Abstract
Study Objectives:
We hypothesized that DNA methylation patterns may contribute to disease severity or the development of hypertension and excessive daytime sleepiness (EDS) in patients with obstructive sleep apnea (OSA).
Methods:
Illumina's (San Diego, CA, USA) DNA methylation 27-K assay was used to identify differentially methylated loci (DML). DNA methylation levels were validated by pyrosequencing. A discovery cohort of 15 patients with OSA and 6 healthy subjects, and a validation cohort of 72 patients with sleep disordered breathing (SDB).
Results:
Microarray analysis identified 636 DMLs in patients with OSA versus healthy subjects, and 327 DMLs in patients with OSA and hypertension versus those without hypertension. In the validation cohort, no significant difference in DNA methylation levels of six selected genes was found between the primary snoring subjects and OSA patients (primary outcome). However, a secondary outcome analysis showed that interleukin-1 receptor 2 (IL1R2) promoter methylation (−114 cytosine followed by guanine dinucleotide sequence [CpG] site) was decreased and IL1R2 protein levels were increased in the patients with SDB with an oxygen desaturation index > 30. Androgen receptor (AR) promoter methylation (−531 CpG site) and AR protein levels were both increased in the patients with SDB with an oxygen desaturation index > 30. Natriuretic peptide receptor 2 (NPR2) promoter methylation (−608/−618 CpG sites) were decreased, whereas levels of both NPR2 and serum C type natriuretic peptide protein were increased in the SDB patients with EDS. Speckled protein 140 (SP140) promoter methylation (−194 CpG site) was increased, and SP140 protein levels were decreased in the patients with SDB and EDS.
Conclusions:
IL1R2 hypomethylation and AR hypermethylation may constitute an important determinant of disease severity, whereas NPR2 hypomethylation and SP140 hypermethylation may provide a biomarker for vulnerability to EDS in OSA.
Commentary:
A commentary on this article appears in this issue on page 723.
Citation:
Chen YC, Chen TW, Su MC, Chen CJ, Chen KD, Liou CW, Tang P, Wang TY, Chang JC, Wang CC, Lin HC, Chin CH, Huang KT, Lin MC, Hsiao CC. Whole genome DNA methylation analysis of obstructive sleep apnea: IL1R2, NPR2, AR, SP140 methylation and clinical phenotype. SLEEP 2016;39(4):743–755.
Keywords: AR, DNA methylation, excessive daytime sleepiness, IL1R2, NPR2, obstructive sleep apnea, oxygen desaturation index, SP140
Significance.
This is the first study to perform a large-scale DNA methylation analysis with replication of the principal finding in OSA patients. The identification of the abnormal DNA methylation marks in OSA provides novel biomarkers for prediction and diagnosis, and affords novel therapeutic targets for the prevention of adverse consequences of OSA. Further investigation is required to clarify whether these changes in peripheral blood mononuclear cells can be translated to neurons, endothelium, or other end-organ tissues, and to clarify the cause-and-effect relationship between the DNA methylation changes and the development of clinical phenotypes of OSA syndrome.
INTRODUCTION
Obstructive sleep apnea (OSA) is an oxidative stress disorder, in which chronic intermittent hypoxia with reoxygenation (IHR) is thought to result in damage similar to that caused by ischemia-reperfusion and to lead to delayed apoptosis of blood immune cells through mechanisms different from sustained hypoxia.1,2 Increases in inflammatory cytokines and adhe sion molecules have been proposed to lead to activation of peripheral blood mononuclear cells, neutrophils, and endothelial cells resulting in various adverse consequences, such as hypertension, ischemic heart disease, stroke, and excessive daytime sleepiness (EDS).3,4 Two thirds of patients with OSA will ultimately have diurnal hypertension, and approximately one half will experience EDS.5,6 Over the past two decades, extensive research to identify genetic determinants of OSA has shown that only a few genetic polymorphisms are independently and consistently associated with apnea-hypopnea index (AHI), even though about 30% to 40% of the variability in apneic activity can be explained by familial factors.7–9
DNA methylation occurring at position 5 of the pyrimidine ring of cytosines in the context of the cytosine followed by guanine dinucleotide sequence (CpG) form the basis of epi-genetic mechanisms modulating gene expressions by inhibition of the binding of transcription factors at the promoter regions. DNA methylation is a heritable, tissue-specific, and reversible gene regulatory process that is highly modified in response to environmental factors. DNA hypermethylation of superoxide dismutase 2 induced by neonatal intermittent hypoxia has been demonstrated to mediate programming of hypoxic sensitivity and the ensuing autonomic dysfunction in adult rats, including increased hypoxic ventilatory response and sleep apnea events.10 Epigenetic programming of gene expression patterns in response to gestational hypoxia has been shown to play a crucial role in the fetal origins of neurological diseases.11 However, little is known about the role of DNA methylation patterns in the development of OSA syndrome and its adverse consequences.
The aim of this study was to improve the understanding of the epigenetic mechanisms that regulate inflammatory responses and other pathways related to chronic IHR in the blood immune cells of patients with OSA by investigating the DNA methylation levels of genes in peripheral blood mono-nuclear cells on a genome-wide scale. We hypothesized that the DNA methylation profiles of peripheral blood mononu-clear cells involved in the inflammatory response to chronic IHR in patients with OSA would be different from those in healthy subjects, and that additional differences would be seen between patients with OSA with and without hypertension or with and without EDS, with the goal of identifying novel epigenetic changes related to disease severity, hypertension, and EDS.
METHODS
Subjects
This study was approved by the Institutional Review Board of Chang Gung Memorial Hospital, Taiwan (certificate number: 99-3517B). The study participants were recruited from the sleep center and health examination center of Kaohsiung Chang Gung Memorial Hospital from January 2012 through December 2014. Written informed consent was obtained from each subject participating in the study, who were all age 20 y or older. The exclusion criteria included ongoing infections, autoimmune disease, use of immunosuppressive agent in the past 6 mo, narcolepsy, severe obesity (body mass index [BMI] ≥ 35 kg/m2), old age (> 65 y), and those with a BMI < 21 kg/m2. The discovery cohort used for the whole-genome DNA methylation microarray experiment included 16 treatment-naïve patients with OSA and eight healthy subjects without habitual snoring. OSA was diagnosed by full-night polysomnography examination at the sleep center of Kaohsiung Chang Gung Memorial Hospital as described previously.12 The eight healthy subjects underwent overnight pulse oximetry (WristOx 3100, Nonin Medical Inc., Minnesota, USA) and showed normal results with no symptoms of sleep disordered breathing (SDB).The validation cohort included 48 treatment-naïve patients with OSA and 24 subjects with primary snoring (PS), all of whom received a diagnosis via full-night PSG examinations. SDB was defined as a history of loud snoring for more than 2 y, and a snoring index of > 20 counts/h. Snoring sampling was performed at 10 to 50 Hz, and all sounds of more than 50% above baseline amplitude lasting for 0.5–5 sec were recorded by Piezo crystal snore sensor (SleepSense, Scientific Laboratory Products, Elgin, USA). Nocturnal hypoxemia was evaluated in terms of the percentage of total minutes of recording time with an oxyhemoglobin saturation < 90% (%time < 90% SaO2), mean SaO2, minimum SaO2, and the number of dips > 4% of basal SaO2%//h (oxygen desaturation index [ODI]). The Epworth Sleepiness Scale (ESS) recorded at the examination was used to measure sleep propensity in every study subject.
Genome-wide DNA Methylation Assay
Venous blood (20 mL) was obtained from all treatment-naïve patients with SDB and healthy subjects. Peripheral blood mononuclear cells were isolated by Ficoll-Hypaque gradient centrifugation (HISTOPAQUE-119, Sigma-Aldrich, Inc., St. Louis, MO, USA), and DNA was extracted using Puregene Core kit (Qiagen, Baltimore, MD, USA). Electropherograms using an Agilent BioAnalyzer with Agilent DNA 12000 chips showed the fragment size to be > 10000 bp. Illumina's Infinium Methylation-27k BeadChip v1.2 (San Diego, CA, USA) was used to detect 27,578 CpG sites genome-wide, located within the promoter regions of 14,495 genes, with the distance to the transcription start site ranging from 0 to 1499 bp. For bisulfite conversion, EZ DNA methylation kit (Zymo Research, Irvine, USA) was used. Approximately 200 ng of each bisulfite-converted genomic DNA sample was applied per BeadChip according to the manufacturer's instructions. During hybridization, the DNA molecules anneal to two different bead types with locus-specific DNA oligomers, with one corresponding to the methylated (C) and the other to the unmethylated (T) state. After extension, the array was fluorescently stained, scanned, and the intensities of the unmethylated and methylated bead types were measured.13
Genome-wide DNA Methylation Data Analysis
We used the Methylation Module in the Illumina Genome Studio V2009.2 to generate the β value for each CpG locus. The β value was calculated as: (intensity of methylated probe) / (intensity of methylated probe + intensity of unmethylated probe). The β values ranged between 0 (least methylated) and 1 (most methylated) and were proportional to the degree of the methylated state of a particular locus. The raw β value was transformed into a M value to achieve better statistical properties,14 which was the log2 ratio of the intensity of methylated probes versus unmethylated probes using the following equation: M value = log 2 (β value / (1 − β value)). A positive M value meant higher intensity from the methylated probes than the unmethylated probes and a negative M value meant the opposite. The significance threshold in M value comparisons was P < 0.005 and a false discovery rate (q) < 0.5.
Principal component analysis plotting showed that almost all of the 24 samples in the discovery cohort clustered together quite well except for samples from one OSA patient and two healthy subjects from the same chip (Figure S1, supplemental material). Considering their significant batch effect on the microarray experiment, we excluded them from analysis, leaving 15 cases and 6 control samples.15 To identify differential methylated CpG sites, M values of the case and control groups were analyzed with the Mann-Whitney U test by Partek Genomics Suite software (Partek Incorporated, Missouri, U.S.A.) to obtain a P value and false discovery rate (q value). Significantly differentially methylated CpG sites with P < 0.005, q < 0.5, at least a 10% difference in their β value (large effect size), and known biological or functional relevance were selected for further verification and validation.16 For the differentially methylated CpG sites, their corresponding gene symbols were used for pathway analysis using MetaCore from Thomson Reuters (New York, USA), which uses hypergeometric tests to examine whether the genes are enriched in any known pathway. The top 10 pathways were selected based on their P values (< 0.005) and q values (< 0.2). All methylation datasets have been deposited in the NCBI Gene Expression Omnibus with the accession number GSE61463.
Measurement of DNA Methylation Levels of Four Selected Gene Promoter Regions by Bisulfite Pyrosequencing in the Discovery and Validation Cohort
Initially, pyrosequencing was applied to verify the DNA methylation levels of 20 CpG sites over 16 gene promoter regions, including IL21R, SP140, ACOT11, CHFR, CXCL9, GIMAP5, IL24, NPR2, AR, IL1R2, KCNH5, KCNA3, VNN3, ANGPTL3, UBASH3A, and GPR21 (based on the aforementioned criteria), in the same 21 samples used in the genome-wide discovery experiment. Among them, seven CpG sites of six genes that showed similar methylation changes to the microarray data and difference in β value ≥ 10% were selected for pyrosequencing validation in the validation cohort (Table S1, supplemental material). The promoter regions of six selected genes, including interleukin-1 receptor 2 (IL1R2), androgen receptor (AR), natriuretic peptide receptor 2 (NPR2), speckled protein 140 (SP140), interleukin-21 receptor (IL21R), and G protein-coupled receptor 21 (GPR21), were amplified. Bisulfite treatment was performed using an EpiTect 96 Bisulfite Kit (Qiagen) and polymerase chain reaction (PCR) amplification was performed using a PyroMark PCR Kit (Qiagen). The PCR conditions were 45 cycles of 95°C for 20 sec, 50°C for 20 sec, and 72°C for 20 sec, followed by 72°C for 5 min. The primer sequences used for PCR amplification and pyrosequencing for these regions are listed in Table S2, supplemental material. The biotin-labeled PCR product was captured by Streptavidin-Sepharose HP (Amersham Pharmacia, New Jersey, USA). Quantitation of cytosine methylation was performed using a PyroMark Q24 system (Qiagen). The amount of C relative to the sum of the amounts of C and T at each CpG site was calculated as a percentage.17 One CpG site (−114, cg17142183) of the IL1R2 gene, one CpG site (−531) of the AR gene, two CpG sites (−608, cg12876594; −618) of the NPR2 gene, one CpG site (−194, cg05564251) of the SP140 gene, one CpG site (−1227, cg02656594) of the IL21R gene, and one CpG site (−438, cg047843150) of the GPR21 gene were assayed. Representative pyrograms of the seven CpG sites assayed are presented in Figure S2, supplemental material.
Measurement of Protein Expression Levels of the Four Selected Genes From Peripheral Blood Mononuclear Cell Samples and Serum Type C Natriuretic Peptide in the Validation Cohort
Peripheral blood mononuclear cells were lysed in radioimmunoprecipitation assay-buffer containing a protease inhibitor cocktail (Sigma-Aldrich). Protein lysate normalized to 20 ng total protein was used to measure IL1R2, AR, NPR2, and Sp140 levels by a commercial enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems, Minneapolis, MN). Briefly, 40-μL dye (Bio-Rad Protein Assay Dye Reagent Concentrate #500-0006, Bio-Rad, California, USA) was added to 10 μL bovine serum albumin with serial dilution to generate a standard curve for total protein concentration by measuring values at Optical Density 595 nm (Figure S3, supplemental material). Accordingly, protein lysate equivalent to 20 ng of total protein was used for each protocol in ELISA analysis. Serum type C natriuretic peptide (CNP) levels were also measured by ELISA (USCN Business Co., China) according to the manufacturer's instructions.
Statistical Analysis
Continuous values were expressed as mean ± standard deviation (SD). The differences between two groups were analyzed using the Student t-test or χ2 test, as appropriate. Subgroup comparisons of continuous variables in the validation cohort were performed using the independent Student t-test, followed by multivariate linear regression analysis to adjust for confounding factors, including age, sex, BMI, comorbidities, smoking, and a history of alcoholism, and to obtain adjusted P values with 95% confidence interval (CI). In the comparisons between the patients with SDB with and without EDS, the AHI was added as a covariate in the linear regression model to examine the independent effect of EDS on methylation or protein levels. The null hypothesis was rejected at P < 0.05. All analyses were performed using SPSS software version 17.0 (SPSS Corp., Chicago, IL). To assist in the interpretation of P values given the number of statistical tests performed, q values were calculated separately for multiple comparisons of the DNA methylation and protein expression levels using the Benjamini-Hochberg test with R Console software (R foundation for Statistical Computing, Vienna).
RESULTS
Demographic Data of the Participants
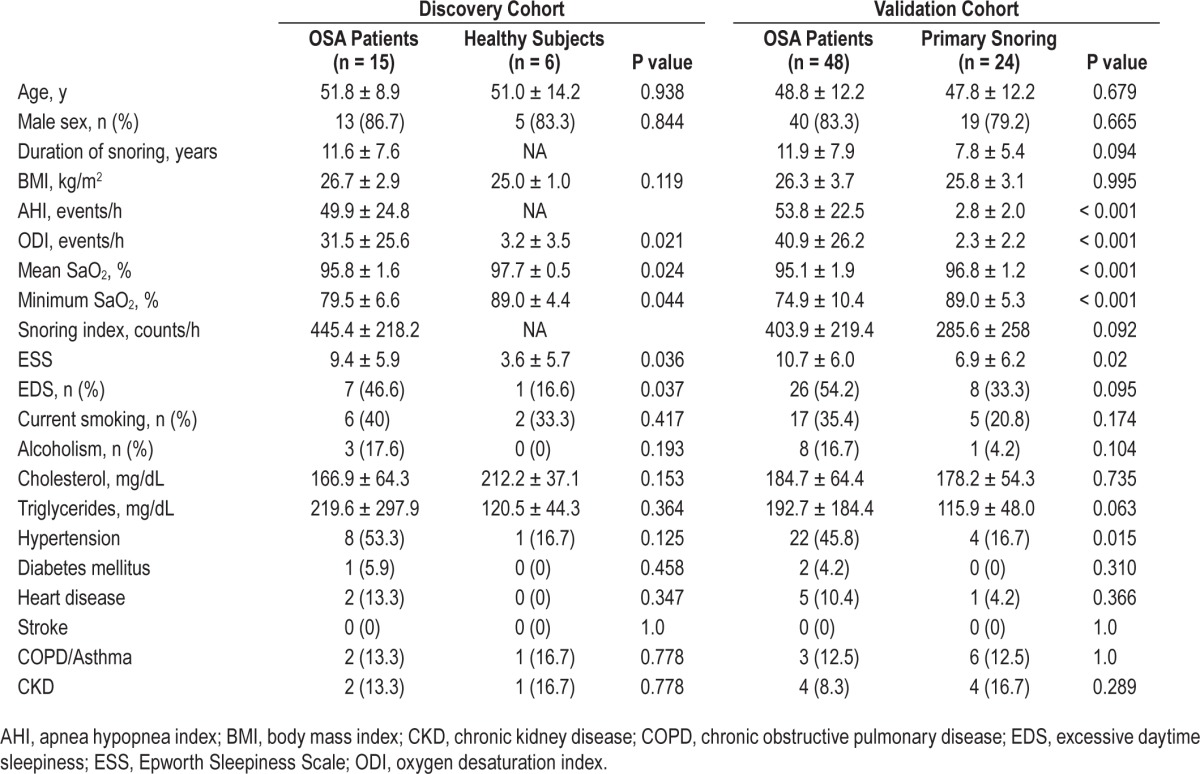
The baseline, sleep, and biochemistry data of both the discovery and validation cohorts are listed in Table 1. The study population comprised residents of Taiwan. Age, BMI, male sex ratio, smoking history, alcoholism history, and comorbidities were all matched between the case and control groups, except that more patients with OSA had hypertension than those with PS in the validation cohort.
Table 1.
Demographic, sleep, and biochemistry data for the participants in both the discovery and validation cohorts.
Whole-Genome DNA Methylation Profiles of the Discovery Cohort
A total of 21 samples were grouped and analyzed in two different comparisons. The first comparison (I) was between normal patients (n = 6) and patients with OSA (n = 15). The second (II) was between patients with OSA with hypertension (n = 8) and those without hypertension (n = 7). For each comparison, 636 and 327 significantly differentially methylated loci (DML) were identified, respectively. Figure 1 shows two-dimensional hierarchical clustering of the patients with OSA versus the healthy subjects (A, 377 hypermethylated DMLs and 259 hypomethylated DMLs, all P < 0.005, all q < 0.5; comparison I), and patients with OSA with hypertension versus those without hypertension (B, 117 hypermethylated DMLs and 210 hypomethylated DMLs; all P < 0.005, all q < 0.5; comparison II). Hypomethylated OSA-related DMLs included ACOT11, IL21R, KCNA3, and NPR2, whereas hypermethylated OSA-related DMLs included IL24, CXCL9, CRTAM, and cycLTR2 (Table 2). Hypermethylated hypertension-related DMLs included LTA, CXCR3, GIMAP5, and SP140, whereas hypomethylated hypertension-related DMLs included TRPM2, VMP, IL1R2, and CCL26 (Table S3, supplemental material). Hypermethylated EDS-related DMLs included F8A1, TNXB, GNL3L, and ALX4, whereas hypomethylated EDS-related DMLs included HSD17B4, PDCD6IP, DHCR24, and HSD3B7 (data not shown). Product names and gene ontology functions of the genes with DML in the comparisons I and II, which meet the objective criteria, are provided in Table S4, supplemental material. The intersection of comparison I and comparison II resulted in three DMLs with correspondently increased methylation levels in both comparisons of OSA versus healthy subjects and hypertension versus no hypertension (SP140, CYP2D7P1, GIMAP5).
Figure 1.
Heatmaps of the differentially methylated loci (DML) for the three comparisons in the discovery cohort. Hierarchical clustering of DML in 21 samples classified into three comparisons: (A) Patients with OSA versus healthy subjects without habitual snoring. (B) Patients with OSA with hypertension versus those without hypertension.
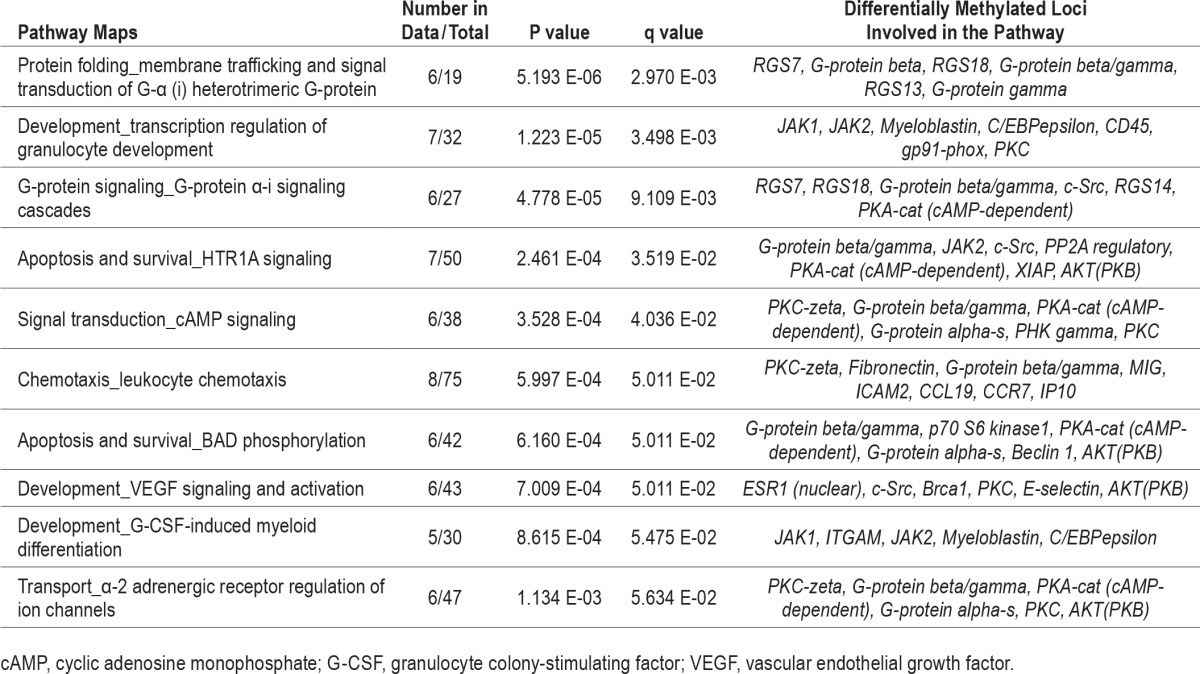
Table 2.
Top 10 pathways enriched in the comparison between patients with OSA and healthy subjects without habitual snoring (comparison I).
Enrichment Pathway Analysis of the Differentially Methylated Genes in the Three Comparisons
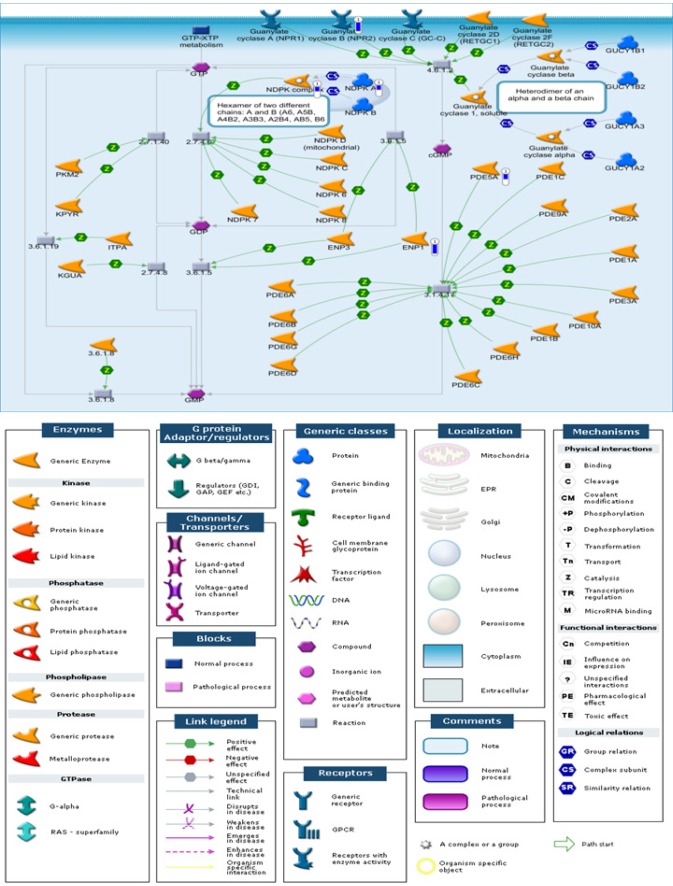
The top-ranking pathways that were enriched in comparison I included membrane trafficking and signal transduction of G-Protein α, transcription regulation of granulocyte development, and G-Protein α-i signaling cascades (Table 3). In addition, the NPR2/cGMP signaling pathway was enriched with five DMLs, all showing hypomethylation (NPR2, PDE5A, ENP1, NDPK complex, NDPK A) in the patients with OSA relative to the healthy subjects (Figure 2). The top-ranking pathways enriched in comparison II included antiapoptotic action of membrane-bound ESR1, transcription of P53 signaling, and immune response of HMGB1 release from the cell (Table S5, supplemental material).
Table 3.
Top differentially methylated loci in the comparison between patients with OSA and healthy subjects without habitual snoring (comparison I).
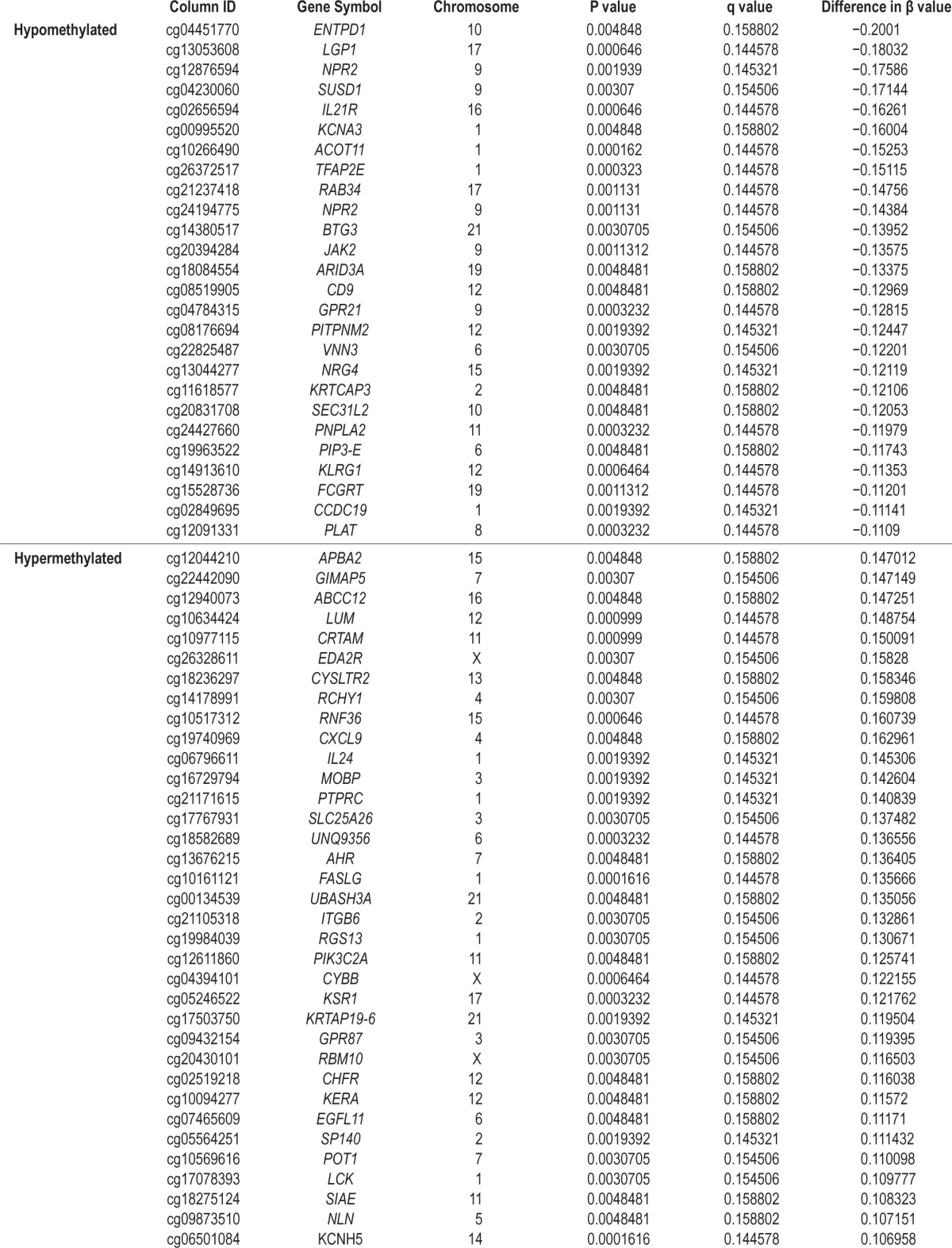
Figure 2.
NPR2 and cGMP signaling pathway enriched in patients with OSA of the discovery cohort. In comparison between patients with OSA and healthy subjects, the significantly hypomethylated genes were highlighted with a blue-colored barometric bar. The changes represent the differences between the median M values of healthy patients and those with OSA. For example, the median M value for healthy patients and those with OSA is 1.27 and 0.37, respectively, for NPR2. Therefore, there is a lower methylation level (−0.9) in patients with OSA, which was shown as a blue bar alongside the NPR2 gene.
Differential DNA Methylation and Corresponding Protein Expression Levels of Four of the Six Selected Genes in the Validation Cohort
DNA methylation levels over the promoter regions of six selected genes with respect to disease severity of OSA and the presence of hypertension or EDS were validated by an independent method of quantification for DNA methylation, pyrosequencing, in the validation cohort of 24 PS subjects and 48 patients with OSA. No significant difference in DNA methylation levels of the six selected genes were found between the PS subjects and the patients with OSA (primary outcome). However, significant correlation between the DNA methylation level and either ODI or ESS was found. Thus, a secondary outcome analysis was performed, and all 72 patients with SDB were reclassified into two groups based on ODI with a cutoff value of 30 events/h, or ESS with a cutoff value of 10.
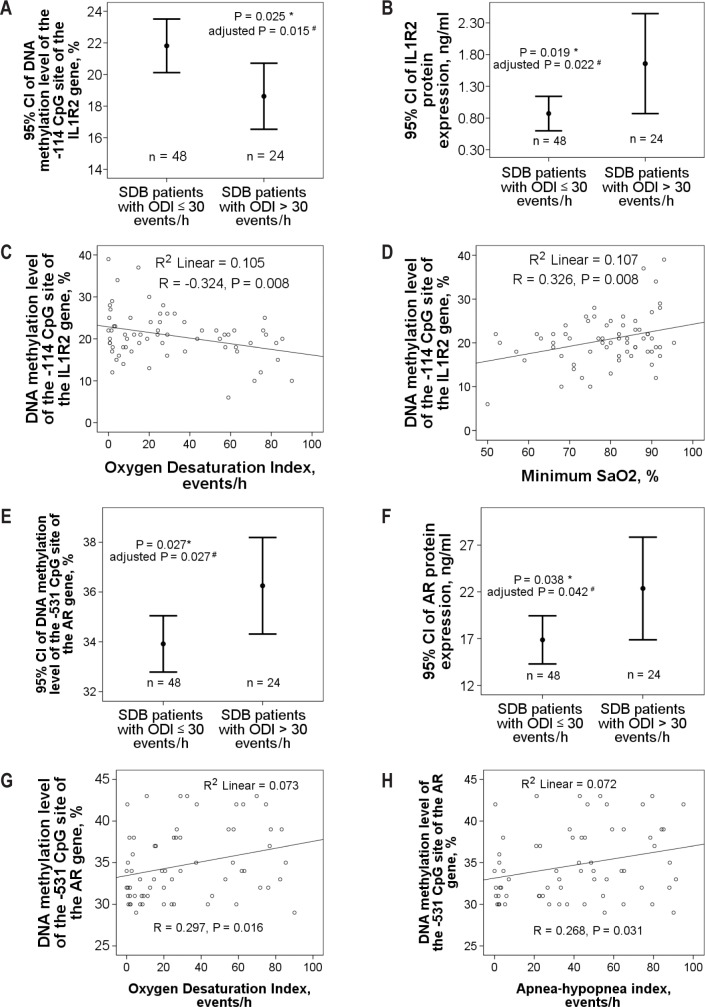
DNA methylation levels over the −114 CpG site of the IL1R2 promoter region were decreased (18.63 ± 4.95% versus 21.81 ± 5.83%, adjusted P = 0.015, 95% CI −7.48 to −0.64, q = 0.067, Figure 3A) and IL1R2 protein expression levels were increased (1.66 ± 1.87 versus 0.87 ± 0.94 ng/mL, adjusted P = 0.022, 95% CI 0.04 to 1.63, q = 0.048, Figure 3B) in the patients SDB with an ODI > 30 as compared to those with an ODI ≤ 30. DNA methylation level over the −114 CpG site of the IL1R2 gene was negatively correlated with ODI (R = −0.324, P = 0.008, Figure 3C) and positively with minimum SaO2 (R = 0.326, P = 0.008, Figure 3D). DNA methylation levels over the −531 CpG site of the AR promoter region (36.25 ± 4.59 versus 33.91 ± 3.85 %, adjusted P = 0.027, 95% CI 0.04 to 4.24, q = 0.067, Figure 3E) and AR protein expression (21.39 ± 12.25 versus 15.69 ± 7.6 ng/mL, adjusted P = 0.042, 95% CI 0.6 to 17.82, q = 0.048, Figure 3F) were both increased in the patients with SDB with an ODI > 30 as compared to those with an ODI 30. DNA methylation level over the −531 CpG site of the AR gene was positively correlated with both ODI (r = 0.297, P = 0.016, Figure 3G) and AHI (R = 0.268, P = 0.031, Figure 3H).
Figure 3.
DNA methylation and protein expression levels of the IL1R2 and AR genes in the validation cohort. (A) DNA methylation levels over −114 CpG site of the IL1R2 promoter region were decreased and (B) IL1R2 protein expression levels were increased in patients with sleep disordered breathing (SDB) with oxygen desaturation index (ODI) > 30 as compared to those with ODI ≤ 30. (C) DNA methylation level over −114 CpG site of the IL1R2 gene was correlated with ODI negatively and (D) minimum SaO2 positively. (E) DNA methylation levels over −531 CpG site of the AR promoter region and (F) AR protein expression were both increased in patients with SDB with ODI > 30 as compared to those with ODI ≤ 30. (G) DNA methylation level over −531 CpG site of the AR gene was positively correlated with both ODI and (H) AHI. *Compared by independent Student t-test. #Adjusted by multivariate linear regression model.
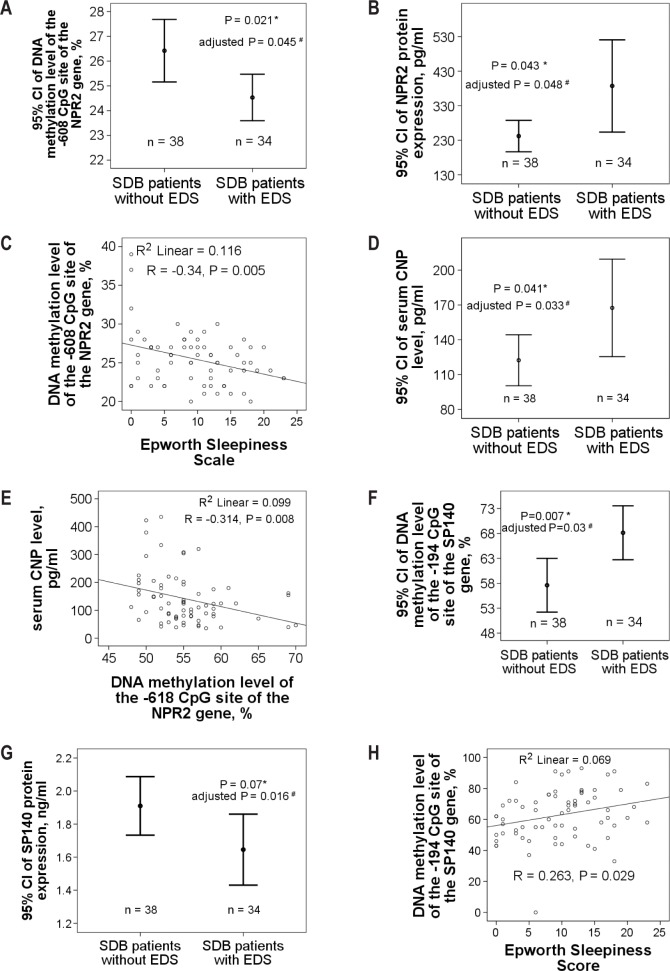
DNA methylation levels over both the −608 CpG site (24.53 ± 2.6% versus 26.42 ± 3.85 %, adjusted P = 0.045, 95% CI −3.12 to −0.08, q = 0.081, Figure 4A) and −618 CpG site (54.13 ± 4.51% versus 56.26 ± 1.26 %, adjusted P = 0.029, 95% CI −5.84 to −0.23, q = 0.067) of the NPR2 promoter region were decreased, whereas the expression level of NPR2 protein was increased (387.13 ± 376.38 versus 242.14 ± 138.31 pg/mL, adjusted P = 0.048, 95% CI 36.9 to 337.3, q = 0.048, Figure 4B) in the patients with SDB and EDS (ESS > 10) as compared to those without EDS. DNA methylation level over the −608 CpG site of the NPR2 gene was negatively correlated with ESS (R = −0.34, P = 0.005, Figure 4C). Because CNP binds to NPR2 to mediate the regulation of endochondral ossification, reproduction, nervous system development, and the maintenance of cardiovascular health,18 we measured serum CNP protein expression levels of the 72 patients with SDB. Serum CNP levels were increased in patients with SDB and EDS as compared to those without EDS (167.47 ± 112.51 versus 122.31 ± 69.6, adjusted P = 0.033, 95% CI 1.85 to 118.86, q = 0.048, Figure 4D). Serum CNP was negatively correlated with the level of DNA methylation over the −618 CpG site of the NPR2 gene (R = −0.314, P = 0.008, Figure 4E), and positively correlated with NPR2 protein expression level (R = 0.284, P = 0.016). DNA methylation level over the −194 CpG site of the SP140 promoter region in patients with SDB and EDS was increased (68.65 ± 15.58% versus 57.72 ± 15.78 %, adjusted P = 0.03, 95% CI 5.0 to 21.02, q = 0.067, Figure 4F), whereas SP140 protein expression levels were decreased (1.66 ± 0.51 versus 1.91 ± 0.6 ng/mL, adjusted P = 0.016, 95% CI −0.66 to −0.07, q = 0.048, Figure 4G) in the patients with SDB and EDS (ESS > 10). DNA methylation level over the −194 CpG site of the SP140 gene was positively correlated with ESS (R = 0.263, P = 0.029, Figure 4H).
Figure 4.
DNA methylation and protein expression levels of the NPR2 and SP140 genes in the validation cohort. (A) DNA methylation levels over both −608 CpG site of the NPR2 promoter region were decreased, (B) whereas NPR2 protein expression levels were increased in patients with sleep disordered breathing (SDB) with excessive daytime sleepiness (EDS) (Epworth Sleepiness Scale [ESS] score > 10) as compared to that in those without EDS. (C) DNA methylation level over −608 CpG site of the NPR2 gene was negatively correlated with ESS. (D) Serum CNP levels were increased in patients with SDB with EDS as compared to that in those without EDS. (E) Serum CNP was negatively correlated with DNA methylation level over −618 CpG site of the NPR2 gene. (F) DNA methylation levels over −194 CpG site of the SP140 promoter region in patients with SDB with EDS were increased, (G) whereas SP140 protein expression levels were decreased in patients with SDB with EDS (ESS > 10) as compared to those without EDS. (H) DNA methylation level over −194 CpG site of the SP140 gene was positively correlated with ESS. *Compared by independent Student t-test. #Adjusted by multivariate linear regression model.
There was no significant difference in the DNA methylation level over either the −1227 CpG site of the IL21R gene or the −438 CpG site of the GPR21 gene between the patients with SDB with an ODI > 30 and those with an ODI ≤ 30, as well as between the patients with SDB and EDS and those without EDS.
DISCUSSION
In this study, we identified and replicated a specific association between ODI and IL1R2 hypomethylation/AR hypermethylation, as well as between EDS and NPR2 hypomethylation/ SP140 hypermethylation in peripheral blood mononuclear cell DNA from patients with SDB. Moreover, we demonstrated their corresponding protein expression changes in three of the four selected genes, indicating a possible repressing effect on gene transcription of these CpG sites. Although some preliminary reports are available on blood DNA methylation changes of several genes in relation to indices of OSA,19,20 this is the first study to perform a large-scale analysis with replication of the principal finding.
Interleukin (IL)1R2, a decoy receptor for IL-1, negatively regulates IL-1 activity by binding IL1β, IL1α, and IL1Ra.21 Bacteremia and anti-inflammatory signals have been reported to enhance IL-1R2 expression, indicating the activation of endogenous pathways of negative regulation of inflammation.22,23 In the current study, we found a negative correlation between methylation levels of the IL1R2 promoter region (−114 CpG site) and ODI in patients with SDB, accompanied by an enhanced IL1R2 protein expression with a higher ODI in the validation cohort. In line with these findings, hypomethylation of the IL1R2 promoter over the same −114 CpG site has been reported to be associated with disease activity of systemic lupus erythematosus and rheumatoid arthritis.24 Additionally, high basal levels of IL1R2 expression have been reported to indicate a lack of response to steroid therapy in patients with autoimmune inner ear disease.23 Furthermore, IL1R2 genetic polymorphisms have recently been reported to be significantly associated with poor sleep maintenance in adults living with human immunodeficiency virus/acquired immunodeficiency syndrome.25 We speculate that DNA methylation changes occurring in the prenatal period or early life may predispose subjects to an epigenotype with decreased methylation and increased expression of the IL1R2 gene, and subsequently a phenotype with more frequent hypoxic events during sleep in adulthood. However, it is equally possible that this may occur in adults as a result of OSA, suggesting that further investigations are required to clarify the cause-and-effect relationship.
Previous studies have shown that inhibition of testosterone action via the 5 alpha-reductase pathway may alleviate breathing instability during sleep, and that testosterone therapy worsens SDB transiently through increasing hyperoxic ventilatory recruitment threshold.26,27 Little is known about the relationship between the AR and OSA, although changes in hypoxia/reoxygenation have been reported to stimulate AR transactivation and sensitization of prostate cancer cells in the presence of androgen.28 Patients with spinal and bulbar muscular atrophy have been shown to have a mutated and gain-of-function AR gene, and increased incidence of OSA.29 In the current study, both AR protein expression and methylation were increased with a higher ODI in patients with SDB. We speculate that an elevated AR expression may predispose subjects to OSA via ventilatory instability, whereas several factors apart from epigenetic regulations may concurrently affect the AR expression. Further studies are required to clarify the effect of AR DNA methylation on sex differences in OSA.
CNP-NPR2 and its signaling pathway connecting to cyclic guanosine monophosphate (cGMP) were found to be involved in the development of OSA and the EDS phenotype in the current study. In addition, both NPR2 and serum CNP expressions were increased, whereas NPR2 methylation was decreased with a higher ESS in the patients with SDB. Consistent with these findings, alterations induced by hyperammonemia on glutamate-nitric oxide-cGMP or NPR2-cGMP pathways in the brain have been reported to result in hepatic encephalopathy characterized by altered sleep-wake patterns and drowsiness.30–32 The second messenger, cGMP, has been reported to possess arousal-promoting activity and to serve as a determinant of wake-promoting output of the circadian system through affecting its downstream protein kinase activity.33,34 Moreover, NPR-2 has been shown to mediate the inhibition of vascular smooth muscle proliferation and migration by CNP in a cGMP-dependent manner.35 The generation of cGMP has been shown to occur through both a nitric oxide synthase (NOS)–soluble guanylate cyclase (sGC) pathway, and NPR1/2-receptor (rGC) pathway.36 Furthermore, phosphodiesterase-5 (PDE5)-mediated cGMP breakdown has been shown to exert a vasoconstrictive effect on coronary resistance vessels through increasing endothelin-1 production.37 The effect of PDE5A inhibitors on improving functional recovery after experimental stroke in rats has been proved.38 However, it has also been reported that the loading-induced CNP/NPR2/cGMP signaling route mediates anabolic events and prevents catabolic activities induced by IL-1β, suggesting additional cytoprotective effects of NPR2.39 Therefore, we speculate that hypomethylation of all of the five DMLs involved in the NPR2 pathway may play a crucial role in the development of the EDS phenotype in OSA. Further investigations are needed to clarify the underlying mechanisms by which the CNP-NPR2-cGMP-PED5A pathway is involved in the development of or resistance to EDS in patients with OSA.
SP140 is an interferon-inducible nuclear leukocyte-specific protein that has been reported to act as an autoantigen in both primary biliary cirrhosis and chronic Kawasaki disease.40,41 Hypermethylation over the −117 CpG site of the SP140 gene has been demonstrated in chronic myelomonocytic leukemia patients with mutant 10-11 translocation oncogene family member 2 and that it results in poor outcomes.42 SP140 has been reported to play a role in interferon response and it has been implicated in innate responses to human immunodeficiency virus type 1.43,44 Decreased protein expression and increased methylation of SP140 over the −194 CpG site were found in SDB patients with EDS in the current study. Previous studies have shown that OSA is associated with altered T helper1:T helper2 balance toward Th1 predominance, leading to interferon-γ secretion, and the interferon type I receptor has been shown to be a determinant of time spent in spontaneous rapid eye movement sleep.45,46 We speculate that SP140 may play a role in the development of EDS via its hypermethylation in the promoter region and possibly under the influence of interferon response, although further investigations are required to elucidate the underlying mechanisms.
There are several limitations to the current study. First, the cause-and-effect relationship could not be determined in this cross-sectional clinical study design, because inherited DNA methylation patterns (epigenotype) may affect the development of disease, and environmental stimuli may cause disease progression through DNA methylation changes. Our preliminary in vitro experiment with peripheral blood mononuclear cell samples from six healthy subjects showed that gene expression and DNA methylation levels of the four selected genes were not altered with 4 d of IHR treatment (7 h of alternative 0% and 21% O2 each day) compared to normoxic conditions (data not shown), indicating that these CpG sites may be the initiators of different phenotypes in OSA but not responders to IHR. However, we acknowledge that we cannot exclude a role for IHR for two reasons: (1) the treatment period was very short (4 d) compared to months/years in patients with OSA; (2) our paradigm for IHR did not mimic exactly cyclical intermittent hypoxia as occurs in humans. Second, DNA methylation and protein expression changes were demonstrated independently in the patients with SDB with different phenotypes (secondary outcome) but not between patients with OSA and PS (primary outcome). Further studies with sufficiently large sample sizes are required for the internal and external validity and the reliability of the results. However, %time < 90% SaO2 has been demonstrated to be the strongest predictor of high-sensitivity C-reactive protein variability in patients with OSA, indicating that the AHI may not reflect the true characteristic of chronic intermittent hypoxia and inflammation.47 Third, gene expression levels of the selected genes were not examined in the peripheral blood mononuclear cell samples of the discovery or validation cohorts because of inadequate RNA samples. However, their protein expressions showed corresponding changes, indicating a potential functional role of these DML in regulating gene expressions. Fourth, hypertension-related DMLs in the discovery cohort, such as IL1R2 and SP140, were shown to be associated with the ODI and EDS, respectively, in the validation cohort. However, these results were not unexpected, because EDS in patients with OSA is a special phenotype, characterized by younger age, higher blood pressure, and more severe hypoxic load.6 We also acknowledge that the study design does not allow an understanding of whether these findings are unique to these outcomes solely in the setting of OSA or whether they represent general differences in those disease states. Fifth, the identified changes in the peripheral blood mononuclear cells may be only partly responsible for the pathogenesis of OSA, and partly mirror differences in other relevant tissues. Further investigation is required to clarify whether these changes can be translated to neurons, endothelium, or other end-organ tissues. Finally, verification and validation of many enriched pathways identified in the discovery phase are ongoing. Among them, the cyclic adenosine monophosphate (cAMP)-protein kinase A signaling-cAMP response element binding protein (CREB) pathway has been reported to play an important role in sleep-wake control, hippocampal neuronal plasticity, and memory processes.48
In summary, we reported a novel association of increased ODI and ESS in adults of Asian origin with aberrant DNA methylation in the promoter regions of the IL1R2, AR, NPR2, and SP140 genes in blood immune cells. The findings extend reports linking IL1R2 and AR with the frequency of hypoxic events in patients with SDB, and provide direct evidence that perturbation of CNP/NPR2/cGMP/PDE5A or SP140 signaling through epigenetic programming may play an important role in the mediation of EDS in OSA.
DISCLOSURE STATEMENT
This was not an industry supported study. This work was supported by grants from the National Science Council, Taiwan (NMRPD1A1331/100-2314-B-182-070, NMRPG8B6191/101-2314-B-182A-130-MY2, & NMRPG8B6192/101-2314-B-182A-130-MY2 to M.C. Lin) and from Chang Gung Memorial Hospital (CMRPG8C1181 to M.C. Lin) Taiwan. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors acknowledge the technical support provided by the Genomic and Proteomic Core Laboratory, and the Internal Medicine Core Facility of the Kaohsiung Chang Gung Memorial Hospital.
ABBREVIATIONS
- AHI
apnea hypopnea index
- AR
androgen receptor
- BMI
body mass index
- CNP
type C natriuretic peptide
- CpG
cytosine followed by guanine dinucleotide sequence
- DML
differentially methylated loci
- EDS
excessive daytime sleepiness
- IHR
intermittent hypoxia with re-oxygenation
- IL1R2
interleukin 1 receptor 2
- NPR2
natriuretic peptide receptor 2
- ODI
oxygen desaturation index
- OSA
obstructive sleep apnea
- %time < 90% SaO2
percentage of total minutes of recording time with oxyhemoglobin saturation < 90%
- PS
primary snoring
- SDB
sleep disordered breathing
- SP140
speckled protein 140
REFERENCES
- 1.Arnardottir ES, Mackiewicz M, Gislason T, Teff KL, Pack AI. Molecular signatures of obstructive sleep apnea in adults: a review and perspective. Sleep. 2009;32:447–70. doi: 10.1093/sleep/32.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavie L. Oxidative stress in obstructive sleep apnea and intermittent hypoxia - revisited - the bad ugly and good: implications to the heart and brain. Sleep Med Rev. 2015;20:27–45. doi: 10.1016/j.smrv.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Dyugovskaya L, Polyakov A, Lavie P, Lavie L. Delayed neutrophil apoptosis in patients with sleep apnea. Am J Respir Crit Care Med. 2008;177:544–54. doi: 10.1164/rccm.200705-675OC. [DOI] [PubMed] [Google Scholar]
- 4.Dyugovskaya L, Polyakov A, Ginsberg D, Lavie P, Lavie L. Molecular pathways of spontaneous and TNF-{alpha}-mediated neutrophil apoptosis under intermittent hypoxia. Am J Respir Cell Mol Biol. 2011;45:154–62. doi: 10.1165/rcmb.2010-0025OC. [DOI] [PubMed] [Google Scholar]
- 5.Mohsenin V. Obstructive sleep apnea and hypertension: a critical review. Curr Hypertens Rept. 2014;16:482. doi: 10.1007/s11906-014-0482-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Zhang C, Jia P, et al. The association between the phenotype of excessive daytime sleepiness and blood pressure in patients with obstructive sleep apnea-hypopnea syndrome. Int J Med Sci. 2014;11:713–20. doi: 10.7150/ijms.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, Tao L, Nie P, et al. Association between 5-HT2A receptor polymorphisms and risk of obstructive sleep apnea and hypopnea syndrome: a systematic review and meta-analysis. Gene. 2013;530:287–94. doi: 10.1016/j.gene.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Varvarigou V, Dahabreh IJ, Malhotra A, Kales SN. A review of genetic association studies of obstructive sleep apnea: field synopsis and meta-analysis. Sleep. 2011;34:1461–8. doi: 10.5665/sleep.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redline S, Tishler PV, Tosteson TD, et al. The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med. 1995;151:682–7. doi: 10.1164/ajrccm/151.3_Pt_1.682. [DOI] [PubMed] [Google Scholar]
- 10.Nanduri J, Makarenko V, Reddy VD, et al. Epigenetic regulation of hypoxic sensing disrupts cardiorespiratory homeostasis. Proc Natl Acad Sci U S A. 2012;109:2515–20. doi: 10.1073/pnas.1120600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Q, Xiong F, Zhang L. Gestational hypoxia and epigenetic programming of brain development disorders. Drug Discov Today. 2014;19:1883–96. doi: 10.1016/j.drudis.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YC, Su MC, Liou CW, et al. Co-upregulation of Toll-like receptors 2 and 6 on peripheral blood cells in patients with obstructive sleep apnea. Sleep Breath. 2015;19:873–82. doi: 10.1007/s11325-014-1116-4. [DOI] [PubMed] [Google Scholar]
- 13.Bibikova M, Le J, Barnes B, et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 14.Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson MD, Kahraman A, Law CW, et al. Statistical methods for detecting differentially methylated loci and regions. Front Genet. 2014;5:324. doi: 10.3389/fgene.2014.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10:949–55. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 17.Chen YC, Hsiao CC, Chen CJ, et al. Aberrant Toll-like receptor 2 promoter methylation in blood cells from patients with pulmonary tuberculosis. J Infect. 2014;69:546–57. doi: 10.1016/j.jinf.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Schulz S. C-type natriuretic peptide and guanylyl cyclase B receptor. Peptides. 2005;26:1024–34. doi: 10.1016/j.peptides.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Bhattacharjee R, Khalyfa A, et al. DNA methylation in inflammatory genes among children with obstructive sleep apnea. Am J Respir Crit Care Med. 2012;185:330–8. doi: 10.1164/rccm.201106-1026OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kheirandish-Gozal L, Khalyfa A, Gozal D, Bhattacharjee R, Wang Y. Endothelial dysfunction in children with obstructive sleep apnea is associated with epigenetic changes in the eNOS gene. Chest. 2013;143:971–7. doi: 10.1378/chest.12-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garlanda C, Riva F, Bonavita E, Mantovani A. Negative regulatory receptors of the IL-1 family. Semin Immunol. 2013;25:408–15. doi: 10.1016/j.smim.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Thompson CM, Park CH, Maier RV, O'Keefe GE. Traumatic injury, early gene expression, and gram-negative bacteremia. Critical Care Med. 2014;42:1397–405. doi: 10.1097/CCM.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vambutas A, DeVoti J, Goldofsky E, Gordon M, Lesser M, Bonagura V. Alternate splicing of interleukin-1 receptor type II (IL1R2) in vitro correlates with clinical glucocorticoid responsiveness in patients with AIED. PloS One. 2009;4:e5293. doi: 10.1371/journal.pone.0005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin SY, Hsieh SC, Lin YC, et al. A whole genome methylation analysis of systemic lupus erythematosus: hypomethylation of the IL10 and IL1R2 promoters is associated with disease activity. Genes Immun. 2012;13:214–20. doi: 10.1038/gene.2011.74. [DOI] [PubMed] [Google Scholar]
- 25.Lee KA, Gay C, Pullinger CR, Hennessy MD, Zak RS, Aouizerat BE. Cytokine polymorphisms are associated with poor sleep maintenance in adults living with human immunodeficiency virus/acquired immunodeficiency syndrome. Sleep. 2014;37:453–63. doi: 10.5665/sleep.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Killick R, Wang D, Hoyos CM, Yee BJ, Grunstein RR, Liu PY. The effects of testosterone on ventilatory responses in men with obstructive sleep apnea: a randomised, placebo-controlled trial. J Sleep Res. 2013;22:331–6. doi: 10.1111/jsr.12027. [DOI] [PubMed] [Google Scholar]
- 27.Chowdhuri S, Bascom A, Mohan D, Diamond MP, Badr MS. Testosterone conversion blockade increases breathing stability in healthy men during NREM sleep. Sleep. 2013;36:1793–8. doi: 10.5665/sleep.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SY, Kim YJ, Gao AC, et al. Hypoxia increases androgen receptor activity in prostate cancer cells. Cancer Res. 2006;66:5121–9. doi: 10.1158/0008-5472.CAN-05-1341. [DOI] [PubMed] [Google Scholar]
- 29.Romigi A, Liguori C, Placidi F, et al. Sleep disorders in spinal and bulbar muscular atrophy (Kennedy's disease): a controlled polysomnographic and self-reported questionnaires study. J Neurol. 2014;261:889–93. doi: 10.1007/s00415-014-7293-z. [DOI] [PubMed] [Google Scholar]
- 30.Konopacka A, Fresko I, Piaskowski S, Albrecht J, Zielinska M. Ammonia affects the activity and expression of soluble and particulate GC in cultured rat astrocytes. Neurochem Int. 2006;48:553–8. doi: 10.1016/j.neuint.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 31.Konopacka A, Zielinska M, Albrecht J. Ammonia inhibits the C-type natriuretic peptide-dependent cyclic GMP synthesis and calcium accumulation in a rat brain endothelial cell line. Neurochem Int. 2008;52:1160–6. doi: 10.1016/j.neuint.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Monfort P, Munoz MD, ElAyadi A, Kosenko E, Felipo V. Effects of hyperammonemia and liver failure on glutamatergic neurotransmission. Metab Brain Dis. 2002;17:237–50. doi: 10.1023/a:1021993431443. [DOI] [PubMed] [Google Scholar]
- 33.Ribeiro AC, Kapas L. The effects of intracerebroventricular application of 8-Br-cGMP and LY-83,583, a guanylyl cyclase inhibitor, on sleep-wake activity in rats. Brain Res. 2005;1049:25–33. doi: 10.1016/j.brainres.2005.04.067. [DOI] [PubMed] [Google Scholar]
- 34.Langmesser S, Franken P, Feil S, Emmenegger Y, Albrecht U, Feil R. cGMP-dependent protein kinase type I is implicated in the regulation of the timing and quality of sleep and wakefulness. PloS One. 2009;4:e4238. doi: 10.1371/journal.pone.0004238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahmutula D, Gardner DG. C-type natriuretic Peptide down-regulates expression of its cognate receptor in rat aortic smooth muscle cells. Endocrinology. 2005;146:4968–74. doi: 10.1210/en.2005-0262. [DOI] [PubMed] [Google Scholar]
- 36.Zhang M, Kass DA. Phosphodiesterases and cardiac cGMP: evolving roles and controversies. Trends Pharmacol Sci. 2011;32:360–5. doi: 10.1016/j.tips.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z, de Beer VJ, Bender SB, et al. Phosphodiesterase-5 activity exerts a coronary vasoconstrictor influence in awake swine that is mediated in part via an increase in endothelin production. Am J Physiol Heart Circ Physiol. 2014;306:H918–27. doi: 10.1152/ajpheart.00331.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menniti FS, Ren J, Sietsma DK, et al. A non-brain penetrant PDE5A inhibitor improves functional recovery after stroke in rats. Restor Neurol Neurosci. 2012;30:283–9. doi: 10.3233/RNN-2012-110187. [DOI] [PubMed] [Google Scholar]
- 39.Peake N, Su N, Ramachandran M, et al. Natriuretic peptide receptors regulate cytoprotective effects in a human ex vivo 3D/bioreactor model. Arthritis Res Ther. 2013;15:R76. doi: 10.1186/ar4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Granito A, Yang WH, Muratori L, et al. PML nuclear body component Sp140 is a novel autoantigen in primary biliary cirrhosis. Am J Gastroenterol. 2010;105:125–31. doi: 10.1038/ajg.2009.596. [DOI] [PubMed] [Google Scholar]
- 41.Reindel R, Bischof J, Kim KY, et al. CD84 is markedly up-regulated in Kawasaki disease arteriopathy. Clin Exper Immunol. 2014;177:203–11. doi: 10.1111/cei.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki J, Taby R, Vasanthakumar A, et al. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics. 2012;7:201–7. doi: 10.4161/epi.7.2.19015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regad T, Chelbi-Alix MK. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–86. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 44.Madani N, Millette R, Platt EJ, et al. Implication of the lymphocytespecific nuclear body protein Sp140 in an innate response to human immunodeficiency virus type 1. J Virol. 2002;76:11133–8. doi: 10.1128/JVI.76.21.11133-11138.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bohnet SG, Traynor TR, Majde JA, Kacsoh B, Krueger JM. Mice deficient in the interferon type I receptor have reduced REM sleep and altered hypothalamic hypocretin, prolactin and 2',5'-oligoadenylate synthetase expression. Brain Res. 2004;1027:117–25. doi: 10.1016/j.brainres.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 46.Tan HL, Gozal D, Wang Y, et al. Alterations in circulating T-cell lymphocyte populations in children with obstructive sleep apnea. Sleep. 2013;36:913–22. doi: 10.5665/sleep.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang XB, Zen HQ, Lin QC, Chen GP, Chen LD, Chen H. TST, as a polysomnographic variable, is superior to the apnea hypopnea index for evaluating intermittent hypoxia in severe obstructive sleep apnea. Eur Arch Otorhinolaryngol. 2014;271:2745–50. doi: 10.1007/s00405-014-3044-0. [DOI] [PubMed] [Google Scholar]
- 48.Kreutzmann JC, Havekes R, Abel T, Meerlo P. Sleep deprivation and hippocampal vulnerability: changes in neuronal plasticity, neurogenesis and cognitive function. Neuroscience. 2015;309:173–90. doi: 10.1016/j.neuroscience.2015.04.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.