Phosphorylation of two highly conserved phosphosites preceding the kinase catalytic subdomain VIII region in the activation T-loop of the MAP kinase ERK1 negatively regulates its protein phosphotransferase activity. Hyperphosphorylation of most protein-serine/threonine kinases in this manner may be a general mechanism for their down-regulation.
Abstract
The catalytic domains of most eukaryotic protein kinases are highly conserved in their primary structures. Their phosphorylation within the well-known activation T-loop, a variable region between protein kinase catalytic subdomains VII and VIII, is a common mechanism for stimulation of their phosphotransferase activities. Extracellular signal–regulated kinase 1 (ERK1), a member of the extensively studied mitogen-activated protein kinase (MAPK) family, serves as a paradigm for regulation of protein kinases in signaling modules. In addition to the well-documented T202 and Y204 stimulatory phosphorylation sites in the activation T-loop of ERK1 and its closest relative, ERK2, three additional flanking phosphosites have been confirmed (T198, T207, and Y210 from ERK1) by high-throughput mass spectrometry. In vitro kinase assays revealed the functional importance of T207 and Y210, but not T198, in negatively regulating ERK1 catalytic activity. The Y210 site could be important for proper conformational arrangement of the active site, and a Y210F mutant could not be recognized by MEK1 for phosphorylation of T202 and Y204 in vitro. Autophosphorylation of T207 reduces the catalytic activity and stability of activated ERK1. We propose that after the activation of ERK1 by MEK1, subsequent slower phosphorylation of the flanking sites results in inhibition of the kinase. Because the T207 and Y210 phosphosites of ERK1 are highly conserved within the eukaryotic protein kinase family, hyperphosphorylation within the kinase activation T-loop may serve as a general mechanism for protein kinase down-regulation after initial activation by their upstream kinases.
INTRODUCTION
Protein kinases are major players in intracellular signal transduction through their catalysis of the reversible phosphorylation of the vast majority of proteins in cells on serine, threonine, and tyrosine residues. Such phosphorylation provides an efficient and effective means to regulate most physiological activities, including metabolism, transcription, DNA replication and repair, cell proliferation, and apoptosis (Krebs, 1993; Hunter, 2000; Pawson and Scott, 2005). Dysregulation of protein phosphorylation is implicated in >400 types of human diseases, including cancer, diabetes, and cardiovascular, neurological, and immunological disorders (Hunter, 1998; Blume-Jensen and Hunter, 2001).
Eukaryotic protein kinases (EPKs) comprise a ubiquitous and broadly expanded family of enzymes (Manning et al., 2002a). In humans, at least 536 protein kinases have been identified, including 484 typical protein kinases that feature one or sometimes two conserved catalytic domains (Manning et al., 2002b). Within the catalytic domain, 12 extremely conserved subdomains were originally defined by based on sequences of 65 protein kinases (Hanks and Hunter, 1995; Hanks et al., 1998). The functional roles of these subdomains were assigned using the results from site-directed mutagenesis (Gibbs and Zoller, 1991) and the crystal structure of the catalytic subunit of cAMP-dependent protein kinase (PKA; Knighton et al., 1991a, b). The activation T-loop is a variable segment that extends from subdomain VII (DFG sequence) to subdomain VIII (APE sequence). In most protein kinases, reversible phosphorylation and dephosphorylation of this segment on one or more phosphosites plays a key role in stimulating their phosphotransferase activity (Taylor and Radzio-Andzelm, 1994; Tayor and Kornev, 2011). Many EPKs are capable of autoactivation through autophosphorylation within their activation loop. Comparisons of active and inactive protein kinase structures indicate that conformational changes of this flexible region via phosphorylation promote the correct domain orientations and stabilize the macromolecule for catalysis (Johnson et al., 1996; Kornev et al., 2006).
Mitogen-activated protein kinases (MAPKs) play fundamental roles in the regulation of a broad diversity of functions, including meiosis, cell cycle progression, and stress responses, from yeast to humans (Schaeffer and Weber, 1999; Raman et al., 2007). The Ras-Raf–MAPK/extracellular signal–regulated kinase (ERK) kinase (MEK)–ERK signaling module has served as a paradigm for the study of intracellular signal transduction and protein kinase regulation (Wortzel and Seger, 2011). The activity of this pathway is commonly up-regulated in various types of cancers (Dhillon et al., 2007). In ERK1/2, dual phosphorylation on a threonine–glutamic acid–tyrosine residue (TEY) motif within the activation loop by the upstream kinases MAPK/ERK-1 and 2 (MEK1/2) was initially described more than 25 yr ago as a critical event in the stimulation of ERK1/2 (Anderson et al., 1990; Ahn et al., 1991; Payne et al., 1991). Phosphosite-specific antibodies targeting the phosphorylated TEY site have been widely used to indirectly monitor the activation state of ERK1/2. However, besides the TEY motif, three flanking phosphorylation sites have been identified by mass spectrometry (MS) in multiple cell lines and tumor samples, as documented in PhosphoSitePlus (www.phosphosite.org) and PhosphoNET (www.phosphonet.ca).
In this study, we investigated the functional roles of the three additional phosphosites flanking the TEY phosphorylation motif in human ERK1. We also explored the possible regulatory mechanisms of these phosphorylation sites by comparing the primary sequences of all of the human typical protein-serine/threonine kinases. The influences of these highly evolutionarily conserved phosphosites on ERK1 conformation and functional activity may reflect a general mechanism for tight regulation of the EPKs, which represent the second largest class of signaling proteins after the G protein–coupled receptors. This may ensure that the cell will permit the signaling to advance to the next stages only when a sufficient portion of the population of a particular protein-serine/threonine kinase is activated over a sustained period.
RESULTS
T207 and Y210 are highly conserved phosphorylation sites in ERK1
To investigate the regulation of protein kinases, we aligned 496 catalytic domain sequences of 484 human protein kinases with information for many of their known experimentally confirmed phosphosites (Supplemental Table S1). Of 1950 phosphosites initially identified, only 27 (1.4%) were known before this study to be inhibitory, and seven of these were located in kinase subdomain I in the GxGxxG motif. Another 303 (15.6%) phosphosites were reported to stimulate protein kinase phosphotransferase activity, and of these, 75% were located within the activation T-loop (Figure 1A). In human ERK1, these included the well-characterized T202 and Y204 phosphosites at aligned amino acid residue positions 150 and 152, respectively. Also found in ERK1 was a phosphorylatable threonine residue at aligned position 155 (T207) and a phosphorylatable tyrosine residue (Y210) at the aligned position 158, located −5 and −2 residues before the subdomain VIII APE sequence. The T207 and Y210 phosphorylation sites in ERK1 were highly conserved across 20 diverse species, as revealed on the PhosphoNET website (www.phosphonet.ca/evolution.aspx?protname=P27361%20T207). In fact, these flanking phosphosites were identified as among the most conserved sites in most human protein-serine/threonine kinases, including MAPKs, cyclin-dependent kinases, protein kinase C’s, and protein kinase B/AKTs (Figure 1B). Of the 393 catalytic domains of human typical protein-serine/threonine kinases, a serine or threonine residue was featured at position 155 in 342, and a tyrosine residue was found at position 158 in 272 (Supplemental Table S1). Of these, phosphorylated residues were reported in the literature at one or both of these positions in 174 of these protein-serine/threonine kinase catalytic domains (Supplemental Table S2).
FIGURE 1:

Phosphorylation sites in human protein kinase catalytic domains. (A) Distribution of experimentally confirmed phosphosites in human protein kinase domains. Phosphosites identified in human protein kinase catalytic domains were mapped on the alignment provided in Supplemental Table S1. The total number of phosphosites was 1950, with 304 activatory sites (dark purple) mostly clustering at activation loop between aligned amino acid residues 139 and 159, and inhibitory sites (red). (B) Distribution of phosphotyrosine (orange), phosphothreonine (green), and phosphoserine (blue) residues in the kinase activation T-loop. The locations of the phosphorylation sites in ERK1 are shown in purple. These include the highly conserved threonine phosphorylation site at the aligned position 155 and tyrosine phosphorylation site at the aligned position 158. The T198 phosphosite of ERK1 is located with the Insert/Gap 8 region at aligned position 148.
Of interest, 100% of human protein-tyrosine kinases featured a tryptophan residue instead of tyrosine residue at aligned position 158, and 98% had a proline residue (alanine and leucine for the exceptions) instead of a threonine residue at aligned position 155. This further supported possible specific roles of these phosphosites in selective protein-serine/threonine kinases.
ERK1 slowly autophosphorylates T207 in vitro
The aforementioned findings compelled us to investigate more carefully the functional relevance of the T207 and Y210 phosphosites in ERK1, along with T198, which was also a highly conserved phosphosite at aligned position 148, which encompassed the highly variable Insert/Gap 8 segment found in the T activation loop of protein kinases. We raised polyclonal antibodies in rabbits against the phospho-T207 and phospho-Y210 sites of ERK1 (T190 and Y193 in ERK2), and subjected the sera to negative purification with phosphotyrosine-agarose columns to deplete generic phosphotyrosine-specific antibodies. The nonphosphospecific antibodies were removed with peptide columns with ligands that corresponded to the unphosphorylated version of these sites, followed by affinity purification with columns that used the original immunizing phosphopeptides as ligands. Unphosphorylated peptides with the corresponding sequences were also added in the antibody incubation solutions to block nonphosphospecific binding. The specificity of each antibody preparation was confirmed by peptide dot blot analyses.
To evaluate whether the phosphorylation of these sites was due to autocatalysis, a kinase-dead (KD) mutant of ERK1 was created by substitution of the Lys-71 residue from the subdomain II AxK motif with an alanine residue (K71A). Purified glutathione S-transferase (GST) fusion proteins of ERK1 wild type (WT) and KD mutant were incubated with or without a constitutively active version of the upstream kinase MEK1 (MEK1-ΔN3EE) in the presence of ATP (Figure 2A). On the one hand, phosphorylation of T207 was detected in WT but not KD ERK1, which confirmed autophosphorylation of the site in vitro. On the other hand, phosphorylation of Y210 was induced by MEK1 with both WT and KD ERK1. We performed a time-course experiment to determine the correlation between these phosphorylation events and ERK1 activation by MEK1 (Figure 2B). Over 60 min of incubation time, T207 phosphorylation increased slowly, whereas the phospho-signal from the TEY site was saturated more quickly after 20 min. Activation of ERK1 by MEK1-ΔN3EE may induce the ability of the kinase to more rapidly autophosphorylate at the T207 site.
FIGURE 2:

Phosphorylation of ERK1 T207 and Y210 in vitro. (A) ERK1-WT and KD phosphorylation by MEK1-ΔN3EE. The reactions were carried out at 30°C for 15 min. (B) Time-course experiment of ERK1-WT phosphorylation. At each time point, an aliquot of the incubation mix was taken and mixed with SDS–PAGE sample buffer to terminate the reaction. The samples were subsequently probed with phosphosite-specific antibodies (ERK1 pT207, PYKSD8 for pY210 and dual phospho-ERK1/2 pTEpY for pT202 and pY204) or the pan-expression ERK1/2-CT (ERK-CT) antibody. Western blots from the region of the migration of ERK1. Similar results were obtained in at least three independent experiments.
Observations from mass spectrometry analyses also indirectly supported autophosphorylation of the T207 site. We prepared samples of trypsin-digested peptides from ERK1-WT and KD that were phosphorylated by MEK1 in vitro and subjected them to MS analysis. In the ERK1-KD sample, a peptide from the activation segment (amino acid residues 190–208) with the TEY motif (T202 and Y204) dually phosphorylated was the only phosphopeptide detected. The unphosphorylated form of this peptide was also detected in the KD sample but not in the WT sample. Surprisingly, the peptide corresponding to the activation loop was not detected at all in the ERK1-WT in two independent experiments. Possible explanations for these negative results are that the additional phosphate groups on this target peptide made it difficult to ionize for MS detection and the resultant fragment had too high a charge-to-mass ratio and traveled too quickly through the mass spectrometer (Xie et al., 2011).
T207 and Y210 play critical roles in ERK1 regulation
To further characterize these phosphosites, we mutated T207 and Y210, as well as another threonine residue (T198) upstream on the N-terminal side of TEY in ERK1 (Figure 3A). Coupled kinase assays with recombinant MEK1-ΔN3EE, ERK1, and myelin basic protein (MBP) as a substrate revealed the functional effects of phosphorylation of T207 and Y210, but not T198, on ERK1 activation and its downstream phosphotransferase activity in vitro (Figure 3, B and C).
FIGURE 3:

Phosphorylation and activity of ERK1 and its mutants. Six ERK1 mutants were created to characterize the functional roles of the three flanking phosphosites near the TEY motif (A). Purified recombinant ERK1 and its mutants were incubated with MEK1-ΔN3EE (orange) or kinase dilution buffer (blue) in presence of 50 μM ATP at 30°C for 15 min. An aliquot of each reaction mix was mixed with 2.5 μg of MBP and incubated for another 2 min. Samples were mixed with SDS–PAGE sample buffer and analyzed by Western blotting using the dual phospho-ERK1/2 pTEpY antibody (B) and phospho-MBP antibody (C). The results are averaged from three to five separate experiments with the SDs indicated by bars. **p < 0.005.
Substitution of Thr-207 to Ala (T207A) markedly increased the autophosphorylation at the TEY phosphosites. Surprisingly, the T207A mutant preserved only ∼20% of the phosphotransferase activity toward MBP when compared with WT. The T207E mutant was phosphorylated by MEK1-ΔN3EE to a similar extent with WT and T207A, but it completely failed to phosphorylate MBP. The T207E phosphosite-mimetic mutant was consistently slightly more inhibitory in its MBP phosphotransferase activity than the T207A mutants (Figure 3C), which further supports an inhibitory role for phosphorylation of the WT ERK1 at this site. These findings demonstrated that the autophosphorylation of T207 can be independent of TEY phosphorylation by MEK1. Furthermore, phosphorylation at the TEY site does not necessarily correlate with ERK1 phosphotransferase activity toward an exogenous substrate.
All three mutants with the Tyr-210 substituted by Phe or Glu (Y210F or Y210E) or Phe in combination with alanine residue replacements of T198 and T207 sites (2AF) were not recognized by MEK1-ΔN3EE for phosphorylation, indicating an important role for this tyrosine residue in providing the proper conformation of the activation T-loop of the kinase for recognition by MEK1.
Mutation at T207 does not affect the specificity of ERK1 toward peptide substrates
To further characterize the effects of T207 phosphorylation on ERK1 phosphotransferase activity, we tested ERK1 wild type (WT), T207A, and T207E on a Kinex kinase substrate peptide microarray, which permitted assessment of the phosphotransferase activity of kinases toward 445 different peptides patterned after optimal substrate consensus sequences for hundreds of different protein kinases. Recombinant ERK1 and its mutants were preactivated by incubation with MEK1, and the MEK1 phosphotransferase activity was subsequently inhibited by adding the compound UO126 at the end of the preincubation. After analyzing the microarray image, we observed no phosphotransferase activity of ERK1-T207E mutant compared with the MEK1/UO126 control field (Supplemental Figure S1). The T207A and WT preparations showed the same selectivity in phosphorylating the substrate peptides on the chip. The strongest phosphorylation detected in both fields was from the same substrate peptide with the sequence (GGSFPLSPGKKGG). The ratio of net signal strength between WT and T207A from this peptide was 10:3. Among the top hits from the T207A mutant, 14 of 16 peptides were also strongly phosphorylated by ERK1 WT. These results are consistent with the in vitro kinase assays described earlier and supported the conclusion that an alanine mutation at T207 of ERK1 did not affect the specificity toward peptide substrates but decreased the overall phosphotransferase activity of the kinase by ∼70%.
Phosphorylation at T207 may reduce the stability of activated ERK1
To investigate this matter further in a physiologically relevant cell-based system, we transfected ERK1-WT, T207A, and T207E constructs with Flag tags into the human embryonic kidney HEK293 cell line. After stimulation of overnight serum-deprived cells with 10% fetal bovine serum (FBS) for 10 min to obtain maximal ERK1/2 activation, we immunoprecipitated Flag-ERK1 from the lysates of the harvested cells. We observed that with similar amounts of the kinase, the T207A mutant had a significantly higher level of TEY phosphorylation than WT (Figure 4A). However, only a weak phospho-TEY signal was detected from the T207E phosphorylation-mimicking mutant. This indicated that the phosphorylation of T207 might interfere with MEK1 phosphorylation and activation of ERK1 in vivo and/or enhance the dephosphorylation of the TEY phosphosites, since the absolute levels of the Flag-ERK1 constructs were comparable. Considering that all three forms of ERK1 were phosphorylated to an equal level by MEK1-ΔN3EE at TEY in vitro, the weaker TEY phosphorylation of ERK1-T207E in vivo was unlikely to be due to decreased phosphorylation by MEK1.
FIGURE 4:

Phosphorylation and activity of ERK1-WT, T207A, and T207E in HEK293 cells. (A) Phosphorylation of TEY motif of ERK1 under serum stimulation. HEK293 cells stably expressing Flag-ERK1 were starved overnight before stimulation with 10% FBS for 10 min. (B) Activity of immunoprecipitated Flag-ERK1. After serum stimulation, Flag-ERK1 was immunoprecipitated by Flag-tag antibody and incubated with 5 μg of MBP and 50 μM ATP at 30°C for 15 min. The samples were subsequently subjected to SDS–PAGE and Western blotting with phosphosite-specific antibodies for the dual phospho-ERK1/2 phosphosite pTEpY and myelin basic protein (pMBP) and the Flag tag (Flag). (C) Each image is representative of three independent experiments, and the averages of the phosphorylation of the pTEpY site and MBP from the three separate experiments are shown with the SDs indicated by bars. **p < 0.005.
The immunoprecipitated ERK1 proteins were also tested for their kinase activity toward MBP (Figure 4B). Despite the stronger phosphorylation of ERK1-T207A at the TEY site after incubated with ATP, neither the Ala nor Glu mutants were able to phosphorylate MBP as a substrate. Substitution of a nonphosphorylatable alanine residue at T207 induced only the autophosphorylation activity of ERK1, indicating that the unphosphorylated threonine residue is important for the kinase to maintain its active conformation. Once phosphorylated at T207, the phosphotransferase activity of the kinase would be suppressed, and this might also serve to increase recognition of the TEY site for dephosphorylation by phosphatases.
Phosphorylation of T207 is regulated by protein phosphatases
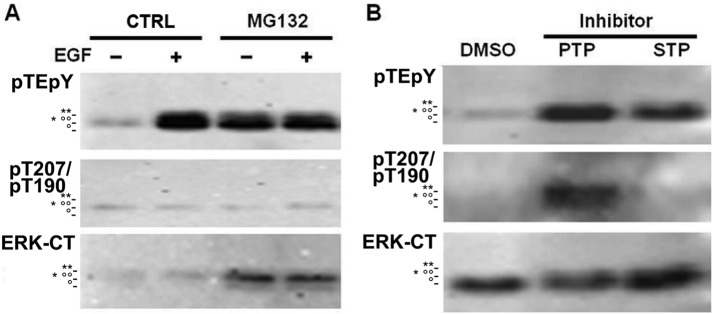
We also examined the phosphorylation of T207 in multiple cell lines with various treatments that activated ERK1/2 (Supplemental Figure S2). None of these conditions was able to produce stable phosphorylation of T207 of ERK1 or T190 of ERK2, which indicated that ERK1 and ERK2 that were phosphorylated on this threonine site were quickly removed from the cells by dephosphorylation and/or degradation. Protein levels of ERK1 and ERK2 have been reported to be negatively regulated by the ubiquitin/proteasome pathway (Lu et al., 2002). However, treatment with a specific proteasome inhibitor, MG132, increased the phosphorylation of only the TEY site and not that of the T207/T188 sites of ERK1/2 in A431 cells (Figure 5A). Given the earlier results that expression of ERK1-T207E did not affect the protein level of ERK1/2 in HEK293 cells, we believe that the T207 phosphorylation of ERK1 was probably not related to enhanced degradation of the kinase in vivo.
FIGURE 5:
Phosphorylation of ERK T207/T190 is regulated by protein phosphatases in A431 cells. (A) A431 cells were treated with 10 μM proteasome inhibitor MG132 for 4 h, followed by stimulation with 100 ng/ml EGF for 5 min. (B) A431 cells were treated with 0.025% dimethyl sulfoxide (CTRL), PTP inhibitors (25 μM PAO and 50 μM Na3VO4), or STP inhibitor (30 mM NaF) for 30 min. The samples were subsequently subjected to SDS–PAGE and Western blotting with antibodies that were dual phospho-ERK1/2 pTEpY phosphosite-specific and ERK1 pT207 phosphosite-specific (which cross-reacts with ERK2 pT190) or the pan-expression ERK1/2-CT (ERK-CT) antibody. The migration positions of phospho-ERK1 (**), phospho-ERK2 (°°), ERK1 (*), and ERK2 (°) on the SDS–PAGE gel are indicated.
To investigate the effect of protein phosphatases on ERK1/2 T207/T190 phosphorylation, we treated A431 cells with protein-tyrosine phosphatase (PTP) or serine/threonine phosphatase (STP) inhibitors (Figure 5B). Both sets of inhibitors were able to elevate the TEY phosphorylation level, but the phospho-T207/T190 signal increased only in PTP inhibitor-treated cells. These PTP inhibitors (phenylarsine oxide [PAO] and Na3VO4) suppress the activity of protein-tyrosine phosphatases and dual-specificity phosphatases/MAPK phosphatases. Because the expression of the phosphomimicking T207E mutant of ERK1 resulted in decreased levels of TEY phosphorylation, we suspect that phosphorylation of T207 might induce the dephosphorylation of ERK1 by MAPK phosphatases.
DISCUSSION
Phosphorylation of the activation T-loop is the most common mechanism for posttranslational kinase stimulation. Protein kinases are dynamically regulated in this flexible segment to control downstream signaling. Under physiological conditions, activation of protein kinases is usually transient, and robust treatments are required to ensure elicitation of the appropriate physiological responses. Various processes, including phosphorylation at inhibitory sites, dephosphorylation at activatory sites, and enhanced proteolysis, have been reported to account for negative regulation of the phosphotransferase activity of different individual protein kinases. However, no common mechanism has yet been proposed for negative regulation of protein-serine/threonine kinases in general, although phosphorylation of several of the cyclin-dependent protein kinases within the subdomain I GxGxxG motif is known to be inhibitory (Gu et al., 1992; Mueller et al., 1995; Welburn et al., 2007).
In this study, we focused on the activation segment of human protein kinases and by careful alignment identified the two most ubiquitous phosphorylation sites in all protein-serine/threonine kinases. At least 35 articles have been published regarding the functional analyses of these two phosphosites, using mutagenesis techniques with 33 protein kinases (Figure 6; Fisher and Morgan, 1994; Alessi et al., 1996; Butch and Guan, 1996; Raingeaud et al., 1996; Konishi et al., 1997; Lawler et al., 1997; Siow et al., 1997; Tassi et al., 1999; Masai et al., 2000; Lee and Chung, 2001; Rutter et al., 2001; Tan et al., 2001; Zhang et al., 2001, 2006; Liu et al., 2002; Huang et al., 2003; Moran et al., 2003; Rafie-Kolpin et al., 2003; Durkin et al., 2004; Arnold et al., 2005; Beullens et al., 2005; Graves et al., 2005; Jung et al., 2005; Kaur et al., 2005; Gagnon et al., 2006; Lu et al., 2006; Stafford et al., 2006; Bunkoczi et al., 2007; Harraz et al., 2007; Mattison et al., 2007; Hashimoto et al., 2008; Tyler et al., 2009; Guo et al., 2010; Jin et al., 2010; Li et al., 2010; Rodgers et al., 2010; Zheng et al., 2010; Gordon et al., 2011). All of the mutants created, including substitutions with nonphosphorylatable residues or phosphomimicking residues, were reported to show decreased activity, and it has been repeatedly interpreted that phosphorylation of these phosphosites may play a critical role in maintaining the active conformation of kinases. Indeed, in Figure 1, these were accordingly represented as stimulatory sites, although we now challenge these conclusions. Our findings indicate that phosphorylation of these two phosphosites may actually repress the catalytic activity of these protein kinases, which is contrary to the interpretations offered in most of these previous studies.
FIGURE 6:
Site-directed mutagenesis of phosphorylation sites in the activation T-loop of protein-serine/threonine kinases. Publications were identified in which site-directed mutagenesis had been performed on protein kinases that featured a phosphorylated threonine residue at aligned position 155 and/or a phosphorylated tyrosine residue at aligned position 158 in their catalytic domains. The effects of mutation of these and flanking phosphosites are shown as gain of function (GoF), loss of function (LoF), or without known effect (NoE) on the phosphotransferase activities of the tested protein kinases. Activatory phosphosites are highlighted in green, and suspected inhibitory phosphosites are highlighted in pink. A more complete list of mutated phosphorylation sites in diverse human proteins is provided in Supplemental Table S3B.
We used glutamic acid substitutions as phosphomimetics to explore the roles of the phosphorylation sites in the ERK1 activation T-loop. The locations of the oxygen atoms in the carboxyl moiety in the side chain of glutamic acid better approximates the positions of oxygen atoms in the phosphate moiety in a phosphorylated serine or threonine residue than they do in the shorter side chain aspartic acid residue. However, it was interesting to learn which acidic amino acid residue was in practice a better phosphomimetic. As shown in Supplemental Table S3B, primarily from the UniProt website, we were able to identify 695 human phosphosites that were demonstrated to be functionally important and for which site-directed mutagenesis had been performed to alter these critical residues. These included 477 activatory phosphosites and 183 inhibitory phosphosites. As summarized in Supplemental Table S3A, for a glutamic acid substitution of a serine, threonine, or tyrosine residue that corresponded to a phosphosite, the activatory or inhibitory effect of the phosphorylation of that site on the protein’s function was successfully mimicked 70.0, 77.6, and 55.6% of the time, respectively. For an aspartic acid substitution of a serine, threonine, or tyrosine residue that corresponded to a phosphosite, the activatory or inhibitory effect of the phosphorylation of that site on the protein’s function was successfully reproduced 89.8, 85.0, and 83.3% of the time, respectively. Based on these literature findings, it would appear that aspartic acid is more often a slightly better mimetic of phosphorylation than glutamic acid. Although we did not test aspartic acid mutants of the ERK1 T207 site, apparently ERK2 T190D mutants behave very much like the ERK1 T207E mutants with respect to enhanced autophosphorylation at the TEY phosphosites and abolition of phosphotransferase activity toward exogenous substrates (Smorodinsky-Atias et al., 2016).
We used ERK1, one of the most fundamental and well-studied kinases, as a model in which to investigate the functional roles of these highly conserved phosphorylation sites in activation T-loop. Based on our data, T207 in ERK1 arose from autophosphorylation, whereas the phosphorylation of Y210 was catalyzed by MEK1. All of the mutants of these two sites (T207A, T207E, Y210F, and Y210E) showed significantly decreased phosphotransferase activity toward MBP regardless of the level of TEY phosphorylation, which is consistent with the results from studies of other protein-serine/threonine kinases with mutation of equivalent amino acid residues (Figure 6). The Y210 mutants were not recognized by MEK1, indicating the importance of this residue in its unphosphorylated form for the maintaining the structure of ERK1 so that it can be targeted by MEK1 and MEK2.
Careful examination of the x-ray crystallographic structure of human ERK1, with and without phosphorylation of the activating Y204 site, revealed that the internal amino acid interactions associated with the side chains in T207 and Y210 are relatively unaffected by the presence of the phosphate moiety on Y204. As shown with the three-dimensional (3D) structure of monophosphorylated ERK1 in Figure 7, T207 appears to interact with K168, D166, and Y210, whereas Y210 may also interact with P169, E237, and W209. In particular, as documented in Supplemental Table S4A, in unphosphorylated ERK1, the oxygen atom (OG1) in the T207 side-chain hydroxyl group was within 3.6 Å of the two oxygen atoms (OD1 and OD2) in the side-chain carboxyl group of D166 and 2.8 Å from the nitrogen atom (NZ) in the side-chain amino group of K168. In T204-phosphorylated ERK1, these atoms pairs were still within 4.0 Å of each other. Similarly, with unphosphorylated ERK1, the oxygen in the hydroxyl moiety of Y210 was within 3.5 Å of the two oxygen atoms (OD1 and OD2) in the side-chain carboxyl group of E237. In T204-phosphorylated ERK1, these atom pairs were also still within 3.9 Å of each other. D166 and K168 in ERK1 correspond to the highly conserved positions D116 and K118 in kinase subdomain VI in the aligned sequences of protein kinase catalytic domains. Site-directed mutagenesis studies with the yeast cAMP-dependent protein kinase demonstrated that the D116 and K118 positions are critical for the phosphotransferase activity (Gibbs and Zoller, 1991). E237 in ERK1 corresponds to highly conserved E185 site in the aligned sequences of protein kinase catalytic domains and is located 5 amino acid residues after subdomain IX. This glutamic acid residue has also been identified as important for protein substrate recognition, particularly for basic amino acid side chains that are located two residues before the phosphoacceptor site in substrates (Gibbs and Zoller, 1991). As shown in Supplemental Figure S3B, these interactions are similarly preserved in both the inactive and T185/Y187 dual-phosphorylated forms of ERK2. Consequently, loss of T207 and Y210 interactions with other amino acid residues in ERK1, and the equivalent T190 and Y193 interactions in ERK2, which would be highly disrupted with their phosphorylation, would also be expected to be inhibitory to the catalytic activities of both of these kinases. Emrick et al. (2006) originally proposed that hydrogen bonding of the T190 in ERK2 to D149 (aligned position D116) might be responsible for maintaining the conformation of the activation loop and preventing kinase autoactivation by autophosphorylation. The highly increased autophosphorylation of ERK1 that we observed with the T207A ERK1 mutant in our study is in keeping with this idea.
FIGURE 7:

Interactions with T207 and Y210 residues in the 3D structure of human ERK1. The x-ray crystallographic structure of T204 phosphorylated human ERK1 (PDB Id 2ZOQ) was originally deduced by Kinoshita et al. (2008) and is rendered with JMol on the RCSB PDB website. The backbone atoms of ERK1 appear with white bonds, and most of the atoms in the tyrosine, tryptophan, and proline side-chain residues are colored orange, green, and yellow, respectively. Most oxygen, nitrogen, and carbon atoms in the other amino acid residue side chains appear as red, blue, and gray, respectively. Distances of atoms in the side chains of T207 and Y210 that were within 5 Å of the atoms of other amino acid side chains are indicated with orange dashed lines. In particular, T207 appears to interact with K168, D166, and Y210, whereas Y210 also interacts with P169, E237, and W209. Supplemental Table S4, A and B, respectively, provides listings of the actual distances between the phosphoacceptor residues in ERK1 and ERK2 with neighboring amino acid residues.
Autophosphorylation of the T190 site in ERK2 was reported to promote cardiac hypertrophy in both mice and humans without affecting the phosphotransferase activity of the kinase (Lorenz et al., 2009; Ruppert et al., 2013). By contrast, our results with ERK1 mutants of the corresponding T207 site indicate that phosphorylation of this site apparently inhibits the phosphotransferase activity of ERK1 toward exogenous substrates. Any mutation of the T207 residue resulted in a dramatic decrease of ERK1 phosphotransferase activity even when the TEY activating motif was phosphorylated.
Both the T207 and Y210 sites were phosphorylated slowly in vitro, and their phosphorylations were usually increased along with TEY phosphorylation and kinase activation. In contrast to our in vitro studies with recombinant protein kinases, in cell lysates, we observed very low levels of T207/T190 phosphorylation even when the ERK1/2 pathways were activated. This might be taken to indicate that these sites might not be appreciably phosphorylated in cells. However, in HEK293 cells that stably expressed the phosphomimicking ERK1-T207E mutant, the phosphorylation of TEY under serum stimulation was lower than with WT and T207A, indicating that the hyperphosphorylated forms of ERK1/2 were quickly removed from these cells. This hypothesis was supported by increased levels of both TEY and T207/T190 phosphorylation of ERK1/2 in PTP inhibitor-treated A431 cells.
Careful inspection of the PhosphoSitePlus website (www.phosphosite.org) for the number of reports by mass spectrometry of the phosphorylation of threonine and serine residues at aligned position 155 and tyrosine residues at aligned position 158 in protein-serine/threonine kinases revealed that phosphorylation at these sites is detected in general at much lower frequency than other phosphosites in the activation T-loops of protein kinases (Supplemental Table S2). This may be related to the difficulty of detecting highly phosphorylated short peptides in large-scale MS studies. As mentioned in the Results section, we experienced difficulty in detecting phosphorylation of the T207 and Y210 sites of ERK1 in vitro by MS. It is also likely that the hyperphosphorylated forms of protein kinases are subjected to increased rates of degradation or dephosphorylation in vivo.
Phosphosite-specific antibodies for the detection of phosphorylation of the TEY site in MAP kinases have been extensively used in thousands of studies to demonstrate the state of activation of these protein kinases. Although such antibodies can reveal the stimulations of signaling pathways that converge on these and other protein kinases, our results reveal that despite the presence of specific phosphorylation of the TEY site, the MAP kinases can still be catalytically inactive. In fact, this could be a very general problem, in view of the high conservation of the ERK1 T207 and Y210 phosphosites in most protein-serine/threonine kinases. At the very least, phosphosite-specific antibodies for stimulatory sites in the T-loop can demonstrate that the target kinase has been recently activated in its history, although not necessarily at the time of the isolation of the kinase.
We conclude that ERK1 slowly autophosphorylates the T207 site and this phosphorylation is markedly stimulated with ERK1 activation, phosphorylation at this site inhibits the phosphotransferase activity of ERK1 toward other substrates, and it is involved in the dephosphorylation and deactivation of ERK1 by MAPK phosphatases. At least a few previous studies also suggested that the phosphorylation of the serine or threonine residues at aligned position 155 might be inhibitory for Chk2 (Schwarz et al., 2003), MARK4 (Timm et al., 2003), and NEK2 (Rellos et al., 2007). We propose this may be an important general mechanism by which protein-serine/threonine kinases negatively regulate their activity after their initial activation. This ensures that only when a sufficient portion of the population of a particular protein-serine/threonine kinase is activated over a sustained period will the cell permit the signaling to advance to the next steps.
For many cellular processes, the subsequent inactivation of protein kinases is just as critical as their initial recruitment. Such processes include, among others, cell cycle progression in response to growth stimuli, relief from checkpoint controls with availability of nutrients and repair of DNA damage (Elledge, 1996; Johnson and Walker, 1999; Kastan and Bartek, 2004), resumption of anabolic reactions and inhibition of catabolic reactions with restoration of ATP levels after buildup of AMP and cAMP, and depression of the immune system after an infection has subsided.
The ability of signaling proteins to autoinactivate is not uncommon. For example, G proteins feature an intrinsic GTPase activity that curtails their actions on effector proteins. Many GTPase-activating proteins act to stimulate the rates of GTP hydrolysis by G proteins. We speculate that some regulatory proteins might act directly on protein kinases to accelerate or depress their rates of autoinactivation by stimulation of phosphorylation of the inhibitory phosphosites upstream of the kinase subdomain VIII APE sequence. Such kinase-inactivating proteins might specifically recognize the phosphorylated region just before the APE region, which in the phosphorylated state would be very conserved among most protein-serine/threonine kinases. These regions also may act as docking sites for members of the large family of dual-specificity protein phosphatases to facilitate their dephosphorylation of the activating phosphorylation sites that are located further downstream in the activation T-loop.
A tyrosine residue at aligned position 158 does not occur in any of the 90 human protein-tyrosine kinases. Previous studies showed that this region near the C-terminus of activation loop is one of the major determinants in conferring the specificity of protein-tyrosine kinases versus protein-serine/threonine kinases (Taylor et al., 1995; Hubbard, 1997). Protein-tyrosine kinases would be more likely than protein-serine/threonine kinases to undergo rapid autophosphorylation and inactivation with a tyrosine residue just before the APE region. Of interest, one-third of the human protein-tyrosine kinases feature a tyrosine residue that is located just four amino acid residues C-terminal to the subdomain VIII APE sequence (aligned position 166), and phosphorylation of this site has been confirmed for two-thirds of these tyrosine residues by mass spectrometry. It is tempting to speculate that this highly conserved tyrosine phosphosite serves as an inactivating autophosphorylation site for protein-tyrosine kinases. Although little is known from experimental studies about the physiological role of this tyrosine phosphorylation site, this residue at T701 in TrkA is known to be inhibitory for at least this kinase (de Pablo et al., 2008).
MATERIALS AND METHODS
Plasmids and commercial antibodies
The plasmids pGEX-2T-GST-ERK1, pGEX-2T-GST-ERK1-K71A, and pGEX-2T-GST-MEK1-ΔN3EE were constructed by David L. Charest from our lab (Charest et al., 1993). The plasmid containing full-length wild-type ERK1 (pGEX-2T-GST-ERK1) was used for mutagenesis (Q5 Site-Directed Mutagenesis Kit; New England BioLabs, Whitby, ON, Canada). ERK1 and its mutants were also subcloned into pcDNA3 vectors with Flag tag for mammalian cell expression. All DNA oligos were synthesized by Integrated DNA Technologies (Coralville, IA). We used the following commercial antibodies for Western blotting or immunoprecipitation (IP): ERK1/2-CT for the C-terminus of ERK1 and ERK2 (NK055-6; Kinexus), dual phospho-ERK1/2 pTEpY (KAP-MA021, discontinued; Stressgen, Victoria, BC, Canada), Flag tag antibody (2368; Cell Signaling Technology, Danvers, MA), and phospho-MBP (05-429; Upstate/EMD Millipore, Billerica, MA).
ERK1 phospho-T207 and phospho-Y210 antibodies
A phosphopeptide corresponding to human ERK1 204-209 (pYVApTRW) was synthesized to raise phosphosite-specific antibodies in rabbits against the ERK1 T207 (ERK2 T188) site, and this was subsequently used to affinity purify the antibodies from the rabbit sera. Non–phosphosite-specific antibodies were depleted by an agarose column coupled with pYVATRW peptide and an agarose column coupled with phosphotyrosine just before affinity purification with immunogenic peptide column. In Western blotting experiments using the ERK1 p207 antibody (PK866; Kinexus), the antibody solution was supplemented with 15 μg/ml pYVATRW peptide to further eliminate the binding to unphosphorylated ERK1. Dot blot studies demonstrated that this antibody preparation immunoreacted similarly with phosphopeptides with the sequences pYVApTRW and YVApTRW. The ERK1 pY210 site was detected with the PYKSD8 antibody (PG005; Kinexus), which was raised in rabbits against the WpYRAPE peptide. The PYKSD8 antibody was subsequently affinity purified from serum with an agarose column to which this peptide was covalently coupled after the serum was initially passaged through a phosphotyrosine-agarose column to deplete generic phosphotyrosine antibodies.
Cell cultures, transfections, and treatments
Human epidermoid carcinoma A431 and human embryonic kidney/HEK293 cells were cultured in DMEM (Life Technologies/Thermo Fisher Scientific, Waltham, MA) and MEM (Life Technologies), respectively, supplemented with 10% FBS (Applied Biological Materials, Richmond, BC, Canada). HEK293 cells were used as transfection hosts of Flag-ERK1 and its mutants. Culture dishes were coated with 2% poly-l-lysine solution (Sigma-Aldrich, Oakville, ON, Canada) for 20 min before each use. At 50–60% confluency, cells were transfected with pcDNA3.0-ERK1 plasmids using Lipofectamine 2000 transfection reagent (Life Technologies). The transfection medium was replaced 6 h after transfection. Cells were cultured for another 48 h before selection with 500 μg/ml G418 for positively transfected clones. Clones stably expressing Flag-ERK1 were tested and expanded for further experiments.
For growth factor treatments, the cells (80% confluency) were maintained in serum-free medium for 16–18 h and then treated separately with one of the following agents: 10 ng/ml human epidermal growth factor (EGF) for 5 min; 100 nM phorbol 12-myristate 13-acetate (PMA) for 10 min; 100 nM tumor necrosis factor-alpha (TNFα) for 10 min; and 10% FBS for 10 min. For the following treatments, cells were previously maintained in 10% FBS: 25 μM arsenite for 2 h, 25 μM PAO for 30 min, 50 μM Na3VO4 for 30 min, 30 mM NaF for 30 min, 10 μM MG132 (Calbiochem/EMD Millipore, Billerica, MA) for 4 h, and 100 ng/ml nocodazole for 16 h. After treatments, cells were usually washed with ice-cold phosphate-buffered saline before scraping into lysis buffer (20 mM 3-(N-morpholino)propanesulfonic acid [MOPS], pH 7.2, 5 mM EDTA, 2 mM ethylene glycol tetraacetic acid [EGTA], 0.5% [vol/vol] Triton X-100, 30 mM NaF, 20 mM Na4P2O7, 1 mM Na3VO4, 40 mM β-glycerophosphate, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 3 mM benzamidine, 5 μM pepstatin A, and 10 μM leupeptin). Cells were briefly sonicated and then subjected to ultracentrifugation at 100,000 × g for 30 min to prepare lysates for Western blotting.
In vitro kinase assay with myelin basic protein
Purified GST-ERK1 was first incubated with constitutively active mutant of MEK1 (MEK1-ΔN3EE) in 5 mM MOPS, pH 7.2, 2.5 mM β-glycerolphosphate, 0.4 mM EDTA, 1 mM EGTA, 5 mM MgCl2, 0.5 mM DTT, and 100 μM ATP at 30°C for 15 min. Purified MBP was added to the mixture and incubated for another 2–5 min before the reaction was terminated. Flag-ERK1 and its mutants were immunoprecipitated from cell lysates with Flag-tag antibody before incubation with MBP and ATP in the same buffer.
Kinase substrate antibody microarray
Kinex kinase substrate microarrays (Kinexus) were printed with 445 kinase substrate peptides as spots in triplicate. Purified ERK1 and its T207A and T207E mutants were preincubated with MEK1-ΔN3EE as described. At the end of the incubation, activated ERK1 was diluted in the same kinase assay buffer with MEK1 inhibitor UO126 to 200 μl. The mixture was then loaded onto a Kinex kinase substrate microarray slide and incubated at 30°C for 2 h. We used Pro-Q Diamond Phosphoprotein Stain (Molecular Probes/Thermo Fisher Scientific, Waltham, MA) to detect phosphorylation on the microarray. Microarray analysis software from ImaGene was used to analyze microarray images. Net signal median was calculated to represent the signal strength of each spot. Standard deviation of triplicates was used to eliminate inconsistent signals. Short lists were generated using a certain threshold percentage when compared with the strongest signal.
Databases and online resources
Information on protein phosphorylation sites was based on data collected from the UniProt (www.uniprot.org/), PhosphoSitePlus (www.phosphosite.org/), and PhosphoNET (www.phosphonet.ca/) databases. Protein structures were from the RCSB protein data bank (PDB; www.rcsb.org/pdb/) and visualized using PDB Protein Workshop. All numbering of amino acids in proteins is based on the unprocessed human forms as reported in UniProt.
Supplementary Material
Acknowledgments
We thank Dirk Winkler, Jane Shi, Hong Zhang, James Hopkins, and the rest of the staff at Kinexus Bioinformatics for their aid in the production of the phosphopeptides and antibodies that were developed for this study. We also thank Leonard Foster at the Centre for High Throughput Biology at the University of British Columbia for performing the mass spectrometry analyses of the ERK1 phosphorylation sites. S.L. was supported by a University Graduate Fellowship from the University of British Columbia and Kinexus. This work was funded by Kinexus.
Abbreviations used:
- DTT
dithiothreitol
- EPK
eukaryotic protein kinase
- ERK
extracellular signal–regulated kinase
- MAPK
mitogen-activated protein kinase
- MBP
myelin basic protein
- MEK
MAPK/ERK kinase
- MS
mass spectrometry
- PAO
phenylarsine oxide.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0527) on January 28, 2016.
REFERENCES
- Ahn NG, Seger R, Bratlien RL, Diltz CD, Tonks NK, Krebs EG. Multiple components in an epidermal growth factor-stimulated protein kinase cascade. In vitro activation of a myelin basic protein/microtubule-associated protein 2 kinase. J Biol Chem. 1991;266:4220–4227. [PubMed] [Google Scholar]
- Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Anderson NG, Maller JL, Tonks NK, Sturgill TW. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;343:651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- Arnold R, Patzak IM, Neuhaus B, Vancauwenbergh S, Veillette A, Van Lint J, Kiefer F. Activation of hematopoietic progenitor kinase 1 involves relocation, autophosphorylation, and transphosphorylation by protein kinase D1. Mol Cell Biol. 2005;25:2364–2383. doi: 10.1128/MCB.25.6.2364-2383.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beullens M, Vancauwenbergh S, Morrice N, Derua R, Ceulemans H, Waelkens E, Bollen M. Substrate specificity and activity regulation of protein kinase MELK. J Biol Chem. 2005;280:40003–40011. doi: 10.1074/jbc.M507274200. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Bunkoczi G, Salah E, Filippakopoulos P, Fedorov O, Müller S, Sobott F, Parker SA, Zhang H, Min W, Turk BE, Knapp S. Structural and functional characterization of the human protein kinase ASK1. Structure. 2007;15:1215–1226. doi: 10.1016/j.str.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butch ER, Guan KL. Characterization of ERK1 activation site mutants and the effect on recognition by MEK1 and MEK2. J Biol Chem. 1996;271:4230–4235. doi: 10.1074/jbc.271.8.4230. [DOI] [PubMed] [Google Scholar]
- Charest DL, Mordret G, Harder KW, Jirik F, Pelech SL. Molecular cloning, expression, and characterization of the human mitogen-activated protein kinase p44erk1. Mol Cell Biol. 1993;13:4679–4690. doi: 10.1128/mcb.13.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pablo Y, Perez-Garcia MJ, Georgieva MV, Sanchis D, Lindqvist N, Soler RM, Comella JX, Llovera M. Tyr-701 is a new regulatory site for neurotrophin receptor TrkA trafficking and function. J Neurochem. 2008;104:124–139. doi: 10.1111/j.1471-4159.2007.05027.x. [DOI] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- Durkin JT, Holskin BP, Kopec KK, Reed MS, Spais CM, Steffy BM, Gessner G, Angeles TS, Pohl J, Ator MA, Meyer SL. Phosphoregulation of mixed-lineage kinase 1 activity by multiple phosphorylation in the activation loop. Biochemistry. 2004;43:16348–16355. doi: 10.1021/bi049866y. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Emrick MA, Lee T, Starkey PJ, Mumby MC, Resing KA, Ahn NG. The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc Natl Acad Sci USA. 2006;103:18101–18106. doi: 10.1073/pnas.0608849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP, Morgan DO. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- Gagnon KB, England R, Delpire E. Characterization of SPAK and OSR1, regulatory kinases of the Na-K-2Cl cotransporter. Mol Cell Biol. 2006;26:689–698. doi: 10.1128/MCB.26.2.689-698.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- Gordon J, Hwang J, Carrier KJ, Jones CA, Kern QL, Moreno CS, Karas RH, Pallas DC. Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-Like kinase Mst3. BMC Biochem. 2011;12:54. doi: 10.1186/1471-2091-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves PR, Winkfield KM, Haystead TA. Regulation of zipper-interacting protein kinase activity in vitro and in vivo by multisite phosphorylation. J Biol Chem. 2005;280:9363–9374. doi: 10.1074/jbc.M412538200. [DOI] [PubMed] [Google Scholar]
- Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ward MD, Tiedebohl JB, Oden YM, Nyalwidhe JO, Semmes OJ. Interdependent phosphorylation within the kinase domain T-loop Regulates CHK2 activity. J Biol Chem. 2010;285:33348–33357. doi: 10.1074/jbc.M110.149609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Harraz MM, Park A, Abbott D, Zhou W, Zhang Y, Engelhardt JF. MKK6 phosphorylation regulates production of superoxide by enhancing Rac GTPase activity. Antioxid Redox Signal. 2007;9:1803–1813. doi: 10.1089/ars.2007.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto YK, Satoh T, Okamoto M, Takemori H. Importance of autophosphorylation at Ser186 in the A-loop of salt inducible kinase 1 for its sustained kinase activity. J Cell Biochem. 2008;104:1724–1739. doi: 10.1002/jcb.21737. [DOI] [PubMed] [Google Scholar]
- Huang WC, Chen JJ, Inoue H, Chen CC. Tyrosine phosphorylation of I-kappa B kinase alpha/beta by protein kinase C-dependent c-Src activation is involved in TNF-alpha-induced cyclooxygenase-2 expression. J Immunol. 2003;170:4767–4775. doi: 10.4049/jimmunol.170.9.4767. [DOI] [PubMed] [Google Scholar]
- Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. The role of tyrosine phosphorylation in cell growth and disease. Harvey Lect. 1998;94:81–119. [PubMed] [Google Scholar]
- Hunter T. Signaling—2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- Jin X, Jin HR, Jung HS, Lee SJ, Lee JH, Lee JJ. An atypical E3 ligase zinc finger protein 91 stabilizes and activates NF-kappaB-inducing kinase via Lys63-linked ubiquitination. J Biol Chem. 2010;285:30539–30547. doi: 10.1074/jbc.M110.129551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Walker CL. Cyclins and cell cycle checkpoints. Annu Rev Pharmacol Toxicol. 1999;39:295–312. doi: 10.1146/annurev.pharmtox.39.1.295. [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Jung HS, Kim DW, Jo YS, Chung HK, Song JH, Park JS, Park KC, Park SH, Hwang JH, Jo KW, Shong M. Regulation of protein kinase B tyrosine phosphorylation by thyroid-specific oncogenic RET/PTC kinases. Mol Endocrinol. 2005;19:2748–2759. doi: 10.1210/me.2005-0122. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kaur R, Liu X, Gjoerup O, Zhang A, Yuan X, Balk SP, Schneider MC, Lu ML. Activation of p21-activated kinase 6 by MAP kinase kinase 6 and p38 MAP kinase. J Biol Chem. 2005;280:3323–3330. doi: 10.1074/jbc.M406701200. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yoshida I, Nakae S, Okita K, Gouda M, Matsubara M, Yokota K, Ishiguro H, Tada T. Structural dissection of human mitogen-activated kinase ERK1. Biochem Biophys Res Commun. 2008;377:1123–1127. doi: 10.1016/j.bbrc.2008.10.127. [DOI] [PubMed] [Google Scholar]
- Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991a;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991b;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- Konishi H, Tanaka M, Takemura Y, Matsuzaki H, Ono Y, Kikkawa U, Nishizuka Y. Activation of protein kinase C by tyrosine phosphorylation in response to H2O2. Proc Natl Acad Sci USA. 1997;94:11233–11237. doi: 10.1073/pnas.94.21.11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs EG. Nobel Lecture. Protein phosphorylation and cellular regulation I. Biosci Rep. 1993;13:127–142. doi: 10.1007/BF01149958. [DOI] [PubMed] [Google Scholar]
- Lawler S, Feng XH, Chen RH, Maruoka EM, Turck CW, Griswold-Prenner I, Derynck R. The type II transforming growth factor-beta receptor autophosphorylates not only on serine and threonine but also on tyrosine residues. J Biol Chem. 1997;272:14850–14859. doi: 10.1074/jbc.272.23.14850. [DOI] [PubMed] [Google Scholar]
- Lee CH, Chung JH. The hCds1 (Chk2)-FHA domain is essential for a chain of phosphorylation events on hCds1 that is induced by ionizing radiation. J Biol Chem. 2001;276:30537–30541. doi: 10.1074/jbc.M104414200. [DOI] [PubMed] [Google Scholar]
- Li X, Moore DJ, Xiong Y, Dawson TM, Dawson VL. Reevaluation of phosphorylation sites in the Parkinson disease-associated leucine-rich repeat kinase 2. J Biol Chem. 2010;285:29569–29576. doi: 10.1074/jbc.M110.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Graham C, Li A, Fisher RJ, Shaw S. Phosphorylation of the protein kinase C-theta activation loop and hydrophobic motif regulates its kinase activity, but only activation loop phosphorylation is critical to in vivo nuclear-factor-kappaB induction. Biochem J. 2002;361:255–265. doi: 10.1042/bj3610255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz K, Schmitt JP, Schmitteckert EM, Lohse MJ. A new type of ERK1/2 autophosphorylation causes cardiac hypertrophy. Nat Med. 2009;15:75–83. doi: 10.1038/nm.1893. [DOI] [PubMed] [Google Scholar]
- Lu TJ, Lai WY, Huang CY, Hsieh WJ, Yu JS, Hsieh YJ, Chang WT, Leu TH, Chang WC, Chuang WJ, et al. Inhibition of cell migration by autophosphorylated mammalian sterile 20-like kinase 3 (MST3) involves paxillin and protein-tyrosine phosphatase-PEST. J Biol Chem. 2006;281:38405–38417. doi: 10.1074/jbc.M605035200. [DOI] [PubMed] [Google Scholar]
- Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell. 2002;9:945–956. doi: 10.1016/s1097-2765(02)00519-1. [DOI] [PubMed] [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002a;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002b;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 bY Cdks. J Biol Chem. 2000;275:29042–29052. doi: 10.1074/jbc.M002713200. [DOI] [PubMed] [Google Scholar]
- Mattison CP, Old WM, Steiner E, Huneycutt BJ, Resing KA, Ahn NG, Winey M. Mps1 activation loop autophosphorylation enhances kinase activity. J Biol Chem. 2007;282:30553–30561. doi: 10.1074/jbc.M707063200. [DOI] [PubMed] [Google Scholar]
- Moran ST, Haider K, Ow Y, Milton P, Chen L, Pillai S. Protein kinase C-associated kinase can activate NFkappaB in both a kinase-dependent and a kinase-independent manner. J Biol Chem. 2003;278:21526–21533. doi: 10.1074/jbc.M301575200. [DOI] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated inhibitory kinase that phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Protein phosphorylation in signaling—50 years and counting. Trends Biochem Sci. 2005;30:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Payne DM, Rossomando AJ, Martino P, Erickson AK, Her JH, Shabanowitz J, Hunt DF, Weber MJ, Sturgill TW. Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase) EMBO J. 1991;10:885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafie-Kolpin M, Han AP, Chen JJ. Autophosphorylation of threonine 485 in the activation loop is essential for attaining eIF2alpha kinase activity of HRI. Biochemistry. 2003;42:6536–6544. doi: 10.1021/bi034005v. [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Whitmarsh AJ, Barrett T, Dérijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- Rellos P, Ivins FJ, Baxter JE, Pike A, Nott TJ, Parkinson DM, Das S, Howell S, Fedorov O, Shen QY, et al. Structure and regulation of the human Nek2 centrosomal kinase. J Biol Chem. 2007;282:6833–6842. doi: 10.1074/jbc.M609721200. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Haas W, Gygi SP, Puigserver P. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab. 2010;11:23–34. doi: 10.1016/j.cmet.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert C, Deiss K, Herrmann S, Vidal M, Oezkur M, Gorski A, Weidemann F, Lohse MJ, Lorenz K. Interference with ERK(Thr188) phosphorylation impairs pathological but not physiological cardiac hypertrophy. Proc Natl Acad Sci USA. 2013;110:7440–7445. doi: 10.1073/pnas.1221999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Michnoff CH, Harper SM, Gardner KH, McKnight SL. PAS kinase: an evolutionarily conserved PAS domain-regulated serine/threonine kinase. Proc Natl Acad Sci USA. 2001;98:8991–8996. doi: 10.1073/pnas.161284798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JK, Lovly CM, Piwnica-Worms H. Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and trans-phosphorylation. Mol Cancer Res. 2003;1:598–609. [PubMed] [Google Scholar]
- Siow YL, Kalmar GB, Sanghera JS, Tai G, Oh SS, Pelech SL. Identification of two essential phosphorylated threonine residues in the catalytic domain of Mekk1. Indirect activation by Pak3 and protein kinase C. J Biol Chem. 1997;272:7586–7594. doi: 10.1074/jbc.272.12.7586. [DOI] [PubMed] [Google Scholar]
- Smorodinsky-Atias K, Goshen-Lago T, Goldberg-Carp A, Melamed D, Shir A, Mooshayef N, Beenstock J, Karamansha Y, Darlyuk-Saadon I, Livnah O, et al. Intrinsically active variants of Erk oncogenically transform cells and disclose unexpected autophosphorylation capability that is independent of TEY phosphorylation. Mol Biol Cell. 2016;27:1026–1039. doi: 10.1091/mbc.E15-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford MJ, Morrice NA, Peggie MW, Cohen P. Interleukin-1 stimulated activation of the COT catalytic subunit through the phosphorylation of Thr290 and Ser62. FEBS Lett. 2006;580:4010–4014. doi: 10.1016/j.febslet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Tan I, Seow KT, Lim L, Leung T. Intermolecular and intramolecular interactions regulate catalytic activity of myotonic dystrophy kinase-related Cdc42-binding kinase alpha. Mol Cell Biol. 2001;21:2767–2778. doi: 10.1128/MCB.21.8.2767-2778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi E, Biesova Z, Di Fiore PP, Gutkind JS, Wong WT. Human JIK, a novel member of the STE20 kinase family that inhibits JNK and is negatively regulated by epidermal growth factor. J Biol Chem. 1999;274:33287–33295. doi: 10.1074/jbc.274.47.33287. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Radzio-Andzelm E. Three protein kinase structures define a common motif. Structure. 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Radzio-Andzelm E, Hunter T. How do protein kinases discriminate between serine/threonine and tyrosine? Structural insights from the insulin receptor protein-tyrosine kinase. FASEB J. 1995;9:1255–1266. doi: 10.1096/fasebj.9.13.7557015. [DOI] [PubMed] [Google Scholar]
- Timm T, Li XY, Biernat J, Jiao J, Mandelkow E, Vandekerckhove J, Mandelkow E-M. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 2003;22:5090–5101. doi: 10.1093/emboj/cdg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler RK, Chu ML, Johnson H, McKenzie EA, Gaskell SJ, Eyers PA. Phosphoregulation of human Mps1 kinase. Biochem J. 2009;417:173–181. doi: 10.1042/BJ20081310. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Tucker JA, Johnson T, Lindert L, Morgan M, Willis A, Noble ME, Endicott JA. How tyrosine 15 phosphorylation inhibits the activity of cyclin-dependent kinase 2-cyclin A. J Biol Chem. 2007;282:3173–3181. doi: 10.1074/jbc.M609151200. [DOI] [PubMed] [Google Scholar]
- Wortzel I, Seger R. The ERK cascade: distinct functions within various subcellular organelles. Genes Cancer. 2011;2:195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Liu T, Qian WJ, Petyuk VA, Smith RD. Liquid chromatography-mass spectrometry-based quantitative proteomics. J Biol Chem. 2011;286:25443–25449. doi: 10.1074/jbc.R110.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Facchinetti V, Wang X, Huang Q, Qin J, Su B. Identification of MEKK2/3 serine phosphorylation site targeted by the Toll-like receptor and stress pathways. EMBO J. 2006;25:97–107. doi: 10.1038/sj.emboj.7600913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Romano PR, Nagamura-Inoue T, Tian B, Dever TE, Mathews MB, Ozato K, Hinnebusch AG. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem. 2001;276:24946–24958. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Peng M, Wang Z, Asara JM, Tyner AL. Protein tyrosine kinase 6 directly phosphorylates AKT and promotes AKT activation in response to epidermal growth factor. Mol Cell Biol. 2010;30:4280–4292. doi: 10.1128/MCB.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.