Significance
PIN-FORMED (PIN) proteins actively transport the plant hormone auxin, of which the directionality, referred to as polarity, steers developmental processes throughout the plant’s lifecycle. The polarity of the PIN localization at the cell membrane is regulated by protein complexes, implying temporary internalization in the cell through vesicles and changes in the activity state. We identified the ROTUNDA3 protein as a regulator of the protein phosphatase 2A-driven PIN recycling and revealed its importance in auxin transport-related plant developmental programs.
Keywords: Arabidopsis, PIN recycling, PP2A, auxin, plant development
Abstract
The shaping of organs in plants depends on the intercellular flow of the phytohormone auxin, of which the directional signaling is determined by the polar subcellular localization of PIN-FORMED (PIN) auxin transport proteins. Phosphorylation dynamics of PIN proteins are affected by the protein phosphatase 2A (PP2A) and the PINOID kinase, which act antagonistically to mediate their apical–basal polar delivery. Here, we identified the ROTUNDA3 (RON3) protein as a regulator of the PP2A phosphatase activity in Arabidopsis thaliana. The RON3 gene was map-based cloned starting from the ron3-1 leaf mutant and found to be a unique, plant-specific gene coding for a protein with high and dispersed proline content. The ron3-1 and ron3-2 mutant phenotypes [i.e., reduced apical dominance, primary root length, lateral root emergence, and growth; increased ectopic stages II, IV, and V lateral root primordia; decreased auxin maxima in indole-3-acetic acid (IAA)-treated root apical meristems; hypergravitropic root growth and response; increased IAA levels in shoot apices; and reduced auxin accumulation in root meristems] support a role for RON3 in auxin biology. The affinity-purified PP2A complex with RON3 as bait suggested that RON3 might act in PIN transporter trafficking. Indeed, pharmacological interference with vesicle trafficking processes revealed that single ron3-2 and double ron3-2 rcn1 mutants have altered PIN polarity and endocytosis in specific cells. Our data indicate that RON3 contributes to auxin-mediated development by playing a role in PIN recycling and polarity establishment through regulation of the PP2A complex activity.
Organ growth is determined by cell numbers produced by meristems and by cell expansion to reach final volume. Plant hormones steer the extent and timing of growth and mediate signals of various types that are transmitted within the cell, between cells, or at a long distance within the plant. The phytohormone auxin is a major regulator of cell division and expansion during plant growth and development. The molecular mechanisms by which auxin controls these essential cellular responses are roughly understood thanks to the recent progress in the identification of auxin receptors and components of auxin signaling, transport, and metabolism (1). Auxin gradients between the cells are generated and maintained by intercellular auxin transport mediated by efflux carriers from the PIN-FORMED (PIN) family (2). PIN proteins contain transmembrane domains and continuously cycle between the basal (rootward) and apical (shootward) plasma membranes and endosomes, allowing rapid and dynamic changes in the PIN localization (3). The sorting of PIN proteins into the apical or basal trafficking pathway depends on the PIN phosphorylation status, which is controlled by the PINOID (PID) protein kinase and phosphatase 2A (PP2A) (4, 5), a heterotrimeric complex consisting of a C-catalytic subunit together with A- and B-regulatory subunits. One of the A-subunit isoforms, ROOTS CURL IN NAPHTHYLPHTHALAMIC ACID1 (RCN1), acts as a key positive regulator of the PP2A activity in seedlings. The rcn1 mutant that lost part of the PP2A activity displays abnormalities related to defective auxin transport, such as altered gravity response and lateral root growth (6, 7).
In an ethyl methanesulfonate-induced collection of Arabidopsis thaliana leaf mutants (8), we identified ROTUNDA3 (RON3) as a proline-rich, plant-specific single-copy gene with a function in auxin-related processes in all organs throughout the plant’s lifecycle. Affinity purification of the PP2A complex with RON3 as bait, and genetic and cell biology analyses support the hypothesis that RON3 affects the cellular dynamics of PIN proteins through interference with the PP2A activity.
Results
RON3 Is a Unique Higher Plant-Specific Gene.
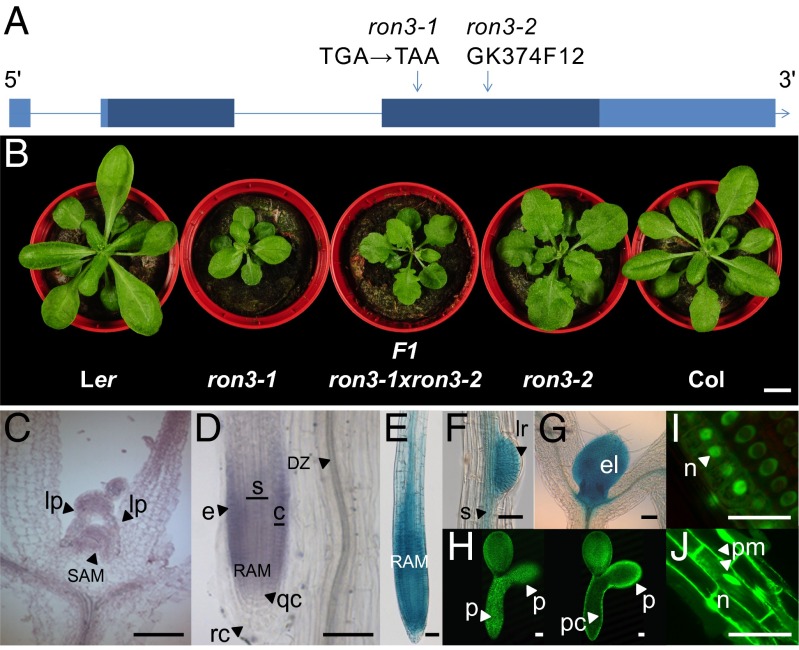
The ron3-1 mutant [Landsberg erecta (Ler)] belongs to the ron class of leaf mutants with large leaf laminas (8). Fine mapping identified a genetic interval of 18 genes (At4g24420–At4g24580) around the RON3 locus (Fig. S1 and Table S1), of which At4g24500 was severely down-regulated in the ron3-1 transcriptome (log fold change = −2.7632) and contained a cytosine to thymine point mutation in the third exon, generating a stop codon in ron3-1 (Fig. 1A). The Gabi-Kat line GK374F12 [designated ron3-2; Columbia (Col)], with a transferred DNA (T-DNA) insertion in the third exon of the At4g24500 gene (Fig. 1A), was crossed with ron3-1. The F1 plants with leaf and root phenotypes similar to those in both parentals confirmed allelism and RON3 as At4g24500 (Fig. 1B). The At4g24500 gene codes for a 319-aa (35-kDa) protein that had previously been identified by the sickle-1 mutant, with a role in splicing and microRNA (miRNA) biogenesis (9). The At4g24500 gene is single copy in Arabidopsis and rice, has two homologs in maize and poplar, and has none in mosses, algae, or nonplant species, suggesting that it is specific for higher plants.
Fig. 1.
RON3 gene structure, expression pattern, and protein localization. (A) RON3 gene structure. Exons are boxed, ORFs are dark blue, UTRs are light blue, introns are shown as lines, and mutations are shown by arrows. (B) Rosette phenotype of Ler, ron3-1 (Ler), F1 ron3-1×ron3-2, ron3-2 (Col), and Col. (C and D) In situ hybridization with RON3 probe of the shoot apex and primary root tip, respectively. (E–H) Marker activity in the RON3::GFP–GUS line in (E) primary root, (F) emerging lateral root, (G) leaf primordia, and (H) heart-stage embryo and (I and J) GFP-RON3 localization in (I) primary root meristem and (J) epidermis. c, Cortex; DZ, differentiation zone; e, epidermis; el, emerging leaf; lp, leaf primordia; lr, lateral root primordium; n, nucleus; p, protoderm; pc, procambium; pm, plasma membrane; qc, quiescent center; RAM, root apical meristem; rc, root cap; s, stele; SAM, shoot apical meristem. (Scale bars: B, 1 cm; C–G, I, and J, 50 µm; H, 250 µm.)
In situ hybridization on young Arabidopsis seedlings showed that RON3 transcripts localized in the shoot apical meristem dome, the emerging leaf primordia, the provascular strands of developing seedlings, and the epidermis and cortex of the meristematic and elongation zones of the primary root tip but absent from the root cap and the differentiated zone (Fig. 1 C and D). Arabidopsis (Col) lines transformed with RON3::GFP–β-glucuronidase (GUS) revealed activity in the primary root tip (Fig. 1E), at initiation sites of lateral roots (Fig. 1F), in the shoot apex (including leaf primordia) (Fig. 1G), and in the protoderm of heart- and torpedo-stage embryos (Fig. 1H), in line with the in situ hybridization expression data. Fusion proteins generated by 35S::GFP-RON3 or 35S::RON3-GFP constructs, restored the ron3-1 mutant phenotype to the WT and localized in the nuclei, excluding the nucleolus, in all cells of the root apical meristem (Fig. 1I). In young cortical cells of the root, nuclear localization and cytosolic and/or membrane associations were observed (Fig. 1J), which were similar to the RCN1::YFP-RCN1 localization pattern (10).
Auxin-Related Phenotypes and Auxin Accumulation in RON3-Perturbed Lines.
Detailed phenotypic analyses of both ron3-1 and ron3-2 mutant alleles were done to gain insight into the RON3 function in growth and development. Mutations in the RON3 gene reduced rosette leaf lamina length, width, and area significantly in ron3-1 (Ler) and ron3-2 (Col) mutants (Fig. S2 A–D). Third leaves were analyzed for cellular parameters in both ron3 alleles, and their reduced area was caused by a reduced total cell number (Fig. S2E). No difference in skoto- or photomorphogenesis was observed in the ron3 mutant alleles, because hypocotyl length was significantly reduced, irrespective of darkness or light quality (Fig. S2F). The ron3-1 and ron3-2 mutants were significantly delayed in flowering time (Fig. S2G), with slightly more rosette leaves. The primary inflorescence length in ron3-1 and ron3-2 was significantly reduced (Fig. S2H), and the secondary inflorescences outgrew the primary inflorescence, a mainly auxin-driven phenomenon called reduced apical dominance.
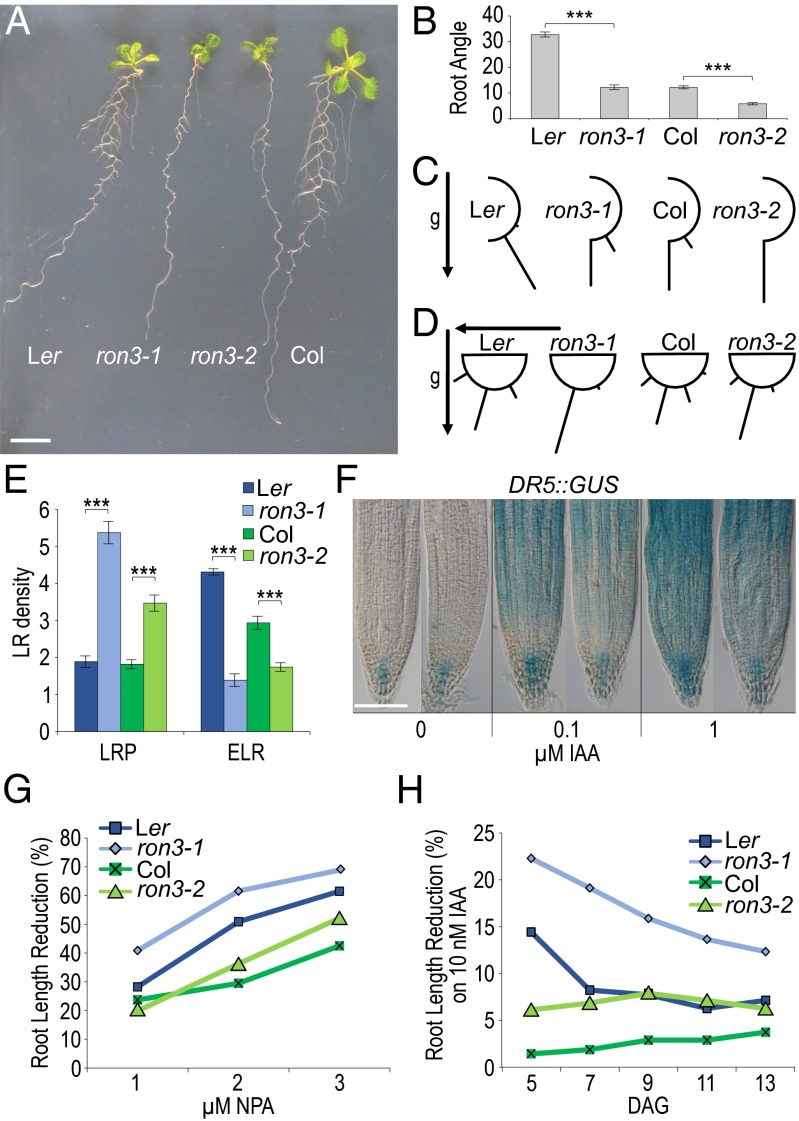
Primary roots of ron3-1 and ron3-2 seedlings showed auxin-related phenotypes, such as reduced and hypergravitropic growth (Fig. 2A and Fig. S3A), which was quantified as significantly reduced gravity deviation angle (Fig. 2B), and horizontal growth index (Fig. S3B), which significantly increased straightness and vertical growth index (Fig. S3 C and D). Hypergravitropic growth and enhanced response to gravitropic stimulus were measured in both ron3 mutant alleles (Fig. 2 C and D), typical for altered auxin transport. Both ron3-1 and ron3-2 seedlings had significantly reduced emerged lateral root density and increased density of stages III-V lateral root primordia (Fig. 2E and Fig. S3E); arrested lateral root primordia were ectopically positioned at the upper part of the root, and a number of emerged lateral roots did not grow out. Primary and lateral root tips of ron3-1 containing the DR5::GUS marker had similar auxin accumulation to that of the WT under native conditions, but on 0.1 and 1 µM indole-3-acetic acid (IAA) treatment, 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid staining was reduced in meristematic and elongation zones of mutant root tips (Fig. 2F and Fig. S3F), suggesting defective cellular import of IAA and/or cell to cell transport. The IAA concentration in the primary root tip of 8-d-old seedlings did not differ between ron3 mutant alleles and their respective WTs but was significantly lower in the ron3-1 remainder of the root and significantly higher in the ron3-1 and ron3-2 shoots (Fig. S3G), in accordance with a defective shoot to root auxin transport. The overexpression line, 35S::RON3_17, had IAA levels similar to those of the ron3 mutants (Fig. S3G), suggesting that a proper stoichiometric balance is required for the RON3 role in auxin transport. Given the increased levels of IAA measured in the shoots and the absence of any differentially expressed auxin biosynthesis-related genes in the microarray data, the hypothesis that auxin biosynthesis was affected by the ron3 mutation was excluded. ron3 mutants grown on increasing concentrations of 1-N-naphthylphthalamic acid had more reduced root lengths than their controls, indicating an enhanced sensitivity to the auxin transport inhibition (Fig. 2G and Fig. S4A), although gravitropism was affected less in the mutants than in the WT (Fig. S5B). Initial root growth on 10 nM IAA was also more reduced in both ron3 mutants, especially ron3-1 (Fig. 2H and Fig. S4C). The defective ERECTA kinase in the Ler ecotype affects auxin distribution and transport only in the shoot apex and not in the root (11); hence, the root-related auxin phenotypes typical for auxin distribution or transport defects in both ron3 alleles provided bona fide functional information.
Fig. 2.
Auxin-related phenotypes in ron3 mutants. (A) Primary and lateral root growth in 7-d-old seedlings grown in vertical position under continuous light. (B) Root angle-θ indicating deviation from perpendicular growth. (C) Gravitropic growth in seedlings 8 DAG in vertical position. Bars give percentages of roots at specific orientations. g, Gravity vector. (D) Gravitropic response in seedlings 4 DAG grown in vertical position followed by a 90° turn; bars indicate percentages of roots at specific orientations. (E) Lateral root (LR) density, total number of lateral root primordia (LRP), and emerged lateral roots (ELRs) per 1 cm primary root. (F) DR5::GUS marker gene activity on 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid staining in WT (Left) and ron3-1 (Right) at different IAA concentrations. (G and H) Sensitivity to 1-N-naphthylphthalamic acid (NPA) and IAA, respectively. ***P < 0.005 (Student’s t test). (Scale bars: A, 1 cm; F, 100 µm.)
Transcriptome analysis of ron3-1 shoot apices revealed that “response to auxin stimulus” was the only prominent gene ontology category in 1,122 down-regulated genes (Fig. S4D), among which were the auxin-inducible AUX/IAA transcriptional repressors. qPCR of ron3 seedlings treated with 20 µM IAA for 2 h showed that representative AUX/IAA genes (i.e., IAA1 and IAA6) were still auxin-inducible, indicating that the auxin signaling machinery was active.
Tandem Affinity Purification of the PP2A Complex with RON3.
Four tandem affinity purifications (TAPs) were done on extracts of Arabidopsis cell suspension cultures and transformed with the 35S::GFP-RON3 and 35S::RON3-GFP constructs that complemented the ron3-1 mutant, showing functionality of the N- or C-tagged RON3 protein. Twenty-one proteins were purified, of which the recurrent proteins are presented in Table 1, the other purified proteins are shown in Table S2, and the MS data in Dataset S1. The high number of purified proteins might indicate that RON3 is involved in multiple cellular processes. Most prominent was the purification of six components of the cytoplasmic PP2A complex (ATBα/PP2AA1/At1g51690, PP2A-4/At3g58500, PP2A-3/At2g42500, ATBβ/At1g17720, PP2AA2/At3g25800, and RCN1/At1g25490) (Table 1) in addition to the ubiquitin-specific proteases UBP12/At5g06600 and UBP13/At3g11910 that were predicted to interact with the ATBβ subunit of PP2A (12). Other copurified proteins were related to histone arginine methylation [protein arginine N-methyltransferase 4A (PRMT4A)/At5g49020 and PRMT4B/At3g06930], protein folding (DnaJ/Hsp40/At3g47650 and ATE1/At1g13690 that stimulates the ATPase activity of DnaK/DnaJ), and DNA damage repair (At5g28740) (Table S2). RON3 might be a substrate of PRMT or work together with PRMT in the regulation of arginine methylation, possibly explaining its previously described nuclear role in splicing and miRNA biogenesis (9) and some of the ron3 phenotypes, such as delay in flowering time (this work and ref. 13).
Table 1.
PP2A components copurified with RON3 as bait in TAPs
| Gene code | Protein name | Experiments |
| AT4G24500 | RON3 | 4 of 4 |
| AT5G06600 | UBP12 | 4 of 4 |
| AT3G11910 | UBP13 | 4 of 4 |
| AT1G51690 | ATBα | 4 of 4 |
| AT1G17720 | ATBβ | 3 of 4 |
| AT3G58500/AT2G42500 | PP2A | 4 of 4 |
| AT1G25490 | RCN1 | 3 of 4 |
| AT3G25800 | PDF1 and PP2AA2 | 3 of 4 |
The PP2A complex was the major protein–protein interaction network with the CORNET prediction software in addition to the PRMT heterodimer. The interaction of RON3 with cytoplasmic and nuclear proteins/complexes correlated with its localization in both compartments (Fig. 1 I and J). The PP2A phosphatase has several substrates, among which the PIN auxin efflux carriers. The role of RON3 in PP2A dephosphorylation of PIN auxin efflux carriers was investigated using a cell biology approach to explain auxin-related developmental phenotypes in the ron3 mutants.
Double Mutants Support a Role for RON3 in Auxin Transport-Related Processes.
The ron3-1 (Ler) auxin-related phenotypes resemble those of the ron1-1 mutant (Ler) (14). The RON1 gene codes for an inositol polyphosphate 1-phosphatase that has a function in inositol triphosphate-mediated Ca2+ signaling that also affects auxin transport and PIN polarity (15). The phenotypes of the ron1-1 ron3-1 double mutant were enhanced compared with those of the two parentals (i.e., smaller mature rosette leaves, more reduced primary root length, severely reduced number of lateral root initiation sites, reduced emerged lateral roots per primary root, reduced lateral root density (Fig. S5 A–D), extremely reduced inflorescence size [13.9 ± 2.0 cm in ron1-1, 9.7 ± 1.3 cm in ron3-1, and 3.6 ± 0.6 cm in ron1-1 ron3-1], reduced apical dominance (Fig. S5E), and significantly enhanced delay in flowering time [36.2 ± 2.9 d after germination (DAG) in ron1-1 ron3-1 compared with 31.6 ± 2.7 DAG in ron1-1 and 23.4 ± 1.4 DAG in ron3-1 parentals]), indicating similar defects in the ron3 and ron1 single mutants and giving additive phenotypes in the ron1-1 ron3-1 double mutant.
The RON3 protein copurified with six components of the PP2A complex, including RCN1. RCN1 codes for the A-regulatory subunit of PP2A, whereas the rcn1 null mutant has an altered sensitivity to the auxin inhibitor 1-N-naphthylphthalamic acid (7). The ron3-2 allele (Col) was crossed with rcn1 [Wassilewskija (WS)] to avoid interference with the er mutation in ron3-1 (Ler). The rosette phenotype of the ron3-2 rcn1 double mutants resembled that of the ron3-2 parental (Col; slightly serrated margin) but not that of rcn1 (Fig. S5F). The length of the ron3-2 rcn1 primary root was significantly shorter than that of the two parentals (Fig. S5G). Lateral root initiation was not affected compared with the WT, but the emerged lateral root number was severely reduced in ron3-2, rcn1, and ron3-2 rcn1 (Fig. S5H); lateral root density of ron3-2 rcn1 was partially restored compared with ron3-2 (Fig. S5I). Apical dominance was severely reduced in ron3-2, because primary inflorescences were overgrown by secondary inflorescences, normal in rcn1, and partially restored in the ron3-2 rcn1 double mutant (Fig. S5J). Hence, ron3 mutation inhibited growth of primary and lateral roots and apical dominance, implying that ron3 is epistatic to rcn1 for these parameters. The ron3 pin double-mutant combinations are probably less informative because of the extensive compensatory functional redundancy among PIN proteins (16).
When the 35S::PID (Col) was combined with the ron3-2 (Col) mutation, no significant phenotypic differences were observed with the 35S::PID overexpressor parental with respect to root morphology (straight short primary root with collapsed tips and perpendicular lateral roots). The ron3-2 mutation might not affect the 35S::PID phenotype, suggesting that there are no genetic interactions or that the very strong 35S::PID phenotypes mask the mild phenotypes of ron3 mutants.
Altered PIN Auxin Transporter Polarity in RON3-Perturbed Lines.
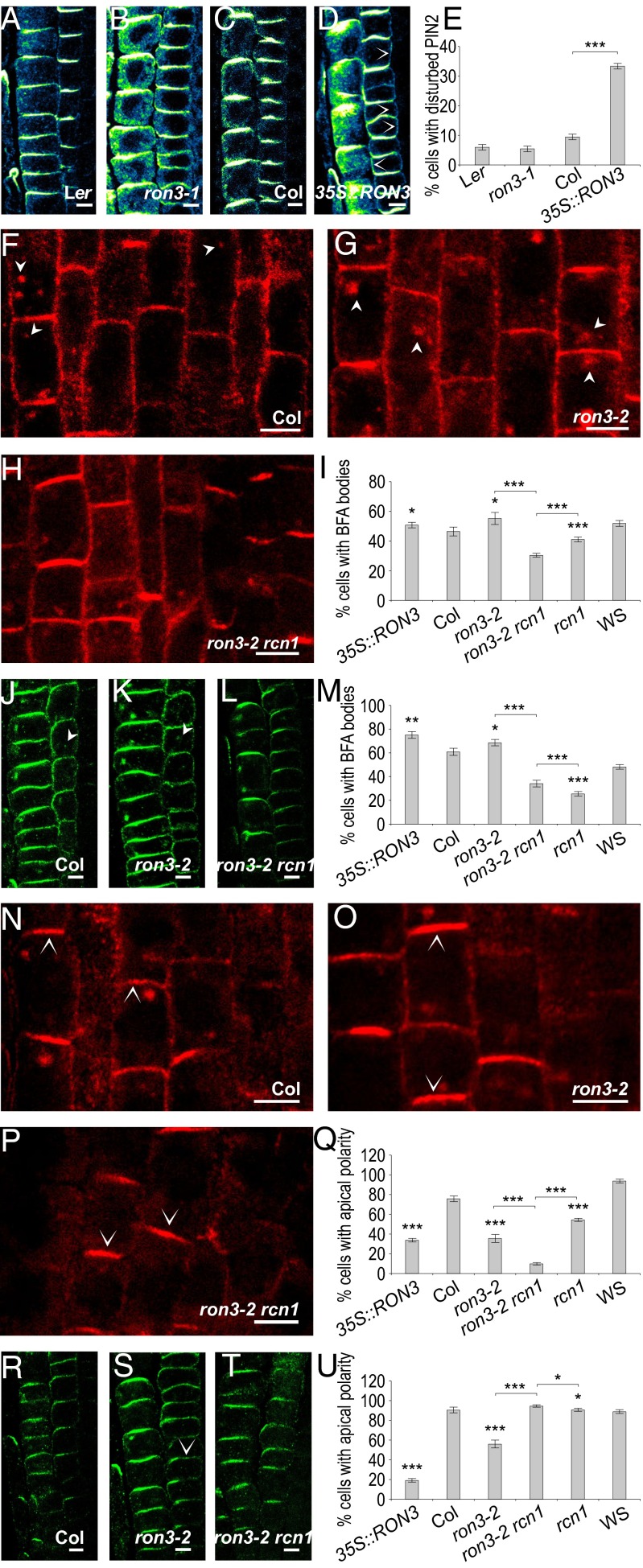
RCN1 and other PP2A subunits that are involved in polar trafficking of PIN auxin transporters (5) were affinity-purified with RON3 as bait. PIN transporters constitutively cycle to and from the plasma membrane and are delivered either basally by the ADP ribosylation factors GTPases and GTPase guanine-nucleotide exchange factor GNOM-dependent mechanism (PIN1) or apically after phosphorylation by the PID kinase, whereas dephosphorylation by the PP2A complex leads preferentially to basal targeting of the PIN proteins (17). Hence, the polar localization and recycling of the PIN1 and PIN2 auxin transporters were investigated by in situ immunodetection in 5-d-old seedlings. No significant alteration of the basal PIN1 localization pattern in the stele (Fig. S6 A–D) or the apical PIN2 pattern in the epidermis (Fig. 3 A–D) was observed in the primary root of either the ron3-1 mutant or the 35::RON3_17 overexpressor compared with their WTs (Ler and Col, respectively). Generally unaffected was also the basal PIN2 in the cortex of the ron3-1 mutant compared with Ler (Fig. 3B compared with Fig. 3A, respectively). However, we observed a weak lateral or apolar PIN2 (Fig. 3D) in a number of young cortical cells in the RON3 overexpressor line (Fig. 3D) in addition to the generally basal PIN2 in the WT (Fig. 3C). We carefully distinguished and counted the cells with disturbed (either the additional lateral or the additional apolar) PIN2 polarity. The percentage was threefold higher than that of cells with basal PIN2 polarity in the RON3 overexpressor compared with Ler (Fig. 3E).
Fig. 3.
Cell biology of ron3-perturbed lines. PIN2 polarity defects. (A–D) Immunolocalization of PIN2 (blue-yellow) in epidermis (Left) and cortex (Right) of roots of Ler, ron3-1, Col, and 35S::RON3_17 and (E) quantification of cortical cells with PIN2 polarity defects (additional lateral or apolar). PIN endocytosis. (F–U) Immunolocalization of PIN1 (red) and PIN2 (green) on Col, ron3-2, and ron3-2 rcn1. (F–M) Imaging and quantification of endocytosis (90 min; 25 µM BFA) in (I) vascular or (M) cortical cells with BFA bodies. (N–U) Imaging and quantification of transcytosis (180 min; 25 µM BFA) in (Q) vascular or (U) cortical cells with apical-only polarity. Arrowheads point to BFA bodies, and open arrowheads in a cell point to the apical or basal membrane with PIN1 or PIN2. Error bars represent SEM. *P < 0.5; **P < 0.05; ***P < 0.005 (Student’s t test). (Scale bars: 5 µm.)
Role of RON3 in PIN Auxin Transporter Internalization.
We examined the internalization step of the PIN intracellular recycling by means of treatments (90 min) with the inhibitor of the basal PIN recycling brefeldin A (BFA) (25 µM) in the lines ron3-2, rcn1, ron3 rcn1, ron3-2, 35S::PID, and 35S::RON3. Under these conditions, basal exocytosis of PIN proteins is inhibited, leading to their concomitant intracellular accumulation (18, 19). We generated Z stacks of confocal images of roots with immunolocalized PIN1 (Fig. 3, red) and PIN2 (Fig. 3, green) after BFA treatment. We distinguished and counted the cells containing one or more clearly visible internalized PIN agglomerates and calculated their percentage in the total number of observed cells per root.
The BFA-induced internalization of PIN1 in the stele of ron3-2 (Fig. 3G) was comparable with that of Col (Fig. 3F). However, in the ron3-2 rcn1 double mutant, PIN1 internalization was reduced (Fig. 3H) compared with both single mutants (Fig. 3 F and G). The overexpressor 35S::RON3 did not show any significant change (Fig. S6E). The rcn1 mutant exhibited significantly reduced internalization of PIN1 (Fig. S6G) compared with its background WS (Fig. S6F). When we introduced the ron3-2 mutation into the 35S::PID (Fig. S6I), we did not observe significant differences compared with the single 35S::PID transgenic (Fig. S6H). Quantification of the cells with BFA-induced intracellular aggregates (Fig. 3I) confirmed that PIN internalization was significantly reduced in the double-mutant ron3-2 rcn1 compared with the ron3-2 and rcn1 parentals. These data support a common, positive role of RON3 and RCN1 in the internalization of the basal PIN1.
In the cortex, basal PIN2 was slightly more internalized in ron3-2 (Fig. 3K) than in Col (Fig. 3J), but in ron3-2 rcn1 (Fig. 3L), the number of cells with obvious internalization aggregates was significantly reduced compared with that in the ron3-2 mutant plants (Fig. 3K). The RON3 overexpressor was apparently unaffected in PIN2 internalization (Fig. S6E). The rcn1 mutation (Fig. S6G) had dramatically decreased intracellular accumulation of PIN2 in the cortex compared with WS (Fig. S6F). The 35S::PID and the double-mutant ron3-2 35S::PID had similar BFA-induced intracellular PIN2 aggregates (Fig. S6 H and I, respectively). Quantification of cortical cells with internalized PIN2 showed significant reduction of the internalization ability of the double-mutant ron3-2 rcn1 compared with its parental ron3-2 and a slight but significant increase compared with rcn1 (Fig. 3M). No major endocytosis alterations were observed for PIN2 in the epidermal cells. In agreement with the suggested positive role on endocytosis in the stele, RON3 together with RCN1 also affect the PIN2 internalization in the cortex.
Role of RON3 in Transcytosis of PIN Auxin Transporters.
Under prolonged BFA treatments (3 h), basal PIN proteins are internalized, lose polarity, and gradually migrate to the top membrane by a mechanism known as transcytosis (19). Moreover, apical PIN proteins are concentrated in the center of the apical membrane, forming a superapical localization domain (19). Similar apicalization of PIN2 in cortex cells was observed in loss-of-function pp2aa and gnom mutants (4, 17) and 35S::PID gain of function (20, 21). Therefore, the role of RON3 in transcytosis of the PIN transporters was investigated.
We monitored the BFA-induced transcytosis of PIN1 (Fig. 3, red) and PIN2 (Fig. 3, green) by immunolocalization and examination of Z-stack confocal images. We noticed qualitative and quantitative variations of polarization between mutants. Phenotypes included the transcytosed apical PIN1 in the stele or PIN2 in the cortex and apical or superapical PIN2 in the epidermis, but also various intensities of apolar, lateral, and basal PIN2 in the cortex. We considered the “apical-only” cells as the apicalization ability measure, and we determined their percentage to the total visible cells per Z stack.
After prolonged BFA application (3 h), PIN1 was much less apicalized in ron3-2 (Fig. 3O) than in Col (Fig. 3N) presenting both apical and basal polarities. Similarly, the rcn1 mutant showed reduced apicalization of PIN1 (Fig. S6L), and the combination of the two single mutations in ron3-2 rcn1 resulted in significantly more reduced apicalization of PIN1 (Fig. 3P). Strong apical PIN1 polarity observed in 35S::PID (Fig. S6M) was not affected by the introduction of the ron3-2 mutation (Fig. S6N). Quantification of the PIN1 transcytosis ability (Fig. 3Q) in the stele of ron3-2 was impaired, and it was even more impaired in the ron3-2 rcn1 double mutant.
Ιn the cortex, 3 h of BFA treatment caused significantly less apicalized and more apolar PIN2 in the ron3-2 mutant (Fig. 3S) than in the Col (Fig. 3R). In the double-mutant ron3-2 rcn1, rcn1 introduction eliminated the disability of the ron3-2 mutant to apicalize PIN2 (Fig. 3T), allowing a PIN2 apical delivery similar to the WT (Fig. 3R). However, overexpression of RON3 caused the most severe effect observed on transcytosis of PIN2 (Fig. S6J). Normal PIN2 transcytosis was observed in the rcn1 mutant (Fig. S6L), comparable with that of the WS (Fig. S6K). Large intracellular PIN2 aggregates were induced without affecting the apical polarity in the cortex of 35S::PID (Fig. S6M) and its double mutant with ron3-2 (Fig. S6N). Quantification of PIN2 apicalization confirmed the significantly reduced PIN2 transcytosis in cortical cells of the ron3-2 mutant, the weak apicalization potential introduced by the RON3 overexpression, and the lack of effects in the remaining genotypes tested (Fig. 3U). The data on internalization and transcytosis indicate that RON3 together with RCN1 play a role in PIN trafficking, presumably promoting endocytosis of basally localized PIN proteins and apical delivery or maintaining localization of already delivered apical PIN cargos.
Because stele cells outnumber the single-layer cortex, a major effect on seedling growth on BFA-containing medium was expected in correlation to the defects in endocytic recycling of polar cargos. Indeed, the fresh weight per seedling was significantly higher in ron3-2 and rcn1 on medium with BFA than that of their WT. The effect was enhanced in ron3-2 rcn1, indicating a synergistic effect between the ron3-2 and rcn1 mutations to resist the inhibitory effects of BFA (Fig. S6O). The seedling growth on BFA-supplemented media correlated well with the reduced levels of internalization and higher preservation of basal PIN1 in the stele.
Discussion
We identified a cytoplasmic function for RON3 as a regulator of the PP2A phosphatase complex activity in the recycling of PIN auxin transporters supported by auxin-related phenotypes in the ron3 mutants, purification of the PP2A complex with RON3 as bait, and altered recycling of the PIN proteins.
RON3 Has a Function in the Dephosphorylation Dynamics of the Cell.
The RON3-mediated purification of six components of the PP2A complex indicated that RON3 modulates the PP2A dephosphorylation machinery, which was supported by auxin transport-related phenotypes shared between ron3-2 and rcn1 (7). Moreover, the localization of RON3 was similar to that of RCN1 (10) in nuclei of the root meristem and both membrane- and nucleus-associated mature cortical and epidermal cells of the primary root. Hence, we hypothesized that the auxin-related phenotypes of ron3 are the consequence of altered PIN phosphorylation status and activity. Additional support came from the resemblance of the ron3 phenotypes to those of ron1, for which defective PIN polarity had been shown (14). The additive phenotypes in the double-mutant ron1 ron3 indicated that RON3 acts in a mode similar to that of RON1, corroborating a role for RON3 in PIN polarity-related processes. Additional confirmation of the implication of RON3 in dephosphorylation processes was provided by the study of the 35S::PID ron3-2 double mutant, in which ron3-2 did not affect the 35S::PID phenotype.
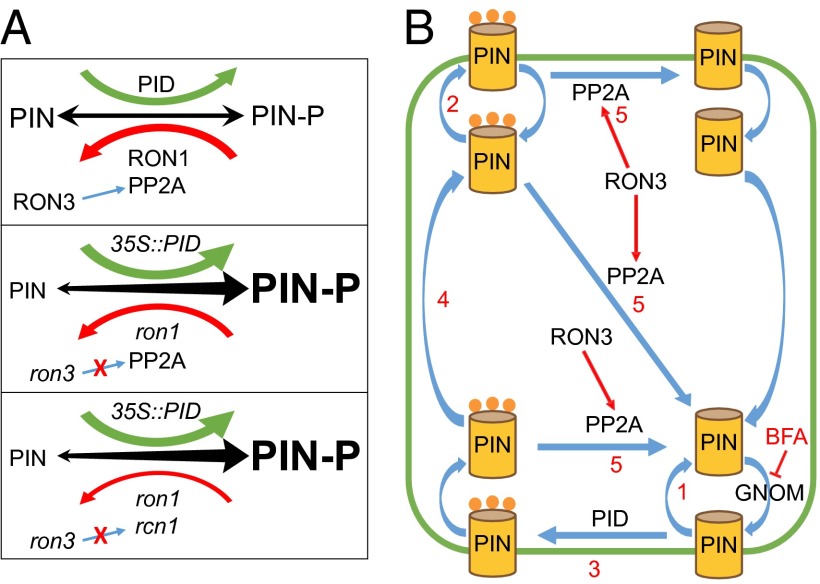
In plant cells PID kinases, RON1/SUPO1 and PP2A complex dephosphatases function in an orchestrated manner to define the phosphorylated status of membrane proteins, such as PIN proteins, and concomitantly, their apical or basal delivery by intracellular trafficking, as represented (Fig. 4A, Top). Our phenotypic and genetic data indicate that RON3 acts positively on the function of the PP2A dephosphorylation machinery. Indeed, single ron3 mutants lack the presumed positive effects on the PP2A complex, resulting in the reduced dephosphorylation rate of the PIN proteins (Fig. 4A, Bottom). In ron1-1 ron3-1 and 35S::PID ron3-2 combinations, dephosphorylation by RON1/SUPO1 and phosphorylation by the PID kinase are affected, so dephosphorylation decreases and phosphorylation increases, respectively (Fig. 4A, Middle). When the rcn1 mutation is coupled with the ron3-2 mutation, again a similar effect of reduced dephosphorylation by the PP2A complex impaired functionality occurs that increases the phosphorylated protein dynamics as PID kinase overexpression (35S::PID) or mutation on RON1 phosphatase (Fig. 4A, Bottom). The PIN recruitment to apical or basal endocytic recycling pathways is expected to be defective and might explain the cell responsiveness to developmental processes or environmental stimuli.
Fig. 4.
Cell biology models. (A) Cell phosphorylation dynamics in (Top) WT plants, (Middle) 35S::PID and ron1 plants, and (Bottom) 35S::PID, ron1, and rcn1 plants. PIN proteins shuffle between phosphorylated and nonphosphorylated state after action of PID kinase or RON1 and PP2A complex phosphatases. Combinations of mutation effects can be speculated. Equilibrium is denoted by arrow thickness and letter size. (B) Model of the RON3 action on the PP2A complex. PIN proteins are recycled either (1) basally in unphosphorylated status or (2) apically when phosphorylated. Although the exact PIN (de)phosphorylation is unclear, we suggest that it happens both in plasma membrane and inside the cell. Basal PIN can be (3) phosphorylated by PID kinase, endocytosed at the basal side, (4) transcytosed, and subsequently, exocytosed apically. (5) Phosphorylated PIN proteins are dephosphorylated by the PP2A complex, of which RCN1 is one component. RON3 assists the PP2A complex action, interacting with several of its components (such as RCN1).
RON3 Function in PIN Internalization and Polarity.
Pharmacological inhibition of exocytosis with BFA showed that the ron3-2 mutation has a positive effect on the basal abundance of PIN1. In control plants, prolonged BFA treatment normally leads to a gradual disappearance of the internalized PIN cargos, loss of any basal polarity, and finally, apicalization of previously basal membrane proteins. The preservation of the basal character of PIN1 after prolonged exocytosis inhibition in the ron3-2 and ron3-2 rcn1 mutants (Fig. 3, O and P, respectively) suggests that RON3 together with the PP2A complex promote PIN internalization and apical delivery. In agreement, we observed that RON3 overexpression preserves internalized PIN cargos and their apolar character but restrains PIN apical polarization (Fig. 3Q and Fig. S6J). We presume that the RON3 protein acts in intermediate steps of transcytosis, supporting dephosphorylation by the PP2A complex and affecting the total cell phosphorylation dynamics and the polar delivery of phosphorylated cargos, such as PIN proteins.
The rcn1 mutation does not have a strong effect on the PIN1 localization. Even after a prolonged inhibition of basal exocytosis (180 min with BFA), only a small increase of PIN1 apicalization was observed that might be justified by a diminished but not abolished PIN1 dephosphorylation (Fig. S6L) (20). Apolar PIN1 indicates that the rcn1 mutation still permits dephosphorylation of PIN proteins. When the ron3-2 mutation was introduced into rcn1, the RON3 regulatory effect on the total PP2A complex was abolished with concomitant effect on the cell’s (de)phosphorylation activity and consequently, trafficking events.
The opposite observations in the internalization and targeting of PIN proteins when the ron3-2 mutation was combined with the rcn1 mutation suggest a different RON3 effect depending on the cargo or cell file. Less endocytosed PIN1 is present in the double mutant, whereas levels of intracellular cortical PIN2 are assisted by the ron3-2 mutation, probably the consequence of the significantly different protein levels of these two proteins. Indeed, the vascular PIN1 is expressed at much higher levels than the cortical PIN2, and the (de)phosphorylation machinery potentials are noticeably different for higher amounts of a protein to be (de)phosphorylated. Another possible explanation could be the phosphorylation dynamics of PIN1 and PIN2. In stele cells, PIN1 is intrinsically basal, indicating a spatially uniform, temporally continuous dephosphorylated status. In contrast, PIN2 is apical in the initial cortical cells but becomes basal in mature cells. This redistribution is mediated by a necessary apical endocytosis and basal exocytosis. Pharmacological or genetic interference with this mechanism could preserve more flexible PIN2 phosphorylation dynamics, in turn leading to more sensitive internalization.
In conclusion, through analysis of auxin-related developmental defects, altered responses to gravistimulation, and cell biology experiments in the ron3 mutants, we showed that, other than its role in the nucleus, the RON3 protein plays a role in the cellular dephosphorylation dynamics (Fig. 4B). We propose that the cytoplasmic fraction of RON3 has a positive interaction with the PP2A complex, influencing endocytosis and transcytosis through influences on the phosphorylation status of PIN proteins.
Materials and Methods
Materials and methods are described at length in SI Materials and Methods.
Plant Material, Media, and Growth Conditions.
The A. thaliana (L.) Heynh. lines are Ler and Col accessions (Nottingham Arabidopsis Seed Collection), ron3-1 and ron1-1 mutants (both Ler) (14), ron3-2 allele (GK374F12 line, homozygous for the T-DNA insertion in At4g24500; Col), rcn1 mutant (WS) (10), 35S::PID, and line PID21 (Col) (20, 21). Media and growth conditions are discussed in SI Materials and Methods.
Map-Based Cloning and Bioinformatics Programs.
For the positional cloning of RON3, an F2 mapping population was derived from an ron3-1 (Ler) × Col cross, and recombinants were used for fine mapping. Details are in SI Materials and Methods.
In Situ Hybridization and Confocal Microscopy.
Shoot apices and root tips of 7-d-old seedlings were used for in situ hybridization and confocal microscopy as described in SI Materials and Methods. Immunodetected proteins were imaged with an LSM 5 Exciter or 710 Microscope (Zeiss).
TAP.
TAP of protein complexes is described in SI Materials and Methods.
Protein Immunodetection.
Whole-mount immunodetection with a liquid handling robot (InsituPro; Intavis) is described in SI Materials and Methods.
Cell Biology.
Categorization (calling) of cells is discussed in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Ghent University Special Research Fund (M.K.), the European Research Council (Project ERC-2011-StG-20101109-PSDP) (to J.F.), and the Körber European Science Foundation (J.F.). S.D.G. is indebted to the Agency for Science and Technology for a predoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Agilent microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE18493).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501343112/-/DCSupplemental.
References
- 1.Perrot-Rechenmann C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb Perspect Biol. 2010;2(5):a001446. doi: 10.1101/cshperspect.a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrášek J, et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312(5775):914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- 3.Dhonukshe P, et al. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol. 2007;17(6):520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 4.Michniewicz M, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130(6):1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Nodzyński T, Pěnčík A, Rolčík J, Friml J. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci USA. 2010;107(2):918–922. doi: 10.1073/pnas.0909460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbers C, DeLong A, Deruére J, Bernasconi P, Söll D. A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 1996;15(9):2115–2124. [PMC free article] [PubMed] [Google Scholar]
- 7.Rashotte AM, DeLong A, Muday GK. Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell. 2001;13(7):1683–1697. doi: 10.1105/TPC.010158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berná G, Robles P, Micol JL. A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics. 1999;152(2):729–742. doi: 10.1093/genetics/152.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan X, et al. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci USA. 2012;109(44):18198–18203. doi: 10.1073/pnas.1216199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blakeslee JJ, et al. Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 2008;146(2):539–553. doi: 10.1104/pp.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M-K, Wilson RL, Palme K, Ditengou FA, Shpak ED. ERECTA family genes regulate auxin transport in the shoot apical meristem and forming leaf primordia. Plant Physiol. 2013;162(4):1978–1991. doi: 10.1104/pp.113.218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisler-Lee J, et al. A predicted interactome for Arabidopsis. Plant Physiol. 2007;145(2):317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu L, Zhang Y, Pei Y, Liu C, Cao X. Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol. 2008;148(1):490–503. doi: 10.1104/pp.108.124727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robles P, et al. The RON1/FRY1/SAL1 gene is required for leaf morphogenesis and venation patterning in Arabidopsis. Plant Physiol. 2010;152(3):1357–1372. doi: 10.1104/pp.109.149369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, et al. Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev Cell. 2011;20(6):855–866. doi: 10.1016/j.devcel.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Vieten A, et al. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132(20):4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- 17.Kleine-Vehn J, et al. PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell. 2009;21(12):3839–3849. doi: 10.1105/tpc.109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geldner N, Friml J, Stierhof Y-D, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413(6854):425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- 19.Kleine-Vehn J, et al. ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr Biol. 2008;18(7):526–531. doi: 10.1016/j.cub.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 20.Friml J, et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306(5697):862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 21.Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development. 2001;128(20):4057–4067. doi: 10.1242/dev.128.20.4057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.