Solo (ARHGEF40) is a RhoA-targeting guanine nucleotide exchange factor that serves to promote stress fibers and precisely organize keratin networks. Solo binds to keratin-8/18 filaments. The interplay between Solo and keratin-8/18 filaments is crucial for tensile force–induced RhoA activation and consequent actin stress fiber reinforcement.
Abstract
Mechanical force–induced cytoskeletal reorganization is essential for cell and tissue remodeling and homeostasis; however, the underlying cellular mechanisms remain elusive. Solo (ARHGEF40) is a RhoA-targeting guanine nucleotide exchange factor (GEF) involved in cyclical stretch–induced human endothelial cell reorientation and convergent extension cell movement in zebrafish gastrula. In this study, we show that Solo binds to keratin-8/keratin-18 (K8/K18) intermediate filaments through multiple sites. Solo overexpression promotes the formation of thick actin stress fibers and keratin bundles, whereas knockdown of Solo, expression of a GEF-inactive mutant of Solo, or inhibition of ROCK suppresses stress fiber formation and leads to disorganized keratin networks, indicating that the Solo-RhoA-ROCK pathway serves to precisely organize keratin networks, as well as to promote stress fibers. Of importance, knockdown of Solo or K18 or overexpression of GEF-inactive or deletion mutants of Solo suppresses tensile force–induced stress fiber reinforcement. Furthermore, knockdown of Solo or K18 suppresses tensile force-induced RhoA activation. These results strongly suggest that the interplay between Solo and K8/K18 filaments plays a crucial role in tensile force–induced RhoA activation and consequent actin cytoskeletal reinforcement.
INTRODUCTION
All of the cells in the body are exposed to mechanical forces. The ability of cells to sense and respond to external forces is essential for numerous pathophysiological processes, such as embryogenesis, organogenesis, tumorigenesis, tissue remodeling, and homeostasis (Wozniak and Chen, 2009; Lecuit et al., 2011). Cells respond to external forces by converting mechanical force signals into biochemical signals, which cause changes in cell activities, including morphogenesis, migration, proliferation, and differentiation (Eyckmans et al., 2011).
Epithelial cells perceive external forces primarily through cell–cell and cell–extracellular matrix (ECM) adhesion sites. At cell–cell adhesions, cadherins and cadherin-associated proteins assemble to form adherence junctions and desmosomes, which anchor cytoplasmic actin filaments and intermediate filaments, respectively (DuFort et al., 2011). At cell–ECM adhesions, integrins and integrin-associated proteins assemble to form focal adhesions and hemidesmosomes, which anchor actin filaments and intermediate filaments, respectively (DuFort et al., 2011). Mechanical forces applied to cells generate tension in the actin filaments and intermediate filaments attached to these cell adhesion sites and induce changes in cytoskeletal organization, such as reinforcement of actin stress fibers, adhesion complexes, and actomyosin contractility, which allows cells to counteract the external forces. Recent studies identified several mechanosensitive proteins, such as α-catenin, p130CAS, and talin, which mediate force–induced cell responses at cell–cell and cell–ECM adhesions (Sawada et al., 2006; del Rio et al., 2009; Moore et al., 2010; Yonemura et al., 2010). However, little is known about how mechanical forces induce cytoskeletal reinforcement (Geiger et al., 2009; Huveneers and de Rooij, 2013).
Rho-family small GTPases are key regulators of actin cytoskeletal reorganization (Jaffe and Hall, 2005). Mechanical force application induces RhoA activation, which promotes stress fiber formation and actomyosin contractility via RhoA effectors such as Rho-associated kinase (ROCK), indicating that RhoA is crucial for force-induced actin remodeling (Zhao et al., 2007; Guilluy et al., 2011; Lessey et al., 2012). Rho-guanine nucleotide exchange factors (Rho-GEFs) activate Rho family GTPases by stimulating the GDP-to-GTP exchange. The Dbl-related Rho-GEFs have two common structural domains: a catalytic Dbl homology (DH) domain and a regulatory pleckstrin homology (PH) domain (Cook et al., 2014). Two RhoA-targeting GEFs, LARG (ARHGEF12) and GEF-H1 (ARHGEF2), have been reported to be involved in tensional force–induced RhoA activation in rat embryonic fibroblasts (Guilluy et al., 2011). We recently showed that Solo (ARHGEF40), a RhoA-targeting GEF (Curtis et al., 2004; Tse et al., 2005), is involved in cyclical stretch–induced reorientation of vascular endothelial cells and their stress fibers (Abiko et al., 2015). Quattro, a zebrafish orthologue of Solo, is involved in convergent extension cell movements of zebrafish gastrula (Daggett et al., 2004). These results suggest that Solo plays a key role in mechanotransduction.
Intermediate filaments are stable but resilient cytoskeletal filaments that provide structural support for cells. Keratins are major intermediate filaments in epithelia and play important roles in the maintenance of cell and tissue integrity against mechanical stresses and mechanoresponsive polarized cell migration (Coulombe and Wong, 2004; Weber et al., 2012). Keratin filaments are anchored to desmosomes and hemidesmosomes, and these keratin-associated cell–cell and cell–ECM adhesion sites sense mechanical forces by changing activity and/or localization of their component proteins (Weber et al., 2012; Osmani et al., 2015). Keratin-8/keratin-18 (K8/K18) filaments mediate ECM rigidity-induced increase in cell stiffness through RhoA/ROCK signaling (Bordeleau et al., 2012). However, it remains unknown how keratin filaments contribute to mechanical force–induced RhoA activation and actin reorganization.
In this study, we show that Solo binds to K8/K18 filaments and is required for the proper organization of actin stress fibers and K8/K18 networks. We also provide evidence that the interplay between Solo and K8/K18 filaments plays a crucial role in mechanical force–induced RhoA activation and stress fiber reinforcement.
RESULTS
Solo binds to K8/K18 filaments
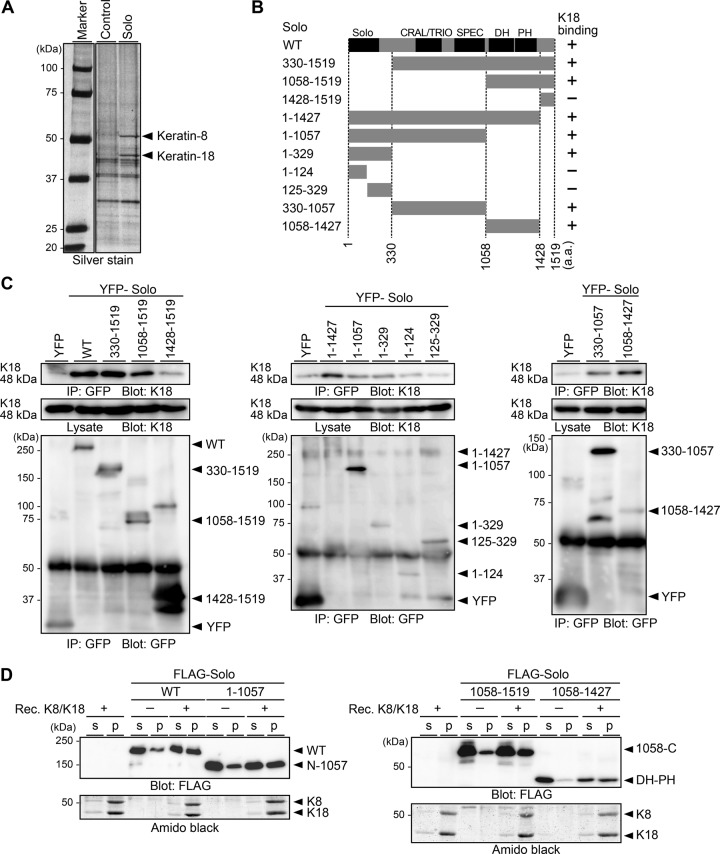
To identify regulator of Solo, we searched for Solo-binding proteins using proteomic analysis. MCF-7 cells stably expressing Halo-tagged Solo or control Halo were lysed, and the lysates were precipitated with Halo ligand–conjugated beads. The coprecipitated proteins were separated by SDS–PAGE and analyzed by mass spectrometry (Figure 1A). K8 and K18 were identified in Halo-Solo coprecipitates but not in control Halo coprecipitates. K8 and K18 are types II and I keratin, respectively, and form heterodimers that further assemble to form intermediate filaments (Loschke et al., 2015). Keratins (such as keratin-1 and -10) were frequently found in proteomics experiments as contaminating proteins from human skin (Global Proteome Machine, www.thegpm.org/crap/index.html), but K8 and K18 are expressed in simple-type epithelia but not in keratinized epidermis. Given these observations, together with the biochemical binding assays described later, it is probable that K8 and K18 are not contaminants but bona fide Solo-binding proteins. Because keratin filaments are involved in mechanotransduction (Weber et al., 2012; Bordeleau et al., 2012; Loschke et al., 2015), we further examined the role of the interaction between Solo and K8/K18 filaments in force-induced cell responses.
FIGURE 1:
Solo binds to K8/K18 IFs. (A) Identification of Solo-binding proteins. Lysates of MCF-7 cells expressing Halo (control) or Halo-Solo were precipitated with Halo-ligand beads. The precipitated proteins were separated by SDS–PAGE, stained with silver, and analyzed by mass spectrometry. (B) Schematic structure of human Solo and its deletion mutants. Conserved domains are denoted by Solo, CRAL/TRIO, SPEC (spectrin repeats), DH, and PH. The K18-binding ability of each construct is indicated in the right column. (C) Coimmunoprecipitation assays. HeLa cells expressing YFP-Solo or its deletion mutants were lysed and immunoprecipitated with an anti-GFP antibody. Immunoprecipitates (IPs) and lysates were analyzed by immunoblotting with an anti-K18 or an anti-GFP antibody. (D) Cosedimentation assays. Recombinant K8 and K18 were polymerized in vitro. Purified FLAG-Solo was incubated with K8/K18 filaments and centrifuged. The supernatant (s) and pellet (p) fractions were analyzed by immunoblotting with an anti-FLAG antibody and Amido black staining.
Solo contains an N-terminal Solo domain (a highly conserved domain in vertebrate Solo proteins), a CRAL/TRIO domain and spectrin repeats in the central region (predicted by National Center for Biotechnology Information Conserved Domain Database analysis), and a C-terminal DH-PH domain (Figure 1B; Daggett et al., 2004; Tse et al., 2005). To determine the keratin-binding region(s) of Solo, we individually expressed yellow fluorescent protein (YFP)–tagged Solo and its deletion mutants (shown in Figure 1B) in HeLa cells and analyzed their K18-binding ability by immunoprecipitation with an anti-GFP antibody, followed by anti-K18 immunoblotting (Figure 1C). K18 coprecipitated with wild-type Solo (Solo-WT) and the deletion mutants 1–329, 330–1057, and 1058–1519, indicating that Solo binds to each of the N-terminal, central, and C-terminal regions independently. K18 bound to the 1058–1427 mutant but not to 1428–1519, indicating that the DH-PH domain is involved in K18 binding. K18 bound to the 1–329 mutant but not to 1–124 or 125–329, suggesting that both the 1–124 and the 125–329 regions are necessary for K18 binding.
We also examined the binding potential of Solo to K8/K18 filaments by in vitro cosedimentation assays. Recombinant K8 and K18 proteins were copolymerized in vitro. Purified FLAG-tagged Solo-WT and its fragments were incubated with K8/K18 filaments and centrifuged (Figure 1D). Substantial amounts of Solo-WT and the fragments 1–1057, 1058–1519, and 1058–1427 were cosedimented with K8/K18 filaments. Together these results suggest that Solo binds to K8/K18 filaments through at least three binding sites, located respectively in the N-terminal, central, and DH-PH regions.
Knockdown of Solo disorganizes actin and keratin cytoskeletons
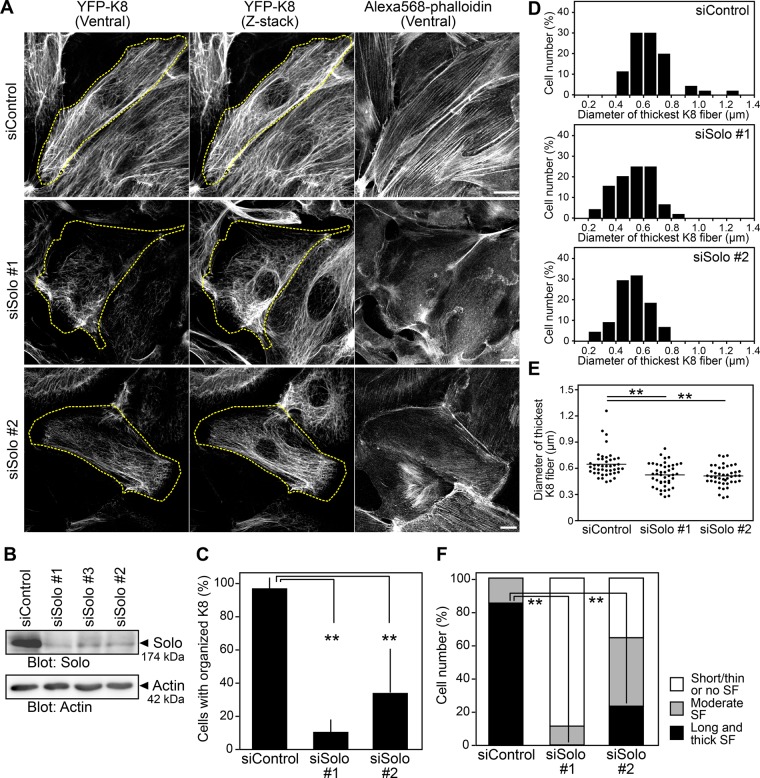
RhoA plays key roles in actin and keratin cytoskeletal organization (Jaffe and Hall, 2005; Waschke et al., 2006). We examined whether Solo knockdown affects the organization of actin and keratin filaments. To visualize K8/K18 filaments, we established Madin–Darby canine kidney (MDCK) cells stably expressing YFP-tagged K8 (MDCK/YFP-K8). We treated the cells with Solo-targeting small interfering RNAs (siRNAs) and analyzed the organizations of K8/K18 and actin filaments using YFP-K8 fluorescence and Alexa Fluor 568–phalloidin staining, respectively. Treatment with Solo-targeting siRNAs decreased the expression of endogenous Solo protein in MDCK/YFP-K8 cells (Figure 2B). Confocal microscopic analysis of the images near the ventral surface of the cell and the z-projection images of the whole cell revealed that control cells formed well-organized K8 networks that continuously distributed over the entire cytoplasmic region, extending from the perinuclear region to the cell margin (Figure 2A). By contrast, Solo-knockdown cells exhibited thinner, shortened, and disorganized K8 filaments whose networks were often distributed disproportionately and discontinuously in the cytoplasm and were lost near the cell peripheral region (Figure 2A). Total internal reflection fluorescence (TIRF) microscopic analysis of YFP-K8 near the ventral surface of the cells also revealed that K8 filaments near the substratum are markedly reduced in Solo-knockdown cells compared with those in control cells (Supplemental Figure S1A). When cells were categorized into those with or without organized K8 networks according to whether or not K8 networks were continuously distributed to the cell margin, the percentage of cells with organized K8 fibers was significantly decreased by Solo knockdown (Figure 2C). We also compared the thickness of K8 fibers in Solo-knockdown cells with that of K8 fibers in control cells. Measurements of the diameter of the thickest K8 fiber in each cell revealed that K8 fibers in Solo-knockdown cells were thinner than those in control cells (Figure 2, D and E). These results indicate that Solo is required for the proper formation and organization of the K8/K18 filament network. We also examined the effects of Solo knockdown on the localizations of β-catenin (a component of adherens junctions) and plakoglobin (a component of desmosomes and adherens junctions; Cowin et al., 1986; Wahl et al., 2000). Compared with control cells, Solo knockdown markedly decreased plakoglobin signals at cell–cell adhesion sites but had no apparent effect on β-catenin signals (Supplemental Figure S1B), which suggests that Solo is involved in the localization of plakoglobin to the cell–cell adhesion sites.
FIGURE 2:
Knockdown of Solo disorganizes actin and keratin cytoskeletons. (A) Confocal microscopic images of K8 filaments and F-actin in control and Solo-knockdown cells. MDCK/YFP-K8 cells were transfected with control or Solo-targeting siRNAs, cultured for 48 h, fixed, and stained with Alexa Fluor 568–phalloidin. The dashed line indicates the cell outline. Scale bars, 20 μm. (B) Effects of Solo knockdown on expression of Solo. MDCK cells were transfected with control or Solo-targeting siRNAs and cultured for 48 h. Cell lysates were analyzed by anti-Solo immunoblotting. (C) Effect of Solo knockdown on the organization of K8 networks. Percentages of cells with organized K8 networks. (D, E) Effect of Solo knockdown on the thickness of K8 fibers. The diameter of the thickest K8 fiber in each cell was measured, and the data are displayed as histograms (D) or dot plots (E). (F) Effect of Solo knockdown on stress fiber formation. Percentages of cells with thick, moderate, or thin stress fibers. In C and F, the data represent the mean ± SD of three independent experiments (at least 10 cells per experiment). **p < 0.01 (one-way ANOVA followed by Dunnett’s test).
Knockdown of Solo also altered F-actin organization. Thick and long stress fibers along the longitudinal axis of the cell were observed in 85% of control cells. By contrast, stress fibers were thinned or absent in most Solo-depleted cells (Figure 2, A and F), suggesting that Solo is required for the formation of thick longitudinal stress fibers.
Expression of Solo or its GEF-inactive mutant affects actin and keratin cytoskeletons
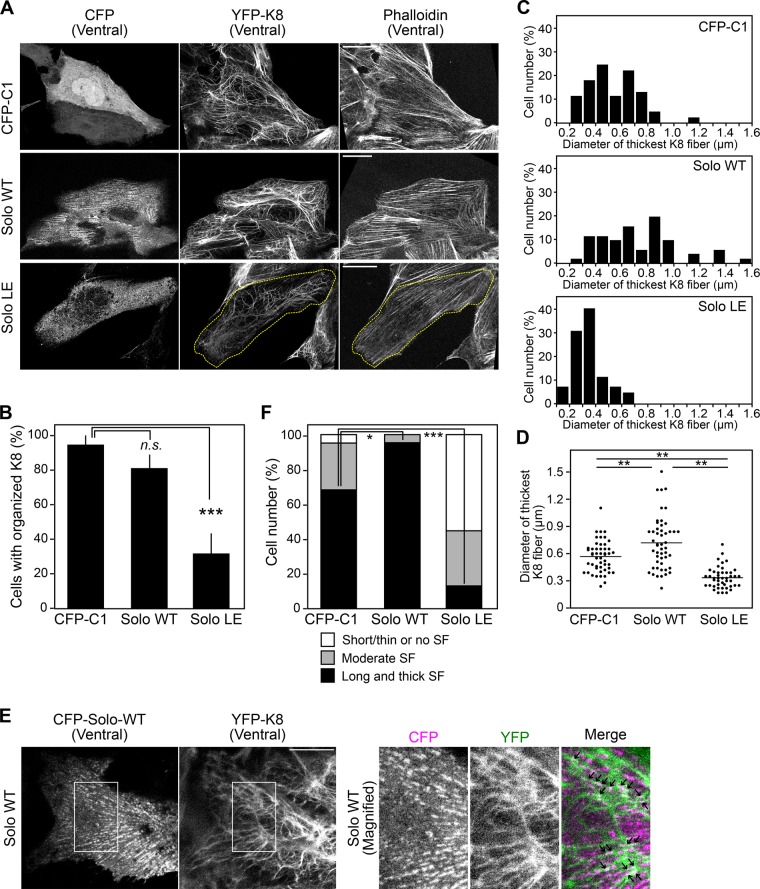
We next examined the effects of overexpression of Solo or its GEF-inactive mutant (Solo-LE), in which Leu-1217 is replaced by Glu, on actin and keratin cytoskeletal organization. The pull-down assays using glutathione-S-transferase (GST)–tagged RhoA-binding domain (RBD) of rhotekin, which binds active RhoA, revealed that expression of Solo-WT, but not that of Solo-LE, leads to RhoA activation (Supplemental Figure S2A), indicating that Solo-LE is a GEF-inactive mutant. MDCK/YFP-K8 cells were transiently transfected with cyan fluorescent protein (CFP), CFP-Solo-WT, or CFP-Solo-LE (Supplemental Figure S2B), and K8 and actin organization was analyzed. Compared with control CFP-expressing cells, expression of Solo-WT led to thick K8 fibers in the basal part of the cell attached to the plate, whereas expression of Solo-LE led to disorganized K8 networks (Figure 3A and Supplemental Figure S2C). The number of cells with organized K8 fibers was significantly decreased by Solo-LE expression (Figure 3B). The diameter of the thickest K8 fiber in each cell was increased by Solo-WT expression but reduced by Solo-LE expression (Figure 3, C and D). High-resolution confocal microscopic analyses revealed that Solo-WT displays punctate localization along the fibrous K8 signals on the ventral side of the cell and partially colocalizes with K8 fibers (Figure 3E). By contrast, Solo-LE was diffusely distributed in the cytoplasm (Figure 3A). In addition, expression of Solo-WT significantly increased the number of cells with thick and long stress fibers, whereas expression of Solo-LE decreased the number of these cells (Figure 3, A and F). These results further indicate that Solo is involved in the proper organization of actin stress fibers and K8 networks and that its GEF activity is required for these functions.
FIGURE 3:
Effects of expression of Solo-WT or its GEF-inactive mutant on actin and keratin cytoskeletons. (A) Fluorescence images of CFP, CFP-Solo (WT or LE), YFP-K8, and F-actin near the ventral surface of the cell. MDCK/YFP-K8 cells were transfected with CFP or CFP-Solo (WT or LE), cultured for 24 h, fixed, stained with Alexa Fluor 568–phalloidin, and analyzed by confocal microscopy. The dashed line in the Solo-LE–expressing cell indicates the cell outline. Scale bars, 20 μm. (B) Effect of Solo knockdown on the organization of K8 networks, analyzed as in Figure 2C. ***p < 0.001; n.s., not significant (one-way ANOVA followed by Dunnett’s test). (C, D) Effect of Solo knockdown on the thickness of K8 fibers, analyzed as in Figure 2, D and E. **p < 0.01 (one-way ANOVA followed by Tukey’s test). (E) Colocalization of Solo-WT and K8 filaments. MDCK/YFP-K8 cells were transfected with CFP-Solo-WT, and fluorescence images of CFP-Solo-WT and YFP-K8 near the ventral surface of the cell were obtained by high-resolution confocal microscopy. Right, magnified images of the white boxes in the left images. Arrows in the merged image indicate colocalization of CFP-Solo-WT and YFP-K8 fibers. Scale bar, 10 μm. (F) Effect of Solo knockdown on stress fiber formation, analyzed as in Figure 2F. *p < 0.05 and ***p < 0.001 (one-way ANOVA followed by Dunnett’s test). In B and F, the data represent the mean ± SD of three independent experiments (at least 13 cells/experiment).
A ROCK inhibitor disorganizes actin and keratin cytoskeletons
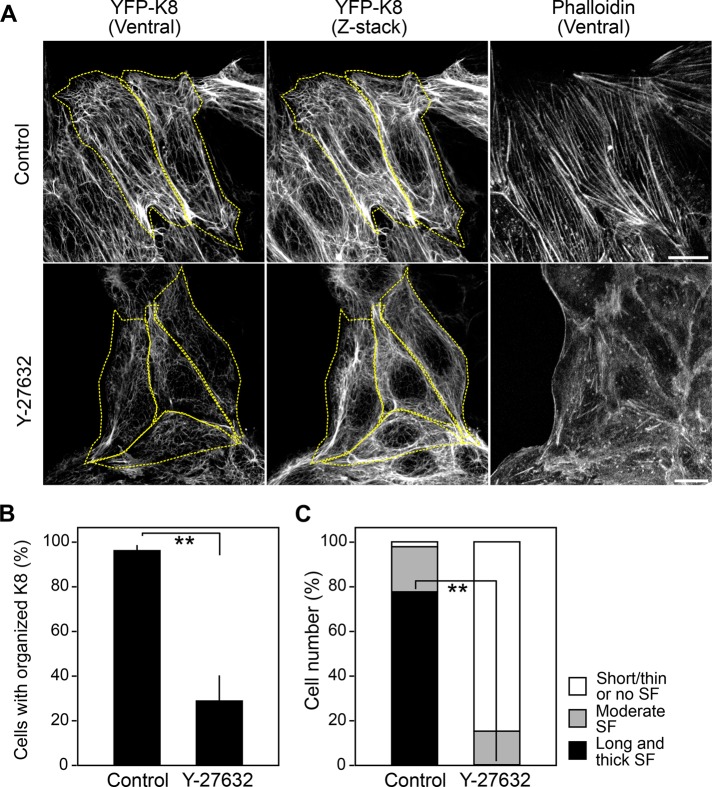
Because Solo is a RhoA-targeting GEF, we examined the role of the Rho-ROCK pathway in actin and keratin cytoskeletal organization. MDCK/YFP-K8 cells were treated with Y-27632, a selective inhibitor of ROCK, and K8 and actin organization was analyzed. Treatment with Y-27632 caused disorganization of K8 networks and suppression of longitudinal actin stress fiber formation (Figure 4, A–C), indicating that the Rho-ROCK pathway is critical for the proper formation and organization of actin stress fibers and K8/K18 networks. The effects of Y-27632 on the organization of K8 fibers and actin stress fibers were similar to those observed by Solo knockdown or Solo-LE overexpression, suggesting that Solo functions in actin and K8/K18 cytoskeletal organization through activation of the Rho-ROCK pathway.
FIGURE 4:
Effects of Y-27632 on actin and keratin organization. (A) Confocal microscopy of YFP-K8 and F-actin. MDCK/YFP-K8 cells were treated with 10 μM Y-27632 or left untreated for 1 h and then fixed and stained with Alexa Fluor 568–phalloidin. The dashed line indicates the cell outline. Scale bars, 20 μm. (B, C) Effects of Solo knockdown on the organization of K8 networks (B) and stress fiber formation (C), analyzed as in Figure 2. The data represent the mean ± SD of three independent experiments (at least 21 cells/experiment). **p < 0.01 (two-tailed paired t test).
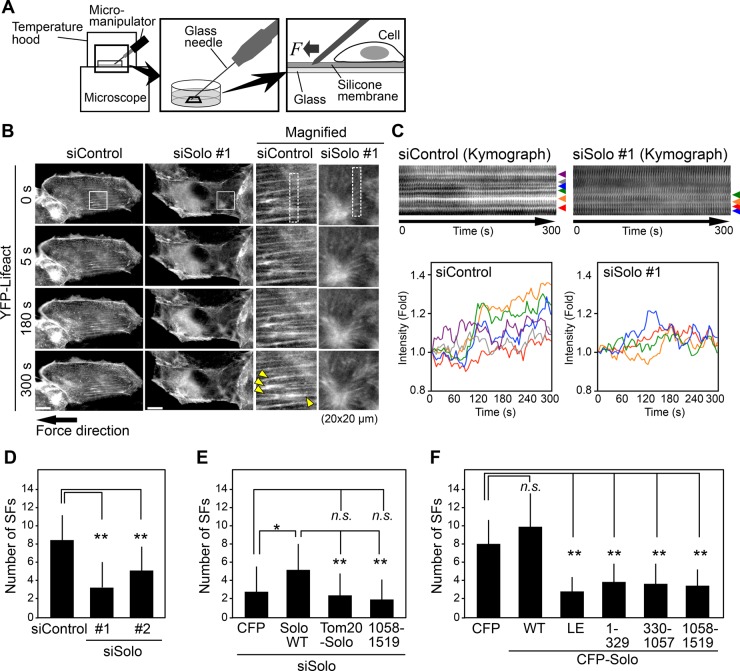
Solo is required for tensional force–induced stress fiber reinforcement
It is generally known that tensional force application reinforces and generates stress fibers in a direction parallel to the force application (Walcott and Sun, 2010; Tojkander et al., 2012). To examine the role of Solo in tensional force-induced stress fiber formation, we established MDCK cells stably expressing YFP-Lifeact (MDCK/YFP-Lifeact) and analyzed changes in actin cytoskeletal organization by live-cell imaging. Cells were cultured on fibronectin (FN)-coated silicone membranes and stretched by inserting a glass needle into the silicone membrane adjacent to the cell and sliding it away from the cell (Figure 5A). This procedure reproducibly caused force-induced stress fiber generation and reinforcement in a direction parallel to the tensile force applied (Figure 5B and Supplemental Movie S1). Kymograph analysis of time-lapse fluorescence images of YFP-Lifeact revealed changes in fluorescence intensity of each stress fiber (Figure 5C). On the basis of these analyses, we quantified force-induced stress fiber reinforcement/formation by counting the number of stress fibers that were reinforced or newly emerged in the direction of the force within 5 min of force application. Because the tensional force application caused the deformation of the silicone membrane, and thereby the cell region adjacent to the site of force application became out of focus, we counted stress fibers only in the cell region at the distance of >10 μm from the site of force application. When MDCK/YFP-Lifeact cells transfected with control or Solo-targeting siRNAs were stretched and analyzed by time-lapse fluorescence microscopy, Solo knockdown significantly reduced the number of stress fibers that were reinforced or emerged by tensional force application (Figure 5, B–D, and Supplemental Movie S1), which indicates that Solo plays a critical role in tensional force–induced stress fiber formation and reinforcement.
FIGURE 5:
Solo is required for tensional force–induced stress fiber formation. (A) Methods of tensional force application and time-lapse observation. (B) Solo knockdown suppresses force-induced stress fiber formation. MDCK/YFP-Lifeact cells were plated on a silicone membrane, transfected with control or Solo-targeting siRNAs, and cultured for 48 h before tensional force was applied. Time-lapse fluorescence images of YFP-Lifeact near the ventral surface of the cell were obtained by confocal microscopy. Right, magnified images of the white boxes in the images on the left. Yellow arrowheads indicate stress fibers that were strengthened or emerged after force application. Scale bars, 20 μm. See also Supplemental Movie S1. (C) Kymographs of the dashed boxes in B. Bottom, changes in fluorescence intensity of each of stress fiber indicated by arrowheads in the kymographs. (D–F) Quantitative analysis of the effects of knockdown of Solo (D), knockdown of Solo followed by expression of Solo-WT or its mutants (E), or expression of Solo or its mutants (F) on force-induced stress fiber formation. The number of stress fibers that were newly generated or reinforced was counted. See also Supplemental Figures S3C and S4B and Supplemental Movies S1–S3. The data shown represent the mean ± SD of 16–19 cells/experiment. *p < 0.05 and **p < 0.01; n.s., not significant (one-way ANOVA followed by Dunnett’s test or Tukey’s test).
We next examined whether the inhibitory effect of Solo-targeting siRNAs on the tensional force-induced stress fiber reinforcement is recovered by expression of siRNA-resistant human Solo-WT or its mutants. MDCK/YFP-Lifeact cells transfected with dog Solo-targeting siRNAs were cultured onto FN-coated dishes and then transfected with CFP-tagged human Solo-WT. Then tensional force was applied as in Figure 5A. Expression of Solo-WT significantly ameliorated the inhibitory effect of Solo knockdown on the force-induced stress fiber reinforcement (Figure 5E, Supplemental Figure S3C, and Supplemental Movie S2), indicating that the inhibitory effect of Solo siRNA treatment is caused by knockdown of endogenous Solo. To examine the importance of the Solo-keratin interaction in tensional force-induced stress fiber formation, we analyzed whether a Solo mutant that is forced to localize to mitochondria could recover the inhibitory effect of Solo knockdown on the force-induced stress fiber formation. We constructed a chimeric protein, Tom20(1-33)-CFP-Solo, which contains a mitochondrial targeting signal of Tom20 (Komatsu et al., 2010) at the N-terminus of CFP-Solo (Supplemental Figure S3A). When expressed in MDCK cells, Tom20(1-33)-CFP-Solo was localized to mitochondria (Supplemental Figure S3B). Expression of Tom20(1-33)-CFP-Solo did not recover the inhibitory effect of Solo knockdown on force-induced stress fiber reinforcement (Figure 5E, Supplemental Figure S3C, and Supplemental Movie S2), which suggests that localization of Solo to K8/K18 filaments is important for tensional force–induced stress fiber reinforcement. In addition, expression of the C-terminal DH-PH–containing fragment of CFP-Solo (1058–1519) also failed to recover the inhibitory effect of Solo knockdown on force-induced stress fiber reinforcement (Figure 5E, Supplemental Figure S3C, and Supplemental Movie S2), which is evidence that the N-terminal region of Solo is required for its function in tensional force–induced stress fiber reinforcement.
Expression of Solo or its mutants affects tensional force–induced stress fiber reinforcement
We further examined the effects of expression of Solo-WT or its mutants on tensional force–induced stress fiber formation. We transfected MDCK/YFP-Lifeact cells with CFP-Solo-WT or its mutants (Supplemental Figure S4A) and applied tensional force as in Figure 5A. Time-lapse fluorescence analysis of YFP-Lifeact revealed that expression of Solo-WT did not significantly affect the number of force-induced stress fibers, but Solo-LE expression significantly decreased the number of these fibers (Figure 5F, Supplemental Figure S4B, and Supplemental Movie S3). These results indicate that the GEF activity of Solo is required for force-induced stress fiber formation and that Solo-LE has a dominant-negative effect against endogenous Solo. Furthermore, expression of the Solo deletion mutants 1–329, 330–1057, and 1058–1519 also decreased the number of force-induced stress fibers (Figure 5F, Supplemental Figure S4B, and Supplemental Movie S3). These results suggest that each fragment acts in a dominant-negative manner against endogenous Solo and that each region of Solo plays a critical role in tensional force–induced Solo activation and stress fiber formation. No obvious change was detected in the localization of CFP-Solo-WT during 5 min after tensional force application under our experimental conditions (Supplemental Movie S4).
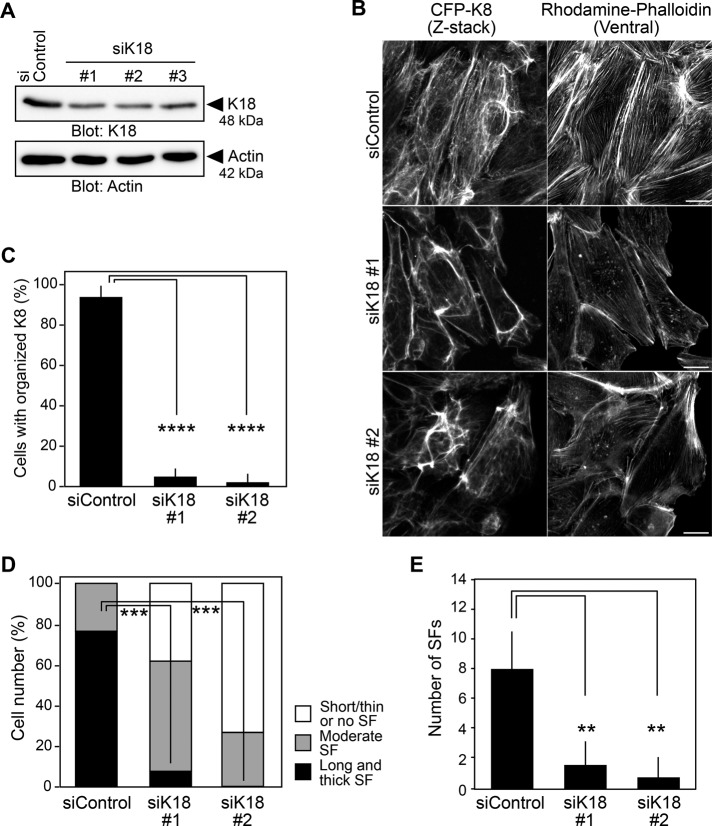
Knockdown of K18 disorganizes actin and keratin cytoskeletons
To examine the role of K8/K18 filaments in actin cytoskeletal organization, we analyzed the effect of K18 knockdown on the organization of actin stress fibers and K8 fibers. Treatment with K18-targeting siRNAs reduced the expression of endogenous K18 protein (Figure 6A). We treated MDCK/CFP-K8 cells with K18-targeting siRNAs and analyzed the organization of K8 and actin fibers by CFP fluorescence and rhodamine–phalloidin staining, respectively. Knockdown of K18 markedly decreased the number of K8 fibers and led to disorganized K8 networks, likely due to reduced formation of K8/K18 filaments (Figure 6, B and C). Phalloidin staining revealed that K18 knockdown suppressed the formation of longitudinal actin stress fibers (Figure 6, B and D). These results indicate that K18 is required for the formation of K8/K18 filaments and that K8/K18 filaments are involved in actin stress fiber formation.
FIGURE 6:
K18 is required for actin filament organization and force-induced stress fiber formation. (A) Effects of K18 knockdown on expression of K18. MDCK cells were transfected with control siRNA or K18-targeting siRNAs and cultured for 48 h. Cell lysates were analyzed by immunoblotting with anti-K18 and anti-actin antibodies. (B) Confocal images of CFP-K8 and F-actin. MDCK/CFP-K8 cells were transfected with control or K18-targeting siRNAs, cultured for 48 h, fixed, and stained with rhodamine–phalloidin. Scale bars, 20 μm. (C, D) Effects of K18 knockdown on the organization of K8 networks (C) and stress fiber formation (D), analyzed as in Figure 2. The data shown represent the mean ± SD of three independent experiments (at least 12 cells/experiment). (E) Effect of K18 knockdown on force-induced stress fiber formation, analyzed as in Figure 5D. See also Supplemental Figure S5 and Supplemental Movie S5. The data shown represent the mean ± SD of 17–21 cells/experiment. **p < 0.01, ***p < 0.001, and ****p < 0.0001 (one-way ANOVA followed by Dunnett’s test).
K18 is required for tensional force–induced stress fiber reinforcement
To understand the role of K8/K18 filaments in the mechanical force–induced cell response, we examined the effects of K18 knockdown on tensional force–induced stress fiber formation. We treated MDCK/YFP-Lifeact cells with control or K18-targeting siRNAs and analyzed tensional force–induced stress fiber formation as shown in Figure 5A. Time-lapse fluorescence analysis showed that K18 knockdown significantly decreased the number of force-induced stress fibers that were reinforced or newly emerged after force application (Figure 6E, Supplemental Figure S5, and Supplemental Movie S5). These results suggest that K8/K18 filaments are required for tensional force–induced stress fiber formation/reinforcement.
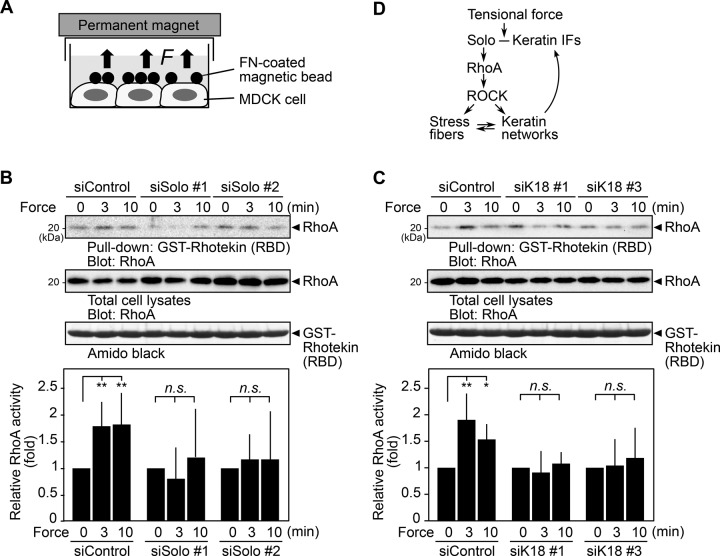
Solo and K18 are required for force-induced RhoA activation
Previous studies showed that tensional force application increases RhoA activity in cells (Zhao et al., 2007; Guilluy et al., 2011). To examine the role of Solo in force-induced RhoA activation, we analyzed the effect of Solo knockdown on tensional force-induced RhoA activation. We cultured MDCK cells treated with control or Solo-targeting siRNAs until they were nearly confluent and serum starved and then applied FN-coated magnetic beads to the cells. Then we applied tensional force to the cells by placing a permanent magnet on top of the culture dish (Figure 7A). The active RhoA pull-down assays using GST-RBD revealed that active RhoA was significantly (1.8- to 1.9-fold) increased 3–10 min after tensional force application in control siRNA cells (Figure 7B). The levels of RhoA activation were similar to those induced by the treatment of cells with 1 μM lysophosphatidic acid (LPA; Supplemental Figure S6). By contrast, treatment of MDCK cells with Solo-targeting siRNAs almost completely suppressed tensional force–induced RhoA activation (Figure 7B), which suggests that Solo plays a crucial role in tensional force–induced RhoA activation in MDCK cells.
FIGURE 7:
Solo and K18 are required for tensional force–induced RhoA activation. (A) Method of tensile force application. The FN-coated magnetic beads were placed on MDCK cells. Tensile force was applied by placing a permanent magnet on the top of the dish. (B, C) Effects of knockdown of Solo (B) or K18 (C) on tensile force–induced RhoA activation. Active RhoA was analyzed by GST pull-down assays using GST-rhotekin (RBD). Relative RhoA activity is shown as the mean ± SD of at least three independent experiments, with the activity at zero time set as 1.0. *p < 0.05 and **p < 0.01; n.s., not significant (one-way ANOVA followed by Dunnett’s test). (D) Model for the role of Solo and keratin intermediate filaments (IFs) in tensional force–induced RhoA activation and stress fiber formation.
Because K18 knockdown disrupted K8/K18 networks and suppressed tensile force–induced stress fiber formation, K8/K18 networks appear to be involved in mechanotransduction. We therefore examined the effect of K18 knockdown on tensional force–induced RhoA activation. We treated MDCK cells with K18-targeting siRNAs and subjected them to force application assays as shown in Figure 7A. The active RhoA pull-down assays revealed that K18 knockdown almost completely suppressed force-induced RhoA activation (Figure 7C), which suggests that K8/K18 networks are required for tensional force–induced RhoA activation.
DISCUSSION
We provided evidence that Solo plays a crucial role in actin and keratin cytoskeletal organization. Knockdown of Solo or treatment with the ROCK inhibitor Y-27632 suppressed stress fiber formation and led to disorganized keratin networks, whereas overexpression of Solo enhanced formation of thick stress fibers and keratin bundles. Because Solo is a RhoA-targeting GEF, these results indicate that Solo promotes stress fiber formation and proper organization of keratin networks through activation of the Rho-ROCK pathway. We also showed that Solo physically associates with K8/K18 filaments and colocalizes with K8/K18 filaments on the ventral side of the cell. Of note, knockdown of Solo or K18 suppressed tensile force–induced RhoA activation and stress fiber formation. The suppressive effect of Solo knockdown on force-induced stress fiber formation was recovered by expression of Solo-WT but not mitochondrial targeting Solo. Furthermore, expression of a GEF-inactive mutant or deletion mutants of Solo suppressed force-induced stress fiber formation. On the basis of these observations, we propose a model for the role of Solo and K8/K18 filaments in mechanical force–induced RhoA activation and stress fiber formation (Figure 7D). In this model, the tensile force induces activation of Solo and its downstream target RhoA. Because depletion of K18 suppresses force-induced RhoA activation, the association with K8/K18 filaments likely plays an important role in force-induced Solo activation (see later discussion). Activated RhoA stimulates downstream effectors, such as ROCK, and then promotes the formation of stress fibers and keratin networks. Because the Solo-RhoA-ROCK pathway serves to organize keratin networks and keratin networks serve to activate Solo, Solo and keratin filaments can generate a positive feedback loop by which local activation of Solo and RhoA and accumulation of actin stress fibers and keratin filaments are attainable at the sites of mechanical force loading (Figure 7D). Local activation of Solo and reinforcements of actin stress fibers and keratin fibers at cell adhesion sites probably contribute to the protection of cells from external forces.
K8 and K18 are the major keratin pair in simple epithelia and play important roles in the maintenance of cell and tissue integrity during mechanical stress (Coulombe and Wong, 2004; Loschke et al., 2015). A recent study showed that knockdown of K8 causes inactivation of RhoA and suppression of stress fiber formation (Bordeleau et al., 2012). In this study, we provided evidence that K8/K18 filaments are required for force-induced RhoA activation and stress fiber formation, indicating that organized K8/K18 networks are necessary for cells to sense and respond to mechanical forces. Because keratin filaments form networks throughout epithelial cells and are anchored to the cell–cell and cell–ECM adhesion sites via desmosomes and hemidesmosomes, it is likely that mechanical force application induces deformation of keratin networks. Thus K8/K18 filaments are likely involved in triggering mechanotransduction by deforming their networks in response to mechanical forces.
Solo, when overexpressed, localizes at cell–cell and cell–ECM adhesion sites (Abiko et al., 2015). This study showed that, in Solo-overexpressing cells, Solo colocalizes with K8 fibers at the ventral side of cells. In addition, knockdown of Solo suppressed force-induced RhoA activation when force was applied via cadherin-coated beads (Abiko et al., 2015) or fibronectin-coated beads (shown in this study). These results suggest that Solo is involved in force-induced RhoA activation and cytoskeletal remodeling at both cell–cell and cell–ECM adhesion sites.
Depletion of Solo or treatment with Y-27632 caused marked disruption of K8/K18 networks, implying a role for the Solo-RhoA-ROCK pathway in maintaining the integrity of keratin networks. However, the mechanisms by which this pathway regulates keratin network organization remain unknown. Previous studies showed that incubation with pemphigus autoantibodies against desmosomal cadherins or overexpression of the desmosomal protein kazrin causes desmosome disassembly by reducing RhoA activity (Waschke et al., 2006; Sevilla et al., 2008), suggesting that RhoA is involved in keratin network organization by maintaining desmosome assembly and/or anchoring keratin filaments to desmosomes. This study showed that Solo knockdown markedly reduces the localization of plakoglobin at cell–cell adhesion sites. Considering that plakoglobin is a component of the desmosome, these results suggest that knockdown of Solo might cause the disorganization of K8/K18 networks, at least in part, by reducing the localization of plakoglobin to desmosomes and thereby suppressing the anchoring of keratin filaments to desmosomes. On the other hand, it has also been reported that keratin filament dynamics are regulated by actin filaments (Kölsch et al., 2009; Windoffer et al., 2011). Therefore it is possible that the Solo-RhoA pathway controls keratin networks via promotion of actin remodeling. It is also conceivable that ROCK directly regulates keratin filament organization by phosphorylation, although phosphorylation of intermediate filaments usually reduces their stability (Omary et al., 2006; Chung et al., 2013). Further studies are required to better understand the mechanism of RhoA-mediated control of keratin network organization.
The mechanism of force-induced Solo activation remains elusive. We showed that Solo has multiple keratin-binding sites and that overexpression of each of the keratin-binding fragments of Solo suppressed force-induced stress fiber formation. This suggests that each fragment of Solo has a dominant-negative effect against endogenous Solo and that Solo binding to keratin filaments via multiple sites is critical for its function in force-induced stress fiber formation. We previously showed that full-length Solo expressed in cells induces RhoA activation but its C-terminal fragment (1058–1519) does not (Abiko et al., 2015), indicating that the N-terminal region is required for its RhoA-GEF activity. Taking into account these findings, it is conceivable that Solo is activated by conformational change, mediated by force-induced deformation of K8/K18 networks, to which Solo binds through multiple sites. It is also possible that Solo is activated by force signal–induced modifications, such as phosphorylation, or association with as-yet-unidentified Solo-activating factors.
This study demonstrates the importance of the interplay between Solo, RhoA, keratin filaments, and actin stress fibers in enabling cells to sense and respond to mechanical forces. Quattro, a zebrafish orthologue of Solo, was shown to play a critical role in convergent extension cell movements during gastrulation of zebrafish embryos (Daggett et al., 2004). Because convergent extension is a process requiring mechanosensing (Lecuit et al., 2011), it is likely that Solo has a physiological function in the transduction of mechanical signals in this organism. Solo is also required for stretch-induced reorientation of human vascular endothelial cells (Abiko et al., 2015), indicating that the function of Solo in mechanotransduction is conserved between zebrafish and humans. A better understanding of the mechanisms of Solo-mediated mechanotransduction in cells and tissues will be important for elucidating the cooperative roles of actin and keratin cytoskeletons in mechanosensing and mechanotransduction.
MATERIALS AND METHODS
Reagents and antibodies
FLAG peptide was purchased from Sigma-Aldrich (St. Louis, MO), recombinant K8 and K18 proteins were purchased from Progen (Darra, Australia), rhodamine-labeled phalloidin was purchased from Wako Pure Chemical Industries (Osaka, Japan), Alexa Fluor 568–labeled phalloidin was purchased from Life Technologies (Grand Island, NY), LPA was purchased from Cayman Chemical (Ann Arbor, MI), and Cytopainter MitoRed Indicator was purchased from Abcam (Cambridge, United Kingdom). Rabbit polyclonal antibodies against human Solo, which also recognized dog Solo, were prepared as previously described (Abiko et al., 2015). Other antibodies were purchased as follows: anti-K18 (DA-7; BioLegend, San Diego, CA) for HeLa cells, anti-K18 (Ks 18.04; Progen, Heidelberg, Deutschland) for MDCK cells, anti-FLAG (M2; Sigma-Aldrich), anti-GFP (A-6455; Life Technologies, Camarillo, CA), anti–β-actin (AC-15; Sigma-Aldrich), anti–β-catenin (Clone 14; BD Biosciences, Franklin Lakes, NJ), anti-plakoglobin (Clone 15; BD Biosciences), and anti-RhoA (sc-418; Santa Cruz Biotechnology, Dallas, TX).
Plasmid construction and siRNAs
The expression plasmid encoding YFP-Solo was constructed as described previously (Abiko et al., 2015). Expression plasmids for CFP-, FLAG-, and Halo-tagged Solo were constructed by inserting the Solo cDNA into the pECFP-C1 (Clontech, Mountain View, CA), pFLAG-C1, and pHaloTag-Hyg-C1 vectors. The pFLAG-C1 vector was constructed by replacing the YFP cDNA with the FLAG epitope sequence in the pEYFP-C1 vector (Clontech). The pHaloTag-Hyg-C1 vector was constructed by replacing the AcGFP cDNA with the Halo cDNA (Promega, Madison, WI) in the pAcGFP1-Hyg-C1 vector (Clontech). The plasmid encoding Solo-LE, in which Leu-1217 in the DH domain was replaced by Glu, was constructed using a site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The plasmids encoding deletion mutants of Solo were constructed by PCR amplification and subcloning into the pECFP-C1 vector. To construct expression plasmids encoding YFP- and CFP-K8, the cDNA encoding human K8 was amplified by PCR using the MegaMam Human Transcriptome Library (Agilent Technologies) and inserted into the pEYFP-C1 and pECFP-C1 vectors, respectively. To construct expression plasmid encoding Tom20(1-33)-CFP-Solo, the cDNA encoding human Tom20(1-33) was amplified by PCR using the MegaMam Human Transcriptome Library and inserted into the CFP-Solo expression plasmid in the pECFP-C1 vector. The primer set for amplifying Tom20(1-33) was designed as previously reported (Komatsu et al., 2010). The siRNAs targeting dog Solo and K18 were designed using the siDirect website (http://sidirect2.rnai.jp) and purchased from Sigma-Aldrich. The sequences of siRNAs are listed in Supplemental Table S1.
Cell culture and transfection
MDCK, HeLa, COS-7, and MCF-7 cells were cultured in DMEM supplemented with 10% fetal bovine serum. MDCK cells stably expressing CFP-K8, YFP-K8, or YFP-Lifeact were established using G418-containing medium for selection. Cells were transfected with plasmids and siRNAs using Lipofectamine LTX and RNAiMAX (Life Technologies), respectively. The siRNAs were used at a final concentration of 50 nM.
Proteomic analysis
MCF-7 cells stably expressing control Halo or Halo-Solo were established using hygromycin B–containing medium for selection. Cells were lysed with lysis buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 8.0, 150 mM NaCl, 1 mM dithiothreitol [DTT], 5% glycerol, 1 mM MgCl2, 1 mM CaCl2, protease inhibitor cocktail [Promega], PhosSTOP phosphatase inhibitor cocktail [Roche Diagnostics, Indianapolis, IN], 10 μg/ml DNase I, and 10 μg/ml RNase A). After sonication, cell lysates were centrifugation at 880 × g for 3 min, and the supernatants were incubated with Halo ligand–conjugated magnetic beads (Promega) at 4°C for 2 h. Beads were sequentially washed with lysis buffer, PBS, and wash buffer (50 mM HEPES, pH 8.0, 5% glycerol, 1 mM MgCl2, 1 mM CaCl2) containing 0.3 and 0.5 M NaCl. Bound proteins were eluted with wash buffer containing 1 M NaCl and subjected to SDS–PAGE. Protein bands were excised and treated with trypsin, and recovered peptides were analyzed by matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry (TOF/TOF 5800 system; AB Sciex, Framingham, MA). Proteins were identified using MS-Fit software (http://prospector.ucsf.edu/prospector/mshome.htm).
Coimmunoprecipitation assay
HeLa cells transfected with YFP-Solo or its mutants were lysed with ice-cold lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM DTT, 0.25 μM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 2 μg/ml pepstatin). Cell lysates were clarified by centrifugation at 18,000 × g for 10 min and incubated 2 h at 4°C with an anti-GFP antibody. Immunoprecipitates were analyzed by immunoblotting with an anti-K18 antibody.
K8/K18 cosedimentation assay
FLAG-Solo or its deletion mutant proteins expressed in COS-7 cells were purified by immunoprecipitation with an anti-FLAG antibody. FLAG-Solo proteins were eluted by incubation with 30 mM Tris-HCl (pH 8.0) containing 200 μg/ml FLAG peptide for 30 min on ice. K8/K18 filaments were assembled using the low-Tris buffer method (Herrmann et al., 2002). Recombinant K8 and K18 proteins were dissolved in 30 mM Tris-HCl (pH 8.0), 9.5 M urea, 2 mM DTT, 2 mM EDTA, and 10 mM methylammonium chloride according to the manufacturer’s protocol. Equimolar K8 and K18 were mixed and dialyzed into the dialysis buffer (10 mM Tris-HCl, pH 8.8, 2 mM DTT) at 4°C by lowering the urea concentration in a stepwise manner (4, 2, and 0 M). Finally, the mixture was dialyzed into low-Tris buffer (2 mM Tris-HCl, pH 8.8, 2 mM DTT) at 4°C overnight. Filament assembly was initiated by addition of an equal volume of 2× assembly buffer (20 mM Tris-HCl, pH 7.0, 2 mM MgCl2) at room temperature for 1 h. Purified FLAG-Solo proteins were incubated with 5 μg of K8/18 filaments on ice for 30 min and then centrifuged at 18,000 × g for 30 min. Equal portions of the supernatants and precipitates were subjected to SDS–PAGE and analyzed by Amido black staining.
Immunofluorescence staining and fluorescence imaging
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 30 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min. After washing with PBS, F-actin was stained with rhodamine- or Alexa Fluor 568–labeled phalloidin. For visualizing mitochondria, cells were incubated with Cytopainter MitoRed Indicator before fixation according to the manufacturer’s protocol. Fluorescence images were obtained using an LSM 510 laser scanning confocal microscope (Carl Zeiss, Jena, Germany) or a high-resolution confocal microscope TCS SP8 (Leica Microsystems, Wetzlar, Germany) equipped with a PL-Apo 63× oil objective lens (numerical aperture [NA] 1.4). TIRF images were obtained using a Leica SR-GSD microscope (Leica Microsystems) equipped with 488-nm lasers, using a 100× oil immersion objective lens. Images were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD).
Time-lapse analysis of force-induced stress fiber formation
Polydimethylsiloxane (PDMS; Silpot 184; Dow Corning, Midland, MI) was prepared by mixing the prepolymer and cross-linker at a 10:1 (vol/vol) ratio. The mixture was spread over a hole (18-mm diameter) in a 35-mm glass-bottomed dish and cured in an oven at 56°C overnight to make a PDMS membrane mat. The PDMS surface was treated with 10% (vol/vol) of (3-aminopropyl)-trimethoxysilane (Sigma-Aldrich) in water for 2 h at room temperature and coated with 2 μg/cm2 FN in PBS at 37°C overnight. MDCK/YFP-Lifeact cells were cultured on the FN-PDMS–coated dishes and transfected with plasmids or siRNAs. After incubation for 48 h, cells were subjected to time-lapse analysis using an LSM 710 confocal microscope equipped with an EC Plan N 40× oil objective lens (NA 1.3), a definite-focus module of AxioObserver, and a temperature hood to keep the stage at 37°C. A glass needle (FemtotipsII; Eppendorf, Hamburg, Germany) was controlled using a Micromanipurator 5171 (Eppendorf). A glass needle was inserted into the PDMS membrane within 5 μm from the cell periphery, held for 30 s, and then moved 2–5 μm away from the cell. Fluorescence and bright-field images were acquired every 5 s for 5 min with a 514 nm argon-ion laser. Image analysis was performed using ImageJ software. Stress fibers that were newly generated or enhanced in the direction of the force within 5 min of force application were counted. Because the cell region adjacent to the site of force application became out of focus, we counted stress fibers in the cell region at the distance of >10 μm from the site of force application. To exclude the random fluorescence changes of Lifeact, only stress fibers whose fluorescence was persistently enhanced for >10 s were counted.
Preparation of magnetic particles and force application
Spherical magnetic beads of 2.8-μm diameter (Dynabeads M-280 Tosylactivated; Life Technologies) were washed with phosphate buffer and incubated with 100 μg/ml FN (Sigma-Aldrich) at 37°C for 24 h. The FN-coated magnetic beads were washed three times with PBS containing 0.1% bovine serum albumin. MDCK cells were cultured on 35-mm dishes, transfected with Solo- or K18-targeting siRNAs, and cultured for 18 h in growth medium. Then cells were cultured at low serum medium (0.1% serum/DMEM) for 8 h and serum starved with medium without serum for 16 h before bead treatment. The FN-coated magnetic beads were sonicated, and 0.25 mg of beads was incubated with cells in a 35-mm dish for 60 min. Force application was conducted as previously reported (Zhao et al., 2007; Guilluy et al., 2011). A ceramic permanent magnet (DynaMag-2; Life Technologies) was used to generate perpendicular, tensile forces on beads attached to the dorsal surface of cells. The manufacturer’s value for the magnetization is an average of 4000 G. For all experiments, the pole face was parallel with and 1 cm from the culture dish surface.
GST-RBD pull-down assay
The active form of RhoA was detected by pull-down assays using the RBD of rhotekin fused to GST, as described previously (Ren and Schwartz, 2000). To analyze the GEF activity of Solo (WT or LE), CHO-K1 cells were transfected with YFP-Solo (WT or LE) or CFP-LARG (as a positive control) and cultured for 14 h in growth medium. Then cells were serum starved for 10 h, and cell lysates were subjected to pull-down assays. To analyze force-induced RhoA activation, tensional force was applied to serum-starved MDCK cells for 3–10 min, and cell lysates were subjected to pull-down assays. To analyze LPA-induced RhoA activation, MDCK cells were cultured onto 35-mm dishes in growth medium for 10 h and serum starved for 18 h. Cells were then stimulated with LPA at a final concentration of 1 μM, and cell lysates were subjected to pull-down assays.
Statistical analysis
Data are expressed as the mean ± SD of more than three independent experiments. Statistical analysis was performed using Prism 4 (GraphPad Software). The p values were calculated using a one-way analysis of variance (ANOVA) followed by Dunnett’s test, Tukey’s test, or a two-tailed paired t test. Statistical significance was set at p < 0.05.
Supplementary Material
Acknowledgments
We thank Masaaki Sato (Tohoku University, Sendai, Japan), Shinji Deguchi, and Tsubasa Matsui (Nagoya Institute of Technology, Nagoya, Japan) for helpful advice, Nahoko Higashitani and Atsushi Higashitani (Tohoku University) for advice on proteomics analysis, Kota Miyasaka and Toshihiko Ogura for TIRF analysis, and Kazuyasu Shoji and Shota Takahashi for technical assistance. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K.O. (Grants 23112005 and 25440076) and K.M. (Grants 24370051 and 26102505) and support from the Mitsubishi Foundation and the Uehara Memorial Foundation to K.M.
Abbreviations used:
- CFP
cyan fluorescent protein
- DH
Dbl homology
- DTT
dithiothreitol
- ECM
extracellular matrix
- FN
fibronectin
- GEF
guanine nucleotide exchange factor
- GST
glutathione-S-transferase
- K8
keratin-8
- K18
keratin-18
- LPA
lysophosphatidic acid
- MDCK
Madin–Darby canine kidney
- PBS
phosphate-buffered saline
- PDMS
polydimethylsiloxane
- PH
pleckstrin homology
- RBD
RhoA-binding domain
- ROCK
Rho-associated kinase
- siRNA
small interfering RNA
- TIRF
total internal reflection fluorescence
- WT
wild type
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-06-0417) on January 28, 2016.
REFERENCES
- Abiko H, Fujiwara S, Ohashi K, Hiatari R, Mashiko T, Sakamoto N, Sato M, Mizuno K. Rho guanine nucleotide exchange factors involved in cyclic-stretch-induced reorientation of vascular endothelial cells. J Cell Sci. 2015;128:1683–1695. doi: 10.1242/jcs.157503. [DOI] [PubMed] [Google Scholar]
- Bordeleau F, Myrand Lapierre ME, Sheng Y, Marceau N. Keratin 8/18 regulation of cell stiffness-extracellular matrix interplay through modulation of Rho-mediated actin cytoskeleton dynamics. PLoS One. 2012;7:e38780. doi: 10.1371/journal.pone.0038780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BM, Rotty JD, Coulombe PA. Networking galore: intermediate filaments and cell migration. Curr Opin Cell Biol. 2013;25:600–612. doi: 10.1016/j.ceb.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe PA, Wong P. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat Cell Biol. 2004;6:699–706. doi: 10.1038/ncb0804-699. [DOI] [PubMed] [Google Scholar]
- Cowin P, Kapprell HP, Franke WW, Tamkun J, Hynes RO. Plakoglobin: a protein common to different kinds of intercellular adhering junctions. Cell. 1986;46:1063–1073. doi: 10.1016/0092-8674(86)90706-3. [DOI] [PubMed] [Google Scholar]
- Curtis C, Hemmeryckx B, Haataja L, Senadheera D, Groffen J, Heisterkamp N. Scambio, a novel guanine nucleotide exchange factor for Rho. Mol Cancer. 2004;3:10. doi: 10.1186/1476-4598-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett DF, Boyd CA, Gautier P, Bryson-Richardson RJ, Thisse C, Thisse B, Amacher SL, Currie PD. Developmentally restricted actin-regulatory molecules control morphogenetic cell movements in the zebrafish gastrula. Curr Biol. 2004;14:1632–1638. doi: 10.1016/j.cub.2004.08.024. [DOI] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyckmans J, Boudou T, Yu X, Chen CS. A hitchhiker’s guide to mechanobiology. Dev Cell. 2011;21:35–47. doi: 10.1016/j.devcel.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Guilluy C, Swaminathan V, Garcia-Mata R, O’Brien ET, Superfine R, Burridge K. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat Cell Biol. 2011;13:722–727. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Wedig T, Porter RM, Lane EB, Aebi U. Characterization of early assembly intermediates of recombinant human keratins. J Struct Biol. 2002;137:82–96. doi: 10.1006/jsbi.2002.4466. [DOI] [PubMed] [Google Scholar]
- Huveneers S, de Rooij J. Mechanosensitive systems at the cadherin-F-actin interface. J Cell Sci. 2013;126:403–413. doi: 10.1242/jcs.109447. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölsch A, Windoffer R, Leube RE. Actin-dependent dynamics of keratin filament precursors. Cell Motil Cytoskeleton. 2009;66:976–985. doi: 10.1002/cm.20395. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Lenne PF, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- Lessey EC, Guilluy C, Burridge K. From mechanical force to RhoA activation. Biochemistry. 2012;51:7420–7432. doi: 10.1021/bi300758e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschke F, Seltmann K, Bouameur JE, Magin TM. Regulation of keratin network organization. Curr Opin Cell Biol. 2015;32C:56–64. doi: 10.1016/j.ceb.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary M, Ku NO, Tao GZ, Toivola D, Liao J. “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem Sci. 2006;31:383394. doi: 10.1016/j.tibs.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Osmani N, Labouesse M. Remodeling of keratin-coupled cell adhesion complexes. Curr Opin Cell Biol. 2015;32:30–38. doi: 10.1016/j.ceb.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Ren XD, Schwartz MA. Determination of GTP loading on Rho. Methods Enzymol. 2000;325:264–272. doi: 10.1016/s0076-6879(00)25448-7. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla LM, Nachat R, Groot KR, Watt FM. Kazrin regulates keratinocyte cytoskeletal networks, intercellular junctions and differentiation. J Cell Sci. 2008;121:3561–3569. doi: 10.1242/jcs.029538. [DOI] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Lappalainen P. Actin stress fibers—assembly, dynamics and biological roles. J Cell Sci. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- Tse SW, Broderick JA, Wei ML, Luo MH, Smith D, McCaffery P, Stamm S, Andreadis A. Identification, expression analysis, genomic organization and cellular location of a novel protein with a RhoGEF domain. Gene. 2005;359:63–72. doi: 10.1016/j.gene.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Wahl JK, 3rd, Nieset JE, Sacco-Bubulya PA, Sadler TM, Johnson KR, Wheelock MJ. The amino- and carboxyl-terminal tails of b-catenin reduce its affinity for desmoglein 2. J Cell Sci. 2000;113:1737–1745. doi: 10.1242/jcs.113.10.1737. [DOI] [PubMed] [Google Scholar]
- Walcott S, Sun SX. A mechanical model of actin stress fiber formation and substrate elasticity sensing in adherent cells. Proc Natl Acad Sci USA. 2010;107:7757–7762. doi: 10.1073/pnas.0912739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschke J, Spindler V, Bruggeman P, Zillikens D, Schmidt G, Drenckhahn D. Inhibition of Rho A activity causes pemphigus skin blistering. J Cell Biol. 2006;175:721–727. doi: 10.1083/jcb.200605125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Beil M, Magin TM, Leube RE. Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J Cell Biol. 2011;194:669–678. doi: 10.1083/jcb.201008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Zhao XH, Laschinger C, Arora P, Szászi K, Kapus A, McCulloch CA. Force activates smooth muscle alpha-actin promoter activity through the Rho signaling pathway. J Cell Sci. 2007;120:1801–1809. doi: 10.1242/jcs.001586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.