GTP) (M12415; Invitrogen) and approximately 3 μM GST-OsYchF1 or GST-OsYchF1-G4′ was added to each reaction. GTP/ATP and Mant-GTP were added in a ratio of 1:1 or 1:10 for competition assays. Signals emitted from the spectrum 400 nm to 500 nm were collected.
The GTPase/ATPase activity was determined by monitoring the release of inorganic phosphate (Pi) during GTP/ATP hydrolysis using the EnzChek Phosphate Assay Kit (E6646; Molecular Probes) (31). In a 165-μL reaction, 0.8 nmol each of the purified GST-OsYchF1-G4′, GST-OsYchF1, or GST protein was incubated with 400 μM GTP/ATP. To test the GTPase/ATPase-activating activities of OsGAP1, 0.4 nmol each of the purified GST-OsGAP1 or GST was mixed with 0.8 nmol each of the purified GST-OsYchF1 or GST-OsYchF1-G4′ before the enzymatic assay.
Plant Materials, Growth Conditions, and Transgenic Plants.
Chemicals for plant growth were from Sigma-Aldrich. The potting soil for growing A. thaliana was from Florgard Vertriebs (Gerhard-Stalling). To construct transgenic A. thaliana lines expressing OsYchF1 or OsYchF1-G4′ the coding region of each gene was subcloned into the binary vector V7 (32) and transformed (33) into the ecotype Col-0. Homozygous lines were used for functional analyses. The same recombinant constructs expressing OsYchF1 or OsYchF1-G4’ were also used to transform tobacco BY-2 cells, using a published transformation protocol (34). A. thaliana lines expression AtYchF1 or AtYchF1-G4′ were constructed by using similar methods, except that AtYchF1 knockdown mutant (TAIR: CS855214; a T-DNA insertion mutant) was used as the host.
Pathogen Inoculation Test.
Five-week-old A. thaliana plants used for pathogen inoculation tests were first grown on MS agar plates for 2 wk before being transferred to Floragard potting soil and cultivated in a growth chamber (22–24 °C; relative humidity 70–80%; light intensity 80–120 μE with a 16-h light–8-h dark cycle). The inoculation of P. syringae pv. tomato DC3000 (Pst DC3000) and the subsequent titer determination of 3-d after inoculation samples were performed by using a plate count method (35).
Abiotic Stress Treatment and Assays.
Abiotic stress treatments of Arabidopsis lines were performed by transferring 10-d-old seedlings onto MS agar plates containing 150 mM NaCl. For stress treatments of BY-2 cells, 150 mM NaCl was applied to 3-d-old suspension cultures with gentle mixing (5 × g) overnight. Chlorophyll contents, degree of lipid peroxidation, cell viability, and reactive oxygen species (ROS) accumulation were assayed as described (11).
Statistical Analysis.
Data were analyzed by using the Statistical Package for Social Sciences (version 15.0). The mean difference was analyzed by using one-way analysis of variance followed by the Tukey’s post hoc test.
Acknowledgments
We thank Ms. Iris Tong for technical help, and Ms. Jee-Yan Chu helped edit this manuscript. This work is supported by Hong Kong Research Grants Council General Research Fund 468013 (to H.-M.L. and K.-B.W.), National Natural Science Foundation of China Funds 91519332 and 31370720 (to Z.C.), and Funding for Extramural Scientists of State Key Laboratory of Agrobiotechnology Grant 2016SKLAB6-2 (to H.-M.L. and Z.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5EE0, 5EE1, 5EE3, and 5EE9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522966113/-/DCSupplemental.
References
- 1.Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: Insights from Arabidopsis and rice mutants. Curr Opin Plant Biol. 2004;7(6):719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294(5545):1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 3.Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 4.Koller-Eichhorn R, et al. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J Biol Chem. 2007;282(27):19928–19937. doi: 10.1074/jbc.M700541200. [DOI] [PubMed] [Google Scholar]
- 5.Gradia DF, et al. Characterization of a novel Obg-like ATPase in the protozoan Trypanosoma cruzi. Int J Parasitol. 2009;39(1):49–58. doi: 10.1016/j.ijpara.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 6.Teplyakov A, et al. Crystal structure of the YchF protein reveals binding sites for GTP and nucleic acid. J Bacteriol. 2003;185(14):4031–4037. doi: 10.1128/JB.185.14.4031-4037.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danese I, et al. The Ton system, an ABC transporter, and a universally conserved GTPase are involved in iron utilization by Brucella melitensis 16M. Infect Immun. 2004;72(10):5783–5790. doi: 10.1128/IAI.72.10.5783-5790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Rubio V, Lieberman MW, Shi ZZ. OLA1, an Obg-like ATPase, suppresses antioxidant response via nontranscriptional mechanisms. Proc Natl Acad Sci USA. 2009;106(36):15356–15361. doi: 10.1073/pnas.0907213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao RF, et al. OLA1 protects cells in heat shock by stabilizing HSP70. Cell Death Dis. 2013;4:e491. doi: 10.1038/cddis.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung MY, Li MW, Yung YL, Wen CQ, Lam HM. The unconventional P-loop NTPase OsYchF1 and its regulator OsGAP1 play opposite roles in salinity stress tolerance. Plant Cell Environ. 2013;36(11):2008–2020. doi: 10.1111/pce.12108. [DOI] [PubMed] [Google Scholar]
- 12.Cheung MY, et al. An ancient P-loop GTPase in rice is regulated by a higher plant-specific regulatory protein. J Biol Chem. 2010;285(48):37359–37369. doi: 10.1074/jbc.M110.172080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong YJ, Kim DE, Patel SS. Nucleotide binding induces conformational changes in Escherichia coli transcription termination factor Rho. J Biol Chem. 2004;279(18):18370–18376. doi: 10.1074/jbc.M309162200. [DOI] [PubMed] [Google Scholar]
- 14.Yung YL, et al. Site-directed mutagenesis shows the significance of interactions with phospholipids and the G-protein OsYchF1 for the physiological functions of the rice GTPase-activating protein 1 (OsGAP1) J Biol Chem. 2015;290(39):23984–23996. doi: 10.1074/jbc.M115.655639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker M, et al. The 70S ribosome modulates the ATPase activity of Escherichia coli YchF. RNA Biol. 2012;9(10):1288–1301. doi: 10.4161/rna.22131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sprang SR. G protein mechanisms: Insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. J Biol Chem. 2000;275(33):25299–25307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- 18.Zurita A, Zhang Y, Pedersen L, Darden T, Birnbaumer L. Obligatory role in GTP hydrolysis for the amide carbonyl oxygen of the Mg(2+)-coordinating Thr of regulatory GTPases. Proc Natl Acad Sci USA. 2010;107(21):9596–9601. doi: 10.1073/pnas.1004803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endicott JA, Noble MEM, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81:587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- 20.Burnstock G. Historical review: ATP as a neurotransmitter. Trends Pharmacol Sci. 2006;27(3):166–176. doi: 10.1016/j.tips.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Chan KH, Lee KM, Wong KB. Interaction between hydrogenase maturation factors HypA and HypB is required for [NiFe]-hydrogenase maturation. PLoS One. 2012;7(2):e32592. doi: 10.1371/journal.pone.0032592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong YH, et al. Assembly of preactivation complex for urease maturation in Helicobacter pylori: Crystal structure of UreF-UreH protein complex. J Biol Chem. 2011;286(50):43241–43249. doi: 10.1074/jbc.M111.296830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski ZMinor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Cheung MY, et al. Constitutive expression of a rice GTPase-activating protein induces defense responses. New Phytol. 2008;179(2):530–545. doi: 10.1111/j.1469-8137.2008.02473.x. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Remans T, et al. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta. 2008;227(6):1343–1349. doi: 10.1007/s00425-008-0706-4. [DOI] [PubMed] [Google Scholar]
- 30.Rohn TT, Nelson LK, Davis AR, Quinn MT. Inhibition of GTP binding to Rac2 by peroxynitrite: Potential role for tyrosine modification. Free Radic Biol Med. 1999;26(9-10):1321–1331. doi: 10.1016/s0891-5849(98)00332-3. [DOI] [PubMed] [Google Scholar]
- 31.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA. 1992;89(11):4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brears T, Liu C, Knight TJ, Coruzzi GM. Ectopic overexpression of asparagine synthetase in transgenic tobacco. Plant Physiol. 1993;103(4):1285–1290. doi: 10.1104/pp.103.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 34.Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell-line as the Hela-cell in the cell biology of higher-plants. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- 35.Katagiri F, Thilmony R, He SY. The Arabidopsis thaliana-Pseudomonas syringae interaction. In: Last R, Chang C, Jander G, Kliebenstein D, McClung R, Millar H, editors. The Arabidopsis Book. Am Soc Plant Biol; Rockville: 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]