Abstract
Synaptic signaling involves the release of neurotransmitter from presynaptic active zones (AZs). Proteins that regulate vesicle exocytosis cluster at AZs, composing the cytomatrix at the active zone (CAZ). At the Drosophila neuromuscular junction (NMJ), the small GTPase Rab3 controls the distribution of CAZ proteins across release sites, thereby regulating the efficacy of individual AZs. Here we identify Rab3-GEF as a second protein that acts in conjunction with Rab3 to control AZ protein composition. At rab3-GEF mutant NMJs, Bruchpilot (Brp) and Ca2+ channels are enriched at a subset of AZs, leaving the remaining sites devoid of key CAZ components in a manner that is indistinguishable from rab3 mutant NMJs. As the Drosophila homologue of mammalian DENN/MADD and Caenorhabditis elegans AEX-3, Rab3-GEF is a guanine nucleotide exchange factor (GEF) for Rab3 that stimulates GDP to GTP exchange. Mechanistic studies reveal that although Rab3 and Rab3-GEF act within the same mechanism to control AZ development, Rab3-GEF is involved in multiple roles. We show that Rab3-GEF is required for transport of Rab3. However, the synaptic phenotype in the rab3-GEF mutant cannot be fully explained by defective transport and loss of GEF activity. A transgenically expressed GTP-locked variant of Rab3 accumulates at the NMJ at wild-type levels and fully rescues the rab3 mutant but is unable to rescue the rab3-GEF mutant. Our results suggest that although Rab3-GEF acts upstream of Rab3 to control Rab3 localization and likely GTP-binding, it also acts downstream to regulate CAZ development, potentially as a Rab3 effector at the synapse.
Keywords: Active Zone, Drosophila, Neuromuscular Junction, Rab3, Synapse
Significance Statement
The release of neurotransmitter at synapses is regulated by proteins that form the presynaptic release machine. We show here that Rab3-GEF is required for controlling the protein composition of release sites in the Drosophila neuromuscular junction (NMJ). Our results indicate that loss of Rab3-GEF results in the development of NMJs that are structurally and functionally indistinguishable from NMJs that lack the protein Rab3. Our studies reveal that Rab3 and Rab3-GEF act within the same molecular mechanism and support a model wherein Rab3-GEF serves as a Rab3 effector to control active zone protein composition. This work provides further insight into a novel molecular mechanism that controls the strength of synapses.
Introduction
At synapses, neurotransmitter exocytosis is regulated by a complex of release machinery proteins. These proteins aggregate at presynaptic active zones (AZs) to form the cytomatrix at the active zone (CAZ), which controls the recruitment, docking, and priming of synaptic vesicles and enhances calcium channel accumulation (Gundelfinger and Fejtova, 2012). CAZ size and composition is variable, correlating with site-specific release efficacy. It is also plastic and can be remodeled to regulate synapse function (Sigrist and Schmitz, 2011; Michel et al., 2015). However, the mechanisms that control CAZ assembly and plasticity remain unclear.
The Drosophila larval neuromuscular junction (NMJ) is composed of hundreds of release sites defined by the apposition of postsynaptic clusters of glutamate receptors (GluRs) with presynaptic accumulations of the CAZ protein Bruchpilot (Brp), the fly ortholog of mammalian and Caenorhabditis elegans CAST/ERC/ELKS (Atwood et al., 1993; Marrus and DiAntonio, 2004; Kittel et al., 2006; Wagh et al., 2006). Several recent studies have striven to define the molecular mechanisms that control Brp aggregation at release sites. The number of AZs that contain Brp is reduced following the disruption of many proteins, including the cell adhesion proteins teneurin and neuroligin (Mosca et al., 2012), postsynaptic actin 57B (Blunk et al., 2014), the serine threonine kinase Unc-51 (Wairkar et al., 2009), the protein phosphatase PP2A (Viquez et al., 2009), and the vesicle proteins synaptotagmin-1 (Paul et al., 2015) and Rab3 (Graf et al., 2009). Of the mutants thus far identified, loss of Rab3 leads to the most severe disruption of Brp localization and distribution across AZs. At rab3 mutant NMJs, two-thirds of AZs are devoid of essential CAZ proteins. Rather, multiple presynaptic components including Brp, calcium channels, and T-bars are concentrated at a minority of sites where they form enlarged aggregates each containing an increased number of CAZ proteins (Graf et al., 2009; Ehmann et al., 2014). This altered distribution of CAZ proteins leads to the formation of a small number of functional sites, each with enhanced release efficacy (Peled and Isacoff, 2011; Graf et al., 2009).
Rab3 is a member of a large family of small GTPases that participates in the tethering of vesicles to target membranes (Bhuin and Roy, 2014). Similar to other small GTPases, Rab3 activity is controlled by the opposing actions of GTPase accelerating proteins (GAPs) and guanine nucleotide exchange factors (GEFs) that determine its GDP- versus GTP-bound state (Cherfils and Zeghouf, 2013). Rab3 function for controlling Brp distribution requires both GTP binding and membrane association (Chen et al., 2015), but not the binding of the effector protein RIM (Rab3 interacting molecule; Graf et al., 2012). However, the mechanism by which Rab3 controls CAZ formation and the proteins with which Rab3 must interact are largely unknown.
Through a genetic screen, we now identify Drosophila Rab3-GEF as an essential regulator of CAZ development at the NMJ. Rab3-GEF is the orthologue of mammalian DENN/MADD/Rab3-GEP and C. elegans AEX-3 (Iwasaki et al., 1997; Wada et al., 1997). We find that loss of rab3-GEF results in the development of NMJs that are morphologically and functionally indistinguishable from rab3 mutant NMJs. Prior work in other organisms indicates that Rab3-GEF acts upstream of Rab3 to control Rab3 activity via GDP to GTP exchange and by linking Rab3 to kinesin for proper axonal trafficking (Wada et al., 1997; Niwa et al., 2008). Consistent with these studies, we show that Rab3 trafficking is defective in the rab3-GEF mutant. However, these functions are not sufficient to explain the rab3-GEF phenotype. Transgenic expression of Rab3Q80L, a GTP-locked variant of Rab3 that rescues the rab3 mutant, enhances the accumulation of Rab3 at rab3-GEF mutant NMJs but still fails to rescue the defective distribution of AZ components. Thus, in addition to its roles regulating Rab3 activity and localization, these findings support the model that Rab3-GEF also functions downstream of Rab3, potentially as a Rab3 effector protein to dock associated vesicles at active zones.
Materials and Methods
Fly stocks
Flies were maintained at 25°C on standard fly food. Wild-type (WT) flies were Canton S (CS) or CS outcrossed to dvglut NMJX-Gal4 (Daniels et al., 2008). The following fly lines were obtained from the Bloomington Stock Center: the P-element line P{EPgy2}Rab3-GEFEY06511, the deficiency lines Df(1)ED7289 and Df(2R)ED2076, the UAS-cacophony-GFP line P(UAS-cac1-EGFP)422A (Kawasaki et al., 2004), and the UAS-YFP-rab3 line P{UASp-YFP.Rab3}4EHP05b (Zhang et al., 2007). The rab3rup mutant (Graf et al., 2009) and the lines containing the UAS-rab3 and UAS-rab3Q80L transgenes (Chen et al., 2015) were described previously.
Cloning of rab3-GEF cDNA
To generate the 6.3 kb full-length rab3-GEF cDNA, we cloned two overlapping cDNA fragments spanning the entire rab3-GEF ORF and then sequentially ligated individual fragments together. In brief, total RNA was extracted from adult Drosophila heads and reverse-transcription (RT) reactions were performed using gene-specific primers to obtain two cDNA fragments: NotI-NheI and NheI-XbaI. NotI and XbaI sites were introduced into the 5′ end and 3′ end, respectively, to facilitate subcloning into the pUAST vector to generate UAS-rab3-GEF.
Generation of rab3-GEF mutants
To generate the collection of EMS mutagenized lines, isogenic males were fed a solution of 5% sucrose and 10 mm methanesulfonic acid ethyl ester (EMS; Sigma-Aldrich) for 12–16 h. These males were mated with FM7a balanced females. <2000 F1 female virgin offspring were individually crossed with FM7a balanced males to create the stock collection used in the anatomical genetic screen.
Deletion of the rab3-GEF gene was generated by imprecise excision of the P-element P{EPgy2}Rab3-GEFEY06511, which is located within an intron near the 3′ end of the rab3-GEF gene. To initiate the excision, homozygous y, w-, P{EPgy2}Rab3-GEFEY06511 females were crossed to y, w-/Y; Xa/CyO; Δ2-3, Sb males carrying the transposase. F1 male progeny y, w-, P{EPgy2}Rab3-GEFEY06511/Y; +/CyO; +/Δ2-3, Sb were collected and crossed to FM7j, B1 females. The F2 progeny were screened for white-eyed females, which were crossed individually to FM7j, B1/Y males to set up stocks that were subsequently screened by PCR to identify those in which DNA surrounding the P-element had been excised. A single mutant with a substantial excision was identified, rab3-GEFSC225, and the following primer pairs were chosen to span the excised region, creating a PCR product that was sequenced to determine the precise nature of the excision: 5′-GCGTCACTTCTCCGATTCCG-3′ and 5′-CCCTTCTGGGTGAAGCACTTGCGG-3′.
Antibody generation
Polyclonal anti sera were generated against a synthetic Rab3-GEF peptide (CDSDRELTSRRDSDQQRLH). The peptide was conjugated to KLH and injected into rabbits for the generation of antiserum (YenZym Antibodies). The antiserum was affinity purified using the peptide conjugated to SulfoLink Coupling Resin (Thermo Scientific). The antibodies were eluted from the affinity column using 0.1 m glycine, pH 2.7. The purified α-Rab3-GEF antibody was used at 1:500.
Immunohistochemistry
Third-instar larvae were dissected in PBS and fixed in either Bouin’s fixative for 5 min or 4% formaldehyde for 20 min. Larvae were washed with PBS containing 0.1% Triton X-100 (PBT) and blocked in 5% NGS in PBT for 30 min, followed by overnight incubation in primary antibodies in 5% NGS in PBT, three washes in PBT, incubation in secondary antibodies in 5% NGS in PBT for 45 min, three final washes in PBT, and equilibration in 70% glycerol in PBS. Samples were mounted in VectaShield (Vector Laboratories). The following primary antibodies were used: mouse α-Brp, 1:250 (Developmental Studies Hybridoma Bank), rabbit α-DGluRIII, 1:2500 (Marrus et al., 2004), and rabbit α-Rab3, 1:1000. Goat Cy3-conjugated secondary antibodies against mouse and rabbit IgG and Cy3- and AlexaFluor 647-conjugated goat α-HRP were used at 1:1000 and were obtained from Jackson ImmunoResearch. AlexaFluor 488-conjugated goat secondary antibody against rabbit IgG and AlexaFluor 647-conjugated secondary antibodies against mouse and rabbit IgG were used at 1:1000 and were obtained from Life Technologies. FITC-conjugated goat α-GFP was used at 1:500 and was obtained from Abcam. Antibodies obtained from the Developmental Studies Hybridoma Bank were developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences of the University of Iowa, Iowa City, IA.
Imaging and analysis
Samples were imaged using a Nikon C2 confocal microscope. All genotypes for an individual experiment were imaged at the same gain and set such that signals from the brightest genotype for a given experiment were not saturating. NM4b NMJs on muscle 4 were analyzed. Images were analyzed using MetaMorph software (Molecular Devices). Statistical analysis was performed using either Student’s t test or ANOVA followed by Bonferroni’s test for comparison of samples within an experimental group. All histograms and measurements are shown as mean±SEM. Statistical analyses are summarized in Table 1. Italicized superscript letters associated with p values correspond to rows in the table.
Table 1.
Statistical Table
| Data structure | Type of test | Power (α=0.05) | |
|---|---|---|---|
| a (Fig. 2B) | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| b (Fig. 2C) | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| c (Fig. 2E) | Normally distributed | Student’s t test | 1 |
| d | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| e | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| f | Normally distributed | Student’s t test | 0.05241 |
| g | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| h | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| i (Fig. 4B) | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| j (Fig. 4C) | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| k | Normally distributed | ANOVA followed by Bonferroni’s test | 0.07662 |
| l | Normally distributed | ANOVA followed by Bonferroni’s test | 0.15701 |
| m (Fig. 5B) | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| n | Normally distributed | ANOVA followed by Bonferroni’s test | 0.15085 |
| o | Normally distributed | ANOVA followed by Bonferroni’s test | 0.13093 |
| p (Fig. 7C) | Normally distributed | Student’s t test | 1 |
| q (Fig. 7D) | Normally distributed | Student’s t test | 1 |
| r (Fig. 7D) | Normally distributed | Student’s t test | 1 |
| s (Fig. 7E) | Normally distributed | Student’s t test | 1 |
| t (Fig. 8B) | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| u (Fig. 8D) | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| v (Fig. 8E) | Normally distributed | ANOVA followed by Bonferroni’s test | 1 |
| w | Normally distributed | Student’s t test | 0.05575 |
| x | Normally distributed | Student’s t test | 0.0937 |
| y | Normally distributed | Student’s t test | 1 |
| z | Normally distributed | Student’s t test | 0.99976 |
| aa | Normally distributed | Student’s t test | 0.0588 |
| bb | Normally distributed | Student’s t test | 0.13625 |
To determine the percentage of GluR clusters apposed by Brp, Brp, and DGluRIII puncta were manually counted, and DGluRIII clusters that were not opposite to a detectable Brp punctum were counted as unapposed DGluRIII clusters. MetaMorph software was used for the quantification of Brp puncta size, Cacophony-GFP cluster size, and Rab3 average intensity. For measurement of Brp area, thresholds were kept constant across all genotypes for a given experiment. Although most Brp puncta were distinct, occasional overlapping puncta were separated with the cut drawing tool. For measurements of Rab3 intensity at the NMJ, the area of the NMJ was first defined by HRP and Brp signal. For measurements of Rab3 intensity in the nerve, the area of a nerve segment was first defined by HRP signal. The average intensity of Rab3 signal within each defined NMJ or nerve segment was then calculated, and the average background intensity was subtracted.
Electrophysiology
Two electrode voltage-clamp recordings were done in muscle six of abdominal segments A3 and A4 of wandering third instar larvae. Electrodes with resistances between 10 and 20 MΩ were used. The cells were held at −70 mV in a modified HL-3 saline solution (Stewart et al., 1994) containing the following (in mm): 70 NaCl, 5 KCl, 10 NaHCO3, 115 sucrose, 5 trehalose, 5 HEPES, 0.4 CaCl, and 10 MgCl. Recordings were done in voltage-clamp mode using an AxoClamp 2B Amplifier (Axon Instruments) low-pass filtered at 1kHz, and digitized at 10 kHz with an Instrutech ITC-18 computer interface using Patchmaster Software (HEKA Electronics). Cells requiring 1 nA or more holding current were discarded. Spontaneous events were recorded for 1 min and the average amplitude and frequency of the spontaneous events was quantified using Mini Analysis Software (Synaptosoft). For stimulated excitatory junctional currents (EJCs), AgCl2 wire in a glass suction electrode was used to stimulate the cut end of the segmental nerve for 1 ms at 1.5× the threshold voltage. A Master 8 stimulator and Isoflex stimulation isolation unit (AMPI) were used to control the duration and amplitude of the stimulation. The elicited currents from 10 successive stimulation protocols were averaged off-line using custom programs written by Josef Trapani (Amherst College) in Igor Pro software (WaveMetrics). For stimulus trains, the nerve was stimulated with 10 trains of five pulses at 20 Hz with 5 s rest intervals between trains. The 10 trains were averaged and the amplitudes of the first and fifth pulse were used to calculate the facilitation index. Histograms and measurements are shown as mean±SEM. Statistical analyses are summarized in Table 1. Italicized superscript letters associated with p values correspond to rows in the table.
Results
rab3-GEF mutants display defective Brp distribution
To identify proteins that control CAZ assembly and active zone composition, we conducted an anatomical genetic screen on a collection of EMS mutagenized Drosophila lines. A set of >2000 homozygous mutant larvae carrying mutagenized X chromosomes were dissected and costained with antibodies against the presynaptic active zone protein Brp and the essential glutamate receptor subunit DGluRIII. Immunostained NMJs were visualized by fluorescence microscopy and assessed for defects in Brp localization and distribution across active zones, including changes in Brp puncta size and number and the apposition of Brp with postsynaptic glutamate receptor clusters. From this screen, we identified two lines, MA18 and MA20, which displayed a prominent Brp distribution phenotype (Fig. 1). At MA18 and MA20 mutant NMJs, gross NMJ morphology and α-DGluRIII staining appears normal. However, Brp aggregates at only a subset of active zones in puncta that are significantly larger than normal, leaving the majority of GluR clusters unapposed to a Brp-positive AZ (Fig. 1B ).
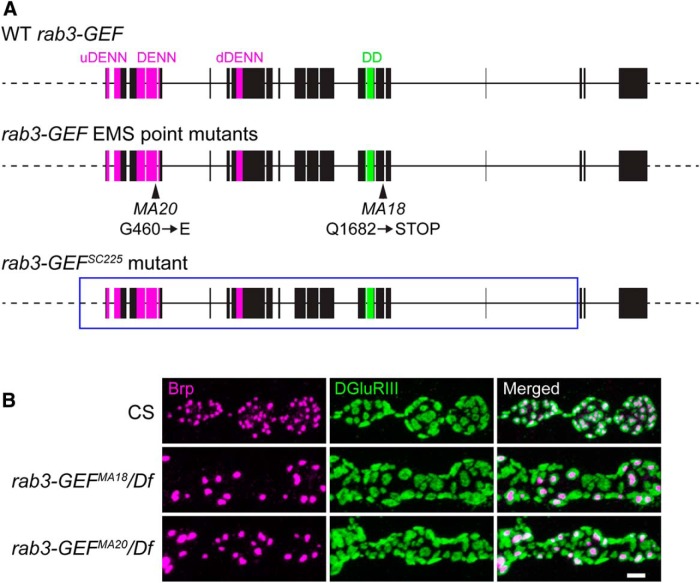
Figure 1.
Bruchpilot distribution is disrupted in rab3-GEF mutant NMJs. A, Schematic representations of the cloned WT Drosophila rab3-GEF gene, the rab3-GEFMA18 and rab3-GEFMA20 mutant genes generated by EMS mutagenesis, and the rab3-GEFSC225 mutant gene generated by P-element excision. Magenta exonic regions correspond to the tripartite DENN domain composed of the uDENN, DENN, and dDENN modules. The green exonic region corresponds to the DD. Arrowheads identify the Q1862STOP and G460E point mutations associated with the rab3-GEFMA18 and rab3-GEFMA20 mutant alleles, respectively. Boxed region of the rab3-GEFSC225 mutant allele denotes the part of the gene that is excised. B, Confocal images of muscle 4 NMJs costained with antibodies against the presynaptic AZ protein Brp (magenta) and the postsynaptic receptor DGluRIII (green) from WT (CS), rab3-GEFMA18 mutant (rab3-GEFMA18/Df(1)ED7289) and rab3-GEFMA20 mutant (rab3-GEFMA20/Df(1)ED7289) third-instar larvae. Scale bar, 2 µm.
We noted that the distribution of Brp in MA18 and MA20 mutant NMJs was remarkably similar to the synaptic phenotype observed in rab3 mutant larvae (Graf et al., 2009). However, ma18 and ma20 are not mutant alleles of rab3 as they map to the X chromosome whereas rab3 is on the second chromosome. To determine the gene responsible for the synaptic phenotype observed in MA18 and MA20 mutant NMJs, we first mapped the mutations by complementation testing. MA18 and MA20 are recessive alleles and transheterozygous larvae carrying both MA18 and MA20 failed to complement one another, suggesting that they are mutant alleles of the same gene. We identified the deficiency chromosome Df(1)ED7289 as failing to complement both MA18 and MA20 (Fig. 1B ). This deficiency deletes 18 predicted genes between 13A5 and 13A12, including rab3-GEF, the Drosophila orthologue of mammalian DENN/MADD/Rab3-GEP and C. elegans AEX-3. In mammals and C. elegans, DENN/MADD/Rab3-GEP and AEX-3 are important regulators of Rab3, acting as GEFs that catalyze GDP to GTP exchange, and disruption of these genes results in defective Rab3 function (Iwasaki et al., 1997; Wada et al., 1997; Oishi et al., 1998; Coppola et al., 2002; Mahoney et al., 2006; Niwa et al., 2008). Due to the similarity between rab3 mutant and MA18 and MA20 mutant NMJs, we tested whether rab3-GEF was responsible for the synaptic phenotype in MA18 and MA20 mutant larvae.
We first analyzed the structure and sequence of the Drosophila rab3-GEF gene. Sequence of the rab3-GEF transcript had been predicted by analysis of incomplete CDS clones but no full-length rab3-GEF cDNA had been identified. To determine experimentally the sequence of the rab3-GEF coding region, we cloned the gene in two overlapping cDNA fragments from mRNA extracted from adult Drosophila heads and ligated the two fragments together to form a 6.3 kb cDNA that spanned the entire rab3-GEF open reading frame. Examination of the full-length cDNA indicates that the Drosophila rab3-GEF transcript codes for a 2115 aa protein that contains the standard domains present in its mammalian and C. elegans orthologues (Schievella et al., 1997; Chow et al., 1998; Levivier et al., 2001; Marat et al., 2011), including an N-terminal tripartite DENN domain composed of the typical uDENN (upstream DENN), DENN, and dDENN (downstream DENN) modules and a single Death domain (DD) near its C-terminal end (Fig. 1A ). Drosophila Rab3-GEF is ∼500–700 amino acids longer than its orthologues due to a modest lengthening of the regions separating domains and the addition of ∼300 amino acids at its C-terminal end following the Death domain.
Sequencing of rab3-GEF in the MA18 and MA20 mutant lines revealed mutations within rab3-GEF coding regions predicted to disrupt or eliminate important domains (Fig. 1A ). rab3-GEFMA18 contains a nonsense mutation that results in a premature stop codon in place of a glutamine residue at position 1682 downstream of the Death Domain. rab3-GEFMA20 contains a missense mutation in which glycine 460 is replaced by glutamate (G460E). Residue 460 is located within the second DENN module of the tripartite DENN domain among a sequence of primarily non-polar amino acids. Although the precise functional deficits of Rab3-GEF in the MA18 and MA20 lines remain unknown, our analysis indicates that rab3-GEFMA18 and rab3-GEFMA20 are mutant alleles of rab3-GEF, and their lack of complementation with each other and Df(1)ED7289 suggests that the observed active zone phenotype in each is due to rab3-GEF dysfunction.
rab3-GEFSC225 phenocopies the active zone phenotype of the rab3 mutant
Because the mutations present in the rab3-GEFMA18 and rab3-GEFMA20 alleles do not result in clear functional nulls, we generated a third rab3-GEF mutant by excision of a transposable P-element located within an intron near the 3′ end of the gene. Imprecise excision of P{EPgy2}Rab3-GEFEY06511 generated rab3-GEFSC225 which contains a large excision that results in the deletion of the majority of the rab3-GEF gene, including the start codon and part of the predicted 5′ UTR (Fig. 1A ). In the rab3-GEFSC225 mutant, the entire coding region is removed except for the last three exons that code for the final 364 amino acids of the protein, which is unlikely to be expressed because there is no start codon. Thus, we hypothesized that rab3-GEFSC225 should represent a genetic and functional null allele of rab3-GEF and utilized it as a basis for our subsequent studies of Rab3-GEF function.
Initial analysis of the MA18 and MA20 mutants indicated that rab3-GEF disruption results in defective Brp distribution across active zones. Examination of Brp and DGluRIII localization in the rab3-GEFSC225 mutant demonstrates that excision of the rab3-GEF gene also results in the concentration of Brp protein at a small fraction of available sites (Fig. 2A ). Quantification of immunostained NMJs reveals that whereas Brp is apposed to nearly all GluR clusters at WT NMJs, in the rab3-GEFSC225 mutant, only one-third of GluR clusters are apposed to a Brp punctum (Fig. 2B ). Furthermore, average Brp punctum size is twice as large in the rab3-GEFSC225 mutant compared with WT (Fig. 2C ). In addition, Brp-GluR apposition and average Brp area are the same in both homozygous rab3-GEFSC225 mutant larvae and transheterozygotes of rab3-GEFSC225 and Df(1)ED7289 (Fig. 2A–C ), indicating that rab3-GEFSC225 behaves as a genetic null. Despite the observed active zone phenotype, rab3-GEFSC225 mutants are both viable and fertile.
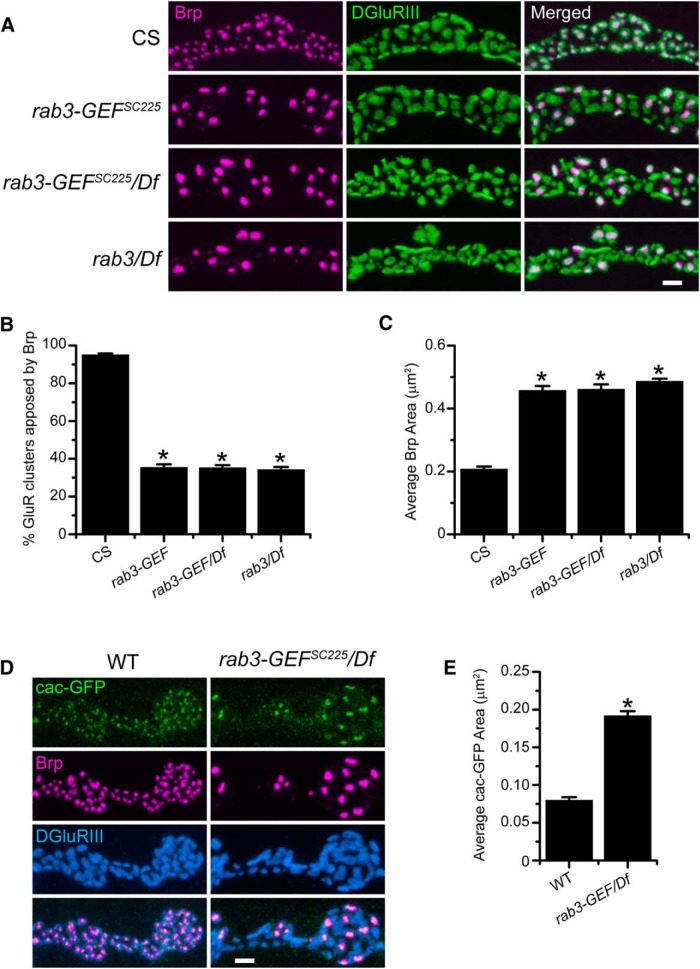
Figure 2.
The altered distribution of Brp in rab3-GEFSC225 mutant NMJs is identical to the rab3 mutant and results in the clustering of Ca2+ channels at a small number of release sites. A, Images of NMJs costained with α-Brp (magenta) and α-DGluRIII (green) from CS, homozygous rab3-GEFSC225 mutant larvae, rab3-GEFSC225/Df(1)ED7289 mutant larvae, and rab3rup/Df(2R)ED2076 mutant larvae. Scale bar, 2 µm. B, C, Histograms show (B) the average percentage of DGluRIII clusters apposed to Brp puncta per NMJ and (C) the average area of individual Brp puncta for the genotypes listed in A. n = 10 NMJs for all genotypes; (B) *p≪0.000001a versus CS; (C) *p≪0.000001b versus CS. D, Images of NMJs expressing cacophony-GFP (green) driven by dvglut NMJX-Gal4 in from WT (dvglut NMJX-Gal4/+; UAS-Cacophony-GFP/+) and rab3-GEFSC225 mutant (dvglutNMJX-Gal4, rab3-GEFSC225/Df(1)ED7289; UAS-Cacophony-GFP/+) larvae. NMJs are costained with α-Brp (magenta) and α-DGluRIII (blue) antibodies. Scale bar, 2 µm. E, Histogram shows the average area of cacophony-GFP clusters for the genotypes listed in D. n = 10 NMJs for both genotypes; *p≪0.000001c versus WT.
In mammals and C. elegans, loss of DENN/MADD/Rab3-GEP and AEX-3 causes defects in Rab3 function (Iwasaki et al., 1997; Yamaguchi et al., 2002; Mahoney et al., 2006; Niwa et al., 2008). To determine whether loss of Rab3-GEF mimics loss of Rab3, we compared Brp distribution between the two mutants and examined the degree of similarity (Fig. 2A–C ). Quantification reveals no difference in Brp localization between rab3/Df and rab3-GEFSC225/Df mutants, both in terms of percentage of GluR clusters apposed to Brp (p=0.601a) and average Brp area (p=0.156b). Interestingly, rab3-GEFSC225 is the first mutant identified to have a Brp distribution phenotype that is indistinguishable from the rab3 mutant.
We also wished to quantify the severity of the Brp distribution phenotype in rab3-GEFMA18 and rab3-GEFMA20 mutant NMJs where the functional disruption of Rab3-GEF is less clear. Whereas the percentage of GluR clusters apposed to Brp in rab3-GEFMA18 and rab3-GEFMA20 mutant NMJs is significantly reduced compared with WT, the reduction is slightly less severe than in the rab3 mutant (CS: 93.8 ± 0.9%; rab3-GEFMA18/Df(1)ED7289: 38.0 ± 1.3%; rab3-GEFMA20/Df(1)ED7289: 39.4 ± 1.6%; rab3rup/Df(2R)ED2076: 31.6 ± 1.2%; n = 10 NMJs for all genotypes; p≪0.000001d for CS vs MA18 and MA20; p=0.00499d for rab3 vs MA18; p=0.000393d for rab3 vs MA20). Conversely, average Brp area is similar in rab3-GEFMA18 and rab3-GEFMA20, and rab3 mutant NMJs (CS: 0.19 ± 0.005 µm2; rab3-GEFMA18/Df(1)ED7289: 0.41 ± 0.014 µm2; rab3-GEFMA20/Df(1)ED7289: 0.44 ± 0.010 µm2; rab3rup/Df(2R)ED2076: 0.43 ± 0.011 µm2; n = 10 NMJs for all genotypes; p≪0.000001e for CS vs MA18 and MA20; p=1.00e for rab3 vs MA18 and MA20). These results suggest that rab3-GEFMA18 and rab3-GEFMA20 behave as strong hypomorphs.
Our results indicate that Brp distribution is indistinguishable between rab3-GEFSC225 and rab3 mutant NMJs. Do other morphological similarities exist? In rab3 mutant NMJs, multiple presynaptic components required for vesicle release are concentrated at the active zones where Brp is aggregated, including calcium channels (Graf et al., 2009). To determine whether presynaptic calcium channels are distributed similarly at rab3-GEFSC225 mutant NMJs, we used the Gal4/UAS system to express a GFP-tagged version of the calcium channel subunit Cacophony in motor neurons (Fig. 2D ). When expressed in WT neurons, cacophony-GFP accumulates opposite DGluRIII at the majority of release sites. Conversely, in rab3-GEFSC225 mutant NMJs, Cacophony-GFP aggregates selectively at Brp-positive active zones, failing to accumulate opposite the majority of DGluRIII clusters (Fig. 2D ). Furthermore, aggregations of cacophony-GFP protein are larger in the rab3-GEFSC225 mutant compared with WT (Fig. 2E ). Thus, in terms of both Brp and calcium-channel distribution, the rab3-GEFSC225 mutant phenocopies the morphological defects observed at rab3 mutant NMJs. These results are consistent with the hypothesis that Rab3 function is compromised in the rab3-GEFSC225 mutant and that the developmental defects observed in the two mutants are mechanistically related.
Rab3-GEF localizes to both the NMJ and cell body
Our morphological analysis indicates that rab3-GEF dysfunction results in defective AZ development. To study how Rab3-GEF functions to control synapse development, we first examined where Rab3-GEF protein localizes within Drosophila neurons. We generated a polyclonal antibody to a protein epitope in the region separating the uDENN and DENN modules of Drosophila Rab3-GEF. This antibody stains WT NMJs in a mottled pattern throughout the synaptic terminal (Fig. 3A ). α-Rab3-GEF and α-Brp staining partially overlap, although regions of greater Rab3-GEF intensity have a tendency to be adjacent to Brp puncta rather than colocalizing with them (Fig. 3A , inset). Because Rab3-GEF may regulate Rab3 function, we also examined the degree of colocalization between Rab3 and Rab3-GEF at the NMJ by expressing YFP-tagged Rab3 in WT neurons with the dvglutNMJX-Gal4 driver (Fig. 3B ). Rab3-GEF and YFP-Rab3 also partially overlap, although the colocalization between YFP-Rab3 and Brp is more prominent, as described previously for endogenous Rab3 (Graf et al., 2009; Chen et al., 2015). In addition, Rab3-GEF localization at the NMJ appears generally unaffected by rab3 disruption. The average intensity of Rab3-GEF staining at rab3 mutant NMJs is similar to WT (CS: 100.0 ± 4.4 a.u.; rab3/Df: 99.1 ± 4.0 a.u.; n = 8 NMJs for both genotypes; p=0.879f).
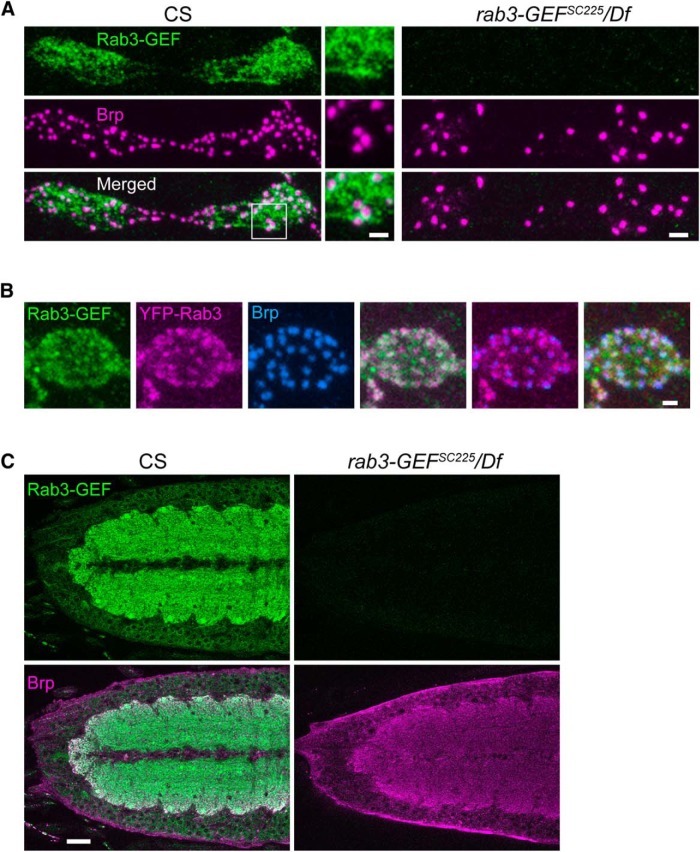
Figure 3.
α-Rab3-GEF immunostaining reveals Rab3-GEF protein at the NMJ and in ventral nerve cord cell bodies and neuropil. A, Images of NMJs immunostained with α-Rab3-GEF (green) and α-Brp (magenta) from CS and rab3-GEFSC225/Df(1)ED7289 mutant larvae. Inset, Magnified view of boxed region, indicating partial overlap of α-Rab3-GEF and α-Brp signal but little colocalization. Scale bars: lower-magnification images, 2 µm; higher-magnification inset, 1 µm. B, Image of a single bouton of a WT NMJ expressing UAS-rab3-YFP (magenta) driven by the dvglutNMJX-Gal4 driver and costained with α-Rab3-GEF (green) and α-Brp (blue). Scale bar, 1 µm. C, Single optical sections of ventral nerve cords from CS and rab3-GEFSC225/Df(1)ED7289 mutant larvae immunostained with α-Rab3-GEF (green) and α-Brp (magenta). In WT brains, Rab3-GEF immunostaining is prominent in the neuropil, as well as in the surrounding neuronal cell bodies visualized by cytosolic Rab3-GEF surrounding immuno-negative nuceli. Scale bar, 20 µm.
Examination of the larval ventral nerve cord (VNC) also reveals prominent α-Rab3-GEF staining in the cell bodies and neuropil of the brain (Fig. 3C ), suggesting that Drosophila Rab3-GEF may function both in neuronal cell bodies and at the synapse. Importantly, α-Rab3-GEF signal is completely absent in rab3-GEFSC225 mutant animals, both at the NMJ and in the brain (Fig. 3A,C ), indicating that the antibody is specific for Rab3-GEF and that Rab3-GEF protein is not expressed in the rab3-GEFSC225 mutant.
We also examined Rab3-GEF protein localization in the hypomorphic rab3-GEFMA18 and rab3-GEFMA20 mutants. Rab3-GEF immunostaining reveals that the MA18 point mutation results in a decrease in Rab3-GEF protein. Average Rab3-GEF intensity is significantly reduced at the NMJ (CS: 100.0 ± 6.8 a.u.; rab3-GEFMA18/Df: 5.9 ± 0.5 a.u; n = 8 NMJs for both genotypes; p≪0.000001g) and within the cell body enriched cortex of the VNC (CS: 100.0 ± 5.3 a.u.; rab3-GEFMA18/Df: 28.6 ± 2.3 a.u.; n = 8 single optical sections for both genotypes; p≪0.000001h). Conversely, in the rab3-GEFMA20 mutant, Rab3-GEF staining is close to normal in VNC cell bodies (CS: 100.0 ± 5.3 a.u.; rab3-GEFMA20/Df: 84.5 ± 4.5 a.u.; n = 8 single optical sections for both genotypes; p=0.04962h) but is nearly absent at NMJs (CS: 100.0 ± 6.8 a.u.; rab3-GEFMA20/Df: 5.0 ± 0.5 a.u; n = 8 NMJs for both genotypes; p≪0.000001g). Because the rab3-GEFMA20 mutant has a strong phenotype, the selective loss of Rab3-GEF at mutant NMJs but not within cell bodies suggests that there may be a local requirement for Rab3-GEF protein at the NMJ.
Transgenic expression of UAS-Rab3-GEF rescues Brp distribution
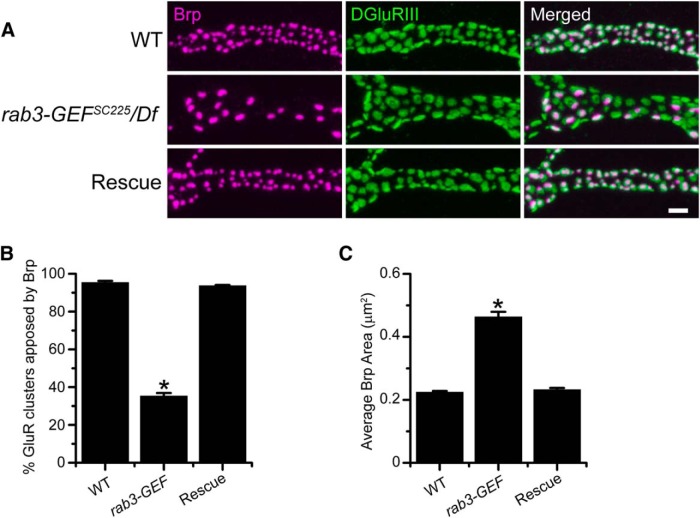
The complete loss of Rab3-GEF protein in the rab3-GEFSC225 mutant suggests that the AZ phenotype is due to the absence of rab3-GEF. However, to ensure that the loss of Rab3-GEF is responsible for the synaptic development defects observed in the rab3-GEFSC225 mutant and to assess cell autonomy, we expressed transgenic rab3-GEF in the rab3-GEFSC225 mutant background using the Gal4/UAS system. Neuronal expression of UAS-rab3-GEF by the dvglutNMJX-Gal4 driver fully rescues the Brp distribution phenotype of the rab3-GEFSC225 mutant (Fig. 4A ). Whereas Brp fails to cluster at >60% of GluR clusters in the rab3-GEFSC225 mutant, the percentage of GluR clusters apposed to Brp is similar to WT when UAS-rab3-GEF is expressed in the rab3-GEFSC225 mutant (Fig. 4B ). Transgenic expression of UAS-rab3-GEF also reduces average Brp puncta size to WT levels (Fig. 4C ). Hence, neuronal expression of Rab3-GEF is required for proper AZ development.
Figure 4.
Neuronal expression of transgenic rab3-GEF rescues Brp distribution in rab3-GEFSC225 mutant NMJs. A, Images of NMJs costained with α-Brp (magenta) and α-DGluRIII (green) from WT (dvglutNMJX-Gal4/+), the rab3-GEFSC225 mutant (dvglutNMJX-Gal4, rab3-GEFSC225/Df(1)ED7289), and rescue larvae corresponding to the rab3-GEFSC225 mutant with neuronal expression of UAS-rab3-GEF (dvglutNMJX-Gal4, rab3-GEFSC225/Df(1)ED7289; UAS-rab3-GEF/+). Scale bar, 2 µm. B, C, Histograms show (B) the average percentage of DGluRIII clusters apposed to Brp puncta per NMJ and (C) the average area of individual Brp puncta for the genotypes listed in A. n = 11 NMJs for all genotypes; (B) *p≪0.000001i versus both WT and rescue larvae; (C) *p≪0.000001j versus both WT and rescue larvae.
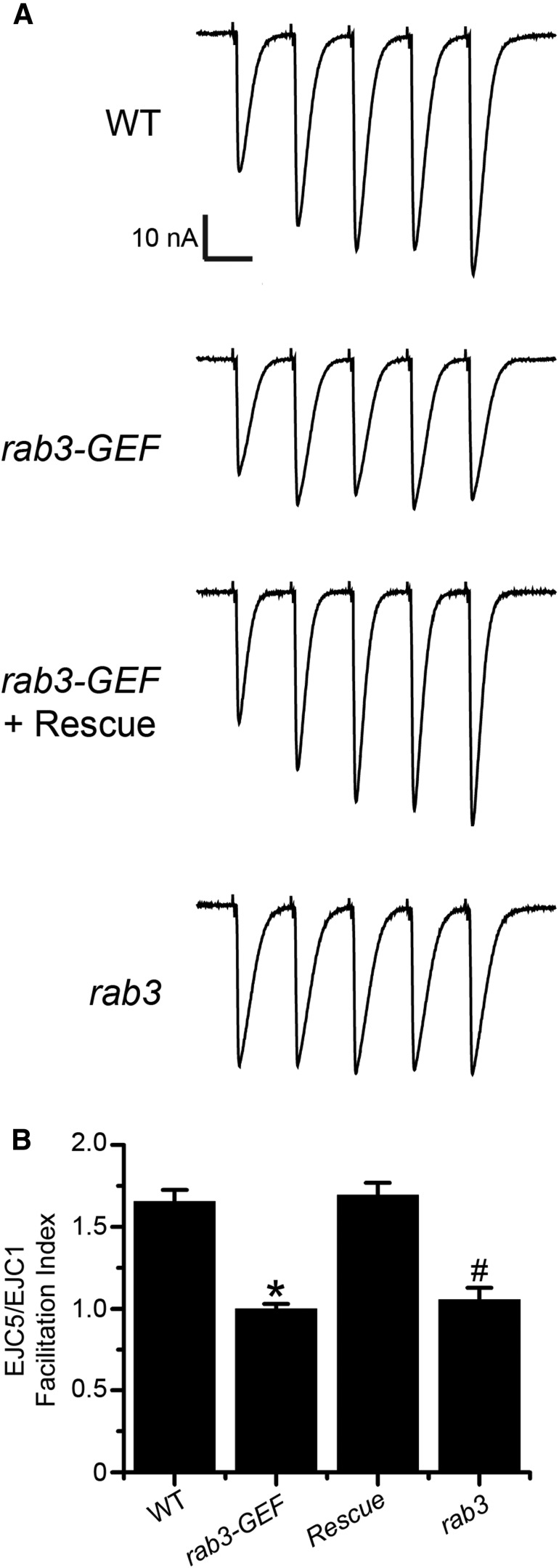
Rab3-GEF is required for normal short-term facilitation
Loss of Rab3-GEF results in the defective distribution of AZ components across release sites in a manner that is similar to rab3 mutant NMJs. Because release efficacy is related to Brp accumulation (Marrus and DiAntonio, 2004; Peled and Isacoff, 2011; Peled et al., 2014), our morphological analysis suggests that rab3-GEFSC225 mutant NMJs are composed primarily of very low probability, Brp-negative sites interspersed by high probability sites where Brp and calcium channels are concentrated. At rab3 mutant NMJs, a similar distribution of AZ components results in defective release during high-frequency stimulation but normal release at low-stimulation frequencies (Graf et al., 2009). To determine the effect of rab3-GEF excision on NMJ function and examine how synaptic function compares between rab3 mutant and rab3-GEFSC225 mutant larvae, we performed voltage-clamp recordings from muscle 6 of segments A3 and A4 at WT, rab3-GEFSC225 mutant, and rab3 mutant NMJs. In all three genotypes, spontaneous miniature EJC amplitude is similar (WT: 0.64 ± 0.03 nA, n = 11; rab3-GEF/Df: 0.61 ± 0.03 nA, n = 13; rab3/Df: 0.66 ± 0.04 nA, n = 12; p=1.00k for rab3-GEF vs WT; p=0.829k for rab3-GEF vs rab3). Furthermore, when evoked by low-frequency stimuli in 0.4 mm Ca2+, EJC amplitude is comparable between the three genotypes (WT: 31.6 ± 3.0 nA, n = 13; rab3-GEF/Df: 38.3 ± 3.7 nA, n = 13; rab3/Df: 35.2 ± 5.4 nA, n = 12; p=0.744l for rab3-GEF vs WT; p=1.00l for rab3-GEF vs rab3). Thus, similar to the rab3 mutant, at low-stimulation frequencies synaptic strength is normal at rab3-GEFSC225 mutant NMJs even though the number (n) of AZs containing key presynaptic components is decreased.
In the rab3 mutant, evoked vesicle release occurs at Brp-positive sites where the probability of release (p) is enhanced due to the augmented concentration of CAZ components (Graf et al., 2009; Peled and Isacoff, 2011). The increased p at these sites maintains neurotransmitter release despite the reduction in n, leading to normal EJC amplitude at low-stimulation frequencies. However, the enhanced release probability of rab3 mutant active zones results in defective short-term facilitation (Graf et al., 2009). At WT NMJs composed primarily of lower probability release sites, high-frequency stimulation in low extracellular calcium results in facilitation. EJC size increases with each subsequent pulse, likely due to a buildup of residual calcium in the axon terminal (Zucker and Regehr, 2002). Conversely, facilitation is attenuated at NMJs with primarily high probability sites, such as is observed in the rab3 mutant (Graf et al., 2009). Is short-term facilitation also defective in the rab3-GEFSC225 mutant? To assay short-term facilitation, we stimulated WT, rab3-GEFSC225 mutant, and rab3 mutant NMJs with a short train of action potentials evoked at a frequency of 20 Hz in 0.4 mm Ca2+. To determine the magnitude of facilitation in each genotype, the facilitation index (FI) was calculated by dividing the amplitude of the fifth EJC by the first. Whereas facilitation is observed at WT NMJs, it is nearly absent in the rab3-GEFSC225 mutant (Fig. 5). Defective facilitation in the rab3-GEFSC225 mutant is equivalent to that observed in the rab3 mutant, resulting in similar FIs between the two genotypes (p=0.448m). Conversely, facilitation is rescued to WT levels when the UAS-rab3-GEF transgene is expressed in rab3-GEFSC225 mutant neurons (Fig. 5). Hence, these results suggest that the functional release sites in rab3-GEFSC225 mutant NMJs have a high p and fire in low calcium concentrations. Additional low p sites are likely not recruited following subsequent stimuli, resulting in reduced facilitation. Our functional and morphological data are consistent, together indicating that rab3-GEFSC225 mutant NMJs contain a reduced number of functional sites that each have a high probability of release. Furthermore, our findings reveal that loss of rab3-GEF phenocopies both the altered distribution of CAZ components and defective function of rab3 mutant NMJs.
Figure 5.
Short-term facilitation is defective in the rab3-GEFSC225 mutant. A Representative EJC traces of a train of five stimuli given at a frequency of 20 Hz in 0.40 mm Ca2+ from WT (dvglutNMJX-Gal4/+), the rab3-GEFSC225 mutant (dvglutNMJX-Gal4, rab3-GEFSC225/Df(1)ED7289), rescue larvae corresponding to the rab3-GEFSC225 mutant with neuronal expression of UAS-rab3-GEF (dvglutNMJX-Gal4, rab3-GEFSC225/Df(1)ED7289;;UAS-rab3-GEF/+), and rab3 mutant larvae (dvglutNMJX-Gal4/+; rab3rup/Df(2R)ED2076). The stimulation artifact has been removed for clarity. B, Histogram showing quantification of the average FI for each of the four genotypes listed in A, calculated by dividing the amplitude of the fifth EJC by the amplitude of the first EJC in a 20 Hz stimulus train. WT, n = 13; rab3-GEFSC225 mutant, n = 12; rab3-GEFSC225 with rescue, n = 14; rab3 mutant, n = 9; *p≪0.000001m versus both WT and rescue larvae; #p=1.83 × 10−6 m versus WT.
Rab3 and Rab3-GEF act within the same pathway to control AZ development
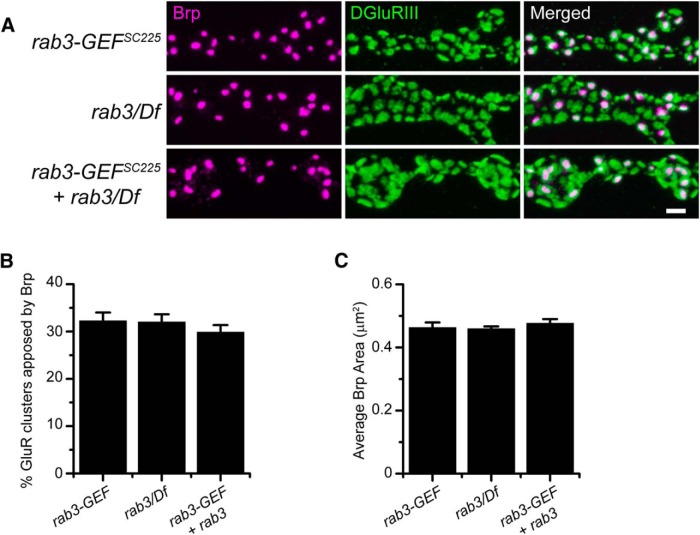
Because loss of Rab3-GEF mimics the loss of Rab3, we hypothesize that the two proteins act via the same molecular pathway to control the distribution of active zone components. To test whether Rab3 and Rab3-GEF act through the same mechanism or separate, parallel pathways, we examined Brp distribution in rab3/rab3-GEF double-mutant larvae. If rab3rup and rab3-GEFSC225 mutant alleles disrupt separate pathways, larvae mutant for both genes should display an enhanced phenotype resulting from an additive effect. Alternatively, if they act through the same pathway, disruption of both genes should result in NMJs that are identical to single knockout of each gene. Examination of α-Brp and α-DGluRIII immunostaining in rab3/rab3-GEF double mutant larvae reveals no difference in Brp distribution across AZs compared with rab3 and rab3-GEFSC225 single mutants (Fig. 6). The percentage of GluR clusters apposed to Brp is similar between the rab3/rab3-GEF double-mutant and each single mutant (p=0.902n for rab3-GEF vs double-mutant; p=1.00n for rab3 vs double-mutant; Fig. 6B ). In addition, average Brp puncta area is equivalent between all three genotypes (p=1.00° for rab3-GEF vs double-mutant; p=0.986° for rab3 vs double-mutant; Fig. 6C ). These results support the hypothesis that Rab3 and Rab3-GEF act within the same molecular pathway to control Brp distribution.
Figure 6.
Simultaneous disruption of both rab3 and rab3-GEF does not enhance the Brp distribution phenotype. A, Images of NMJs costained with α-Brp (magenta) and α-DGluRIII (green) from rab3-GEFSC225 mutant, rab3rup/Df(2R)ED2076 mutant, and rab3-GEFSC225; rab3rup/Df(2R)ED2076 double-mutant larvae. Scale bar, 2 µm. B, C, Histograms show (B) the average percentage of DGluRIII clusters apposed to Brp puncta per NMJ and (C) the average area of individual Brp puncta for the genotypes listed in A. n = 10 NMJs for all genotypes.
rab3-GEF mutation results in defective Rab3 trafficking
How does Rab3-GEF function to control NMJ development? Prior work in mammals and C. elegans indicates that DENN/MADD/Rab3-GEP and AEX-3 can act upstream of Rab3 to control Rab3 function via two separate mechanisms. Orthologues of Drosophila Rab3-GEF catalyze GTP to GDP exchange to increase the population of active, GTP-bound Rab3 (Oishi et al., 1998; Coppola et al., 2002; Mahoney et al., 2006). In addition, DENN/MADD/Rab3-GEP and AEX-3 are required for the anterograde axonal trafficking of Rab3 from the cell body to distal synapses, acting as a linker between GTP-bound Rab3 and the kinesin family proteins KIF1A and KIF1Bβ (Iwasaki et al., 1997; Niwa et al., 2008). However, Rab3-GEF may also act downstream of Rab3 function. Studies of synaptic vesicle exocytosis in mammalian neurons indicate that DENN/MADD/Rab3-GEP acts via an unidentified postdocking mechanism to control vesicle fusion (Yamaguchi et al., 2002). Furthermore, because Rab3-GEF has been identified as a Rab3 effector during axonal trafficking, it may also act as a Rab3 effector at the AZ itself.
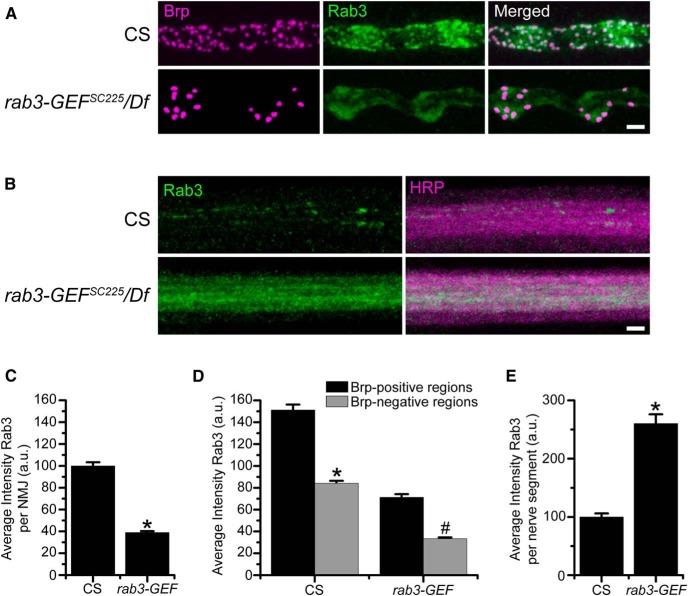
To determine how Rab3-GEF functions in Drosophila neurons to control active zone protein composition, we first asked whether Drosophila Rab3-GEF is required for the trafficking of Rab3 to NMJs by analyzing the localization of Rab3 in the rab3-GEFSC225 mutant. As described previously (Graf et al., 2009), Rab3 localizes throughout WT NMJs in a pattern suggestive of synaptic vesicle proteins but with aggregation at active zones visualized by colocalization with Brp. However, Rab3 accumulation at the NMJ is disrupted in the rab3-GEF mutant (Fig. 7A ). The average intensity of α-Rab3 signal at the NMJ is reduced by approximately 65% in the rab3-GEFSC225 mutant compared with WT (Fig. 7C ). Interestingly, although the pattern of Rab3 localization at the NMJ is more diffuse in the rab3-GEFSC225 mutant and Rab3 fails to aggregate in discrete puncta that colocalize with Brp, it still localizes preferentially to Brp-positive areas of the NMJ (Fig.7A ). In both WT and rab3-GEFSC225 mutant NMJs average Rab3 intensity is significantly greater when measured at Brp-positive regions compared with Brp-negative regions (Fig. 7D ).
Figure 7.
Rab3-GEF is required for proper trafficking of Rab3. A, Images of NMJs costained with α-Brp (magenta) and α-Rab3 (green) from CS and rab3-GEFSC225/Df(1)ED7289 mutant larvae. Scale bar, 2 µm. B, Images of motor nerve segments immunostained with α-Rab3 (green) and α-HRP (magenta) from from CS and rab3-GEFSC225/Df(1)ED7289 mutant larvae. Scale bar, 2 µm. C, Histogram shows the average intensity of α-Rab3 signal throughout the entire NMJ for the genotypes listed in A. n = 10 NMJs for both genotypes; *p≪0.000001p versus CS. D, Histogram shows the average intensity of α-Rab3 signal in Brp-positive and Brp-negative regions of NMJs for the genotypes listed in A. n = 10 NMJs for both genotypes; *p≪0.000001q versus α-Rab3 intensity at the Brp-positive regions in WT NMJs; #p≪0.000001r versus α-Rab3 intensity at the Brp-positive regions in rab3-GEF mutant NMJs. E, Histogram shows the average intensity of α-Rab3 signal per nerve segment as defined by α-HRP staining for CS (n = 24 nerve segments) and rab3-GEFSC225/Df(1)ED7289 mutant larvae (n = 12 nerve segments); *p≪0.000001s versus CS.
Reduced accumulation at the NMJ could result from defective transport of Rab3 from the cell body to the distal axon. To analyze whether axonal trafficking of Rab3 is compromised in the rab3-GEFSC225 mutant, we examined the localization and intensity of α-Rab3 signal in peripheral nerves. In WT nerves, Rab3 is observed in individual axons, often localizing in discrete clusters. However, in the rab3-GEFSC225 mutant, Rab3 protein accumulates throughout entire axon (Fig. 7B ). The buildup of Rab3 in peripheral axons results in a significant increase in average Rab3 intensity per nerve segment (Fig. 7E ). Protein aggregation in axons is a defining characteristic of defective axon transport in Drosophila larvae (Hurd and Saxton, 1996). Thus, the accumulation of Rab3 in peripheral nerves indicates that Rab3-GEF is required for effective transport of Rab3 in the fly.
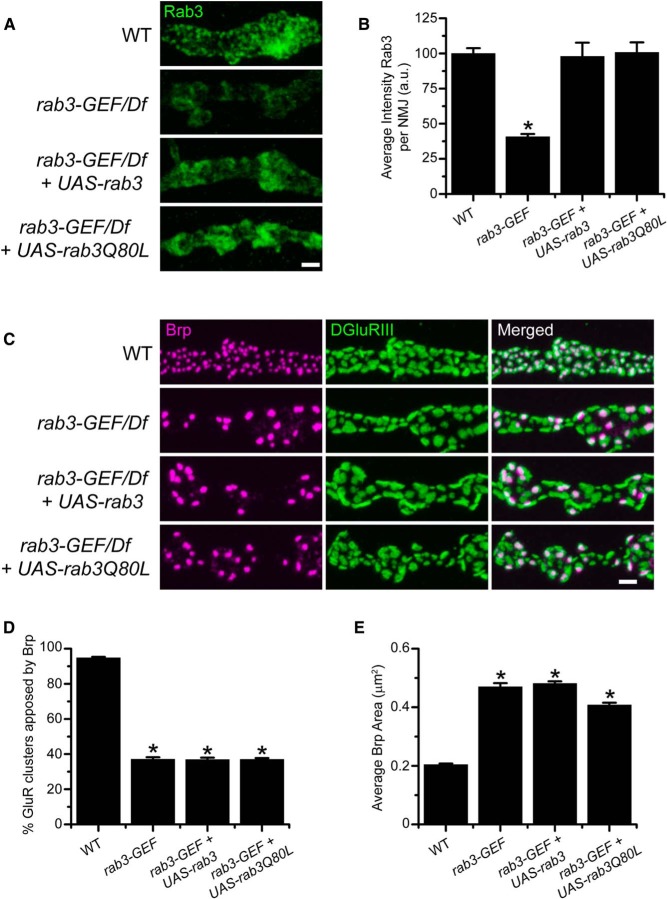
We next tested whether defective Brp distribution results simply from reduced levels of Rab3 protein at the NMJ. Even though the axonal transport of Rab3 is compromised in the rab3-GEFSC225 mutant, Rab3 protein still accumulates at mutant NMJs, albeit at lower levels (Fig. 7), indicating that Rab3 is able to reach the axon terminal in the absence of Rab3-GEF. Therefore, to increase Rab3 protein levels at the NMJ, we enhanced rab3 expression, using the dvglutNMJX-Gal4 neuronal driver to express WT transgenic UAS-rab3 in the rab3-GEFSC225 mutant background. Following transgenic expression of UAS-rab3, Rab3 protein levels in the rab3-GEFSC225 mutant are significantly increased (Fig. 8A ). Average intensity of α-Rab3 signal at the NMJ is equivalent between WT NMJs and rab3-GEFSC225 mutant NMJs expressing UAS-rab3 (Fig. 8A,B ). However, increased Rab3 expression does not rescue Brp distribution in rab3-GEFSC225 mutant NMJs (Fig. 8C ). Analysis of α-Brp and α-DGluRIII staining in rab3-GEFSC225 mutant larvae reveals that UAS-rab3 expression has no effect on the percentage of GluR clusters apposed to Brp or average Brp puncta area (Fig. 8D,E ). Thus, defective AZ development in the rab3-GEFSC225 mutant is not solely due to the lack of Rab3 protein at the NMJ.
Figure 8.
Transgenic expression of WT Rab3 or a GTP-locked variant of Rab3 increases Rab3 accumulation at the NMJ but fails to rescue Brp distribution in the rab3-GEFSC225 mutant. A, Images of NMJs immunostained with α-Rab3 (green) from WT (dvglutNMJX-Gal4/+), the rab3-GEFSC225 mutant (dvglutNMJX-Gal4, rab3-GEFSC225/Df(1)ED7289), the rab3-GEFSC225 mutant expressing UAS-rab3 (dvglutNMJX-Gal4, rab3-GEFSC225/Df(1)ED7289;; UAS-rab3/+), and the rab3-GEFSC225 mutant expressing UAS-rab3Q80L (dvglutNMJX-Gal4, rab3-GEFSC225/Df(1)ED7289;; UAS-rab3Q80L/+). Scale bar, 2 µm. B, Histogram shows the average intensity of α-Rab3 signal throughout the entire NMJ for the genotypes listed in A. n = 10 NMJs for all genotypes; *p= 6.7 × 10−7t versus WT, 1.35 × 10−6t versus the rab3-GEF mutant expressing UAS-rab3, and 5.12 × 10−7t versus the rab3-GEF mutant expressing UAS-rab3Q80L. C, Images of NMJs costained with α-Brp (magenta) and α-DGluRIII (green) from the genotypes listed in A. Scale bar, 2 µm. D, E, Histograms show (D) the average percentage of DGluRIII clusters apposed to Brp puncta per NMJ and (E) the average area of individual Brp puncta for the genotypes listed in A. n = 10 NMJs for all genotypes; (D) *p≪0.000001u versus WT; (E) *p≪0.000001v versus WT.
Expression of a GTP-locked variant of Rab3 fails to rescue rab3-GEFSC225 mutant NMJs
Because overexpression of WT Rab3 fails to rescue the synaptic phenotype of rab3-GEFSC225 mutant NMJS, Rab3-GEF must have additional required functions for active zone development. Enhanced expression of WT Rab3 may not rescue AZ development due to defective guanine nucleotide cycling of both endogenous and transgenically expressed Rab3. Rab3 cycles between an active GTP-bound state and an inactive GDP-bound state. Following the binding of GTP, Rab3 hydrolyzes the guanine nucleotide to GDP, inactivating itself in a process that is regulated by Rab3-GAP (GTPase accelerating protein; Cherfils and Zeghouf, 2013). GEF proteins act as exchange factors that catalyze the release of GDP so that a new GTP molecule can bind (Bos et al., 2007). Hence, defective GEF activity in rab3-GEFSC225 mutant neurons may result in a buildup of GDP-bound Rab3 and a comparable reduction in the GTP-bound population due to an inability of Rab3 to release GDP following hydrolysis.
Because Rab3 must be GTP-bound to control Brp distribution across active zones (Chen et al., 2015), defective guanine nucleotide exchange in the rab3-GEFSC225 mutant may lead to the observed synaptic phenotype. Therefore, we tested whether expression of Rab3Q80L, a GTP-locked variant of Rab3, could rescue Brp distribution in rab3-GEFSC225 mutant neurons. The Q80L mutation inhibits the GTPase activity of Rab3 even in the presence of Rab3-GAP, resulting in a constitutively active variant (Brondyk et al., 1993). We previously showed that expression of Rab3Q80L rescues both the morphological and functional deficits of rab3 mutant NMJs, indicating that it is functional in the presence of Rab3-GEF (Chen et al., 2015). To determine whether GTP-locked Rab3 rescues rab3-GEF mutant NMJs, we assayed Brp distribution in rab3-GEFSC225 mutant neurons expressing UAS-rab3Q80L. Driving UAS-rab3Q80L with DVgult-Gal4 in rab3-GEFSC225 mutant larvae increases the accumulation of Rab3 protein at the NMJ (Fig. 8A ). Average intensity of α-Rab3 staining at the NMJ is similar between WT animals and rab3-GEFSC225 mutant larvae expressing either the UAS-rab3 or UAS-rab3Q80L variants of transgenic Rab3 (Fig. 8B ). However, expression of UAS-rab3Q80L fails to rescue the Brp-GluR apposition phenotype of the rab3-GEFSC225 mutant (Fig. 8C ). The percentage of GluR clusters apposed to Brp does not increase following UAS-rab3Q80L expression in the rab3-GEFSC225 mutant (Fig. 8D ). Interestingly, driving transgenic Rab3Q80L in the rab3-GEFSC225 mutant does modestly but significantly decrease average Brp puncta area (p=3.07 × 10−5v). Although this may correspond to a mild rescue of Brp puncta size, previous analysis indicates that Rab3Q80L expression also reduces average Brp puncta area when driven in WT NMJs (Chen et al., 2015), suggesting that Rab3Q80L may have a gain of function affect that decreases Brp cluster size regardless of the presence or absence of Rab3-GEF.
It is possible that Rab3-GEF is required for Rab3Q80L to bind GTP. While this may explain the inability of Rab3Q80L to rescue Brp distribution in the rab3-GEFSC225 null mutant, we would predict that Rab3Q80L should be GTP-bound when driven in the rab3-GEFMA18 and rab3-GEFMA20 hypomorphic alleles that have unaltered death domains and retain partial function. However, similar to our analysis of rab3-GEFSC225 mutant NMJs, neuronal expression of UAS-rab3Q80L fails to rescue the percentage of GluR clusters apposed to Brp when driven in either the rab3-GEFMA18 mutant (rab3-GEFMA18/Df: 36.1 ± 0.7%; UAS-rab3Q80L expressed in rab3-GEFMA18/Df: 35.7 ± 1.4%: n = 8 NMJs for both genotypes; p=0.814w) or the rab3-GEFMA20 mutant (rab3-GEFMA20/Df: 40.1 ± 2.0%; UAS-rab3Q80L expressed in rab3-GEFMA18/Df: 38.3 ± 1.8%; n = 8 NMJs for both genotypes; p=0.523x). Also similar to the rab3-GEFSC225 mutant, a moderate decrease is observed in average Brp puncta size following Rab3Q80L expression in the the rab3-GEFMA18 mutant (rab3-GEFMA18/Df: 0.44 ± 0.007 µm2; UAS-rab3Q80L expressed in rab3-GEFMA18/Df: 0.32 ± 0.010 µm2; n = 8 NMJs for both genotypes; p=1.17 × 10−7y) and in the rab3-GEFMA20 mutant (rab3-GEFMA20/Df: 0.42 ± 0.011 µm2; UAS-rab3Q80L expressed in rab3-GEFMA18/Df: 0.34 ± 0.009 µm2; n = 8 NMJs for both genotypes; p=3.96 × 10−5z). Hence, Rab3Q80L is unable to rescue the rab3-GEF apposition phenotype even in the presence of genetic hypomoprhs of Rab3-GEF that retain some activity. Because Rab3-GEF levels are significantly reduced at rab3-GEFMA18 and rab3-GEFMA20 mutant NMJs, it is possible that GEF activity is required locally at the NMJ to maintain the GTP-bound state of Rab3Q80L. However, Rab3-GEF protein is present in the cell bodies of rab3-GEFMA18 and rab3-GEFMA20 mutant neurons at reduced or nearly normal levels, respectively, and may thus be available to convert Rab3Q80L to a GTP-bound state prior to being trafficked to the NMJ.
Together, these results indicate that even though expression of Rab3Q80L increases Rab3 accumulation at the NMJ and should enhance the population of GTP-bound Rab3, it is unable to rescue Brp distribution in multiple mutant alleles of rab3-GEF. Thus, our mechanistic studies suggest that although Rab3-GEF may act upstream of Rab3 to control Rab3 trafficking and activation in Drosophila neurons, it may also play an additional role that is downstream of Rab3. Because the rab3 and rab3-GEF mutant phenotypes are identical, this additional function is likely associated with Rab3 itself. It also likely requires the combined action of both Rab3 and Rab3-GEF as enhancing Rab3-GEF expression in the rab3 mutant fails to rescue either the percentage of GluR clusters apposed to Brp (rab3 mutant: 33.7 ± 0.9%; UAS-rab3-GEF expressed in rab3 mutant: 34.2 ± 1.1%; n = 10 NMJs for both genotypes; p=0.77393aa) or average Brp area (rab3 mutant: 0.39 ± 0.005 µm2; UAS-rab3-GEF expressed in rab3 mutant: 0.38 ± 0.015 µm2; n = 10 NMJs for both genotypes; p=0.381bb). We hypothesize that Rab3-GEF may act as an effector of Rab3 in the process of docking Rab3-associated vesicles at release sites to control protein composition at AZs.
Discussion
We show that Rab3-GEF controls CAZ assembly and the distribution of AZ components across release sites at the Drosophila NMJ. In the rab3-GEFSC225 mutant, Brp and calcium channels fail to cluster at two-thirds of available sites. Rather, CAZ components are concentrated at a minority of sites where they aggregate in enlarged clusters. rab3-GEFSC225 mutant NMJs function normally when exposed to low-stimulation frequencies. However, high-frequency stimulation reveals defective short-term facilitation in the rab3-GEFSC225 mutant, a hallmark of NMJs composed primarily of sites exhibiting high-release probability characteristics. Thus, both morphological and functional studies suggest that rab3-GEF disruption results in the formation of a small number of functional sites with enhanced release efficacy.rab3-GEFSC225 mutant NMJs have a synaptic phenotype that is indistinguishable from NMJs in the rab3 mutant (Graf et al., 2009), suggesting that they function together to control AZ composition. Prior studies of the mammalian and C. elegans orthologues DENN/MADD/Rab3-GEP/AEX-3 indicate that Drosophila Rab3-GEF likely functions as a guanine nucleotide exchange factor for Rab3, as well as a Rab3 effector that participates in the axonal transport of Rab3 to synapses (Iwasaki et al., 1997; Wada et al., 1997; Oishi et al., 1998; Coppola et al., 2002; Mahoney et al., 2006; Niwa et al., 2008). Therefore, it is possible that Rab3 itself is dysfunctional in the mutant. In support of this, we show that loss of Rab3-GEF results in defective trafficking of Rab3 and reduced accumulation of Rab3 at NMJs. However, mechanistic studies reveal that the rab3-GEF synaptic phenotype cannot be fully explained by loss of Rab3 function. Attempts to rescue the rab3-GEFSC225 synaptic phenotype through transgenic expression of Rab3 to enhance its accumulation at the NMJ fail to rescue Brp distribution, potentially due to loss of GEF activity and a reduction in GTP-bound Rab3 in the rab3-GEF mutant. However, expression of Rab3Q80L, a GTP-locked variant, also fails to rescue the rab3-GEFSC225 mutant phenotype even though it should enhance the GTP-bound population of Rab3 and suppress the effects caused by loss of GEF activity. Thus, our results support the model that in addition to its roles in Rab3 transport and guanine nucleotide exchange, Rab3-GEF is also required downstream of Rab3 function. We hypothesize that Rab3-GEF acts as a Rab3 effector to dock Rab3-associated vesicles at the synapse for proper CAZ assembly.
Control of CAZ assembly by Rab3
Double-knockout of both rab3 and rab3-GEF fails to enhance the NMJ phenotype, indicating that the two proteins likely act within the same molecular pathway to control AZ development. How does Rab3 control CAZ assembly? Prior mutational studies suggest that Drosophila Rab3 acts to localize CAZ proteins via a mechanism that is typical of Rab proteins. GTP-binding and residues required for effector interaction and membrane association are essential for proper Rab3 function at the NMJ (Chen et al., 2015). The requirement of the Rab3 C-terminal membrane association domain suggests that Rab3 may control Brp distribution via a standard vesicle-docking mechanism. However, RIM is not required for Rab3 to control Brp distribution (Graf et al., 2012). Both the type of vesicle and the effector proteins involved in its tethering to target membranes remain unknown.
Mammalian Rab3 is associated with neurotransmitter-filled synaptic vesicles, Piccolo–Bassoon transport vesicles (PTVs), and other types of secretory vesicles (Darchen and Goud, 2000; Schlüter et al., 2002; Shapira et al., 2003). The role of Rab3 in synaptic vesicle dynamics has been extensively studied (Wang et al., 1997; Schlüter et al., 2006). We have previously shown that general alterations in synaptic vesicle release caused by increasing or decreasing neuronal excitability do not affect Brp distribution in WT or rab3 mutant NMJs (Graf et al., 2009), so it is unlikely that Rab3 controls Brp distribution via a synaptic vesicle associated mechanism. Interestingly, recent studies of the synaptic vesicle protein synaptotagmin-1 (Syt) reveal that Syt disruption also results in Brp localization defects, although the morphological phenotype resulting from Syt knockdown is different than that observed in rab3 and rab3-GEF mutant NMJs (Paul et al., 2015). The fact that both Rab3 and Syt are required for proper Brp localization suggests that a linkage may exist between neurotransmitter release and CAZ assembly; however, the mechanism of this potential linkage is unclear. A simpler explanation for Rab3 function in the control of CAZ assembly involves the docking of transport vesicles homologous to mammalian PTVs, which contain a pre-assembled complex of AZ proteins for insertion at nascent release sites during synapse development (Zhai et al., 2001). Such a model is consistent with previous studies that suggest that Drosophila Rab3 acts to nucleate the formation of new Brp clusters at AZs (Graf et al., 2009). However, an analogous population of transport vesicles has not been identified in Drosophila neurons, and further work will be required to determine whether such vesicles are involved in this mechanism of Rab3 action to control Brp distribution.
Analysis of Rab3-GEF function for controlling AZ composition
We now show that Rab3-GEF is an essential player in this Rab3-associated mechanism to control CAZ assembly. Previous work in other organisms indicates that Rab3-GEF acts upstream of Rab3 to regulate Rab3 function. Consistent with studies in mammals and C. elegans (Iwasaki et al., 1997; Niwa et al., 2008), we show that Drosophila Rab3-GEF is required for the axonal trafficking of Rab3. Rab3 protein accumulates in axon shafts, leading to decreased levels at rab3-GEFSC225 mutant NMJs. Disrupted localization of Rab3 and its associated vesicles may lead to defective Brp distribution in the rab3-GEFSC225 mutant. However, enhanced expression of both transgenic WT Rab3 and Rab3Q80L fail to rescue the rab3-GEFSC225 mutant phenotype even though the accumulation of Rab3 is increased at NMJs to endogenous levels. These results suggest that disrupted Brp distribution in the rab3-GEFSC225 mutant is not solely due to defective Rab3 trafficking. Conversely, although enhanced expression of Rab3 may increase the overall accumulation of Rab3 at the terminal, Rab3-GEF may also be required for more discrete localization of Rab3 within the NMJ, which may not be rescued by Rab3 overexpression. The localization pattern of Rab3 within the NMJ in rab3-GEFSC225 mutants expressing transgenic Rab3 differs from endogenous Rab3; however, this may also be due to the fact that CAZ protein distribution is defective at these NMJs.
A second potential mechanism of Rab3-GEF action is via its role as a GEF. The mammalian and C. elegans orthologues of Drosophila Rab3-GEF act as GEFs to catalyze GDP to GTP exchange (Wada et al., 1997; Mahoney et al., 2006). It has previously been shown that GTP-binding is required for Rab3 to control Brp distribution as GTP-binding defective variants of Rab3 fail to rescue the rab3 mutant phenotype (Chen et al., 2015). Thus, rab3-GEF knockout may disrupt Rab3 function in Drosophila neurons by reducing the GTP-bound population of Rab3. However, our studies indicate that the rab3-GEF synaptic phenotype cannot solely be due to loss of GEF activity or defective Rab3 localization. Enhancing the population of GTP-bound Rab3 by expression of Rab3Q80L, a hydrolysis defective variant, fails to rescue the rab3-GEFSC225 mutant phenotype. Rab3Q80L accumulates at the NMJ at levels similar to endogenous Rab3. Furthermore, previous analysis indicates that Rab3Q80L can rescue the morphological and physiological phenotypes of the rab3 mutant, indicating that it is functional when Rab3-GEF is present. Hence, the lack of rescue following Rab3Q80L expression in the rab3-GEF mutant suggests that the synaptic phenotype is not simply due to loss of GEF activity. Additional lines of evidence support this finding. GEF activity has been mapped to the death domain in the mammalian and C. elegans orthologues of Rab3-GEF. However, rab3-GEFMA18 and rab3-GEFMA20 mutants both have synaptic phenotypes nearly as severe as the rab3-GEFSC225 mutant, even though the death domain is intact in the MA18 and MA20 mutant alleles. Furthermore, studies in mice reveal the presence of significant amounts of GTP-bound Rab3 despite DENN/MADD disruption (Yamaguchi et al., 2002; Niwa et al., 2008), potentially due to the presence of other proteins that can act as GEFs for Rab3.
It is possible that Rab3Q80L itself requires the nucleotide exchange activity of Rab3-GEF to transition into a GTP-bound state, which could explain the lack of rescue when Rab3Q80L is expressed in the rab3-GEFSC225 mutant. However, Rab3Q80L also fails to rescue the synaptic phenotype when driven in the rab3-GEFMA18 and rab3-GEFMA20 hypomorphs that retain some activity. This may be due to a general decrease in Rab3-GEF protein throughout rab3-GEFMA18 mutant neurons. However, Rab3-GEF protein is present at nearly normal levels in the cell bodies of rab3-GEFMA20 mutant neurons even though it is absent at rab3-GEFMA20 mutant NMJs. Although it is possible that Rab3Q80L requires GEF activity in the NMJ to maintain its active state, the presence of Rab3-GEF in rab3-GEFMA20 cell bodies suggests that activation of Rab3Q80L in the rab3-GEFMA20 mutant could occur prior to trafficking to the NMJ. In mammalian neurons, axonal transport of Rab3 is most effective when Rab3 is in a GTP-bound state (Niwa et al., 2008), suggesting that GEF activity generally occurs in the cell body, as well as at the synapse. Thus, together, our results suggest that Rab3-GEF plays other roles in the control of AZ development in addition to its functions in Rab3 trafficking and as a GEF.
How might Rab3-GEF act in addition to its previously identified roles? Because the rab3-GEF and rab3 mutant phenotypes are indistinguishable from each other and from the double-mutant, this additional function must be within the Rab3 pathway. We hypothesize that Rab3-GEF also acts downstream of Rab3 to control AZ composition. Rab3-GEF is a Rab3 effector protein, linking Rab3 to kinesin for axonal trafficking (Niwa et al., 2008). Rab3-GEF may also act as a Rab3 effector at the NMJ itself, playing a direct role in the docking of Rab3-associated vesicles at the plasma membrane. Conversely, Rab3-GEF may be required for an unidentified postdocking step as has been suggested by studies of synaptic vesicle exocytosis in mouse neurons lacking Rab3-GEP (Yamaguchi et al., 2002). Additional work will be required to determine the mechanism by which Rab3 and Rab3-GEF act to control CAZ protein composition at active zones.
Synthesis
The decision was a result of the Reviewing Editor Vivian Budnik and the peer reviewers coming together and discussing their recommendations until a consensus was reached. A fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision is listed below. The following reviewers agreed to reveal their identity: J. Troy Littleton, Dion Dickman
The authors have addressed prior comments by reviewers.
While no additional experiments are required, authors might consider suggestions by reviewer 2.
Reviewer 1
The authors have addressed my prior concerns by showing Rab3-GEF localization and levels in the mutants, an the localizaiton of Rab3-GEF in rab3 mutants. The lack of rescue of the Rab3 mutant phenotype by Rab3-GEF is also a nice addition. Overall, a solid study of a new player in Rab3 active zone biology.
Reviewer 2
The authors conduct a rigorous genetic analysis of Rab3-GEF function in Drosophila, elegantly demonstrating that mutations in Rab3-GEF effectively phenocopy mutants in Rab3. This point is rigorously shown and makes up the bulk of the study. While this discovery itself is not necessarily surprising, the thoroughness of their analysis, clarity of their findings, and importance in understanding this layer of regulation (transport and regulation of Rab3 by Rab3-GEF) should be of considerable interest to the field.
Important questions remain regarding the mechanistic relationship between Rab3 and Rab3-GEF, and the enigma of why a constitutively active Rab3 does not rescue Rab3-GEF mutants. The authors do a commendable job of attempting to provide insight into these difficult questions, proposing the idea that in addition to Rab3-GEF regulating Rab3 activity, it also may have separate roles in active zone regulation, perhaps "downstream" of Rab3.
The authors have already performed a lot of the obvious experiments to provide insight into these questions, so I would not demand that additional ones, as most would require work that itself would be an entire new study. However, it would be of interest to know the answer to these experiments:
1. The authors have already done the rescue experiments of overexpressing Rab3 and constitutively active Rab3 in Rab3GEF mutant backgrounds and did not see rescue. However, have the authors attempted to rescue active zone organization by overexpressing Rab3-GEF in a Rab3 mutant alone and perhaps overexpressing both Rab3-GEF and constitutively active Rab3 in Rab3 mutants? This may provide insight into whether Rab3 expression or activity is needed for the proposed downstream requirements of Rab3-GEF.
2. Another open question is whether the GTP exchange activity of Rab3-GEF is needed for its functions in Rab3 transport and/or downstream functions in active zone organization. Although some insights are made by using the MA20 allele, a more specific way to address this would be to specifically mutate the GEF activity in a Rab3-GEF cDNA and then attempt to rescue Rab3-GEF mutants (and monitor Rab3 transport with and without expression of constitutively active Rab3) with this transgene.
Nonetheless, this study raises interested questions about the role and regulation of Rab3 and Rab3-GEF that should be of interest to many readers.
References
- Atwood HL, Govind CK, Wu CF (1993) Differential ultrastructure of synaptic terminals on ventral longitudinal abdominal muscles in Drosophila larvae. J Neurobiol 24:1008-1024. 10.1002/neu.480240803 [DOI] [PubMed] [Google Scholar]
- Bhuin T, Roy JK (2014) Rab proteins: the key regulators of intracellular vesicle transport. Exp Cell Res 328:1-19. 10.1016/j.yexcr.2014.07.027 [DOI] [PubMed] [Google Scholar]
- Blunk AD, Akbergenova Y, Cho RW, Lee J, Walldorf U, Xu K, Zhong G, Zhuang X, Littleton JT (2014) Postsynaptic actin regulates active zone spacing and glutamate receptor apposition at the Drosophila neuromuscular junction. Mol Cell Neurosci 61:241-254. 10.1016/j.mcn.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129:865-877. 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Brondyk WH, McKiernan CJ, Burstein ES, Macara IG (1993) Mutants of Rab3A analogous to oncogenic Ras mutants: sensitivity to Rab3A-GTPase activating protein and Rab3A-guanine nucleotide releasing factor. J Biol Chem 268:9410-9415. [PubMed] [Google Scholar]
- Chen S, Gendelman HK, Roche JP, Alsharif P, Graf ER (2015) Mutational analysis of Rab3 function for controlling active zone protein composition at the Drosophila neuromuscular junction. PLoS One 10:e0136938. 10.1371/journal.pone.0136938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93:269-309. 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- Chow VT, Lim KM, Lim D (1998) The human DENN gene: genomic organization, alternative splicing, and localization to chromosome 11p11.21–p11.22. Genome 41:543-552. [DOI] [PubMed] [Google Scholar]
- Coppola T, Perret-Menoud V, Gattesco S, Magnin S, Pombo I, Blank U, Regazzi R (2002) The death domain of Rab3 guanine nucleotide exchange protein in GDP/GTP exchange activity in living cells. Biochem J 362:273-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RW, Gelfand MV, Collins CA, DiAntonio A (2008) Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol 508:131-152. 10.1002/cne.21670 [DOI] [PubMed] [Google Scholar]
- Darchen F, Goud B (2000) Multiple aspects of Rab protein action in the secretory pathway: focus on Rab3 and Rab6. Biochimie 82:375-384. [DOI] [PubMed] [Google Scholar]
- Ehmann N, van de Linde S, Alon A, Ljaschenko D, Keung XZ, Holm T, Rings A, DiAntonio A, Hallermann S, Ashery U, Heckmann M, Sauer M, Kittel RJ (2014) Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat Commun 5:4650. 10.1038/ncomms5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Daniels RW, Burgess RW, Schwarz TL, DiAntonio A (2009) Rab3 dynamically controls protein composition at active zones. Neuron 64:663-677. 10.1016/j.neuron.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Valakh V, Wright CM, Wu C, Liu Z, Zhang YQ, DiAntonio A (2012) RIM promotes calcium channel accumulation at active zones of the Drosophila neuromuscular junction. J Neurosci 32:16586-16596. 10.1523/JNEUROSCI.0965-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger ED, Fejtova A (2012) Molecular organization and plasticity of the cytomatrix at the active zone. Curr Opin Neurobiol 22:423-430. 10.1016/j.conb.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Hurd DD, Saxton WM (1996) Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics 144:1075-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K, Staunton J, Saifee O, Nonet M, Thomas JH (1997) aex-3 encodes a novel regulator of presynaptic activity in C. elegans. Neuron 18:613-622. [DOI] [PubMed] [Google Scholar]
- Kawasaki F, Zou B, Xu X, Ordway RW (2004) Active zone localization of presynaptic calcium channels encoded by the cacophony locus of Drosophila. J Neurosci 24:282-285. 10.1523/JNEUROSCI.3553-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, Hell SW, Buchner E, Heckmann M, Sigrist SJ (2006) Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312:1051-1054. 10.1126/science.1126308 [DOI] [PubMed] [Google Scholar]
- Levivier E, Goud B, Souchet M, Calmels TP, Mornon JP, Callebaut I (2001) uDENN, DENN, and dDENN: indissociable domains in Rab and MAP kinases signaling pathways. Biochem Biophys Res Commun 287:688-695. 10.1006/bbrc.2001.5652 [DOI] [PubMed] [Google Scholar]
- Mahoney TR, Liu Q, Itoh T, Luo S, Hadwiger G, Vincent R, Wang ZW, Fukuda M, Nonet ML (2006) Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol Biol Cell 17:2617-2625. 10.1091/mbc.E05-12-1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat AL, Dokainish H, McPherson PS (2011) DENN domain proteins: regulators of Rab GTPases. J Biol Chem 286:13791-13800. 10.1074/jbc.R110.217067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrus SB, DiAntonio A (2004) Preferential localization of glutamate receptors opposite sites of high presynaptic release. Curr Biol 14:924-931. 10.1016/j.cub.2004.05.047 [DOI] [PubMed] [Google Scholar]
- Marrus SB, Portman SL, Allen MJ, Moffat KG, DiAntonio A (2004) Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J Neurosci 24:1406-1415. 10.1523/JNEUROSCI.1575-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel K, Müller JA, Opri̦soreanu AM, Schoch S (2015) The presynaptic active zone: a dynamic scaffold that regulates synaptic efficacy. Exp Cell Res 335:157-164. 10.1016/j.yexcr.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Mosca TJ, Hong W, Dani VS, Favaloro V, Luo L (2012) Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature 484:237-241. 10.1038/nature10923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S, Tanaka Y, Hirokawa N (2008) KIF1Bβ- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat Cell Biol 10:1269-1279. 10.1038/ncb1785 [DOI] [PubMed] [Google Scholar]
- Oishi H, Sasaki T, Nagano F, Ikeda W, Ohya T, Wada M, Ide N, Nakanishi H, Takai Y (1998) Localization of the Rab3 small G protein regulators in nerve terminals and their involvement in Ca2+-dependent exocytosis. J Biol Chem 273:34580-34585. [DOI] [PubMed] [Google Scholar]
- Paul MM, Pauli M, Ehmann N, Hallermann S, Sauer M, Kittel RJ, Heckmann M (2015) Bruchpilot and Synaptotagmin collaborate to drive rapid glutamate release and active zone differentiation. Front Cell Neurosci 9:29. 10.3389/fncel.2015.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled ES, Isacoff EY (2011) Optical quantal analysis of synaptic transmission in wild-type and rab3-mutant Drosophila motor axons. Nat Neurosci 14:519-526. 10.1038/nn.2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled ES, Newman ZL, Isacoff EY (2014) Evoked and spontaneous transmission favored by distinct sets of synapses. Curr Biol 24:484-493. 10.1016/j.cub.2014.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schievella AR, Chen JH, Graham JR, Lin LL (1997) MADD, a novel death domain protein that interacts with the type 1 tumor necrosis factor receptor and activates mitogen-activated protein kinase. J Biol Chem 272:12069-12075. [DOI] [PubMed] [Google Scholar]
- Schlüter OM, Khvotchev M, Jahn R, Südhof TC (2002) Localization versus function of Rab3 proteins: evidence for a common regulatory role in controlling fusion. J Biol Chem 277:40919-40929. 10.1074/jbc.M203704200 [DOI] [PubMed] [Google Scholar]
- Schlüter OM, Basu J, Südhof TC, Rosenmund C (2006) Rab3 superprimes synaptic vesicles for release: implications for short-term synaptic plasticity. J Neurosci 26:1239-1246. 10.1523/JNEUROSCI.3553-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, Zhai RG, Dresbach T, Bresler T, Torres VI, Gundelfinger ED, Ziv NE, Garner CC (2003) Unitary assembly of presynaptic active zones from Piccolo–Bassoon transport vesicles. Neuron 38:237-252. [DOI] [PubMed] [Google Scholar]
- Sigrist SJ, Schmitz D (2011) Structural and functional plasticity of the cytoplasmic active zone. Curr Opin Neurobiol 21:144-150. 10.1016/j.conb.2010.08.012 [DOI] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF (1994) Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A 175:179-191. [DOI] [PubMed] [Google Scholar]
- Viquez NM, Füger P, Valakh V, Daniels RW, Rasse TM, DiAntonio A (2009) PP2A and GSK-3beta act antagonistically to regulate active zone development. J Neurosci 29:11484-11494. 10.1523/JNEUROSCI.5584-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Nakanishi H, Satoh A, Hirano H, Obaishi H, Matsuura Y, Takai Y (1997) Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J Biol Chem 272:3875-3878. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E (2006) Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49:833-844. 10.1016/j.neuron.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Wairkar YP, Toda H, Mochizuki H, Furukubo-Tokunaga K, Tomoda T, Diantonio A (2009) Unc-51 controls active zone density and protein composition by downregulating ERK signaling. J Neurosci 29:517-528. 10.1523/JNEUROSCI.3848-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Südhof TC (1997) Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature 388:593-598. 10.1038/41580 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Tanaka M, Mizoguchi A, Hirata Y, Ishizaki H, Kaneko K, Miyoshi J, Takai Y (2002) A GDP/GTP exchange protein for the Rab3 small G protein family up-regulates a postdocking step of synaptic exocytosis in central synapses. Proc Natl Acad Sci U S A 99:14536-14541. 10.1073/pnas.212511399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Vardinon-Friedman H, Cases-Langhoff C, Becker B, Gundelfinger ED, Ziv NE, Garner CC (2001) Assembling the presynaptic active zone: a characterization of an active one precursor vesicle. Neuron 29:131-143. [DOI] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP (2007) Thirty-one flavors of Drosophila rab proteins. Genetics 176:1307-1322. 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker RS, Regehr WG (2002) Short-term synaptic plasticity. Annu Rev Physiol 64:355-405. 10.1146/annurev.physiol.64.092501.114547 [DOI] [PubMed] [Google Scholar]