Abstract
Background
Chickens and ducks are major hosts of Newcastle disease virus (NDV) with distinct responses to infection. However, whereas ducks are generally asymptomatic or exhibit only mild symptoms following NDV infection and are thus regarded as potential long-term reservoirs of the virus, chickens exhibit severe clinical lesions, transient infections and even death due to NDV infection. These differences may in part result from the host innate immune response to NDV infection.
Methods
To better understand the host innate immune response to NDV infection in avian species, by using the quantitative real-time polymerase chain reaction method we examined the messenger RNA expression levels of immune-related genes in chicken embryonic fibroblasts (CEFs) and duck embryonic fibroblasts (DEFs) when infected with NDV of different pathogenicities.
Results
Gene expression profiles showed that the expression of IL-1beta, TNF-α-like factor (LITAF) and interferon (IFN)-beta was upregulated in both CEFs and DEFs infected with SS-10 and NH-10 viruses or treated with polyinosinic:polycytidylic acid [poly(I:C)], as well as that expression levels were greater in CEFs than in DEFs. The expression of TLR3, TLR7, IL-6, IFN-alpha, IFN-gamma, MHC-I and MHC-II, except for IL-8, were also greater in CEFs than in DEFs in response to infection to both viruses or treatment with poly(I:C). However, unlike moderate virulent NH-10, highly virulent SS-10 induced greater pattern recognition receptors and cytokines, except for IFNs, in CEFs and DEFs.
Conclusion
Results show distinct expression patterns of cytokines, Toll-like receptors and IFNs associated with inflammatory immune responses to NDV between species and by virulence.
Background
Caused by Newcastle disease virus (NDV), Newcastle disease (ND) is one of the most critical diseases in poultry and wild birds, largely due to its high morbidity and mortality, as well as its worldwide distribution and threat of considerable economic losses to avian industries [1]. NDV is a negative-sense, single-stranded and enveloped RNA virus with approximately 15.2 kb genome composed of six genes encoding at least six structural proteins and additional non-structural proteins—namely, hemagglutinin–neuraminidase (HN), nucleoprotein (NP), fusion (F), phosphoprotein (P), matrix (M), RNA-dependent RNA polymerase (L) [2], V, and possibly the W protein, produced by RNA editing of the P coding region [3, 4]. With a wide range of hosts, NDV is known to infect at least 250 bird species through either experimental or natural routes [1]. Given the cleavage site of the F protein and due to the severity of disease, NDV strains in 1-d-old specific pathogen-free (SPF) chickens are categorized as highly virulent (i.e., velogenic), intermediate virulent (i.e., mesogenic) or nonvirulent (i.e., lentogenic) [1, 5] according to the Intracerebral Pathogenicity Index.
Wild waterfowl and shorebirds can act as reservoir hosts, as well as hosts by which viruses with pandemic potential are known to be effectively transmitted to other avian species, and have thus gained attention with the emergence and perpetuation of virulent NDV through serial passage in susceptible animals [6–9]. Even so, few studies have addressed viral pathogenesis and host innate immune response in avian species, thereby leaving gaps in the knowledge of NDV in avian hosts. In particular, chickens and ducks respond to virulent NDV infections differently, and many cases have demonstrated that infection with a specific virulent NDV strain may cause lesions and even death in chickens, whereas a duck infected with the same virus would be asymptomatic and rarely die due to the infection [10, 11]. Moreover, NDV shedding in infected chickens is transient and involves the host’s rapid clearance [12, 13], whereas infected ducks exhibit intermittent, prolonged shedding [11]. Another difference is chickens’ greater probability than ducks of an earlier, stronger humoral immune response to NDV infection [14]. Furthermore, though previous studies have shown that NDV replicates preferentially in both specifies’ respiratory systems and lymphoid tissues, including the lungs, spleen, thymus and bursa of Fabricius [10, 11], only in ducks does NDV’s distribution remain limited to lymphoid tissues [15]. Perhaps more significantly, though having adapted efficient replication in chickens, NDV does not always replicate in ducks, yet depends on its adaptation to different hosts and vice versa. However, to our knowledge, very few studies have compared the viral pathogenesis of or host innate immune responses to the same NDV in chicken and duck embryonic fibroblasts.
At the cellular level, a host’s recognition of viruses is mediated by Toll-like receptors (TLRs), such as TLR3 and TLR7, which recognize viral components and activate intracellular signal transduction pathways. Those processes result in the production of antiviral cytokines such as type I interferons (IFNs) and proinflammatory cytokines and chemokine, including IL-6 and IL-1beta, as well as major histocompatibility complexes (MHC) that support host defenses against clearance of viruses [16]. MHCs of classes I and II exhibit an antigen presentation associated with cell-mediated immunity (CMI) that plays an important role in defending T lymphocytes (e.g., cytokine-secreting CD4+ T helper cells and CD8+ cytotoxic T lymphocytes) against viral infection and is essential for viral clearance [17, 18]. Previous studies have reported that in MHC class I and II molecules, pattern recognition receptor (PRRs) and antiviral cytokines were involved in the host innate immune response of avian species, including chickens and ducks, when infected with NDV [19–21]. Nevertheless, very few studies have compared the induction and role of MHC class I and II molecules, PRRs and antiviral cytokines in avian embryo fibroblasts when infected with NDVs of different pathogenicities.
For this study, we selected a model of chicken embryo fibroblasts (CEFs) and duck embryo fibroblasts (DEFs) to observe host innate immune responses in vitro following infection with NDVs of different pathogenicities. To better understand the host immune responses and mechanisms supporting the different pathogeneses of NDV infection of two the highly relevant avian species of chickens and ducks, we compared the expression of cytokines and PRRs, including TLRs and proinflammatory and antiviral cytokines, in response to NDV infection, all with the quantitative real-time polymerase chain reaction (qRT-PCR) method. With that same method, we also examined cell-mediated immune responses and MHC class I and II molecules in CEFs and DEFs.
Methods
Ethics statement
This experiment was conducted with the approval of the South China Agricultural University Experimental Animal Welfare Ethics Committee (permit no. 2015–08).
Cell lines and virus
CEFs and DEFs were obtained from 10-d-old SPF chicken embryos and 11-d-old Pekin duck embryos (South China Agricultural University, Guangzhou, China) as previously described [22]. The CEFs and DEFs were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10 % fetal bovine serum (FBS) (Gibco), 100 U/mL penicillin and 100 ug/mL streptomycin at 37 °C with 5 % CO2 until cell density reached approximately 80 % confluence. The two NDV strains used were Duck/CH/GD/SS/10 (SS-10) and Duck/CH/GD/NH/10 (NH-10), genotypes VII and IX, respectively, both duck-adapted viruses characterized well in the Key Laboratory of Animal Disease Control and Prevention of the Ministry of Agriculture, College of Veterinary Medicine [11]. The viruses were inoculated into the allantoic cavity of 10-d-old SPF-embryonated chicken eggs at 37 °C for 3 d according to the standard procedures of the Office International Des Epizooties [1]. Fresh allantoic fluid was collected and stored at −80 °C until used, and virus titers were quantified by plaque assay with both CEFs and DEFs [23].
Growth characteristics of the two NDVs in CEFs and DEFs
To determine the multicycle growth kinetics of SS-10 and NH-10, CEFs and DEFs in triplicate wells of six-well plates were infected with either virus at a multiplicity of infection (moi) of 1. Following 1 h of adsorption, the cells were washed and covered with DMEM containing 2 % heat-inactivated FBS at 37 °C and 5 % CO2. Cell culture supernatant samples were collected and replaced with an equal volume of fresh medium at 6, 12, 24, 36, 48 and 60 h post-infection (p.i.). Virus titers were quantified by a plaque assay on CEFs as previously described [23].
Virus infection
CEFs and DEFs were seeded 16 h prior to infection in triplicate wells of six-well plates at a cell density of approximately 1.7 × 105 cells/well. The cells were infected with genotype VII SS-10 and genotype IX NH-10, both predominant duck-origin genotypes of NDV strains circulating in Guangdong Province, at an moi of 1 and incubated at 37 °C in a humidified atmosphere containing 5 % CO2 for 1 h. Afterward, the growth medium was replaced with DMEM supplemented with 2 % heat-inactivated FBS. Mock-infected cells were regarded as negative controls. CEFs and DEFs were treated with the double-stranded RNA (dsRNA) analog poly(I:C) (Sigma–Aldrich) at a concentration of 20 ug/mL and used as positive controls. At 0, 6, 24 and 36 h p.i., cell monolayers were harvested and stored at −80 °C for RNA extraction.
RNA extraction and cDNA synthesis
Total RNA was extracted from the infected and negative control embryo fibroblast cells harvested at each time point using the RNeasy Mini RNA Purification Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Viral RNA was extracted from culture supernatants by using an RNA extraction kit (Takara, Japan). RNA in each sample was quantified using an Ultrospec 2000 mass spectrophotometer (Pharmacia Biotech, Uppsala, Sweden). Approximately 2 ug RNA from each sample was treated with DNase to remove genomic DNA and was later reverse-transcribed to cDNA using the SuperScript® III First-Strand Synthesis System (Clontech) according to the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The qRT-PCR method was applied in a final volume of 25 uL using a QuantiFast SYBR Green PCR Master Mix Kit (Qiagen) with specific primers designed with Oligo 7 software (http://www.oligo.net/) based on published sequences [24]. Primers were developed for IL-1beta, lipopolysaccharide-induced TNF-α-factor (LITAF), IL-2, IL-6, IFN types I and II (IFN-alpha, IFN-beta and IFN-gamma), MHC class I and II molecules, TLR3, TLR7 and IL-8, all derived from published sequences. Predicted product sizes are shown in Table 1. All qRT-PCR assays were conducted using the ABI Prism 7500 Fast Real-Time PCR System (Applied Biosystems), which involved predegeneration for 2 min at 50 °C and initial denaturation for 30 s at 95 °C, followed by 40 cycles of 5 s at 94 °C and 34 s at 60 °C, as well as a melt-curve analysis to confirm the specificity of the SYBR green PCR signal. A one-step qRT-PCR assay of viral RNA using NDV P/V/W gene-specific primers was performed as previously described. Cycle threshold (CT) values were converted to viral gene copy numbers following a standard curve generated using cDNA. To rule out genomic contamination, control qRT-PCRs were performed in the absence of reverse transcriptase. Amplified products were run on a gel and extracted using a QIAEX II DNA gel extraction kit (Qiagen, Germany) according to the manufacturer’s instructions. To validate assays, purified products were subcloned into pMD19-T with a TA cloning kit (Clontech, Japan) and sequenced for verification using M13 forward and reverse primers.
Table 1.
Primer sequences for quantitative real-time polymerase chain reaction
| RNA target | Forward primer (5' → 3') | Reverse primer (5' → 3') | Product size (bp) | GenBank accession no. |
|---|---|---|---|---|
| Chicken | ||||
| GAPDH | CCTCTCTGGCAAAGTCCAAG | CATCTGCCCATTTGATGTTG | 200 | NM_204305 |
| TLR3 | ACAATGGCAGATTGTAGTCACCT | GCACAATCCTGGTTTCAGTTTAG | 123 | NM_001011691 |
| TLR7 | TGTGATGTGGAAGCCTTTGA | ATTATCTTTGGGCCCCAGTC | 218 | DQ780342 |
| IL-1β | GCTCTACATGTCGTGTGTGATGAG | TGTCGATGTCCCGCATGA | 80 | NM204524 |
| IL-6 | CCTGTTCGCCTTTCAGACCT | GGGATGACCACTTCATCGGG | 171 | EU170468 |
| IL-8 | ATTCAAGATGTGAAGCTGAC | AGGATCTGCAATTAACATGAGG | 196 | DQ393272 |
| LITAF | CCGCCCAGTTCAGATGAGTT | GCAACAACCAGCTATGCACC | 130 | AY765397 |
| IFN-a | ATGCCACCTTCTCTCACGAC | AGGCGCTGTAATCGTTGTCT | 387 | EU367971 |
| IFN-r | TGAGCCAGATTGTTTCGATG | CTTGGCCAGGTCCATGATA | 248 | DQ906156 |
| MHC-I | AAGAAGGGGAAGGGCTACAA | AAGCAGTGCAGGCAAAGAAT | 222 | NM001031338 |
| MHC-II | CTCGAGGTCATGATCAGCAA | TGTAAACGTCTCCCCTTTGG | 312 | DQ008588 |
| Duck | ||||
| GAPDH | ATGTTCGTGATGGGTGTGAA | CTGTCTTCGTGTGTGGCTGT | 176 | AY436595 |
| TLR3 | GAGTTTCACACAGGATGTTTAC | GTGAGATTTGTTCCTTGCAG | 200 | NM_001310782 |
| TLR7 | CCTTTCCCAGAGAGCATTCA | TCAAGAAATATCAAGATAATCACATCA | 154 | AY940195 |
| IL-1β | TCGACATCAACCAGAAGTGC | GAGCTTGTAGCCCTTGATGC | 185 | DQ393268 |
| IL-6 | TTCGACGAGGAGAAATGCTT | CCTTATCGTCGTTGCCAGAT | 150 | AB191038 |
| IL-8 | AAGTTCATCCACCCTAAATC | GCATCAGAATTGAGCTGAGC | 174 | AB236334 |
| LITAF | ACAGGACAGCCTATGCCAAC | CATCTGAACTGGGCGGTCAT | 96 | EU375296 |
| IFN-a | TCCTCCAACACCTCTTCGAC | GGGCTGTAGGTGTGGTTCTG | 232 | EF053034 |
| IFN-r | GCTGATGGCAATCCTGTTTT | GGATTTTCAAGCCAGTCAGC | 247 | AJ012254 |
| MHC-I | GAAGGAAGAGACTTCATTGCCTTGG | CTCTCCTCTCCAGTACGTCCTTCC | 196 | AB115246 |
| MHC-II | CCACCTTTACCAGCTTCGAG | CCGTTCTTCATCCAGGTGAT | 229 | AY905539 |
| Chicken and Duck IFN-β | CCTCAACCAGATCCAGCATT | GGATGAGGCTGTGAGAGGAG | 259 | AY831397 |
Data and statistical analysis
The cDNA sample of each CEF and DEF was tested in triplicate. Relative expression levels were calculated according to the 2-△△CT method, which involved using glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) as the endogenous control to normalize the level of target gene expression [25]. Data were expressed as M ± SD. Growth characteristics of each group were analyzed using an unpaired Student’s t-test, and differences between the means of the CEF and DEF target genes were evaluated using two-way analysis of variance (ANOVA) followed by Duncan’s test. All p values less than .05 were considered to be statistically significant. Statistical analyses for M, two-way ANOVA and SD were conducted by using Prism 6 (GraphPad Software, Inc., San Diego, CA, USA).
Results
Replication kinetics of CEFs and DEFs infect with SS-10 and NH-10
The multicycle growth kinetics and replication magnitude of SS-10 and NH-10 were determined in CEFs and DEFs by using a plaque assay. CEFs and DEFs were inoculated with each virus at a moi of 1, and cell supernatants were harvested at the time points indicated. As shown in Fig. 1a and b, SS-10 replicated more efficiently and had a significantly higher titer in CEFs and DEFs than NH-10 at each time point, though both viruses achieved similar maximum titers at 36 h p.i. On the whole, the virus titers of both strains were higher in CEFs than in DEFs during the 60 h of testing (Fig. 1a and b). Additionally, CEFs and DEFs were infected with SS-10 and NH-10 at a moi of 1 over a period of 36 h. Normalized to the endogenous control, viral P/V/W gene RNA accumulation in DEFs was consistently less than that in the corresponding CEFs for two NDV isolates (Fig. 1c and d).
Fig. 1.
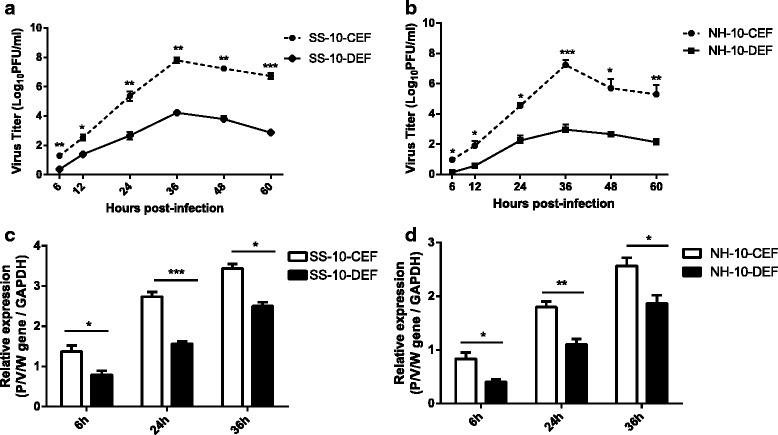
Comparison of multicycle growth kinetics and progeny virus outputs of SS-10 and NH-10 strains on chicken embryo fibroblasts (CEFs) and duck embryo fibroblasts (DEFs). CEFs and DEFs were inoculated with SS-10 (a) and NH-10 (b) at a moi of 1, and cell supernatant was sampled at the time points indicated. Virus titers in the cell supernatant were determined in CEFs via plaque assay. SS-10 (c) and NH-10 (d) at a moi of 1 conferred higher levels of accumulation of P/V/W gene RNA in CEFs than in DEFs. Values represent averages of the results from three independent experiments with standard error bars. *p < .05, **p < .01, ***p < .001, as determined by an unpaired Student’s t-test
Expression of TLR genes in NDV-infected CEFs and DEFs
We compared the expression levels of TLR genes TLR3 and TLR7 in CEFs and DEFs infected with SS-10 and NH-10 at 6, 24 and 36 h p.i. Compared to those in mock-infected samples, the expression levels of TLR3 and TLR7 in CEFs were upregulated at 6 h p.i. and peaked at 24 h p.i., with the exception of TLR7’s expression level induced by SS-10 at 36 h p.i. Thereafter, the levels decreased slightly at 36 h p.i. when either infected with both viruses or treated with poly(I:C), as shown in Fig. 2a and b. In DEFs, the expression levels of TLR3 and TLR7 induced by both viruses and positive control poly(I:C) clearly exhibited patterns of expression. For TLR3, the expression level was downregulated at 6 and 24 h p.i. (0.85- and 0.75-fold, respectively) and upregulated at 36 h p.i. (1.46-fold) in response to SS-10 infection, whereas the expression level of TLR3 was upregulated at 6 h p.i. and downregulated at 24 and 36 h p.i. following infection with NH-10 or after stimulation with poly(I:C) (Fig. 2a). Meanwhile, the expression level of TLR7 was upregulated throughout the period of infection, except at 36 h p.i. induced by NH-10, in response to infection with both viruses or treatment with poly(I:C) (Fig. 2b). Notably, the expression levels of TLR genes TLR3 and TLR7 in CEFs induced by both viruses and poly(I:C) were greater than in DEFs throughout the testing period, whereas TLR7 was slightly increased in DEFs at 24 h p.i. following stimulation with dsRNA analogs (poly(I:C)).
Fig. 2.
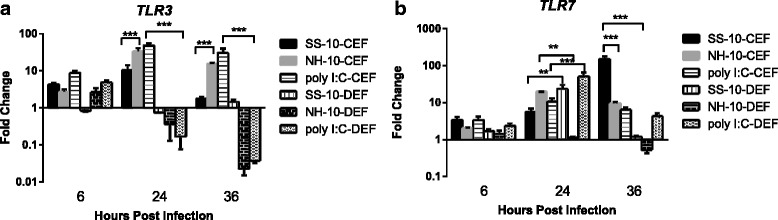
The expression of TLRs TLR3 (a) and TLR7 (b) in chicken embryo fibroblasts (CEFs) and duck embryo fibroblasts (DEFs) following infection with SS-10 and NH-10 or treatment with poly(I:C) at 6, 24 and 36 h p.i. Data represent the mean fold change expression of either CEFs and DEFs compared with mock-infected controls after normalization to the expression of the housekeeping gene glyceraldehyde-3-phosphate-dehydrogenase. Significance was analyzed with two-way analyses of variance between the SS-10 and NH-10 groups in CEFs and DEFs at each time point (*p < .05, *p < .01, *p < .001). Error bars represent SD
Differential expression of proinflammatory cytokines and chemokines in CEFs and DEFs infected with NDV of different pathogenicities
To compare proinflammatory and Th1-associated cytokines and chemokines in CEFs and DEFs infected by SS-10 and NH-10, cytokines and chemokines such as IL-1beta, IL-6, IL-8 and LITAF were measured early during either NDV infection or treatment with poly(I:C). Compared to uninfected cells, CEFs and DEFs exhibited upregulated expression levels of IL-1beta and LITAF in response to infection with SS-10 and NH-10 or treatment with poly(I:C) during the testing period (Fig. 3a and d), though the expression levels of IL-1beta and LITAF showed different expression patterns at each time point for both infected CEFs and DEFs (Fig. 3a and d). In CEFs, the expression of both IL-1beta and LITAF was upregulated when induced by infection with SS-10 and NH-10 at all time points and peaked at 24 h p.i. (22.87- and 12.66-fold versus 319.48- and 21.13-fold, respectively), but was greater for SS-10 than NH-10. Albeit also upregulated, IL-1beta expression in DEFs was induced at a slightly weaker level and peaked at 36 h p.i. (17.93- and 7.88-fold, respectively), induced by both viruses. LITAF expression in DEFs was also induced 6.93- and 4.84-fold at 6 h p.i., respectively, decreased slightly at 24 h p.i. (3.71- and 4.25-fold, respectively) and peaked at 36 h p.i. (12.33- and 6.86-fold, respectively), when infected with SS-10 and NH-10, yet was induced at a far lower level than with CEFs during the observation period (Fig. 3d). The expression level of IL-6 was upregulated in CEFs during the period of infection, peaking at 36 h p.i. and 8 h p.i. (88.22-, 7.60- and 21.21-fold, respectively) in response to infection with SS-10 and NH-10 or treatment with poly(I:C) compared to uninfected CEFs. However, it was downregulated at 6 and 24 h p.i. and maintained baseline level at 36 h p.i. in DEFs induced by SS-10. It was moreover upregulated 2.49-fold at 6 h p.i. and downregulated between 24 and 36 h p.i. (0.56- and 0.34-fold, respectively) in DEFs induced by NH-10 (Fig. 3b). Remarkably, SS-10 induced the expression of proinflammatory cytokine IL-6 to a greater extent than NH-10 during the testing period, whereas it was not statistically significant in DEFs (Fig. 3b).
Fig. 3.
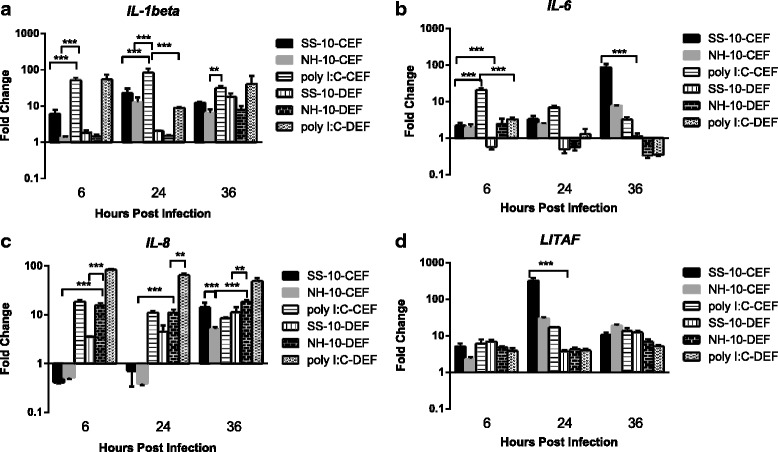
Proinflammatory cytokines IL-1beta (a), IL-6 (b), LITAF (c) and chemokine IL-8 (d) expression in chicken embryo fibroblasts (CEFs) and duck embryo fibroblasts (DEFs) following infection with SS-10 and NH-10 or treatment with poly(I:C) at 6, 24 and 36 h p.i. Data represent the mean fold change expression of either CEFs and DEFs compared with mock-infected controls after normalization to the expression of the housekeeping gene glyceraldehyde-3-phosphate-dehydrogenase. Significance was analyzed with two-way analyses of variance between the SS-10 and NH-10 groups in CEFs and DEFs at each time point (*p < .05, **p < .01, ***p < .001). Error bars represent SD
The expression level of chemokine IL-8 was suppressed at 6 and 24 h p.i., yet upregulated at 36 h p.i. in CEFs when induced by SS-10 and NH-10. However, the expression level of IL-8 showed a different pattern of increase at 6 h p.i. in DEFs and maintained the same tendency at 24 and 36 h p.i. when infected with SS-10 and NH-10, but was higher than that of CEFs at each time point. By contrast, the expression level of IL-8 was upregulated when treated with poly(I:C) in CEFs and DEFs during the testing period, but was higher in DEFs than in CEFs (Fig. 3c).
In sum, these results indicate that the expression levels of proinflammatory cytokines and chemokines IL-1beta, IL-6, IL-8 and LITAF in CEFs and DEFs showed different patterns following infection with NDVs of different pathogenicities.
Differential expression of antiviral cytokines in CEFs and DEFs in NDV-infected embryo fibroblast cells
As is well known, the IFN system is the most important host defense mechanism during infection with viral pathogens, for it controls and surpasses viral replication and modulates innate immune responses. In that context, in this study we compared the induction of IFN type I and II (IFN-alpha, IFN-beta and IFN-gamma) responses in CEFs and DEFs triggered by the SS-10 and NH-10 of different pathogenicities at the early stage of infection and that exhibited a similar species-dependent immune response. IFN-alpha, IFN-beta and IFN-gamma expression was upregulated in CEFs throughout the experiment period and peaked at 24 h p.i. (126.96- and 535.49-fold, 77.32- and 474.78-fold and 152.19- and 409.92-fold, respectively) induced by SS-10 and NH-10 when compared to mock-infected controls, yet was greater for NH-10 than SS-10 (Fig. 4a, b and c). In DEFs, the expression level of IFN-alpha was upregulated at 6 h p.i. (2.21- and 16.36-fold, respectively), gradually decreased to baseline level at 24 h p.i. (1.72- and 1.14-fold, respectively) and decreased further at 36 h p.i. (1.62- and 0.55-fold, respectively) after infection with SS-10 and NH-10 (Fig. 4a). The expression level of IFN-beta was upregulated during the tested period and peaked at both 6 and 24 h p.i. (7.09- and 25.28-fold, respectively) induced by both viruses (Fig. 4b). The expression level of IFN-gamma was downregulated for the duration of the study induced by SS-10; however, it was upregulated at 6 h p.i. (1.83-fold) and then downregulated at the other time points induced by NH-10 (Fig. 4c). In brief, these results demonstrate that the induction of rapid and robust type I and II IFNs in CEFs is far higher than in DEFs following challenge with virulent NDV and is lower for SS-10 than NH-10.
Fig. 4.
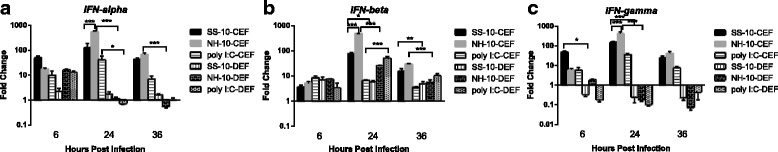
Type I and II interferon (IFN)-alpha (a), IFN-beta (b) and IFN-gamma (c) expression in chicken embryo fibroblasts (CEFs) and duck embryo fibroblasts (DEFs) following infection with SS-10 and NH-10 or treatment with poly(I:C) at 6, 24 and 36 h p.i. Data represent the mean fold change expression of either CEFs and DEFs compared with mock-infected controls after normalization to the expression of the housekeeping gene glyceraldehyde-3-phosphate-dehydrogenase. Significance was analyzed with two-way analyses of variance between the SS-10 and NH-10 groups in CEFs and DEFs at each time point (*p < .05, **p < .01, ***p < .001). Error bars represent SD
MHC class I and II molecule expression preference by embryo fibroblast cells
To compare the expression of MHC class I and II molecules, we examined their relative expression in CEFs and DEFs by qRT-PCR during early-phase NDV infection. MHC class I molecule expression was upregulated in CEFs and DEFs induced by SS-10 and NH-10 or treatment with poly(I:C) during the period of infection, except at 6 and 24 h p.i. in DEFs in response to infection with SS-10 (Fig. 5a). Importantly, the expression level of MHC class I induced by SS-10 was higher than NH-10 in CEFs, yet lower for SS-10 than NH-10 in DEFs at all time points (Fig. 5a). The expression level of MHC class II molecules was downregulated when induced by SS-10 and NH-10 or following stimulation with poly(I:C) for the duration of the study, whereas it was upregulated in CEFs at all time points when treated with poly(I:C) compared to mock-infected controls (Fig. 5b). Most importantly, MHC class I and II molecule expression in CEFs and DEFs showed different expression patterns that were associated with different pathogenicities against ND.
Fig. 5.
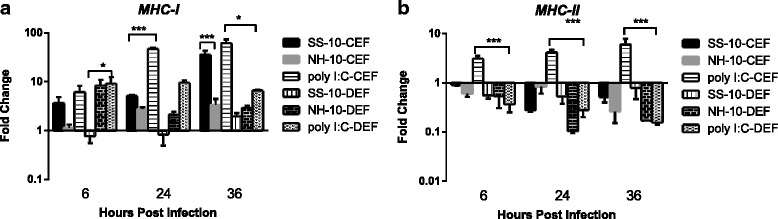
MHC I (a) and II (b) expression in chicken embryo fibroblasts (CEFs) and duck embryo fibroblasts (DEFs) following infection with SS-10 and NH-10 or treatment with poly(I:C) at 6, 24 and 36 h p.i. Data represent the mean fold change expression of either CEFs and DEFs compared with mock-infected control after normalization to the expression of the housekeeping gene glyceraldehyde-3-phosphate-dehydrogenase. Significance was analyzed with two-way analyses of variance between the SS-10 and NH-10 groups in CEFs and DEFs at each time point (*p < .05, **p < .01, ***p < .001). Error bars represent SD
Discussion
ND is a highly devastating viral disease in avian species that results high mortality and morbidity [1]. A wide variety of birds infected with NDV have been reported, though different species have exhibited different pathogenicities following infection with specific NDVs [26, 27]. Moreover, various NDV strains induce different host innate immune responses in specific animals [20, 21]. In this study, according to the results of replication kinetics in CEFs and DEFs when infected by virulent NDVs, the titers in CEFs were higher than in DEFs at each time point. However, the reason for this varying replication ability between the two species remains unknown, as does the role that the difference of disease severity plays in host pathogen immune responses to NDV infection.
Studies have demonstrated that NDV infection in immune cells—for instance, peripheral blood mononuclear cells and macrophages—results in extremely robust proinflammatory and antiviral cytokine induction both in vivo and in vitro [14, 20, 28, 29]. The expression of cytokines such as LITAF, IL-1beta, IL-8 and IL-6 in splenic leukocytes, macrophages and lymphoid tissues in chickens, ducks, geese and pigeons immediately are distinct in response to NDV infection [19–21, 30]. Our results show that proinflammatory cytokines IL-1beta, IL-6, chemokine IL-8, antiviral cytokines IFNs and PRRs such as TLR3 and TLR7, as well as MHC class I and II molecules, show different expression patterns, whereas LITAF is indistinct between the two species. The production of higher inflammatory immune responses to CEFs furthermore contrasts that of DEFs, which might at least partially explain the high morbidity and mortality of these birds following virulent NDV infection. Positive control stimulation with poly(I:C) in embryo fibroblast cultures also shows that differences in species are specific to the NDV. The increased production of proinflammatory cytokines and the severity of the cytopathic effect in CEFs when compared with DEFs following NDV infection might provide a plausible explanation for retinoic acid–inducible gene I (RIG-I) absence in CEFs, a viral RNA sensor that plays a crucial role in IFN-mediated antiviral immunity responses [31].
Our study has moreover shown an elevated induction of type I and II IFNs in CEFs and a weak production of type I and II IFNs in DEFs in response to NDV infection, which suggests the relative susceptibility of CEFs to NDV infection over DEFs, as consistent with previous observations of pathogenicity variation in different birds [11, 27]. The infection of CEFs and DEFs with SS-10 resulted in the weak induction of type I IFN compared to NH-10, likely due to cysteine-rich C terminus deletion in its V protein, which is critical for blocking IFN induction in embryo fibroblast cells [32]. The interaction of V and laboratory of genetics and physiology 2 or melanoma differentiation-associated gene 5 required for targeting STAT1 for degradation results in the inhibition of IFN signaling in chicken cells and Vero cells [32–34]. Rue et al. have shown that highly virulent NDV induces higher host innate immune responses compared with avirulent NDV in chicken spleens [19]. In our study, we found that CEFs induce significantly higher levels of IFN than DEFs following virus infection when compared with the expression levels of type I and II IFNs (Fig. 4). Studies have shown that the infection of chickens with virulent NDV resulted in a weak induction of IFNs that correlated with a longer shedding period, higher virus titers and greater disease severity [11, 35]. According to the above results, we speculate that the higher overall induction of IFNs by CEFs following infection with virulent NDV reflects what happens at the level of the organism, meaning shorter shedding and more rapid viral clearance in chickens and and lower virus replication and weaker viral clearance in ducks, as well as a longer shedding period.
We also found that the IL-6 mRNA transcript was upregulated in CEFs with both viruses and in treatment with polyI:C. By contrast, it was downregulated in DEFs with virulent NDV infection at 24 h p.i. (Fig. 3b). IL-6 mediates the limit and containment of NDV replication in the spleen of infected chickens during the early phase of infection, namely through the activation of host innate immune mechanisms such as macrophages and TLRs, which can contribute to pathological damage observed in NDV-infected chickens [19, 36]. Studies have shown that TLR3 plays a fundamental role in the expression of proinflammatory cytokines such as IL-6 and IL-1beta in fibroblasts or classical dendritic cells derived from TLR3-deficient mice after infection with NDV [37]. TLR3-deficient mice exhibited prolonged survival accompanied with reduced proinflammatory cytokines IL-6 and IFNs when infected with the Sendai virus, an enveloped animal virus of the family Paramyxoviridae similar to NDV [37]. Based on our results, there is a positive correlation in CEFs and DEFs infected with NDV in terms of the expression level of IL-6 and TLR3, which suggests the fundamental role of IL-6 in NDV pathogenesis.
TLRs such as TLR3 and TLR7 play an essential role in producing inflammatory cytokines and IFNs, as well as in activating host innate immune responses by triggering pathogen-associated molecular patterns, including the nucleic acid of RNA viruses such as NDV in mammals, insects and domestic poultry [38]. Yilmaz et al. [39] have reported that chicken TLR3 and TLR7 were highly expressed in the kidneys, liver, heart, spleen, intestines, lungs and oviduct, whereas TLR3 mRNA in ducks was only highly expressed in the spleen and lungs, moderately expressed in the intestines, liver and kidneys, poorly expressed in the heart, brain, bursa, and skin and not expressed whatsoever in muscle tissue [40]. Duck TLR7 mRNA was moreover highly expressed in the spleen, lungs and bursa, poorly expressed in the kidneys and liver, and not expressed whatsoever in the heart and brain, which is distinct from the expression patterns of TLR3 and TLR7 in chickens [41]. Our results reveal distinct expression patterns for TLR3 and TLR7 in CEFs and DEFs when exposed to NDV infection or treatment with polyI:C (Fig. 2a and b). The observed difference in TLR3 and TLR7 expression may be due to differences in the genome of tissues of chickens and ducks, or else the presence of resident cells that express TLR3 and TLR7 receptors absent in chickens.
Conclusions
In sum, our results reveal differences in the mRNA expression levels of TLRs, proinflammatory cytokines, IFNs and other immune-related genes between CEFs and DEFs in response to infection with two NDVs with different virulence or treatment with poly(I:C). Our findings also highlight that differential modulation of the host response by NDV strains of different virulence could be an important aspect of NDV pathogenesis. However, to further evaluate virus-specific differences in avian species, more comparative studies need to assess differences in host innate immune responses in avian species following NDV infection.
Acknowledgment
This study was mainly funded by grants from the National Natural Science Foundation of China (No. 31372412), the Chinese Special Fund for Agro-scientific Research in the Public Interest (No.201303033), and the Specialized Research Fund for Doctoral Program of Higher Education of China (No. 20124404110016).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YFK, TR participated in the design of the experiments and performed the experiments. MSF, XQZ, BX conceived of the study, and participated in its design and coordination and helped to draft the manuscript. PG YLL TR contributed reagents and performed the statistical analysis. YFK Wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Yinfeng Kang, Email: kangyinfeng2008@hotmail.com.
Minsha Feng, Email: 592334789@qq.com.
Xiaqiong Zhao, Email: 251033114@qq.com.
Xu Dai, Email: 545628515@qq.com.
Bin Xiang, Email: 273166125@qq.com.
Pei Gao, Email: 275743800@qq.com.
Yulian Li, Email: 511800192@qq.com.
Yanling Li, Email: 964816052@qq.com.
Tao Ren, Phone: 02085283054, Email: rentao6868@126.com.
References
- 1.OIE . OIE terrestrial manual 2012: manual of diagnostic tests and vaccines for terrestrial animals. Paris: World Organisation for Animal Health; 2012. Newcastle disease.Chapter 2.3.14; pp. 576–89. [Google Scholar]
- 2.Chambers P, Millar NS, Bingham RW, Emmerson PT. Molecular cloning of complementary DNA to Newcastle disease virus, and nucleotide sequence analysis of the junction between the genes encoding the haemagglutinin-neuraminidase and the large protein. J Gen Virol. 1986;67:475–86. doi: 10.1099/0022-1317-67-3-475. [DOI] [PubMed] [Google Scholar]
- 3.Chambers P, Samson A. Non-structural proteins in Newcastle disease virus-infected cells. J Gen Virol. 1982;58:1–12. doi: 10.1099/0022-1317-58-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Steward M, Vipond IB, Millar NS, Emmerson PT. RNA editing in Newcastle disease virus. J Gen Virol. 1993;74:2539–48. doi: 10.1099/0022-1317-74-12-2539. [DOI] [PubMed] [Google Scholar]
- 5.Alexander D, Senne D. Diseases of poultry. 12. Ames: Blackwell Publishing; 2008. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections, p 75–98. [Google Scholar]
- 6.Dortmans JC, Rottier PJ, Koch G, Peeters BP. Passaging of a Newcastle disease virus pigeon variant in chickens results in selection of viruses with mutations in the polymerase complex enhancing virus replication and virulence. J Gen Virol. 2011;92:336–45. doi: 10.1099/vir.0.026344-0. [DOI] [PubMed] [Google Scholar]
- 7.Kommers GD, King DJ, Seal BS, Brown CC. Pathogenesis of chicken-passaged Newcastle disease viruses isolated from chickens and wild and exotic birds. Avian Dis. 2003;47:319–29. doi: 10.1637/0005-2086(2003)047[0319:POCNDV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Kommers GD, King DJ, Seal BS, Brown CC. Virulence of six heterogeneous-origin Newcastle disease virus isolates before and after sequential passages in domestic chickens. Avian Pathol. 2003;32:81–93. doi: 10.1080/0307945021000070750. [DOI] [PubMed] [Google Scholar]
- 9.Shengqing Y, Kishida N, Ito H, Kida H, Otsuki K, Kawaoka Y, Ito T. Generation of velogenic Newcastle disease viruses from a nonpathogenic waterfowl isolate by passaging in chickens. Virology. 2002;301:206–11. doi: 10.1006/viro.2002.1539. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, Wang X, Zhao C, Liu D, Hu Y, Zhao J, Zhang G. Phylogenetic and pathotypical analysis of two virulent Newcastle disease viruses isolated from domestic ducks in China. PLoS One. 2011;6:e25000. doi: 10.1371/journal.pone.0025000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang Y, Li Y, Yuan R, Li X, Sun M, Wang Z, Feng M, Jiao P, Ren T. Phylogenetic relationships and pathogenicity variation of two Newcastle disease viruses isolated from domestic ducks in Southern China. Virol J. 2014;11:147. doi: 10.1186/1743-422X-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller PJ, Estevez C, Yu Q, Suarez DL, King DJ. Comparison of viral shedding following vaccination with inactivated and live Newcastle disease vaccines formulated with wild-type and recombinant viruses. Avian Dis. 2009;53:39–49. doi: 10.1637/8407-071208-Reg.1. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Chai T, Wang Z, Song C, Cao H, Liu J, Zhang X, Wang W, Yao M, Miao Z. Occurrence and transmission of Newcastle disease virus aerosol originating from infected chickens under experimental conditions. Vet Microbiol. 2009;136:226–32. doi: 10.1016/j.vetmic.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Kapczynski DR, Afonso CL, Miller PJ. Immune responses of poultry to Newcastle disease virus. Dev Comp Immunol. 2013;41:447–53. doi: 10.1016/j.dci.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Anis Z, Morita T, Azuma K, Ito H, Ito T, Shimada A. Comparative study on the pathogenesis of the generated 9a5b Newcastle disease virus mutant isolate between chickens and waterfowl. Vet Pathol. 2013;50:638–47. doi: 10.1177/0300985812467470. [DOI] [PubMed] [Google Scholar]
- 16.Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa MT, Engele M, Sieling PA, Barnes PF, Rollinghoff M, Bolcskei PL, Wagner M, et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–7. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 17.Weiss S, Bogen B. MHC class II-restricted presentation of intracellular antigen. Cell. 1991;64:767–76. doi: 10.1016/0092-8674(91)90506-T. [DOI] [PubMed] [Google Scholar]
- 18.Lennon-Dumenil A-M, Bakker AH, Wolf-Bryant P, Ploegh HL, Lagaudriere-Gesbert C. A closer look at proteolysis and MHC-class-II-restricted antigen presentation. Curr Opin Immunol. 2002;14:15–21. doi: 10.1016/S0952-7915(01)00293-X. [DOI] [PubMed] [Google Scholar]
- 19.Rue CA, Susta L, Cornax I, Brown CC, Kapczynski DR, Suarez DL, King DJ, Miller PJ, Afonso CL. Virulent Newcastle disease virus elicits a strong innate immune response in chickens. J Gen Virol. 2011;92:931–9. doi: 10.1099/vir.0.025486-0. [DOI] [PubMed] [Google Scholar]
- 20.Kang Y, Li Y, Yuan R, Feng M, Xiang B, Sun M, Li Y, Xie P, Tan Y, Ren T. Host innate immune responses of ducks infected with Newcastle disease viruses of different pathogenicities. Front Microbiol. 2015;6:1283. doi: 10.3389/fmicb.2015.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasoli M, Yeap SK, Tan SW, Moeini H, Ideris A, Bejo MH, Alitheen NB, Kaiser P, Omar AR. Alteration in lymphocyte responses, cytokine and chemokine profiles in chickens infected with genotype VII and VIII velogenic Newcastle disease virus. Comp Immunol Microbiol Infect Dis. 2014;37:11–21. doi: 10.1016/j.cimid.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Vaheri A, Ruoslahti E, Sarvas M, Nurminen M. Mitogenic effect by lipopolysaccharide and pokeweed lectin on density-inhibited chick embryo fibroblasts. J Exp Med. 1973;138:1356–64. doi: 10.1084/jem.138.6.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khattar SK, Samal S, Devico AL, Collins PL, Samal SK. Newcastle disease virus expressing human immunodeficiency virus type 1 envelope glycoprotein induces strong mucosal and serum antibody responses in Guinea pigs. J Virol. 2011;85:10529–41. doi: 10.1128/JVI.05050-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Adams SC, Xing Z, Li J, Cardona CJ. Immune-related gene expression in response to H11N9 low pathogenic avian influenza virus infection in chicken and Pekin duck peripheral blood mononuclear cells. Mol Immunol. 2009;46:1744–9. doi: 10.1016/j.molimm.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 < sup > − ΔΔCT</sup > Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Kaleta EF, Baldauf C. Newcastle Disease in Free-Living and Pet Birds. In Newcastle Disease. Edited by Alexander DJ. Boston, MA: Springer US; 1988;197–246.
- 27.Kang Y, Xiang B, Yuan R, Zhao X, Feng M, Gao P, Li Y, Li Y, Ning Z, Ren T. Phylogenetic and pathotypic characterization of Newcastle disease viruses circulating in South China and transmission in different birds. Front Microbiol. 2016;7:119. doi: 10.3389/fmicb.2016.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoss A, Zwarthoff EC, Zawatzky R. Differential expression of interferon alpha and beta induced with Newcastle disease virus in mouse macrophage cultures. J Gen Virol. 1989;70:575–89. doi: 10.1099/0022-1317-70-3-575. [DOI] [PubMed] [Google Scholar]
- 29.Zawatzky R, Wurmbaeck H, Falk W, Homfeld A. Endogenous interferon specifically regulates Newcastle disease virus-induced cytokine gene expression in mouse macrophages. J Virol. 1991;65:4839–46. doi: 10.1128/jvi.65.9.4839-4846.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y, Ding N, Ding SS, Yu S, Meng C, Chen H, Qiu X, Zhang S, Yu Y, Zhan Y, Ding C. Goose RIG-I functions in innate immunity against Newcastle disease virus infections. Mol Immunol. 2013;53:321–7. doi: 10.1016/j.molimm.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Karpala AJ, Stewart C, McKay J, Lowenthal JW, Bean AG. Characterization of chicken Mda5 activity: regulation of IFN-beta in the absence of RIG-I functionality. J Immunol. 2011;186:5397–405. doi: 10.4049/jimmunol.1003712. [DOI] [PubMed] [Google Scholar]
- 32.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A. 2004;101:17264–9. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Z, Krishnamurthy S, Panda A, Samal SK. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J Virol. 2003;77:8676–85. doi: 10.1128/JVI.77.16.8676-8685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Childs K, Randall R, Goodbourn S. Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. J Virol. 2012;86:3411–21. doi: 10.1128/JVI.06405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park MS, Garcia-Sastre A, Cros JF, Basler CF, Palese P. Newcastle disease virus V protein is a determinant of host range restriction. J Virol. 2003;77:9522–32. doi: 10.1128/JVI.77.17.9522-9532.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 37.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–7. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 38.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–45. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 39.Yilmaz A, Shen S, Adelson DL, Xavier S, Zhu JJ. Identification and sequence analysis of chicken Toll-like receptors. Immunogenetics. 2005;56:743–53. doi: 10.1007/s00251-004-0740-8. [DOI] [PubMed] [Google Scholar]
- 40.Jiao PR, Wei LM, Cheng YQ, Yuan RY, Han F, Liang J, Liu WL, Ren T, Xin CA, Liao M. Molecular cloning, characterization, and expression analysis of the Muscovy duck Toll-like receptor 3 (MdTLR3) gene. Poult Sci. 2012;91:2475–81. doi: 10.3382/ps.2012-02394. [DOI] [PubMed] [Google Scholar]
- 41.MacDonald MR, Xia J, Smith AL, Magor KE. The duck toll like receptor 7: genomic organization, expression and function. Mol Immunol. 2008;45:2055–61. doi: 10.1016/j.molimm.2007.10.018. [DOI] [PubMed] [Google Scholar]


