A. EXECUTIVE SUMMARY
The impact of advances in sleep and circadian sciences over the last 20 years on medicine, health, and public safety has been limited in part by the lack of availability of objective tools capable of quantifying sleep and circadian function in point-of-care (p-o-c) settings. This whitepaper is a product of a workshop that was designed to bring together thought-leaders in biomarker development, experts in sleep-circadian biology and sleep disorders to identify barriers and opportunities informing the future development of p-o-c diagnostic tools. The workshop entitled, “Developing Biomarker Arrays Predicting Sleep and Circadian-Coupled Risks to Health,” was held in Bethesda April 27–28 2015, and was jointly sponsored by the National Heart Lung and Blood Institute, National Institute on Aging and the Sleep Research Society (hereafter referred to as the biomarker workshop, (http://www.nhlbi.nih.gov/research/reports). The Sleep Research Society supported a number of early career investigators to attend the workshop. They contributed to the writing of this whitepaper. A biomarker is a “biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, condition or disease.”1,2 For the purpose of this whitepaper, “biomarkers” include quantifiable molecules and chemical properties of easily accessible biological samples (e.g., blood, urine, saliva). An ultimate goal is the development of robust and practical approaches for p-o-c or contact implementation in population-based research and most importantly, for clinical applications to enhance sleep and circadian health.
Biomarkers to assess current alertness status, sleep health and circadian function are needed for: research to further understanding of sleep and circadian health science, p-o-c diagnosis of sleep and circadian disorders, for prognosis and to evaluate the risk of associated heart, lung, blood, and aging diseases and disorders, and to assess the adequacy of therapy. The ideal biomarker would show high sensitivity (correctly identify the state and degree of acute sleep loss, and possibly even duration that such a status has been ongoing), specificity (correctly identify the presence/absence of a chronic sleep deficiency, such as would be useful in an annual primary care medical visit). However, the field is currently without any viable biomarkers measurable in easily accessible bio-specimens. The availability of objective platforms capable of quantifying sleep and circadian function will ultimately determine whether advances in understanding sleep and circadian biology can be applied to improve health and disease management and reduce risks to health and public safety.
In parallel with the development of p-o-c biomarkers, which take years to develop, there is urgent need to enhance and validate biosensors, mobile and wearable technologies that can be used in population phenotyping to accurately track sleep and circadian physiology and behavior. Such wearable technologies may facilitate the phenotyping needed to validate biomarkers, but in addition, will provide valuable information that will be useful for improving discovery, clinical diagnostics and intervention, as well as self-management of sleep for wellness.
Together these efforts to develop biomarkers and biosensors will position the field for participation in the Million Vet Program (http://www.research.va.gov/MVP/) and the Precision Medicine Initiatives (http://www.nih.gov/precision-medicine-initiative-cohort-program). With diagnostic tools for p-o-c measurement of sleep and circadian function, medical care and physician practices addressing sleep and circadian disorders and risk for metabolic and other diseases, will be vastly improved, translating into lower health care costs and a healthier population.
B. THE NEED AND USE CASES FOR SLEEP-CIRCADIAN BIOMARKERS
A marker of acute sleep loss that tracks alertness and predicts impaired behavioral function and performance relevant to fitness for duty might be used for roadside screening to assess sleep loss as a risk to performance and safe operation of a vehicle. Biomarker detection, sensitive enough to detect a single night of sleep loss would need to be developed with consideration of individual differences, including age and sex. Further, baseline data would likely be required to establish a reliable comparison for subsequent detection of change. Individuals vary in vulnerability to sleep loss and research is needed to elucidate benchmarks of normality.
A marker of chronic sleep loss that predicts risks to health reflecting long term deficiency in duration, timing, or quality of sleep might be used in conjunction with the annual physical examination by the primary care provider. There is a need to develop clinical definitions and phenotypic measures relevant to predicting sleep health-related outcomes.
Biomarkers, as diagnostic aids to facilitate pre-test triage for probable sleep apnea identification, and to help personalize/ optimize therapeutic strategy, identify comorbid health risks, and prognostic outcome stratification, would be of great value in patient care. Sleep apnea biomarkers would help improve risk stratification algorithms and elucidate domains of potential comorbidity overlap (heart, lung, and blood disorders), and ultimately inform the selection of targeted/personalized treatments.
In addition to duration and sleep disordered breathing, the timing of sleep is important for optimal restorative, physiological, and homeostatic benefits of sleep. A biomarker that could assess the appropriateness of the timing of the sleep period in circadian time could be used by primary care providers to differentiate insomnia, circadian phase disorders, shiftwork disorder, and extreme chronotypes. Circadian phase bio-markers could also enable the personalization of medical therapy with time-of-day dosing for oncologic and other pharmacotherapies. Surgical outcomes with potential links to circadian physiology such as cardioprotection from remote limb ischemic preconditioning, transplant tissue matching, vaccination, and metabolic disease management would also benefit from such biomarkers.
Sleep deficiency can be built up, not only by acute sleep deprivation, where individuals get by on little or no sleep for a night or two, but by an accrual of smaller deficits that are cumulative, such as 2 or 4 hours of lost sleep, per night3 over multiple nights, or even longer. Current tools to measure acute and chronic sleep patterns and their adverse consequences are either highly subjective, such as self-reported questionnaires, or involve complex laboratory measurements. The former is subject to imprecision and variation due to both human fallibility as well as disincentive for accurate reporting, for example in employment-based consequences of sleep reporting for transportation or aviation professionals. Laboratory measurements such as EEG based assessment of sleepiness or objective performance measures are expensive and complex, not amenable to general use and there is no conclusive evidence that they provide gold standard measurements by which assessment of the state of chronic sleep loss can be determined. In addition, a longer term sleep deficit may not be recognized by the individual, as there is evidence for a dissociation of subjective sleepiness from objective indices of ability to stay on task and maintain optimal vigilant performance.3 We know that there are individual differences in response to sleep loss at a behavioral level of assessment4,5 and this is highly heritable.6,7
A recent joint consensus statement of the American Academy of Sleep Medicine and Sleep Research Society recommended adults 18–60 years of age get 7 or more hours of sleep per night.8 The guidelines were based on the preponderance of evidence that failure to get enough sleep on a regular basis is associated with several adverse health outcomes including the development of obesity, and risk for depression, diabetes, hypertension, stroke, cardiovascular disease, and mortality.9–13 The term inadequate sleep may be used in reference to insufficient duration, quality and/or timing of sleep.
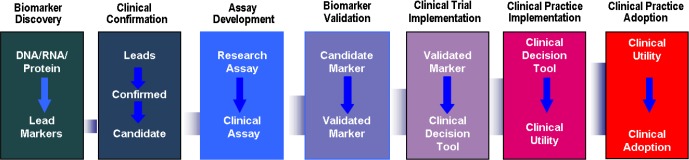
There are now fairly standard approaches to developing bio-markers (see Figure 1). First, it is important to utilize broad discovery approaches not only in humans but also in model systems, e.g., rats, mice, Drosophila. It is unknown at the outset which particular approach will be most valuable, hence all the “omic” approaches should be applied—transcriptomics, metabolomics, proteomics, lipidomics, etc.—to determine which platform or combination will ideally identify sleep associated markers. Initial studies could be done in a fairly small sample of subjects (20–40) who are very well phenotyped and characterized. A molecular signature that is found across different species adds confidence that the candidate biomarker is robust. When such a signature is identified, the next step is to validate the reproducibility of the specific signature in larger human cohorts (preferably prospectively, but also from existing) with the appropriate samples and phenotype information. This approach is applied in iterative fashion to refine and perfect the biomarker or biomarker panel.
Figure 1.
The Iterative process of biomarker development, illustrating the path for biomarkers from laboratory concept on the left, to clinical adoption, on the right. The connecting band represents the growing number of subjects required at each step in development, from biomarker discovery, leading to clinical adoption.
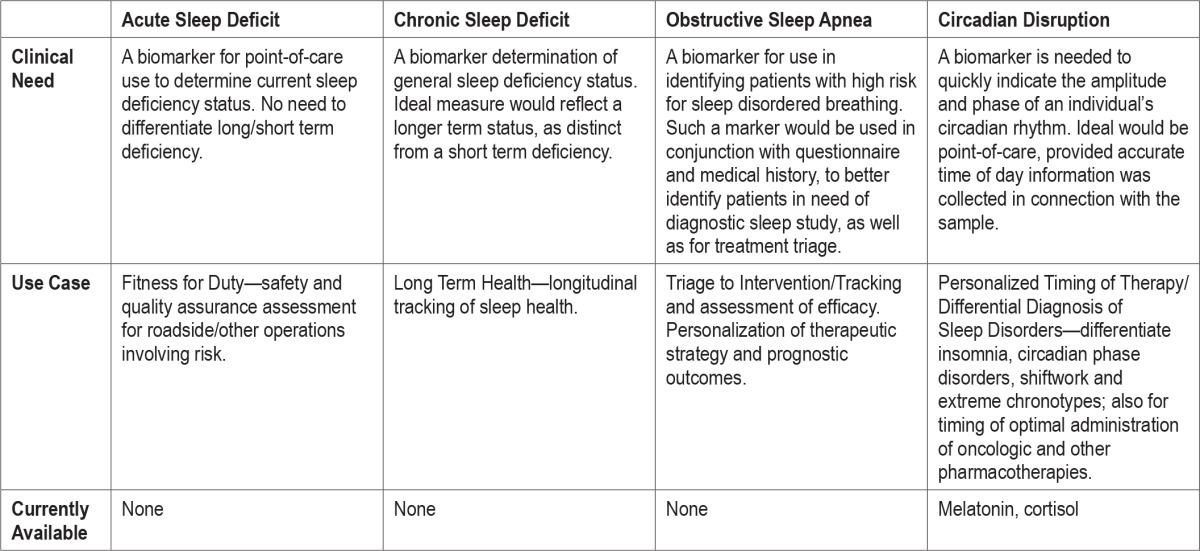
At the biomarker workshop, breakout groups were formed for the purpose of discussing various sleep-circadian use cases. The availability of biomarkers, needs and use cases for bio-markers are summarized in Table 1. The current state of the field for each area of biomarker development is described in the sections that follow, along with current opportunities and finally, recommendations for how to move forward in the most efficient way.
Table 1.
Summary of need for a sleep and/or circadian biomarker: use cases, currently available markers.
C. USE CASES
1. Development of a Biomarker of Acute Sleep Deficit
1a. Current Work
The fundamental amount of sleep that may be required between persons varies by age, and it is unclear whether the operational criteria for identifying acute sleep loss should be considered in light of an individual's inherent sleep need, or based upon a universal threshold. Given that sleep need may vary across the lifespan,14 a minimum threshold for acutely inadequate sleep for research purposes, would be less than or equal to 6 hours. Recognizing that with more severe acute sleep loss (i.e., total acute sleep loss), consideration of inter-individual differences in sleep need are a less pressing concern in establishing operational definitions of what constitutes inadequate sleep. Regardless of these challenges, the use of acute sleep loss as a research paradigm in humans and animals has yielded potentially promising candidate biomarkers, although as yet, there are none that can fulfil the demands of the use cases.
The term “acute” sleep loss is used here to refer to sleep loss that occurs in the 1 to 3 day range, whereas “chronic” refers to runs of more than 3 days of insufficient sleep that may occur repeatedly over many weeks, months or years (see chronic sleep deficit section, C2). Routine short sleep duration appears to be a particularly appropriate model of chronic sleep insufficiency. The estimated prevalence of short sleep duration (< 6 h per night) across the population is 30%.15 Individual differences in what constitutes sufficient sleep and acute sleep loss are, however, likely to exist. This underscores the need for objective molecular assessments of consequences that will contribute to a personalized assessment.
Based on the well-established relationship between sleep loss and immune function; many have tested, in controlled hypothesis-driven research, whether a biomarker exits in immune sub-cell populations (such as neutrophils, monoctyes, lymphocytes) or inflammatory markers found in plasma, saliva, or urine.16 Several markers that have been found to be dysregulated in response to acute sleep loss; for example inter-leukin (IL)-6 in plasma/serum, saliva, or expressed by monocytes,17–24 IL-1β or IL-1 receptor antagonist,25,26 tumor necrosis factor (TNF) alpha or receptors,17,19,29,27,28 chemokine receptor CXCR2 expression,29 cellular adhesion molecules,30 natural killer cells,31 and salivary amylase,32 among others. Methods that identify candidate markers in invertebrate and vertebrate species, and use these findings to replicate markers in humans, are particularly efficient.
The wide-spread use of exploratory methodologies, using omics based technology for biomarker discovery is largely based on the idea that a molecular profile (i.e. “signature”) or a single biomarker may best identify acute sleep loss.33 Transcriptomics and metabolomics have been most commonly assessed for biomarker discovery in biofluids for acute sleep loss.34 There have been few proteomics studies and they have been limited to brain tissue from murine sleep loss models.35,36 In humans, studies have reported a reduced diurnal amplitude of several transcripts in individuals identified as neurobehaviorally resistant to sleep loss, compared to those classified as sensitive to sleep loss.37 Acute sleep loss has been further shown to increase inflammation-related transcripts23,38 and cause differential expression of metabolites related to lipid metabolism, neurotransmitters, vitamin B3, and gut metabolism, as measured in plasma.39–40 The challenge is complex; and the effects of total sleep deprivation on the transcriptome have been shown to be sensitive to a chronic sleep debt prior to the acute sleep deprivation.38
1b. Immediate Opportunities
There are several immediate opportunities for implementation of potentially high-yield strategies in acute sleep loss biomarker research. Since many of the candidate biomarkers that have been identified using hypothesis-driven approaches (such as those related to inflammation and immune function) may also be aberrant in other pathological states besides sleep loss, and thus ultimately have lower probability of demonstrating sufficient specificity as a biomarker for sleep loss in isolation, the search should move beyond those established candidates. Validation of other novel biomarkers such as those related to phospholipid metabolism that have been recently identified41 using metabolomics, may provide important leads in the search for an optimal biomarker. However, we recognize it is quite possible that no one molecule will sufficiently demonstrate the required sensitivity and specificity to serve as a biomarker for acute sleep loss, and thus combinations of biomarkers (i.e., signatures), that may span several functional domains, may ultimately be required.
2. Development of a Biomarker of Chronic Sleep Deficit
2a. Current Work
Insufficient sleep affects immune function and restricting sleep for several nights leads to increases in neutrophils, monocytes and lymphocytes, and alteration in their circadian rhythmicity.16,42 Genome wide blood transcript analysis has identified changes in the abundance of mRNAs of genes related to oxidative stress, metabolism, and immune function in response to one week of insufficient sleep.38 Exploratory metabolomics have found that sleep reduction by approximately a third for 8 days led to altered metabolic biomarkers, with the clearest effects being elevated plasma amino acids.43 The effects are small but the findings provide useful information for future studies of chronic sleep loss. Furthermore, diurnal transcript profiles have been shown to associate with neurocognitive resilience to total sleep loss, which is an important consideration in evaluating the utility of a potential biomarker. From a behavioral standpoint, the degree to which an individual is susceptible or resistant to the cognitive effects of chronic sleep loss may be task dependent,7,44,45 and further research is needed to identify a biomarker that might also correspond with cognitive processing speed or other similar parameters of relevance necessary to safely operate vehicles.
Metabolomics approaches have successfully identified metabolites perturbed under controlled experimental models of both total sleep deprivation and partial sleep restriction, with a notable role for lipids.40 This latter study identified two potential markers as common to sleep restriction in humans and animals which may be useful for translational biomarker work. Potential protein markers have also been reported, with particular interest in salivary assessment. Amylase protein was proposed as a possible biomarker of sleep loss almost a decade ago. Saliva peptides have more recently been identified from fatigue studies which may be applicable to sleep loss.46 However, as salivary amylase is driven by increased sympathetic activity, it may be altered under many circumstances, and therefore lack the necessary specificity of a good biomarker.
A problem common to all these approaches is that systems specific responses to sleep loss have not been well characterized, and their dynamics during several days of sleep loss alternating with one or two recovery sleep episodes, have not been documented. An ideal marker of chronic sleep loss would need to stand out over the circadian influences that might also be characteristic of the biomarker or constellation of bio-markers, under study. Specificity or the degree to which a bio-marker exclusively represents chronic sleep loss is important for predictability, but thus far, all of the sleep-related candidate biomarkers appear to be pleiotropic in nature. For instance, adenosine increases with time spent awake, but functions in vasodilation and energy metabolism as well. Cytokines and neurotrophins are additional examples of multifunctional molecules (e.g., innate immunity and inflammation) that are known sleep-related biomarkers. Weighting pleiotropic biomarkers in proportion to their contribution as predictors of chronic sleep loss could allow inclusion and limit their potential bias of depicting alternative processes. Use of multiple sleep loss biomarkers may limit false detection and increase the predictability in a biomarker panel. For use in the clinic the sleep loss biomarker should ideally be accessible using blood, urine, or saliva. While it would be most efficient to have a biomarker panel that could be performed in a single assay type, e.g., DNA, RNA, or protein; a range of panels could be processed independently and subsequently integrated in the analysis phase to provide a single metric or set of linked metrics.
2b. Immediate Opportunities
Despite the many challenges to identifying and validating sleep loss biomarkers, several recent advancements in science have provided the field with important tools by which to overcome some of the road blocks and facilitate sleep loss biomarker profiling. A first immediate opportunity is capitalizing on the development, and increased sensitivity and affordability of omics technologies which greatly expand the number of variables that can be gleaned from just one sample. For example, common microarray techniques provide genome-wide transcriptional coverage of well-characterized genes, gene candidates, and splice variants. Parallel to gene expression analyses, proteomics and metabolomics offer new opportunities to examine in a comprehensive way the dynamic impact chronic sleep loss has on molecular processes. Advanced mass spec (e.g., Tandem, and MALDI-TOF) approaches provide different information than array data or ELISAs. Other exciting research directions at the forefront of science include epigenetics and micro-RNAs as signaling molecules. The idea of state- dependent non-coding RNAs, histone modifications, micro-vesicle or exosome packaging, transport, recognition and docking are all virtually uncharted areas that could yield key biomarkers of chronic sleep loss. Many of these processes are activity-dependent, but their relationship to sleep and responses to sleep loss are just starting to be elucidated.
3. Development of a Biomarker of Obstructive Sleep Apnea (OSA)
3a. Current Work
An ideal biomarker of OSA would be highly sensitive and specific for OSA-induced end-organ dysfunction, include genetic and behavioral risk factors, and provide information related to prognosis and response to treatment.47 Identification of such an OSA biomarker signature would have significant real-world effects. For example, we would be able to identify “vulnerable” and “resilient” patients, allowing us to target therapeutic interventions or offer latitude with watchful waiting. Thus, studies assessing biomarkers in the context of well-defined and validated clinical algorithms are needed.
Biomarkers can also be a physiological signal-based signature such as blood pressure48 or snoring.49 Over the last 15 years, a substantial number of studies have tackled the identification of an ideal biomarker for OSA using exhaled breath condensate, salivary, serum and urinary molecules. Although no simple and clinically useful biomarker signature for OSA exists today, considerable progress has been made50–62 to identify and detect the presence of OSA and OSA-related consequences. While underlying genetic risk factors are not yet established, gene expression arrays reveal a number of genes that may have discriminatory ability to recognize OSA in children.63 Furthermore, whole-genome expression measurement of peripheral blood leukocytes was performed at baseline and following continuous positive airway pressure (CPAP) therapy in a small cohort of adult patients with OSA; although the diagnostic ability of such approach was not specifically examined, the investigators found evidence supporting the presence of transcriptional suppression in cancer-related pathways.64 Similarly, urine is a highly desirable biological fluid for diagnostic testing because of its ease of collection and widespread use in clinical laboratories. Urinary proteomic analyses have been used to identify candidate biomarkers for a broad range of human disorders that have included renal disease, diabetes, atherosclerosis, Alzheimer's disease and cancer.65–67 In a cohort of children, 12 candidate urinary biomarkers were identified that could reliably detect OSA.68 The latter study underscores the feasibility of urine to report pathophysiology in OSA and further suggests that a similar approach may be informative for identifying OSA-associated comorbidities.69 The use of unique sub-population clusters such as patients with resistant hypertension and OSA, have resulted in specific blood-based biomarkers that can predict favorable or ineffective response to PAP.70
It is important to stress that the measures derived from lab-based sleep studies or home-based multichannel recordings are poor predictors of OSA-associated morbidities. In other words, two patients with similar PSG-defined OSA severity may present with markedly different clinical phenotypes— one will manifest substantial end-organ morbidities related to the presence of OSA, while any such morbid features will be absent in the other. The phenotypic variance in the clinical morbidity of OSA has therefore prompted exploration of bio-markers that would enable the identification of the more “vulnerable” or more “resilient” patients, who would more likely benefit from timely and targeted therapeutic interventions or may be allowed increased latitude regarding their treatment options. Thus, studies assessing the possibility of incorporation of morbidity biomarkers into well-defined and validated clinical algorithms become very attractive. The discovery of an ideal biomarker for OSA-associated morbidities also offers the opportunity to provide information related to prognosis and response to treatment.71 Ideal biomarkers should be highly sensitive and specific for OSA-induced end-organ dysfunction, should be involved in a causal pathway, and temporal changes in such biomarkers in the context of OSA treatment should reliably predict improvements in the specific end-organ outcomes.
In the context of use cases for biomarker development, a prominent challenge derives from the complex interactions between sleep apnea and obesity, the latter being present in approximately two-thirds of all patients. Thus, the risk exists whereby the discovery process of biomarkers may yield what essentially would correspond to markers of obesity rather than markers of sleep apnea. Thus, it is important to sample broadly, so that lean and non-obese populations are included and can contribute to isolation of OSA-specific markers.
3b. Immediate Opportunities
To successfully achieve such a challenging task, coordinated efforts are needed to link a carefully constructed phenotype to all the omics available today. Once we agree as a community what this phenotype is, we may be able to identify a clean and robust signature in a select, small group of OSA and control subjects. In practice, we would need to collect a medical history (age, family history, co-morbidities, etc.) as well as specimens (blood, urine, saliva and breath volatiles/condensate) and standardized PSG variables and longitudinally correlate them to morbidities and mortality. Animal models of OSA would be essential to identify potential mechanisms at the tissue level that underlies cancer, neurological, cardiovascular, and metabolic disease. Comprehensive data could then be used to risk stratify the long term prognosis and treatment response into a weighed risk score based on what is clinically relevant and what is mechanistically informative. Ultimately we need a unique OSA biomarker signature that is context-relevant, simple and easy to use. Ideally, clinicians would use a disease score to (1) determine prognosis (end-organ dysfunction vs. irreversibility) (2) gauge treatment response at the molecular and cellular level, and (3) understand how OSA contributes to other chronic diseases.
We propose 3 major areas of research to develop OSA bio-markers that include both discovery and validation steps. The first is the development of a diagnostic biomarker or biomarker signature (biological candidates and current gold standard PSG) that reliably discriminate between OSA and healthy individuals, between symptomatic individuals with and without OSA, or alternatively, high-risk populations such as commercial drivers. Identifying an OSA biomarker signature will improve overall health by decreasing the burden of oxidative stress and chronic inflammation. In addition, such a biomarker will help identify potential individuals with excessive daytime sleepiness for PSG screening, and thereby help to prevent sleep-related accidents, such as motor vehicle accidents. The second is prognosis biomarkers, which targets symptoms such as sleepiness, (morbidity)—develop a biomarker signature that consistently and accurately identifies which patients with OSA are at risk for developing end-organ morbidities and which patients already have end-organ morbidities. For example, identifying unique biomarker signatures of excessive daytime sleepiness would be clearly desirable.72–76 Third is treatment adherence/outcome biomarkers, to develop a biomarker signature in treated patients that reliably discerns those that are disease free (vs. those that have residual OSA), and detects continued presence of OSA in adherent patients (to alter treatment). Such a biomarker will decrease healthcare-related costs by eliminating or delaying the need for treatment based on accurate prognostication.
There are many examples of work that can be done immediately. First, we can develop and perform a general population screen using the recording of snoring and breathing patterns on mobile devices such as cell phones. Second, we can work with current non-sleep cohorts and add sleep questionnaires, sleep studies and bank additional blood for future studies. Third, we need to agree as a community what is an OSA phenotype to better select subjects for in-depth, discovery omics. Fourth, using PSGs and biological samples we can start to risk stratify all the variables within the PSG (not only AHI) and develop a weighted PSG “biomarker.” Lastly, we can perform gene sequencing in infants born with apnea.
4. Development of a Biomarker of Circadian Disruption
4a. Current Work
Currently, the diagnosis and treatment of circadian rhythm sleep-wake disorders is dependent primarily on either self-report, or the use of activity monitoring devices. Actigraphy, which consists of an accelerometer that is usually worn on the non-dominant wrist, can be helpful to provide objective measurements of activity and rest periods. Actigraphy has been validated as a means of measuring sleep and wake77 and recently was demonstrated to be approximately 80% accurate in distinguishing sleep and wake when compared to polysomnography (PSG).78 Newer devices also are capable of measuring both the intensity and wavelength of light exposure, which can provide additional data to aid in the diagnosis and management of circadian disorders. However, both actigraphy and sleep logs have the limitation that they are a reflection primarily of the sleep-wake cycle, which can be heavily influenced by social obligations and work schedules, so may not provide accurate representation of the internal biological rhythm.
Measurement of melatonin, the circadian hormone produced by the pineal, is one way that circadian phase and amplitude have been measured. Under normal conditions, the onset of melatonin secretion in dim light (DLMO) generally occurs 2–3 hours prior to the habitual sleep time, and melatonin secretion peaks in the middle of the night.79 Melatonin can be measured in plasma, however this is impractical in the out-patient setting, as samples need to be collected every 30 to 60 minutes for at least 4–5 hours prior to the predicted sleep onset time, most practically through an indwelling venous catheter. As an alternative to obtaining frequent blood samples, melatonin can also be collected in the saliva. Salivary melatonin has been validated as a comparable measure to plasma, and can accurately be obtained by patients using sampling kits at home.80 However again this requires multiple samples be collected over a 4 to 5 or more, hour time window.81 If underlying sleep-wake patterns are variable, this window may need to be expanded up to 24–36 hours. In cases such as these where the predicted DLMO is unclear, urine samples may be more useful, as the primary metabolite of melatonin, 6-sulfatoxymelatonin (aMT6s) is secreted in the urine. Urine samples can be collected at scheduled intervals over a 24 to 48 hour period and provide an estimate of timing and amount of daily melatonin production.79 However, urine collection provides much less precision regarding overall circadian phase when compared to saliva or blood sampling.
Cortisol also has a circadian pattern of release and can be measured in blood or saliva. However, similar to melatonin, accurate phase assessment is dependent on obtaining multiple blood samples over time, making it less practical as an outpatient assessment of circadian phase. In addition, cortisol levels can also be heavily influenced by external factors such as acute stressors. Estimated core body temperature is another marker of circadian timing. However, traditional methods of data collection to approximate core temperature, involving rectal probes or telemetry using swallowed transmitter pills, are not practical. Newer techniques are being developed using wrist- mounted thermometer to measure the daily fluctuations in temperature. While body temperature normally falls as an individual is falling asleep, reaching a nadir, and then beginning to rise ∼2 hours prior to waking, skin temperature, on the other hand, begins to increase prior to bedtime, and drops just after awakening.82 Validation studies have shown good correlation between the evening temperature increase measured at the wrist and DLMO suggesting that this may be another less invasive means of measuring circadian phase timing.83 However, further validation studies are needed in which the timing of sleep is altered relative to the melatonin and body temperature rhythms. Under conditions when sleep timing is not at the usual nocturnal placement, such as shift-work, it may be more difficult to predict circadian phase.
Individual differences in vulnerability to sleep loss appear to be associated with circadian rhythmicity, such that circadian rhythmicity is not only reduced during sleep deprivation,38 but the reduction of circadian amplitude is dependent on vulnerability to sleep loss, with more resistant individuals showing more robust suppression of circadian rhythmicity.37 Lipid homeostasis is heavily influenced by circadian rhythms and several recent studies have used lipid-based omics methods to investigate these systems. One study of 20 healthy young male subjects who had their blood sampled every 4 hours, found that, in a given individual, about 18% of the lipids detected exhibited circadian variation, mostly di- and triglycerides that peaked around waking.84 Omic investigations have found circadian rhythms in metabolic phenotypes including lipid-mediated energy storage, transport and signaling, however, the rhythms are quite variable across subjects,84,85 making circadian biomarker discovery a challenging, but rich area for phenotypic exploration.86 Likewise, approximately 6% to 9% of the human blood transcriptome displays circadian rhythmicity38,87 and could therefore potentially be used to develop a one-sample biomarker for circadian phase. However, this rhythmicity is greatly affected by the timing of sleep,87 posing a significant challenge for biomarker development.
As for the development of biomarkers for acute and chronic sleep loss, integration of clinical and preclinical work will be important. For example, comparisons of rhythmicity in the transcriptome in the brain and other organs of rodents to rhythmicity in the human blood transcriptome may aid the development of indicators of circadian phase in the brain and other organs.88
4b. Immediate Opportunities
There are immediate opportunities to advance the study of circadian biomarkers in both the omics and wearable domains. In the omics domain, there is a rapid growth in publicly available datasets with omics data (genomics, proteomics, transcriptomics, metabolomics, etc.) through NIH data repositories, such as the cardiovascular cohorts, and other large scale biobank efforts. To the extent that the samples are time-stamped for when they were collected, there are opportunities for data mining in order to identify signatures of circadian phase and amplitude that could be validated in other datasets. A systems biology approach will be needed to integrate data across omics levels.
There are also immediate opportunities to utilize wearable technology to develop circadian biomarkers. The sleep and circadian fields have pioneered work in this area through the use of actigraphy over the past several decades. There is a rapid growth in the types of biosensors available, with heart rate and temperature routinely collected with several available devices. The widespread use of smartphones compliments the use of biosensors through the use of apps. Such technology could be used to track activities, food intake and other factors that can influence circadian rhythms. There is substantial investment by a number of companies in this space so there is considerable opportunity to partner with industry in these efforts. Given that these data are collected in real-world settings, development of sensor technology will need to be paralleled by improved analytic methods that can account for masking influences and identify the endogenous circadian rhythm in the midst of substantial noise.
D. LONG TERM NEEDS, STRATEGIES AND RECOMMENDATIONS FOR SLEEP AND BIOMARKERS SCIENCE
The workshop participants agreed that a systems biology approach will be needed to integrate data across acute sleep loss, chronic sleep loss and sleep disordered breathing and circadian phase. Insofar as sleep deficiency presents a biological stress or burden to the organism, it is reasonable to expect that the consequences will vary in part based on genetic and epigenetic systems vulnerabilities. Long-term strategies for developing biomarkers in the sleep and circadian field will need to include several levels of investigation, applying new biomarker measurement technologies.
Although considerable limitations exist in translatability, animal models are still very useful given the possibility of highly controlled and manipulated environments available to examine the impact of sleep loss on biomarkers, not possible to do in humans. Additionally, there is very high conservation between animal and human metabolism, lipids, and signaling pathways, and animal models are cost-effective and capable of high throughput testing compared to human sleep research. Basic animal models and basic human physiology phenotyping are fundamental to the phenotyping discovery process.
Basic human physiology research conducted under highly controlled protocols is needed in order to deeply phenotype and generate a short list of candidate biomarkers, which then inform larger-scale population studies to validated candidate biomarkers. This work must be done in concert with sensitivity to genetic and epigenetic influences on the measures under study. Research must demonstrate reproducibility within individuals, on different occasions, where sleep and circadian factors are held constant. Also important, the research should include conventional non-biochemical markers of acute and chronic sleep loss, such as behavioral and cognitive performance and observable physiologies as these will be needed in iterative validation to help construct an index of sleep loss provided by the biomarker panel.
Biomarker research on sleep and circadian health will require interdisciplinary teams of sleep and circadian scientists, clinicians, biomarker experts, quantitative scientists as well as stakeholders from the military and public health and safety domains. Several recommendations emerged from the breakout group discussions, as well as from the convened group discussions of reports. A few consensus recommendations emerged from the discussions:
Network Capacity—The establishment of a collaborative national sleep-circadian network. This was one of the central recommendations of the Institute of Medicine Report,89 and other countries, such as Canada, have recently established such a mechanism. A collaborative national network would have interdisciplinary involvement, help to integrate and link the biomarker, big data, novel therapeutic development and training pipeline. In addition, such a mechanism would enable more cohesive international collaboration and scientific, clinical-translational advancement. A national sleep-circadian network would embrace “team science” and help to foster communication among interdisciplinary scientists and bridge the basic-clinical-translational divide, to expedite the integration of omics and other new technologies, and facilitate an iterative approach to biomarker development. It would facilitate the involvement of experts in biomarker development in sleep-circadian research.
Integrated National Training and Workforce Pipeline Development—Development of interdisciplinary training programs that bring sleep scientists, biomarker researchers and data science experts together. Encourage integrative interdisciplinary team research approaches in training programs in order to capitalize on the opportunities to advance science and improve public health.
Near-term biomarker discovery—Foster biomarker development through secondary analysis programs and incorporation of sleep markers into existing and new cohorts using mobile approaches. Facilitate the identification and advancement of diagnostic biomarkers. Secondary analyses of well-characterized datasets enhanced with sleep/circadian phenotypes would open the door to a new array of hypothesis testing, risk stratification and ultimately new opportunities for the precision medicine initiative. Access to cohorts with specimens time-stamped for sleep and circadian information should be enhanced through a public index that could be accessed through an increased network capacity.
Big Data and Data Analysis Innovation—Collaboration is critical to moving big data initiatives forward and data management and sharing technologies are key. Time information as well as multi-dimensional methods for integrating biological and behavioral data are necessary for the advancement of sleep-circadian modeling, therapeutic development, and translation. The availability of circadian time-stamped biosamples is a major barrier to biomarker development. Adding time stamps to index samples collected in all ongoing cohort studies could potentially increase the tissue resource suitable for sleep/ circadian biomarker research. However, since existing cohorts are not phenotyped with regard to circadian phase, the elucidation of circadian biomarkers will be primarily dependent on prospective tissue collection (urine, saliva, blood, epithelial cells) combined with wearable sleep/circadian devices and omic analyses.
Novel Technologies and Therapeutics—There is a need to foster the development and validation of “wearable” devices, m-Health applications, and instruments suitable for population-based and big data approaches used in developing predictive models and biomarker platforms assessing sleep and circadian health. The public pressure evident by the popularity of devices to track activity and sleep demonstrates the need to quickly validate the cost-effective implementation of miniaturized sleep tracking devices in clinical-translational research.
CITATION
Mullington JM, Abbott SM, Carroll JE, Davis CJ, Dijk DJ, Dinges DF, Gehrman PR, Ginsburg GS, Gozal D, Haack M, Lim DC, Macrea M, Pack AI, Plante DT, Teske JA, Zee PC. Developing biomarker arrays predicting sleep and circadian-coupled risks to health. SLEEP 2016;39(4):727–736.
DISCLOSURE STATEMENT
This is not an industry supported study. Dr. Dijk has received research support from GW Pharma, Lilly, and Janssen and has consulted for Servier and Shire. Dr. Gehrman has received research support from Merck and has consulted for Johnson & Johnson. The other authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The National Heart Lung and Blood Institute (NHLBI) and the National Institute of Aging (NIA) jointly sponsored the meeting on which this whitepaper is based in partnership with the Sleep Research Society. The authors acknowledge the contributions of Drs. Michael Twery (NHLBI), Miroslaw Mackiewicz (NIA), and Karen Teff of the National Institutes of Diabetes and Digestive and Kidney Diseases through the scientific discussion of programmatic opportunities and meeting organization under the aegis of the National Institutes of Health. The authors also acknowledge the biomarker and additional sleep and circadian experts who participated in the meeting and breakout groups including:
Ronald G. Crystal, Weill Cornell Medical College
Karyn A. Esser, University of Kentucky
Joshua J. Gooley, Duke-National University of Singapore Graduate Medical School
Alfred Hero, The University of Michigan
John B. Hogenesch, University of Pennsylvania
M.Mahmood Hussain, SUNY Downstate Medical Center
Rima Kaddurah-Daouk, Duke University School of Medicine
Satchindananda Panda, The Salk Institute
Peipei Ping, UCLA School of Medicine
Virend Somers, Mayo Clinic
Russell Tracy, The University of Vermont
Martin Young, University of Alabama School of Medicine
REFERENCES
- 1.Gozal D. Serum, urine, and breath-related biomarkers in the diagnosis of obstructive sleep apnea in children: is it for real? Curr Opin Pulmonary Med. 2012;18:561–7. doi: 10.1097/MCP.0b013e328358be2d. [DOI] [PubMed] [Google Scholar]
- 2.Wong TK. The search on an ideal disease marker for childhood obstructive sleep apnea syndrome. Sleep. 2011;34:133–4. doi: 10.1093/sleep/34.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Dongen HP, Maislin G, Mullington JM, Dinges DF. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep. 2003;26:117–26. doi: 10.1093/sleep/26.2.117. [DOI] [PubMed] [Google Scholar]
- 4.Van Dongen HP, Baynard MD, Maislin G, Dinges DF. Systematic interindividual differences in neurobehavioral impairment from sleep loss: evidence of trait-like differential vulnerability. Sleep. 2004;27:423–33. [PubMed] [Google Scholar]
- 5.Van Dongen HPA, Maislin G, Dinges DF. Dealing with inter-individual differences in the temporal dynamics of fatigue and performance: importance and techniques. Aviat Space Environ Med. 2004;75:A147–54. [PubMed] [Google Scholar]
- 6.Kuna ST, Maislin G, Pack FM, et al. Heritability of performance deficit accumulation during acute sleep deprivation in twins. Sleep. 2012;35:1223–33. doi: 10.5665/sleep.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler JF, Arnold RD, Phillips JB, Turnmire AE. Predicting individual differences in response to sleep loss: application of current techniques. Aviat Space Environ Med. 2013;84:927–37. doi: 10.3357/asem.3581.2013. [DOI] [PubMed] [Google Scholar]
- 8.Watson NF, Badr M, Belenky G, et al. Recommended amount of sleep for a healthy adult: a joint consensus statement of the American Academy of Sleep Medicine and Sleep Research Society. Sleep. 2015;38:843–4. doi: 10.5665/sleep.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gangwisch JE. A review of evidence for the link between sleep duration and hypertension. Am J Hypertens. 2014;27:1235–42. doi: 10.1093/ajh/hpu071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangwisch JE, Malaspina D, Posner K, et al. Insomnia and sleep duration as mediators of the relationship between depression and hypertension incidence. Am J Hypertens. 2010;23:62–9. doi: 10.1038/ajh.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullington JM, Haack M, Toth M, Serrador J, Meier-Ewert HK. Cardiovascular, inflammatory, and metabolic consequences of sleep deprivation. Progr Cardiovasc Dis. 2009;51:294–302. doi: 10.1016/j.pcad.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parthasarathy S, Vasquez MM, Halonen M, et al. Persistent insomnia is associated with mortality risk. Am J Med. 2015;128:268–75. doi: 10.1016/j.amjmed.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiegel K, Knutson K, Leproult R, Tasali E, Van Cauter E. Sleep loss: a novel risk factor for insulin resistance and Type 2 diabetes. J Appl Physiol. 2005;99:2008–19. doi: 10.1152/japplphysiol.00660.2005. [DOI] [PubMed] [Google Scholar]
- 14.Ohayon MM, Carskadon MA, Guilleminault C, Vitiello MV. Meta-analysis of quantitative sleep parameters from childhood to old age in healthy individuals: developing normative sleep values across the human lifespan. Sleep. 2004;27:1255–73. doi: 10.1093/sleep/27.7.1255. [DOI] [PubMed] [Google Scholar]
- 15.Chen JC, Espeland MA, Brunner RL, et al. Sleep duration, cognitive decline, and dementia risk in older women. Alzheimers Dement. 2016;12:21–33. doi: 10.1016/j.jalz.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullington JM, Simpson NS, Meier-Ewert HK, Haack M. Sleep loss and inflammation. Best Pract Res Clin Endocrinol Metab. 2010;24:775–84. doi: 10.1016/j.beem.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shearer WT, Reuben JM, Mullington JM, et al. Soluble TNF-α receptor 1 and IL-6 plasma levels in humans subjected to the sleep deprivation model of spaceflight. J Allergy Clin Immunol. 2001;107:165–70. doi: 10.1067/mai.2001.112270. [DOI] [PubMed] [Google Scholar]
- 18.Vgontzas AN, Papanicolaou DA, Bixler EO, et al. Circadian interleukin-6 secretion and quantity and depth of sleep. J Clin Endocrinol Metab. 1999;84:2603–7. doi: 10.1210/jcem.84.8.5894. [DOI] [PubMed] [Google Scholar]
- 19.Irwin MR, Wang MG, Campomayor CO, Collado-Hidalgo A, Cole S. Sleep deprivation and activation of morning levels of cellular and genomic markers of inflammation. Arch Intern Med. 2006;166:1756–62. doi: 10.1001/archinte.166.16.1756. [DOI] [PubMed] [Google Scholar]
- 20.Irwin MR, Witarama T, Caudill M, Olmstead R, Breen EC. Sleep loss activates cellular inflammation and signal transducer and activator of transcription (STAT) family proteins in humans. Brain Behav Immun. 2015;47:86–92. doi: 10.1016/j.bbi.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimitrov S, Lange T, Benedict C, et al. Sleep enhances IL-6 trans-signaling in humans. FASEB J. 2006;20:2174–6. doi: 10.1096/fj.06-5754fje. [DOI] [PubMed] [Google Scholar]
- 22.Frey DJ, Fleshner M, Wright KP., Jr The effects of 40 hours of total sleep deprivation on inflammatory markers in healthy young adults. Brain Behav Immun. 2007;21:1050–7. doi: 10.1016/j.bbi.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Thimgan MS, Gottschalk L, Toedebusch C, et al. Cross-translational studies in human and drosophila identify markers of sleep loss. Plos One. 2013;8:e61016. doi: 10.1371/journal.pone.0061016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faraut B, Nakib S, Drogou C, et al. Napping reverses the salivary interleukin-6 and urinary norepinephrine changes induced by sleep restriction. J Clin Endocrinol Metab. 2015;100:E416–26. doi: 10.1210/jc.2014-2566. [DOI] [PubMed] [Google Scholar]
- 25.Moldofsky H, Lue FA, Davidson JR, Gorczynski R. Effects of sleep deprivation on human immune functions. FASEB J. 1989;3:1972–7. doi: 10.1096/fasebj.3.8.2785942. [DOI] [PubMed] [Google Scholar]
- 26.Dinges DF, Douglas SD, Hamarman S, Zaugg L, Kapoor S. Sleep deprivation and human immune function. Adv Neuroimmunol. 1995;5:97–110. doi: 10.1016/0960-5428(95)00002-j. [DOI] [PubMed] [Google Scholar]
- 27.Irwin MR, Carrillo C, Olmstead R. Sleep loss activates cellular markers of inflammation: sex differences. Brain Behav Immun. 2010;24:54–7. doi: 10.1016/j.bbi.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimitrov S, Besedovsky L, Born J, Lange T. Differential acute effects of sleep on spontaneous and stimulated production of tumor necrosis factor in men. Brain Behav Immun. 2015;47:201–10. doi: 10.1016/j.bbi.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Christoffersson G, Vagesjo E, Pettersson US, et al. Acute sleep deprivation in healthy young men: impact on population diversity and function of circulating neutrophils. Brain Behav Immun. 2014;41:162–72. doi: 10.1016/j.bbi.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Redwine L, Dang J, Irwin M. Cellular adhesion molecule expression, nocturnal sleep, and partial night sleep deprivation. Brain Behav Immun. 2004;18:333–40. doi: 10.1016/j.bbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Ozturk L, Pelin Z, Karadeniz D, Kaynak H, Cakar L, Gozukirmizi E. Effects of 48 hours sleep deprivation on human immune profile. Sleep Research Online. 1999;2:107–11. [PubMed] [Google Scholar]
- 32.Seugnet L, Boero J, Gottschalk L, Duntley SP, Shaw PJ. Identification of a biomarker for sleep drive in flies and humans. Proc Natl Acad Sci U S A. 2006;103:19913–8. doi: 10.1073/pnas.0609463104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Schantz M, Skene DJ. Telling biological time from a blood sample: current capabilities and future potential. Ann Clin Biochem. 2015;52:699–701. doi: 10.1177/0004563215597943. [DOI] [PubMed] [Google Scholar]
- 34.Goel N. ‘Omics’ approaches for sleep and circadian rhythm research: biomarkers for identifying differential vulnerability to sleep loss. Curr Sleep Med Rep. 2015;1:38–46. [Google Scholar]
- 35.Pawlyk AC, Ferber M, Shah A, Pack AI, Naidoo N. Proteomic analysis of the effects and interactions of sleep deprivation and aging in mouse cerebral cortex. J Neurochem. 2007;103:2301–13. doi: 10.1111/j.1471-4159.2007.04949.x. [DOI] [PubMed] [Google Scholar]
- 36.Naidoo N. Potential of proteomics as a bioanalytic technique for quantifying sleepiness. J Clin Sleep Med. 2011;7:S28–S30. doi: 10.5664/JCSM.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnardottir ES, Nikonova EV, Shockley KR, et al. Blood-gene expression reveals reduced circadian rhythmicity in individuals resistant to sleep deprivation. Sleep. 2014;37:1589–U172. doi: 10.5665/sleep.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moller-Levet CS, Archer SN, Bucca G, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci U S A. 2013;110:E1132–41. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies SK, Ang JE, Revell VL, et al. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci U S A. 2014;111:10761–6. doi: 10.1073/pnas.1402663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weljie AM, Meerlo P, Goel N, et al. Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci U S A. 2015;112:2569–74. doi: 10.1073/pnas.1417432112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chua EC-P, Shui G, Cazenave-Gassiot A, Wenk MR, Gooley JJ. Changes in plasma lipids during exposure to total sleep deprivation. Sleep. 2015;38:1683–91. doi: 10.5665/sleep.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Besedovsky L, Lange T, Born J. Sleep and immune function. Pflugers Arch. 2012;463:121–37. doi: 10.1007/s00424-011-1044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell LN, Kilkus JM, Booth JN, Bromley LE, Imperial JG, Penev PD. Effects of sleep restriction on the human plasma metabolome. Physiol Behav. 2013;122:25–31. doi: 10.1016/j.physbeh.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson ML, Gunzelmann G, Whitney P, et al. Deconstructing and reconstructing cognitive performance in sleep deprivation. Sleep Med Rev. 2013;17:215–25. doi: 10.1016/j.smrv.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rupp TL, Wesensten NJ, Balkin TJ. Trait-like vulnerability to total and partial sleep loss. Sleep. 2012;35:1163–72. doi: 10.5665/sleep.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michael DJ, Daugherty S, Santos A, Ruby BC, Kalns JE. Fatigue biomarker index: an objective salivary measure of fatigue level. Accid Anal Prev. 2012;45:68–73. doi: 10.1016/j.aap.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 47.Montesi SB, Bajwa EK, Malhotra A. Biomarkers of sleep apnea. Chest. 2012;142:239–45. doi: 10.1378/chest.11-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai M, Stockbridge N, Temple R. Blood pressure as an example of a biomarker that functions as a surrogate. AAPS J. 2006;8:E146–52. doi: 10.1208/aapsj080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams CJ, Hu FB, Patel SR, Mantzoros CS. Sleep duration and snoring in relation to biomarkers of cardiovascular disease risk among women with type 2 diabetes. Diabetes Care. 2007;30:1233–40. doi: 10.2337/dc06-2107. [DOI] [PubMed] [Google Scholar]
- 50.Jullian-Desayes I, Joyeux-Faure M, Tamisier R, et al. Impact of obstructive sleep apnea treatment by continuous positive airway pressure on cardiometabolic biomarkers: a systematic review from sham CPAP randomized controlled trials. Sleep Med Rev. 2015;21:23–38. doi: 10.1016/j.smrv.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Altinas N, Mutlu LC, Akkoyun DC, et al. Effect of CPAP on new endothelial dysfunction marker, endocan, in people with obstructive sleep apnea. Angiology. 2015 Jun; doi: 10.1177/0003319715590558. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stradling JR, Schwarz EI, Schlatzer C, et al. Biomarkers of oxidative stress following continuous positive airway pressure withdrawal: data from two randomised trials. Eur Respir J. 2015;46:1065–71. doi: 10.1183/09031936.00023215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu A, Lamendola C, Ariel D, et al. Usefulness of fetuin-a to predict risk for cardiovascular disease among patients with obstructive sleep apnea. Am J Cardiol. 2015;116:219–24. doi: 10.1016/j.amjcard.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kheirandish-Gozal L, Gileles-Hillel A, Alonso-Alvarez M, et al. Effects of adenotonsillectomy on plasma inflammatory biomarkers in obese children with obstructive sleep apnea: a community-based study. Int J Obes. 2015;39:1094–100. doi: 10.1038/ijo.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canto GDL, Pacheco-Pereira C, Aydinoz S, Major PW, Flores-Mir C, Gozal D. Biomarkers associated with obstructive sleep apnea: a scoping review. Sleep Med Rev. 2015;23:28–45. doi: 10.1016/j.smrv.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Araújo Freitas IG, de Bruin PF, Bittencourt L, de Bruin VM, Tufik S. What can blood biomarkers tell us about cardiovascular risk in obstructive sleep apnea? Sleep Breath. 2015;19:755–68. doi: 10.1007/s11325-015-1143-9. [DOI] [PubMed] [Google Scholar]
- 57.Arnardottir ES, Lim DC, Keenan BT, et al. Effects of obesity on the association between long-term sleep apnea treatment and changes in interleukin-6 levels: the Icelandic Sleep Apnea Cohort. J Sleep Res. 2015;24:148–59. doi: 10.1111/jsr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Luca Canto G, Pacheco-Pereira C, Aydinoz S, Major PW, Flores-Mir C, Gozal D. Diagnostic capability of biological markers in assessment of obstructive sleep apnea: a systematic review and meta-analysis. J Clin Sleep Med. 2015;11:27–36. doi: 10.5664/jcsm.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kosacka M, Brzecka A, Piesiak P, Korzeniewska A, Jankowska R. Soluble Ligand CD40 and uric acid as markers of atheromatosis in patients with obstructive sleep apnea. Inflammatory Bowel Dis. 2015;839:55–60. doi: 10.1007/5584_2014_44. [DOI] [PubMed] [Google Scholar]
- 60.Maeder MT, Strobel W, Christ M, et al. Comprehensive biomarker profiling in patients with obstructive sleep apnea. Clin Biochem. 2015;48:340–6. doi: 10.1016/j.clinbiochem.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Stradling JR, Craig S, Kohler M, Nicoll D, Ayers L, Nunn AJ. Markers of inflammation: data from the MOSAIC randomised trial of CPAP for minimally symptomatic OSA. Thorax. 2015;70:181. doi: 10.1136/thoraxjnl-2014-205958. [DOI] [PubMed] [Google Scholar]
- 62.Kunos L, Bikov A, Lazar Z, et al. Evening and morning exhaled volatile compound patterns are different in obstructive sleep apnoea assessed with electronic nose. Sleep Breath. 2015;19:247–53. doi: 10.1007/s11325-014-1003-z. [DOI] [PubMed] [Google Scholar]
- 63.Tan HL, Kheirandish-Gozal L, Gozal D. The promise of translational and personalised approaches for paediatric obstructive sleep apnoea: an ‘Omics’ perspective. Thorax. 2014;69:450–6. doi: 10.1136/thoraxjnl-2013-204640. [DOI] [PubMed] [Google Scholar]
- 64.Gharib SA, Seiger AN, Hayes AL, Mehra R, Patel SR. Treatment of obstructive sleep apnea alters cancer-associated transcriptional signatures in circulating leukocytes. Sleep. 2014;37:709–U309. doi: 10.5665/sleep.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kentsis A. Challenges and opportunities for discovery of disease biomarkers using urine proteomics. Pediatr Int. 2011;53:1–6. doi: 10.1111/j.1442-200X.2010.03253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zurbig P, Jerums G, Hovind P, et al. Urinary proteomics for early diagnosis in diabetic nephropathy. Diabetes. 2012;61:3304–13. doi: 10.2337/db12-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zimmerli LU, Schiffer E, Zurbig P, et al. Urinary proteomic biomarkers on coronary artery disease. Mol Cell Proteomics. 2008;7:290–8. doi: 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- 68.Gozal D, Jortani S, Snow AB, et al. Two-dimensional differential in-gel electrophoresis proteomic approaches reveal urine candidate biomarkers in pediatric obstructive sleep apnea. Am J Respir Crit Care Med. 2009;180:1253–61. doi: 10.1164/rccm.200905-0765OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kheirandish-Gozal L, Mcmanus CJT, Kellermann GH, Samiei A, Gozal D. Urinary neurotransmitters are selectively altered in children with obstructive sleep apnea and predict cognitive morbidity. Chest. 2013;143:1576–83. doi: 10.1378/chest.12-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanchez-de-la-Torre M, Khalyfa A, Sanchez-de-la-Torre A, et al. Precision medicine in patients with resistant hypertension and obstructive sleep apnea blood pressure response to continuous positive airway pressure treatment. J Am Coll Cardiol. 2015;66:1023–32. doi: 10.1016/j.jacc.2015.06.1315. [DOI] [PubMed] [Google Scholar]
- 71.Montesi SB, Bajwa EK, Malhotra A. Biomarkers of sleep apnea. Chest. 2012;142:239–45. doi: 10.1378/chest.11-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thimgan MS, Toedebusch C, McLeland J, Duntley SP, Shaw PJ. Excessive daytime sleepiness is associated with changes in salivary inflammatory genes transcripts. Mediators Inflamm. 2015;2015:539627. doi: 10.1155/2015/539627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paik MJ, Kim DK, Nguyen DT, et al. Correlation of daytime sleepiness with urine metabolites in patients with obstructive sleep apnea. Sleep Breath. 2014;18:517–23. doi: 10.1007/s11325-013-0913-5. [DOI] [PubMed] [Google Scholar]
- 74.Uysal A, Liendo C, McCarty DE, et al. Nocturnal hypoxemia biomarker predicts sleepiness in patients with severe obstructive sleep apnea. Sleep Breath. 2014;18:77–84. doi: 10.1007/s11325-013-0851-2. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez-de-la-Torre M, Barcelo A, Pierola J, et al. Plasma levels of neuropeptides and metabolic hormones, and sleepiness in obstructive sleep apnea. Respir Med. 2011;105:1954–60. doi: 10.1016/j.rmed.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 76.Bravo MD, Serpero LD, Barcelo A, Barbe F, Agusti A, Gozal D. Inflammatory proteins in patients with obstructive sleep apnea with and without daytime sleepiness. Sleep Breath. 2007;11:177–85. doi: 10.1007/s11325-007-0100-7. [DOI] [PubMed] [Google Scholar]
- 77.Ancoli-Israel S, Cole R, Alessi C, Chambers M, Moorcroft W, Pollak CP. The role of actigraphy in the study of sleep and circadian rhythms. Sleep. 2003;26:342–92. doi: 10.1093/sleep/26.3.342. [DOI] [PubMed] [Google Scholar]
- 78.Marino M, Li Y, Rueschman MN, et al. Measuring sleep: accuracy, sensitivity, and specificity of wrist actigraphy compared to polysomnography. Sleep. 2013;36:1747–55. doi: 10.5665/sleep.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benloucif S, Burgess HJ, Klerman EB, et al. Measuring melatonin in humans. J Clin Sleep Med. 2008;4:66–9. [PMC free article] [PubMed] [Google Scholar]
- 80.Burgess HJ, Wyatt JK, Park M, Fogg LF. Home circadian phase assessments with measures of compliance yield accurate dim light melatonin onsets. Sleep. 2015;38:889–97. doi: 10.5665/sleep.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voultsios A, Kennaway DJ, Dawson D. Salivary melatonin as a circadian phase marker: validation and comparison to plasma melatonin. J Biol Rhythms. 1997;12:457–66. doi: 10.1177/074873049701200507. [DOI] [PubMed] [Google Scholar]
- 82.Sarabia J, Rol M, Mendiola P, Madrid J. Circadian rhythm of wrist temperature in normal-living subjects A candidate of new index of the circadian system. Physiol Behav. 2008;95:570–80. doi: 10.1016/j.physbeh.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 83.Bonmati-Carrion M, Middleton B, Revell V, Skene D, Rol M, Madrid J. Circadian phase asessment by ambulatory monitoring in humans: correlation with dim light melatonin onset. Chronobiol Int. 2014;31:37–51. doi: 10.3109/07420528.2013.820740. [DOI] [PubMed] [Google Scholar]
- 84.Chua EC-P, Shui G, Lee IT-G, et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci U S A. 2013;110:14468–73. doi: 10.1073/pnas.1222647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kasukawa T, Sugimoto M, Hida A, et al. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci U S A. 2012;109:15036–41. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gooley JJ, Chua EC. Diurnal regulation of lipid metabolism and applications of circadian lipidomics. J Genet Genomics. 2014;41:231–50. doi: 10.1016/j.jgg.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 87.Archer SN, Laing EE, Moller-Levet CS, et al. Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc Natl Acad Sci U S A. 2014;111:E682–91. doi: 10.1073/pnas.1316335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laing EE, Johnston JD, Moller-Levet CS, et al. Exploiting human and mouse transcriptomic data: identification of circadian genes and pathways influencing health. Bioessays. 2015;37:544–56. doi: 10.1002/bies.201400193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colten HR, Altevogt BM, editors. Sleep disorders and sleep deprivation: an unmet public health problem. Washington, DC: The National Academies Press; 2006. [PubMed] [Google Scholar]