Significance
This study examines how the high-energy, dome-shaped infectious form of the bacteriophage T4 baseplate assembles as opposed to how it assembles in the low-energy, star-shaped form that occurs after infection. Normal expectations would be that a molecular assembly occurs as a result of loss of energy. However, a virus has to be poised in a high-energy form to fight its way into a host. Our investigations of T4 have now shown how bacteriophage T4 can assemble into a high-energy form and how the structure of the components directs the sequential conformational changes that gain access to the host, an Escherichia coli bacterium.
Keywords: bacteriophage T4, cryo-EM reconstruction, baseplate assembly, conformational changes, near-atomic resolution
Abstract
Bacteriophage T4 consists of a head for protecting its genome and a sheathed tail for inserting its genome into a host. The tail terminates with a multiprotein baseplate that changes its conformation from a “high-energy” dome-shaped to a “low-energy” star-shaped structure during infection. Although these two structures represent different minima in the total energy landscape of the baseplate assembly, as the dome-shaped structure readily changes to the star-shaped structure when the virus infects a host bacterium, the dome-shaped structure must have more energy than the star-shaped structure. Here we describe the electron microscopy structure of a 3.3-MDa in vitro-assembled star-shaped baseplate with a resolution of 3.8 Å. This structure, together with other genetic and structural data, shows why the high-energy baseplate is formed in the presence of the central hub and how the baseplate changes to the low-energy structure, via two steps during infection. Thus, the presence of the central hub is required to initiate the assembly of metastable, high-energy structures. If the high-energy structure is formed and stabilized faster than the low-energy structure, there will be insufficient components to assemble the low-energy structure.
Most bacteriophages have a tail. At the distal end of the tail there is usually a baseplate that is decorated by some fibers (1). The baseplate initiates infection when the tail fibers bind to a host cell. Signals are transmitted from the tail fibers via the baseplate to the tail that then trigger the ejection of the phage genome from the head into the host cell through the tail tube. Two evolutionary related structures, of pyocin (2, 3) and of the type VI secretion system (4, 5), are found in bacteria as defense systems to kill competing bacteria. These structures are remarkably similar to the tail baseplate structure of bacteriophages, suggesting that tail baseplate-like structures are effective organelles for infecting bacteria (6, 7).
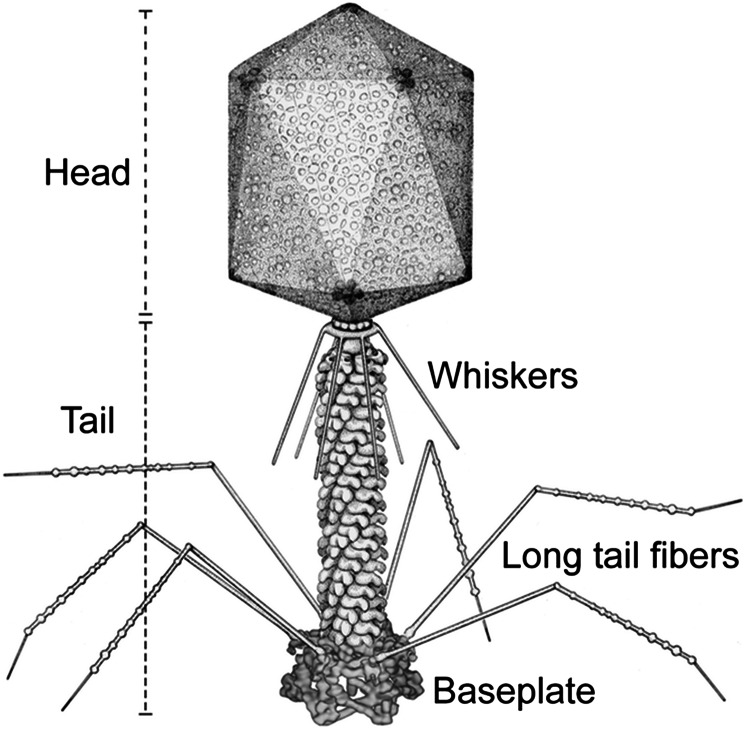
T4 is a member of the Myoviridae family of bacteriophages. These phages have a sheath around the tail tube that contracts during infection (Fig. 1) (8). T4 has a complex baseplate that is essential for assuring a highly efficient infection mechanism (8). After recognition of an Escherichia coli host cell by some of the six long-tail fibers (LTF), the short-tail fibers (STF) that are a part of the baseplate, bind irreversibly to the cell. This process is accompanied by a large conformational change in the baseplate from a “high-energy” dome- to a “low-energy” star-shaped structure (9, 10), although each of these structures represent an energy minimum in the energy landscape of the baseplate assembly. This change triggers contraction of the tail sheath, driving the tail tube into the outer host cell membrane and further across the periplasmic space to the inner membrane. The genomic DNA is then ejected into the host’s cytoplasm. Hence, the baseplate serves as the nerve center for transmitting signals from the tail fibers to the head for the release of DNA into the host.
Fig. 1.
Schematic diagram of bacteriophage T4. Bacteriophage T4 has a contractile tail and a complex baseplate. Six long-tail fibers are attached to the upper part of the baseplate and six short-tail fibers are folded under the baseplate before infection. Reproduced with permission from ref. 50, copyright American Society for Microbiology.
The hexagonal dome-shaped T4 baseplate assembles from six wedges surrounding a central hub (8). A total of 134 protein subunits from 15 different proteins form the ∼6.5-MDa baseplate (Table S1) (8, 11). The structure of the T4 baseplate has been studied extensively by cryoelectron microscopy (cryo-EM) of the whole virus and X-ray crystallography of individual proteins (12). Cryo-EM maps of the baseplate when in the dome- (9) and star-shaped (10) conformations were previously reported to 12 Å and 16 Å resolution, respectively. The dome-shaped baseplate was found to be ∼520 Å in diameter and ∼270 Å high, whereas the diameter and height of the star-shaped baseplate were ∼610 Å and ∼120 Å, respectively.
Table S1.
List of the baseplate proteins, their position and interacting protein partners in the tail and their putative role
| Protein | No. of residue | Oligomeric state in solution | Position and interacting protein partner(s) | Putative role |
| gp5 | 575 | Trimer | Hub | Lysis of host cell membrane |
| gp5.4, gp27 | ||||
| gp5.4 | 97 | Monomer | Distal end of the tail | Puncturing of host cell membrane |
| gp5 | ||||
| gp6 | 660 | Dimer | Wedge | Inter wedge interaction and wedge to hub interaction |
| gp7, gp8, gp25, gp27, gp53 | ||||
| gp7 | 1032 | Monomer | Wedge | Connecting the wedge proteins |
| gp6, gp8, gp9, gp10, gp53 | ||||
| gp8 | 334 | Dimer | Wedge | Raising the baseplate periphery |
| gp6, gp7, gp10 | ||||
| gp9 | 288 | Trimer | Wedge | Attachment of LTF |
| gp7, LTF | ||||
| gp10 | 602 | Trimer | Wedge | Attachment of STF |
| gp7, gp8, gp11, gp12 | ||||
| gp11 | 219 | Trimer | Baseplate periphery | Bending and attachment of STF |
| gp10, gp12 | ||||
| gp12 (STF) | 527 | Trimer | Baseplate periphery | Irreversible binding to host cell receptor |
| gp10, gp11 | ||||
| gp25 | 132 | Monomer | Wedge | Initiation of the tail sheath polymerization |
| gp6, gp18, gp48–gp54, gp53 | ||||
| gp27 | 391 | Trimer | Hub | Wedge to hub interaction |
| gp5, gp6, gp48–gp54 | ||||
| gp29 | 590 | ND | Hub | Tail length determination |
| gp3, gp19, gp48–gp54 | ||||
| gp48 | 364 | ND | Hub | Termination of baseplate assembly |
| gp19, gp25, gp27, gp29, gp54 | ||||
| gp53 | 196 | Monomer | Wedge | Holding of inter wedge junction |
| gp6, gp7, gp25 | ||||
| gp54 | 320 | ND | Hub | Initiation of the tail tube polymerization |
| gp19, gp27, gp29, gp48 |
LTF, long-tail fiber; STF, short tail fiber; ND, not determined.
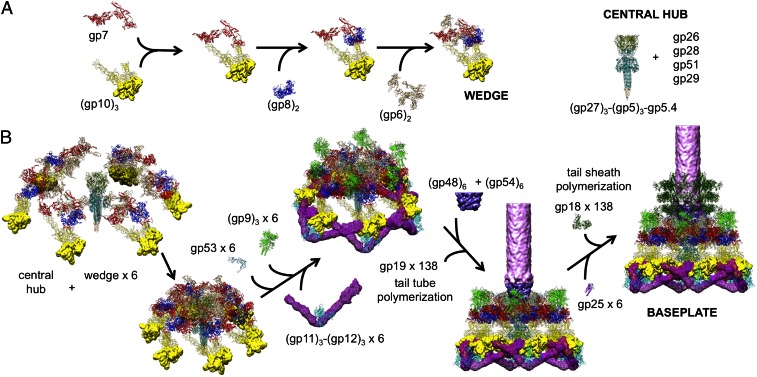
The assembly of a wedge had been shown to follow a strictly ordered sequence. First, an initial complex is formed by a monomer of gp7 and a trimer of gp10, followed sequentially by binding of a dimer of gp8 and a dimer of gp6 to the complex (13, 14). In the absence of a central hub, at least five proteins (gp7, gp10, gp8, gp6, and gp53) are required for assembly of wedges in vitro into a star-shaped, low-energy baseplate-like structure (14). Assembly of the high-energy, dome-shaped structure requires the presence of the central hub. However, how the sequential wedge assembly events are regulated remained unknown. In particular, the question remained how the high-energy dome-shaped baseplate could assemble.
We report here a 3.8-Å resolution 3D cryo-EM map of a ∼3.3-MDa in vitro-assembled star-shaped, hubless, baseplate-like complex (Fig. 2). The component proteins of this in vitro-assembled baseplate were gp7, gp10, gp8, gp6, and gp53. We show that gp7 provides the primary control of the sequential assembly events that regulate the conformational changes of the baseplate during assembly and during infection. We also show that interaction between gp6 and gp27 in associating the wedges around the central hub is the critical nucleation step to form the high-energy dome-shaped baseplate. Furthermore, we describe that the transition of the baseplate from the dome-shaped to the star-shaped conformations probably occurs in two steps.
Fig. 2.
Cryo-EM 3D reconstruction of the in vitro-assembled hubless baseplate. (A) The cryo-EM density showing the various proteins colored according to the index at the bottom left. (B) Ribbon representation of the protein structures in a single wedge using the same color code. Domains II and III of gp10 are shown as cryo-EM density.
Results
The electron density map of the in vitro-assembled star-shaped baseplate complex made it possible to build the polypeptide chains of gp7, gp53, and N-terminal parts of gp6 and gp10 whose structures had not been previously determined. In addition, fitting of the previously determined X-ray structures of gp8 (15), and C-terminal parts of gp6 (16) and gp10 (17), produced a model of the entire hubless baseplate-like complex. Fitting of all available structures, including the newly determined structures and the hub proteins [gp5 (18), gp5.4, gp6 (16), gp7, gp8 (15), gp9 (19), gp10 (17), gp11, gp12, gp18 (20, 21), gp25, gp27 (18), and gp53] into the earlier reconstructions of the T4 tail when in the dome- and star-shaped conformations, showed how these proteins rearrange themselves during infection.
Structure of gp7.
Gp7 (1,032 residues) is an elongated molecule that can be divided into six domains connected by flexible linkers (Fig. 3). Domain I (residues 1–106) has an Ig-like fold and belongs to the fibronectin type III superfamily. Domain II (residues 107–171 and 538–623) is a four-stranded, antiparallel β-sheet surrounded by five short α-helices. Domain III (residues 172–537) is inserted into domain II and has a β-propeller-like fold. The closest structural relative of this domain [with a DALI Z-score of 13.2 (22)] is phytase [Protein Data Bank (PDB) ID code 2POO]. The two structures can be superimposed with rmsd of 3.6 Å between 231 equivalenced Cα atoms. Domain III has a 24-Å-long loop (residues 454–475) that is important for interwedge interactions with gp8. Domain II is connected to domain IV via a long linker (residues 624–694), which has an extended part (residues 624–638) and a helix-turn-helix motif (residues 639–694). Domain IV (residues 695–762 and 863–894) consists of a four-helix bundle and a three-stranded β-sheet. Domain V (residues 763–862) is an insertion into domain IV and consists of an eight-stranded β-barrel followed by two short α-helices. Domain VI is connected to domain IV through a 20-residue-long linker (residues 909–928) that adopts a β-strand conformation in the baseplate wedge. This linker is important for binding the gp8 dimer and forms β-sheets together with β-strands in the gp8 molecule. Domain VI (residues 921–1001) forms a ribbon-like structure that interacts with N-terminal region of gp10. The flexibility and complexity of the gp7 structure explains why all attempts to crystallize this protein were unsuccessful.
Fig. 3.
Ribbon diagrams of the newly determined protein structures gp7, gp10, gp53, and gp6. Each polypeptide chain is rainbow-colored from blue at the N terminus to red at the C terminus. Protein domains are indicated by roman numerals. Domains II and III of gp10 are shown as cryo-EM density. Helices of gp7 and gp6, involved in the formation of the three-helix coiled-coils located at the interface between wedges, are indicated by red arrows. Also shown diagrammatically is the ring of gp6 dimers that surround the gp27 hub in the dome-shaped baseplate. Each gp6 dimer has a different color. The amino end of each gp6 monomer makes a trimeric coiled-coil with a part of gp7. The wedge boundaries are indicated by a black outline. Each wedge contains one gp6 dimer formed by association of the gp6 C-terminal regions. Contact between the N-terminal regions of gp6 is outlined by a rectangle, whereas the contact between the C-terminal regions of gp6 is outlined by a circle.
Structure of gp10.
Gp10 is a 602-residue-long trimeric protein consisting of four domains (Fig. 3). Each domain forms a homotrimer. However, the threefold symmetry axes of the individual domains are not collinear and change their relative orientations during the conformational rearrangement of the baseplate.
The N-terminal domain I (residues 1–143) of gp10 has 21% sequence identity with the N-terminal region of gp9 (19) (PDB ID code 1S2E). The N-terminal domain is located in the region of the baseplate, which was previously assigned as domain A of gp7 in the low-resolution cryo-EM study (10). Residues 1–39 form a trimeric coiled-coil region, which interacts with the C-terminal domain VI of gp7. Residues 40–143 of gp10 fold into a nine-stranded β-barrel that forms a triangular prism with the threefold-related chains.
Domains II and III of gp10 are located in the baseplate’s periphery, which is less well-resolved than the inner part of the baseplate. As a result, the resolution for these domains is limited to ∼5 Å. Domain II of gp10 has a prism-like structure and is an adaptor for binding the STFs, gp12. Domain III of gp10 is a three-finger-like domain, providing a site for the attachment of gp11 to the baseplate. The crystal structure of the gp10 C-terminal domain IV (residues 406–602) had been determined from a truncated and protease-resistant fragment (17). This crystal structure was fitted into the density and was found to have a real-space correlation of 0.85 with the cryo-EM map (23). Domain IV of gp10 attaches to domain III of gp7 in the baseplate; in the star-shaped baseplate it has additional contacts with gp9.
Structure of gp6.
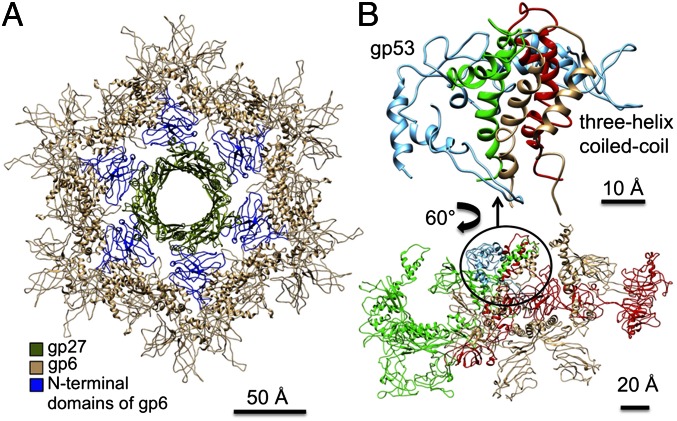
Gp6 is a 660-residue-long dimeric protein (Fig. 3). In the dome-shaped baseplate, six gp6 dimers interact with each other and form a ring around the central hub. The crystal structure of a dimer of the gp6 C-terminal part (residues 340–660) had been previously determined (16) and fits well into the cryo-EM density of the in vitro-assembled baseplate with a real space correlation of 0.86 between the cryo-EM density and crystal structure. The structure of the N-terminal part of gp6 consists of a helix-turn-helix motif (residues 26–79) and two spatially separated domains. Domain I (residues 80–170 and 278–316) has a core with a lipoxygenase homology-like fold, whereas domain II (residues 171–279), which is an insertion into domain I, has an Ig-like fold. The region of gp6, which includes the helix-turn-helix motif and three helices in domain I (residues 80–103 and 319–355) is similar to the region 645–753 of gp7. These gp6 and gp7 regions can be superimposed with a rmsd of 3.5 Å between equivalenced Cα atoms. In the baseplate, the N-terminal regions of two gp6 chains and the similar region of gp7 interact and form a three-helix coiled-coil structure.
There are two chains of gp6 in each wedge; these have different environments and different conformations of their N-terminal parts (Fig. 3). There are two types of contacts between the gp6 chains in the baseplate. One of these contacts is between the C-terminal regions, similar to what was observed in the crystal structure of gp6 (16). The other contacts are between the N-terminal parts of two gp6 chains different to those that make contact between the C-terminal regions. Because the assembly of the wedges into the baseplate is dependent on the presence of gp53 (13, 14), and because the structure of the baseplate reported here shows that N-terminal contacts of gp6 are closely associated with gp53, it is reasonable to assume that the N-terminal contacts are between gp6 chains in neighboring wedges and that the C-terminal contacts are between gp6 chains within a wedge. This result is different to what had previously been assumed (16).
Structure of gp53.
Gp53 is a 196-residue-long monomeric, V-shaped protein (Fig. 3). The protein has a core with a LysM-like fold (24) from which two “arms” extend. Each arm is formed by two α-helices, with residues 1–44 and 172–192 forming one arm, and residues 89–103 and 155–168 forming the other arm. In the baseplate, gp53 embraces a three-helix coiled-coil consisting of two gp6 chains and one gp7 chain, thus joining wedges into the baseplate. A small 67-residue protein gpX with a similar LysM-like core, but lacking the arms, was found in the baseplate of phage P2 (25). This protein is, therefore, likely to have a similar function and a common evolutionary origin as gp53. However, the presence of the arms in gp53 suggests that this protein is more efficient as an interwedge clamp, compared with its P2 ortholog.
Discussion
Wedge Assembly.
Wedge assembly starts with formation of an initial complex between gp7 and gp10, followed by binding of gp8 and then gp6 (Fig. 4A and Movie S1) (13, 14). Gp7 serves as a backbone for attachment of other proteins. The gp7 molecule is elongated and presumably rather flexible, considering its six domains are connected by unsupported linkers. The structure shows that when gp10 binds to gp7, in the first step in wedge assembly, domains I and IV of gp10 bind to domains VI and III of gp7, respectively, to form a ring-like structure, which reduces the number of degrees of freedom of the flexible gp7 molecule. Binding of gp10 to gp7 also results in stretching the 20-residue-long linker between domains IV and VI of gp7, required for subsequent binding of a gp8 dimer. The residues 894–902 of this linker are involved in parallel β-strand interactions with the residues 323–334 of one gp8 chain, whereas the residues 906–917 are involved in antiparallel β-strand interactions with the residues 323–334 of another gp8 chain. Presumably, binding of the rigid gp8 dimer would further reduce the flexibility of gp7 and restrict movements of domains I, IV, V, and VI of gp7. As a result, the helix-turn-helix motif of gp7 adopts an orientation favorable for subsequent binding of gp6, which binds to domain IV and the helix-turn-helix motif of gp7 to complete the assembly of a wedge. In the assembled structure of the baseplate, the gp10, gp8, and gp6 proteins interact extensively with the backbone gp7, but the contact surfaces between gp10, gp8, and gp6 within a wedge are only small. These observations show that the flexibility of gp7 is the key for controlling the sequential assembly.
Fig. 4.
Assembly of a baseplate based on present and earlier results. (A) Wedge assembly. Gp10, gp8, and gp6 bind sequentially to the gp7 backbone protein. The central hub of the baseplate is assembled independently. (B) Baseplate and tail assembly. Six wedges assemble around the central hub to form a baseplate. Gp53 binds adjacent wedges together. Subsequently, gp9 and the gp11–gp12 complex bind to the baseplate, further stabilizing the dome-shaped configuration. Then, gp48 and gp54 bind to the top of the central hub and initiate polymerization of the tail tube. Gp25 attaches to the gp48–gp54 complex, initiating polymerization of the tail sheath. For clarity, only three rings of the tail sheath are shown.
Baseplate Assembly.
How is it possible to assemble the high-energy dome-shaped baseplate initially as opposed to the low-energy star-shaped baseplate?
After the individual wedges have assembled, six wedges assemble around the central hub (12) to form the dome-shaped baseplate (Fig. 4B and Movie S2). The present structure has shown the complete structure of gp6, which when fitted into the cryo-EM density of the dome-shaped baseplate shows that gp6 binds tightly around the central hub protein gp27 (Fig. 5A). Thus, the nucleation of the baseplate assembly from individual wedges is dependent on the interaction of gp6 with gp27. However, in the star-shaped baseplate structure the tight ring of six gp6 dimers around gp27 has expanded and lost contacts with gp27. Thus, the initiation of baseplate assembly is the association of gp6 with gp27, from wedges that are probably assembled as in the dome-shaped baseplate. In the absence of gp27, as was the case for the in vitro-assembled baseplate reported here, gp6 can assemble only as a star-shaped baseplate. Thus, the route to the low-energy star-shaped wedge is via the initially assembled dome-shaped wedge.
Fig. 5.
Position of proteins around the central hub. (A) Interactions of gp6 with the central hub protein gp27. The N-terminal domains of gp6 that interact with gp27 (olive) to associate the wedges around the central hub to assemble the dome-shaped baseplate are highlighted in blue. (B) Interactions of adjacent wedges in the baseplate. The helix-turn-helix motifs of gp7 (red) and the two gp6 chains (green and light brown) from adjacent wedges are held together by gp53 (light blue) in the baseplate. An enlargement of the interacting region is shown.
In the assembled dome-shaped baseplate the interface between the wedges and the hub is formed mainly by the N-terminal domains of gp6 and by gp27 (Fig. 5A). Gp6 forms a ring around the central hub, and gp53 attaches to the trimeric coiled-coil formed by gp6 and gp7 (Fig. 5B), presumably stabilizing the hexameric association of the wedges in the dome-shaped configuration.
Fitting of the protein structures reported here into the lower-resolution structure of the dome-shaped baseplate shows that the domains of each protein are rigid units that do not significantly change their structure between the dome- and star-shaped conformations of the baseplate. In the final assembled baseplate, gp9 is bound to domain III of gp7, further stabilizing the baseplate. Pulse-chase analysis (26) suggested that gp9 attaches to the baseplate followed by the simultaneous attachment of gp11 and of the STFs, gp12. In the dome-shaped baseplate, the STFs are kinked and folded underneath the baseplate, across three adjacent wedges. Because gp11 attaches to the kinked region of the STFs, gp11 might help to bend the STFs and to locate them under the baseplate (Fig. 4B).
Tail Assembly.
The tail tube (gp19) and tail sheath (gp18) polymerization are initiated by the proteins at the center of the dome-shaped baseplate. After the wedges have assembled into the hexameric baseplate, six copies of gp48 and six copies of gp54 bind onto the central hub (27, 28) and probably form hexameric rings on top of gp27 (Fig. 4B). Gp54 is predicted to have a domain with a fold similar to the tail-tube protein. Thus, the bound gp54 in association with the tape measure protein, gp29, can initiate the polymerization of gp19 to form the tail tube (27, 28).
The gp25 protein then attaches to gp53 and gp6, further strengthening the interwedge interactions (29). It also binds to the gp48–gp54 complex reinforcing interactions between the hub and wedges (29, 30). Fitting of the gp25 structure (PDB ID code 4HRZ) into the earlier reported dome-shaped baseplate (9) shows that gp25 interacts extensively with the gp48–gp54 complex, suggesting that these proteins are important for attachment of gp25 to the baseplate. Gp25 has a structural similarity to the C-terminal domain of the tail sheath protein, gp18. Thus, it is likely that gp25 initiates the polymerization of the tail sheath.
Infection Mechanism.
Phage T4 uses the LTFs to recognize a LPS and/or OmpC receptor on the surface of the host cell (8). Because phages have been observed that have baseplates, which have changed to star-shaped structure, but that still have extended sheaths (31), presumably, the initial step of infection is the change of the dome-shaped structure to an intermediate structure in which the STFs are released. Subsequently, attachment of the STFs to cell-surface molecules triggers the baseplate to change its conformation to the final star shape, which in turn causes the sheath to contract and the tail tube to puncture the outer cell membrane. Thus, it is likely that the conformational changes of the baseplate occur in two steps—namely, first from the dome-shaped structure to an intermediate star-shaped structure and then from the intermediate structure to the final star-shaped structure.
The LTFs are attached to the baseplate via an adaptor protein gp9 (32). The fit of the gp7 and gp9 structures into the 16-Å resolution cryo-EM density of the dome-shaped baseplate (19) shows that domain III of gp7 interacts with density that represents the N-terminal part of gp9 (Movie S2). Thus, a flexible region of gp9 contacts gp7, allowing the LTFs to have multiple conformations in solution. When the LTFs attach to the cell, the Brownian motion of the phage particle may cause these flexible regions (31) to pull on domain III of gp7. This domain has extensive interactions with the gp8 dimer in the adjacent wedge, and with the C-terminal domain IV of gp10 in the same wedge. The rigid gp8 dimer in turn interacts extensively with the C-terminal domain VI of gp7, which is attached to the N-terminal domain I of gp10. Therefore, the interaction of gp9 with domain III of gp7 is likely to disturb interactions among gp7, gp8, and gp10 at the vertices of the dome-shaped baseplate and trigger the periphery of the baseplate to change to a star-shaped conformation (Movie S3). As a result, the STFs attached to gp10 might unfold from the baseplate and point toward the cell surface with their C-terminal domain, whereas the N-terminal part of the STFs remain attached to domain II of gp10, after release of gp11 (Fig. 6). During the conformational change from dome-shaped to a star-shaped intermediate structure, the gp11 molecules, attached to domain III of the gp10 molecules, move closer to the phage head and push on the end of the LTFs close to the baseplate. The interaction of gp11 with the LTFs probably causes the parts of the LTFs closest to the baseplate to rotate with respect to the baseplate as seen in the star-shaped baseplate. Thus, LTFs attached to the cell surface are used as levers to bring the phage closer to the cell surface. On completion of these movements, the baseplate will have the structure of the intermediate state that has a star-shaped periphery (gp7, gp8, gp9, gp10, gp11, and gp12), although the inner ring of gp6-gp53-gp25 molecules probably remains attached to the hub (Fig. 6).
Fig. 6.
Conformational changes of the baseplate during infection. Domains II and III of gp10, gp48, gp54, and the proximal part of the tail tube are presented as electron density. The proximal end of LTFs and STFs are presented as rods. Three rings of the tail sheath are shown. The identity of each protein is indicated in the index. (A) The dome-shaped baseplate. (Right) The inner ring of gp6-gp53-gp25 and part of gp7. The ring is attached to the central hub. (B) The final star-shaped baseplate with three rings of the contracted tail sheath. (Right) The inner ring of gp6-gp53-gp25 and part of gp7. The ring is expanded and detached from the central hub. The intermediate star-shaped baseplate presumably would have star-shaped periphery (B, Center) but the inner ring of gp6-gp53-gp25 and part of gp7 remains attached to the central hub (A, Right).
The movement of the phage on the surface of E. coli will be restricted because the LTFs are attached to the cell and because steric clashes with gp11 would restrict the movement of the LTFs in the star-shaped baseplate. Random Brownian motion is likely to occasionally bring the phage closer to the cell surface, allowing the STFs to search for suitable receptors. Once the phage is firmly fixed to the host’s surface, Brownian motion of the phage may result in the STFs pulling on the baseplate, triggering the signal for the contraction of the tail sheath. A likely pathway for signal transmission from STF to the central part of the baseplate would be via gp10, gp8, gp7, and, finally, to the ring of gp6. Comparison of the gp6 ring in the dome-shaped and star-shaped structures shows that the inner diameter of the gp6 ring expands from 71 to 101 Å. Thus, the signal initiated by the interaction of the STFs with the E. coli surface causes gp6 to detach from the central hub; in turn, this would cause movement of the gp53 and gp25 molecules, attached to the heterotrimeric (gp6)2(gp7) coiled-coil (Fig. 6 and Movie S3). This movement of gp25 would nudge the C-terminal domain of the tail sheath protein, gp18, thus initiating contraction of the metastable tail sheath (Movie S3).
Assembly of High-Energy Biological Structures.
Biological machines require energy that can be provided by catalyzing a chemical reaction of high-energy molecules such as ATP, or by using the energy stored when macromolecules fold and assemble. Phage baseplates are an example of the latter. The current analysis of the T4 baseplate shows how the energy required for infection can in part be stored in the dome-shaped baseplate; it also shows that assembly of the dome-shaped baseplate requires the presence of a template (the central hub) that traps wedges by selecting wedges when they are briefly in a high-energy conformation.
Methods
Electron Microscopy.
Crystals of hubless baseplates that diffracted X-rays to 4.2 Å resolution (12, 33) were dissolved in 50 mM Tris, 100 mM NaCl buffer at pH 8 to a concentration of 2 µg/mL. Aliquots of 3 µL of sample were loaded on glow-discharged C-Flat grids (CF-1.2/1.3-4C-50, CF-2/2-4C-50). These grids were then blotted for 2 s and flash frozen in liquid ethane using the Gatan CP3 plunge freezer. The grids were viewed in an FEI Titan Krios microscope operated at 300 kV. Images were recorded using a K2 Summit detector at a calibrated magnification of 38,168 yielding a pixel size of 0.655 Å. A total dose of 35 e−/Å2 and an exposure time of 7.6 s were used to collect 38 movie frames with defocus ranging from 0.5 to 3.5 µm. Fully automated data collection was operated in counting mode using the MSI-Template application in LEGINON (34). The dosef_gpu_driftcorr software (35) integrated in the Appion pipeline (36) was used to perform frame alignments to correct stage drift and beam-induced motion. A frame offset of seven and a B-factor of 1,000 pixels squared were used to align the movie frames.
A total of 1,725 images were collected; from these, 67,920 particles were boxed manually using the EMAN2 package (37). Contrast transfer function parameters were estimated using CTFFIND3 (38). The 2D and 3D classifications were performed using RELION (39). An initial model and a mask were generated using EMAN2. In total, 45,607 particles were used in the final reconstruction. The 3D reconstruction was performed with RELION, and further refinement was performed using the JSPR software package (40).
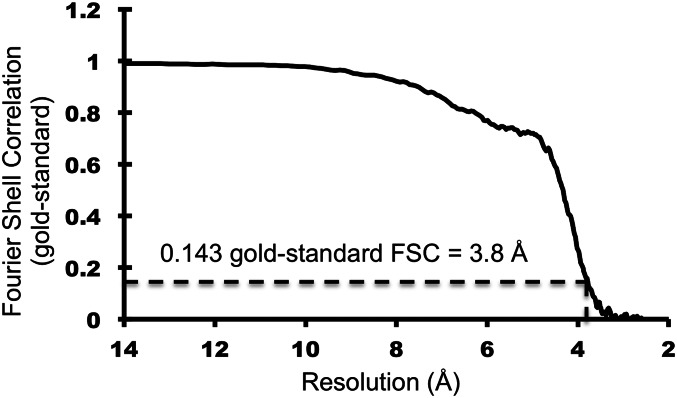
The electron density map was subjected to postprocessing by JSPR for map sharpening by applying a negative B-factor of 100 Å2. The Fourier shell correlation curve was calculated for an electron density map in which the solvent regions were set to a constant value (41). The resolution of the final map was estimated to be 3.8 Å (Figs. S1 and S2), based on the gold-standard Fourier shell correlation (FSC) criterion of 0.143 (42). The absolute hand of the map was determined by fitting the various known crystal structure baseplate components into the density map.
Fig. S1.
Gold-standard FSC curve. Resolution corresponding to 0.143 FSC cutoff is 3.8 Å. The cryo-EM 3D reconstruction map for the in vitro-assembled, hubless baseplate-like structure reported in this study is estimated as 3.8 Å resolution.
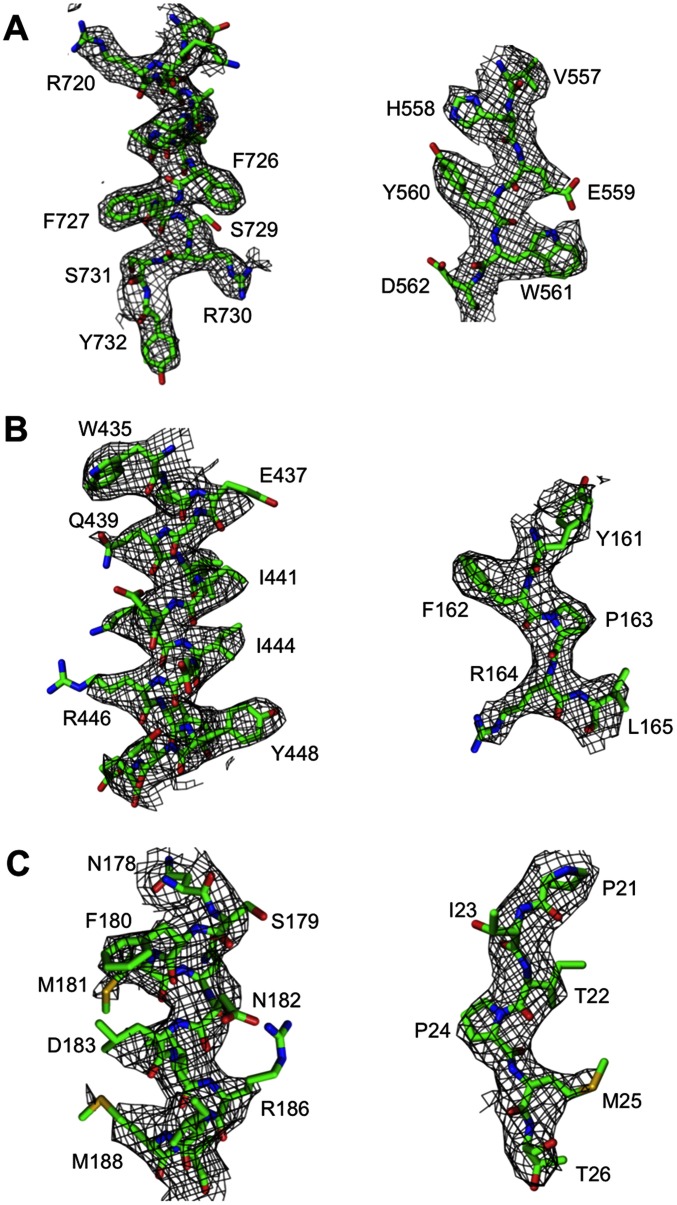
Fig. S2.
Representative areas of the density map around (A) gp7, (B) gp6, and (C) gp53.
Model Building.
Crystal structures of complete gp8, partial gp6, and gp10 were fitted into the density map using CHIMERA (23). Models of the other wedge proteins were built using COOT (43). The model of the entire wedge was subjected to real-space refinement (44) in PHENIX (45) to 3.8 Å resolution. The quality of the cryo-EM density was significantly better near the center of the baseplate, which made it possible to build side chains for gp6, gp7, and gp53. However, gp10 was built as main chain. Figures were prepared using the CHIMERA (23) and the PyMOL (46). Movies were prepared using CHIMERA (23).
Fitting of Protein Structures into Previous Cryo-EM Reconstructions.
The crystal structures of gp9 [(19), PDB ID code 1S2E], gp11 [(47), PDB ID code 1EL6], gp27–gp5 [(18), PDB ID code 1K28], gp8 [(15), PDB ID code 1N80], gp12 N terminus [(48), PDB ID code 1OCY], gp10 C terminus [(17), PDB ID code 2FKK], gp6 C terminus [(16), PDB ID code 3H2T], gp18 [(20), PDB ID code 3FO8; (21), PDB ID codes 3J2N and 3J2M], gp25 (PDB ID code 4HRZ) and gp5.4 (PDB ID code 4KU0) were fitted into earlier cryo-EM maps (accession codes EMD-1048, EMD-1086, and EMD-1126). Models of gp7, gp10 N terminus, gp6 N terminus, and gp53 from this study were fitted into the maps of the dome-shaped (9, 49) and the star-shaped (10) baseplates using the CHIMERA program (23).
Supplementary Material
Acknowledgments
We thank Sheryl Kelly for help in the preparation of this manuscript. This work was supported by NIH Grant AI081726 (to M.G.R.). Some of the work presented here was conducted at the National Resource for Automated Molecular Microscopy, which is supported by National Institute of General Medical Sciences Grant GM103310 from the NIH.
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryo-EM reconstruction map of the in vitro-assembled, star-shaped, hubless, T4 baseplate-like complex has been deposited in the Electron Microscopy Data Bank, www.emdatabank.org/ (EMDB accession code EMD-8064). The coordinates of a wedge (gp7-gp10-gp8-gp6-gp53) have been deposited in the Protein Data Bank, www.rcsb.org/ (PDB accession code 5HX2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601654113/-/DCSupplemental.
References
- 1.Calendar R, editor. The Bacteriophages. Oxford Univ Press; New York: 2006. [Google Scholar]
- 2.Kageyama M. Studies of a pyocin. I. Physical and chemical properties. J Biochem. 1964;55:49–53. doi: 10.1093/oxfordjournals.jbchem.a127839. [DOI] [PubMed] [Google Scholar]
- 3.Ishii SI, Nishi Y, Egami F. The fine structure of a pyocin. J Mol Biol. 1965;13(2):428–431. doi: 10.1016/s0022-2836(65)80107-3. [DOI] [PubMed] [Google Scholar]
- 4.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7(1):25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483(7388):182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge P, et al. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat Struct Mol Biol. 2015;22(5):377–382. doi: 10.1038/nsmb.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudryashev M, et al. Structure of the type VI secretion system contractile sheath. Cell. 2015;160(5):952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karam JD, editor. Molecular Biology of Bacteriophage T4. Am Soc Microbiology; Washington, DC: 1994. [Google Scholar]
- 9.Kostyuchenko VA, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10(9):688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- 10.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118(4):419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Leiman PG, et al. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap ML, Rossmann MG. Structure and function of bacteriophage T4. Future Microbiol. 2014;9(12):1319–1327. doi: 10.2217/fmb.14.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikuchi Y, King J. Genetic control of bacteriophage T4 baseplate morphogenesis. I. Sequential assembly of the major precursor, in vivo and in vitro. J Mol Biol. 1975;99(4):645–672. doi: 10.1016/s0022-2836(75)80178-1. [DOI] [PubMed] [Google Scholar]
- 14.Yap ML, Mio K, Leiman PG, Kanamaru S, Arisaka F. The baseplate wedges of bacteriophage T4 spontaneously assemble into hubless baseplate-like structure in vitro. J Mol Biol. 2010;395(2):349–360. doi: 10.1016/j.jmb.2009.10.071. [DOI] [PubMed] [Google Scholar]
- 15.Leiman PG, et al. Structure and location of gene product 8 in the bacteriophage T4 baseplate. J Mol Biol. 2003;328(4):821–833. doi: 10.1016/s0022-2836(03)00366-8. [DOI] [PubMed] [Google Scholar]
- 16.Aksyuk AA, Leiman PG, Shneider MM, Mesyanzhinov VV, Rossmann MG. The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate. Structure. 2009;17(6):800–808. doi: 10.1016/j.str.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Leiman PG, Shneider MM, Mesyanzhinov VV, Rossmann MG. Evolution of bacteriophage tails: Structure of T4 gene product 10. J Mol Biol. 2006;358(3):912–921. doi: 10.1016/j.jmb.2006.02.058. [DOI] [PubMed] [Google Scholar]
- 18.Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415(6871):553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 19.Kostyuchenko VA, et al. The structure of bacteriophage T4 gene product 9: the trigger for tail contraction. Structure. 1999;7(10):1213–1222. doi: 10.1016/s0969-2126(00)80055-6. [DOI] [PubMed] [Google Scholar]
- 20.Aksyuk AA, et al. The tail sheath structure of bacteriophage T4: A molecular machine for infecting bacteria. EMBO J. 2009;28(7):821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fokine A, et al. The molecular architecture of the bacteriophage T4 neck. J Mol Biol. 2013;425(10):1731–1744. doi: 10.1016/j.jmb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299(4):1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- 25.Maxwell KL, et al. Structural and functional studies of gpX of Escherichia coli phage P2 reveal a widespread role for LysM domains in the baseplates of contractile-tailed phages. J Bacteriol. 2013;195(24):5461–5468. doi: 10.1128/JB.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson PL, Coombs DH. Pulse-chase analysis of the in vivo assembly of the bacteriophage T4 tail. J Mol Biol. 2000;297(1):99–117. doi: 10.1006/jmbi.2000.3551. [DOI] [PubMed] [Google Scholar]
- 27.King J. Bacteriophage T4 tail assembly: Four steps in core formation. J Mol Biol. 1971;58(3):693–709. doi: 10.1016/0022-2836(71)90034-9. [DOI] [PubMed] [Google Scholar]
- 28.Meezan E, Wood WB. The sequence of gene product interaction in bacteriophage T4 tail core assembly. J Mol Biol. 1971;58(3):685–692. doi: 10.1016/0022-2836(71)90033-7. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi Y, King J. Genetic control of bacteriophage T4 baseplate morphogenesis. II. Mutants unable to form the central part of the baseplate. J Mol Biol. 1975;99(4):673–694. doi: 10.1016/s0022-2836(75)80179-3. [DOI] [PubMed] [Google Scholar]
- 30.Watts NRM, Coombs DH. Analysis of near-neighbor contacts in bacteriophage T4 wedges and hubless baseplates by using a cleavable chemical cross-linker. J Virol. 1989;63(6):2427–2436. doi: 10.1128/jvi.63.6.2427-2436.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu B, Margolin W, Molineux IJ, Liu J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci USA. 2015;112(35):E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urig MA, Brown SM, Tedesco P, Wood WB. Attachment of tail fibers in bacteriophage T4 assembly. Identification of the baseplate protein to which tail fibers attach. J Mol Biol. 1983;169(2):427–437. doi: 10.1016/s0022-2836(83)80059-x. [DOI] [PubMed] [Google Scholar]
- 33.Yap ML, et al. Structure of the 3.3MDa, in vitro assembled, hubless bacteriophage T4 baseplate. J Struct Biol. 2014;187(2):95–102. doi: 10.1016/j.jsb.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 39.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo F, Jiang W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol Biol. 2014;1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henderson R, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20(2):205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Afonine PV, Headd JJ, Terwilliger TC, Adams PD. New tool: phenix.real_space_refine. Computational Crystallography Newsletter. 2013;4:43–44. [Google Scholar]
- 45.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrödinger LLC. 2015 The PyMOL Molecular Graphics System, version 1.8. Available at www.pymol.org/
- 47.Leiman PG, et al. Structure of bacteriophage T4 gene product 11, the interface between the baseplate and short tail fibers. J Mol Biol. 2000;301(4):975–985. doi: 10.1006/jmbi.2000.3989. [DOI] [PubMed] [Google Scholar]
- 48.Thomassen E, et al. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J Mol Biol. 2003;331(2):361–373. doi: 10.1016/s0022-2836(03)00755-1. [DOI] [PubMed] [Google Scholar]
- 49.Kostyuchenko VA, et al. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12(9):810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- 50.Eiserling FA, Black LW. Pathways in T4 morphogenesis. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Am Soc Microbiology; Washington, DC: 1994. pp. 209–212. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.