Abstract
Until recently the set of “model” species used commonly for cell biology was limited to a small number of well-understood organisms, and developing a new model was prohibitively expensive or time-consuming. With the current rapid advances in technology, in particular low-cost high-throughput sequencing, it is now possible to develop molecular resources fairly rapidly. Wider sampling of biological diversity can only accelerate progress in addressing cellular mechanisms and shed light on how they are adapted to varied physiological contexts. Here we illustrate how historical knowledge and new technologies can reveal the potential of nonconventional organisms, and we suggest guidelines for selecting new experimental models. We also present examples of nonstandard marine metazoan model species that have made important contributions to our understanding of biological processes.
In scientific investigation the fortunate choice of animal often suffices to resolve general questions of the greatest importance.
Claude Bernard, Introduction a l’étude de la médecine expérimentale
This sentiment, expressed by Claude Bernard in 1865 (Bernard et al., 1865, p. 27 [translation from French by the authors]), was echoed 60 years later by August Krogh: “For such a large number of problems there will be some animal of choice, or a few such animals, on which it can be most conveniently studied” (Krogh, 1929, p. 202). It is as true today in the era of genomics as it was in those days, that choosing experimental organisms on the basis of particular physiological features or practical suitability for a given technique is often the key to unlocking a biological question.
Individual species used for scientific investigation, in particular those used repeatedly, are commonly referred to as “models.” In practice, the term is generally used more restrictively to refer only to those organisms that have been heavily studied and that are tractable to genetic and/or molecular analysis, obvious examples being the fruit fly, mouse, yeast, Arabidopsis, nematode, zebrafish, and Xenopus. Such heavily studied species having many resources may be described as “traditional,” “conventional,” “standard,” “canonical,” “favored,” “well-established,” or “dominant,” whereas organisms studied by a small number of labs and having fewer molecular tools may be called “emerging,” “historical,” “unusual,” “nonstandard,” “marginal,” or “understudied.” It is worth pointing out that the adjective “emerging,” does not imply recent introduction into the laboratory: many of the “emerging models” have been studied since the 19th century; in this usage, “emerging” indicates a recent increase in molecular tools and methodologies, speed of scientific progress, or the number of laboratories working with a particular organism.
The “traditional” model organisms are very well understood through accumulated knowledge and intense study and have proven broad utility for research in many different fields, but they are unable to cover the full range of biological enquiry. This is because, as Claude Bernard implied 150 years ago, many biological processes are absent, masked, or not accessible in these organisms, and only a tiny fraction of existing molecular and taxonomic biodiversity is represented (Abzhanov et al., 2008; Bolker, 2012; Sullivan, 2015; Warren, 2015). In the past, there was no good alternative to explore this diversity: developing a new organism as a model was time-consuming and costly.
Today, many of the limitations in developing new model organisms are disappearing. With the advent of molecular methods, in particular low-cost high-throughput sequencing and easier approaches to genetic, epigenetic, and functional analysis without the need for conventional genetics, it is now possible to develop genomic resources and adapt analytical methods for new models fairly rapidly. These molecular resources can then allow rapid progress in addressing research questions that are intractable using current models and permit the exploration and development of new biotechnologies based on the unique biological characteristics of a particular species. In this new era, cross-talk between communities exploiting living organisms for applied aims and for basic research is facilitated. Organisms already cultured or collected for commercial purposes can be readily tested for their use in the laboratory, while better understanding of particular biological processes or traits in laboratory models can open up translational research avenues. Moreover, models of interest to multiple disciplines (basic biology, industry, medicine, teaching) acquire synergistic added value, as each increase in knowledge or resources will benefit all sectors that use it.
Importantly, “non-model” organisms have an excellent track record contributing to major discoveries in cell biology. Table 1 presents a nonexhaustive list to highlight some of the most prominent examples, many of which have led to Nobel Prize–winning discoveries. Notably, these discoveries were made before the new molecular technologies became available. These examples illustrate the potential for virtually any organism to become a “model,” and developing new models, while still not straightforward, is less daunting since the advent of new molecular technologies.
TABLE 1:
Examples of contributions from marine model organisms.
| Model species or group | Key biological features and breakthroughs | Awards | Key references |
|---|---|---|---|
| Sea urchin (Arbacia punctulata, Strongylocentrotus purpuratus,a Lytechinus variegatus, Paracentrotus lividus) | • Rapid, synchronous development and “biochemical” quantities of the easy-to-handle sea urchin embryos make them a key model for cell and developmental biology. | 2001 Nobel Prize in Physiology or Medicine: identification of the key mitotic protein cyclin | Dorée and Hunt, 2002; Davidson, 2009 |
| • Circa 1900, Boveri proposed the chromosome theory of inheritance and discovered centrosomes in sea urchins. | |||
| • Important models for studying mechanisms of cell cycle and transcriptional regulation. | |||
| Starfish (e.g., Patiria pectinifera, Patiria miniataa, Marthasterias glacialis) | • Concept of “maturation (M-phase) promoting factor” was established by cytoplasmic transfer experiments in amphibian and starfish oocytes, providing the foundation for much of cell cycle research. | Kanatani et al., 1969; Kishimoto and Kanatani, 1976 | |
| • Starfish were among the first organisms in which the meiosis-inducing hormone was identified. | |||
| Clam (Spisula solidissima and other bivalve mollusks, e.g., mussel, oyster) | • Extremely large number of oocytes allows establishment of cell-free systems that recapitulate cell cycle transitions, which has led to significant advances in the understanding of the cell cycle and translational control. | 2001 Nobel Prize in Physiology or Medicine: cyclins | Sudakin et al., 1995 |
| 2004 Nobel Prize in Chemistry: discovery of ubiquitin-mediated protein degradation system | |||
| Sea hares/slugs (Aplysia californicaa, other Aplysia species) | • The nervous system is composed of a small number of large cells, many of which are invariant and identifiable, rendering sea slugs an ideal model to understand the physiological basis of learning and memory. | 2000 Nobel Prize in Physiology or Medicine: discoveries concerning signal transduction in the nervous system | Carew and Kandel, 1973 |
| Squid (Loligo spp.) | • Squids feature a giant axon (up to 1 mm in diameter) in which voltage clamp electrodes can be inserted, allowing electrophysiology studies. | 1963 Nobel Prize in Physiology or Medicine: discovery of the ionic mechanism of the action potential | Vale et al., 1985; Schwiening, 2012 |
| • Observations of axonal transport led to the discovery of kinesin, the first microtubule motor protein. | |||
| Sea squirts (Ciona intestinalisa, Ciona savigny,a Phallusia mammillataa, Halocynthia roretzia, Botryllus schlosseria, Styela partita) | • Owing to their copious gametes and easy culture methods, sea squirts (ascidians) are a historical model for basic cell and developmental biology. | Nishida and Sawada, 2001; Brozovic et al., 2016 | |
| • In 1905, observations of the reorganization and partitioning of the pigmented myoplasm led Conklin to propose the concept of maternal determinants and the role of asymmetric division in specifying cell fates. | |||
| Hydrozoan jellyfish (Aequorea victoria, Clytia hemisphaerica) | • Hydrozoans have been used to study bioluminescence and for traditional experimental embryology. | 2008 Nobel Prize in Chemistry: discovery of GFP and the intracellular calcium sensor aequorin | Zimmer, 2009 |
| • Laboratory model hydrozoans have provided evidence for the evolutionarily ancient and conserved roles of signaling pathways in embryo polarity, development, and oocyte maturation. | |||
| Ragworm (Platynereis dumerilii) | • This organism has a short generation time and synchronous and stereotypic development of thousands of transparent embryos. | Tosches et al., 2014 | |
| • Research has addressed diverse questions in development, evolution, and neurobiology concerning phototaxis, introns, microRNA, the control of diel vertical migration via melatonin, and nervous system cell types. |
This table is far from exhaustive and omits many laboratory models with huge potential such as the amphipod crustacean Parhyale hawaiensis, the larvacean Oikopleura dioica, and important fish models such as medaka (Oryzias latipes) and puffer fish (Takifugu rubripes).
aGenome available publicly in January 2016.
It is not a coincidence that the examples in Table 1 are all marine organisms, because these organisms are, for historical and practical reasons, often suitable for development as experimental models. Since the 19th century, marine stations around the world have provided access to biological material covering a spectacular range of biodiversity; all animal phyla are present in the sea, several of them uniquely so. Marine stations have thus pushed forward research into all aspects of biology and ecology, thereby generating a large body of knowledge concerning where to find organisms, particularities of their physiology and life cycles, and how to manipulate them in the laboratory. This knowledge is of enormous importance in guiding the efficient choice and development of new models. On the practical side, many marine organisms produce vast quantities of freely accessible eggs, embryos, and larvae that are naturally transparent and therefore ideal for microscopy; and many, too, lack the robust protective cell walls, shells, cuticles, and exoskeletons that are common in terrestrial organisms and render the latter less amenable to experimental manipulation.
The motivation to find a new model organism is usually driven by a particular research challenge; existing models may not be suitable for the approaches envisaged to address particular biological processes; or an unexplored species or taxonomic group may have characteristics that are unique, exaggerated, or especially accessible to analysis. The rapidly decreasing start-up costs and technical investment needed to establish molecular and technical resources for any organism now make it economically feasible for a single laboratory or small consortium to test a number of possible new model species, then choose to develop and explore one (or more) of these for specific research aims. However, “lower” costs are not necessarily negligible: developing a new model can require establishing laboratory cultures and determining whether the biology of the organism is suitable for the desired experiments, and this effort can be quite labor- and material-intensive.
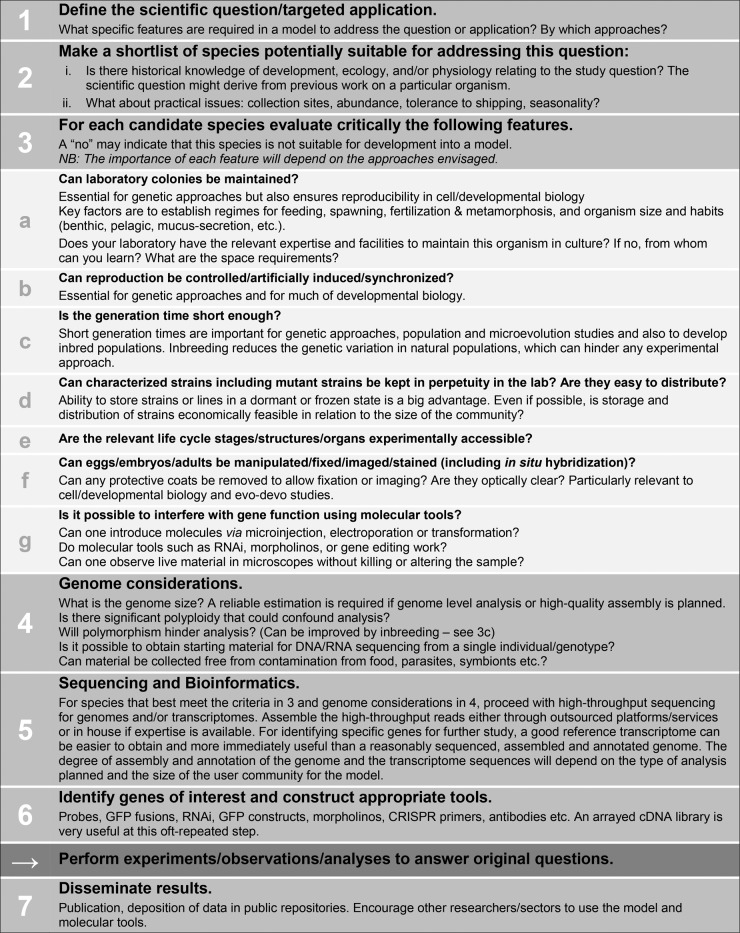
We have used our own experience developing models to outline some of the factors to be considered before developing resources for new models (Figure 1). We identify decision points that can help researchers to develop new models efficiently or to abandon those organisms that do not meet key criteria. We emphasize that prior biological knowledge is essential in the selection of potential models and that existing expertise can significantly decrease the financial and time costs for developing a new model. We have included in the outline the development of genomics resources such as a transcriptome or genome sequences, facilitated by high-throughput sequencing approaches. These resources are essential for many approaches in modern biology (such as comparative transcriptomics and promoter analysis) and greatly accelerate others, including genetics, biochemistry/proteomics, and gene function studies. Our own expertise is marine organisms, but our criteria will be useful when considering any potential new model, and in principle, any new tissue and cell cultures.
FIGURE 1:
Considerations and workflow for developing a new model organism.
We emphasize that the considerations listed in Figure 1 are a thinking aid, not a recipe to be followed to the letter. Many potential models may return a “no” to the criteria queried in part 3 of Figure 1 but nevertheless prove valuable. This is demonstrated by the first five models in Table 1, none of which meet every criterion. The main aim of this figure is to alert researchers to potential bottlenecks involved in developing a new organism as a model and to weigh them against potential benefits.
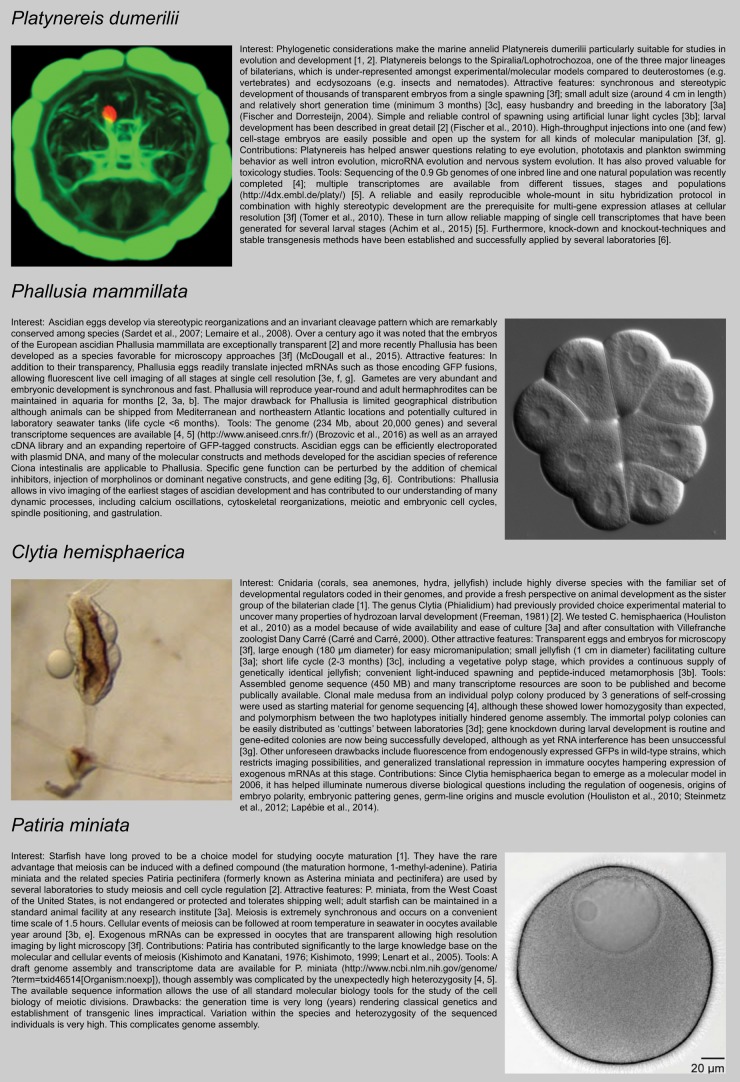
Four model species for which we have firsthand experience are presented in Figure 2 (Kishimoto and Kanatani, 1976; Kishimoto, 1999; Freeman, 1981; Carré and Carré, 2000; Fischer and Dorresteijn, 2004; Fischer et al., 2010; Lenart et al., 2005; Sardet et al., 2007; Lemaire et al., 2008; Houliston et al., 2010; Tomer et al., 2010; Steinmetz et al., 2012; Lapébie et al., 2014; Achim et al., 2015; McDougal et al., 2015; Brozovic et al., 2016). We have for each case matched the relevant features of these species with the considerations listed in Figure 1. For the annelid Platynereis dumerilii and the jellyfish Clytia hemisphaerica, for instance, paying attention to these criteria paid off and expectations have largely been fulfilled. Furthermore, some unexpected advantages emerged (e.g., vegetatively propagating Clytia polyp colonies being extremely convenient for maintenance of wild-type and gene-edited strains, Phallusia eggs readily translating exogenous mRNA even before fertilization), but also some unforeseen drawbacks.
FIGURE 2:
Case histories for four marine model animal species. Numbers in square brackets correspond to the points listed in Figure Figure 1.
As demonstrated by the examples in Table 1, nonstandard model organisms provide tremendous opportunities for increasing our understanding of biological processes, and their study has resulted in the development of a range of new technologies that have benefited academic research, health care, and the biotechnology industry. Particular scientific and technological advances are not necessarily anticipated directly, but can emerge progressively once sufficient resources and know-how have accumulated to allow in-depth analyses. We hope that this overview will provide a stimulus to turn from the beaten path of standard models and explore new avenues: the time is ripe to do so!
Acknowledgments
This work was supported by the European Commission FP7 Research Infrastructure Project, the European Marine Biological Resource Centre (ref. no. 262280), the European Marine Biological Resource Centre-France infrastructure project to the contributors from France, and European Molecular Biology Laboratory core funding. We thank Alex McDougall and Philippe Dru (Sorbonne Universités, UPMC Université Paris 06, CNRS, UMR 7009, Laboratoire de Biologie du Développement de Villefranche-sur-Mer, Observatoire Océanologique de Villefranche-sur-Mer, Villefranche-sur-Mer, France), Mark Cock (Station Biologique de Roscoff, Roscoff, France), and Olivier Thomas (Institut de Chimie de Nice UMR 7272, Universitè Nice Sophia Antipolis, Nice, France) for useful comments.
Footnotes
REFERENCES
- Abzhanov A, Extavour CG, Groover A, Hodges SA, Hoekstra HE, Kramer EM, Monteiro A. Are we there yet? Tracking the development of new model systems. Trends Genet. 2008;24:353–360. doi: 10.1016/j.tig.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Achim K, Pettit J-B, Saraiva LR, Gavriouchkina D, Larsson T, Arendt D, Marioni JC. Single-cell expression profiling and spatial mapping into tissue of origin. Nat Biotech. 2015;33:503–509. doi: 10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- Bernard C, Baillière JB, Fernández Carril A, Calleja y Sánchez J. Introduction a l’étude de la médecine expérimentale. Paris: J.-B. Baillière et Fils; 1865. [Google Scholar]
- Bolker J. Model organisms: there’s more to life than rats and flies. Nature. 2012;491:31–33. doi: 10.1038/491031a. [DOI] [PubMed] [Google Scholar]
- Brozovic M, Martin C, Dantec C, Dauga D, Mendez M, Simion P, Percher M, Laporte B, Scornavacca C, Di Gregorio A, et al. ANISEED 2015: a digital framework for the comparative developmental biology of ascidians. Nucleic Acids Res. 2016;44:D808–D818. doi: 10.1093/nar/gkv966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew TJ, Kandel ER. Acquisition and retention of long-term habituation in Aplysia: correlation of behavioral and cellular processes. Science. 1973;182:1158–1160. doi: 10.1126/science.182.4117.1158. [DOI] [PubMed] [Google Scholar]
- Carré D, Carré C. Origin of germ cells, sex determination, and sex inversion in medusae of the genus Clytia (Hydrozoa, leptomedusae): the influence of temperature. J Exp Zool. 2000;287:233–242. doi: 10.1002/1097-010x(20000801)287:3<233::aid-jez5>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Network design principles from the sea urchin embryo. Curr Opin Genet Dev. 2009;19:535–540. doi: 10.1016/j.gde.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorée M, Hunt T. From Cdc2 to Cdk1: when did the cell cycle kinase join its cyclin partner. J Cell Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- Fischer A, Dorresteijn A. The polychaete Platynereis dumerilii (Annelida): a laboratory animal with spiralian cleavage, lifelong segment proliferation and a mixed benthic/pelagic life cycle. BioEssays. 2004;26:314–325. doi: 10.1002/bies.10409. [DOI] [PubMed] [Google Scholar]
- Fischer AH, Henrich T, Arendt D. The normal development of Platynereis dumerilii (Nereididae, Annelida) Front Zool. 2010;7:31. doi: 10.1186/1742-9994-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman G. The cleavage initiation site establishes the posterior pole of the hydrozoan embryo. Wilhelm Rouxs Arch Dev Biol. 1981;190:123–125. doi: 10.1007/BF00848406. [DOI] [PubMed] [Google Scholar]
- Houliston E, Momose T, Manuel M. Clytia hemisphaerica: a jellyfish cousin joins the laboratory. Trends Genet. 2010;26:159–167. doi: 10.1016/j.tig.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Kanatani H, Shirai H, Nakanishi K, Kurokawa T. Isolation and identification of meiosis inducing substance in starfish Asterias amurensis. Nature. 1969;221:273–274. doi: 10.1038/221273a0. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. Activation of MPF at meiosis reinitiation in starfish oocytes. Dev Biol. 1999;214:1–8. doi: 10.1006/dbio.1999.9393. [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Kanatani H. Cytoplasmic factor responsible for germinal vesicle breakdown and meiotic maturation in starfish oocyte. Nature. 1976;260:321–322. doi: 10.1038/260321a0. [DOI] [PubMed] [Google Scholar]
- Krogh A. The progress of physiology. Science. 1929;70:200–204. doi: 10.1126/science.70.1809.200. [DOI] [PubMed] [Google Scholar]
- Lapébie P, Ruggiero A, Barreau C, Chevalier S, Chang P, Dru P, Houliston E, Momose T. Differential responses to Wnt and PCP disruption predict expression and developmental function of conserved and novel genes in a cnidarian. PLoS Genet. 2014;10:e1004590. doi: 10.1371/journal.pgen.1004590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire P, Smith WC, Nishida H. Ascidians and the plasticity of the chordate developmental program. Curr Biol. 2008;18:R620–R631. doi: 10.1016/j.cub.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P, Bacher CP, Daigle N, Hand AR, Eils R, Terasaki M, Ellenberg J. A contractile nuclear actin network drives chromosome congression in oocytes. Nature. 2005;436:812–818. doi: 10.1038/nature03810. [DOI] [PubMed] [Google Scholar]
- McDougall A, Chenevert J, Pruliere G, Costache V, Hebras C, Salez G, Dumollard R. Centrosomes and spindles in ascidian embryos and eggs. Methods Cell Biol. 2015;129:317–339. doi: 10.1016/bs.mcb.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Nishida H, Sawada K. macho-1 encodes a localized mRNA in ascidian eggs that specifies muscle fate during embryogenesis. Nature. 2001;409:724–729. doi: 10.1038/35055568. [DOI] [PubMed] [Google Scholar]
- Sardet C, Paix A, Prodon F, Dru P, Chenevert J. From oocyte to 16-cell stage: cytoplasmic and cortical reorganizations that pattern the ascidian embryo. Dev Dyn. 2007;236:1716–1731. doi: 10.1002/dvdy.21136. [DOI] [PubMed] [Google Scholar]
- Schwiening CJ. A brief historical perspective: Hodgkin and Huxley. J Physiol. 2012;590:2571–2575. doi: 10.1113/jphysiol.2012.230458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz PRH, Kraus JEM, Larroux C, Hammel JU, Amon-Hassenzahl A, Houliston E, Wörheide G, Nickel M, Degnan BM, Technau U. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W. The Institute for the Study of Non-model Organisms and other fantasies. Mol Biol Cell. 2015;26:387–389. doi: 10.1091/mbc.E14-03-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomer R, Denes AS, Tessmar-Raible K, Arendt D. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell. 2010;142:800–809. doi: 10.1016/j.cell.2010.07.043. [DOI] [PubMed] [Google Scholar]
- Tosches MA, Bucher D, Vopalensky P, Arendt D. Melatonin signaling controls circadian swimming behavior in marine zooplankton. Cell. 2014;159:46–57. doi: 10.1016/j.cell.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. In praise of other model organisms. J Cell Biol. 2015;208:387–389. doi: 10.1083/jcb.201412145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M. GFP: from jellyfish to the Nobel Prize and beyond. Chem Soc Rev. 2009;38:2823–2832. doi: 10.1039/b904023d. [DOI] [PubMed] [Google Scholar]