Abstract

High-throughput biology has contributed a wealth of data on chemicals, including natural products (NPs). Recently, attention was drawn to certain, predominantly synthetic, compounds that are responsible for disproportionate percentages of hits but are false actives. Spurious bioassay interference led to their designation as pan-assay interference compounds (PAINS). NPs lack comparable scrutiny, which this study aims to rectify. Systematic mining of 80+ years of the phytochemistry and biology literature, using the NAPRALERT database, revealed that only 39 compounds represent the NPs most reported by occurrence, activity, and distinct activity. Over 50% are not explained by phenomena known for synthetic libraries, and all had manifold ascribed bioactivities, designating them as invalid metabolic panaceas (IMPs). Cumulative distributions of ∼200,000 NPs uncovered that NP research follows power-law characteristics typical for behavioral phenomena. Projection into occurrence–bioactivity–effort space produces the hyperbolic black hole of NPs, where IMPs populate the high-effort base.
Introduction
The advent of high-throughput screening (HTS) and the subsequent development of a plethora of compatible biological assays have led to a staggering amount of bioactivity data. Beyond the inherent difficulty of managing high volumes of data, the validity of the hits and assays has to be questioned. Some compounds, in commercial or privately assembled chemical libraries, were shown to be responsible for a disproportionate fraction of the hits in these screens.1 Moreover, many of these hits often appeared to be acting as panaceas, i.e., they showed activity in several disparate assays (frequent hitters), suggesting that they could be lead structures for drug development for several different diseases. In most cases, these were false positives, and extensive efforts have been devoted to understand the mechanisms involved. The designation of some of those compounds as PAINS (pan-assay interference compounds)2 and, more broadly, as promiscuous inhibitors3 adequately reflects how this select group of chemicals has led, and may continue to lead, to wasted effort and resources in futile development programs.
Evidence for Panacea Natural Products
A suspicion that these nonspecific inhibition phenomena might not be restricted to synthetic chemical libraries but might extend to natural product (NP) programs is the driving force for the present investigation. Evidence for panacea NPs came from various aspects of our research programs. One source of evidence relates to the widely observed challenges associated with identifying highly selective bioactive principles in complex NP extracts. Examples include our own efforts to advance interdisciplinary drug discovery, e.g., the development of anti-TB leads from NPs,4 as well as botanical dietary supplement research programs, e.g., efforts to advance the rationalized pharmacognosy of black cohosh (Actaea racemosa).5 Indeed, insight is growing that the active principles in many crude NP therapies are polyfactorial agents (multiagent, multitarget).6−8 This challenges the long-held paradigm of bioassay-guided fractionation as the standard discovery process for NP bioactives and instigates the development of new approaches such as biochemometrics,9 databases and metabolic networks,10 or machine learning.11 An extensive list of unconventional approaches can be found in a timely review compiled by Wolfender et al.12
A second indication that panacea NPs exist came from the observation of an inverse correlation between the purity and anti-TB bioactivity of ursolic acid,13 leading to the establishment of (quantitative) purity–activity relationships ([q]PARs) as a means of hit validation.13,14 A subsequent global literature search of the biological profile of ursolic acid using the NAPRALERT database (unpublished data) confirmed the earlier notion13 that panacea-like properties have been ascribed to this near-ubiquitous plant NP.
A third source of evidence for panacea NPs originated from an extensive meta-analysis of the primary literature on bioactive NPs, evaluating the status quo of methodology used in the analysis and purification of natural products:15 the AnaPurNa study uncovered a variety of analytical and conceptional parameters that can impact the validity of a hit. Developed further in the AnaPurNa study, the several factors that were identified as impacting the NP valid lead discovery process (e.g., purity, metabolomic sources, analytical methodology, and bioassay specificity) are distinctly different from the mechanisms of biological promiscuity discussed in the present perspective. However, both the previous and present findings ring true with the points raised by Cordell7 about ecopharmacognosy and the multiple challenges of NPs as treatments or tools for drug supply and discovery.
A fourth source of evidence involves a broader body of our own work that led to the establishment of the concept of residual complexity (RC) (see http://go.uic.edu/residualcomplexity).15,16 RC refers to the convolution of major and minor chemical species in NP preparations and other materials that originate from (bio)synthetic reaction mixtures, i.e., the designated actives, impurities, and biogenetic congeners or side products. The RC concept is particularly significant in instances where minor (residual) constituents cannot be neglected and are actually key to the explanation of biological outcomes achievable with NPs. This applies regardless of whether the residual agent is already present (static) or formed over time (dynamic), e.g., during the biological evaluation.
Meta-Analysis with the NAPRALERT Database
The above points provided ample rationale to propose that a holistic analysis of the world literature on bioactive NPs was required to detect additional key parameters and identify global principles that are important to the success of the (drug) discovery process. The primary resource for undertaking this otherwise Herculean ab initio task was the natural products alert (NAPRALERT) relational database. The ChEMBL17 and PubChem18,19 databases were used as secondary sources for structural and bioassay data.
Invalid Metabolic Panaceas
In its overall outcome, this study recognized multiple factors driving NP-based discovery. In particular, we observed that certain NPs, designated invalid metabolic panaceas (IMPs), interfere with this process. IMPs are familiar to most researchers in the field, but they are not necessarily well-understood metabolites. Some of the identified IMPs were recognized by the PAINS filters, others were proven to be aggregators or to show the characteristics of both groups, while still others fit neither of these categories.
IMPs extend the established principle of promiscuous molecules such as PAINS rather than being a subset thereof. This is supported by the outcome that for some of the IMPs no known promiscuous characteristic, other than observed promiscuity itself, could be found. Like PAINS, IMPs tend to divert major research effort and scientific focus away from potentially more promising molecules. The present perspective both identifies IMPs and provides potential routes for avoiding unproductive effort in NP-based research programs.
Opening the NAPRALERT Window
Bioassay Interference with Natural Products
The insight that certain compounds could “trick” bioassays (Figure 1, middle) became actively disseminated at the end of the 1990s with the description of the promiscuous effects of some chemical substances in HTS assays.20,21 Systematic recognition of aggregation as a major cause of artifacts in bioassays commenced with the ground-breaking work of the Shoichet group and their 2002 publication.3 The authors applied a variety of orthogonal methods to demonstrate that certain compounds could act as false positives in bioassays by forming aggregates binding to protein targets in the aqueous media that are predominantly used in bioassays.22 The definition of PAINS entered the picture of bioassay artifacts in 2010: the Baell group2 recognized PAINS as compounds that bear problematic substructures that escape traditional substructures filters, while still being identified as hits in assays that were specially crafted to reduce aggregation artifacts as unveiled by the Shoichet group.
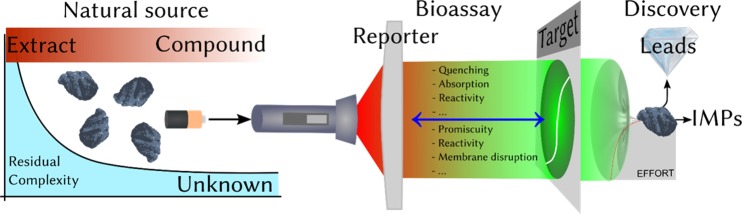
Figure 1.
Relationship among the source, the bioassay, and the interpretation of data from the NP discovery pathway is complex by nature.
Some PAINS possess substructures that were considered to be characteristic of aggregating compounds, despite the adapted bioassays.23 Moreover, new compounds that cannot be avoided by PAINS removal strategies continue to be discovered,24 indicating that other forms of promiscuity exist in both natural and synthetic molecules. Other mechanisms by which compounds will act as promiscuous agents have been summarized.25 The most common process of bioassay interference is related to fluorescence, a frequent read-out in HTS assays.26 Additional means of interference include precipitation of an analyte as a cause for false negatives, light scattering leading to false positives in UV/visible read-out assays, and membrane disruption causing issues in whole-cell assays.
All of these bioassay-related means of generating anomalous results in compound libraries apply equally to NP-based drug discovery. Compared to synthetic libraries, the NP approach to drug discovery has a more prominent analytical dimension, at least historically, due to the need for isolation, purification, and identification. The increasing recognition of the impact of multiple reaction products and purity in combinatorial libraries27,28 has its parallel in the combinatorial biosynthesis of NPs. In fact, as residual complexity (RC) is found almost ubiquitously in NPs, there is a need for the careful analysis of at least static RC, especially when considering the historically poor attention to analyte purity and the inherent complexity of biological matrices.15
In the case of NP lead discovery, the relationship between the agent and the bioassay, consisting of the reporter and the target, is rather complex (Figure 1). This is a result of three main factors: (a) the RC (purity) of the NP sample, which arises from its natural source and is the main energizer of the discovery process (Figure 1, left); (b) a variety of known potential interferences that can occur between the agent and the reporter components of the bioassay (Figure 1, middle); and (c) the nature and/or complexity of the actual biological target (Figure 1, middle right). All three factors contribute to the current status of NP-based discovery in terms of effort spent, leads identified, and IMPs encountered during the process.
Initial Observations
The present work began with two basic enquiries regarding the long-term distribution of NPs in the literature and the global effort expended on them.
-
(i)
Which metabolites are the most reported in the literature?
-
(ii)
Which metabolites are the most reported as showing biological activities?
The NAPRALERT database29 was used throughout this project. Housed at the University of Illinois at Chicago for over 45 years, it covers more than 80 years of predominantly phytochemical natural product research, including both chemical and biological information. More importantly, it is the most complete and comprehensive applicable database available. It includes almost all phytochemicals reported for the covered years as well as their biological activities and source organisms. A preliminary study showed that the distribution of both groups of highly reported compounds followed a rather specific, but not mathematically uncommon, pattern. This revealed, with the confidence of thousands of supporting primary references collected in the database, that some metabolites were heavily over-represented. The present meta-analysis represents the logical extension of this initial result and is aimed at learning more about these highly reported compounds, often with similar structures, and the reasons for their over-representation in the scientific literature.
The Origins of NAPRALERT Information
It is important to understand the origin of the information deposited in the database, in order to appreciate the significance of the meta-analysis results. NP-based research programs are often driven by interest in identifying bioactive metabolites. The organism provides the library of metabolites that may be investigated. The choice of organelle(s) or plant part(s) along with the extraction method(s) provides the first level of metabolite selection. In bioassay-guided fractionation schemes, the researcher is interested only in identifying and isolating metabolites that display bioactivity. A variation of bioassay-guided fractionation seeks to identify and isolate novel structures associated with selected bioactivities. Metabolites with promising bioactivities may be reisolated from the sources, or isolated from other sources, in order to continue bioactivity investigations. Persistent bioactivity studies are desirable to develop promising drug leads and are facilitated by databases that report both chemical and biological activity data.
Some chemistry-driven research programs are specifically interested in novel metabolites regardless of their bioactivity. In addition, some metabolomic studies have been performed to identify and isolate a large number of both known and previously unknown compounds from natural sources. Chemotaxonomic and endophyte-targeted investigations provide other motivations for metabolite identification and isolation. Accordingly, the information available in NAPRALERT is similar to, but not the same as, the HTS campaigns that led to identification of panacea compounds.
Another influential aspect is that the limited bioassay information, typically found in individual NP publications, makes it difficult to perceive bigger patterns unless a very large number of publications is studied. Even larger NP discovery campaigns have had the inherent handicap of a relative paucity of biological information, resulting in a limited ability to recognize global trends. This limitation was one of the key motivations for the founder of NAPRALERT and his colleagues to embark on the long journey of creating this unique resource.
On the other hand, the curators of experimental HTS data are faced with the treatment of thousands to millions of entries for each campaign. Therefore, it is more likely that HTS-driven initiatives can produce awareness of the existence of nuisance compounds that show strong connections to producing unexpected or undesired outcomes.
The Next Step beyond Identifying Metabolic Panaceas
Recently, an increasing number of studies have aimed at unravelling the mechanisms of panacea compounds. The most prevalent manifestations of assay interference are aggregation, precipitation, instability, chemical reactivity, optical opacity (absorption, diffusion), oxidation/reduction, fluorescence quenching, and RC (impurities). While some of these phenomena can lead to false negatives, most possess substantial disrupting potential by producing either false positives or yielding results that appear to be incoherent when comparing them with results from orthogonal (bio)assays. While some studies have started to investigate these phenomena in NPs,30,31 it appears that the NP literature has not yet embraced these concepts as being essential for a more targeted discovery process and/or acknowledged the nuisance character of certain hits. We felt that a study specifically designed to identify and evaluate the suspicious characteristics of certain NPs was in order. To this end, this Perspective also seeks to raise awareness by summarizing recent work on nuisance mechanisms that are also applicable to NPs.
The Challenges Associated with Bioassays
Several mechanisms are known to underlie unwanted interferences between screened materials and bioassays. The following section begins with a summary of NP-specific parameters that can add another (undesired) dimension to the interpretation of biological outcomes. The subsequent survey of in vitro interference mechanisms also brings to mind that the continued predominance of in vitro screening over in vivo assessment does not come without its challenges. On the basis of their over two decades of experience in the discovery of chemopreventive agents, Kinghorn and co-workers32 pointed out that the difficulty of finding promising NP leads may be correlated with the trend of eliminating pharmacological in vivo models, in favor of higher throughput in vitro assays. It is an intriguing question to ask, whether the early significant NP discoveries that resulted from in vivo primary screening efforts (e.g., the 1960s to early 1980s NCI campaign yielding taxol, camptothecin, maytansine, dolastatins, bryostatins, etc.) would also have been made in programs driven by in vitro assays. Collectively, this may generate impulses for a future comparative assessment of the overall effectiveness of the two paradigmatic approaches, which could add a useful dimension to the present discussion.
The Complexity of Natural Sources
As documented by the highly comprehensive work of Newman, Cragg, and their co-workers (refs (33) and (34) and references therein), NPs are a vital source of drugs and/or molecular scaffolds for drugs. It can be perceived as unfortunate, or the natural challenge, that this enormous potential is confounded by complex issues of sourcing, purification, and assay perturbation.15,31,35 The source organism’s metabolic matrix is usually complex already, containing compounds produced for, e.g., metabolic purposes, defense, or interspecies communication. Metabolites are typically products or substrates of enzymes that can have homologues in target organisms. It should be no surprise if at least some metabolites could actually have an effect on these homologous enzymes, thus giving rise to interesting and even unexpected activities. Similar considerations apply for primordial molecules that might appear commonly across distant taxa; as Sandor and Mehdi inferred in 1979, “steroids are very ancient bioregulators, which evolved prior to the appearance of eucaryotes or were even possibly synthesized abiotically”.36 This idea was later reinforced in 1993 by Agarwal in his review of steroid hormones receptors in microbes and plants.37 Examples include mammalian steroid hormones that are known to also be present in plants (e.g., progesterone in walnut leaves)38 and 3-O-sulfation as a shared means of steroidal metabolism in plants (e.g., Adonis aleppica) and mammals.38
Purity and Residual Complexity
Whereas purity is central to the definition of pharmaceutical quality, purity of assayed metabolites or fractions is often overlooked or assessed unreliably.39,40 For NPs in particular, many commercially available metabolites are of moderate purity, typically in the range of 90–95% declared purity, and only infrequently assessed by independent methods.40 For metabolites obtained by bioassay-guided fractionation, the problem of residual complexity (RC) is inherent. In static RC, the residual components are chemically stable and do not change over time.13 In dynamic RC, not only does the concentration of the metabolite change, but also new chemical entities appear in the sample over time and as a function of environmental conditions, e.g., the bioassay.41 While this concept originated with NPs, it can apply to synthetic compounds as well, where each sample carries its synthetic history rather than a biogenetic heritage. Whereas the identified (often major) component may be benign, an impurity that is part of either static or dynamic RC may be the active component or the interfering troublemaker. Fortunately, awareness of the role of purity and the potential of quantitative 1H NMR (qHNMR) as a versatile, orthogonal analytical method has recently increased in the scientific community in general and in this journal in particular.42
Aggregating Metabolites
Performing an extensive study of the behavior of aggregating compounds,1,3,22,43−50 the Shoichet group has gathered clear evidence that some compounds have the ability to sequester proteins from the assay, thus likely leading to false-positives. Conversely, as the free concentration of the molecule of interest may be only minute and possibly below the critical aggregating concentration, the same basic mechanism can also lead to a reduction of apparent activity or false negatives.51 Thus, any observed puzzling bell-shaped concentration–activity relationships may be related to aggregation phenomena.49 Notably, even some commercial drugs have displayed aggregation potential.43,48,50 During an HTS campaign, Feng et al. found that 95% of the actives were aggregators.52 A study of 14 selected compounds that are present in abundance in traditional Chinese medicine (TCM) preparations detected 10 with aggregating potential. This indicates that caution is required when interpreting in vitro assay results with both these compounds and their source TCMs in general.50
PAINS
An acronym for pan-assay interference compounds, PAINS are a collection of problematic substructures unveiled in a 2010 groundbreaking paper by Baell and Holloway.2 The authors compiled previously published20,53,54 and new guidelines in the form of a set of Sybil Line Notation filters. These filters have been integrated in more generic tools such as the FAF-Drug55 and the Eli Lilly set of rules.56 While the PAINS substructures may not always be problematic and the kiss of death for a compound containing them, it is important to be aware of their existence. In any event, it is vital to verify that the bioactivity of a compound is authentic before designating it as a lead or, alternatively, removing it from consideration.
Optical and Fluorescence Effects
Fluorescence detection is one of the favorite reporter mechanisms in bioassays, as it is usually highly sensitive. However, certain compounds are fluorescent by themselves57 or have the ability to quench fluorescence through diverse mechanisms.58 In fact, one of the most widely used reporters, firefly luciferase, has been shown to be inhibited by almost 60% of the compounds in cell-based screening campaigns (see ref (25) and references therein). On the other hand, other compounds may show intrinsic fluorescence, which, if not taken into account, may compromise the reading. Optical interferences that occur with colored extracts and compounds that can impede optical detection of activity can also create detection issues for fluorescence- or absorbance-based assays.
Chelation, Metals, and Redox (Re-)Activity
Chelation of metals has also proven to be a source of spurious inhibition.2 Some commonly used bioassays are sensitive to certain chelators.59 In the case of cell-based assays, chelation can sequester vital ions, thus reducing cell viability. When working with enzymes that contain a metal cofactor, chelating compounds can also impede the assay. On the other hand, some metals themselves can lead to the formation of reactive species or production of hydrogen peroxide, and/or they can elicit other unexpected inhibition.60
Another form of assay interference is observed with compounds that can covalently bind to or otherwise modify the target. Some compounds may oxidize susceptible enzymes or intermediaries used in the bioassays. While this phenomenon is a known in vivo mechanism to regulate enzyme activity,61 it is usually unwanted in a controlled bioassay environment. In other cases, the interfering compounds may be involved in the generation of hydrogen peroxide when reducing agents are used.62 Phenolic compounds are abundant redox-active metabolites and should be held under scrutiny. For example, catechol moieties in polyphenols have been shown to form quinones and/or radicals through redox cycling, even without enzymatic catalysis.63 While this mechanism potentially applies to a large number of compounds, a generalized conclusion about bioassay interference or even lack of drug lead potential cannot be made, as is evident from the large number of drugs with the catechol motif.
Surfactants and Membrane Perturbation
Several prominent groups of NPs, such as saponins and certain fatty acids and their derivatives, have surfactant-like properties. Regarding fatty acids, Balunas et al. studied the effects of 11 fatty acids on enzymatic and cell-based bioassays.64 While the reported effect on cell-based assays is low, the effect on enzymatic assays is significant. Linoleic acid has shown in vitro estrogenic activity65 and in vitro binding to human δ opioid receptors.66 However, as noticed by the authors of the last paper, the same compound was identified as a noncompetitive inhibitor for three independent targets, making conformational changes a more plausible explanation of the observed effect.67 As aggregator compounds are sensitive to detergent concentration, it is possible that some NPs trigger the destabilization of aggregates. This could restore an activity that had disappeared as a result of the introduction of the aggregating compound in the assay, i.e., acting as antiaggregators and producing double false positives. Such a case would, of course, make the assay even harder to interpret, especially if the aggregating and destabilizing agents and/or characteristics are unknown, as is often the case in early stage NP programs working with multicomponent mixtures. Moreover, in assays involving cells or reconstituted membranes, compounds showing surfactant properties may show disturbing results if their effect on membranes is not assessed or is not the subject of the assay itself.68,69
All of the above effects, alone and/or in combination, have proven to be major issues impacting biological screening. A study by Jadhav et al. showed that 93% of the hits were nonspecific due to a combination of these kinds of effects.1 Comprehensive reviews of some of these interferences have been compiled by Thorne et al.25 and Sink et al.23 The principles of affinity, efficacy, potency, and mass action are not discussed here, as they have been covered by Borgert et al. in their review on endocrine active substances.70
Material and Methods
NAPRALERT
The detailed description of the design of this relational database used to collect NP research data has been published71 and was followed-up by a more recent summary of its capabilities.29 The database is accessible via STN and a web interface at https://www.napralert.org, which has been redesigned as of October 2015. While NAPRALERT continues to be run on its original MSSQL/.NET platform, it is currently being rewritten using modern technologies that will provide easier access to data and complex requests. Meanwhile, the data used for this study has been exported from MSSQL format to a series of CSV files, which were used as raw data for the analyses.
Data Analysis
While many tools are able to cope with moderate amounts of data (up to millions of entries), Python (https://www.python.org) was chosen for its ease of use, status as Free software, optimized data-analysis libraries, and the ease of incremental development and interactive data-mining. Pandas (http://pandas.pydata.org) and Scipy (https://www.scipy.org) libraries were used for data-analysis. Bokeh (http://bokeh.pydata.org) provided interactive graphics during the data exploration phase. Matplotlib (https://www.matplotlib.org) was used to generate the static graphics that were further processed with Inkscape (https://www.inkscape.org). Blender (http://www.blender.org) was used for 3D artwork. The incremental development and interactive mining of NAPRALERT’s raw data was made possible by using the Jupyter Notebook (https://www.jupyter.org) software, allowing for work in a web-browser with remote access. The Jupyter environment was running in a Docker instance (https://www.docker.com) using the scipyserver container (https://github.com/ipython/docker-notebook). Long-tail fitting utilized the power-law Python package.72
Research on PubChem data was performed manually using the Web site: https://pubchem.ncbi.nlm.nih.gov/. Search data was downloaded as CSV files and treated using shell scripts and the Python infrastructure described above. The ratio of actives over total reported in the confirmatory assays was then calculated. Data was grouped by targets to avoid inflation of the scores by assay repetitions/duplication.
A local PostgreSQL instance of the ChEMBL database (version 20) was used to automatically gather the structures of the compounds.
Defining the Bottom of the Black Hole of Natural Products
Beginning with NAPRALERT data, this study focused on a merged set of top-scoring NPs in three categories. (i) Occurrence: the top-20 metabolites according to their described occurrence in organisms, i.e., frequency of a report as a constituent of any organism. (ii) Activities: the top-20 metabolites tested for bioactivity and/or designated as bioactive, including those reported as a bioactive principle and/or marker. (iii) Distinct activity: the top-20 metabolites regarding their assigned unique biological activities, determined as a measure of the number of distinct targets they have been assayed for.
Occurrences (O)
In order to determine the number of organisms for which a metabolite has been reported, a relatively large set of search criteria was used (see Table S1, Supporting Information). These criteria were modeled to accommodate the ways by which the presence of a metabolite is usually reported in publications. Presently, NAPRALERT contains organism-specific information on 189,740 metabolites from 43,578 organisms. The database contains additional information on metabolites that, by nature of the study material or as a result of the style of the report, were not attributable precisely to a single organism. On average, NAPRALERT has 11 metabolites per organism, with a maximum of 795 NPs reported for a single organism (Nicotiana tabacum; see Table S2, Supporting Information). The Tables S3 and S4 in the Supporting Information contain data on the top-20 organisms and families, respectively, in terms of distinct metabolites. For the purpose of assessing occurrence, the names of the organisms were used as reported by the authors. NAPRALERT contains a synonym dereplication system that is currently being reworked to cope with recent nomenclature updates and to link it to taxonomy databases.
The distribution of metabolites across all investigated organisms documented in Table S2 in the Supporting Information reveals that, on average, a NP is described in 37 organisms. Considering that, at the same time, more than 50% of the NPs are described only once, this already shows the relatively strong tendencies toward the two extremes in the occurrence reporting of NPs.
Activities (A)
In the PubChem database (accessed April 18, 2015), of the 68,280,771 compounds described, 2,082,979 have been tested for activity. Thus, only 3% of the reported compounds have associated bioactivity results. By comparison, of the 189,740 metabolites entered into NAPRALERT (accessed on the same date), 50,379 have been evaluated biologically. This activity coverage of 27% is almost 10 times that for PubChem, demonstrating NAPRALERT’s information richness for bioactive NPs. The aim of PubChem is not to provide activity data, and many of the compounds it covers were not synthesized with drug discovery in mind. However, recently, the effort to acquire more data has become a very active process, with support of other databases and private entities sharing their own data. This represents an advantage over NAPRALERT bioassay data in the coverage of non-NP-related sources.
One of the characteristics of NP research is the dominant role of bioassay-guided fractionation, which, if followed rigorously, would lead exclusively to bioactive compounds, at least in theory. However, another significant characteristic of NP research is the quest for new molecular entities, particularly new structural types. Moreover, NP science also includes metabolomics investigations and chemotaxonomy studies that are not necessarily bioactivity driven, hence yielding the occurrence of a reasonable proportion of compounds with no reported bioactivity. It is noteworthy that >75% of the compounds with assigned bioactivity in NAPRALERT have been reported to occur in only one or two organisms (see Table S5, Supporting Information).
Table 1 shows that the extent of the biological evaluation of 96% of all NPs recorded in NAPRALERT is limited, to no more than 10 reported activities per metabolite. Whereas NAPRALERT includes the entire breadth of (mostly plant related) NP publications, and despite linkage with pharmacological data being one of its mainstays, this resource still cannot be expected to completely cover the biological activities of all included metabolites. In order to compensate for this limitation, the present study utilized other available bioassay databases. These can be readily accessed through public application programming interfaces (APIs) and/or linked together with cross-referencing systems such as UniChem.73
Table 1. Number of Activity Tests Reported for NPs Included in NAPRALERT in Five Categories of Frequency of Evaluation.
| no. of reported activities | 0 | 1–10 | 11–100 | 101–1000 | >1000 |
|---|---|---|---|---|---|
| compounds (%) | 139,361 (73) | 44,219 (23) | 5798 (3.0) | 355 (<1.0) | 7 (<0.01) |
Factors Affecting (Reported) Activities
Under certain instances, over the course of their investigation, metabolites and their activity data can fade or even disappear from the NAPRALERT radar. To date and by design, the database relies on editorial work involving systematic but manual selection of articles for inclusion and encoding their content. There are three main reasons for the fading or disappearance of a metabolite from the inclusion efforts:
-
(i)
The compound becomes commercially available or is synthesized successfully. As a result, it is no longer being related to an organism. However, such compounds will still remain visible if these publications occur in the surveyed NP journals.
-
(ii)
Compounds that are transferred between laboratories, e.g., as a gift or inside collaborative teams, are frequently used for biological purposes without being linked to their origin.
-
(iii)
The compounds are used in studies that are published in non-NP-related journals, which are too numerous to be part of the NAPRALERT encoding efforts.
The first point highlights the multiple effects that obscure the precise origin of a compound, especially if it comes (originally) from natural sources or is semisynthetic. Collectively, the frequency of occurrence of all three instances, (i)–(iii), combined can be estimated to be in the 20–30% range. While being limited in journal coverage relative to NAPRALERT, our previous AnaPurNa study15 was designed to distinguish isolated from synthesized compounds, purchased materials, and gifts from colleagues. The breadth of the AnaPurNa study is sufficient to conclude that a significant proportion of NPs escapes the systematic NAPRALERT survey by mechanisms (i)–(iii). This affects up to one-third of NPs (unpublished data from the analysis of raw AnaPurNa data15).
Another important general trend that we have observed as part of our NAPRALERT work, as well as during the AnaPurNa project, is that publications with a strong focus on biological effects tend to omit chemical quality/grade, lot number, or manufacturer/source information on the investigated NPs. This appears to be counterintuitive when considering the parallel trend toward increasingly stringent editorial and documentation guidelines (e.g., Good Laboratory Practices). However, three factors are of considerable importance for a better understanding of reported NP activities: the limitations posed by the relatively frequent lack of information on (a) positive identification of the NP, (b) its purity, and (c) possible variations in the RC of the investigated material.16
Upon closer inspection, despite being related to minor components, the RC issue can be of major importance. First, even in instances where purity is assessed (≪1% of all NP studies;15 more common for commercial compounds), there is usually no information about what exactly constitutes the missing few (X) percent of the “100 – X% pure” materials. Moreover, the historically predominant (UV-)HPLC assays exhibit an acknowledged limitation regarding their universality and selectivity. More importantly, while the typical range of 2–5% can seem a low value for an impurity, this value should be put in relation to the actual amount that gets into the assay. For example, a molar impurity of 2% of a sample applied at 10 μM is still present at 200 nM, clearly a concentration of potential pharmacological relevance. This quantitative side of the purity coin also has its qualitative counterpart: as isolation procedures, source organisms, synthetic routes, or suppliers change, the RC profile is likely to change as well, even if the NP shows the same labeled purity value. Instances falling under the umbrella of both static and dynamic RC were also discussed by Baell et al. in the PAINS context,24 demonstrating that these issues are not limited to the NP world.
Distinct Activities (D)
This set is derived from the activities set (A) by filtering out the duplicate bioassay targets. These duplicates are usually not replicates, as it is difficult to correlate different activity levels when the assay is not normalized or when the purity of the NP is not assessed. Comparison of the sets of distinct activities (D) vs tested activities (A) exhibits an average duplicate ratio of 3.2, with a maximum of 6.0.
Overlapping the Three Sets: Occurrences, Activities, and Distinct Activities
Merging the three top-20 sets of metabolites that are most occurring (O), most tested for activity (A), and with the most distinct (D) activities yielded 39 metabolites (Table 2). A Venn diagram of this overlap is displayed in Figure 2. While the activity (A) and distinct activity (D) sets are similar (12 overlapping metabolites, equivalent to 60% similarity), the occurrence (O) set is clearly separated from both other sets. This implies that the (chemical) occurrences and number of (biological) activities tested in the entire group of 39 metabolites (39/60 = 65% total overlap) may not be highly correlated.
Table 2. Top Reported Compounds for Each of the Three Categories: Occurrences (O), Activities (A), and Distinct Activities (D)a.
| no. | compound | O | rank | A | rank | D | rank | Agg | PAINS | % actives |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | quercetin | 4115 | 2 | 3004 | 1 | 686 | 1 | * | * | 52.4 |
| 2 | gossypol | 495 | 112 | 2642 | 2 | 433 | 3 | * | * | 41.3 |
| 3 | β-sitosterol | 7640 | 1 | 805 | 14 | 201 | 29 | ± | 5.6b | |
| 4 | genistein | 431 | 139 | 1630 | 3 | 468 | 2 | * | 18.6 | |
| 5 | rutin | 2889 | 4 | 1025 | 6 | 355 | 5 | * | 14.3 | |
| 6 | kaempferol | 2531 | 6 | 939 | 9 | 313 | 9 | * | 25.1 | |
| 7 | berberine | 1365 | 33 | 1258 | 5 | 319 | 8 | 5.5 | ||
| 8 | curcumin | 106 | 657 | 1347 | 4 | 371 | 4 | * | 18.0 | |
| 9 | apigenin | 1533 | 27 | 937 | 10 | 325 | 7 | * | 30.4 | |
| 10 | (+)-catechin | 910 | 50 | 998 | 8 | 341 | 6 | * | 8.6 | |
| 11 | luteolin | 1903 | 13 | 758 | 18 | 246 | 14 | * | * | 35.8 |
| 12 | caffeic acid | 1581 | 25 | 770 | 17 | 238 | 16 | ± | * | 15.9 |
| 13 | (−)-epicatechin | 764 | 67 | 772 | 16 | 271 | 12 | * | 9.3 | |
| 14 | resveratrol | 209 | 306 | 874 | 11 | 296 | 10 | 23.9 | ||
| 15 | glycyrrhizin | 189 | 352 | 809 | 13 | 294 | 11 | ± | 4.9 | |
| 16 | gallic acid | 1154 | 39 | 790 | 15 | 198 | 30 | * | 34.6 | |
| 17 | EGCG | 141 | 494 | 813 | 12 | 248 | 13 | * | 35.4 | |
| 18 | ursolic acid | 1623 | 18 | 563 | 30 | 172 | 38 | ± | 13.5 | |
| 19 | taxol | 555 | 100 | 1009 | 7 | 158 | 47 | 18.5 | ||
| 20 | eugenol | 723 | 72 | 720 | 20 | 191 | 32 | 2.8 | ||
| 21 | (+)-tetrandrine | 72 | 1009 | 734 | 19 | 245 | 15 | * | 6.2 | |
| 22 | myricetin | 666 | 84 | 581 | 28 | 223 | 20 | * | * | 40.4 |
| 23 | stigmasterol | 2857 | 5 | 272 | 109 | 81 | 148 | ± | 0 | |
| 24 | α-pinene | 3007 | 3 | 224 | 135 | 78 | 156 | 0 | ||
| 25 | capsaicin | 63 | 1137 | 636 | 23 | 235 | 17 | ± | 6.5 | |
| 26 | ginsenoside Rb-1 | 454 | 130 | 504 | 39 | 228 | 18 | 0 | ||
| 27 | ginsenoside Rg-1 | 470 | 123 | 463 | 44 | 228 | 19 | N/Ab | ||
| 28 | limonene | 2313 | 8 | 295 | 95 | 98 | 99 | 6.7b | ||
| 29 | isoquercitrin | 2128 | 10 | 258 | 118 | 117 | 80 | * | 17.3 | |
| 30 | daucosterol | 1995 | 11 | 281 | 102 | 103 | 92 | 50b | ||
| 31 | 1,8-cineol | 1931 | 12 | 344 | 69 | 92 | 118 | 1.3 | ||
| 32 | lupeol | 1827 | 15 | 310 | 85 | 104 | 90 | * | 100b | |
| 33 | palmitic acid | 2145 | 9 | 129 | 277 | 76 | 159 | 20.4 | ||
| 34 | linalool | 1849 | 14 | 282 | 101 | 61 | 214 | 0 | ||
| 35 | β-pinene | 2351 | 7 | 132 | 273 | 46 | 318 | 0 | ||
| 36 | linoleic acid | 1608 | 20 | 203 | 154 | 82 | 139 | ± | 18.8 | |
| 37 | oleic acid | 1617 | 19 | 149 | 228 | 75 | 162 | ± | 8.9 | |
| 38 | p-cymene | 1734 | 16 | 113 | 327 | 34 | 472 | 0 | ||
| 39 | myrcene | 1665 | 17 | 95 | 377 | 41 | 376 | 1.7 |
The Agg column denotes if the metabolite itself (*) or a close analogue (±) has been reported as aggregating. The PAINS column indicates if the metabolite is recognized by the PAINS filters. The % actives column shows the percentage of confirmatory assays from PubChem, in which the NP has been reported as active. EGCG means epigallocatechin gallate.
Denotes metabolites with less than 50 assays reported in PubChem.
Figure 2.
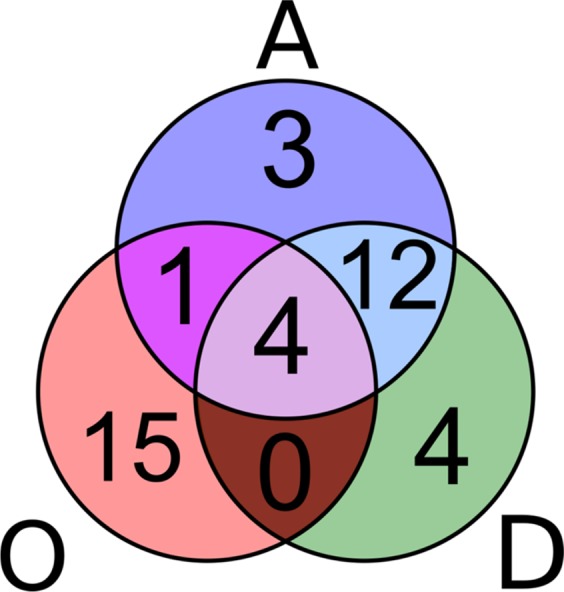
Venn diagram of the three considered sets of the top 39 metabolites: most occurring (O), most reported as tested for activity (A), and most distinct (D) activities. The activities/distinct activities set are highly overlapping, whereas the occurrence set tends to be isolated from these.
At first glance, 39 metabolites may seem to be a vanishingly small number compared to the almost 200,000 NPs contained in NAPRALERT. However, Table 3 shows that, while these metabolites represent <0.002% of the database, they account for 5–8% (2500- to 4000-fold; depending on group O vs A vs D) of the total reports in the database.
Table 3. Percentage of the Top-20 Metabolites of Each Set (O/A/D) Relative to the Total NAPRALERT Databasea.
Possibly, the most important insight from recognizing these 39 metabolites through NAPRALERT data mining is that this group of metabolites likely contains the most prominent IMPs produced by nature. Evidence that some are indeed IMPs will be presented in the following discussion. Notably, all three cumulative distributions (O/A/D) follow power-law functions rather than showing the Gaussian behavior of statistical chance. This is an additional preliminary indicator that discovery serendipity or chance arise from non-Gaussian events.
As fully explained in the next section, the 39 metabolites of the merged set are located at the very bottom of a hyperbolic body, designated as the black hole of NPs: this shape is formed when plotting the cumulative 2D power-law distribution of all NPs in 3D bioactivity and occurrence distribution space. Thus, the distribution analysis of the NP literature reveals a characteristic behavior of NP research in which a significant amount of effort is expended on a tiny fraction of chemical diversity and with little production of valuable drug leads.
The Big Picture: The Holistic Distribution of Natural Products
Scattered Distributions and Cumulative Sums
The scatter plot of the distribution of occurrences (O) and activities (A) displayed in Figure 3 shows that no visible correlation exists between these two parameters for these merged sets. This confirms the implication from the Venn diagram (Figure 2) that something other than occurrence is driving the reporting of activities. Comparing the two complete O and A distributions through their Spearman rank correlation gives a positive score <0.3 (p < 0.01) on the scale −1 to 1, whereas the activity (A) and distinct activity (D) sets are more correlated, with a score of 0.994 (p < 0.01). Contrary to the Pearson correlation, the Spearman rank correlation reacts not only to a linear correlation between ranks but also a monotonic relationship. Thus, by giving an indication of the similarity of the ordering, it also has the advantage of being valid when it is performed on data sets that are not normally distributed (non-Gaussian). While the occurrence of a given metabolite in an organism by default cannot predict the number of its bioactivities, the observed non-null correlation between O and A may indicate that this relationship is still (perceived as) a factor that has some influence. However, the interpretation of correlation values must be done with caution, as the distribution of the underlying data points is clearly asymmetrical, with most of the data points being in the tail of low citations per metabolite. Moreover, the high number of points involved in the present study artificially decreases the p value, thus rendering these numbers to be interpreted with caution rather than designating them as directly representing tendencies and/or indications of similarities.
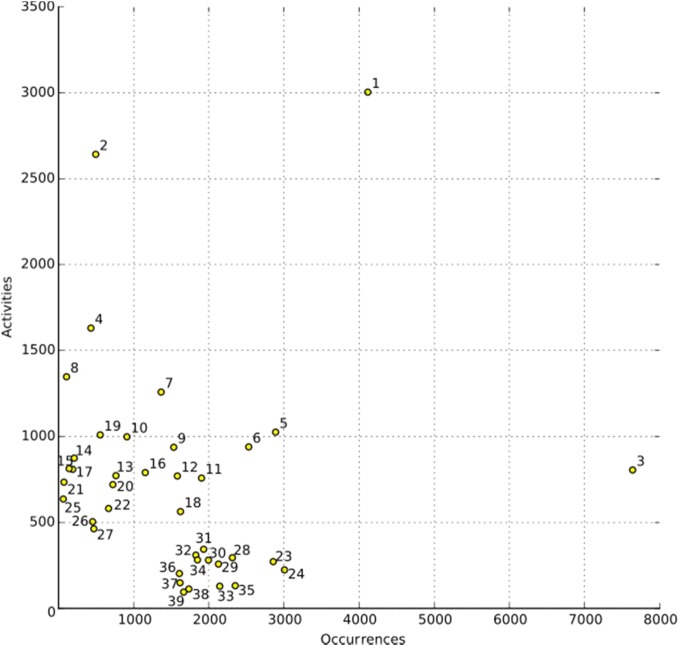
Figure 3.
Scatter plot for the merged sets of the 39 metabolites that are highly occurring (O) and have the most reported activities (A) of all nearly 200,000 NPs included in NAPRALERT with annotations matching the metabolite numbers in Table 2. While not showing a clear correlation between these two sets (A vs O), some metabolites are clearly outliers (1, 2, 3), and two major groups emerge for metabolites highly occurring and metabolites with a high number of activities reported.
Upon examination of the cumulative sum plots of each individual distribution (O vs A vs D), shown in Figure 4, two striking conclusions are apparent. First, a major portion of the compounds is present only once, or a handful of times, in each data set. Second, only a very limited number of metabolites represents a large number of citations. These features imply that, when all three sets are considered concurrently, they are likely to be long-tail-distributed. These types of distributions are characterized by having a non-negligible part of their populations outside of the range that would otherwise be expected to fall within a Gaussian-type distribution. The A/O/D distributions have apparent long-tail characteristics. This has two major consequences: first, data sets of this nature make it nearly impossible to predict the behavior or the importance of new or known elements. This results from the fact that an unexpected single element can have an influence that overwhelms all of the already known elements. Second, most of the classical statistical tools cannot be applied to these types of distributions.74−76This also means that the statistical models most widely used in pharmaceutical research, and engrained into the general modes of scientific questioning, do not apply for long-tail distributions, including those of bioactive NPs.
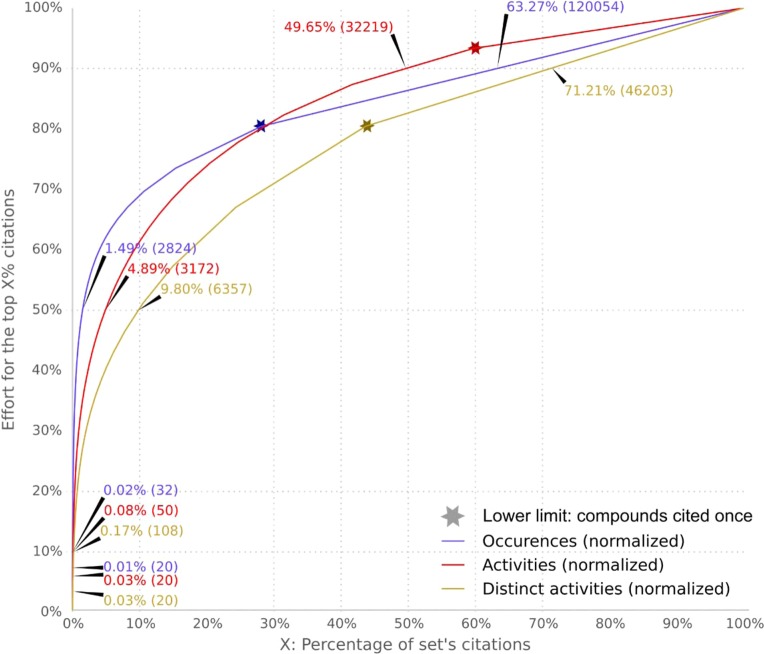
Figure 4.
Cumulative sums of the distributions of the three top-20 sets of NPs: occurrences (O) in blue, activities (A) in red, and distinct activities (D) in brown. The arrows show the percentage of citations at each given point, as well as the number of citations (in parentheses) that represent the top-20 NPs (bottom left), as well as the top 10% (lower left), top 50% (middle), and top 90% (upper right) of all NPs. The stars indicated the beginning of the single citation per compound zone, which continues on the right, ending at 100%.
Natural Product Research Follows Power-Laws
Looking more closely at the distributions of the number of citations in each of the three sets (O/A/D), it became evident that the distributions follow a power-law rule (Table 4 and Figure 5). Comparing the different distributions proposed by the power-law package (log-normal, power-law, truncated power-law, exponential, stretched_exponential), the truncated power-law was always the one with the best fit. Such a distribution is characterized by truncated power-law equation (eq 1)
| 1 |
where xmin is the truncation value, Λ is the scaling factor, and α is the exponent.
Table 4. Parameters of the Power-Law Distributions Function in Equation 1 of the Three Sets of NPs.
| α | Λ | xmin | |
|---|---|---|---|
| occurrences (O) | 1.96 | 2.58 × 10–4 | 6 |
| activities (A) | 2.28 | 2.14 × 10–4 | 11 |
| distinct activities (D) | 2.10 | 2.87 × 10–3 | 10 |
Figure 5.
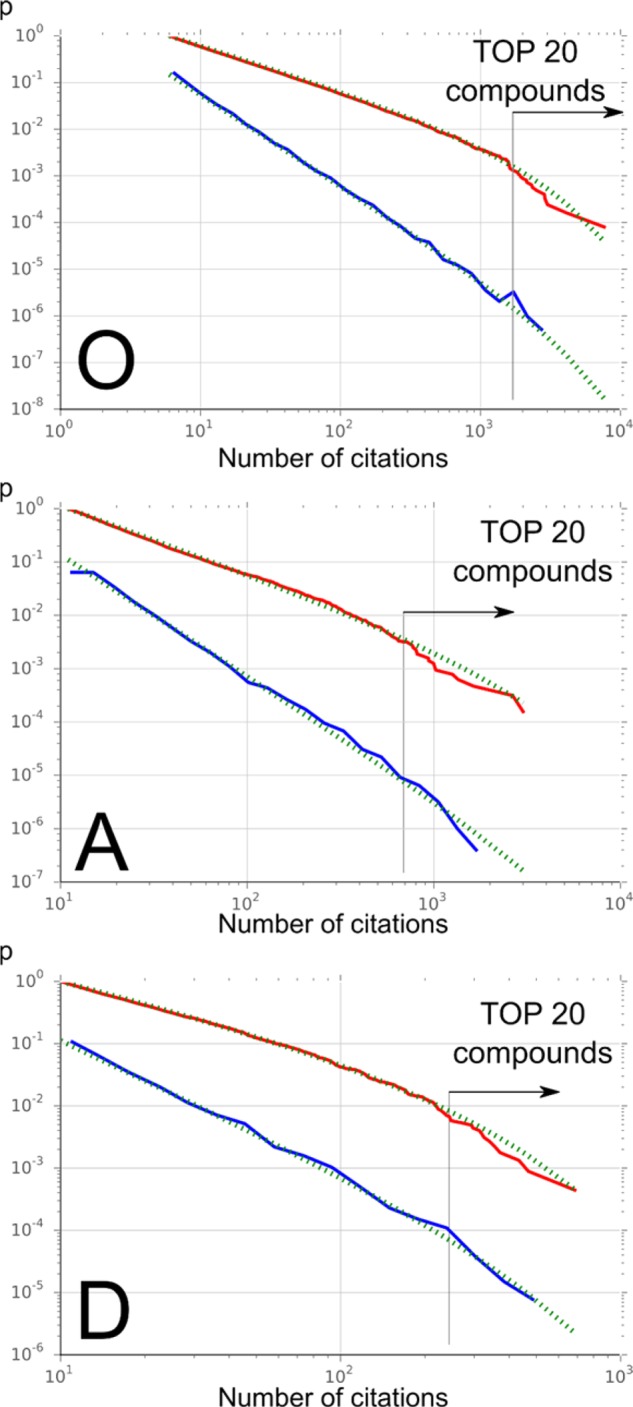
Truncated power-law fitting of the distributions (blue) and cumulative sums (red) of the three sets: occurrences (O), activities (A), and distinct activities (D). These graphics represent the cumulative complementary density functions, representing the probabilities (y-axis) of obtaining a given value (x-axis). They clearly show that low-citation compounds (left) are more likely to happen than high-citations ones (right). Dotted green lines are the truncated power-law fitting according to eq 1.
Figure 5 displays the fit of a truncated power-law distribution for the three sets considered (Figure 4). While the fit is good for most of the distribution, the high-citation part of the distribution is noisier. The same applies to the low end, which is hidden due to truncation, which is a consequence of the greater sampling errors of the number of citations for low-ranking compounds vs those of the rest of the distribution. The different fitting parameters for eq 1 are described in Table 4. Many power-law-type distributions could fit this data, as they all share similar characteristics. However, the fact that compounds with a small citation number could be hampered by data entry errors, unresolved synonyms, or wrong structural elucidation makes this part of the distribution more prone to error. On the other hand, the high-citation side of the distribution could also hide undescribed metabolites because the criteria used for identification and the completeness of the identification processes may not be sufficient to discern them with sufficient certainty from analogues.77
Power-law distributions, truncated or not, are found in many natural or human behavioral phenomena including linguistics, astronomy, demography, and, remarkably, citation analysis.75 These kinds of distributions are usually seen in resource-limited events, as exemplified by the finite number of words in the vocabulary of all languages. In the case of NP-based research, financial resources, human effort, popularity, comfort factor, and sampling of biological sources are likely the main finite factors contributing to the power-law nature of the discovery process. This popularity factor was hypothesized as being responsible for the low level of new kinase targets and poor selectivity of assayed drugs by Fedorov et al.78 Zipf came up with a similar power-law regarding the distribution of words in written language. In his 1949 book, Human Behavior and the Principle of Least Effort,79 he hypothesized that the tendency of choosing the path of least resistance may be one of the main causes for such a distribution. Compared with other distributions and intrinsic characteristics of statistical correlations, the mathematics behind the accurate fitting of power-law distributions is still highly debated.72,75,76 This includes the question of whether accurate fitting is possible at all in these cases. From a statistical perspective, some of the mathematical properties normally applied to the commonly used distributions are considered to be difficult to describe for power-law functions. For example, for α < 3, their variance is not finite, nor is their mean for α < 2.
The importance of power-law distribution is also discussed in domains for which prediction tools based on standard statistical distributions fail to be resilient to extreme events. This resilience explains why power-law distributions are often perceived as causing frustration: their ability to cope with extreme values or events is paired with their characteristic to be of minimal use as predictive tools, which is the actual intent of most fitting applications. In other words, and from a global perspective, this exemplifies how generalization and prediction often show counterintuitive and/or counterproductive behavior.
The Most-Studied Natural Products Are a Subset of All Metabolites
When NAPRALERT was initially compiled, compounds widely designated as primary metabolites were (intentionally) excluded. This reflects the general notion that these housekeeping metabolites are neither reported nor studied by most NP chemists. This gap between biochemistry and NP research has been recognized and discussed in detail by Firn and Jones.80−82 These authors collected evidence for an array of hypotheses including how the primary vs secondary metabolites dichotomy makes little sense, and how metabolic pathways, and not just metabolites or enzymes, could have been chosen by means of natural selection. While experimental evidence in support of their plausible hypotheses may be difficult to obtain, Firn and Jones’ evolutionary rationales, the outcomes of the present study, and the general experience of NP discovery research point in the same direction. The borderline between those NPs that can be considered to be potential or verified leads vs those NPs that should be ignored for this purpose is likely an infinitely thin membrane or simply nonexistent.
This generates the thought-provoking question: What is the best way to qualify ubiquitous NPs such as two of the top-four IMPs (Table 2), β-sitosterol and quercetin? As Firn and Jones deduced, quercetin-type flavonoids or carotenoids are often put into the secondary metabolite category, but when their biological pathways are knocked-out, the producing organisms are no longer viable. This happens either because these compounds were involved in direct support of the organism or the impacted pathways were a crucial step leading to a vital metabolite.81,82 Evidently, from the perspective of a target system or organism, the ignored primary or household metabolites may well play a role beyond their basic integration into metabolism.82 One such link could relate to the solubility of the metabolite(s) considered to be the active principle, as proposed by Choi et al. with the natural deep-eutectic solvents (NADES) concept,83 in which some naturally occurring compound mixtures may help to solubilize other constituents of the organism.
A relatively unmapped territory in the field of HTS is data that compares hits from synthetic libraries with those from libraries containing (only) NPs or NP-like compounds. As the theoretical chemical space of 30 or fewer heavy atoms is more than 50 orders of magnitude bigger than the number of actually reported compounds, NP or not, Hert et al. have questioned why hits are seen at all.84 The authors suggest that libraries fit for screening should contain many more biological-like scaffolds than is usually the case. Their suggestion followed the rationale that the biogenic bias of using molecules already known to play a biological role, i.e., being bioactives, is more likely to lead to success, even if the actual bioactivity is unknown. This explanation seems even more plausible when considering that biologically evolved small molecules, which are mostly made by proteins, have already successfully passed the same set of evolutionary filters that affect living organisms.
Considering the problematic nature of certain metabolites in HTS campaigns, the flipside of the coin is the final soul-searching question of the present study: How do organisms cope with promiscuous and over-represented molecules such as the IMPs? This question extends beyond the producing organisms, considering that several of the IMPs identified from the three merged sets of most reported NPs (Figure 2 and Table 2) are likely consumed by living organisms on a daily basis.
Why Are the IMPS so Prevalent?
Addressing this question reverts back to the Introduction and the discussion of aggregation as a recently recognized phenomenon with huge potential impact on bioassays. Currently published data on the aggregation properties of the 39 metabolites identified via merging of the three top-20 sets (Table 2 and Figure 2) reveal two things: first, many of these metabolites are, in fact, problematic as potential aggregators; second, another important group has little to no reported activity. Of the top-39 metabolites, 10 are aggregators, 11 are PAINS, and four are both. Of the top-20 compounds of just the activity (A) set, eight (including all of the top-4 that show aggregating behavior) plus two more might be potential aggregators according to Aggregator Advisor. Moreover, nine of the top-20 most active NPs exhibit PAINS substructures, of which three are also aggregators. This means that 14 out of 20 (70%) may be problematic metabolites and true IMPs that require more scrutiny in any program involving biological assessment. This figure could be even higher as, to our best knowledge, some of these metabolites have not been investigated for aggregation. Of the four all-intersecting metabolites (Figure 2) and, thus, most prominent IMPs, rutin (5) is the only compound that has not been reported as an aggregator.
For metabolites with distinct activities (set D), the situation is similar, also producing a striking fit for what could be expected of promiscuous compounds. Of these 20 metabolites, nine show aggregating behavior, two more may well be aggregators due to similarities with known aggregators, and nine are PAINS (of which four are also aggregators). This means that, again, 14 of 20 compounds (70%) in set D are identified as problematic when applying only these two criteria.
Bioassay Interference of Prominent IMPs
A compilation of PubChem confirmatory assay data85−92 on the four most prominent IMPs is presented in Table 5. It shows that they were all tested as being active in a series of experiments that can be classified as highly specific.
Table 5. PubChem-Based Review of Eight High-Specificity Assays in Which the Four Most Prominent IMPs Identified in This Study Showed Activity.
| PubChem ID | assay | active (out of 4) |
|---|---|---|
| AID_399341 | Antioxidant activity assessed as superoxide-scavenging activity by the nitrite method | 4 |
| AID_455702 | Inhibition of Clostridium perfringens neuraminidase | 4 |
| AID_455703 | Noncompetitive inhibition of recombinant influenza A virus rvH1N1 A/Bervig_Mission/1/18 neuraminidase | 4 |
| AID_399340 | Inhibition of xanthine oxidase assessed as decrease in uric acid production by spectrophotometry | 3 |
| AID_293298 | Antioxidant activity assessed as inhibition of superoxide production by xanthine/xanthine oxidase method | 3 |
| AID_366284 | Inhibition of Influenza A Jinan/15/90 H3N2 virus neuraminidase activity by MUN-ANA substrate based fluorometric assay | 2 |
| AID_366285 | Inhibition of Influenza A PR/8/34 H1N1 virus neuraminidase activity by MUN-ANA substrate based fluorometric assay | 1 |
| AID_366286 | Inhibition of Influenza A Jiangsu/10/2003 virus neuraminidase activity by MUN-ANA substrate based fluorometric assay | 1 |
Whereas the two antioxidant activities are not surprising, the high hit rate for the neuraminidase activities can be perceived as being suspicious. Upon close inspection, quercetin (1) is known to interfere with the 2′-O-(4-methylumbelliferyl)-N-acetylneuraminic acid (MUN-ANA) used in these assays.58 This established interference and the close structural similarity of the four top IMPs raise concerns about the validity of the lead character of these NPs for the reported targets.
The broader relevance of this interference mechanism is documented in several publications on fluorescence-based assays. One study showed quenching of BSA auto fluorescence as being an underlying mechanism.93 Another showed that 1 exhibits fluorescence when internalized into cells,94 and the authors hypothesized that noncovalent binding to some proteins was involved. A study of two very commonly investigated NPs, curcumin (8) and 1, demonstrated that these IMPs were capable of quenching the thioflavine T reported in β-amyloid aggregation inhibition assays.95 Finally, small-molecule aggregates were shown to inhibit amyloid aggregation in vitro, thus probably impacting the validity of conclusions drawn from these assays.47 These documented interferences exemplify the highly counterproductive twists that can occur in the logical chain between the agent and the reporter component(s) of the bioassay (Figure 1, middle).
Regarding NPs that could impede the reporter of the assay, probably the most striking example relates to luciferase activators/inhibitors. While none of the metabolites of the three sets have been tested on this target yet, common IMPs are showing activities in three different counterscreens (Table 6).96−98
Table 6. NPs Active on the Luciferin/Luciferase Counterscreening Assays in PubChem.
| compound | luciferase perturbing assay |
|---|---|
| resveratrol (14) | AID_411 |
| genistein (4) | AID_624030 |
| genistein (4), luteolin (11), resveratrol (14) | AID_588342 |
Another broader conclusion from the recognition of many flavonoids as IMPs is that, in general, this class of NPs should be studied carefully because they tend to form aggregates and/or disrupt assays.49 Their observed activities on several nuclear receptors99 may alternatively be viewed as a sign of suspicion. Moreover, quercetin (1), genistein (4), rutin (5), kaempferol (6), apigenin (9), luteolin (11), and myricetin (22) are also recognized as membrane disruptors, being able to increase or decrease membrane fluidity depending on the individual structure and compound concentration.100,101 Some of these NPs are known for effects on MDR mechanisms, lipid membrane permeability and structure, as well as fluorophore distribution in certain assays.100 Moreover, it has been shown that some of these NPs have the ability to produce false positives in MTT-based cell-viability assays and that adequate washing may reduce the interferences.102,103 Additional details and references about issues related to MTT-based assays can be found in the comprehensive review by Fallarero et al.104 The need to overcome some of the issues associated with tetrazolium salt-based assays is reflected by the NCIs’ efforts to develop alternative cell-viability screening assays.105
Finally, fatty acids such as linoleic (36) and oleic acid (37) are another group of prominent IMPs, known for their noncompetitive inhibitor characteristics on three of the receptors, as evaluated by Ingkaninan et al.67 A typical warning flag alerting to a more in-depth (literature) analysis is the effect of unsaturated fatty acids on cellular assays vs noncellular assays.64 Palmitic acid (33) did not influence the tested targets, but still exhibited a high rate of activity in PubChem confirmatory assays.
As exemplified for both the flavonoid and fatty acid portion of the IMPs, it is important that all known interference factors are taken into account prior to making conclusions about the validity of a hit and/or claims about their activity. It should also be kept in mind that, even in the absence of interferences, positive in vitro bioassay outcomes of the IMPs identified here may or may not be predictive of an in vivo effect. This is supported by NAPRALERT. One means of avoiding such pitfalls in the long run is thorough literature searches and the use of publicly accessible databases. This allows for activities related to potential interaction with one of the assay’s ingredients to be examined, or for other physicochemical properties that may interfere in an assay.
Navigating the Black Hole and Recognizing Traps
Several strategies have been proposed to address the key challenges of NP drug discovery presented in the Introduction that lead to invalid hits. One takes into account the responsibility of a small number of (sub)structures for a disproportionately large fraction of the hits.25 Another considers the disruptive properties of promiscuous compounds that are aggregators and/or PAINS, as recognized by the groups of Shoichet23 and Baell,2 respectively.
The disruptive factors that characterize IMPs include both of these concepts, as well as two additional phenomena described in the present study. The first phenomenon is the power-law behavior of the cumulative distribution of bioactive NPs. The second is the hyperbolic shape that results from mapping these cumulative distributions in 3D occurrence–bioactivity–effort space, resembling the black hole of NPs (see The Big Picture: The Holistic Distribution of Natural Products).
Now that the traps and the topology of the terrain have been defined, the following discussion seeks to outline potential strategies for enhancing navigation in and around the black hole.
Searching for PAINS
One important tactic for addressing the challenges posed by compound promiscuity and PAINS is the use of orthogonal assays,25 which are based on different reporters and/or different detection mechanisms.
Detecting and Avoiding Aggregation
Assays capable of detecting aggregating behavior have been developed through the use of NMR,106 dynamic light scattering, transmission electronic microscopy, or detergents.22,107 The addition of small amounts of certain detergents in assays has been shown to reduce effectively or even eliminate aggregation in most cases.108 For detergent-intolerant assays, centrifugation or addition of serum proteins may help to reduce protein–aggregate interactions,108,109 with the caveat that the final concentration of the assayed compound may be more difficult to determine precisely. An online tool compiled by the Shoichet group, Aggregator Advisor (http://advisor.bkslab.org/search), is capable of predicting the likelihood of a given structure belonging to this class of nuisance compounds. Shoichet’s group disseminates their considerable experience in this area on their Web site at http://www.bkslab.org/take-away.php. This resource was built from testing >70,000 compounds for detergent-mediated activity in an AmpC β-lactamase assay.
Fluorescence Issues
While quenching issues can usually be solved only by changing the detection method, a compound’s fluorescence impact can be lowered by the use of red-shifted fluorophores,110 which are rare among NPs, or by using ratiometric or time-resolved fluorescence approaches.111 Further references regarding these effects can be found in the review by Thorne et al.25
Redox Issues
The redox related issues are diverse, and several assays have been developed to help with their identification. One must keep in mind that these activities may be wanted in some assays. Electrochemical methods can be applied as described by Liu et al.112 While more classical colorimetric assays detecting formation of hydrogen peroxide have been developed,61,62 special care must be taken, as some compounds may interfere either through their color or through unexpected reactions with the intermediates.
Reactivity Issues
A knowledge base for compounds reacting with thiols was assembled by Dahlin et al., who recognized the critical impact of these reactive agents during their quest for inhibitors of histone acetyltransferase Rtt109: only three out of 1500 active hits could be confirmed as actual leads.113 Extrapolating from this experience, it is likely that other target- or assay-specific HTS studies might also pinpoint unexpected promiscuous compounds that bear a high risk of being pursued as leads. Involving the meta-analysis of published data, another predictive strategy to address promiscuity by reactivity has been developed recently by Hu et al. using PubChem confirmatory bioassay data.114 Interestingly, these authors have shown distribution curves that closely resemble the power-law characteristics uncovered in the present study. In a similar meta-approach, Nissink et al. chose data mining and binomial experiments as tools to map frequent-hitter behavior in published data sets.115
Bioassay Issues
Using another strategy geared toward identifying components of bioassays as the root of erroneous bioactivity recognition, the groups of Fallarero and Agarwal have compiled advisory evidence and references that provide an invaluable resource for the development of NP HTS campaigns.104,116 The former group also advocated the routine assessment of all test compounds by fluorescence and UV/vis spectroscopy, dynamic light scattering (DLS), and label-free detection methods such as electrochemical approaches in case they exhibit UV/vis absorbing, diffusing, or fluorescent properties.
Prevention by Prediction
Another line of defense against being waylaid by promiscuity, PAINS, and IMPs is the use of databases and molecular substructure filters capable of putting warnings on assayed compounds. However, this approach requires structural information, which is typically unavailable during bioguided fractionation procedures in NP programs. This once more emphasizes the value of rapid dereplication,12 especially when it is performed as early as possible in the fractionation process and with a focus on known problematic compounds. At the same time, it should not be overlooked that available dereplication schemes are less rigorous than full structure elucidation protocols. It is known that elucidation procedures fail more than occasionally, mostly due to the insufficient use of analytical orthogonality (see ref (77) for a comprehensive overview of failed NP leads) and/or as a result of inadequate reporting of 1H NMR spectroscopic data (see ref (117) and references therein).
As far as predictive tools are concerned, the free FAF-Drugs3 (http://fafdrugs3.mti.univ-paris-diderot.fr) is a very useful tool that can calculate important molecular characteristics and immediately includes useful filters such as for PAINS, aggregation data (the Shoichet laboratory Web site provides very comprehensive coverage), and the Eli Lilly MedChem rules.56 Mixtures remain difficult to resolve, as they can show enhanced, reduced, or nulled effects compared to those of the individual components. These effects can be due to synergistic or antagonistic action on the target,118−120 solubility effects,83 or impact on aggregation.46 Evidently, NP-driven programs are notoriously plagued with the mixture problem, as discussed above with regard to RC.
Whether in the form of filters, rules, or predictions, the NPs chemist should always be aware of how and for what specific purpose these controls have been defined. This awareness is critical, as it enables the researcher to recognize when control(s) mask positive events in the data (i.e., the one-off, true hits) and/or obscure their negative counterparts (a potential role for true IMPs, PAINS, and other promiscuous compounds). The ability to avoid wasting a positive event, e.g., by letting it be collected into the solvent waste during final LC purification, while spending endless efforts on chasing a negative event with its spurious activities, requires awareness of this unresolved dichotomy.
By lack of devoted efforts, the majority of compounds are not studied much more beyond their initial discovery. Bringing friends to help search for the keys under the street light is unlikely to increase the discovery rate, and we are convinced that this analogy applies to NPs as well. However, in order for serendipitous discoveries to occur, one must be receptive, prepared, and accept the occurrence of unexpected events.
Conclusions and Outlook
At the risk of oversimplifying a complex matter, “yes” is still a reasonable simplistic answer to the title question. The present study provided clear evidence for the existence of IMPs and for their ability to interfere with the NP-based drug discovery process, using various meanings of the term interference when it is applied to bioassay-driven approaches. Located in the same region of the black hole of NPs, where the density of effort is very high, IMPs are direct neighbors of true leads. Taleb described these sought-after marvels of drug discovery as positive black swans.74 By following a power-law distribution, true leads are like black swans: they are neither predictable nor readily distinguished from IMPs at the early discovery stage. The recognition of IMPs presented in this work builds on the large body of NP literature encoded into NAPRALERT. From a holistic perspective, this also leads to the conclusion that orthogonality applied to both biology and chemistry is essential to both IMP recognition and avoidance.
Reflections on IMPs and the Black Hole of Natural Products
While the evidence collected so far is insufficient to assign IMPs the role of consistently negative black swans, the analogy is at least thought provoking. The special role of the top-39 compounds identified in this study and their potential IMP status beg two immediate questions: Shall the compounds be completely eliminated from the list of potential lead compounds, or (in the Boolean sense) is the IMPs character of any given compound actually a signature of its unique, yet unrecognized, role in nature?
Figure 6 summarizes the key findings of the present study and provides a visual impression about the ambivalence of the compound–abundance–bioactivity space of NPs. One key conclusion is that a relatively small group of molecules can indeed be defined that are invalid metabolic panaceas, IMPs. Located at the bottom of the 3D hyperbolic space, i.e., the black hole of NPs (Figure 6C), the IMPs are neighbors but antonyms of true lead compounds. The shape of the black hole is essentially identical for all three investigated parameters (O/A/D; see Defining the Bottom of the Black Hole of Natural Products). Moreover, the black hole provides the sense of the extremely high effort (density) expended on relatively few NPs, whereas the majority of NPs remain vastly underexplored, both chemically and biologically.
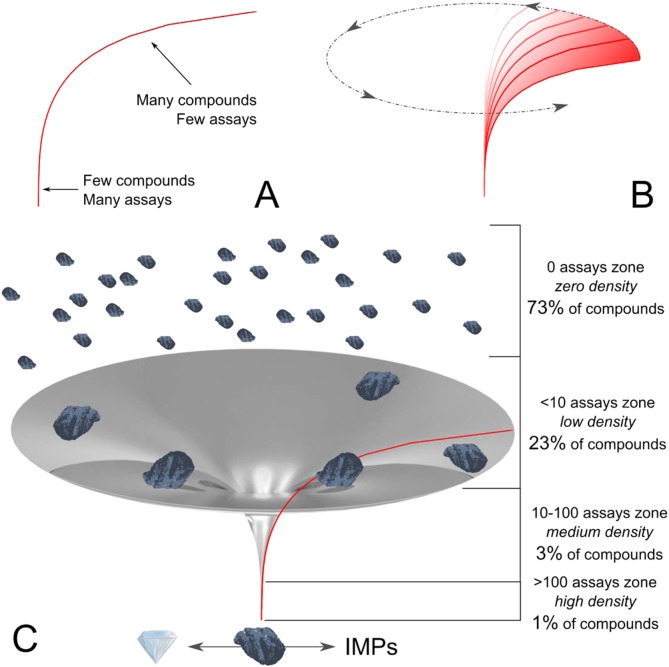
Figure 6.
Cumulative abundances of the reporting of the occurrences, activities, and distinct activities all follow the same principal power-law distribution. A typical curve is shown in A, indicating the two major regions of overattention to a few and lack of effort on many NPs. Distributing this NP–abundance–bioactivity space, which was built on the base of NAPRALERT’s nearly 200,000 compounds, into the third dimension (B) generates a hyperbolic structure that resembles a well-known corpus in astrophysics and is, therefore, termed the black hole of NPs. Panel C shows its various zones that categorize all NPs by their attached biological knowledge and abundance of the test parameter (O/A/D; see main text). Similar to a stellar black hole, density (representing research effort) increases dramatically toward the bottom (with infinite effort not being a scientific option). In distinction to its true counterpart, the black hole of NPs has a (virtual) outlet toward the bottom (C), which release either precious hits or IMPs.
As detailed in the above section, The Big Picture: The Holistic Distribution of Natural Products, the hyperbolic shape follows power-law functions, which are principles found to govern a breadth of natural and social phenomena. The authors interpret this analogy as a hint by nature that a common and possibly invariable law drives human ability, curiosity, and discovery equally.
Considering the undeniable abundance of success stories of NP-based drug discovery,33 there clearly are diamonds (true hits) to be discovered. One interesting instance is that of taxol (19), which is contained in the top-39 compounds in Table 2. This is mainly the result of 19 generating massive and broad interest in the research community, which led to a large number of reports (high count in the A category; see Table 2) within a rather focused window of biological activity. While clearly representing a valid drug (lead), the placement of 19 on the list of potential IMPs may furthermore imply that the compound also has interference qualities, which were uncovered while performing random searches for alternative uses of the compound. Continuing this interpretation would even generate the scenario that a valid hit receives a false-positive promiscuity label if it is tested only in a sufficient number of invalid assays before being assayed in the (otherwise decisive) test. Conversely, the designation of a compound as an IMP has a dynamic component that results from the potentially volatile, power-law driven scientific interest in it, which, in turn, reflects the behavior of IMPs as proverbial imps. Again, it remains to be shown whether true hits can emerge from unpredictable events or as the direct result of a systematic and truly targeted approach.
Nevertheless, caution even applies when using such compounds as positive controls in bioassays. Their ability to interfere with many reporters (e.g., fluorophores, oxidation dependent chromophores) as well as with the targets (aggregation, nonspecific binding) increases the likelihood of their activity scores not being comparable to those of their real targets. Moreover, assays that suffer from sensitivity to these interferences will likely only enrich compounds or fractions that bear the same issues.
The Bigger Data Approach
The recognition of IMPs, PAINS, and other promiscuous molecules requires bigger picture approaches, looking at relatively large amounts of data including experimental, HTS, and the broader literature. Relational databases, in particular those collecting and sometimes editing (published) meta-information, are the prime tools for the meta-analysis part of such undertakings. NAPRALERT was uniquely positioned to serve the present study due to its comprehensiveness and design. This was evident from the long-term involvement of one of the authors (J.G.) with NAPRALERT and copious personal communications of several of the authors with its founder, the late Dr. Norman Farnsworth. Collectively, there are clear indications that, from its early days, NAPRALERT was designed to encode published information about bioactive NPs in a unique comprehensive fashion: with very broad coverage (high journal diversity), using a multidisciplinary approach (aimed at collaborative pharmaceutical research), with linkage to the original primary articles (physical collection), and such that its ultimate utility increases over time beyond projected linear growth of information content/value. Hence, NAPRALERT inherently can address more general questions that otherwise would be beyond the capacity even of large academic research programs.
The linkage of NAPRALERT output with other databases, while representing an important tool during the present study, also illustrates the importance of the availability and accessibility (public licensing) of comprehensive software solutions for data analysis. Ideally, such tools are backed by dedicated, global user communities and documentation, as is the case for PubChem and ChEMBL used here. When coupled with public databases, which are becoming increasingly available, the mining of bigger data becomes feasible even for the less computer-initiated researcher and, thereby, can provide new means of answering important scientific questions related to drug discovery.
However, it is equally important to realize that the treatment of huge amounts of data always presents the risk of being subject to sourcing bias, noncurated data artifacts, or simple misunderstanding of parameters. One reason is that the manual curation of the breadth and depth of published results can quickly produce demands beyond human capacity. For both the producers and the consumers of such data, the importance of awareness for the inherent risks of invalid data or analyses cannot be overemphasized. The key role of data quality and compilation practices also explains why efforts for finding the most advanced forms of the description of metadata (e.g., biological profiles), chemical structures,121,122 spectra (e.g., access to raw data), and interpreted analytical data (e.g., NMR tables117) are more critical than ever.
More manageable, interface-ready, and reproducible forms of dissemination are essential for the ability of future researchers to cope with the tremendous amount of research data produced every day. A new generation of bioinformatic tools is required to enable recognition of global patterns behind research outcomes. Such tools will also be needed to confirm (or reject) the present outcomes of power-law principles that produce the black hole of NPs and/or the identification of IMPs as a new class of compounds that produce red flags in NP drug discovery programs.
Nature’s Dichotomy of Prioritization
The (drug) discovery process is filled with decision making. While it is frequently termed prioritization, emphasizing its analogue nature, decisions are inevitably binary, especially from the perspective of a single element (e.g., candidate molecule, NP extract, or fraction). Materials are studied because their bioactivity levels can achieve a certain threshold. Biological profiles receive favorable evaluation, or they do not; serendipity strikes, or it does not; etc. Despite wide awareness of these relationships, this reflection serves as a reminder that dichotomous paths bear the same risks as binary decisions: single-point errors can lead to total disorientation, like in a maze.
The same is true for virtual software filters or real filters used in the discovery process, such as bioassay-guided fractionation and HTS of pure compounds: the results are indicators rather than definitive answers. However, dichotomy trumps in another way: the complementary nature of two existing, but radically different, approaches to NP-based drug discovery. One follows the rational prioritization of crude NP extracts, e.g., via bioassay guidance or by other means. The contrasting approach involves the systematic mining of single chemical entities (pure NPs), which can result from various forms of purification campaigns and often are not (perceived as being) very targeted. Both approaches can actually benefit from each other: the former, for example, by allowing more efficient prioritization using information available from pure compounds, and the latter, by inspiring a search for chemical novelty in materials with interesting biological profiles. This provides rationale for consideration of another dimension of the meaning of “targeted” in (NP) drug discovery programs.
The findings of the present study indicate that harnessing the uninterrupted power and potential of NPs31,33,35 will not only benefit from both approaches, but also gain from, or even require, the advancement of experimental approaches. This includes the following three exemplary avenues: (i) the development of more integrated approaches to the prioritization of actives; (ii) a more in-depth biological assessment and interpretation of bioassay data; and/or (iii) an early stage validation of compounds designated as being bioactive, especially when involving broader terms such as antifungal, antimicrobial, or the like.123 Collectively, quality control of early hits and leads would enhance the validity and prioritization of true leads.
Finally, the identification of IMPs as a group of ubiquitous nuisance compounds that attract an overproportional amount of experimental attention has the potential to spark further thoughts that may eventually lead to a paradigmatic shift in NP-based drug discovery and related fields of research. While IMPs do not necessarily represent futile compounds, their true role in nature is likely understood poorly or not at all. The present study makes it a plausible hypothesis that IMPs are actually part of the power-law functionality that continues to be responsible for the generation of confusion across a breadth of sciences, including the pharmaceutical disciplines. This hypothesis is visualized by the NP black hole, a 3D space model of occurrence, bioactivity, and research effort, in which IMPs populate the high-density bottom. To this end, the recognition of the existence of both IMPs and the NP black hole, gleaned from 80+ years of reported NPs research, may inspire future progress in the field.
Acknowledgments
We are pleased to acknowledge the numerous helpful discussions of general drug discovery topics with our UIC colleagues, Drs. Scott G. Franzblau and David C. Lankin. Furthermore, we are grateful to Drs. David Newman and Gordon Cragg (NCI, Frederick, MD) for their valuable input and helpful information regarding the NCI NP drug discovery program. The authors gratefully acknowledge support of this work by grant U41 AT008706 from NCCIH and ODS, as well as support of their biomedical research on natural products and related training efforts through the following grants from NCCIH (formerly NCCAM), ODS, NIDCR, and NIAID, all of the NIH: P50 AT000155, RC2 AT005899, R44 AT004534, R43 AT001758, R01 DE021040, R21 AI093919, T32 AT007533, T32 DE018381, and R21 AI82847. We also appreciate the creative advice of Sheila Miguez, Aaron Lav, and Jenny Tong (Pumping Station: ONE, Chicago, IL).
Glossary
Abbreviations Used
- A
activities set
- Agg
aggregation
- D
distinct activities set
- HTS
high-throughput screening
- IMPs
invalid metabolic panaceas
- NADES
natural deep-eutectic solvents
- NCI
National Cancer Institute
- NP
natural product
- O
occurrences set
- PAINS
pan-assay interference compounds
- qPARs
quantitative purity–activity relationships
- RC
residual complexity
- anti-TB
anti-tuberculosis
- TCM
traditional Chinese medicine
Biographies
Jonathan Bisson obtained an M.S. degree in structural biochemistry and started his phytochemistry journey under the mentorship of Dr. Vincent Dumontet at the Institut de Chimie des Substances Naturelles, Gif-sur-Yvette, France. He then obtained a Ph.D. in Science, Technology and Health from the University of Bordeaux, France, under the mentorship of Dr. Pierre Waffo-Téguo, specializing in methodology at the chemistry–biology interface and practicing liquid–liquid chromatography, NMR spectroscopy, and hyphenated techniques. In 2013, he joined the University of Illinois at Chicago, where he is currently a Postdoctoral Research Associate, developing new methods and IT-based tools for natural products research in interdisciplinary programs, mainly through NMR and chromatographic approaches. Recently, he has been involved in the redesign of the NAPRALERT database.
James B. McAlpine obtained a Ph.D. from the University of New England, Armidale, New South Wales, Australia. Postdoctoral work at Northwestern University Medical School, studying the biosynthesis and mode of action of macrolide antibiotics, followed. In 1972, he joined Abbott Laboratories and worked on macrolides, aminoglycosides, and quinolones before heading up their natural product discovery project from 1981 to 1996; he discovered Tiacumicin B, the API of Difficid. He joined Phytera Inc. as VP Chemistry in 1996, discovering drugs from manipulated plant cell cultures, and in 2002, he joined Ecopia Bio Sciences as VP Chemistry and Discovery using genomics to discover novel secondary metabolites. He has authored or coauthored 120+ papers, is inventor on 50 U.S. patents, and joined UIC as Adjunct Research Professor in 2011.
J. Brent Friesen received his Ph.D. in Natural Products Chemistry from the University of Minnesota, focusing on the biosynthesis of pyridine alkaloids in tobacco. He has spent 10 years in Africa teaching organic chemistry in N’Djamena, Chad, and studying native plants used in Chadian traditional medicines. Currently a Professor of Chemistry at Dominican University, River Forest (IL), he holds an appointment as Adjunct Research Professor at University of Illinois at Chicago. His research encompasses the use of innovative NMR applications in undergraduate laboratories and research as well as the chromatography of bioactive natural products. He has participated in the international countercurrent separation community since 2003 and published articles both independently and in collaboration with the Pauli group at the University of Illinois at Chicago.
Shao-Nong Chen completed a B.S. degree in Organic Chemistry from Lanzhou University, China, as well as an assistantship in the Lanzhou Institute of Chemical Physics (CAS), and then pursued his interests in natural products chemistry, obtaining a Ph.D. under the joint mentorship of Drs. Yao-Zu Chen, Lanzhou University, and Han-Dong Sun, Kunming Institute of Botany (KIB, CAS). After 2 years of postdoctoral training with Dr. Guo-Wei Qin at Shanghai Institute of Materia Medica (SIMM), he joined Dr. Sydney Hecht’s group at the University of Virginia. He moved to UIC in 2000, where he currently is an Assistant Research Professor, working on botanical standardization and integrity in the UIC/NIH Botanical Center, as well as on method development for the analysis of bioactive natural products in interdisciplinary programs.
James Graham received his Ph.D. in Pharmacognosy in 2001, under the tutelage of Dr. Norman Farnsworth at the University of Illinois at Chicago (UIC), conducting ethnobotanical fieldwork in remote areas of Amazonian Peru. He received an NIH postdoctoral Fellowship at Florida International University in the area of Tropical Botanical Medicine. He was also a Technical Officer at the World Health Organization as part of the Traditional Medicine team. Currently, he is a Research Assistant Professor in the Department of Medicinal Chemistry and Pharmacognosy, College of Pharmacy, at UIC, and has served as editor of the NAPRALERT database since 2011. In addition, he is a Research Associate in Botany at the Field Museum in Chicago, and continues botanical exploration of remote rainforests of Peru.
Guido F. Pauli is a pharmacist by training and holds a doctoral degree in natural products chemistry and pharmacognosy. As Professor and University Scholar at UIC, Chicago (IL), he is principal investigator and collaborator in interdisciplinary natural product-centered research projects as well as director of the NAPRALERT database and the newly formed Center for Natural Products Technologies. His main interests are in the metabolome analysis of complex natural products, bioactive principles, herbal dietary supplements, anti-TB drug discovery, and dental applications of natural agents. Scholarly activities include international collaborations and guest professorships, the education of the next generation of pharmacognosists, and service on funding agency panels, pharmacopoeial expert committees, and in professional societies. His portfolio comprises 150+ peer-reviewed journal articles, international seminars and conference presentations, four book chapters, patents, as well as journal editorial and board functions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.5b01009.
Tables of criteria for occurring compounds, descriptive statistics of compounds by plant and distribution of occurrences of compounds, most studied plants in NAPRALERT, most studied families in NAPRALERT, descriptive statistics of the distribution of the three sets (A/D/O), and analysis of PubChem confirmatory assays data (PDF)
Compressed archive containing part of the Python code and compound lists as CSV files, with SMILES codes, PubChemID, and ChEMBLID data (ZIP)
The authors declare no competing financial interest.
Notes
This perspective represents part 28 of the series on Residual Complexity and Bioactivity (http://go.uic.edu/residualcomplexity).
Dedication
Dedicated to the late Dr. Norman R. Farnsworth, creator of the NAPRALERT database and spiritus rector, as well as his wife, Priscilla Farnsworth, a continuous supporter of pharmacognosy research, on the occasion of what would have been Norman’s 85th birthday.
Supplementary Material
References
- Jadhav A.; Ferreira R. S.; Klumpp C.; Mott B. T.; Austin C. P.; Inglese J.; Thomas C. J.; Maloney D. J.; Shoichet B. K.; Simeonov A. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J. Med. Chem. 2010, 53 (1), 37–51. 10.1021/jm901070c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J. B.; Holloway G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53 (7), 2719–2740. 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- McGovern S. L.; Caselli E.; Grigorieff N.; Shoichet B. K. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002, 45 (8), 1712–1722. 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- Pauli G. F.; Case R.; Inui T.; Wang Y.; Cho S.; Fischer N. H.; Franzblau S. New perspectives on natural products in TB drug research. Life Sci. 2005, 78 (5), 485–494. 10.1016/j.lfs.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Qiu F.; McAlpine J. B.; Krause E. C.; Chen S.-N.; Pauli G. F.. Pharmacognosy of black cohosh: the phytochemical and biological profile of a major botanical dietary supplement. In Progress in the Chemistry of Organic Natural Products 99; Kinghorn A. D., Falk H., Kobayashi J., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp 1–68. [DOI] [PubMed] [Google Scholar]
- Wagner H. Synergy research: approaching a new generation of phytopharmaceuticals. Fitoterapia 2011, 82 (1), 34–37. 10.1016/j.fitote.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Cordell G. A. Ecopharmacognosy and the responsibilities of natural product research to sustainability. Phytochem. Lett. 2015, 11, 332–346. 10.1016/j.phytol.2014.09.003. [DOI] [Google Scholar]
- Gertsch J. Botanical drugs, synergy, and network pharmacology: forth and back to intelligent mixtures. Planta Med. 2011, 77 (11), 1086–1098. 10.1055/s-0030-1270904. [DOI] [PubMed] [Google Scholar]
- Inui T.; Wang Y.; Pro S. M.; Franzblau S. G.; Pauli G. F. Unbiased evaluation of bioactive secondary metabolites in complex matrices. Fitoterapia 2012, 83, 1218–1225. 10.1016/j.fitote.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouslimani A.; Sanchez L. M.; Garg N.; Dorrestein P. C. Mass spectrometry of natural products: current, emerging and future technologies. Nat. Prod. Rep. 2014, 31 (6), 718–729. 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagas-Paula D.; Oliveira T.; Zhang T.; Edrada-Ebel R.; Da Costa F. Prediction of anti-inflammatory plants and discovery of their biomarkers by machine learning algorithms and metabolomic studies. Planta Med. 2015, 81 (06), 450–458. 10.1055/s-0034-1396206. [DOI] [PubMed] [Google Scholar]
- Wolfender J.-L.; Marti G.; Thomas A.; Bertrand S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2014, 1382, 136–164. 10.1016/j.chroma.2014.10.091. [DOI] [PubMed] [Google Scholar]
- Jaki B. U.; Franzblau S. G.; Chadwick L. R.; Lankin D. C.; Zhang F.; Wang Y.; Pauli G. F. Purity–activity relationships of natural products: the case of anti-TB active ursolic acid. J. Nat. Prod. 2008, 71 (10), 1742–1748. 10.1021/np800329j. [DOI] [PubMed] [Google Scholar]
- Qiu F.; Cai G.; Jaki B. U.; Lankin D. C.; Franzblau S. G.; Pauli G. F. Quantitative purity–activity relationships of natural products: the case of anti-tuberculosis active triterpenes from Oplopanax horridus. J. Nat. Prod. 2013, 76 (3), 413–419. 10.1021/np3007809. [DOI] [PubMed] [Google Scholar]
- Pauli G. F.; Chen S.-N.; Friesen J. B.; McAlpine J. B.; Jaki B. U. Analysis and purification of bioactive natural products: the AnaPurNa study. J. Nat. Prod. 2012, 75 (6), 1243–1255. 10.1021/np300066q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli G. F. Residual Complexity of Bioactive Natural Products. http://go.uic.edu/residualcomplexity.
- Bento A. P.; Gaulton A.; Hersey A.; Bellis L. J.; Chambers J.; Davies M.; Krüger F. A.; Light Y.; Mak L.; McGlinchey S.; Nowotka M.; Papadatos G.; Santos R.; Overington J. P. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014, 42 (D1), D1083–D1090. 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton E. E.; Wang Y.; Thiessen P. A.; Bryant S. H. PubChem: integrated platform of small molecules and biological activities. Annu. Rep. Comput. Chem. 2008, 4, 217–241. 10.1016/S1574-1400(08)00012-1. [DOI] [Google Scholar]
- Wang Y.; Xiao J.; Suzek T. O.; Zhang J.; Wang J.; Zhou Z.; Han L.; Karapetyan K.; Dracheva S.; Shoemaker B. A.; Bolton E.; Gindulyte A.; Bryant S. H. PubChem’s bioassay database. Nucleic Acids Res. 2012, 40, D400–D412. 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishton G. M. Reactive compounds and in vitro false positives in HTS. Drug Discovery Today 1997, 2 (9), 382–384. 10.1016/S1359-6446(97)01083-0. [DOI] [Google Scholar]
- Walters W. P.; Murcko A. A.; Murcko M. A. Recognizing molecules with drug-like properties. Curr. Opin. Chem. Biol. 1999, 3 (4), 384–387. 10.1016/S1367-5931(99)80058-1. [DOI] [PubMed] [Google Scholar]
- McGovern S. L.; Helfand B. T.; Feng B.; Shoichet B. K. A specific mechanism of nonspecific inhibition. J. Med. Chem. 2003, 46 (20), 4265–4272. 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- Sink R.; Gobec S.; Pecar S.; Zega A. False positives in the early stages of drug discovery. Curr. Med. Chem. 2010, 17 (34), 4231–4255. 10.2174/092986710793348545. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Ferrins L.; Falk H.; Nikolakopoulos G. PAINS: relevance to tool compound discovery and fragment-based screening. Aust. J. Chem. 2013, 66 (12), 1483–1494. 10.1071/CH13551. [DOI] [Google Scholar]
- Thorne N.; Auld D. S.; Inglese J. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol. 2010, 14 (3), 315–324. 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J.; Johnson R. L.; Simeonov A.; Xia M.; Zheng W.; Austin C. P.; Auld D. S. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007, 3 (8), 466–479. 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- Letot E.; Koch G.; Falchetto R.; Bovermann G.; Oberer L.; Roth H.-J. Quality control in combinatorial chemistry: determinations of amounts and comparison of the “purity” of LC–MS-purified samples by NMR, LC–UV and CLND. J. Comb. Chem. 2005, 7 (3), 364–371. 10.1021/cc049850r. [DOI] [PubMed] [Google Scholar]
- Rizzo V.; Pinciroli V. Quantitative NMR in synthetic and combinatorial chemistry. J. Pharm. Biomed. Anal. 2005, 38 (5), 851–857. 10.1016/j.jpba.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Graham J. G.; Farnsworth N. R.. The NAPRALERT database as an aid for discovery of novel bioactive compounds. In Comprehensive Natural Products II; Liu H.-W., Mander L., Eds.; Elsevier: Amsterdam, 2010; pp 81–94. [Google Scholar]
- Koehn F. E.; Carter G. T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discovery 2005, 4 (3), 206–220. 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- Butler M. S. The role of natural product chemistry in drug discovery. J. Nat. Prod. 2004, 67 (12), 2141–2153. 10.1021/np040106y. [DOI] [PubMed] [Google Scholar]
- Kinghorn A. D.; Pan L.; Fletcher J. N.; Chai H. The relevance of higher plants in lead compound discovery programs. J. Nat. Prod. 2011, 74 (6), 1539–1555. 10.1021/np200391c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J.; Cragg G. M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75 (3), 311–335. 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg G. M.; Grothaus P. G.; Newman D. J. Impact of natural products on developing new anti-cancer agents. Chem. Rev. 2009, 109 (7), 3012–3043. 10.1021/cr900019j. [DOI] [PubMed] [Google Scholar]
- Beutler J. A.Natural products as a foundation for drug discovery. In Current Protocols in Pharmacology; Enna S. J., Williams M., Barret J. F., Ferkany J. W., Kenakin T., Porsolt R. D., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, 2009. [Google Scholar]
- Sandor T.; Mehdi A. Z.. Hormones and Evolution; Academic Press: New York, 1979; Vol. 1, pp 1–72. [Google Scholar]
- Agarwal M. K. Receptors for mammalian steroid hormones in microbes and plants. FEBS Lett. 1993, 322 (3), 207–210. 10.1016/0014-5793(93)81570-P. [DOI] [PubMed] [Google Scholar]
- Pauli G. F.; Friesen J. B.; Gödecke T.; Farnsworth N. R.; Glodny B. Occurrence of progesterone and related animal steroids in two higher plants. J. Nat. Prod. 2010, 73 (3), 338–345. 10.1021/np9007415. [DOI] [PubMed] [Google Scholar]
- Lemoff A.; Yan B. Dual detection approach to a more accurate measure of relative purity in high-throughput characterization of compound collections. J. Comb. Chem. 2008, 10 (5), 746–751. 10.1021/cc800100g. [DOI] [PubMed] [Google Scholar]
- Popa-Burke I.; Novick S.; Lane C. A.; Hogan R.; Torres-Saavedra P.; Hardy B.; Ray B.; Lindsay M.; Paulus I.; Miller L. The effect of initial purity on the stability of solutions in storage. J. Biomol. Screening 2014, 19 (2), 308–316. 10.1177/1087057113492201. [DOI] [PubMed] [Google Scholar]
- Simmler C.; Hajirahimkhan A.; Lankin D. C.; Bolton J.; Jones T.; Soejarto D. D.; Chen S.-N.; Pauli G. F. Dynamic residual complexity of the isoliquiritigenin-liquiritigenin interconversion during bioassays. J. Agric. Food Chem. 2013, 61 (9), 2146–2157. 10.1021/jf304445p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli G. F.; Chen S.-N.; Simmler C.; Lankin D. C.; Gödecke T.; Jaki B. U.; Friesen J. B.; McAlpine J. B.; Napolitano J. G. Importance of purity evaluation and the potential of quantitative 1H NMR as a purity assay. J. Med. Chem. 2014, 57 (22), 9220–9231. 10.1021/jm500734a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler J.; McGovern S. L.; Doman T. N.; Shoichet B. K. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem. 2003, 46 (21), 4477–4486. 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- Feng B. Y.; Shelat A.; Doman T. N.; Guy R. K.; Shoichet B. K. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 2005, 1 (3), 146–148. 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- Shoichet B. K. Screening in a spirit haunted world. Drug Discovery Today 2006, 11 (13–14), 607–615. 10.1016/j.drudis.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. Y.; Shoichet B. K. Synergy and antagonism of promiscuous inhibition in multiple-compound mixtures. J. Med. Chem. 2006, 49 (7), 2151–2154. 10.1021/jm060029z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. Y.; Toyama B. H.; Wille H.; Colby D. W.; Collins S. R.; May B. C. H.; Prusiner S. B.; Weissman J.; Shoichet B. K. Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol. 2008, 4 (3), 197–199. 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen S. C.; Doak A. K.; Wassam P.; Shoichet M. S.; Shoichet B. K. Colloidal aggregation affects the efficacy of anticancer drugs in cell culture. ACS Chem. Biol. 2012, 7 (8), 1429–1435. 10.1021/cb300189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen S. C.; Doak A. K.; Ganesh A. N.; Nedyalkova L.; McLaughlin C. K.; Shoichet B. K.; Shoichet M. S. Colloidal drug formulations can explain “bell-shaped” concentration–response curves. ACS Chem. Biol. 2014, 9 (3), 777–784. 10.1021/cb4007584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D.; Doak A. K.; Nedyalkova L.; Shoichet B. K. Colloidal aggregation and the in vitro activity of traditional chinese medicines. ACS Chem. Biol. 2015, 10 (4), 978–988. 10.1021/cb5009487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjala L.; Tammela P. Aggregating behavior of phenolic compounds — a source of false bioassay results?. Molecules 2012, 17 (9), 10774–10790. 10.3390/molecules170910774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. Y.; Simeonov A.; Jadhav A.; Babaoglu K.; Inglese J.; Shoichet B. K.; Austin C. P. A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem. 2007, 50 (10), 2385–2390. 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- Walters W. P.; Murcko M. A. Prediction of “drug-likeness.. Adv. Drug Delivery Rev. 2002, 54 (3), 255–271. 10.1016/S0169-409X(02)00003-0. [DOI] [PubMed] [Google Scholar]
- Verheij H. J. Leadlikeness and structural diversity of synthetic screening libraries. Mol. Diversity 2006, 10 (3), 377–388. 10.1007/s11030-006-9040-6. [DOI] [PubMed] [Google Scholar]
- Lagorce D.; Sperandio O.; Galons H.; Miteva M. A.; Villoutreix B. O. FAF-Drugs2: free ADME/Tox filtering tool to assist drug discovery and chemical biology projects. BMC Bioinf. 2008, 9 (1), 396. 10.1186/1471-2105-9-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns R. F.; Watson I. A. Rules for identifying potentially reactive or promiscuous compounds. J. Med. Chem. 2012, 55 (22), 9763–9772. 10.1021/jm301008n. [DOI] [PubMed] [Google Scholar]
- Simeonov A.; Jadhav A.; Thomas C. J.; Wang Y.; Huang R.; Southall N. T.; Shinn P.; Smith J.; Austin C. P.; Auld D. S.; Inglese J. Fluorescence spectroscopic profiling of compound libraries. J. Med. Chem. 2008, 51 (8), 2363–2371. 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- Kongkamnerd J.; Milani A.; Cattoli G.; Terregino C.; Capua I.; Beneduce L.; Gallotta A.; Pengo P.; Fassina G.; Monthakantirat O.; Umehara K.; De-Eknamkul W.; Miertus S. The quenching effect of flavonoids on 4-methylumbelliferone, a potential pitfall in fluorimetric neuraminidase inhibition assays. J. Biomol. Screening 2011, 16 (7), 755–764. 10.1177/1087057111409221. [DOI] [PubMed] [Google Scholar]
- Schorpp K.; Rothenaigner I.; Salmina E.; Reinshagen J.; Low T.; Brenke J. K.; Gopalakrishnan J.; Tetko I. V.; Gul S.; Hadian K. Identification of small-molecule frequent hitters from AlphaScreen high-throughput screens. J. Biomol. Screening 2014, 19 (5), 715–726. 10.1177/1087057113516861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann J. C.; Chen Y.; Wartchow C.; Menke J.; Gao L.; Gleason S. K.; Haynes N.-E.; Scott N.; Petersen A.; Gabriel S.; Vu B.; George K. M.; Narayanan A.; Li S. H.; Qian H.; Beatini N.; Niu L.; Gan Q.-F. Metal impurities cause false positives in high-throughput screening campaigns. ACS Med. Chem. Lett. 2013, 4 (2), 197–200. 10.1021/ml3003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lor L. A.; Schneck J.; Mcnulty D. E.; Diaz E.; Brandt M.; Thrall S. H.; Schwartz B. A simple assay for detection of small-molecule redox activity. J. Biomol. Screening 2007, 12 (6), 881–890. 10.1177/1087057107304113. [DOI] [PubMed] [Google Scholar]
- Johnston P. A.; Soares K. M.; Shinde S. N.; Foster C. A.; Shun T. Y.; Takyi H. K.; Wipf P.; Lazo J. S. Development of a 384-well colorimetric assay to quantify hydrogen peroxide generated by the redox cycling of compounds in the presence of reducing agents. Assay Drug Dev. Technol. 2008, 6 (4), 505–518. 10.1089/adt.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.; Gordonov T.; Liu Y.; Bentley W. E.; Payne G. F. Reverse engineering to suggest biologically relevant redox activities of phenolic materials. ACS Chem. Biol. 2013, 8 (4), 716–724. 10.1021/cb300605s. [DOI] [PubMed] [Google Scholar]
- Balunas M. J.; Su B.; Landini S.; Brueggemeier R. W.; Kinghorn A. D. Interference by naturally occurring fatty acids in a noncellular enzyme-based aromatase bioassay. J. Nat. Prod. 2006, 69 (4), 700–703. 10.1021/np050513p. [DOI] [PubMed] [Google Scholar]
- Liu J.; Burdette J. E.; Sun Y.; Deng S.; Schlecht S. M.; Zheng W.; Nikolic D.; Mahady G.; van Breemen R. B.; Fong H. H. S.; Pezzuto J. M.; Bolton J. L.; Farnsworth N. R. Isolation of linoleic acid as an estrogenic compound from the fruits of Vitex agnus-castus l. (chaste-berry). Phytomedicine 2004, 11 (1), 18–23. 10.1078/0944-7113-00331. [DOI] [PubMed] [Google Scholar]
- Tarawneh A. H.; León F.; Radwan M. M.; Wang X.; Dale O. R.; Husni A. S.; Rosa L. H.; Cutler S. J. Fatty acids with in vitro binding affinity for human opioid receptors from the fungus Emericella nidulans. J. Agric. Food Chem. 2013, 61 (44), 10476–10480. 10.1021/jf4019849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingkaninan K.; von Frijtag Drabbe Künzel J. K.; Ijzerman A. P.; Verpoorte R. Interference of linoleic acid fraction in some receptor binding assays. J. Nat. Prod. 1999, 62 (6), 912–914. 10.1021/np9805490. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ferrer A.; Laveda F.; Garcia-Carmona F. Substrate-dependent activation of latent potato leaf polyphenol oxidase by anionic surfactants. J. Agric. Food Chem. 1993, 41 (10), 1583–1586. 10.1021/jf00034a010. [DOI] [Google Scholar]
- D’Auria S.; Cesare N. D.; Gryczynski I.; Rossi M.; Lakowicz J. R. On the effect of sodium dodecyl sulfate on the structure of β-galactosidase from Escherichia coli. a fluorescence study. J. Biochem. 2001, 130 (1), 13–18. 10.1093/oxfordjournals.jbchem.a002951. [DOI] [PubMed] [Google Scholar]
- Borgert C. J.; Baker S. P.; Matthews J. C. Potency matters: thresholds govern endocrine activity. Regul. Toxicol. Pharmacol. 2013, 67 (1), 83–88. 10.1016/j.yrtph.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Loub W. D.; Farnsworth N. R.; Soejarto D. D.; Quinn M. L. NAPRALERT: computer handling of natural product research data. J. Chem. Inf. Model. 1985, 25 (2), 99–103. 10.1021/ci00046a009. [DOI] [PubMed] [Google Scholar]
- Alstott J.; Bullmore E.; Plenz D. Powerlaw: a python package for analysis of heavy-tailed distributions. PLoS One 2014, 9 (1), e85777. 10.1371/journal.pone.0085777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J.; Davies M.; Gaulton A.; Hersey A.; Velankar S.; Petryszak R.; Hastings J.; Bellis L.; McGlinchey S.; Overington J. P. UniChem: a unified chemical structure cross-referencing and identifier tracking system. J. Cheminf. 2013, 5 (1), 3. 10.1186/1758-2946-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleb N. N.The Black Swan: The Impact of the Highly Improbable Fragility, 2nd ed.; Random House: New York, 2010. [Google Scholar]
- Clauset A.; Shalizi C.; Newman M. Power-law distributions in empirical data. SIAM Rev. 2009, 51 (4), 661–703. 10.1137/070710111. [DOI] [Google Scholar]
- Newman M. E. J. Power laws, Pareto distributions and Zipf’s law. Contemp. Phys. 2005, 46 (5), 323–351. 10.1080/00107510500052444. [DOI] [Google Scholar]
- Nicolaou K. C.; Snyder S. A. Chasing molecules that were never there: misassigned natural products and the role of chemical synthesis in modern structure elucidation. Angew. Chem., Int. Ed. 2005, 44 (7), 1012–1044. 10.1002/anie.200460864. [DOI] [PubMed] [Google Scholar]
- Fedorov O.; Müller S.; Knapp S. The (un)targeted cancer kinome. Nat. Chem. Biol. 2010, 6 (3), 166–169. 10.1038/nchembio.297. [DOI] [PubMed] [Google Scholar]
- Zipf G. K.Human Behavior and the Principle of Least Effort; Addison-Wesley Press: Cambridge, MA, 1949; Vol. XI. [Google Scholar]
- Firn R. D.; Jones C. G. The evolution of secondary metabolism – a unifying model. Mol. Microbiol. 2000, 37 (5), 989–994. 10.1046/j.1365-2958.2000.02098.x. [DOI] [PubMed] [Google Scholar]
- Firn R. D.; Jones C. G. Natural products — a simple model to explain chemical diversity. Nat. Prod. Rep. 2003, 20 (4), 382–391. 10.1039/b208815k. [DOI] [PubMed] [Google Scholar]
- Firn R. D.; Jones C. G. A Darwinian view of metabolism: molecular properties determine fitness. J. Exp. Bot. 2009, 60 (3), 719–726. 10.1093/jxb/erp002. [DOI] [PubMed] [Google Scholar]
- Choi Y. H.; van Spronsen J.; Dai Y.; Verberne M.; Hollmann F.; Arends I. W. C. E.; Witkamp G.-J.; Verpoorte R. Are natural deep eutectic solvents the missing link in understanding cellular metabolism and physiology?. Plant Physiol. 2011, 156 (4), 1701–1705. 10.1104/pp.111.178426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hert J.; Irwin J. J.; Laggner C.; Keiser M. J.; Shoichet B. K. Quantifying biogenic bias in screening libraries. Nat. Chem. Biol. 2009, 5 (7), 479–483. 10.1038/nchembio.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antioxidant activity assessed as superoxide-scavenging activity by nitrite method, ChEMBL (547370) 399341; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=399341.
- Inhibition of Clostridium perfringens neuraminidase, ChEMBL (606760) 455702; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=455702.
- Noncompetitive inhibition of recombinant influenza A virus rvH1N1 A/Bervig_Mission/1/18 neuraminidase, ChEMBL (606761) 455703; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=455703.
- Inhibition of xanthine oxidase assessed as decrease in uric acid production by spectrophotometry, ChEMBL (547369) 399340; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=399340.
- Antioxidant activity assessed as inhibition of superoxide production by xanthine/xanthine oxidase method, ChEMBL (441324) 293298; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=293298.
- Inhibition of Influenza A Jinan/15/90 H3N2 virus neuraminidase activity by MUN-ANA substrate based fluorimetric assay, ChEMBL (514313) 366284; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=366284.
- Inhibition of Influenza A PR/8/34 H1N1 virus neuraminidase activity by MUN-ANA substrate based fluorimetric assay, ChEMBL (514314) 366285; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=366285.
- Inhibition of Influenza A Jiangsu/10/2003 virus neuraminidase activity by MUN-ANA substrate based fluorimetric assay, ChEMBL (514315) 366286; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=366286.
- Papadopoulou A.; Green R. J.; Frazier R. A. Interaction of flavonoids with bovine serum albumin: a fluorescence quenching study. J. Agric. Food Chem. 2005, 53 (1), 158–163. 10.1021/jf048693g. [DOI] [PubMed] [Google Scholar]
- Nifli A.-P.; Theodoropoulos P. A.; Munier S.; Castagnino C.; Roussakis E.; Katerinopoulos H. E.; Vercauteren J.; Castanas E. Quercetin exhibits a specific fluorescence in cellular milieu: a valuable tool for the study of its intracellular distribution. J. Agric. Food Chem. 2007, 55 (8), 2873–2878. 10.1021/jf0632637. [DOI] [PubMed] [Google Scholar]
- Hudson S. A.; Ecroyd H.; Kee T. W.; Carver J. A. The thioflavin T fluorescence assay for amyloid fibril detection can be biased by the presence of exogenous compounds. FEBS J. 2009, 276 (20), 5960–5972. 10.1111/j.1742-4658.2009.07307.x. [DOI] [PubMed] [Google Scholar]
- qHTS Assay for Inhibitors of Firefly Luciferase, NCGC 411; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=411.
- Biochemical firefly luciferase enzyme assay for NPC, NCGC 624030; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=624030.
- qHTS profiling assay for firefly luciferase inhibitor/activator using purifed enzyme and Km concentrations of substrates (counterscreen for miR-21 project), NCGC 588342; National Center for Biotechnology Information: Bethesda, MD. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=588342.
- Avior Y.; Bomze D.; Ramon O.; Nahmias Y. Flavonoids as dietary regulators of nuclear receptor activity. Food Funct. 2013, 4 (6), 831. 10.1039/c3fo60063g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich A. B. Flavonoid-membrane interactions: possible consequences for biological effects of some polyphenolic compounds. Acta Pharmacol. Sin. 2006, 27 (1), 27–40. 10.1111/j.1745-7254.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Thakur P.; Herold K. F.; Hobart E. A.; Ramsey N. B.; Periole X.; de Jong D. H.; Zwama M.; Yilmaz D.; Hall K.; Maretzky T.; Hemmings H. C.; Blobel C.; Marrink S. J.; Koçer A.; Sack J. T.; Andersen O. S. Phytochemicals perturb membranes and promiscuously alter protein function. ACS Chem. Biol. 2014, 9 (8), 1788–1798. 10.1021/cb500086e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggisser R.; von Daeniken K.; Jundt G.; Schaffner W.; Tullberg-Reinert H. Interference of plant extracts, phytoestrogens and antioxidants with the MTT tetrazolium assay. Planta Med. 2002, 68 (5), 445–448. 10.1055/s-2002-32073. [DOI] [PubMed] [Google Scholar]
- Shoemaker M.; Cohen I.; Campbell M. Reduction of MTT by aqueous herbal extracts in the absence of cells. J. Ethnopharmacol. 2004, 93 (2–3), 381–384. 10.1016/j.jep.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Fallarero A.; Hanski L.; Vuorela P. How to translate a bioassay into a screening assay for natural products: general considerations and implementation of antimicrobial screens. Planta Med. 2014, 80 (14), 1182–1199. 10.1055/s-0034-1383061. [DOI] [PubMed] [Google Scholar]
- Boyd M. R.The NCI in vitro anticancer drug discovery screen. In Anticancer Drug Development Guide; Humana Press: Totowa, NJ, 1997; pp 23–42. [Google Scholar]
- LaPlante S. R.; Aubry N.; Bolger G.; Bonneau P.; Carson R.; Coulombe R.; Sturino C.; Beaulieu P. L. Monitoring drug self-aggregation and potential for promiscuity in off-target in vitro pharmacology screens by a practical NMR strategy. J. Med. Chem. 2013, 56 (17), 7073–7083. 10.1021/jm4008714. [DOI] [PubMed] [Google Scholar]
- Ryan A. J.; Gray N. M.; Lowe P. N.; Chung C. Effect of detergent on “promiscuous” inhibitors. J. Med. Chem. 2003, 46 (16), 3448–3451. 10.1021/jm0340896. [DOI] [PubMed] [Google Scholar]
- Feng B. Y.; Shoichet B. K. A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 2006, 1 (2), 550–553. 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassano M. F.; Doak A. K.; Roth B. L.; Shoichet B. K. Colloidal aggregation causes inhibition of G protein-coupled receptors. J. Med. Chem. 2013, 56 (6), 2406–2414. 10.1021/jm301749y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks P.; Gosselin M.; Prystay L. Impact of a red-shifted dye label for high throughput fluorescence polarization assays of G protein-coupled receptors. J. Biomol. Screening 2000, 5 (5), 329–334. 10.1177/108705710000500504. [DOI] [PubMed] [Google Scholar]
- Gribbon P.; Sewing A. Fluorescence readouts in HTS: no gain without pain?. Drug Discovery Today 2003, 8 (22), 1035–1043. 10.1016/S1359-6446(03)02895-2. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Kim E.; White I. M.; Bentley W. E.; Payne G. F. Information processing through a bio-based redox capacitor: signatures for redox-cycling. Bioelectrochemistry 2014, 98, 94–102. 10.1016/j.bioelechem.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Dahlin J. L.; Nissink J. W. M.; Strasser J. M.; Francis S.; Higgins L.; Zhou H.; Zhang Z.; Walters M. A. PAINS in the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. J. Med. Chem. 2015, 58 (5), 2091–2113. 10.1021/jm5019093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Bajorath J. What is the likelihood of an active compound to be promiscuous? systematic assessment of compound promiscuity on the basis of PubChem confirmatory bioassay data. AAPS J. 2013, 15 (3), 808–815. 10.1208/s12248-013-9488-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissink J. W. M.; Blackburn S. Quantification of frequent-hitter behavior based on historical high-throughput screening data. Future Med. Chem. 2014, 6 (10), 1113–1126. 10.4155/fmc.14.72. [DOI] [PubMed] [Google Scholar]
- Agarwal A.; D’Souza P.; Johnson T. S.; Dethe S. M.; Chandrasekaran C. Use of in vitro bioassays for assessing botanicals. Curr. Opin. Biotechnol. 2014, 25, 39–44. 10.1016/j.copbio.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Pauli G. F.; Chen S.-N.; Lankin D. C.; Bisson J.; Case R. J.; Chadwick L. R.; Gödecke T.; Inui T.; Krunic A.; Jaki B. U.; McAlpine J. B.; Mo S.; Napolitano J. G.; Orjala J.; Lehtivarjo J.; Korhonen S.-P.; Niemitz M. Essential parameters for structural analysis and dereplication by 1H NMR spectroscopy. J. Nat. Prod. 2014, 77 (6), 1473–1487. 10.1021/np5002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbaum M. C. A method for testing for synergy with any number of agents. J. Infect. Dis. 1978, 137 (2), 122–130. 10.1093/infdis/137.2.122. [DOI] [PubMed] [Google Scholar]
- Sucher N. J. Searching for synergy in silico, in vitro and in vivo. Synergy 2014, 1 (1), 30–43. 10.1016/j.synres.2014.07.004. [DOI] [Google Scholar]
- Qin C.; Tan K. L.; Zhang C. L.; Tan C. Y.; Chen Y. Z.; Jiang Y. Y. What does it take to synergistically combine sub-potent natural products into drug-level potent combinations?. PLoS One 2012, 7 (11), e49969. 10.1371/journal.pone.0049969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles S. J.; Day N. E.; Murray-Rust P.; Rzepa H. S.; Zhang Y. Enhancement of the chemical semantic web through the use of InChI identifiers. Org. Biomol. Chem. 2005, 3 (10), 1832. 10.1039/b502828k. [DOI] [PubMed] [Google Scholar]
- Southan C. InChI in the wild: an assessment of InChIkey searching in Google. J. Cheminf. 2013, 5, 10. 10.1186/1758-2946-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot M.; Jeanmart S. Pan assay interference compounds (PAINS) and other promiscuous compounds in antifungal research. J. Med. Chem. 2015, 10.1021/acs.jmedchem.5b00361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.