Abstract
Atopic dermatitis (AD) is a chronic multifactorial inflammatory skin disease. The pathogenesis of AD remains unclear, but the disease results from dysfunctions of skin barrier and immune response, where both genetic and environmental factors play a key role. Recent studies demonstrate the substantial evidences that show a strong genetic association with AD. As for example, AD patients have a positive family history and have a concordance rate in twins. Moreover, several candidate genes have now been suspected that play a central role in the genetic background of AD. In last decade advanced procedures similar to genome-wide association (GWA) and single nucleotide polymorphism (SNP) have been applied on different population and now it has been clarified that AD is significantly associated with genes of innate/adaptive immune systems, human leukocyte antigens (HLA), cytokines, chemokines, drug-metabolizing genes or various other genes. In this review, we will highlight the recent advancements in the molecular genetics of AD, especially on possible functional relevance of genetic variants discovered to date.
Keywords: Atopic dermatitis, molecular genetics, immune genes, cytokine, chemokine, drug-metabolizing genes
Introduction
Atopic dermatitis (AD) is a very frequent multifactorial chronic inflammatory skin disorder and is characterized by xerosis, pruritus and erythematous lesions with increased transepidermal water loss. (1) Despite the power of molecular approaches and persistent investigative efforts, AD remains an enigmatic disorder and the agent (or agents) triggering this skin disorder remains to be completely identified. AD is thought to be associated with the dysfunctioning of skin barrier and Th2 cell adaptive immune responses to common environmental allergens. It is a disease with complex genetic and environmental susceptibility factors. Although it is likely that many genetic loci are involved, the association of filaggrin (FLG) null mutations with AD has provided a major step forward in our understanding of disease pathogenesis. (2) FLG is expressed in keratinocytes and is thought to have a role in skin barrier function, cutaneous pH, and hydration. (3) Several clinical phenotypes of AD have now been discovered, but they exhibit great variations in disease severity among affected individuals. (4) Moreover, some cases have the atopic march, while others are susceptible to skin infections such as staphylococcus aureus, eczema herpeticum and malassezia. (4) Approximately 80% of AD patients have raised serum of immunoglobulin E (IgE) and/or immediate skin test reactivity to allergens. (5–7) Advances in genomic medicine have improved our understanding of the human genome’s contribution to health and disease. Genome wide association studies (GWAS) are proved to be a powerful method for identifying disease susceptibility genes for common human diseases and has begun to reveal underlying cellular pathways and point to new therapeutic approaches. (8) This review focuses on the recent advancement of the molecular genetics of atopic dermatitis, especially on the genes located within and around the susceptible loci and their possible roles in pathogenesis of atopic dermatitis. Before beginning of discussion on the molecular genetics, we will highlight the factors that influence on the skin barrier abnormalities.
Factors influence on skin barrier dysfunctions
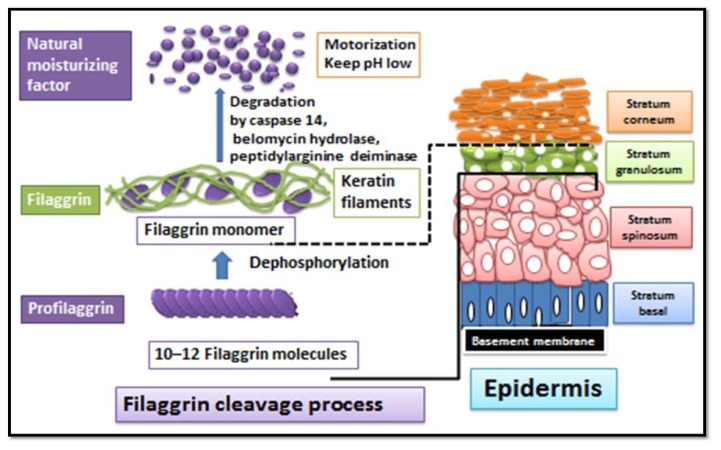
It has now been well established that FLG derived natural moisturizing factors (NMFs) are important in maintenance of epidermal water and low acidity thus preserving the barrier function of outermost stratum corneum. (9) FLG is involved in the development of keratinocytes to maintain epidermal integrity and it is an important marker of keratinocytes differentiation. During keratinocytes differentiation, profilaggrin is dephosphorylated and degraded into 10–12 FLG molecules, which condense in the cytoskeleton of keratin to form an intensive protein-lipid matrix. Consequently, these FLG monomers are degraded into NMFs, which are important to maintain skin water, a low pH or maintaining the barrier function of the stratum corneum (Fig. 1).
Figure 1. Cleavage of profilaggrin into natural moisturizing factors via degradation of filagrgrin molecules.
Filagrgrin is involved in the development of keratinocytes to maintain epidermal integrity and it is an important marker of keratinocytes differentiation. (after Kabashima, 2013 (9)).
Recently, it has been reported that intragenic copy number variation (20–24 copies in one person) within a FLG gene contributes to the risk of AD with a dose-dependent effect. (10) Profilaggrin may prevent the extracellular secretion of lamellar bodies, as contain lipids, and may block or decrease the degradation of FLG, which results in reduce in the production of NMFs. Reduced production of acidic metabolites of FLG results an increase of skin surface pH or activating neutral pH dependent kallikreins with downstream influences on skin barrier function. (11, 12) An increased pH may also decrease the β-glucocerebrosidase activity and sharing lipid-processing defect may delay epidermal barrier recovery. (11–13) Also, the elevated pH takes place a decrease in acidic sphingomyelinase catalytic activity and causes degradation of β-glucocerebrosidase and acidic sphingomyelinase. (11–13) These enzymes playing main role in ceramide synthesis. Ceramides are essential lipid moieties that are included in preserving epidermal permeability barriers. Thereby, a change in epidermal surface pH may regulate homeostasis of the epidermal permeability barrier, as well as stratum corneum integrity and cohesion. The following are the factors influence on FLG expression in AD. (A) Loss-of-function mutations of FLG result in decreased or totally absent levels of FLG. (14–16) The prevalence of FLG mutation in Northern European subjects about 10% whereas in Chinese, Japanese, and Korean subjects range from 3% to 6%, (16)however, in certain Northern European populations with AD, the prevalence of FLG mutations is reach to 25%: 50%. (2) Consistently focusing in the research of AD for more than 20 year on genetic factors, recent strong role of a genetically predetermined skin barrier disturbance came to a large extent from the discovery the linkage between the development of AD and loss-of-function mutations in the gene encoding FLG. (14–16) (B) Expression of epidermal FLG is down regulated by cytokines IL-4, IL-13, IL-17A, IL-22, IL-25, IL-31, TNF-α in AD patients. (17–23) (C) Effect of environmental factors which lead to FLG proteolysis such as low ambient humidity, skin irritants, pruritus / excoriations / mechanical damage, age and water. (24, 25) Later will be reviewed the genetic variation of FLG.
Study on single nucleotide polymorphism of the claudin-1 gene (CLDN1) in AD patients demonstrated that tight junctions contribute to the barrier dysfunction and immune dysregulation in AD patients and this may be mediated in part by reductions in tight junction protein claudin-1. (26) In addition, it is also reported that activation of toll like receptor (TLR)-2 plays a role on tight junction barrier via the protein kinase in the tight junction biogenesis. (27) Moreover, IL-4 and IL-13 also promote keratinocyte CLDN-1 expression. (27) Thus TLR-2 activation enhanced expression of CLDN-1 in keratinocytes of AD patients. In addition, TLR-2 lacking mice showed delayed or incomplete barrier recovery. (28) These genetic modifications of TLR-2 might lead to barrier dysfunctions and predispose in AD patients.
Evidences of the genetics role in atopic dermatitis
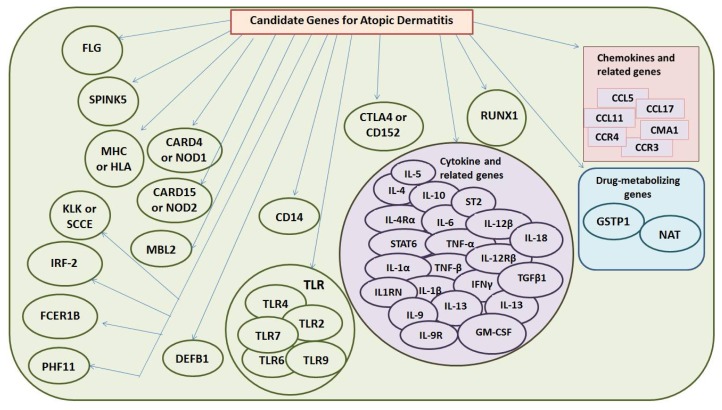
Recent advancements in clinical experience and molecular reseach on atopic dermatitis have strongly been influenced by genetically alterations. (29) All identified major candidate genes for AD pathogenesis has been listed in Figure 2. Studies on twin have provided the role of a genetic background with a concordance rate of 72–86 % in identical twins and 21–23 % in dizygotic twins, indicating high heritability of AD. (30, 31) In addition multiple studies on affected individuals and their families have a positive family history of AD. However the heritability of AD was determined at 72 % by study on Norwegian twin. (31) Recently, a total of 19 susceptibility loci have been estimated at a genome-wide level of significance. Contribution of such these genetic factors may affect diverse phenotypes of AD among individuals. Hence the disease seems to be caused by genetic factor that lead to an immune system deregulation. However deficiencies in innate and adaptive immunity based on, endogenous factor as genetic predisposition result in skin barrier dysfunction and exogenous factor as hyper reactivity to environmental stimuli and susceptibility to skin infections which influence the course and severity of AD. In fact the pathogenesis of AD has been attributed to a complex and multifactorial interactions of the environment and host susceptibility genes, altered skin barrier function, the immune system, and pruritus. (6)
Figure 2. Identified candidate genes in atopic dermatitis.
FLG, Filaggrin; SPINK5, Serine Protease Inhibitor Kazal-Type 5 ; MHC, major histocompatibility complex; HLA, human leukocyte antigen; CARD4, Caspase recruitment domain-containing protein 4; NOD, nucleotide binding oligomerization domain protein; CD, cluster of differentiation; MBL2, mannose binding lectin-2; TLR, toll like receptor; DEFB1, human defernsin 1; IL, interleukin; IL-4Rα, interleukin 4 receptor alpha; STAT6, signal transducer and activator of transcription-6; TNF, tumor necrosis factor; IFNγ, interferon gamma; IL1RN, interleukin 1 receptor antagonist; ST2, suppression of tumorigenicity-2; TGF, transforming growth factor; GM-CSF, granulocyte macrophage colony stimulating factor; CCL, chemokine C-motif ligand; CCR, chemokine C motif receptor; CMA1; mast cell chymase 1; GSTP1, clutathionine S-transferase P1; NAT, N-acetyl transferase; CTLA4, cytotoxic T-lymphocyte associated antigen-4; KLK, kallikrein; RUNX1, runt-related transcription factor 1; TRF-2, interferon regulatory factor 2; FCER1B, high affinity IgE receptor beta chain; PHF11, plant homeodomain zink finger 11 protein 11.
Molecular genetic tools of atopic dermatitis
In progress of biotechnology and molecular biology, more candidate genes have now been determined to be associated with AD. According to the available knowledge, the genes that are related to the structural abnormalities of the epidermis and immune dysregulation play a pivotal role in the etiology of AD. (32) Human genetic variants are either common or rare, common variants, known as polymorphisms and are defined as variants with a minor allele frequency of at least 1% within the population. (30, 31) Single nucleotide polymorphisms (SNP) are the most common uses for determining of genetic variations within individuals. (33) Recently GWAS comprehensively surveys associations between SNP in common diseases. (34) Genome-wide linkage analysis had been used in different populations to date, and multiple candidate regions on multiple chromosomes had been associated with AD. (35–39) The linkage regions vary in different populations, and there is no extensive overlap among studies. Some candidate regions are close to functional genes linked to the various phenotypes of AD. Systemic, well-powered, genome-wide surveys using GWAS and immunochip analyses have estimated the relationship between SNP and susceptibility to AD. (31, 39, 40) It is well established that common loss of function variants of FLG are a major predisposing factor for AD. As for example, the FLG mutations are involved in AD and are featured in some review articles. (31, 42–45) The AD associated major candidate genes and their chromosomal location or genetic variations have been summarized in Table 1. The following genes play a central role in the pathogenesis of AD and now have been offered a several novel potential therapeutic opportunities for AD.
Table 1.
Atopic dermatitis associated candidate genes and their chromosomal location and genetic variation in different population.
| Gene Symbol | Gene name | Chromosomal location | Genetic variation | Population | References |
|---|---|---|---|---|---|
| FLG | Filaggrin | 1q21 | R501X-2282del4 | Caucasian Northern-American European and Asian ancestry, Chinese German |
(Palmer et al. 2006) (Brown and McLean 2012; Hu et al. 2009; Brown et al. 2008; Ekelund et al. 2008; Barker et al. 2007; Morar et al. 2007; Weidinger et al. 2005) |
| S2377X (FLG2) Q294X (TCHHL1) R501X, 2282del4, E2554X, R2447X, 1249insG, R826X, 2767insT, E2422X P478S |
Irish Japanese African Americans Chinese |
(Ekelund et al. 2008) (Marenholz et al. 2006) (Brown and McLean 2012) (Osawa et al. 2010) (Margolis et al. 2014; Winge et al. 2011) (Polcari et al. 2014) (Kim et al. 2009) |
|||
|
| |||||
| SPINK5 (LEKTI) | Serine Protease Inhibitor Kazal-Type 5 and Lymphoepithelial Kazal-Type-Related Inhibitor | 5q31 | 1258G > A IV12-26C > T IVS12-10A > G IVS14 + 19 G > A IVS 13-50 G > A 1103A > G 1156 G > A 1188T > C 1258G > A |
Japanese | (Kato et al. 2003; Nishio et al. 2003) |
|
| |||||
| MHC (HLA) | The major histocompatibility complex or human leukocyte antigen | 6p21 | A24 allele of HLA | Korean | (Lee et al. 2001) |
|
TAP1, Val333Ile Gly637Asp |
Tunisians | (Ismail et al. 1997) | |||
|
TAP2, Ile379Val Thr565Ala Ala665Thr Gln687Stop |
Korean | (Braff et al. 2005) | |||
|
| |||||
| Innate Immune genes | |||||
|
| |||||
| CARD4 (NOD1) | Caspase recruitment domain-containing protein 4 | 7p15-p14 | rs2736726 (A > G) rs2075817 (A > G) rs2975632 (C > T) rs3030207 (A > G) rs2075818 (C > G) rs2235099 (C > T) rs2075821 (A > G) rs2075822 (C > T) rs2907749 (A > G) rs2907718 (C > T) rs5743368 (A > G) | German | (Weidinger et al. 2005) |
|
| |||||
| CARD15 (NOD2) | Caspase recruitment domain-containing protein 15 | 16q21 | 2104C > T 2722 G > C 802T > C 534 G > C rs1077861 (intron 10A > T) 2863 G > A 4278A > G 60A > G |
German | (Weidinger et al. 2005; Kabesch et al. 2003) |
|
| |||||
| CD14 | Monocyte differentiation antigen | 5q22-q32 | 159C > T 1145 G > A, 1359 G > T and −550C > T |
Chinese German Singaporean Chinese |
(Lange et al. 2005) (Sengler et al. 2003) (Liang et al. 2006) |
|
| |||||
| (MBL2) | Mannose-binding lectin | 10q11.2-q21 | Gly54 Asp | Japanese | (Hashimoto et al. 2005) |
|
| |||||
| TLR2 | Toll-like receptor 2 | 4q32 | R753Q A-16934T 2258G/A, 896A/G (rs4696480 (T > A), rs3804099 (T > C), rs3804100 (T > C), rs5713708 (G > A) |
German Italian Italian Ukrainian German |
(Niebuhr et al. 2010; Mrabet-Dahbi et al. 2008; Niebuhr et al. 2008) (Salpietro et al. 2011) (Salpietro et al. 2011) (Levchenko et al. 2013) (Potaczek et al. 2011; Oh et al. 2009) |
|
| |||||
| TLR4 | Toll-like receptor 4 | 9q32-q33 | rs4986790 (A > G) rs4986791 (C > T) (rs2770150 (T > C), rs6478317 (A > G), rs1927911 (C > T), rs2149356 (C > T), rs4986790 (A > G), rs4986791 (C > T), rs7873784 (G > C), rs1927906 (A > G) A-896G |
Italian Ukrainian German |
(Salpietro et al. 2011) (Levchenko et al. 2013) (Oh et al. 2009; Weidinger et al. 2006) |
| German | (Weidinger et al. 2006) | ||||
|
| |||||
| TLR6 | Toll-like receptor 6 | 4p14 | Ser249Pro rs5743810 (T> C) T597C C1350T |
German Dutch |
(Hoffjan et al. 2005) |
|
| |||||
| TLR9 | Toll-like receptor 9 | 3p21.3 | C-1237T | German | (Novak et al. 2007) |
|
| |||||
| DEFB1 | Human β-defensin 1 | 8p23 | 2266T>C 1241T>G |
Korean | (Kim et al. 2009) |
| 692 A>G 1654 A>G 20 G>A 5′-UTR 44 C>G 5′-UTR 52 G>A 5′-UTR |
Egyptian Mexican Brazilian |
(Mohamed et al. 2009) (Prado-Montes et al. 2007) (Segat et al. 2010) |
|||
|
| |||||
| Cytokines and related genes | |||||
|
| |||||
| IL-4 | Interleukin 4 | 5q31–33 | −590 C/T 1098G/T 589C>T −33 C/T |
Egyptian Czech Japanese Chinese Czech Macedonian Candian Egyptian |
(Hussein et al. 2014) (Kayserova et al. 2012) (Walley et al. 2001) (Chang et al. 2006) (Kayserova et al. 2012) (Stavric et al. 2012) (He et al. 2003) (Hussein et al. 2014) |
|
| |||||
| IL-4R α | Interleukin 4 receptor alpha | 16p11.2-12.1 | −3112C > T, −1803T > C, −327C > A, −326A > C −186G > A |
Japanese | (Hosomi et al. 2004) |
| 1727G > A 1199C > A, 1242T > G, 1507C > T 1727G > A I50 V |
British, Japanese Chinese |
(Callard et al. 2002) (Chang et al. 2006; Oiso et al. 2000) |
|||
| Q576R 3223C/T |
Egyptian | (Hussein et al. 2014) | |||
| Japanese | (Hosomi et al. 2004) | ||||
|
| |||||
| STAT6 | signal transducer and activator of transcription | 12q13.3–q14.1 | 2964 G > A 2892 C/T 1315-GT repeat in exon 1 |
Egyptian | (Hussein et al. 2014) |
| Chinese Japanese |
(Chang et al. 2006) (Tamura et al. 2003) |
||||
|
| |||||
| IL-10 | Interleukin 10 | 1q31-q32 | 1082A > G 819T > C 592A > C 1082A/G, −819C/T, −592A/C --7616AGG |
Macedonian Chinese German Czech |
(Stavric et al. 2012) (Chang et al. 2006) (Hosomi et al. 2004) (Kayserova et al. 2012) |
|
| |||||
| IL-6 | Interleukin 6 | 7p21 | −174C > G −922A > G |
German | (Hosomi et al. 2004) |
| −174C/G nt565A/G |
Czech | (Kayserova et al. 2012) | |||
|
| |||||
| TNF-α | Tumor necrosis factor alpha | 6p21.3 | −308 G > A 1031T > C, −863C > A, −857C > T, −308G > A −238G > A |
British Chinese |
(Rafatpanah et al. 2003) (Chang et al. 2006) |
| AG cdn25 | Macedonians | (Stavric et al. 2012) | |||
|
| |||||
| TNF-β | Tumor necrosis factor beta | 6p21.3 | 238G > A 308G > A |
German | (Hosomi et al. 2004) |
|
| |||||
| IL-1 α | Interleukin 1 alpha | 2q14 | −899T > C | Macedonians | (Stavric et al. 2012) |
|
| |||||
| IFNγ | Interferon gamma | 12q14 | 874 A>T | Macedonians | (Stavric et al. 2012) |
|
| |||||
| IL-1β | Interleukin 1 beta | 2q14 | 511C > T, 3953T > C, 3953T > C 1418T > C 315T > C |
German British Macedonian |
(Hosomi et al. 2004) (Rafatpanah et al. 2003) (Stavric et al. 2012) |
|
| |||||
| IL1RN | Interleukin 1 receptor antagonist | 2q14.2 | variable number of tandem repeat in intron 2 | German | (Hosomi et al. 2004) |
|
| |||||
| IL1RL1 (ST2) | Interleukin 1 receptor-like 1 (Suppression of tumorigenicity 2) | 2q12 | 26999G > A 2992C > T, 5283G > A, 5860C > A, 11147C > T, 744C > A 27639A > G |
Japanese | (Shimizu et al. 2005) |
|
| |||||
| IL-5 | Interleukin 5 | 5q31.1 | 703C > T | Japanese | (Yamamoto et al. 2003) |
|
| |||||
| IL-12β | Interleukin 12beta | 5q31.1-q33.1 | 1188A > C 4237 G > A, 4496A > G 4510G > A |
Japanese Chinese |
(Tsunemi et al. 2002) (Chang et al. 2006) |
|
| |||||
| IL-12Rβ | Interleukin 12 receptor beta | 19p13.1 | 111A > T 2C > T, 4443C > T 5970 G > C 17183T > C 17369C > T 25748T > C 27637A > T |
Japanese | (Takahashi et al. 2005) |
|
| |||||
| IL-13 | Interleukin 13 | 5q31 | 1111C > T 1024C > T 704A > C 1103C > T Arg144Gln Arg 130 Gln 1111C > T, 1293C > T, Arg144Gln |
Chinese Japanese German Japanese German Canadian German Japanese |
(Chang et al 2006) (Tsunemi et al. 2002) (Hummelshoj et al. 2003) (Tsunemi et al. 2004) (Folster-Holst et al. 2005) (He et al. 2003) (Hummelshoj et al. 2003) (Tsunemi et al. 2002) |
|
| |||||
| IL-18 | Interleukin 18 | 11q22.2-q22.3 | 132A > G, 133C > G 137G > C, 113T > G 127C > T |
German | (Novak et al. 2005) |
|
| |||||
| TGF β1 | Transforming growth factor beta 1 | 19q13.2 | 1 915 G > C 869T > C 590C > T |
British | (Arkwright et al. 2001) |
|
| |||||
| GM-CSF | Granulocyte macrophage colony-stimulating factor | 5q31.1 | 677A > C 1916T > C |
British | (Rafatpanah et al. 2003) |
| 3606T > C 3928C > T |
Japanese Canadian |
(Saeki et al. 2006) (He et al. 2003) |
|||
|
| |||||
| IL-9 | Interleukin 9 | 5q31–35 | 4091G>A | Korean | (Namkung et al. 2011) |
|
| |||||
| IL-9R | Interleukin 9 receptor | Xq/Yq | 1737C>T | Korean | (Namkung et al. 2011) |
|
| |||||
| Chemokines and related genes | |||||
|
| |||||
| CCL5 (RANTES) | Chemokine (C-Cmotif) ligand 5 (Regulated upon activation, normally T cell expressed +secreted ) | 17q11.2-q12 | 28C>G 403 G>A 2518A > G |
German Japanese Hungarian |
(Nickel et al. 2000) (Bai et al. 2005) (Kozma et al. 2002) |
|
| |||||
| CCL11 | chemokine(C-Cmotif) ligand 11 (eotaxin 1) | 17q21.1-q21.2 2 | 426C > T, 384A > G |
Japanese | (Wakugawa et al. 2002) |
|
| |||||
| CCL17 (TARC) | chemokine(C-Cmotif) ligand 17 (Thymus and activation-regulated chemokine) | 16q13 | 431C > T | Japanese | (Gangur and Oppenheim 2000) |
|
| |||||
| CCR3 | Chemokine (C-Cmotif) receptor3 | 3p21.3 | 1052T > C | American | (Tsunemi et al. 2003) |
|
| |||||
| CCR4 | Chemokine (C-Cmotif) receptor 4 | 3p24 | 1014C > T | Japanese | (Tsunemi et al. 2004) |
|
| |||||
| CMA1 | Mast cell chymase 1 | 14q11.2 | (−1903A > G | Japanese Italian |
(Iwanaga et al. 2004) (Weidinger et al. 2005) |
|
| |||||
| Drug-metabolizing genes | |||||
|
| |||||
| GSTP1 | Glutathione S-transferase pi | 11q13 | 1404A > G 2294C > T |
Russian | (Safronova et al. 2003) |
|
| |||||
| NAT | N-acetyl transferase | 8p23.1-p21.3 | 481C > T 590G > A 857G > A |
Russian Caucasian |
(Makarova et al. 2005) (Ueda et al. 2003) |
|
| |||||
| Other genes | |||||
|
| |||||
| CTLA4 (CD152) | Cytotoxic T-lymphocyte-associated antigen-4 | 2q33 | 49A > G | Chinese | (Yang et al. 2004) |
|
| |||||
| KLK (SCCE) | Kallikrein (stratum corneum chymotryptic enzyme) | 19q13.33 | AACCAACC variant of the KLK7 (SCCE) | British | (Vasilopoulos et al. 2004) |
|
| |||||
| RUNX1 | Runt-related transcription factor 1 binding site between solute carrier | rs734232 | Japanese | (Hosomi et al. 2004) | |
|
| |||||
| IRF-2 | Interferon regulatory factor 2 | 4q35 | 829C>T 830C>T 684C>T 467G>A 921G>A), 10-bp deletion in 3′ UTR 1739[ATCCC]8>6 |
Japanese | (Nishio et al. 2001) |
|
| |||||
| FCER1B | High affinity IgE receptor beta chain | 11q13 | RsaIin2, RsaIex7 | British | (Cox et al. 1998) |
|
| |||||
| PHF11 | Plant homeodomain Zink finger 11 protein | 13q14 | intron3 T>C 3UTR G>A |
Australian | (Jang et al. 2005) |
1- Filaggrin Gene
FLG gene is located on chromosome 1q21 in a region called the epidermal differentiation complex. It is one of genes that code for S100-fused-like proteins (SFTP) which harbor several proteins e.g. profilaggrin (FLG), hornerin (HRNR), FLG-2 (FLG2), repetin (RPTN), cornulin (CRNN), trichohyalin (TCHH), and trichohyalin-like 1(TCHHL1). (41) These genes are very similar together in structure and function, and locate in close proximity to each other in the epidermal differentiation complex. (13,41,42) On the basis of previous illustrations with FLG, it has been postulated that a stop-gain (null) mutation in exon 3 of any of the SFTP genes may be result in reduced or absent protein production. (41–44) Genetic associations of FLG mutations with AD were confirmed and further verified by many independent groups using numerous cohorts of different populations e.g. American, (45) Caucasian and Northern-American AD, (2) European and Asian ancestry, (43,46–51) Chinese, (52) German, (53) Irish, (43) and Japanese. (54) More than 40 FLG loss-of-function mutations have been described in Europeans and Asians. (43) While in African-American children with AD revealed a total of 289 variants in FLG, 107 variants in FLG2, 339 variants in HRNR, 4 variants in RPTN, 37 variants in CRNN, 88 variants in TCHH, and 14 variants in TCHHL1. In addition three novel identified FLG stop-gain mutations, Q570X, R3409X and S3707X, were investigated only once. S2392X and S2377X in FLG2 were indicated 1 and 16 times, respectively. However, none of these variants could be detected in other members of African-American PEER children cohort. In TCHHL1, the variant Q294X was noted twice. (43–45, 55, 56) These findings are in agreement with those of Winge et al., who also failed to detect common FLG loss-of-function mutations in people of African ancestry with AD. (55) Their report is from the largest whole-exome sequencing study of African Americans with AD suggesting that S2377X (FLG2) and Q294X (TCHHL1) variants may not be clinically important with respect to incident AD and it seems unlikely that FLG stop-gain mutations have a prominent role with respect to incident AD in African Americans children. (55,56) Whereas, the recent demonstration that FLG2 mutations increase the persistence of AD in African Americans (57) further suggests a pathogenic role for FLG2 mutations (rs12568784 and rs16833974) and supports the sequencing of FLG2in African Americans with IV with or without AD. (44) Polcari data was demonstrated a prevalence of filaggrin mutations including R501X, 2282del4, E2554X, R2447X, 1249insG, R826X, 2767insT, and E2422X in the African American population that exceeds previously published data, although the overall prevalence is still lower than in other populations. (58) In addition other study was added that the FLG P478S polymorphism alone and combined with other factors influences free fatty acids levels and increases the susceptibility to AD among Chinese population. (59)
2- SPINK5 and LEKTI gene
The Serine Protease Inhibitor Kazal-Type 5 (SPINK5) gene located on chromosome 5q32 within genomic region has been linked to AD and encodes a 15-domain protease inhibitor Lymphoepithelial Kazal-Type-Related Inhibitor (LEKTI) which is expressed in epithelial and mucosal surfaces and in the thymus. In fact Serine protease enzymes play an important role in skin barrier homeostasis, including SC desquamation, lipid barrier construction and cornified cell envelope. While, SPINK5 participates to the regulation of proteolysis in keratinocyte differentiation and the generation of normal epithelium, and LEKTI had been found to be involved in maintaining the permeability of normal skin. SPINK5 polymorphisms are associated with the incidence and severity of AD among Chainese, (60, 61) athough no association was confirmed with AD in German. (62) Otherwise, a recent study was suggested that the E420K LEKTI variant is a risk factor for AD through an increase in the TSLP expression. (63) Subsequently, recent studies found a significant association between (1258G > A) SPINK5 polymorphism and AD in Japanese, (64, 65) and in German. (66) In individuals affected by AD, a significant maternal over-transmission of the risk allele to their children was demonstrated Walley et al. (67) Folster-Holset al., studied 8 SNPs in different regions of the SPINK5 gene, including 4 non-synonymous SNPs leading to an amino acid change (Asp106Asn (G316A), Asn368Ser (1103A > G) and Asp386Asn (1156G > A), and, Gly463Gly (A1389G), Val553Val (C1659T), Leu756Leu (C2358T) and Gly804Gly C2412T) and Glu 825Asp (C2475T)). (62) None of the SNPs were associated with an increased risk of Ad. Kato et al. examined associations between 8 SNPs (IV12-26C > T, IVS12-10A > G, IVS14 + 19 G > A, IVS 13-50 G > A, Asn 368Ser (1103A > G), Asp 386Asn (1156 G > A), His 396His (1188T > C), Glu 420Lys (1258G > A)) of the SPINK5 gene and AD in a Japanese population and found a positive association of 7 SPINK5 SNPs (except for 1156G > A) with AD. (64, 65) In addition a study of Six SNPs rs17718511, rs17860502, KN0001820, rs60978485, rs17718737, and rs1422985 in the SPINK5 gene provides evidence for a significant interaction between the SPINK5 gene that may contribute to AD susceptibility among Korean. (68)
3- MHC (or HLA) genes
The major histocompatibility complex (MHC) (also known as human leukocyte antigen (HLA) complex) is classified into three classes. Human leukocyte antigen class-I (HLA-I) sub-classified as A, B, and C, are produce by all human cells except erythrocytes and trophoblasts The HLA group of genes located at chromosome 6p21.3. Eleven HLA-A (1, 2, 3, 11, 24, 26, 29, 30, 31, 33 and 66) and twenty seven HLA-B (7, 8, 13, 14, 16, 27, 35, 37, 38, 39, 46, 48, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 67, 71 and 75) alleles are frequently found in Koreans. Among these, only the twenty four alleles of HLA were significantly associated with AD. (69) The 333 Val and 637 Gly alleles of the transporter for antigen presentation 1 (TAP1) gene were significantly associated with susceptibility to AD among Tunisians population, (70) while allelic frequencies of the TAP1 gene polymorphisms were not associated with the disease. (69) The 565Ala and 665Thr alleles of the TAP2 gene may be associated with susceptibility to AD in a Korean population. (69)
4- Innate Immune system genes
As components of innate immunity, antimicrobial peptides (AMPs) produced by keratinocytes play a crucial role in the clearance of microbial pathogens and in preserving epidermal barrier effectiveness. Consequently deficiencies in these AMPs play roles in the pathogenesis of AD. Mainly AMPs include proteins from the β-defensin family (hBD-1, 2, and 3), cathelicidins, psoriasin, and ribonuclease (RNase). (71) Mutations in PRRs s and AMPs are play roles in the initiation and exacerbation of AD. However pattern-recognition receptors (PRRs) help protect organisms from microbial pathogens such as Staphylococcus aureus (S. aureus) and Malassezia furfur (M. furfur). Generally, in AD patients, AMPs expression is not reduced, but significantly varies according to the type of AMPs. (5) Recent progress has revealed that innate immune responses are initiated by pattern-recognition receptors (PRR) a family of proteins that enhance certain cytokine gene transcription in response to various pathogenic ligands and control acquired immune responses such as Th1 responses. (72,73) Mutations in PRRs such as toll-like receptors (TLRs) and nucleotide-binding oligomerization domain like receptors (NLRs), as well as mutations in AMPs are associated with susceptibility to skin infections and play key roles in the initiation and aggravation of AD. (74,75) The following are the genes associated innate immune system and have a role in AD pathogenesis.
4 (a) - CARD4 (or NOD1) gene
Caspase recruitment domain – containing protein (CARD) 4 is located at chromosome 7p15-p14 and about 54.49 kb in length. Eleven CARD4 or nucleotide – binding oligomerization domain protein (NOD)1 polymorphisms, such as rs2736726 (A > G), rs2075817 (A > G), rs2975632 (C > T), rs3030207 (A > G), rs2075818 (C > G), rs2235099 (C > T), rs2075821 (A > G), rs2075822 (C > T), rs2907749 (A > G), rs2907718 (C > T), rs5743368 (A > G), were investigated in a German population. Genotypes AA at rs2736726 and GG at rs2075817 were associated with AD. It has been also observed that haplotype rs 2736726 A- rs2075817 G -rs2975632 T- rs3030207 A-rs2075818 Crs2235099 C- rs2075821 G - rs2075822 Trs2907749 A- rs2907718 C- rs5743368 G is weakly associated with atopic eczema. (76)
4 (b) - CARD15 (or NOD2) gene
CARD15 is located at chromosome 16q21 and about 39.45 kb in length. No significant associations between AD and any polymorphisms (2104C > T, 2722 G > C, 802T > C, 534 G > C, rs1077861 (intron 10A > T), 2863 G > A, 4278A > G, -60A > G) or haplotype of CARD15 were observed. (77) Associations of three SNPs (2104C > T, 2722 G > C and 3020iC) with AD in children have been reported. Children with the C allele of 2722 G > C SNP had a 1.85-fold risk of developing AD in German. (78)
4 (c) - Monocyte differentiation antigen (or CD14) gene
CD14 gene is located at chromosome 5q22-q32 and about 1.95 kb in length. In a small study, children with the CT genotype of the CD14–159C > T SNP had a significantly lower prevalence of AD at three years of age compared with those with the genotypes CC and TT combined, (79) although the CD14-159C > T SNP was not associated with an increased risk of AD in German children. (80) No significant difference was found in the genotype frequencies of -159C > T, -1145 G > A, 1359 G > T and -550C > T SNPs between AD patients and controls. (81)
4 (d) - MBL2 gene
The mannose-binding lectin (MBL2) gene is located at 10q11.2-q21 chromosome and about 6.32 kb in length. Recently, three variants at codons 52, 54, and 57 of exon 1 of the MBL2 gene have been identified. The MBL2 Gly54 Asp SNP was not associated with an increased risk of AD in a Japanese population. (82)
4 (e) - TLR2, TLR4, TLR6 and TLR 9 genes
Toll-like receptor (TLR)-2, TLR4, TLR6 and TLR9 are located at chromosome 4q32, 9q32-q33, 4p14, and 3p21.3, respectively. The TLR2 R753Q polymorphism modulates the innate and adaptive immunity through control of cytokine production (IL-2, IL-6, IL-8, and IL-12), and via changing TLR2 and CD36 expression in AD cases [83–85]. TLR2 rs5743708 (A > G) polymorphisms were demonstrated among Italian, Ukrainian and German children with severe AD, (86, 87) while AD patients exhibit a higher frequency of the TLR4 polymorphisms rs4986790 (A > G) among Italian and among Ukrainian, (86, 87) and rs4986791 (C > T) among German [88, 89]. However, it has been found that common TLR2 (rs4696480 (T > A), rs3804099 (T > C), rs3804100 (T > C), rs5713708 (G > A) or TLR4(rs2770150 (T > C), rs6478317 (A > G), rs1927911 (C > T), rs2149356 (C > T), rs4986790 (A > G), rs4986791 (C > T), rs7873784 (G > C), rs1927906 (A > G)) variants or haplotypes were not associated with an increased risk of AD in another German population. The TLR4 A-896 G polymorphism was associated with severe AD patients. (90) There was no association between the TLR6 rs5743810 (T> C) polymorphism and risk for AD. While T597C TLR6, C 1350T TLR6 were associated with AD in German and Dutch. (91, 92) Moreover, C-1237T TLR9 promoter polymorphism had been found in AD patients. (93)
4 (f) - DEFB1 gene
The human β-defensin 1 (DEFB1) gene is located at 8p23.2266T/C and 1241T/GSNPs of DEFB1 gene had been demonstrated to be associated with AD among Korean. (94) While 692 A>G and 1654 A>GSNPs of DEFB1 gene had been demonstrated to be associated with AD among Egyptian, (107) and among Mexican populations. (95, 96) As controversial findings have been achieved, a study had been not confirmed that the role of −20 G/A (rs11362), −44 C/G (rs1800972), and −52 G/A (rs1799946) at 5′-UTR of DEFB1 gene in the development of AD among Brazilian population. (97)
5- Adaptive immune system genes
Alterations in the adaptive immune system are also play key role with AD. Where AD is going through a biphases, the first phase is prevailed by T helper type 2 (Th2) cytokines that later turns to the second phase a more chronic Th1-dominated eczematous phase. The effective elevation of IgE in atopic disease by B cells depends on support by Th2 cells, which mainly produce interleukin-4 (IL-4), IL-5, IL-9 and IL-13. Adaptive immune genes are including cytokine genes and chemokine.
5(a) - Cytokines and related genes
Cytokines have been classified into two subgroups according to their function: Th1 cytokines, mainly interleukin (IL) 2, IL12, interferon (IFN)γ, and tumor necrosis factor (TNF)α, which activate the cellular machinery of the immune system; and Th2 (IL4, IL5, IL6, IL10 and IL13) cytokines, which activate the humeral machinery. Some cytokines, such as IL-1α, IL-2, and TGF-βwere found to be decreased in AD, (98, 99) while other such as IFN-γ, IL-12 and GM-CSF were elevated in chronic AD. Several data have been shown the association between cytokine polymorphism and AD. Some reports show positive association of certain cytokine polymorphisms with AD, while others are controversial. (100)
5(a-1) - IL-4 gene
Interleukin (IL)-4 is a glycoprotein encoded by the IL-4 gene which has been mapped to chromosome 5q31–33 and about 9.01 kb in length. Promoter polymorphisms of IL-4-590C/T were significantly associated with atopic AD in Egyptian population, (101) Czech population, (102) and Japanese population. (67) On contrast, IL-4-590C/T SNP of IL-4 was not associated with AD in Chinese, (103) Egyptian (101) and Czech population. (102) IL-4-1098G/T polymorphism was significantly associated with atopic AD in Czech population, (102) while no association in Macedonians. (104) No association was found between AD and −33C/T,IL-4 polymorphism in Chinese, (103) Czech (102) and Macedonians population. (104) In Caucasians the T allele of IL-4-589C>T SNP was significantly associated with the development of AD at 24 months of age. (105)
5(a-2) - IL-4Rα gene
Interleukin 4 receptor alpha gene (IL-4Rα) is located at chromosome 16p11.2-12.1 and about 50.86 kb in length. Several IL4R polymorphisms (−3112C > T, −1803T > C, −327C > A, −326A > C and −186G > A) have been found to associated with AD in Japanese. (106) Seven polymorphisms (223C > G > T > A, 1199C > A, 1291C > T, 1307T > C, 1727G > A, 2356C > T) and a silent 1242T > G have been demonstrated to have functional significance. Caucasian children with the rare homozygous 1727G > A polymorphism had a higher prevalence of flexural eczema in the first 6 months compared with the heterozygote and the wild type homozygote genotypes combined in British. (107) It has been demonstrated that the 1727G > A SNP (Gln551Arg) was significantly associated with AD in another Japanese population. (108) In contrast no association between (1199C > A, 1242T > G, 1507C > T and 1727G > A) of IL4R polymorphism and AD in a Chinese population. (103) In addition in Egyptian population the IL-4Rα I50 V and Q576R, polymorphisms were significantly associated with the development of AD. (101) Another polymorphism 3223C/T IL4Rα was associated with AD in Japanese. (106)
5(a-3) - STAT6 gene
The Signal transducer and activator of transcription (STAT)6 gene have been mapped to chromosome 12q13.3–q14.1, and about 16.79 in length. There was no association between AD risk and the 2964 G > A SNP of the STAT6 gene while the 1315-GT repeat allele heterozygosity of the dinucleotide repeat in exon 1 (13-, 14-, 15- and 16-GT repeat alleles) was significantly associated with allergic disease including AD in Japanese. (109) However, the short tandem repeat in exon 1 was not associated with AD risk in Chinese. (103) In Egyptian STAT6 2964 G/A and 2892 C/T polymorphisms were significantly associated with the development of AD. (101)
5(a-4) - IL-10 gene
IL-10 gene is located at chromosome 1q31-q32 and about 4.89 kb in length. The −1082A > G, −819T > C and −592A > C SNPs of the IL10 gene did not contribute to the development of AD in Macedonian, and in German. (104,106) Also, −7616AGG promoter, −6365C > G and −3526A > T, −795G>A, −1328 C>T, −2127 G>C, 3976 A>G and – 4311T>C SNPs of the IL10 gene were not associated with AD in German. (106,109) Whereas-1082A/G, −819C/T, and −592A/C were significantly associated with atopic AD in Czech population. (102)
5(a-5) - IL-6 gene
IL-6 is located at chromosome 7p21 and about 6.12 kb in length. No association was found between the −174C > G SNP of the IL6 gene and AD. (106) As well as the −174C > G and −922A > G SNPs were not found. (106) Whereas −174C/G and nt565A/G were found that significantly associated with atopic AD in a Czech population. (102)
5(a-6) - TNF-α gene
Tumor necrosis factor (TNF)-α gene is located at chromosome 6p21.3. No significant association was found between −308 G > A SNP of the TNFα gene and AD in English. (110) Neither −1031T > C, −863C > A, −857C > T, −308G >A nor −238G > A SNPs of the TNFα gene was associated with AD in a Chinese population. (103) Whereas, no association was not found between AG cdn25 and AD in population of Macedonians. (104)
5(a-7) - TNF-β gene
Tumor necrosis factor (TNF)-β is located at chromosome 6p21.3. No association was found between AD and −238G >A or −308G > A SNPs of the TNFβ gene in a German. (106)
5(a-8)- IL-1α gene
Interleukin (IL)-1α gene is located at chromosome 2q14 and about 11.48 kb in length. The −899T > C SNP of the IL1A gene was not associated with AD in Macedonians. (104)
5(a-9) - IFNγ gene
Interferon gamma (IFNγ) gene is located at chromosome 12q14 and about 16.25 kb in length. Study on a Chinese population was demonstrated no association between short tandem repeats at the first intron of IFNγ gene and AD. (103) In addition, no association was found between 874 A>T IFNγ gene and AD in population of Macedonians. (104)
5(a-10) - IL-β gene
IL-1β is located at chromosome 2q14 and about 7.16 kb in length. No association was found between either the −511C > T, 3953T > C, 3953T > C, −1418T > C or the 315T > C SNPs of the IL1B gene and AD in American, German, English and Macedonians. (104,106,110)
5(a-11) - IL1RN gene
Interleukin 1 receptor antagonist (IL1RN) gene is located at chromosome 2q14.2 and about 34.70 kb in length. The polymorphism in intron 2 of the IL1RN gene is caused by a variable copy number of an 86-bp sequence. The 4-repeat (IL1RN *1) and 2-repeat (IL1RN *2) alleles are most common, while the other alleles occur at a combined frequency of less than 5%. No association was found between the variable number of tandem repeat polymorphisms in intron 2 of the IL1RN gene and AD. (106)
5(a-12) - IL1RL1 (or ST2) gene
Interleukin 1 receptor-like 1 (IL1RL1) is located at chromosome 2q12 and 40.54 kb in length. A significant association between AD and the −26999G > A or SNP of the suppression of tumorigenicity (ST)2 gene was found in a Japanese population. (111) On other hand, 2992C > T, 5283G > A, 5860C > A, 11147C > T, 744C > A and −27639A > G SNPs were not 5q31.1 associated with AD risk. (111)
5(a-13) - IL-5 gene
IL-5 is located at chromosome and about 2.08 kb in length. The −703C > T SNP of IL5 was not significantly associated with AD in Japanese. (112)
5(a-14) - IL-12 β gene
IL-12 β is located at chromosome 5q31.1-q33.1 and about 15.69 kb in length. The AA genotype of IL12B 1188A > C SNP was associated with decreased risk of AD in a Japanese population [113]. The 4237 G > A, 4496A > G and 4510G > A SNPs of the IL12B gene were not associated to the development of AD. (103) In addition 30 untranslated region of the IL-12B gene was associated with the AD phenotype. (113)
5(a-15) - IL-12R β
Interleukin 12 receptor beta (IL-12R β) is located at chromosome 19p13.1 and about 39.94 kb in length. Among eight SNPs (−111A > T, −2C > T, 4443C > T, 5970 G > C, 17183T > C, 17369C > T, 25748T > C and 27637A > T), the TT genotype of the −111A >T SNP and the TT genotype of the −2C > T SNP were significantly associated with an increased risk of AD in a Japanese population. (114)
5(a-16) - IL-13 gene
IL-13 is located at chromosome 5q31 and about 4.85 kb in direct. No association between the −1111C > T SNP of IL13 and AD in Chinese population (103) and Japanese population. (115) While significant association between the −1024C > T SNP of the IL13 gene and AD was confirmed in German. (116) In the Japanese population there was no significant association between two SNPs of 704A > C and 1103C > T whereas the Arg allele of Arg144Gln SNP was significantly associated with an increased risk of AD in Japanese. (115) As well as the Arg 130 Gln polymorphism was confirmed in Canadian, (117) German, (116) and Japanese. (115) The A allele of the Arg144Gln SNP was associated with AD in German. (118) In Caucasians, haplotypes consisting of IL13 Arg144Gln with AD were associated with AD. None of the three SNPs (−1111C > T, 1293C > T, and Arg144Gln) were associated with AD during the first year. (106)
5(a-17)- IL-18 gene
IL-18 is located at chromosome 11q22.2-q22.3 and about 21.61 kb in length. Among five SNPs (−132A > G, −133C > G, −137G > C, −113T > G and 127C > T), only the C allele of the −137G > C SNP was associated with an increased risk of AD. (119)
5(a-18) - TGF-β1 gene
Transforming growth factor β1 (TGF-β1) is located at chromosome 19q13.2 and about 52.34 kb in length. The C allele of the TGF-β1 915 G > C SNP was associated with an increased risk of AD in British, whereas there was no significant difference in the frequencies of the 869T > C genotypes [120]. No association between AD and the −590C > T SNP was not found. (106)
5(a-19) - GM-CSF gene
Granulocyte macrophage colony-stimulating factor (GM-CSF) is located at chromosome 5q31.1 and about 2.38 kb in length. The A allele of the −677A > C SNP in the promoter region of the GM-CSF gene was associated with an increased risk of AD in British. (110) Although −1916T > C SNP was significantly associated with an increased risk of AD, there was a strong linkage disequilibrium existed between the −677A > C and −1916T > C SNPs. (110) The 3606T > C and 3928C > T SNPs of the GM-CSF gene was not associated with susceptibility to AD in Japanese. (121) While a strong linkage disequilibrium between 3606T > C and 3928C > T SNPs of the GM-CSF with AD in Canadian. (117)
5(a-20) - IL-9 gene
IL-9 is located at chromosome 5q31–35 and about 4 kb in length. 4091G>A of IL-9 gene polymorphism was associated with an increased susceptibility to Korean AD. (122)
5(a-21) - IL-9R gene
Interleukin 9 receptor (IL-9R) is located at chromosome Xq/Yq. 1737C/T gene polymorphism seems to be associated with an increased risk for developing non-allergic Korean AD. (122)
6 (b) - Chemokines and related genes
Chemokines are classified into four classes on the basis of their protein structure: CXC (16 chemokines), CC (28 chemokines), C (2 chemokines), and CX3C chemokines (1 chemokines). (123) The CC and CXC chemokines are belong into inflammatory chemokines while the C and CX3C chemokines are belong to immune chemokines. Some chemokines, such as CCL5 (RANTES), CCL11 (exotoxin), CCL2 (MCP-1), CCL13 (MCP-4), CCL7 (MCP-3), CCL8 (MCP-2), and macrophage inflammatory protein (MIP)-1α CCL3, lead to cellular activation and inflammatory mediator secreted by basophils and eosinophil’s. Several receptors have been shown to bind the chemokines: CCR Receptors for CC chemokine; CXCR receptors for CXC chemokines; XCR1 receptor for C and CX3CR1 receptor for CX3C chemokines. (123) At least three chemokine receptors have been demonstrated to mediate the recruitment of Th2 cells: CCR3, the receptor for CCL5 (RANTES), CCL11 (exotoxin), CCL2 (MCP-1), and CCL13 (MCP-4), which is also secreted in eosinophils and basophils, CCR4, the receptor for CCL17 (TARC), CCL22 (MDC) and CCR8, the receptor for CCL1 (I-309). Enhanced levels of both CCL5 (RANTES) and CCL11 (exotoxin 1) have been determined in the sera of AD patients, (124) with CCL5 (RANTES) signifying a crucial correlation with both total serum IgE levels and eosinophil numbers. Exotoxin 1 also has shown a high pattern of gene expression with AD patients. (125)
5(b-1) - CCL5 gene (or RANTES)
Chemokine (C-Cmotif) ligand 5 (CCL5) gene is located at chromosome 17q11.2–q12 and about 9.01 kb in length. Two polymorphisms in the CCL5 (RANTES) promoter region (−28CG and −403 GA) increase CCL5 (RANTES) expression in humans. (126) Indeed, these two (−28CG and −403 GA) CCL5 polymorphisms have been associated with susceptibility to AD patients. (127) In addition, the −401 G > A polymorphism has been shown a significantly frequency in AD patients among German population. In contrast, other study was not confirmed such as these association between (−28C > G, −403 G > A and −2518A > G) CCL5 polymorphisms with AD patients among Hungarian population. (128)
5(b-2) - CCL11 (or Exotoxin 1) gene
Chemokine(C-Cmotif) ligand 11 (CCL11) is located at chromosome 17q21.1–q21.2 2 and about 66 kb in length. In spite of multiple polymorphisms have been identified in the gene, only the two polymorphisms (−426C > T, −384A > G) were associated with serum IgE levels in Japanese AD. (129)
5(b-3)- CCL17 (or TARC) gene
The Chemokine(C-Cmotif) ligand 17 (CCL17) or thymus and activation-regulated chemokine (TARC) levels expression of AD patients were associated with disease activity. (130) In spite of some CCL17 polymorphisms are candidates as a genetic factor in AD, no correlation between AD and the −431C > T SNP of the CCL17 gene was found in a Japanese population. (131)
5(b-4)- CCR3 gene
Chemokine (C-Cmotif) receptor3 (CCR3) gene is located at chromosome 3p21.3 and about 143.70 kb in length. As mentioned before, CCR3 is a receptor for different chemokines which are play important role in AD pathogenesis. Hence, the biological activities of CCR3 suggest that polymorphisms of CCR3 may be an increased risk for AD. There was no significant difference in genotype frequencies of 51T > C SNP of the CCR3 gene between AD patients and controls among Japanese. (131)
5(b-5)- CCR4 gene
Chemokine (C-Cmotif) receptor-4 (CCR4) gene is located at chromosome 3p24 and about 3.36 kb in length. One study was not demonstrated a significant association between the 1014C > T CCR4 polymorphism and AD patients. (132)
5(b-6)- CMA1 gene
Mast cell chymase 1 (CMA1) gene is located at chromosome 14q11.2 and about 2.91 kb in length. A family-based association study in Caucasians revealed a significant association of this polymorphism with total IgE levels in patients with self-reported AD. (133) A significant association between the CMA1-1903A > G polymorphism and AD was observed by Weidinger et al. (134) It may be speculated whether this DNA variant alters the expression of chymase. It has also been shown that CMA1 is increased in chronic atopic eczema skin lesions (135) and a potential role of chymase in the promotion of skin barrier defects and cutaneous neovascularization has been suggested. (136)
6- Drug-metabolizing genes
The metabolism of xenobiotic involves oxidation, reduction, and hydrolysis (phase I) and conjugation (phase II) reactions.
6(a-1)- GST genes
Certain genes within the glutathione S-transferase (GST) M, GSTT and GSTP subfamilies (GSTM1, GSTT1and GSTP1) are polymorphic in humans and the levels of individual enzymes expressed can be influenced by induction and genetic polymorphism. The GSTM1, GSTT1and GSTP1genes are located on chromosomes 1p13.3, 22q11.23 and 11q13, respectively. The 1404A > G (Ile105Val) and 2294C > T (Ala114 Val) SNPs of the GSTP1 gene were associated with a significantly increased risk of AD in Russian. (137)
6(a-2)- NAT-2 gene
N-acetyl transferase (NAT)2 gene is located at chromosome 8p23.1-p21.3 and about 9.97 kb in length. N-acetylation is an important genetic polymorphic pathway in the biotransformation of one or more single-based mutations in the NAT2 gene known to cause low expression levels of functional NAT2 enzyme. 481C > T (synonymous mutation) and 590G > A SNPs were not correlated with susceptible to AD in Russian. (138) Moreover, 481C > T, 590 G >A or 857G > A, were not associated with an increased risk of AD in Caucasian. (139)
7- Other genes associated with atopic dermatitis
7(a-1)- CTLA-4 (or CD152) gene
Cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) gene is located at chromosome 2q33 and about 5.55 kb in length. CTLA −4 is associated to an increased the risk of autoimmune diseases. While, the 49A > G SNP of CTLA-4 gene was not associated with AD in Chinese. (140)
7(a-2)- KLK ( or SCCE) gene
Kallikrein (KLK) or stratum corneum chymotrypsin enzyme (KLK) gene is located at chromosome 19q13.33 and about 7.57 kb in length. A significant genetic association was found between the rare AACCAACC variant of the KLK7 gene and AD in British. (141)
7(a-3)- RUNX1 gene
Runt-related transcription factor 1 binding site between solute carriers (RUNX1) gene is located at chromosome 21q22.3. There was no significant allelic association between the RUNX1 polymorphism (rs734232) and AD in a small Japanese adult population. (142)
7(a-4)- IRF2 gene
Interferon regulatory factor 2 (IRF2) gene is located at chromosome 4q35. IRF-2--829C>T, −830C>T, −684C>T, −467G>A, one silent mutation in exon 9 (921G>A), and a 10-bp deletion in the 3′ untranslated region (1739[ATCCC]8>6) polymorphisms were significantly associated with atopic AD in Japanese. (143)
7(a-5)- FCER1B gene
High affinity IgE receptor beta chain (FCER1β) is located at chromosome 11q13. RsaIin2, RsaIex7 polymorphisms were significantly associated with atopic AD in British. (144)
7(a-6)- PHF11 gene
Plant homeodomain Zink finger 11 (PHF11) gene is Located at 13q14. T/C intron3, G/A 3UTR polymorphisms were significantly associated with atopic AD in Australian. (145)
Conclusions
Immune system plays a key role in the pathogensis of atopic dermatitis, therefore it is important to design an appropriate epidemiological investigation of immune system polymorphism for atopic dermatitis patients. Continued advances in molecular genetics and in high-throughput of genotyping methods will facilitate the analysis of multiple polymorphisms within genes and the analysis of multiple genes pathways. The effects of polymorphisms are best represented by their haplotypes. Data from multiple polymorphisms within a gene can be combined to create haplotypes and the set of multiple alleles on a single chromosome. Polymorphisms, even those not significantly associated with AD, should be considered as potentially important public health issues. In addition, it is important to keep in mind that a susceptibility factor in one population may not be a factor in another. There are differences in the prevalence rates of immune system polymorphisms among populations. In a population where the frequency of an “at-risk” genotype in a given polymorphism is too low, the “at-risk” allele or “at-risk” genotype can be rare to assess its associated risk.
Acknowledgements
This work was supported by funds from College of Medicine, Qassim University.
References
- 1.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–94. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 2.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 3.Sandilands A, Sutherland C, Irvine AD, McLean WH. Filaggrin in the frontline: role in skin barrier function and disease. J Cell Sci. 2009;122:1285–94. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tokura Y. Extrinsic and intrinsic types of atopic dermatitis. J Dermatol Sci. 2010;58:1–7. doi: 10.1016/j.jdermsci.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Leung DY. New insights into atopic dermatitis: role of skin barrier and immune dysregulation. Allergol Int. 2013;62:151–61. doi: 10.2332/allergolint.13-RAI-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akdis CA, Akdis M, Bieber T European Academy of Allergology; Clinical Immunology/American Academy of Allergy, Asthma and Immunology/PRACTALL Consensus Group. Diagnosis and treatment of atopic dermatitis in children and adults: European Academy of Allergology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology/ PRACTALL Consensus Report. Allergy. 2006;61:969–87. doi: 10.1111/j.1398-9995.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 7.Lyons JJ, Milner JD, Stone KD. Atopic Dermatitis in Children: Clinical Features, Pathophysiology, and Treatment. Immunol Allergy Clin North Am. 2015;35:161–83. doi: 10.1016/j.iac.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng W, Novak N. Recent developments in atopic dermatitis. Curr Opin Allergy Clin Immunol. 2014;14:417–22. doi: 10.1097/ACI.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 9.Kabashima K. New concept of the pathogenesis of atopic dermatitis: interplay among the barrier, allergy, and pruritus as a trinity. J Dermatol Sci. 2013;70:3–11. doi: 10.1016/j.jdermsci.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Brown SJ, Kroboth K, Sandilands A, Campbell LE, Pohler E, Kezic S, et al. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. J Invest Dermatol. 2012;132:98–104. doi: 10.1038/jid.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hachem JP, Crumrine D, Fluhr J, Brown BE, Feingold KR, Elias PM. pH directly regulates epidermal permeability barrier homeostasis, and stratum corneum integrity/cohesion. J Invest Dermatol. 2003;121:345–53. doi: 10.1046/j.1523-1747.2003.12365.x. [DOI] [PubMed] [Google Scholar]
- 12.Hachem JP, Man MQ, Crumrine D, Uchida Y, Brown BE, Rogiers V, et al. Sustained serine proteases activity by prolonged increase in pH leads to degradation of lipid processing enzymes and profound alterations of barrier function and stratum corneum integrity. J Invest Dermatol. 2005;125:510–20. doi: 10.1111/j.0022-202X.2005.23838.x. [DOI] [PubMed] [Google Scholar]
- 13.Elias PM, Steinhoff M. Outside-to-inside (and now back to “outside”) pathogenic mechanisms in atopic dermatitis. J Invest Dermatol. 2008;128:1067–70. doi: 10.1038/jid.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heimall J, Spergel JM. Filaggrin mutations and atopy: consequences for future therapeutics. Expert Rev Clin Immunol. 2012;8:189–97. doi: 10.1586/eci.11.100. [DOI] [PubMed] [Google Scholar]
- 15.Ziyab AH, Karmaus W, Holloway JW, Zhang H, Ewart S, Arshad SH. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J Eur Acad Dermatol Venereol. 2013;27:e420–3. doi: 10.1111/jdv.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thyssen JP, Godoy-Gijon E, Elias PM. Ichthyosis vulgaris: the filaggrin mutation disease. Br J Dermatol. 2013;168:1155–1166. doi: 10.1111/bjd.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, DeBenedetto A, Schneider L, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2009;124:R7–12. doi: 10.1016/j.jaci.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Cornelissen C, Marquardt Y, Czaja K, Wenzel J, Frank J, Luscher-Firzlaff J, et al. IL-31 regulates differentiation and filaggrin expression in human organotypic skin models. J Allergy Clin Immunol. 2012;129:426–33. doi: 10.1016/j.jaci.2011.10.042. [DOI] [PubMed] [Google Scholar]
- 19.Deleuran M, Hvid M, Kemp K, Christensen GB, Deleuran B, Vestergaard C. IL-25 induces both inflammation and skin barrier dysfunction in atopic dermatitis. Chem Immunol Allergy. 2012;96:45–9. doi: 10.1159/000331871. [DOI] [PubMed] [Google Scholar]
- 20.Gutowska-Owsiak D, Schaupp AL, Salimi M, Taylor S, Ogg GS. Interleukin-22 downregulates filaggrin expression and affects expression of profilaggrin processing enzymes. Br J Dermatol. 2011;165:492–8. doi: 10.1111/j.1365-2133.2011.10400.x. [DOI] [PubMed] [Google Scholar]
- 21.Gutowska-Owsiak D, Schaupp AL, Salimi M, Selvakumar TA, McPherson T, Taylor S, et al. IL-17 downregulates filaggrin and affects keratinocyte expression of genes associated with cellular adhesion. Exp Dermatol. 2012;21:104–10. doi: 10.1111/j.1600-0625.2011.01412.x. [DOI] [PubMed] [Google Scholar]
- 22.Hvid M, Johansen C, Deleuran B, Kemp K, Deleuran M, Vestergaard C. Regulation of caspase 14 expression in keratinocytes by inflammatory cytokines-a possible link between reduced skin barrier function and inflammation? Exp Dermatol. 2011;20:633–6. doi: 10.1111/j.1600-0625.2011.01280.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim BE, Howell MD, Guttman-Yassky E, Gilleaudeau PM, Cardinale IR, Boguniewicz M, et al. TNF-α downregulates filaggrin and loricrin through c-Jun N-terminal kinase: role for TNF-α antagonists to improve skin barrier. J Invest Dermatol. 2011;131:1272–9. doi: 10.1038/jid.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angelova-Fischer I, Dapic I, Hoek AK, Jakasa I, Fischer TW, Zillikens D, et al. Skin barrier integrity and natural moisturizing factor levels after cumulative dermal exposure to alkaline agents in atopic dermatitis. Acta Derm Venereol. 2014;94:640–4. doi: 10.2340/00015555-1815. [DOI] [PubMed] [Google Scholar]
- 25.Rinnerthaler M, Duschl J, Steinbacher P, Salzmann M, Bischof J, Schuller M, et al. Age-related changes in the composition of the cornified envelope in human skin. Exp Dermatol. 2013;22:329–35. doi: 10.1111/exd.12135. [DOI] [PubMed] [Google Scholar]
- 26.De Benedetto A, Rafaels NM, McGirt LY, Ivanov AI, Georas SN, Cheadle C, et al. Tight junction defects in patients with atopic dermatitis. J Allergy Clin Immunol. 2011;127:773–86. doi: 10.1016/j.jaci.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuki T, Yoshida H, Akazawa Y, Komiya A, Sugiyama Y, Inoue S. Activation of TLR2 enhances tight junction barrier in epidermal keratinocytes. J Immunol. 2011;187:3230–7. doi: 10.4049/jimmunol.1100058. [DOI] [PubMed] [Google Scholar]
- 28.Kuo IH, Carpenter-Mendini A, Yoshida T, McGirt LY, Ivanov AI, Barnes KC, et al. Activation of epidermal toll-like receptor 2 enhances tight junction function: implications for atopic dermatitis and skin barrier repair. J Invest Dermatol. 2013;133:988–98. doi: 10.1038/jid.2012.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ring J, Mohrenschlager M, Weidinger S. Molecular genetics of atopic eczema. Chem Immunol Allergy. 2012;96:24–9. doi: 10.1159/000331807. [DOI] [PubMed] [Google Scholar]
- 30.Cookson WO. The genetics of atopic dermatitis: strategies, candidate genes, and genome screens. J Am Acad Dermatol. 2001;45:S7–9. doi: 10.1067/mjd.2001.117026. [DOI] [PubMed] [Google Scholar]
- 31.Strachan DP, Wong HJ, Spector TD. Concordance and interrelationship of atopic diseases and markers of allergic sensitization among adult female twins. J Allergy Clin Immunol. 2001;108:901–7. doi: 10.1067/mai.2001.119408. [DOI] [PubMed] [Google Scholar]
- 32.Fiset PO, Leung DY, Hamid Q. Immunopathology of atopic dermatitis. J Allergy Clin Immunol. 2006;118:287–90. doi: 10.1016/j.jaci.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 33.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–51. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 34.Thomsen SF, Ulrik CS, Kyvik KO, Hjelmborg JV, Skadhauge LR, Steffensen I, et al. Importance of genetic factors in the etiology of atopic dermatitis: a twin study. Allergy Asthma Proc. 2007;28:535–9. doi: 10.2500/aap2007.28.3041. [DOI] [PubMed] [Google Scholar]
- 35.Haagerup A, Bjerke T, Schiotz PO, Dahl R, Binderup HG, Tan Q, Kruse TA. A topic dermatitis - a total genome-scan for susceptibility genes. Acta Derm Venereol. 2004;84:346–52. doi: 10.1080/00015550410034426. [DOI] [PubMed] [Google Scholar]
- 36.Kurz T, Altmueller J, Strauch K, Ruschendorf F, Heinzmann A, Moffatt MF, et al. A genome-wide screen on the genetics of atopy in a multiethnic European population reveals a majoratopy locus on chromosome 3q21.3. Allergy. 2005;60:192–9. doi: 10.1111/j.1398-9995.2005.00646.x. [DOI] [PubMed] [Google Scholar]
- 37.Enomoto H, Noguchi E, Iijima S, Takahashi T, Hayakawa K, Ito M, et al. Single nucleotide polymorphism-based genome-wide linkage analysis in Japanese atopic dermatitis families. BMC Dermatol. 2007;7:5. doi: 10.1186/1471-5945-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guilloud-Bataille M, Bouzigon E, Annesi-Maesano I, Bousquet J, Charpin D, Gormand F, et al. Evidence for linkage of a new region (11p14) to eczema and allergic diseases. Hum Genet. 2008;122:605–14. doi: 10.1007/s00439-007-0439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christensen U, Moller-Larsen S, Nyegaard M, Haagerup A, Hedemand A, Brasch-Andersen C, et al. Linkage of atopic dermatitis to chromosomes 4q22, 3p24 and 3q21. Hum Genet. 2009;126:549–57. doi: 10.1007/s00439-009-0692-z. [DOI] [PubMed] [Google Scholar]
- 40.Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43:690–4. doi: 10.1038/ng.851. [DOI] [PubMed] [Google Scholar]
- 41.Henry J, Toulza E, Hsu CY, Pellerin L, Balica S, Mazereeuw-Hautier J, et al. Update on the epidermal differentiation complex. Front Biosci. 2012;17:1517–32. doi: 10.2741/4001. [DOI] [PubMed] [Google Scholar]
- 42.Marenholz I, Rivera VA, Esparza-Gordillo J, Bauerfeind A, Lee-Kirsch MA, Ciechanowicz A, et al. Association screening in the Epidermal Differentiation Complex (EDC) identifies an SPRR3 repeat number variant as a risk factor for eczema. J Invest Dermatol. 2011;131:1644–9. doi: 10.1038/jid.2011.90. [DOI] [PubMed] [Google Scholar]
- 43.Brown SJ, McLean WH. One remarkable molecule: filaggrin. J Invest Dermatol. 2012;132:751–62. doi: 10.1038/jid.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margolis DJ, Kim B, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, et al. Thymic stromal lymphopoietin variation, filaggrin loss of function, and the persistence of atopic dermatitis. JAMA Dermatol. 2014;150:254–9. doi: 10.1001/jamadermatol.2013.7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margolis DJ, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, Campbell LE, et al. The persistence of atopic dermatitis and filaggrin (FLG) mutations in a US longitudinal cohort. J Allergy Clin Immunol. 2012;130:912–7. doi: 10.1016/j.jaci.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu CK, Akiyama M, Nemoto-Hasebe I, Nomura T, Sandilands A, Chao SC, et al. Analysis of Taiwanese ichthyosis vulgaris families further demonstrates differences in FLG mutations between European and Asian populations. Br J Dermatol. 2009;161:448–51. doi: 10.1111/j.1365-2133.2009.09112.x. [DOI] [PubMed] [Google Scholar]
- 47.Barker JN, Palmer CN, Zhao Y, Liao H, Hull PR, Lee SP, et al. Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J Invest Dermatol. 2007;127:564–7. doi: 10.1038/sj.jid.5700587. [DOI] [PubMed] [Google Scholar]
- 48.Weidinger S, Illig T, Baurecht H, Irvine AD, Rodriguez E, Diaz-Lacava A, et al. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol. 2006;118:214–9. doi: 10.1016/j.jaci.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Morar N, Cookson WO, Harper JI, Moffatt MF. Filaggrin mutations in children with severe atopic dermatitis. J Invest Dermatol. 2007;127:1667–72. doi: 10.1038/sj.jid.5700739. [DOI] [PubMed] [Google Scholar]
- 50.Brown SJ, Relton CL, Liao H, Zhao Y, Sandilands A, Wilson IJ, et al. Filaggrin null mutations and childhood atopic eczema: a population-based case-control study. J Allergy Clin Immunol. 2008;121:940–6. doi: 10.1016/j.jaci.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ekelund E, Lieden A, Link J, Lee SP, D’Amato M, Palmer CN, et al. Loss-of-function variants of the filaggrin gene are associated with atopic eczema and associated phenotypes in Swedish families. Acta Derm Venereol. 2008;88:15–9. doi: 10.2340/00015555-0383. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Guo Y, Wang W, Shi M, Chen X, Yao Z. Mutations in the filaggrin gene in Han Chinese patients with atopic dermatitis. Allergy. 2011;66:420–7. doi: 10.1111/j.1398-9995.2010.02493.x. [DOI] [PubMed] [Google Scholar]
- 53.Marenholz I, Nickel R, Rüschendorf F, Schulz F, Esparza-Gordillo J, Kerscher T, et al. Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J Allergy Clin Immunol. 2006;118:866–71. doi: 10.1016/j.jaci.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 54.Osawa R, Konno S, Akiyama M, Nemoto-Hasebe I, Nomura T, Nomura Y, et al. Japanese-specific filaggrin gene mutations in Japanese patients suffering from atopic eczema and asthma. J Invest Dermatol. 2010;130:2834–6. doi: 10.1038/jid.2010.218. [DOI] [PubMed] [Google Scholar]
- 55.Winge MC, Bilcha KD, Lieden A, Shibeshi D, Sandilands A, Wahlgren CF, et al. Novel filaggrin mutation but no other loss-of-function variants found in Ethiopian patients with atopic dermatitis. Br J Dermatol. 2011;165:1074–80. doi: 10.1111/j.1365-2133.2011.10475.x. [DOI] [PubMed] [Google Scholar]
- 56.Thawer-Esmail F, Jakasa I, Todd G, Wen Y, Brown SJ, Kroboth K, et al. South African amaXhosa patients with atopic dermatitis have decreased levels of filaggrin breakdown products but no loss-of-function mutations in filaggrin. J Allergy Clin Immunol. 2014;133:280–2. doi: 10.1016/j.jaci.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margolis DJ, Gupta J, Apter AJ, Ganguly T, Hoffstad O, Papadopoulos M, et al. Filaggrin-2 variation is associated with more persistent atopic dermatitis in African American subjects. J Allergy Clin Immunol. 2014;133:784–9. doi: 10.1016/j.jaci.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polcari I, Becker L, Stein SL, Smith MS, Paller AS. Filaggrin gene mutations in African Americans with both ichthyosis vulgaris and atopic dermatitis. Pediatr Dermatol. 2014;31:489–92. doi: 10.1111/pde.12355. [DOI] [PubMed] [Google Scholar]
- 59.Kim SY, Yang SW, Kim HL, Kim SH, Kim SJ, Park SM, et al. Association between P478S polymorphism of the filaggrin gene & atopic dermatitis. Indian J Med Res. 2013;138:922–7. [PMC free article] [PubMed] [Google Scholar]
- 60.Lan CC, Tu HP, Wu CS, Ko YC, Yu HS, Lu YW, et al. Distinct SPINK5 and IL-31 polymorphisms are associated with atopic eczema and non-atopic and dermatitis in Taiwanese nursing population. Exp Dermatol. 2011;20:975–9. doi: 10.1111/j.1600-0625.2011.01374.x. [DOI] [PubMed] [Google Scholar]
- 61.Zhao LP, Di Z, Zhang L, Wang L, Ma L, Lv Y, et al. Association of SPINK5 gene polymorphisms with atopic dermatitis in Northeast China. J Eur Acad Dermatol Venereol. 2012;26:572–7. doi: 10.1111/j.1468-3083.2011.04120.x. [DOI] [PubMed] [Google Scholar]
- 62.Folster-Holst R, Stoll M, Koch WA, Hampe J, Christophers E, Schreiber S. Lack of association of SPINK5 polymorphisms with nonsyndromic atopic dermatitis in the population of Northern Germany. Br J Dermatol. 2005;152:1365–7. doi: 10.1111/j.1365-2133.2005.06602.x. [DOI] [PubMed] [Google Scholar]
- 63.Fortugno P, Furio L, Teson M, Berretti M, El Hachem M, Zambruno G, et al. The 420K LEKTI variant alters LEKTI proteolytic activation and results in protease deregulation: implications for atopic dermatitis. Hum Mol Genet. 2012;21:4187–200. doi: 10.1093/hmg/dds243. [DOI] [PubMed] [Google Scholar]
- 64.Nishio Y, Noguchi E, Shibasaki M, Kamioka M, Ichikawa E, Ichikawa K, et al. Association between polymorphisms in the SPINK5 gene and atopic dermatitis in the Japanese. Genes Immun. 2003;4:515–7. doi: 10.1038/sj.gene.6363889. [DOI] [PubMed] [Google Scholar]
- 65.Kato A, Fukai K, Oiso N, Hosomi N, Murakami T, Ishii M. Association of SPINK5 gene polymorphisms with atopic dermatitis in the Japanese population. Br J Dermatol. 2003;148:665–9. doi: 10.1046/j.1365-2133.2003.05243.x. [DOI] [PubMed] [Google Scholar]
- 66.Kabesch M, Carr D, Weiland SK, von Mutius E. Association between polymorphisms in serine protease inhibitor, kazal type 5 and asthma phenotypes in a large German population sample. Clin Exp Allergy. 2004;34:340–5. doi: 10.1111/j.1365-2222.2004.01860.x. [DOI] [PubMed] [Google Scholar]
- 67.Walley AJ, Chavanas S, Moffatt MF, Esnouf RM, Ubhi B, Lawrence R, et al. Gene polymorphism in Netherton and common atopic disease. Nat Genet. 2001;29:175–8. doi: 10.1038/ng728. [DOI] [PubMed] [Google Scholar]
- 68.Namkung JH, Lee JE, Kim E, Byun JY, Kim S, Shin ES, et al. Hint for association of single nucleotide polymorphisms and haplotype in SPINK5 gene with atopic dermatitis in Koreans. Exp Dermatol. 2010;19:1048–53. doi: 10.1111/j.1600-0625.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- 69.Lee HJ, Ha SJ, Han H, Kim JW. Distribution of HLA-A, B alleles and polymorphisms of TAP and LMP genes in Korean patients with atopic dermatitis. Clin Exp Allergy. 2001;31:1867–74. doi: 10.1046/j.1365-2222.2001.01042.x. [DOI] [PubMed] [Google Scholar]
- 70.Ismail A, Bousaffara R, Kaziz J, Zili J, el Kamel A, Tahar Sfar M, et al. Polymorphism in transporter antigen peptides gene (TAP1) associated with atopy in Tunisians. J Allergy Clin Immunol. 1997;99:216–23. doi: 10.1016/s0091-6749(97)70099-x. [DOI] [PubMed] [Google Scholar]
- 71.Braff MH, Di Nardo A, Gallo RL. Keratinocytes store the antimicrobial peptide cathelicidin in lamellar bodies. J Invest Dermatol. 2005;124:394–400. doi: 10.1111/j.0022-202X.2004.23443.x. [DOI] [PubMed] [Google Scholar]
- 72.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 73.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–95. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 74.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 75.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–45. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 76.Weidinger S, Klopp N, Rummler L, Wagenpfeil S, Novak N, Baurecht HJ, et al. Association of NOD1 polymorphisms with atopic eczema and related phenotypes. J Allergy Clin Immunol. 2005;116:177–84. doi: 10.1016/j.jaci.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 77.Weidinger S, Klopp N, Rummler L, Wagenpfeil S, Baurecht HJ, Gauger A, et al. Association of CARD15 polymorphisms with atopy-related traits in a population-based cohort of Caucasian adults. Clin Exp Allergy. 2005;35:866–72. doi: 10.1111/j.1365-2222.2005.02269.x. [DOI] [PubMed] [Google Scholar]
- 78.Kabesch M, Peters W, Carr D, Leupold W, Weiland SK, von Mutius E. Association between polymorphisms in caspase recruitment domain containing protein 15 and allergy in two German populations. J Allergy Clin Immunol. 2003;111:813–7. doi: 10.1067/mai.2003.1336. [DOI] [PubMed] [Google Scholar]
- 79.Lange J, Heinzmann A, Zehle C, Kopp M. CT genotype of promotor polymorphism C159T in the CD14 gene is associated with lower prevalence of atopic dermatitis and lower IL-13 production. Pediatr Allergy Immunol. 2005;16:456–7. doi: 10.1111/j.1399-3038.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 80.Sengler C, Haider A, Sommerfeld C, Lau S, Baldini M, Martinez F, et al. German Multicenter Allergy Study Group: Evaluation of the CD14 C-159 T polymorphism in the German Multicenter Allergy Study cohort. Clin Exp Allergy. 2003;33:166–9. doi: 10.1046/j.1365-2222.2003.01549.x. [DOI] [PubMed] [Google Scholar]
- 81.Liang XH, Cheung W, Heng CK, Liu JJ, Li CW, Lim B, Wang de Y. CD14 promoter polymorphisms have no functional significance and are not associated with atopic phenotypes. Pharmacogenet Genomics. 2006;16:229–36. doi: 10.1097/01.fpc.0000197466.14340.0f. [DOI] [PubMed] [Google Scholar]
- 82.Hashimoto S, Nakamura K, Oyama N, Kaneko F, Fujita T, Tsunemi Y, et al. Mannose-binding lectin (MBL) single nucleotide polymorphism is not associated with atopic dermatitis in Japanese patients. J Dermatol. 2005;32:1038–40. doi: 10.1111/j.1346-8138.2005.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 83.Mrabet-Dahbi S, Dalpke AH, Niebuhr M, Frey M, Draing C, Brand S, et al. The Toll-like receptor 2 R753Q mutation modifies cytokine production and Toll-like receptor expression in atopic dermatitis. J Allergy Clin Immunol. 2008;121:1013–9. doi: 10.1016/j.jaci.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 84.Niebuhr M, Langnickel J, Draing C, Renz H, Kapp A, Werfel T. Dysregulation of toll-like receptor-2 (TLR-2)-induced effects in monocytes from patients with atopic dermatitis: impact of the TLR-2 R753Q polymorphism. Allergy. 2008;63:728–34. doi: 10.1111/j.1398-9995.2008.01721.x. [DOI] [PubMed] [Google Scholar]
- 85.Niebuhr M, Langnickel J, Sigel S, Werfel T. Dysregulation of CD36 upon TLR-2 stimulation in monocytes from patients with atopic dermatitis and the TLR2 R753Q polymorphism. Exp Dermatol. 2010;19:e296–8. doi: 10.1111/j.1600-0625.2009.00989.x. [DOI] [PubMed] [Google Scholar]
- 86.Salpietro C, Rigoli L, Miraglia Del Giudice M, Cuppari C, Di Bella C, Salpietro A, et al. TLR2 and TLR4 gene polymorphisms and atopic dermatitis in Italian children: a multicenter study. Int J Immunopathol Pharmacol. 2011;24:33–40. doi: 10.1177/03946320110240S408. [DOI] [PubMed] [Google Scholar]
- 87.Levchenko LIu, Izmailova OV, Shlykova OA, Kaidashev IP. Polymorphism 896A/G of TLR4 gene rather than 1196C/T and 2258G/A of TLR2 gene determines severe and complicated course of atopic dermatitis in children. Tsitol Genet. 2013;47:46–53. [PubMed] [Google Scholar]
- 88.Oh DY, Schumann RR, Hamann L, Neumann K, Worm M, Heine G. Association of the toll-like receptor 2 A-16934T promoter polymorphism with severe atopic dermatitis. Allergy. 2009;64:1608–15. doi: 10.1111/j.1398-9995.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 89.Potaczek DP, Nastalek M, Okumura K, Wojas-Pelc A, Undas A, Nishiyama C. An association of TLR2–16934A >T polymorphism and severity/phenotype of atopic dermatitis. J Eur Acad Dermatol Venereol. 2011;25:715–21. doi: 10.1111/j.1468-3083.2010.03812.x. [DOI] [PubMed] [Google Scholar]
- 90.Weidinger S, Novak N, Klopp N, Baurecht H, Wagenpfeil S, Rummler L, et al. Lack of association between Toll-like receptor 2 and Toll-like receptor 4 polymorphisms and atopic eczema. J Allergy Clin Immunol. 2006;118:277–9. doi: 10.1016/j.jaci.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 91.Hoffjan S, Stemmler S, Parwez Q, Petrasch-Parwez E, Arinir U, Rohde G, et al. Evaluation ofthe toll-like receptor 6 Ser249Pro polymorphism in patients with asthma, atopic dermatitis and chronic obstructive pulmonary disease. BMC Med Genet. 2005;6:34. doi: 10.1186/1471-2350-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miedema KG, Tissing WJ, Te Poele EM, Kamps WA, Alizadeh BZ, Kerkhof M, et al. Polymorphisms in the TLR6 gene associated with the inverse association between childhood acute lymphoblastic leukemia and atopic disease. Leukemia. 2012;26:1203–10. doi: 10.1038/leu.2011.341. [DOI] [PubMed] [Google Scholar]
- 93.Novak N, Yu CF, Bussmann C, Maintz L, Peng WM, Hart J, et al. Putative association of a TLR9 promoter polymorphism with atopic eczema. Allergy. 2007;62:766–72. doi: 10.1111/j.1398-9995.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 94.Kim E, Lee JE, Namkung JH, Kim PS, Kim S, Shin ES, et al. Single nucleotide polymorphisms and the haplotype in the DEFB1 gene are associated with atopic dermatitis in a Korean population. J Dermatol Sci. 2009;54:25–30. doi: 10.1016/j.jdermsci.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 95.Mohamed HG, Abbas A, El-Kabarity RH, Diab HM. Association of beta-defensin 1 single nucleotide polymorphism with atopic dermatitis. Egypt J Immunol. 2009;16:125–38. [PubMed] [Google Scholar]
- 96.Prado-Montes de Oca E, García-Vargas A, Lozano-Inocencio R, Gallegos-Arreola MP, Sandoval-Ramírez L, Davalos-Rodriguez NO, et al. Association of beta-defensin 1 single nucleotide polymorphisms with atopic dermatitis. Int Arch Allergy Immunol. 2007;142:211–8. doi: 10.1159/000097023. [DOI] [PubMed] [Google Scholar]
- 97.Segat L, Guimarães RL, Brandão LA, Rocha CR, Zanin V, Trevisiol C, et al. Beta defensin-1 gene (DEFB1) polymorphisms are not associated with atopic dermatitis in children and adolescents from northeast Brazil (Recife, Pernambuco) Int J Dermatol. 2010;49:653–7. doi: 10.1111/j.1365-4632.2009.04343.x. [DOI] [PubMed] [Google Scholar]
- 98.Yamanaka K, Mizutani H. The role of cytokines/chemokines in the pathogenesis of atopic dermatitis. Curr Probl Dermatol. 2011;41:80–92. doi: 10.1159/000323299. [DOI] [PubMed] [Google Scholar]
- 99.Rigotti E, Piacentini GL, Ress M, Pigozzi R, Boner AL, Peroni DG. Transforming growth factor-beta and interleukin-10 in breast milk and development of atopic diseases in infants. Clin Exp Allergy. 2006;36:614–8. doi: 10.1111/j.1365-2222.2006.02483.x. [DOI] [PubMed] [Google Scholar]
- 100.Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. 2010;125:16–29. doi: 10.1016/j.jaci.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hussein YM, Shalaby SM, Nassar A, Alzahrani SS, Alharbi AS, Nouh M. Association between genes encoding components of the IL-4/IL-4 receptor pathway and dermatitis in children. Gene. 2014;545:276–81. doi: 10.1016/j.gene.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 102.Kayserova J, Sismova K, Zentsova-Jaresova I, Katina S, Vernerova E, Polouckova A, et al. A prospective study in children with a severe form of atopic dermatitis: clinical outcome in relation to cytokine gene polymorphisms. J Investig Allergol Clin Immunol. 2012;22:92–101. [PubMed] [Google Scholar]
- 103.Chang YT, Lee WR, Yu CW, Liu HN, Lin MW, Huang CH, et al. No association of cytokine gene polymorphisms in Chinese patients with atopic dermatitis. Clin Exp Dermatol. 2006;31:419–23. doi: 10.1111/j.1365-2230.2006.02124.x. [DOI] [PubMed] [Google Scholar]
- 104.Stavric K, Peova S, Trajkov D, Spiroski M. Gene polymorphisms of 22 cytokines in Macedonian children with atopic dermatitis. Iran J Allergy Asthma Immunol. 2012;11:37–50. [PubMed] [Google Scholar]
- 105.He JQ, Chan-Yeung M, Becker AB, Dimich-Ward H, Ferguson AC, Manfreda J, et al. Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes Immun. 2003;4:385–9. doi: 10.1038/sj.gene.6363985. [DOI] [PubMed] [Google Scholar]
- 106.Hosomi N, Fukai K, Oiso N, Kato A, Ishii M, Kunimoto H, et al. Polymorphisms in the promoter of the interleukin-4 receptor alpha chain gene are associated with atopic dermatitis in Japan. J Invest Dermatol. 2004;122:843–5. doi: 10.1111/j.0022-202X.2004.22338.x. [DOI] [PubMed] [Google Scholar]
- 107.Callard RE, Hamvas R, Chatterton C, Blanco C, Pembrey M, Jones R, et al. Avon Longitudinal Study of Parents and Children. An interaction between the IL-4R alpha gene and infection is associated with atopic eczema in young children. Clin Exp Allergy. 2002;32:990–3. doi: 10.1046/j.1365-2222.2002.01414.x. [DOI] [PubMed] [Google Scholar]
- 108.Oiso N, Fukai K, Ishii M. Interleukin 4 receptor alpha chain polymorphism Gln551Arg is associated with adult atopic dermatitis in Japan. Br J Dermatol. 2000;142:1003–6. doi: 10.1046/j.1365-2133.2000.03485.x. [DOI] [PubMed] [Google Scholar]
- 109.Tamura K, Suzuki M, Arakawa H, Tokuyama K, Morikawa A. Linkage and association studies of STAT6 gene polymorphisms and allergic diseases. Int Arch Allergy Immunol. 2003;131:33–8. doi: 10.1159/000070432. [DOI] [PubMed] [Google Scholar]
- 110.Rafatpanah H, Bennett E, Pravica V, McCoy MJ, David TJ, Hutchinson IV, et al. Association between novel GM-CSF gene polymorphisms and the frequency and severity of atopic dermatitis. J Allergy Clin Immunol. 2003;112:593–8. doi: 10.1016/s0091-6749(03)01797-4. [DOI] [PubMed] [Google Scholar]
- 111.Shimizu M, Matsuda A, Yanagisawa K, Hirota T, Akahoshi M, Inomata N, et al. Functional SNPs in the distal promoter of the ST2 gene are associated with atopic dermatitis. Hum Mol Genet. 2005;14:2919–27. doi: 10.1093/hmg/ddi323. [DOI] [PubMed] [Google Scholar]
- 112.Yamamoto N, Sugiura H, anaka K, Uehara M. Heterogeneity of interleukin 5 genetic background in atopic dermatitis patients: significant difference between those with blood eosinophilia and normal eosinophil levels. J Dermatol Sci. 2003;33:121–6. doi: 10.1016/s0923-1811(03)00149-x. [DOI] [PubMed] [Google Scholar]
- 113.Tsunemi Y, Saeki H, Nakamura K, Sekiya T, Hirai K, Fujita H, et al. Interleukin-12 p40 gene (IL12B) 3′-untranslated region polymorphism is associated with susceptibility to atopic dermatitis and psoriasis vulgaris. J Dermatol Sci. 2002;30:161–6. doi: 10.1016/s0923-1811(02)00072-5. [DOI] [PubMed] [Google Scholar]
- 114.Takahashi N, Akahoshi M, Matsuda A, Ebe K, Inomata N, Obara K, et al. Association of the IL12RB1 promoter polymorphisms with increased risk of atopic dermatitis and other allergic phenotypes. Hum Mol Genet. 2005;14:3149–59. doi: 10.1093/hmg/ddi347. [DOI] [PubMed] [Google Scholar]
- 115.Tsunemi Y, Saeki H, Nakamura K, Sekiya T, Hirai K, Kakinuma T, et al. Interleukin-13 gene polymorphism G4257A is associated with atopic dermatitis in Japanese patients. J Dermatol Sci. 2002;30:100–7. doi: 10.1016/s0923-1811(02)00065-8. [DOI] [PubMed] [Google Scholar]
- 116.Hummelshoj T, Bodtger U, Datta P, Malling HJ, Oturai A, Poulsen LK, et al. Association between an interleukin-13 promoter polymorphism and atopy. Eur J Immunogenet. 2003;30:355–9. doi: 10.1046/j.1365-2370.2003.00416.x. [DOI] [PubMed] [Google Scholar]
- 117.He JQ, Ruan J, Chan-Yeung M, Becker AB, Dimich-Ward H, Pare PD, et al. Polymorphisms of the GM-CSF genes and the development of atopic diseases in at-risk children. Chest. 2003;123:438S. doi: 10.1378/chest.123.3_suppl.438s. [DOI] [PubMed] [Google Scholar]
- 118.Liu X, Nickel R, Beyer K, Wahn U, Ehrlich E, Freidhoff LR, et al. An IL13 coding region variant is associated with a high total serum IgE level and atopic dermatitis in the German multicenter atopy study (MAS-90) J Allergy Clin Immunol. 2000;106:167–70. doi: 10.1067/mai.2000.107935. [DOI] [PubMed] [Google Scholar]
- 119.Novak N, Kruse S, Potreck J, Maintz L, Jenneck C, Weidinger S, et al. Single nucleotide polymorphisms of the IL18 gene are associated with atopic eczema. J Allergy Clin Immunol. 2005;115:828–33. doi: 10.1016/j.jaci.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 120.Arkwright PD, Chase JM, Babbage S, Pravica V, David TJ, Hutchinson IV. Atopic dermatitis is associated with a low-producer transforming growth factor beta (1) cytokine genotype. J Allergy Clin Immunol. 2001;108:281–4. doi: 10.1067/mai.2001.117259. [DOI] [PubMed] [Google Scholar]
- 121.Saeki H, Tsunemi Y, Asano N, Nakamura K, Sekiya T, Hirai K, et al. Analysis of GM-CSF gene polymorphisms (3606T/C and 3928C/T) in Japanese patients with atopic dermatitis. Clin Exp Dermatol. 2006;31:278–80. doi: 10.1111/j.1365-2230.2005.02052.x. [DOI] [PubMed] [Google Scholar]
- 122.Namkung JH, Lee JE, Kim E, Park GT, Yang HS, Jang HY, et al. An association between IL-9 and IL-9 receptor gene polymorphisms and atopic dermatitis in a Korean population. J Dermatol Sci. 2011;62:16–21. doi: 10.1016/j.jdermsci.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 123.IUIS/WHO Subcommittee on Chemokine Nomenclature. Chemokine/chemokine receptor nomenclature. Cytokine. 2003;21:48–9. [Google Scholar]
- 124.Yawalkar N, Uguccioni M, Scharer J, Braunwalder J, Karlen S, Dewald B, et al. Enhanced expression of eotaxin and CCR3 in atopic dermatitis. J Invest Dermatol. 1999;113:43–8. doi: 10.1046/j.1523-1747.1999.00619.x. [DOI] [PubMed] [Google Scholar]
- 125.Park CW, Lee BH, Han HJ, Lee CH, Ahn HK. Tacrolimus decreases the expression of eotaxin, CCR3, RANTES and interleukin-5 in atopic dermatitis. Br J Dermatol. 2005;152:1173–81. doi: 10.1111/j.1365-2133.2005.06474.x. [DOI] [PubMed] [Google Scholar]
- 126.Nickel RG, Casolaro V, Wahn U, Beyer K, Barnes KC, Plunkett BS, et al. Atopic dermatitis is associated with a functional mutation in the promoter of the C-C chemokine RANTES. J Immunol. 2000;164:1612–6. doi: 10.4049/jimmunol.164.3.1612. [DOI] [PubMed] [Google Scholar]
- 127.Bai B, Tanaka K, Tazawa T, Yamamoto N, Sugiura H. Association between RANTES promoter polymorphism -401A and enhanced RANTES production in atopic dermatitis patients. J Dermatol Sci. 2005;39:189–91. doi: 10.1016/j.jdermsci.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 128.Kozma GT, Falus A, Bojszko A, Krikovszky D, Szabo T, Nagy A, et al. Lack of association between atopic eczema/dermatitis syndrome and polymorphisms in the promoter region of RANTES and regulatory region of MCP-1. Allergy. 2002;57:160–3. doi: 10.1034/j.1398-9995.2002.1s3361.x. [DOI] [PubMed] [Google Scholar]
- 129.Wakugawa M, Torii H, Tamaki K. Eotaxin gene single nucleotide polymorphisms in the promoter and exon regions are not associated with susceptibility to atopic dermatitis, but two of them in the promoter region are associated with serum IgE levels in patients with atopic dermatitis. J Dermatol Sci. 2002;29:222–8. doi: 10.1016/s0923-1811(02)00038-5. [DOI] [PubMed] [Google Scholar]
- 130.Gangur V, Oppenheim JJ. Are chemokines essential or secondary participants in allergic responses? Ann Allergy Asthma Immunol. 2000;84:569–79. doi: 10.1016/S1081-1206(10)62403-9. [DOI] [PubMed] [Google Scholar]
- 131.Tsunemi Y, Sekiya T, Saeki H, Hirai K, Ohta K, Nakamura K, et al. Lack of association of CCR3 single nucleotide polymorphism with atopic dermatitis in Japanese population. J Dermatol Sci. 2003;33:130–3. doi: 10.1016/s0923-1811(03)00178-6. [DOI] [PubMed] [Google Scholar]
- 132.Tsunemi Y, Sekiya T, Saeki H, Hirai K, Ohta K, Nakamura K, et al. Lack of association of CCR4 single nucleotide polymorphism with atopic dermatitis in Japanese patients. Acta Derm Venereol. 2004;84:187–90. doi: 10.1080/00015550410025859. [DOI] [PubMed] [Google Scholar]
- 133.Iwanaga T, McEuen A, Walls AF, Clough JB, Keith TP, Rorke S, et al. Polymorphism of the mast cell chymase gene (CMA1) promoter region: lack of association with asthma but association with serum total immunoglobulin E levels in adult atopic dermatitis. Clin Exp Allergy. 2004;34:1037–42. doi: 10.1111/j.1365-2222.2004.02000.x. [DOI] [PubMed] [Google Scholar]
- 134.Weidinger S, Rummler L, Klopp N, Wagenpfeil S, Baurecht HJ, Fischer G, et al. Association study of mast cell chymase polymorphisms with atopy. Allergy. 2005;60:1256–61. doi: 10.1111/j.1398-9995.2005.00879.x. [DOI] [PubMed] [Google Scholar]
- 135.Badertscher K, Bronnimann M, Karlen S, Braathen LR, Yawalkar N. Mast cell chymase is increased in chronic atopic dermatitis but not in psoriasis. Arch Dermatol Res. 2005;296:503–6. doi: 10.1007/s00403-005-0542-3. [DOI] [PubMed] [Google Scholar]
- 136.Groneberg DA, Bester C, Grutzkau A, Serowka F, Fischer A, Henz BM, et al. Mast cells and vasculature in atopic dermatitis-potential stimulus of neoangiogenesis. Allergy. 2005;60:90–7. doi: 10.1111/j.1398-9995.2004.00628.x. [DOI] [PubMed] [Google Scholar]
- 137.Safronova OG, Vavilin VA, Lyapunova AA, Makarova SI, Lyakhovich VV, Kaznacheeva LF, et al. Relationship between glutathione S-transferase P1 polymorphism and bronchial asthma and atopic dermatitis. Bull Exp Biol Med. 2003;136:73–5. doi: 10.1023/a:1026001216033. [DOI] [PubMed] [Google Scholar]
- 138.Makarova SI, Dodunova EM, Ivanova GG, Vavilin VA, Kaznacheeva LF, Lyakhovich VV. Polymorphism of arylamine-N-acetyltransferase 2 gene is associated with the risk of atopic dermatitis. Bull Exp Biol Med. 2005;139:662–4. doi: 10.1007/s10517-005-0371-6. [DOI] [PubMed] [Google Scholar]
- 139.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–11. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 140.Yang KD, Liu CA, Chang JC, Chuang H, Ou CY, Hsu TY, Wang CL. Polymorphism of the immune-braking gene CTLA-4 (+49) involved in gender discrepancy of serum total IgE levels and allergic diseases. Clin Exp Allergy. 2004;34:32–7. doi: 10.1111/j.1365-2222.2004.01776.x. [DOI] [PubMed] [Google Scholar]
- 141.Vasilopoulos Y, Cork MJ, Murphy R, Williams HC, Robinson DA, Duff GW, et al. Genetic association between an AACC insertion in the 3′UTR of the stratum corneum chymotryptic enzyme gene and atopic dermatitis. J Invest Dermatol. 2004;123:62–6. doi: 10.1111/j.0022-202X.2004.22708.x. [DOI] [PubMed] [Google Scholar]
- 142.Hosomi N, Fukai K, Oiso N, Kato A, Fukui M, Ishii M. No association between atopic dermatitis and the SLC9A3R1-NAT9 RUNX1 binding site polymorphism in Japanese patients. Clin Exp Dermatol. 2005;30:192–3. doi: 10.1111/j.1365-2230.2005.01720.x. [DOI] [PubMed] [Google Scholar]
- 143.Nishio Y, Noguchi E, Ito S, Ichikawa E, Umebayashi Y, Otsuka F, et al. Mutation and association analysis of the interferon regulatory factor 2 gene (IRF2) with atopic dermatitis. J Hum Genet. 2001;46:664–7. doi: 10.1007/s100380170018. [DOI] [PubMed] [Google Scholar]
- 144.Cox HE, Moffatt MF, Faux JA, Walley AJ, Coleman R, Trembath RC, et al. Association of atopic dermatitis to the beta subunit of the high affinity immunoglobulin E receptor. Br J Dermatol. 1998;138:182–7. doi: 10.1046/j.1365-2133.1998.02108.x. [DOI] [PubMed] [Google Scholar]
- 145.Jang N, Stewart G, Jones G. Polymorphisms within the PHF11 gene at chromosome 13q14 are associated with childhood atopic dermatitis. Genes Immun. 2005;6:262–4. doi: 10.1038/sj.gene.6364169. [DOI] [PubMed] [Google Scholar]