SUMMARY
Mammalian genomes are organized into megabase-scale topologically associated domains (TADs). We demonstrate that disruption of TADs can rewire long-range regulatory architecture and result in pathogenic phenotypes. We show that distinct human limb malformations are caused by deletions, inversions, or duplications altering the structure of the TAD-spanning WNT6/IHH/EPHA4/PAX3 locus. Using CRISPR/Cas genome editing, we generated mice with corresponding rearrangements. Both in mouse limb tissue and patient-derived fibroblasts, disease-relevant structural changes cause ectopic interactions between promoters and non-coding DNA, and a cluster of limb enhancers normally associated with Epha4 is misplaced relative to TAD boundaries and drives ectopic limb expression of another gene in the locus. This rewiring occurred only if the variant disrupted a CTCF-associated boundary domain. Our results demonstrate the functional importance of TADs for orchestrating gene expression via genome architecture and indicate criteria for predicting the pathogenicity of human structural variants, particularly in non-coding regions of the human genome.
INTRODUCTION
Approximately 5% of the human genome is structurally variable in the normal population, which includes deletions and duplications (collectively referred to as copy number variants, CNVs), as well as inversions, and translocations. Structural variations have received considerable attention as a major cause for genetic disease, promoting the search for CNVs as a standard diagnostic procedure in conditions such as intellectual disability and congenital malformations (Stankiewicz and Lupski, 2010; Swaminathan et al., 2012). The pathogenicity of many CNVs can be explained by their effect on gene dosage. In contrast, it is difficult to predict the consequences of balanced rearrangements, such as inversions, or the functional impact of CNVs that are limited to non-coding DNA. Such variants have the potential to disrupt the integrity of the genome, causing changes in the regulatory architecture that lead to pathogenic alterations of gene expression levels and patterns (Haraksingh and Snyder, 2013; Spielmann and Mundlos, 2013). However, the lack of a comprehensive understanding of the large-scale functional organization of the regulatory genome is a major limitation in predicting their potential pathogenicity.
New methods for enhancer identification and analysis of chromosome conformation have enabled substantial progress towards elucidating genome-wide regulatory interactions. ChIP-seq performed directly on ex vivo tissues can reveal the location of distant-acting tissue-specific enhancer sequences at genomic scale (Visel et al., 2009a). In parallel, sequencing-based studies of DNA:DNA interactions have provided insight into the general conformation of the genome in living cells, as well as interactions between promoters and distant-acting transcriptional enhancers in specific cell types (Lieberman-Aiden et al., 2009). These data also show that enhancers can control multiple genes, frequently over hundreds of kb away from their target. Only a fraction of enhancers contact the nearest promoter whereas most skip one or more genes (de Laat and Duboule, 2013). How the selective interaction of enhancers with their respective target genes is achieved remains largely unknown but the organization of the genome in domains of interaction that are shielded from each other by boundaries appears to be critical. Genome-wide interaction studies by chromosome conformation capture-based approaches such as Hi-C and 5C show that the genome is partitioned into megabase-scale topologically associated domains (TADs) (Dixon et al., 2012; Nora et al., 2012). These domains have been proposed to represent regulatory units within which enhancers and promoters can interact. They are separated by boundary regions that often contain CTCF binding sites or housekeeping genes representing de facto insulators that block interactions across adjacent TADs (Dixon et al., 2012). The importance of TAD structures is further supported by the finding that TAD boundaries appear to be largely static across different species and cell types. This suggests the existence of a preformed and stable topology that organizes the physical proximity between enhancers and their target genes. However, the observation that TADs exist regardless of transcriptional status has also raised questions regarding their role in cell- and tissue-specific regulatory processes (de Laat and Duboule, 2013). Furthermore, it has remained unclear if alterations in TAD structure, as they may occur in genomic rearrangements, can contribute to disease etiology.
In the present study, we analyze the potential value of annotated TAD boundaries for understanding how structural variation in the human genome elicits pathogenic phenotypes. Focusing on families with rare limb malformations, we identified several rearrangements in the extended WNT6/IHH/EPHA4/PAX3 region and re-engineered them in mice. Through a series of 4C-seq experiments and expression studies in mouse limb tissue and human patient-derived cells, we show that the rearrangements disrupt the normal topology of protein-coding genes and their enhancers relative to TAD boundaries, resulting in inappropriate interactions and misexpression. Our results highlight the utility of considering the three-dimensional architecture of the genome for predicting the consequences of structural variation and reinforce that this approach may be useful for the analysis of structural variants in a wide spectrum of human disease phenotypes.
RESULTS
Disruptions of TAD structure at the EPHA4 locus are associated with limb phenotypes
The EPHA4 gene resides within a large gene desert flanked by a gene-dense region on the centromeric side and the PAX3 gene on the telomeric side. Hi-C data show that the region is organized into three adjacent TADs, the largest encompassing EPHA4 (Figure 1A) (Dixon et al., 2012). Studying the genetic causes of rare limb malformations, we identified a series of structural variants at the EPHA4 locus that potentially interfere with the integrity of this region. In mice, Epha4 is expressed during limb development and required for normal innervation of the limb, but inactivation of Epha4 does not cause changes in the limb skeleton (Helmbacher et al., 2000).
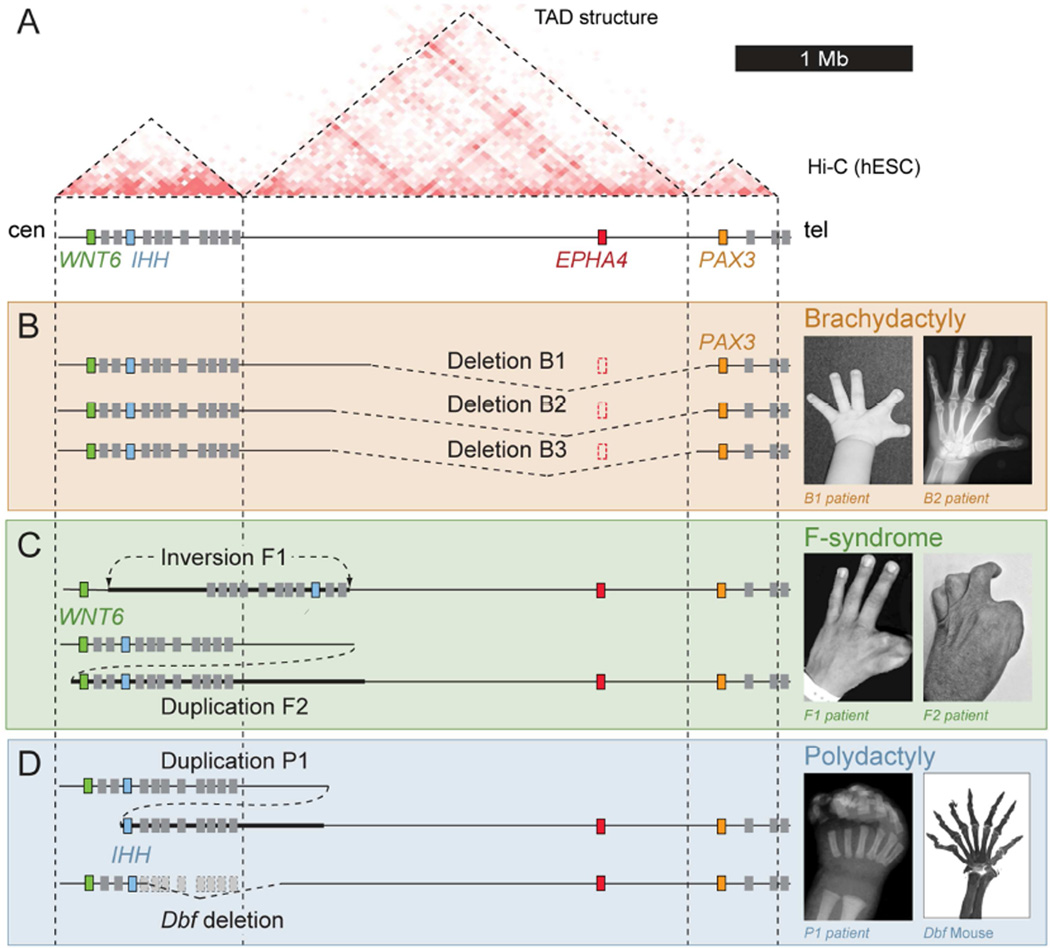
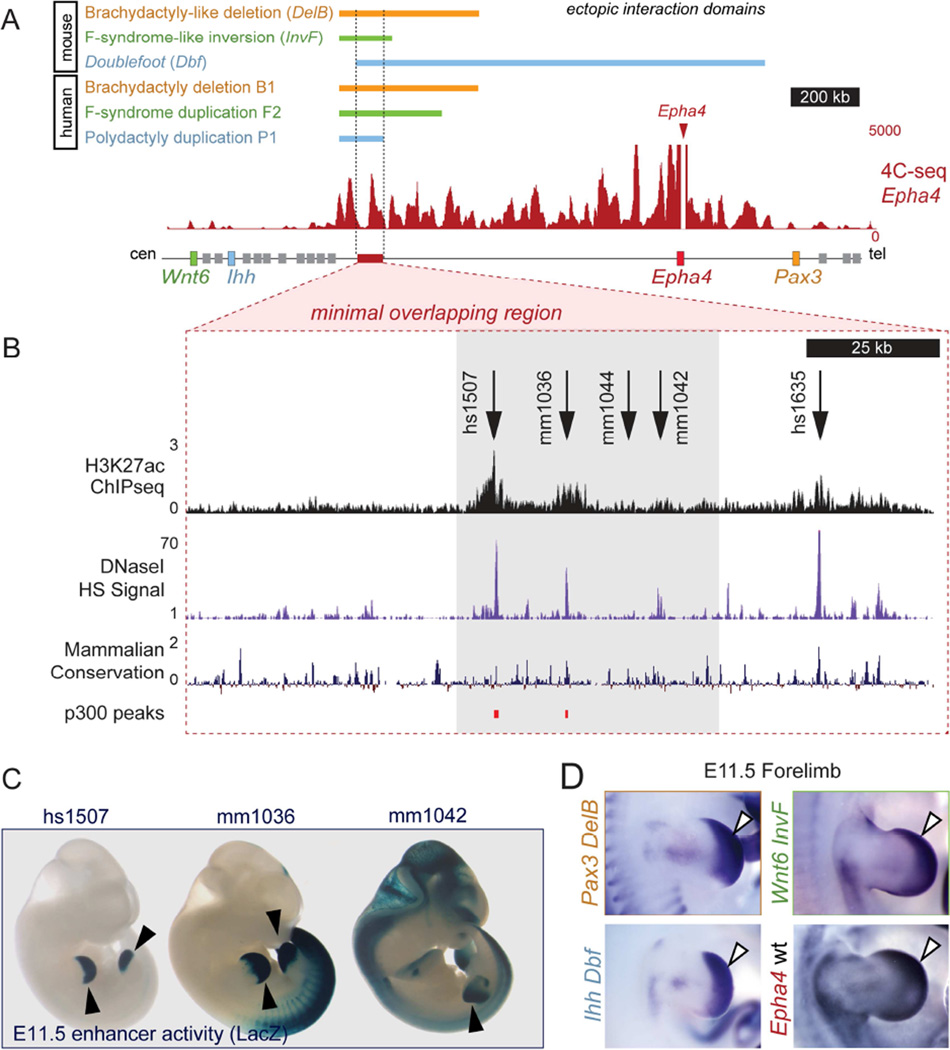
Figure 1. Different limb pathogenic structural variations in human and mouse map to the EPHA4 TAD.
(A) Hi-C profile around the EPHA4 locus in human ES cells (Dixon et al., 2012). Dashed lines indicate the EPHA4 TAD and boundaries. Cen, centromeric; tel, telomeric. (B–D) Schematic of structural variants (left) and associated phenotypes (right). (B) Brachydactyly-associated deletions in families B1, B2 and B3. Note thumb and index finger shortening with partial webbing in a child (B1 patient) and adult (B2 patient). (C) F-syndrome-associated inversion in family F1 and duplication in family F2. Note similar phenotypes of index/thumb syndactyly. (D) Polydactyly-associated duplication (P1) and deletion in the doublefoot (Dbf) mouse mutant. Radiograph of patient hand and skeletal preparation of Dbf/+ mouse show similar 7-digit polydactyly. See also Figure S1.
First, we investigated a dominantly inherited novel type of brachydactyly in three unrelated families, characterized by short digits predominantly on the preaxial (radial) side resulting in stub thumbs, short index fingers and a cutaneous web between the first and second fingers (Figure 1B, Figure S1). High resolution array comparative genome hybridization (CGH) revealed heterozygous deletions of 1.75–1.9 Mb on chromosome 2q35–36 in all three affected families. All three deletions include the EPHA4 gene along with a large portion of its surrounding TAD and extend into the non-coding part of the adjacent PAX3 TAD, thereby removing the predicted boundary between the EPHA4 and PAX3 TADs.
Second, we studied the molecular cause of F-syndrome, a limb malformation syndrome characterized by severe and complex syndactyly, often involving the first and second fingers, and polydactyly of the feet (Figure 1C) (Grosse, 1969). F-syndrome had previously been mapped to this chromosomal region (2q36), but its genetic cause remained unknown (Camera et al., 1995; Thiele et al., 2004). We used whole-exome sequencing to detect mutations in genes located in the linkage interval but were not able to identify any potentially pathogenic changes. To search for non-coding mutations and structural variations, we used whole-genome sequencing. We detected a ~1.1 Mb heterozygous inversion in family F1 and a ~1.4 Mb heterozygous duplication, arranged in direct tandem orientation, in family F2. The telomeric breakpoints were located 1.4 Mb away from the EPHA4 gene within the gene desert in the case of the inversion, and 1.2 Mb in the case of the duplication. The centromeric breakpoints were located centromeric and telomeric of WNT6 in the duplication and inversion, respectively (Figure 1C). Of note, both rearrangements bring the centromeric portion of the EPHA4-containing TAD into close proximity of the WNT6 gene.
Third, we studied a family that carries a heterozygous ~900 kb duplication in chromosomal region 2q35 that results in severe polysyndactyly and craniofacial abnormalities (Figure 1D) (Yuksel-Apak et al., 2012). The phenotype is reminiscent of the doublefoot (Dbf) mouse mutant, which also features massive polysyndactyly and was shown to be caused by a ~600 kb deletion affecting the same region (Babbs et al., 2008). Of note, both the human and the mouse alleles bring the IHH/Ihh gene in proximity to the centromeric portion of the EPHA4-containing TAD.
Chromatin interaction landscape of the extended WNT6/IHH/EPHA4/PAX3 region
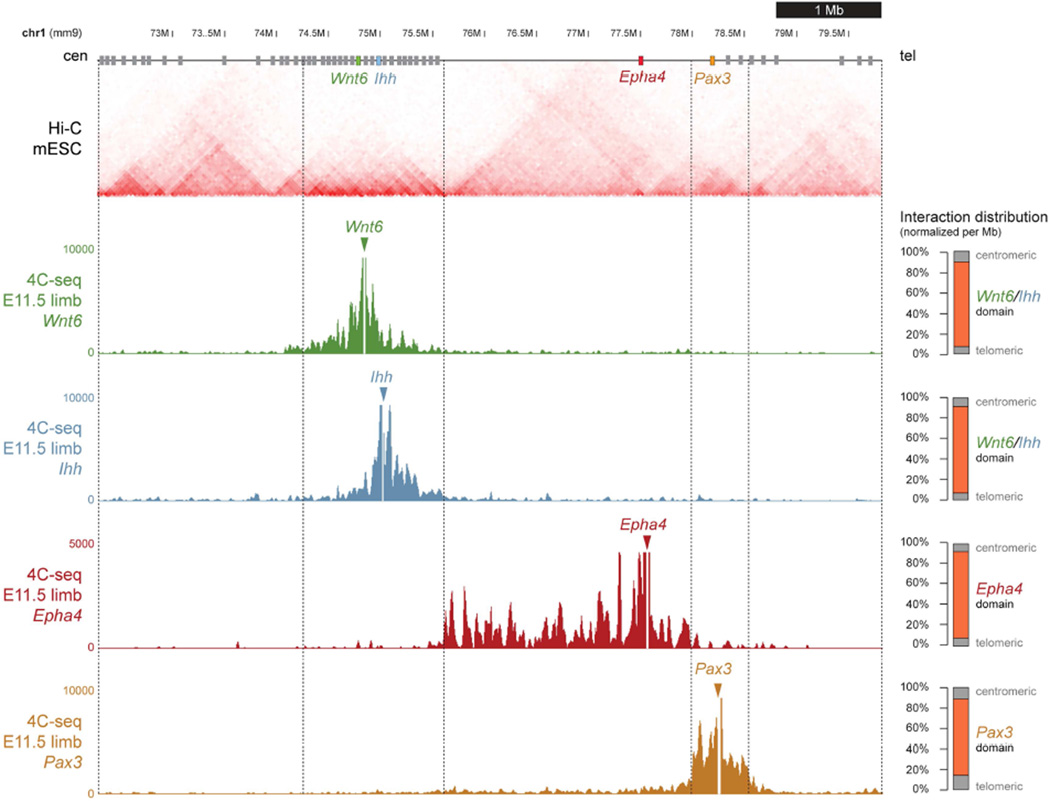
To elucidate the genetic basis of these birth defects, we sought to examine the regulatory landscape at this locus in more detail. In addition to EPHA4, we focused on the IHH, WNT6, and PAX3 genes due to their location near breakpoints in patients and their involvement in other developmental processes (Geetha-Loganathan et al., 2010; Goulding et al., 1994; St-Jacques et al., 1999). In each of the human disease alleles, at least one of the predicted TAD boundaries would be disrupted or its position changed relative to the four genes highlighted above (Figure 1). As illustrated in Figures 1 and 2, Hi-C data from human and mouse cells show a very similar TAD structure (Dixon et al., 2012), indicating that mice could serve as a model system for these diseases. To test if the TAD structure observed in ES cells by Hi-C is consistent with the chromatin conformation in the developing limb, we performed 4C-seq experiments in E11.5 mouse limb buds using the promoters of Epha4, as well as Ihh, Wnt6 and Pax3 as baits. For each promoter we observed interaction domains that were compatible with the Hi-C predicted TADs (Figure 2). These results confirm that the general TAD organization identified by Hi-C around the Epha4 locus is consistent with the interaction landscape during limb development obtained with 4C-seq.
Figure 2. TAD organization around the Wnt6/Ihh/Epha4/Pax3 locus.
Hi-C profile derived from mouse ES cells (Dixon et al., 2012). Predicted TAD boundaries are indicated by vertical dashed lines. 4C-seq profiles from E11.5 mouse distal limbs with viewpoints on Wnt6, Ihh, Epha4, and Pax3. Y-axis indicates number of normalized reads. 4C-seq interactions sharply drop at the Hi-C-predicted boundaries. Percentage of contacts/Mb within TADs is shown on right. Orange bars represent contacts within corresponding TAD. Grey bars represent contacts located in the respective next two centromeric TADs and two telomeric TADs.
CRISPR-mediated re-engineering of human disease alleles in mice
To study the effects of the structural variants observed in human patients in an experimentally accessible in vivo system, we generated mice with genome rearrangements recapitulating the human disease alleles. Using a protocol adapted for the introduction of large structural variants (Kraft et al., 2015), we cotransfected pairs of single guide RNAs (sgRNAs) into mouse embryonic stem cells (ESCs) to induce double strand breaks at desired positions.
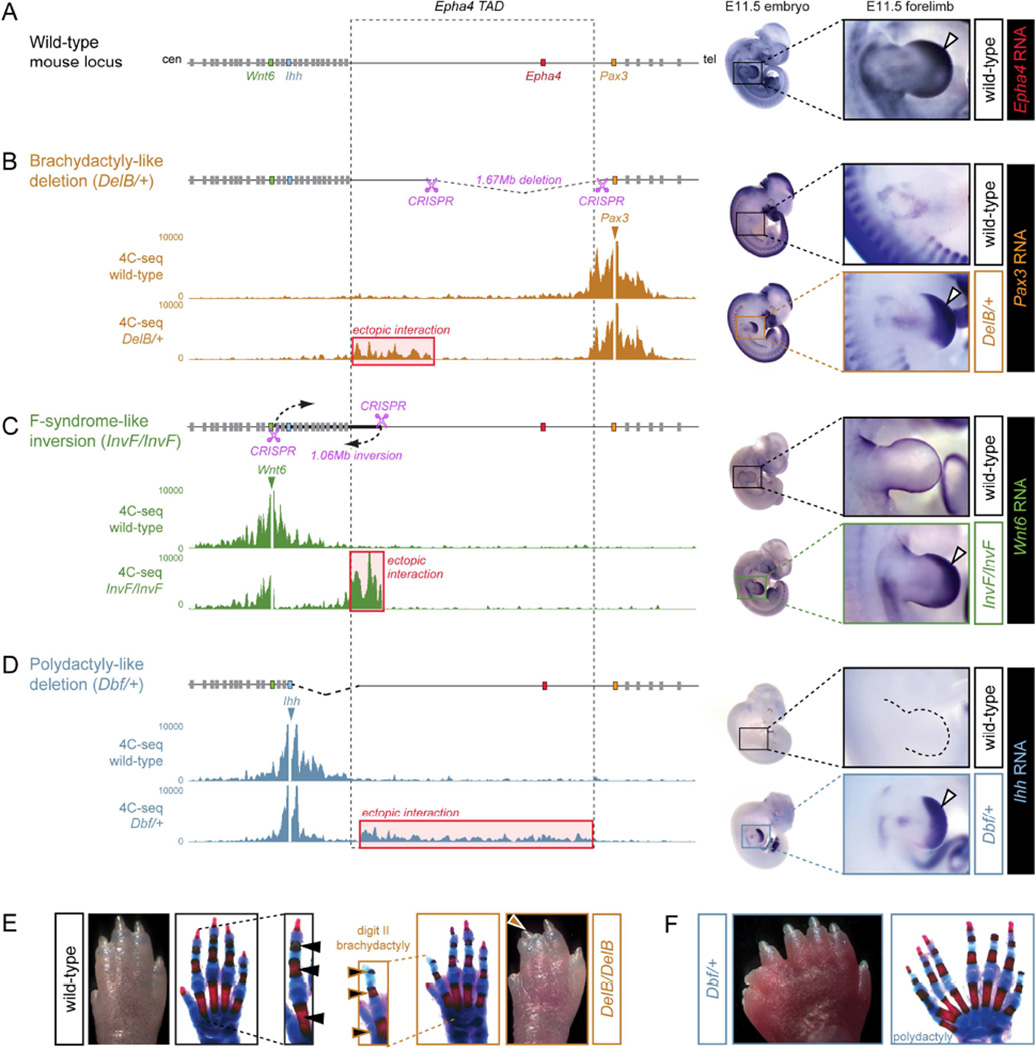
To generate a mouse model for the human brachydactyly cases, mouse line DelB was created from ESC clones carrying a corresponding CRISPR-induced deletion (Figure 3A/B). Heterozygous DelB/+ mice showed shortening of the second and third digits due to hypoplastic phalanges which was most pronounced in the middle phalanx of the second digit (Figure S2). This phenotype increased in severity when mice were bred to homozygosity, leading to severe shortening of the second digits due to very short middle phalanges, syndactyly between the second and third digits and a deviation of these digits towards the radial side (Figure 3E, DelB/DelB). The phenotype is very similar to the human malformation which is also characterized by short thumb and index finger due to short or missing middle phalanges and partial syndactyly (Figure 1B). Thus, mutant mice with a deletion corresponding to the human disease alleles recapitulated the phenotype observed in patients. In addition to these digit malformations, homozygous DelB/DelB mice also showed the known phenotype resulting from Epha4 inactivation, i.e. a resultant hopping gait due to gross motor dysfunction (Helmbacher et al., 2000).
Figure 3. Ectopic interaction of Wnt6, Ihh and Pax3 with the Epha4 TAD and misexpression in mouse models.
(A) Schematic of the locus. Dashed lines indicate Epha4 TAD and boundaries. In situ hybridization (right) shows Epha4 expression in distal limbs of an E11.5 wild-type embryo (white arrow). (B) Brachydactyly-like deletion generated by CRISPR/Cas (pink scissors). 4C-seq analysis with Pax3 as viewpoint in E11.5 wild-type and heterozygous mutant distal limbs shows ectopic interaction with the centromeric part of the Epha4 TAD in the mutant (red box). Pax3 shows ectopic distal limb expression in mutants (white arrow). (C) F-syndrome inversion generated by CRISPR/Cas (pink scissors). 4C-seq analysis with Wnt6 as viewpoint shows ectopic interaction with the centromeric part of the Epha4 TAD in the mutant (red box). Wnt6 shows ectopic distal limb expression in mutant limbs (white arrow). (D) Dbf deletion. 4C-seq analysis with Ihh as viewpoint shows ectopic interaction with most of the Epha4 TAD in mutants (red box). Ihh shows ectopic distal limb expression in mutants (white arrow). (E–F) Autopod and corresponding skeletal preparations of wild-type and mutant mice at postnatal day 3. Insets in (E) show digit II. Note reduction of digit II in the homozygous brachydactyly-like deletion and massive polydactyly in Dbf/+. See also Figures S2, S3 and S4.
To create a mouse model of structural variants found in human F-syndrome patients, we reproduced the inversion observed in family F1 in mice. We obtained heterozygous and homozygous ESC clones carrying a 1.06 Mb CRISPR-induced inversion with breakpoints at comparable positions in the mouse locus (Figure 3C). Heterozygous as well as homozygous newborns generated via tetraploid aggregation died shortly after birth of unknown cause and did not show overt limb phenotypes or other morphological defects (data not shown). Finally, we re-examined the previously described doublefoot (Dbf) mutant mouse strain due to the parallels in phenotype and genomic rearrangement with the human polydactyly patients (Figure 3D) (Yuksel-Apak et al., 2012). Dbf/+ mice have 6–9 digits per limb in a mirror image position with loss of anterior-posterior differences and no thumb-equivalent biphalangeal digit I (Figure 3F). Patients with polydactyly resulting from duplication P1 have a very similar limb phenotype consisting of severe polydactyly, complete fusion of digits (syndactyly), and a mirror configuration of the digits (Figure 1D). In two of three cases the mouse phenotypes closely resemble the human congenital limb defects, further supporting the utility of these mouse models.
Structural changes cause misexpression of developmental genes resembling the endogenous Epha4 pattern
The general nature of the structural variations and the resemblance of phenotypes resulting from inversion and duplication (F-syndrome) or duplication and deletion (polydactyly/Dbf) raise the possibility that these phenotypes are caused by convergent alterations in gene regulation. In the case of Dbf mice, ectopic expression of Ihh in the embryonic limb was previously described (Babbs et al., 2008). To examine the new CRISPR-engineered lines for aberrant expression, we performed RNA-seq experiments in E11.5 limbs of wild-type, DelB/+ (brachydactyly-like deletion), InvF/InvF (F-syndrome-like inversion), and Dbf/+ (polydactyly) embryos. We analyzed the chromosomal region around the Wnt6/Ihh/Epha4/Pax3 locus (chr1:73000000–79000000), for altered levels of gene expression related to the corresponding structural variation. We detected a significant upregulation of Pax3 in DelB/+ limbs, of Wnt6 in InvF/InvF limbs, and of Ihh in the Dbf/+ limbs, whereas all other surrounding genes were unaltered or showed only marginal increases in expression levels (Figure S3). As expected, Epha4, which is contained in the brachydactyly (DelB) deletion, and all the genes located within the Dbf deletion, were down-regulated. Based on these results we analyzed the expression patterns of Pax3, Wnt6 and Ihh, in the respective mouse mutants by in situ hybridization at E11.5 and compared them to the wild-type Epha4 expression pattern.
Epha4 is expressed in a distinct pattern in the developing limb, mainly in the distal mesoderm with predominance to the anterior side (Figure 3A, right). At the same developmental stage, Pax3 is also expressed in the limb bud, but restricted to migrating muscle cells, evident as faint staining in the proximal limb, and absent from the developing hand plate (Figure 3B, top right). DelB/+ (brachydactyly-like deletion) mice showed strong misexpression of Pax3 in the distal anterior part of the autopod, in a pattern resembling endogenous Epha4 expression (Figure 3B, bottom right). Wnt6 is normally expressed in the limb bud ectoderm, but not the distal mesoderm where Epha4 is expressed (Figure 3C, top right). In InvF/InvF (F-syndrome-like inversion) mice, we observed strong misexpression of Wnt6 in the distal limb autopod mesenchyme, in a similar pattern to Epha4 in wild-type controls and Pax3 in DelB/+ mice (Figure 3C, bottom right). The same pattern of misexpression was observed for heterozygous InvF/+ mice (not shown). Finally, Ihh is not expressed at all in the limb bud autopod at E11.5 (Figure 3D, top right). Misexpression of Ihh in the distal limb bud of Dbf mutants was previously demonstrated (Babbs et al., 2008) and comparison at E11.5 revealed a striking resemblance of the expression pattern with Epha4 in wild-type embryos (Figure 3D, bottom right). Taken together, these results indicate that genes near the chromosomal breakpoints are misexpressed in all three mouse lines. In all cases, the acquired expression domain closely resembles the endogenous expression pattern of Epha4, suggesting that regulatory sequences normally controlling Epha4 may play a role in the pathogenesis of the human limb phenotypes.
4C-seq reveals ectopic interaction of misexpressed genes with the Epha4 TAD
To examine whether the structural variants result in aberrant chromatin interactions that may explain the ectopic expression domains observed in mutant mice, we performed 4C-seq on distal E11.5 limbs. We analyzed the different mutants and stage-matched wild-type embryos, using the promoters of Pax3, Wnt6 and Ihh as baits (Figure 3B–D). For the deletions (DelB and Dbf), heterozygous animals were examined in order to minimize the possibility of deleterious effects that otherwise may result from homozygous deletion of genes. For the copy number-balanced inversion (InvF), we examined homozygous animals because no genes are deleted and the absence of the wild-type allele simplifies the interpretation of the 4C-seq data. In wild-type distal limbs, 4C-seq experiments showed minimal interaction of Pax3, Wnt6 and Ihh with non-coding sequences in the Epha4 TAD. In contrast, all three genes showed substantial interaction with the Epha4 TAD in the mutants. In the DelB/+ (brachydactyly-like deletion) mice, Pax3 showed a novel interaction domain of ~800 kb, spanning the remaining part of the Epha4 TAD flanking the centromeric breakpoint (Figure 3B). In InvF/InvF (F-syndrome-like inversion) we detected strong interaction of Wnt6 with a ~300 kb region that corresponds to the centromeric part of the Epha4 TAD, brought to the vicinity of Wnt6 through the inversion (Figure 3C). For better visualization, we mapped the 4C-seq data to a reference sequence that includes the inversion as present in the mice (Figure S4). Comparison with 4C-seq data from wild-type embryos showed that the interaction levels substantially exceed interactions with equidistant sequences in wild-type limb buds. Finally, the Dbf/+ deletion showed extensive interactions of Ihh with sequences throughout the entire Epha4 TAD (Figure 3D). Thus, all rearrangements resulted in novel interactions within the Epha4 TAD and a fusion of adjacent TADs. Remarkably, all novel interactions in the fused TADs respected the adjacent boundaries, i.e. Ihh and Wnt6 did not show interaction with the Pax3 domain and vice versa.
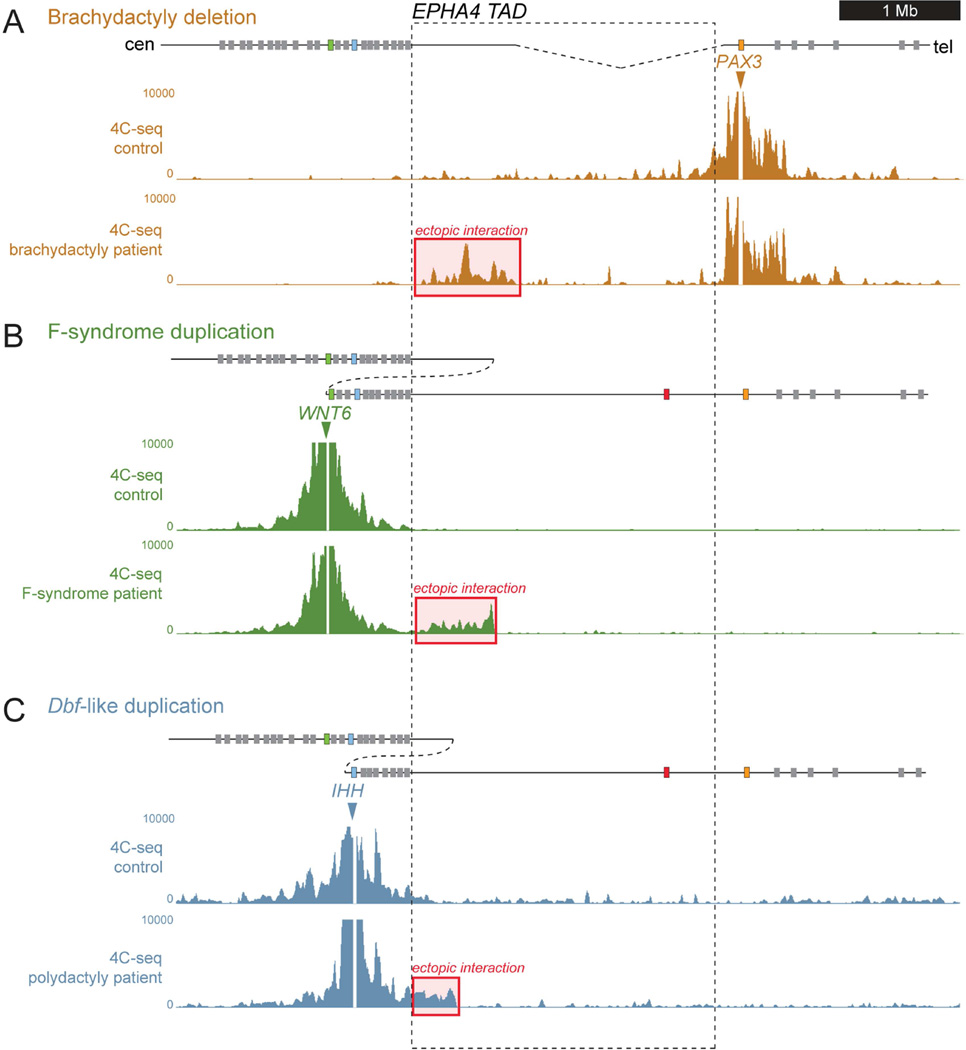
Disrupted TADs result in ectopic interaction in patient cells
Previous studies showed that TADs are highly stable across species and cell lines (Dixon et al., 2012), raising the possibility that patient-derived samples can provide direct insight into regulatory aberrations that affect early embryonic development. To test this paradigm, we applied 4C-seq to human adult fibroblasts (HAFs) and compared the results to data from the mutant mouse strains. We processed HAFs from a brachydactyly patient of family B1, a patient with F-syndrome from family F2 (cells from F1 were not available), and a polydactyly patient (P1) with the duplication. We compared patient samples to HAFs from age-, sex- and passage-matched healthy control donors (Figure 4). All controls showed a 4C-seq profile that was highly similar to that observed in mouse limbs (Figure 2) and human Hi-C data (Figure 1, top). Similar to DelB mice, the human brachydactyly-associated deletion resulted in aberrant contact of the PAX3 promoter region with the centromeric part of the EPHA4 TAD (Figure 4A). Likewise, the F-syndrome-associated duplication showed an ectopic interaction domain in the centromeric regions of the EPHA4 TAD, closely resembling the interaction domain gained in InvF mice (Figure 4B). Finally, the polydactyly-associated human duplication resulted in an overlapping, smaller interaction domain in the most centromeric region of the EPHA4 TAD (Figure 4C). While individual reads could not be unambiguously traced to one of the two copies present in the duplication allele, one plausible explanation for this observation are ectopic interactions between the telomeric copies of Wnt6/Ihh and the centromeric copy of the duplicated regions of the EPHA4 TAD.
Figure 4. 4C-seq analysis in patient-derived human adult fibroblasts.
Schematic representation of the locus and the structural variations are shown on top. 4C-seq profiles from human adult fibroblasts from healthy controls and patients with heterozygous structural variations are shown below. Red boxes indicate ectopic interaction. The EPHA4 TAD is indicated by dashed lines. (A) Patient with brachydactyly- associated deletion. 4C-seq analysis with PAX3 as viewpoint shows extensive ectopic interaction with the EPHA4 TAD (red box). (B) Patient with the F-syndrome-associated duplication. 4C-seq profile with WNT6 as viewpoint shows ectopic interaction with the EPHA4 TAD (red box). (C) Patient with the polydactyly-associated duplication. 4C-seq profile with IHH as viewpoint shows ectopic interaction with the centromeric region of the EPHA4 TAD (red box).
De novo interaction between distal limb enhancers and ectopic target genes upon TAD reorganization
Comparisons between the 4C-seq profiles obtained from the different mutant mouse tissues and patient cells revealed a minimal common region of ~150 kb within the EPHA4 TAD (Figure 5A). 4C-seq analysis of distal wild-type mouse limbs at E11.5 showed that this region frequently interacts with the EPHA4 promoter during normal development across 1.66 Mb of the intervening gene desert (Figure 5A). To identify enhancers with regulatory activity during limb development, we screened public ChIP-seq data for enhancer-associated chromatin marks, DNase HS data from equivalent developmental stages (E11.5) for open chromatin, and sequence conservation in this region (Figure 5B). To examine the strongest candidate sequences identified through this screen in more detail, we studied the in vivo activity pattern of five candidate enhancers. Two of these regions were already studied and documented in the VISTA enhancer browser database (Visel et al., 2007). The remaining three were tested for enhancer activity using a transgenic mouse LacZ enhancer reporter system.
Figure 5. A cluster of limb enhancers is located in the interacting region of the EphA4 TAD.
(A) Color bars indicate the fragment of the Epha4 TAD showing increased interaction with target genes in different mouse mutant limbs or human patient fibroblasts. The 4C-seq profile of Epha4 in E11.5 distal mouse limbs is shown for comparison. Red box indicates region of minimal overlap analyzed for enhancer activity. (B) Close-up view of the chromosomal region analyzed. DNase HS, H3K27ac and p300 ChIP-seq from E11.5 limbs and conservation tracks were used to predict enhancers. Arrows indicate putative enhancer regions. Gray box indicates the location of a cluster of enhancers shown below (C). (C) Transgenic enhancer reporter (LacZ) staining at E11.5. (D) Pattern of forelimb expression of Pax3 (DelB), Wnt6 (InvF), and Ihh (Dbf) in mutants and endogenous pattern of expression of Epha4 by in situ hybridization at E11.5. Note the similarity of endogenous Epha4 and ectopic Pax3, Wnt6, and Ihh expression domains (D, white arrows) with the enhancer activity patterns (C, black arrows). See also Figures S5 and S6.
At E11.5, 4 of 5 regions showed reproducible LacZ reporter activity in the developing limb. Three of these enhancers, clustered in a 30 kb region, showed a high degree of spatial overlap with the endogenous expression pattern of Epha4, as well as the ectopic expression domains of Pax3, Ihh, and Wnt6 gained in DelB, Dbf, and InvF mice, respectively (Figure 5C/D, Figure S5).
To confirm that the interactions in this region previously observed in mouse wild-type and mutant limbs involve these enhancers, we performed 4C-seq using the enhancer cluster as bait. The interaction profile revealed that this region interacts frequently with Epha4 during development in wild-type distal limbs (Figure S6, top). Next we tested if these enhancers had contact with the promoters of Pax3, Wnt6 or Ihh in the distal limbs of mutants compared to wild-type controls (Figure S6, bottom). This analysis confirmed that in all three cases, as a consequence of the different structural variations, new interactions are established between limb enhancers located inside the Epha4 TAD and genes outside the domain.
Boundaries of the EPHA4 TAD determine the pathogenicity of structural variants
All structural variants examined do not only change the arrangement of genes, enhancers, and predicted TAD boundaries relative to each other, but also result in changes in the distance between enhancers and their possible target genes. To investigate if the observed ectopic interactions are caused by disruption of boundary elements or merely by distance effects, we examined the role of the putative boundaries at either side of the Epha4 TAD in additional mouse mutants. Regions with boundary-like properties were suggested by the Hi-C data (Dixon et al., 2012). The analysis of CTCF ChIP-Seq data in human and mouse cell lines and tissues (ENCODE Project Consortium, 2004) showed an absence of CTCF binding sites within the Epha4 TAD, and the presence of several CTCF peaks at each boundary region (Figure 6A). To investigate the possible role of the border regions flanking the Epha4 TAD, we generated DelBS and DbfS mutant mice carrying deletions similar to the DelB (brachydactyly-like) and Dbf (polydactyly-like) mutants, except that the region containing the predicted boundary element was left intact (Figure 6B/C). Animals carrying these deletions had normal limbs and did not show any other abnormalities. Moreover, in situ hybridization for Ihh and Pax3 showed that these genes were not misexpressed and had retained their endogenous pattern of expression (Figures 6B/C, right). To compare the interaction profiles in the absence or presence of boundary elements, we performed 4C-seq experiments using Ihh or Pax3 as baits in E11.5 distal limbs. This analysis revealed that the ectopic interactions of the corresponding gene with the Epha4 TAD observed in DelB and Dbf mice is reduced in DelBS and DbfS mice (Figures 6B/C, left). Vice versa, 4C-seq using the enhancer cluster as bait also showed reduced interaction with the Ihh or Pax3 promoter (Figure S7). Thus, the presence of the boundary elements was sufficient to prevent inappropriate cross-TAD chromatin interactions, ectopic expression of non-target genes, and the morphological phenotypes resulting from this misexpression.
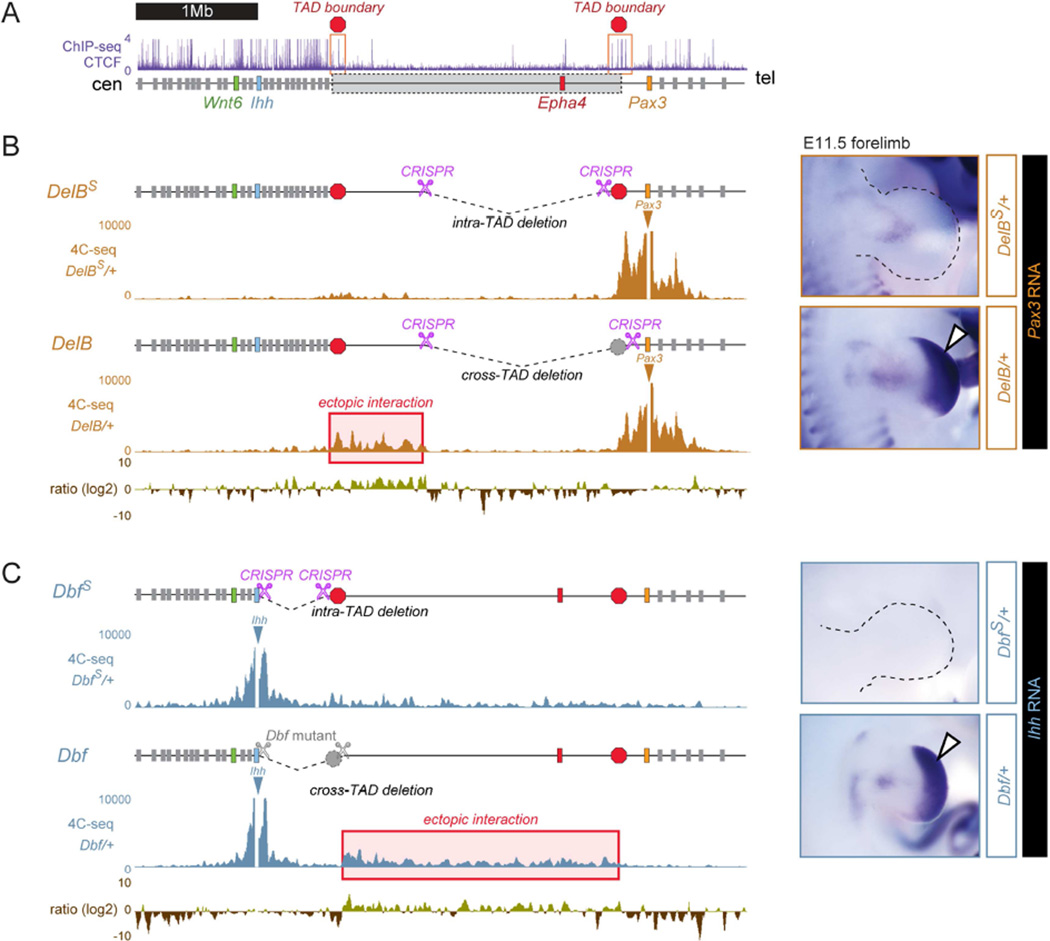
Figure 6. Boundary elements at both sides of the Epha4 TAD prevent ectopic expression of neighboring genes.
(A) CTCF ChIP-seq track in E14.5 mouse limbs (ENCODE/LICR). Red boxes and octagons mark clusters of CTCF peaks located at the boundary of the Epha4 TAD. Grey box indicates Epha4 TAD. 4C-seq profiles were generated from distal limb buds at E11.5. All data was obtained from heterozygous animals. Aberrant interactions are indicated by red boxes. Pink scissors indicate CRISPR/Cas-induced breakpoints in each deletion. (B) A deletion (DelBs) excluding the boundary region and CTCF cluster at the telomeric side of the Epha4 TAD (red octagon) was generated and compared to the brachydactyly-like deletion (DelB, including the CTCF cluster). Log2 ratio of the 4C-seq signal of DelB/DelBS shows increased interaction with the Epha4 TAD in the DelB deletion when compared to DelBs deletion (red box). Pax3 (right) shows normal expression of DelBs /+ deletion mice, in contrast to Pax3 misexpression in DelB/+ mice (white arrow). (C) A deletion (DbfS) excluding the boundary region and CTCF cluster at the centromeric side of the Epha4 TAD (red octagon) was generated and compared to the doublefoot deletion (Dbf, including the CTCF cluster). Log2 ratio of the 4C-seq signal of Dbf/DbfS shows increased interaction with the Epha4 TAD in the Dbf deletion when compared to DbfS deletion (red box). Ihh (right) shows absence of limb expression in Dbfs /+ deletion mice, in contrast to Ihh misexpression in Dbf/+ deletion mice (white arrow). See also Figure S7.
DISCUSSION
Structural variations are common in the human genome, but often difficult to interpret. Their inherent complexity urges for model systems in which the human situation can be faithfully recapitulated and studied. Here we use an adapted CRISPR/Cas genome editing protocol to reproduce human rearrangements in mice (Kraft et al., 2015). Using this approach, we re-engineered 3 human malformation associated rearrangements and investigated their effect on chromatin higher-order structures and gene function.
Disruption of TADs results in aberrant DNA domain topology and gene misexpression
TADs are stable units of genomic architecture that have been proposed to partition the genome into large regulatory units (Dixon et al., 2012). To investigate the effect of structural variants on TAD integrity, we examined limbs from the different mutant mouse strains using 4C-seq, which provides higher resolution than Hi-C. In all cases, we observed ectopic interaction of Pax3, Wnt6 or Ihh with the Epha4 TAD. The extent of interaction varied across the mutants, but included in all cases a minimal overlapping region of 150 kb (chr1:75694480–75848058, mm9). Based on the hypothesis that this region might contain regulatory elements that drive the misexpression of Pax3, Wnt6 and Ihh, we screened the region for enhancers and identified a cluster of regulatory elements driving limb expression. The pattern driven by these enhancers overlaps with the endogenous limb expression of Epha4 and is very similar to the misexpression domains of Pax3, Wnt6 and Ihh observed in the mutant strains. Taken together, these data suggest that the target genes near the breakpoints were adopted by Epha4 enhancers which, in turn, results in their misexpression (Figure 7). In DelB/+ mice we did not detect any other regulated genes. In both, the InvF/InvF and the Dbf/+ mice, only one gene, Cyp27a and Fev, respectively, showed significant up-regulation at overall marginal expression levels. Given the function of these genes in cholesterol metabolism (Cyp27a) and the central serotonin system (Fev), a contribution to the phenotype seems unlikely. Ectopic expression of Wnt6 can cause limb malformations in the chick via its anti-chondrogenic effect (Geetha-Loganathan et al., 2010) and misexpression of hedgehog proteins can induce polydactyly via the disruption of the anterior-posterior GLI3 gradient (Lettice et al., 2002). While the mechanisms by which ectopic expression of Pax3 may affect skeletal morphology remain to be established, the observed misexpression domains in combination with the morphogenetic potential of Wnt6 and hedgehog proteins offer a plausible molecular explanation for at least two of the human phenotypes observed.
Figure 7. A model for pathogenicity of structural variations.
Wild-type conformation shows the structure of two adjacent TADs. The activity of the enhancer (E) is restricted to gene 1 (G1) located inside the TAD. The TADs are separated by a boundary element (black). The black boxes represent the rearranged regions. The box with dotted lines represents the duplicated region. In the inversion, the enhancer is moved out of TADa and placed in the vicinity of gene 2 (G2). The boundary is now on the right side of E. This results in interaction of E with G2, but prevents its original interaction with G1. In the duplication, E is placed next to the duplicated G2’ resulting in interaction and misexpression. If the deletion removes the boundary and parts of TADb (black box), E is able to interact with G1 and G2, resulting in misexpression of G2. If the deletion leaves the boundary intact (DeletionS, black box), the TAD structure remains intact and E interacts only with G1.
Our 4C-seq data using the Epha4 enhancers as a viewpoint (Figure S6) also show that the regions of ectopic interaction cover many other genes besides the identified targets Pax3, Ihh and Wnt6. Nevertheless, expression analysis by RNA-seq showed no substantial upregulation of these genes, indicating that either enhancer-promoter distance or other unknown factors contribute to the receptiveness of a promotor to respond to the enhancer. In a Drosophila in vitro system, housekeeping and developmental promoters can respond to different classes of enhancers (Zabidi et al., 2014). It is possible that similar intrinsic specificities help to guide enhancer-promotor in vertebrate genomes. Here, the activated genes are all developmental genes expressed during limb development indicating that there may be a preference towards genes that are poised to get activated in this tissue.
Conservation of TAD structure across species, tissues and rearrangements
Comparison of TADs across different mouse and human cell types suggests that their boundaries are largely conserved (Dixon et al., 2012). We hypothesized that this conservation allows for the analysis of disrupted regulatory interactions that occur in vivo during early embryonic development, using patient samples collected long after limb morphogenesis has ended. To test this approach, we performed 4C-seq in human adult fibroblasts from patients with the brachydactyly-associated deletion, the F-syndrome-associated duplication and the polydactyly-associated duplication (Yuksel-Apak et al., 2012). Fibroblasts from healthy control individuals showed interaction domains highly similar to wild-type developmental mouse limb buds, whereas patient fibroblasts recapitulated the aberrant interactions observed in the respective mutant mouse strains. The observed chromatin interactions appear to be independent from gene expression levels, since EPHA4 and PAX3 are expressed at robust levels, WNT6 at very low levels, and IHH transcript is not detectable in HAFs (data not shown). While the ectopic interaction as well as the overall configuration was similar between human fibroblasts and mouse limb tissue, the distribution of peaks within the TADs was different. This is likely due to the different transcriptional activity and differentiation status of the analyzed samples (Nora et al., 2012). Regardless of such variation, our data indicate overall strong congruence between the developing mouse tissue and human-derived fibroblasts in the overall configuration of TADs. These results demonstrate the potential of patient-derived chromatin interaction data for gaining insight into the pathology of transient processes that occur during embryonic development.
Perturbation of TAD structure results in the formation of new TADs
All structural variants examined in this study result in aberrant interactions of a regulatory unit with ectopic-target genes that normally do not take place because enhancers interact only with their respective target promoters. TADs have been proposed to play an important role in establishing appropriate enhancer-promoter interactions by providing a structural scaffold that limits the distance and direction over which enhancers operate. However, it remains unclear if the observed partitioning of the genome is cause or consequence of pervasive enhancer-promoter interactions, and what functional role boundary regions between the TADs have (de Laat and Duboule, 2013). The perturbation of TADs by means of chromosomal rearrangements can provide insight into the fundamental mechanisms that drive this process. In vitro experiments at the Xist locus, for example, demonstrated reorganization of a TAD and spill-over of activity upon deletion of a 58 kb element corresponding to the boundary region (Nora et al., 2012). In the present study, variants that delete TAD boundaries result in interaction across the domains with no apparent new boundary formation and thus represent an apparently seamless fusion of neighboring TADs (Figure 7). The interactions within these apparent new TADs, as well as their exact boundaries and their impact on the overall three-dimensional architecture of the locus will have to be resolved by more quantitative analysis methods. However, the 4C-seq data obtained in this study suggest that the new TADs are defined by the next adjacent boundaries and that their functionality is not impaired. In the duplications the boundaries are not removed but the duplicated copy is flanked by an additional boundary, resulting in ectopic interaction within the newly formed TAD. Accordingly, our 4C-seq results show increased frequency of interaction of IHH (polydactyly family) and WNT6 (F-syndrome family) with centromeric parts of the EPHA4 domain (Figure 7, Duplication). In the case of the inversion the EPHA4 enhancer cluster and the adjacent boundary is moved next to WNT6. This results in the ectopic interaction with WNT6 and the formation of a new TAD which is now confined by the former EPHA4 centromeric boundary (Figure 7, Inversion). Similar results were obtained at the Tfap2/Bmp2 locus showing that inversions can result in reorganization of TAD structure by shifting boundaries (Tsujimura et al., 2015). Thus, a boundary element can be inverted or moved to a different chromosomal region without losing its functionality. However, the minimal region to determine boundary functionality is still unknown and therefore the consequences of deleting only the boundary elements remains to be tested.
Boundary structures are important for TAD integrity
We experimentally challenged the assumption that TAD boundary elements are functional and relevant for disease pathogenesis by creating deletions that leave the proposed boundary regions on either side of the Epha4 TAD intact. Both regions contained a cluster of binding sites for CTCF, a factor involved in boundary formation (Dixon et al., 2012; Van Bortle et al., 2014). We engineered additional variants of the Dbf as well as the brachydactyly-associated rearrangements, this time leaving the predicted boundary regions undeleted. No phenotypes and no misexpression of either Ihh or Pax3 were observed. Furthermore, 4C-seq experiments in these mice showed decreased frequency of interaction of the target genes Pax3 and Ihh with the Epha4 domain. Thus, leaving the proposed boundary regions intact diminishes all molecular phenotypes and averts morphological aberrations by preventing ectopic interactions.
Distance between regulatory elements and their target genes may be another determining factor. At the HoxD locus, for example, duplications within the TAD that result in an increase in the distance between promoter and enhancers were shown to result in an impairment of activation (Montavon et al., 2012). While we cannot rule out that distance effects contribute to the attenuation of molecular phenotypes, the difference between the deletion sizes is 100 kb for the Dbf/DbfS alleles and 200 kb for the DelB/DelBS alleles, corresponding to 17% and 12% of the total deletion size, respectively. It appears unlikely that these minor differences in distance alone are sufficient to explain the reversion of molecular phenotypes to near-wild-type levels, given similar deletion size differences of ~200kb that are present across the brachydactyly families (B1, B2 and B3) and have no apparent effect on the phenotype.
A framework for interpreting human structural variation
Depending on their size and position, structural variants may disrupt higher order genomic organization. In this study we present a conceptual framework for the interpretation of such variants using genome-wide chromatin interaction data sets. Our results reinforce the notion that the pathogenicity of a substantial proportion of human disease-associated deletions results from ectopic enhancer-promoter interaction causing gene misexpression due to the elimination of annotated boundaries (Ibn-Salem et al., 2014). The aberrant chromatin interactions observed at the EPHA4 locus in the present study exemplify how the disruption of TAD structure by eliminating or interfering with boundary elements can lead to the functional rewiring of gene-enhancer interactions (schematically shown in Figure 7). This model also illustrates how different types of large-scale structural changes can converge to give rise to the same phenotype. We showed that duplication and an inversion, as observed in the F-syndrome cases, or duplication and a deletion, as in the polydactyly family/mouse mutant, result in nearly identical molecular changes and morphological defects. In both cases the target gene interacts with the same non-coding genomic region and exhibits similar ectopic expression domains, despite the fundamentally different nature of the underlying structural mutation. In the deletions, a boundary is removed from the genome, permitting contact of the enhancer with genes outside of the TAD. In contrast, in the inversion and the duplications the enhancer is placed next to the new target gene. In the latter rearrangements the boundaries are left intact, but their new position no longer restricts contact of the enhancer with the target gene (Figure 7). The effect of such rearrangements is a gain of function via misexpression of one or several target genes. On the other hand, rearrangements of comparable size that do not interfere with TAD boundaries can be without consequence, as shown for the DelBS and DbfS mutants. Thus, considering overall TAD structure and in particular the integrity of the boundaries and their position relative to genes and enhancers appears critical when predicting ectopic and potentially pathogenic enhancer-promoter interactions. As this situation depends on the presence of several factors including available enhancers and receptive genes that can give rise to phenotypes when misexpressed, other pathogenic mechanisms such as loss or gain of gene function have to be considered when interpreting structural variations. Notably, such predictions can be experimentally tested even if the affected tissue is not available. As shown in this study, normal and abnormal TAD structure and enhancer-promoter interactions may be preserved across developmental tissues and adult human cells such as fibroblasts. Hence, 4C-seq in patient cells can yield diagnostically valuable information to predict effects of structural variants on gene regulation and thus disease etiology. While the present study focused on one locus and one set of related morphological phenotypes, TAD data for the entire human and mouse genome is becoming available at increasing resolution (Jin et al., 2013; Rao et al., 2014). These data should also help to interpret rearrangements in regions with higher gene density. Thus, the general principles uncovered in this study, and the resulting approaches for the interpretation of structural variation in human phenotypes are expected to be applicable to other genomic loci and a wide spectrum of genetic conditions caused by structural variants.
EXPERIMENTAL PROCEDURES
Human material
Venous blood samples and skin biopsies and were obtained from the patients and controls by standard procedures. Written informed consent was obtained from all individuals studied to participate in this study. This study was approved by the Charité Universitätsmedizin Berlin ethics committee.
Identification of human structural variations
All experiments were done with genomic DNA extracted from blood. Brachydactyly deletions were identified using array comparative genome hybridization (array CGH). F-syndrome associated duplication and inversion were identified using next generation sequencing (NGS, see supplemental information). Breakpoints for each structural variation were identified by breakpoint spanning PCR and Sanger sequencing.
Generation of transgenic animals using CRISPR/Cas
Mouse ES cells carrying structural variations were created using a CRISPR/Cas based protocol (Kraft et al., 2015)(see supplemental information). The size and position of the human structural variations (hg19) were converted to the mouse genome (mm9) using the USCS liftOver tool. CRISPR guides were designed using the CRISPR design tool based on the algorithm described in Hsu et al. (Hsu et al., 2013) to place guide sequences within close proximity of the predicted breakpoints (see Table S1). To minimize off target effects, guide sequences were chosen to have a quality score above 85%.
Embryos and live animals from ES cells were generated by tetraploid complementation (Artus and Hadjantonakis, 2011). For each structural variation at least two independent clones were aggregated. Genotyping was performed by PCR analysis. Guide primers, genotyping primers and breakpoint coordinates are summarized in Table S1.
RNA-seq
E11.5 distal limbs were microdissected from wild-type or mutant embryos. RNA was isolated from tissue samples using the RNeasy Mini Kit (Qiagen). Samples were sequenced using Illumina HiSeq technology according to standard protocols.
In vivo enhancer validation
Putative enhancer regions were selected based on public tracks for H3K27Ac Chip-Seq (Cotney et al., 2012), DNase HS, p300 Chip-Seq data from 11.5 limbs (Visel et al., 2009b), and conservation available at USCS genome browser (http://http://genome.ucsc.edu/). Selected regions were amplified by PCR from mouse genomic DNA and cloned into a Hsp68-promoter-LacZ reporter vector as previously described (Visel et al., 2009b)(see Table S2). Transgenic embryos were generated and tested for LacZ reporter activity at E11.5. All animal work performed at Lawrence Berkeley National Laboratory was reviewed and approved by the institutional Animal Welfare and Research Committee (AWRC).
In situ hybridization and skeletal preparations
In situ hybridization was performed according to standard protocols. Probes from Pax3, Ihh, Wnt6 and Epha4 were generated by PCR amplification using E11.5 mouse limb cDNA. For skeletal preparation, specimens were stained according to standard Alcian blue/Alizarin red protocols.
All animal procedures were in accordance with institutional, state, and government regulations (Berlin: LAGeSo).
4C-Seq
4C-seq libraries were generated from microdissected tissues or cells as described previously (van de Werken et al., 2012). BglII or HindIII (6-bp cutters) were used as primary restriction enzymes. Csp6I or DpnII were used as secondary restriction enzymes. For each viewpoint, a total of 1.6 mg of each library was amplified by PCR (primer sequence in Table S3). Samples were sequenced with Ilumina Hi-Seq technology according to standard protocols.
Supplementary Material
HIGHLIGHTS.
-
◦
Disruptions of TADs lead to de novo enhancer-promoter interactions and misexpression
-
◦
Misexpression occurs when CTCF-associated TAD boundary elements are disrupted.
-
◦
Structural variations disrupting TAD structures can cause malformation syndromes
-
◦
Different phenotypes can result from one enhancer acting on different target genes
Acknowledgments
D.G.L. is supported by the Fundación Alfonso Martín Escudero. This research was supported by grants from the Deutsche Forschungsgemeinschaft, from the Berlin Institute for Health, and the Max Planck Foundation to SM. M.O. was supported by a Swiss National Science Foundation (SNSF) fellowship. AV was supported by NIH grants R01HG003988, U54HG006997 and U01DE024427. Research conducted at the E.O. Lawrence Berkeley National Laboratory was performed under Department of Energy Contract DE-AC02-05CH11231, University of California. We thank Daniel Ibrahim and Guillaume Andrey for comments on the manuscript. We also thank Nicole Rösener, Asita Stiege, Karol Macura, Nadine Lehmann, Anne Heß and Christin Franke for technical support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All data have been deposited at GEO (GSE66383).
REFERENCES
- Artus J, Hadjantonakis AK. Generation of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Methods in molecular biology. 2011;693:37–56. doi: 10.1007/978-1-60761-974-1_3. [DOI] [PubMed] [Google Scholar]
- Babbs C, Furniss D, Morriss-Kay GM, Wilkie AO. Polydactyly in the mouse mutant Doublefoot involves altered Gli3 processing and is caused by a large deletion in cis to Indian hedgehog. Mechanisms of development. 2008;125:517–526. doi: 10.1016/j.mod.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camera G, Camera A, Pozzolo S, Costa M, Mantero R. F-syndrome (F-form of acro-pectoro-vertebral dysplasia): report on a second family. American journal of medical genetics. 1995;57:472–475. doi: 10.1002/ajmg.1320570322. [DOI] [PubMed] [Google Scholar]
- Cotney J, Leng J, Oh S, DeMare LE, Reilly SK, Gerstein MB, Noonan JP. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome research. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Geetha-Loganathan P, Nimmagadda S, Christ B, Huang R, Scaal M. Ectodermal Wnt6 is an early negative regulator of limb chondrogenesis in the chicken embryo. BMC developmental biology. 2010;10:32. doi: 10.1186/1471-213X-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–971. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- Grosse FRHJ, Opitz JM. The F-form of acropectorovertebral dysplasia: the F-syndrome. Birth Defects Orig Artic. 1969:48–63. [Google Scholar]
- Haraksingh RR, Snyder MP. Impacts of variation in the human genome on gene regulation. Journal of molecular biology. 2013;425:3970–3977. doi: 10.1016/j.jmb.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Helmbacher F, Schneider-Maunoury S, Topilko P, Tiret L, Charnay P. Targeting of the EphA4 tyrosine kinase receptor affects dorsal/ventral pathfinding of limb motor axons. Development. 2000;127:3313–3324. doi: 10.1242/dev.127.15.3313. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibn-Salem J, Kohler S, Love MI, Chung HR, Huang N, Hurles ME, Haendel M, Washington NL, Smedley D, Mungall CJ, et al. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome biology. 2014;15:423. doi: 10.1186/s13059-014-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft K, Geuer S, Will AJ, Chan WL, Paliou C, Borschiwer M, Harabula I, Wittler L, Franke M, Ibrahim DM, et al. Deletions, Inversions, Duplications: Engineering of Structural Variants using CRISPR/Cas in Mice. Cell reports. 2015 doi: 10.1016/j.celrep.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Thevenet L, Duboule D. Impact of copy number variations (CNVs) on long611 range gene regulation at the HoxD locus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20204–20211. doi: 10.1073/pnas.1217659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014 doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann M, Mundlos S. Structural variations, the regulatory landscape of the genome and their alteration in human disease. BioEssays : news and reviews in molecular, cellular and developmental biology. 2013;35:533–543. doi: 10.1002/bies.201200178. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes & development. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annual review of medicine. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Swaminathan GJ, Bragin E, Chatzimichali EA, Corpas M, Bevan AP, Wright CF, Carter NP, Hurles ME, Firth HV. DECIPHER: web-based, community resource for clinical interpretation of rare variants in developmental disorders. Human molecular genetics. 2012;21:R37–R44. doi: 10.1093/hmg/dds362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele H, McCann C, van't Padje S, Schwabe GC, Hennies HC, Camera G, Opitz J, Laxova R, Mundlos S, Nurnberg P. Acropectorovertebral dysgenesis (F syndrome) maps to chromosome 2q36. Journal of medical genetics. 2004;41:213–218. doi: 10.1136/jmg.2003.014894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura T, Klein FA, Langenfeld K, Glaser J, Huber W, Spitz F. A discrete transition zone organizes the topological and regulatory autonomy of the adjacent tfap2c and bmp7 genes. PLoS genetics. 2015;11:e1004897. doi: 10.1371/journal.pgen.1004897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Nichols MH, Li L, Ong CT, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome biology. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, de Vree PJ, Splinter E, Holwerda SJ, Klous P, de Wit E, de Laat W. 4C technology: protocols and data analysis. Methods in enzymology. 2012;513:89–112. doi: 10.1016/B978-0-12-391938-0.00004-5. [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009a;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009b;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic acids research. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuksel-Apak M, Bogershausen N, Pawlik B, Li Y, Apak S, Uyguner O, Milz E, Nurnberg G, Karaman B, Gulgoren A, et al. A large duplication involving the IHH locus mimics acrocallosal syndrome. European journal of human genetics : EJHG. 2012;20:639–644. doi: 10.1038/ejhg.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer--core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2014 doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.