Abstract
Recent estimates suggest that 1 in 12 of the global population suffers from diabetes mellitus. Approximately 40 % of those affected will go on to develop diabetes-related chronic kidney disease or diabetic nephropathy (DN). DN is a major cause of disability and premature death. Existing tests for prognostic purposes are limited and can be invasive, and interventions to delay progression are challenging. MicroRNAs (miRNAs) are a recently described class of molecular regulators found ubiquitously in human tissues and bodily fluids, where they are highly stable. Alterations in miRNA expression profiles have been observed in numerous diseases. Blood and tissue miRNAs are already established cancer biomarkers, and cardiovascular, metabolic and immune disease miRNA biomarkers are under development. Urinary miRNAs represent a potential novel source of non-invasive biomarkers for kidney diseases, including DN. In addition, recent data suggest that miRNAs may have therapeutic applications. Here, we review the utility of miRNAs as biomarkers for the early detection and progression of DN, assess emerging data on miRNAs implicated in DN pathology and discuss how the data from both fields may contribute to the development of novel therapeutic agents.
Keywords: Diabetic nephropathy, MicroRNA, Biomarker, Therapeutics, Fibrosis, Non-coding RNA
Introduction
Diabetic nephropathy (DN) is a complication of type 1 (T1DM) and type 2 diabetes mellitus (T2DM), the incidence of which is increasing, with an estimated 9 % of the adult population affected globally [1]. DN develops in approximately 40 % of patients with T1DM. DN is also common in those with T2DM, who are also at risk of renal dysfunction from other pathologies including ischaemic nephropathy. DN is the leading cause of renal failure requiring renal replacement therapy worldwide [2], but effective methods to identify and halt progression of pathophysiological changes of DN remain elusive.
Numerous risk factors for the development of DN have been postulated, including ethnicity and inherited genetic differences [3, 4]. Downstream contributors include hyperglycaemia and insulin resistance, and aberrant haemodynamics leading to intraglomerular hypertension and hyperfiltration.
Hyperglycaemia is associated with the generation of advanced glycation end products, renin-angiotensin system activation, increased cytokine production (most notably pro-fibrotic transforming growth factor (TGF)-β), reactive oxygen species and protein kinase C activity [5, 6]. Insulin resistance is associated with loss of endothelial and vascular modulation via nitric oxide and NF-κB beta pathways, with defects in podocyte-specific insulin signalling recently implicated in early DN [7]. The histological consequences of such insults include mesangial hyperexpansion, nodular glomerulosclerosis and tubulointerstitial fibrosis [8], the degree of which correlates to loss of glomerular filtration rate and, ultimately, renal failure [9].
MicroRNAs
MicroRNAs (MiRNAs) are ubiquitous endogenous, non-coding, single-stranded (ss)RNA transcripts, most frequently of 19–25 nucleotides in length, that act as post-transcriptional regulators of gene expression by blocking protein translation and/or inducing messenger RNA (mRNA) degradation. It is currently estimated that miRNAs regulate the expression of at least 60 % of all protein coding genes, and alterations in miRNA expression profiles have been observed in numerous pathological processes. Consequently, there is much current interest in miRNAs both as novel biomarkers and as potential targets for therapeutic intervention.
The first miRNA, lin-4, was identified in 1993 in the nematode Caenorhabditis elegans [10], the significance of this finding becoming clearer when a second miRNA, let-7, was also detected [11], and lin-4 and let-7 were found to be highly conserved in eukaryotes, suggesting an important functional role [12, 13]. Over 1000 human miRNAs have now been identified through bioinformatic and molecular cloning approaches, although functional validation has not yet been established in every case.
Biogenesis and Functional Mechanism of MicroRNA-Mediated Translational Repression
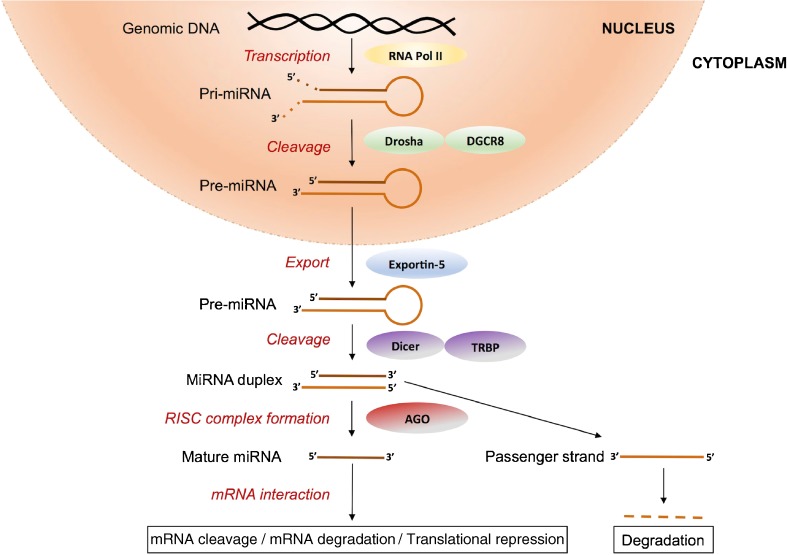
As shown in Fig. 1, canonical miRNA biogenesis begins with nuclear transcription, most frequently by RNA polymerase II, into transcripts known as primary miRNAs (pri-miRNAs). Pri-miRNAs vary in length, in some cases spanning kilobases, and have a distinctive stem-loop structure. They are cleaved within the nucleus into precursor miRNAs (pre-miRNAs) by a multiprotein complex that includes the ribonuclease III (RNase III) enzyme drosha and the cofactor Di George syndrome critical region gene 8 (DGCR8). DGCR8 binds to pri-miRNAs at a specific distance from the ssRNA-double-stranded (dsRNA) junction at the base of the stem-loop [14] and directs drosha to cleave the pri-miRNA 11 nucleotides from the ssRNA-dsRNA junction to form a 70–100 bp pre-miRNA [15]. Pre-miRNAs have a distinctive hairpin structure that is recognised by exportin-5, a dsRNA-binding protein that facilitates egression of pre-miRNAs from nucleus to cytoplasm. Subsequent cytoplasmic modification involves pre-miRNA cleavage to form mature ds miRNAs in a miRNA/miRNA* duplex consisting of two ss miRNAs referred to as the guide strand (miRNA) and the passenger strand (miRNA*). Pre-miRNA cleavage is facilitated by dicer, another RNase III enzyme within a large multiprotein complex that, in this case, contains the TAR RNA-binding protein (TRBP), which binds dsRNA and guides dicer to the correct cleavage site.
Fig. 1.
MicroRNA biogenesis and repression of gene expression
The function of miRNAs in silencing gene expression follows incorporation into the RNA-induced silencing complex (RISC), which includes the argonaute (AGO) proteins, after which the miRNA* is typically degraded. The RISC-bound miRNA then associates with the 3′-untranslated region (3′-UTR) of its target mRNA via classical Watson-Crick base pairing. Perfect sequence complementarity between miRNA and target mRNA 3′-UTR results in mRNA cleavage by AGO2, and imperfect complementarity results in translational repression and/or degradation of the mRNA target [16].
Utility of MicroRNAs as Diabetic Nephropathy Biomarkers
Existing Diabetic Nephropathy Biomarkers
Current DN diagnosis and monitoring of disease progression rely heavily on the detection of urinary microalbuminuria. However, not all patients with microalbuminuria progress to overt proteinuria and nephropathy, complicating prognosis. In addition, tissue damage and induction of inflammation have already occurred by the time that microalbuminuria is detectable. Furthermore, microalbuminuria is not specific to DN but is merely a hallmark of glomerular, and more specifically podocyte, dysfunction. Biopsy is the present diagnostic and prognostic test for intrinsic renal disease, but this highly invasive and expensive procedure has a 3 % risk of major complications.
There is therefore powerful incentive for research teams and industrial collaborators to develop improved prognostic and diagnostic tests for DN based on new biomarkers that provide earlier warning, more specific information on disease progression and, ideally, response to treatment. Pragmatism must be exercised in any biomarker screening programme, with downstream applicability borne in mind throughout. Consideration of test cost, speed and feasibility of entry into existing treatment pathways is essential to optimise use of resources. Nevertheless, the identification of differences in miRNA expression that do not have utility for biomarker analyses may still provide important information on disease mechanism, DN pathology and the identification of potential targets for therapy. The latter areas will be discussed later in this review.
Disease-Specific MicroRNA Signatures and Tissue Specificity
Expression profiling of miRNAs was first carried out in studies on carcinogenesis. Calin and colleagues showed that specific miRNAs were downregulated in circulating mononuclear cells from chronic lymphocytic leukaemia patients and identified a 13-miRNA expression profile associated with prognostic factors and disease progression [17, 18]. Tissue-specific miRNA expression has been reported, with perhaps the best-known example being the discovery that miR-122 expression issue was restricted to the mouse liver [19]. Mapping of miRNA expression in human tissues has since been reported, including profiles from cells in the different nephron regions [20].
Circulating MicroRNAs
Detection of circulating miRNAs offers a quicker, potentially automatable alternative method for DN diagnosis to biopsy and tissue analysis. Extracellular miRNAs were first reported in cell culture medium in an in vitro study that identified exosomal miRNAs that were transferred between mast cells, suggesting that cell-cell communication could be facilitated by fluid-borne miRNAs [21].
Significant differences in biological fluid miRNA expression profiles have now been observed in numerous disease states. In 2008, miRNAs were first detected in plasma and serum [22, 23]. Mitchell and colleagues identified circulating stable, readily detectable miRNAs in the blood of healthy human donors and patients with metastatic prostate cancer [22]. Detection of miR-141 in serum differentiated these two groups, underlining the potential of miRNAs for use as high-throughput diagnostic biomarkers [22]. Similarly, Lawrie and co-workers demonstrated the diagnostic potential of a miRNA panel composed of miR-21, miR-155 and miR-210 in diffuse large B cell lymphoma patients [23]. As reviewed by Allegra and colleagues, blood miRNAs are already established cancer biomarkers, and cardiovascular, metabolic and immune disease miRNA biomarkers are under development [24].
Stability is a key consideration when evaluating the use of any potential biomarker. Characterisation of blood-borne miRNAs by centrifugation and size exclusion chromatography identified two populations of circulating miRNAs: extracellular vesicle-associated (EVA-) and non-vesicle-associated (NVA-) miRNAs [25•], and much work has since been devoted to identifying the factors stabilising fluid-borne miRNAs, as well as the functions of these transcripts.
In human plasma, association of extracellular miRNAs with AGO proteins results in the formation of ribonucleoprotein complexes with the RISC that stabilise these transcripts (Fig. 1), and specific association of selected miRNAs with AGO2 has been demonstrated in plasma [26]. This association protects miRNAs from degradation in RNase-rich biological fluids such as blood [25•, 26, 27].
The role of extracellular vesicles as miRNA transporters that mediate signalling has been proposed [28]. Studies by Wang et al. correlated an increase in detectable NVA-miRNAs during serum-deprived cellular stress, and suggested that their secretion might drive cell regulatory mechanisms via miRNA-regulated cell-cell signalling [29]. The role of high-density lipoproteins in transporting functional NVA-miRNAs has also been posited [30].
Urinary MicroRNAs
The use of urinary miRNAs as disease biomarkers provides the additional advantages of non-invasive testing for which samples can be collected remotely and mailed to the test laboratory, as well as the possibility of a rapid point-of-care test. Recent in-depth studies have analysed the stability of urinary miRNAs [31–33]. Extremes of pH, prolonged storage at room temperature, multiple freeze-thaw cycles and subjection to RNase activity have all been used in these assessments, and we have recently demonstrated that urinary miRNAs are stabilised by association with AGO2 and exosomes [33]. Robust techniques for detection of miRNAs in urine and in urinary sediment have now been established [31–33], but more rapid and higher-throughput protocols are likely to be required for routine clinical testing.
Differential urinary miRNA expression profiles have been reported in kidney disease. One study showed decreased detection of miR-21 and miR-29, and increased miR-93 detection, in urine samples from IgA nephropathy patients compared to controls [32]. Argyropoulos et al. identified a panel of 27 differentially regulated urinary miRNAs that varied with DN progression [34].
The definition of the relative contributions of EVA- and NVA-miRNA populations to the total miRNA complement of each body fluid, the miRNA composition of these populations, and the functionality of miRNAs in each population is presently the subject of much ongoing research. It has been reported that serum and salivary miRNAs reside primarily in exosomes [35]. However, studies on plasma [25•, 26] as well as seminal fluid, dendritic cells, mast cells and ovarian cancer cells [36] contend that the majority of extracellular miRNAs are NVA. Furthermore, it has been suggested that the numbers of EVA-miRNAs are insufficient to mediate signalling [36]. Our recent work has provided definitive evidence of association of EVA urinary miR-16 and miR-192 with exosomes and NVA miR-16 and miR-192 with AGO2 [33].
MicroRNAs in Fibrosis and Diabetic Nephropathy Pathogenesis
MiR-192 and Fibrosis
As mentioned above, TGF-β is a key cytokine with fundamental importance in DN due to its roles in fibrosis and scarring. TGF-β signalling is regulated by miR-21, which can in turn regulate mature miR-21 expression [37], while our analysis of the TGF-β1 3′-UTR has shown evidence of post-transcriptional regulation by miR-744 [38].
An early step in fibrogenesis in DN involves the repression of E-cadherin by TGF-β1. Kato and colleagues observed that, in the early stages of renal injury, mouse mesangial cells treated with TGF-β1 showed upregulated expression of miR-192 and collagen alpha-2(I) [39]. Conversely, studies from this laboratory found decreased miR-192 expression in advanced-stage human DN renal biopsy samples accompanied by low estimated glomerular filtration rate and tubulointerstitial fibrosis [40]. In cultured human proximal tubular cells, we showed a similar TGF-β1-driven downregulation of miR-192 expression, while forced expression of this transcript repressed ZEB1 and ZEB2 expression, thereby derepressing E-cadherin and exerting an anti-fibrotic effect [40]. Further studies in diabetic apoE mice by Wang et al. identified a similar pattern of TGF-β1-mediated downregulation of miR-192 leading to E-cadherin inhibition [41]. We have since reviewed the pleiotropic roles of miR-192 in the kidney, with both anti- and pro-fibrotic effects that are apparently cell-type dependent [42].
MiR-21, miR-200 and miR-29 Families in Diabetic Nephropathy
One of the most abundant miRNAs in human tissues, miR-21, has been studied extensively and implicated in the pathogenesis of a number of malignancies. Similarities of these pathologies with fibrogenesis highlight miR-21 and its mRNA targets as possible candidates for DN progression, and renal miR-21 knockdown suppressed TGF-β1 signalling in a mouse model of T2DM [43•]. Increased miR-21 expression has also been identified in renal transplant patients with fibrotic kidney disease and in the urine of fibrotic patients with IgA nephropathy [32].
Lin and co-workers have recently reported functional interactions of miR-21 and TGF-β signalling by confirming that SMAD7, an inhibitor of TGF-β signalling, is a direct target of miR-21 and targets TGF-β by repressing SMAD7 expression, thus preventing rat renal tubular epithelial cell proliferation in an in vitro DN model [44].
The miR-200 family of miR-200a, miR-200b, miR-200c, miR-429 and miR-141 has been ascribed roles in maintaining epithelial differentiation, suggesting anti-fibrotic functions in DN [45]. Downregulated expression of these transcripts by TGF-β has been shown in cancer cell epithelial-to-mesenchymal transition (EMT) [46]. Conversely, upregulated miR-200b, miR-200c and miR-192 expression was observed in TGF-β-treated mouse mesangial cells [47].
Anti-fibrotic effects in DN have also been reported for miR-29 family members miR-29a, miR-29b and miR-29c. For example, diabetic mice expressing a miR-29a transgene showed improved renal function and better podocyte viability compared to wild-type diabetic mice [48]. Knockdown of miR-29a promoted histone deacetylase activity that lead to podocyte apoptosis, proteinuria and subsequent renal dysfunction [48].
Above, we have summarised studies on expression changes in DN and other renal diseases for the most widely studied miRNAs to date. The identification of miRNAs from biomarker screening studies and from disease model studies both have potential for use as therapeutic targets.
MicroRNAs as Therapeutic Targets
The majority of therapeutic studies investigating miRNAs have so far focussed on cancer, and miRNAs that regulate oncogene expression are often known as oncomiRs. Therapies targeting repression of single oncogenes using small molecule inhibitors have had limited therapeutic response due to the complexity of carcinogenesis, and this complexity is likely to be mirrored in other multifactorial diseases such as DN.
Targeting oncomiRs has the potential benefit of affecting the expression of multiple mRNAs that are oncomiR targets and therefore may target multiple pathways. For example, miR-34a has multiple target mRNAs that include proto-oncogene c-Met, cell cycle regulator cyclin-dependent kinase 4 and B cell lymphoma 2, an anti-apoptotic oncogenic protein [49]. A miR-34 mimic has been used in mouse models of lung cancer resulting in amelioration of disease progression correlated with a repression of all three targets [50]. Clearly, the possibility of deleterious off-target effects must also be considered.
Recent advances in the treatment of hepatitis C virus (HCV) infection represent the best example to date of a miRNA-based therapy. Chimpanzees treated with a miR-122 antagonist conferred long-lasting repression in HCV viraemia [51].
The need for more successful early-stage treatment options for DN patients remains. As discussed above, many studies are now investigating the potential of miRNAs as DN biomarkers. Identification of these miRNAs and other candidates selected on the basis of function and/or GWAS study outputs will allow investigation of their utility as targets to intervene in disease progression.
Several studies have manipulated miRNA expression in in vivo diabetic models. For example, in streptozotocin-treated diabetic mice, miR-192 was downregulated by a locked nucleic acid-modified anti-miRNA in the renal cortex to improve renal fibrosis symptoms [52, 53]. Similarly, knockdown of miR-29c resulted in the prevention of DN progression in db/db mice [54]. In a rat remnant kidney model of renal fibrosis, low-dose treatment with anti-cancer agent paclitaxel improved renal function, inhibited Smad2/3 activation and downregulated miR-192 expression [55].
Unlike liver-specific expression of miR-122, there are no renal-specific miRNAs, but ready uptake from the circulation by renal proximal tubular epithelial cells facilitates targeted delivery to the kidney. Ultrasound-microbubble-mediated gene transfer has been used to deliver plasmids to mammalian kidneys [56]. Developed first as ultrasound contrast agents, microbubbles have since been described as ‘theranostic’ agents, with uses in diagnostics and now in therapeutics, depending on the ultrasound parameters used [57]. High-intensity ultrasound causes microbubble oscillation, resulting in ‘inertial cavitation’ and release of the therapeutic agent(s) attached to, or contained within, the microbubbles [57]. Microbubble technology has been used to knockdown miR-21 expression in db/db mice, ameliorating microalbuminuria, renal fibrosis and inflammation [43•].
Conclusion
Emerging miRNA analyses continue to show promise for these transcripts as both biomarkers and as therapies in renal pathologies [58]. However, considerably more work will be required to consolidate and translate these findings. Biomarker studies of appropriate size and power will be required to augment the studies carried out to date, and animal model studies on mechanism represent the first stage on the long road to therapy. Nevertheless, the identification of miRNAs as novel targets for the improvement of DN patient outcomes provides hope for significant future clinical developments.
Acknowledgments
The Wales Kidney Research Unit is funded by Health and Care Research Wales. The authors also acknowledge support from Kidney Research UK Project Grant Award RP44/2014 to Timothy Bowen. This is a publication of independent research funded by the National Institute for Health Research (NIHR) Invention for Innovation (i4i) Programme (Grant Reference no. II-LA-0712-20003). The principal investigator for the grant is Timothy Bowen. The views expressed in this paper are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health, UK.
Compliance with Ethical Standards
Conflict of Interest
Kate Simpson, Alexa Wonnacott, and Donald J. Fraser declare that they have no conflict of interest.
Timothy Bowen reports grants from Health and Care Research Wales, Kidney Research UK and Knowledge Economy Skills Scholarship. In addition, Dr. Bowen has a pending patent with Cardiff University.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Microvascular Complications—Nephropathy
Contributor Information
Kate Simpson, Email: simpsonka1@cf.ac.uk.
Alexa Wonnacott, Email: wonnacottac@cf.ac.uk.
Donald J. Fraser, Email: fraserdj@cf.ac.uk
Timothy Bowen, Phone: 44-29-2074-8389, Email: bowent@cf.ac.uk.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
- 1.World Health Organisation . Global status on noncommunicable diseases. Geneva: World Health Organisation; 2014. [Google Scholar]
- 2.Jha V, Garcia-Garcia G, Iseki K, Li Z, Naicker S, Plattner B, et al. Chronic kidney disease: global dimension and perspectives. Lancet. 2013;382(9888):260–72. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 3.Cowie CC, Port FK, Wolfe RA, Savage PJ, Moll PP, Hawthorne VM. Disparities in incidence of diabetic end-stage renal disease according to race and type of diabetes. N Engl J Med. 1989;321(16):1074–9. doi: 10.1056/NEJM198910193211603. [DOI] [PubMed] [Google Scholar]
- 4.Regele F, Jelencsics K, Shiffman D, Pare G, McQueen MJ, Mann JF, et al. Genome-wide studies to identify risk factors for kidney disease with a focus on patients with diabetes. Nephrol Dial Transplant. 2015;30 Suppl 4:iv26–34. doi: 10.1093/ndt/gfv087. [DOI] [PubMed] [Google Scholar]
- 5.Makita Z, Radoff S, Rayfield EJ, Yang Z, Skolnik E, Delaney V, et al. Advanced glycosylation end products in patients with diabetic nephropathy. N Engl J Med. 1991;325(12):836–42. doi: 10.1056/NEJM199109193251202. [DOI] [PubMed] [Google Scholar]
- 6.Schrijvers BF, De Vriese AS, Flyvbjerg A. From hyperglycemia to diabetic kidney disease: the role of metabolic, hemodynamic, intracellular factors and growth factors/cytokines. Endocr Rev. 2004;25(6):971–1010. doi: 10.1210/er.2003-0018. [DOI] [PubMed] [Google Scholar]
- 7.Coward RJ, Saleem MA. Podocytes as a target of insulin. Curr Diabetes Rev. 2011;7(1):22–7. doi: 10.2174/157339911794273883. [DOI] [PubMed] [Google Scholar]
- 8.Kolset SO, Reinholt FP, Jenssen T. Diabetic nephropathy and extracellular matrix. J Histochem Cytochem. 2012;60(12):976–86. doi: 10.1369/0022155412465073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Iturbe B, Johnson RJ, Herrera-Acosta J. Tubulointerstitial damage and progression of renal failure. Kidney Int Suppl. 2005;99:S82–6. doi: 10.1111/j.1523-1755.2005.09915.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros C. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 11.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 12.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 15.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 17.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–9. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 20.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 24.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41(6):1897–912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 25.•.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beltrami C, Clayton A, Phillips AO, Fraser DJ, Bowen T. Analysis of urinary microRNAs in chronic kidney disease. Biochem Soc Trans. 2012;40(4):875–9. doi: 10.1042/BST20120090. [DOI] [PubMed] [Google Scholar]
- 28.Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr. 2007;1(3):156–8. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38(20):7248–59. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharkey JW, Antoine DJ, Park BK. Validation of the isolation and quantification of kidney enriched miRNAs for use as biomarkers. Biomarkers. 2012;17(3):231–9. doi: 10.3109/1354750X.2012.657246. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC. Urinary miR-21, miR-29, and miR-93: novel biomarkers of fibrosis. Am J Nephrol. 2012;36(5):412–8. doi: 10.1159/000343452. [DOI] [PubMed] [Google Scholar]
- 33.Beltrami C, Clayton A, Newbury LJ, Corish C, Jenkins RH, Phillips AO, et al. Stabilization of urinary microRNAs by association with exosomes and argonaute 2 protein. Non-Coding RNA. 2015;1(2):151–65. doi: 10.3390/ncrna1020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argyropoulos C, Wang K, McClarty S, Huang D, Bernardo J, Ellis D, et al. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0054662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111(41):14888–93. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin J, Jenkins RH, Bennagi R, Krupa A, Phillips AO, Bowen T, et al. Post-transcriptional regulation of transforming growth factor beta-1 by microRNA-744. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0025044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104(9):3432–7. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of microRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol. 2010;21(3):438–47. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B, Herman-Edelstein M, Koh P, Burns W, Jandeleit-Dahm K, Watson A, et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes. 2010;59(7):1794–802. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkins RH, Martin J, Phillips AO, Bowen T, Fraser DJ. Pleiotropy of microRNA-192 in the kidney. Biochem Soc Trans. 2012;40(4):762–7. doi: 10.1042/BST20120085. [DOI] [PubMed] [Google Scholar]
- 43.•.Zhong X, Chung AC, Chen HY, Dong Y, Meng XM, Li R, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56(3):663–74. doi: 10.1007/s00125-012-2804-x. [DOI] [PubMed] [Google Scholar]
- 44.Lin L, Gan H, Zhang H, Tang W, Sun Y, Tang X, et al. MicroRNA21 inhibits SMAD7 expression through a target sequence in the 3′ untranslated region and inhibits proliferation of renal tubular epithelial cells. Mol Med Rep. 2014;10(2):707–12. doi: 10.3892/mmr.2014.2312. [DOI] [PubMed] [Google Scholar]
- 45.Bracken CP, Khew-Goodall Y, Goodall GJ. Network-based approaches to understand the roles of miR-200 and other microRNAs in cancer. Cancer Res. 2015;75(13):2594–9. doi: 10.1158/0008-5472.CAN-15-0287. [DOI] [PubMed] [Google Scholar]
- 46.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato M, Arce L, Wang M, Putta S, Lanting L, Natarajan R. A microRNA circuit mediates transforming growth factor-beta1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011;80(4):358–68. doi: 10.1038/ki.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin CL, Lee PH, Hsu YC, Lei CC, Ko JY, Chuang PC, et al. MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J Am Soc Nephrol. 2014;25(8):1698–709. doi: 10.1681/ASN.2013050527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4(8) doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–30. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Putta S, Lanting L, Sun G, Lawson G, Kato M, Natarajan R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2012;23(3):458–69. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato M, Natarajan R. MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann N Y Acad Sci. 2015 doi: 10.1111/nyas.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long J, Wang Y, Wang W, Chang BH, Danesh FR. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem. 2011;286(13):11837–48. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun L, Zhang D, Liu F, Xiang X, Ling G, Xiao L, et al. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. J Pathol. 2011;225(3):364–77. doi: 10.1002/path.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deelman LE, Declèves AE, Rychak JJ, Sharma K. Targeted renal therapies through microbubbles and ultrasound. Adv Drug Deliv Rev. 2010;62(14):1369–77. doi: 10.1016/j.addr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sirsi S, Borden M. Microbubble compositions, properties and biomedical applications. Bubble Sci Eng Technol. 2009;1(1-2):3–17. doi: 10.1179/175889709X446507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bowen T, Jenkins RH, Fraser DJ. MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J Pathol. 2013;229(2):274–85. doi:10.1002/path.4119. [DOI] [PubMed]