SUMMARY
Erh1, the fission yeast homolog of Enhancer of rudimentary, is implicated in meiotic mRNA elimination during vegetative growth, but its function is poorly understood. We show that Erh1 and the RNA-binding protein Mmi1 form a stoichiometric complex called EMC (Erh1-Mmi1 complex), to promote meiotic mRNA decay and facultative heterochromatin assembly. To perform these functions, EMC associates with two distinct complexes, MTREC (Mtl1-Red1 core) and CCR4-NOT. Whereas MTREC facilitates assembly of heterochromatin islands coating meiotic genes silenced by the nuclear exosome, CCR4-NOT promotes RNAi-dependent heterochromatin domain (HOOD) formation at EMC-target loci. CCR4-NOT also assembles HOODs at retrotransposons and regulated genes containing cryptic introns. We find that CCR4-NOT facilitates HOOD assembly through its association with the conserved Pir2/ARS2 protein, and also maintains rDNA integrity and silencing by promoting heterochromatin formation. Our results reveal connections among Erh1, CCR4-NOT, Pir2/ARS2 and RNAi, which target heterochromatin to regulate gene expression and to protect genome integrity.
Graphical Abstract
INTRODUCTION
Gametes are produced from germ cells when they undergo meiosis, a complex differentiation process that relies on developmental signaling pathways to direct cell division and reductional chromosome segregation. The fission yeast Schizosaccharomyces pombe is an excellent system for studying meiosis (Yamamoto, 1996a). In fission yeast, cells respond to nitrogen starvation by switching from the mitotic cell cycle to meiotic sexual differentiation, which in turn triggers pheromone production and response, cell conjugation and meiosis to occur in sequential order. Completion of meiosis in diploid cells requires Mei2, a master regulator of meiosis. Mei2 is normally targeted for degradation in vegetative cells through phosphorylation by the Pat1 kinase. However, the expression of Mei3, which is a pseudo-substrate for the Pat1 kinase, inhibits Pat1 activity in conjugated cells, resulting in Mei2 stabilization. Mei2 then binds to meiRNA, a meiosis-specific non-coding RNA encoded by the sme2+ gene, to form a nuclear dot. This Mei2-meiRNA ribonucleoprotein complex is believed to facilitate the transcription of various meiosis-specific genes that are essential to complete meiosis in diploid cells (Yamamoto, 1996b).
Recent advances have now dispelled the long held belief that meiotic gene transcription is active only in meiotic S. pombe cells. Meiotic gene transcription does occur in vegetative cells, but meiotic mRNAs are selectively eliminated from mitotically dividing cells to prevent their inappropriate expression (Harigaya et al., 2007; Yamamoto, 1996a). This mRNA elimination system specifically degrades transcripts that contain a DSR (determinant of selective removal) element and a polyadenylated tail (Harigaya et al., 2006; Yamanaka et al., 2010). The DSR core sequence (UCAAAC or UUAAAC) in meiotic mRNAs is recognized by the YTH domain-containing protein Mmi1, which in turn engages various proteins such as MTREC, composed of the Zn-finger protein Red1 and the Mtr4-like RNA helicase Mtl1, as well as MTREC-associated factors including the serine and proline-rich protein Pir1/Iss10, the Zn-finger protein Red5, the nuclear poly(A)-binding protein Pab2, the canonical poly(A) polymerase Pla1 and the nuclear exosome, to degrade target mRNAs (Chen et al., 2011; Egan et al., 2014; Lee et al., 2013; St-Andre et al., 2010; Sugiyama and Sugioka-Sugiyama, 2011; Sugiyama et al., 2013; Yamanaka et al., 2010; Yamashita et al., 2012; Yamashita et al., 2013; Zhou et al., 2015).
In addition to elimination of meiotic mRNAs during vegetative growth, DSR-dependent mRNA degradation machinery also promotes the assembly of facultative heterochromatin at discrete regions in the S. pombe genome (Hiriart et al., 2012; Tashiro et al., 2013; Yamanaka et al., 2013; Zofall et al., 2012). These regions include RNAi-independent heterochromatin islands formed at meiotic genes (e.g., mei4+ and ssm4+), and RNAi-dependent HOODs, which are heterochromatin domains formed at meiotic genes (e.g., mug5+ and mcp3+), genes regulated by environmental cues (e.g., pho1+), and retrotransposons (Cam et al., 2005; Shah et al., 2014; Yamanaka et al., 2013; Zofall et al., 2012). HOODs are particularly evident under specific growth conditions or when the nuclear exosome subunit Rrp6 is deleted (Yamanaka et al., 2013). The assembly of heterochromatin islands and HOODs is coordinated by distinct MTREC-associated factors. For example, Pir1 assembles heterochromatin islands, whereas Pla1 and Pab2 direct the formation of HOODs (Lee et al., 2013; Yamanaka et al., 2013). However, the detailed mechanism by which the RNA elimination system directs heterochromatin assembly at different sites in the genome is not yet fully established.
The S. pombe protein Erh1 (Enhancer of rudimentary homolog 1) is essential for the suppression of meiotic mRNAs during vegetative growth and for normal cell growth (Krzyzanowski et al., 2012; Yamashita et al., 2013). The gene was first identified in Drosophila as a mutation that enhances the wing phenotype of hypomorphic rudimentary mutations, and was named enhancer of rudimentary (Wojcik et al., 1994). The encoded product ERH is a small protein, and highly conserved orthologs are found in most eukaryotes (Weng and Luo, 2013). ERH has been shown to be critical for cell growth and is implicated in multiple processes such as CENP-E splicing in humans (Weng and Luo, 2013).
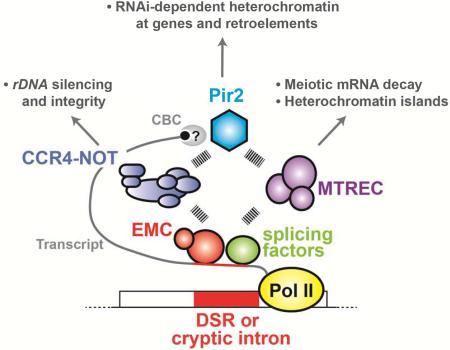
Here we investigate the role of Erh1 in meiotic mRNA degradation and heterochromatin assembly. Interestingly, we find Erh1 and Mmi1 form a tight complex, which we term EMC (Erh1-Mmi1 complex). EMC promotes meiotic mRNA decay and heterochromatin assembly in distinct ways. In addition to directing MTREC-mediated mRNA decay and the formation of heterochromatin islands at meiotic genes silenced by the exosome, EMC engages CCR4-NOT for RNAi-dependent assembly of HOODs. CCR4-NOT also promotes rDNA silencing and integrity, and triggers RNAi-mediated heterochromatin assembly at genes and retrotransposons containing cryptic introns. Importantly, our analyses suggest that CCR4-NOT cooperates with Pir2/Ars2, which is known to associate with the nuclear Cap-binding complex, to facilitate small RNA production and heterochromatin formation. These results provide important insights into pathways governing RNA decay and facultative heterochromatin assembly in different parts of the genome.
RESULTS
Erh1 colocalizes with nuclear RNA degradation machinery
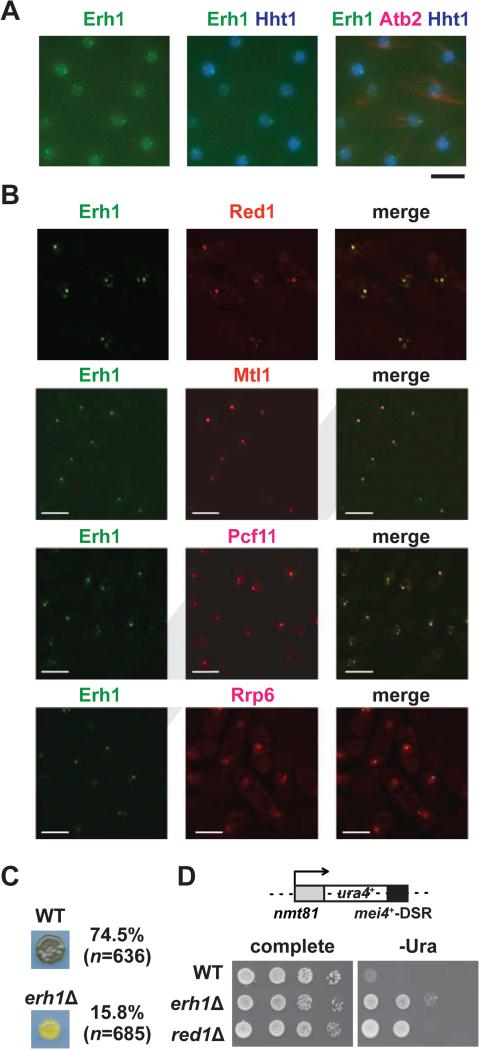
Erh1 localizes to the nucleus (Krzyzanowski et al., 2012), but its in-depth characterization has been lacking. In vegetative cells, we found that GFP-tagged Erh1 was evenly distributed in chromatin regions, which were visualized by co-expressing CFP-tagged histone H3 (Hht1); additionally, Erh1 formed nuclear foci that were often located at the edges of chromatin domains (Figure 1A). Moreover, we found that Erh1 colocalized with foci formed by known nuclear factors involved in meiotic mRNA decay, including Red1, Mtl1 and Rrp6, as well as with Pcf11, a cleavage and polyadenylation specificity factor (Figure 1B). Thus, we find that Erh1 colocalizes with RNA elimination factors to cleavage bodies in vegetative cells.
Figure 1. Characterization of Erh1.
(A) Erh1 forms nuclear foci. Representative images of vegetative fission yeast cells expressing Erh1-GFP (Erh1), Hht1 (histone H3 h3.1)-CFP and mCherry-Atb2 (tubulin alpha2; used to confirm mitotic interphase) are shown. Bar, 5 μm. (B) Erh1 colocalizes with Red1, Mtl1, Pcf11 and Rrp6. Deconvolved images of vegetative fission yeast cells expressing Erh1-GFP (Erh1) with either Red1-tdTomato (Red1), Mtl1-tdTomato (Mtl1), Pcf11-mCherry (Pcf11) or Rrp6-mCherry (Rrp6) are shown. Bars, 5 μm. (C) Cells from a parental WT or erh1Δ homothallic culture were spotted onto minimal (PMG) plates and incubated at 26°C for 3 days. The presence or absence of asci was determined by iodine staining, and the mating efficiencies are noted; “n” indicates the number of cells counted in each strain. (D) Erh1 is required for DSR-dependent mRNA decay. The ura4+-DSR construct is shown (top). WT, erh1Δ, and red1Δ cells carrying the ura4+-DSR were spotted onto complete or uracil-lacking (-ura) minimal plates in the absence of thiamine and then were incubated at 30°C for 3 days (bottom). See also Figure S1.
Loss of Erh1 causes growth defect and mating deficiency
To further understand the functions of Erh1, we generated and characterized an erh1Δ strain. The erh1Δ strain carrying the ade6-M210 or ade6-M216 allele displayed pink and pale pink colony color on adenine-limiting medium, as compared to red and pink colonies formed by WT cells carrying ade6-M210 or ade6-M216, respectively (Figures S1A). In addition, erh1Δ cells displayed a growth defect and cold sensitivity (Figures S1B). These phenotypes consistently segregated with erh1 (Figures S1A and S1B). We next tested if the growth defect may result from the ectopic expression of meiotic mRNAs. Since the lethality of mmi1Δ can be rescued by the deletion of the meiotic gene mei4+ (Harigaya et al., 2006), we asked whether erh1Δ growth defects were also suppressed by mei4Δ. Interestingly, the growth defects observed in erh1Δ at both 30°C and 19°C were suppressed by mei4Δ. In contrast, mei4Δ failed to rescue the growth defect or the cold sensitivity caused by red1Δ (Figure S1C). This result indicates that Mei4 shows a specific genetic interaction with Erh1, but not with Red1, even though both Erh1 and Red1 are required for suppression of mei4 mRNA (Sugiyama and Sugioka-Sugiyama, 2011; Yamashita et al., 2013).
We also noticed that erh1Δ cells displayed a severe mating defect. When meiosis is induced, homothallic (h90) wild-type cells sporulate and form asci. These asci can be detected by exposure to iodine vapor, which stains the starch-like compound in the spore wall. However, erh1Δ cells showed decreased intensity of iodine staining and a reduced mating efficiency (74.5% for WT and 15.8% for erh1Δ) (Figure 1C). Interestingly, we also observed a weak iodine staining in non-switching erh1Δ cells (Figure S1D). Since we did not observe haploid meiosis in non-switching erh1Δ cells by microscopy (data not shown), the weak iodine staining implies that erh1Δ may affect meiotic gene expression and result in ectopic starch accumulation in haploid cells during nitrogen starvation.
Erh1 is required for DSR-mediated mRNA decay
Four meiotic mRNAs containing the DSR sequence (mei4, rec8, ssm4 and spo5) accumulate in erh1Δ cells (Yamashita et al., 2013), suggesting that DSR-dependent mRNA decay requires Erh1. To confirm that Erh1 is indeed required for DSR-dependent mRNA decay, we combined erh1Δ with the ura4+-DSR construct (Harigaya et al., 2006), which comprises the ura4+ gene fused to the mei4+ DSR region (Figure 1D, top). We found that erh1Δ as well as red1Δ carrying ura4+-DSR grew on medium lacking uracil (Figure 1D, bottom), indicating that Erh1 is essential for DSR-directed mRNA degradation in vegetative cells.
Erh1 suppresses meiotic genes in vegetative cells
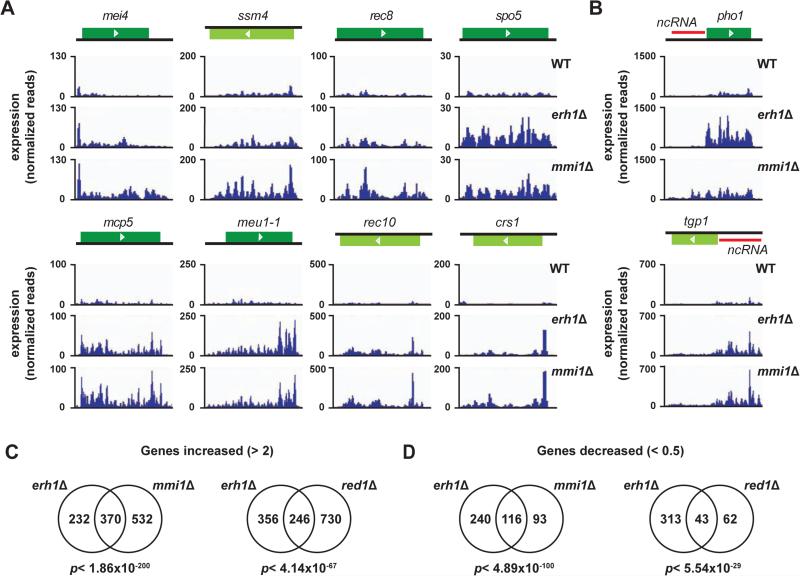
To gain insight into the target transcripts of Erh1, we analyzed the expression profile of vegetative WT and erh1Δ strains using RNA-Seq. We identified 602 transcripts that were upregulated >2-fold in vegetative erh1Δ relative to WT cells (Table S1). Of the 602 transcripts, 223 transcripts were previously shown to increase during nitrogen starvation or meiosis (Wilhelm et al., 2008). For example, DSR-containing mRNAs such as mei4 and ssm4 were increased in erh1Δ (Figure 2A), which is consistent with a previous report (Yamashita et al., 2013). We classified the 223 transcripts into 6 categories based on their expression profiles (Mata et al., 2002), and found that many of the upregulated genes are “early” or “middle” meiotic genes (Figure S2A and Table S1). Consistent with this meiotic gene classification, GO (gene ontology) analysis indicated that genes involved in meiotic nuclear division or meiotic cell cycle progression were upregulated in erh1Δ (Figure S2B and Table S1). In addition to meiosis-related genes, various types of non-coding RNAs (ncRNAs) also accumulated in erh1Δ (Figure S2C and Table S1). These included ncRNAs whose expression is regulated by the environmental growth conditions or developmental signals. For example, the loss of Erh1 caused upregulation of long ncRNA upstream of the phosphate-responsive pho1+ and tgp1+ genes (Figure 2B), which are regulated by RNA processing factors including Mmi1 and MTREC-associated factors (Ard et al., 2014; Chen et al., 2011; Lee et al., 2013; Shah et al., 2014). These results demonstrate that Erh1 is required for the suppression of meiotic genes and many ncRNAs.
Figure 2. Erh1 shares target transcripts with Mmi1 and Red1.
(A) DSR-containing meiotic mRNA levels in erh1Δ and mmi1Δ. RNA-Seq data of 8 representative meiotic mRNAs in WT, erh1Δ and mmi1Δ are shown. (B) Expression levels of pho1+ and tgp1+. Red bars indicate non-coding RNAs. (C and D) The numbers of genes with increased >2-fold (C) or decreased <0.5-fold (D) expression levels in erh1Δ, mmi1Δ or red1Δ cells are presented in Venn diagrams. For each comparison, the same set of 7019 genes was examined. The statistical significance (p-value) of the overlap between each of the two groups is shown under the diagrams. See also Figure S2; Tables S1 and S2.
We also identified 356 transcripts with lower steady-state levels (<0.5-fold reduction) in erh1Δ than in WT (Table S2). Only 13 were meiotic genes (Figure S2D), in contrast to the large fraction of meiotic genes found among upregulated transcripts. Only a few meiosis-related genes were decreased in erh1Δ (Figure S2E). Instead, most of the decreased transcripts, 290 of 356 (81%), represented various ncRNAs, including antisense RNAs, intergenic RNAs, tRNAs, and 5S rRNA (Figure S2F).
Erh1 shares target transcripts with Mmi1 and Red1
We addressed whether Erh1, Red1 and Mmi1 target a common set of transcripts by looking for any overlap among the Red1, Mmi1 and Erh1 regulons. We determined gene expression levels in erh1Δ relative to WT, and compared the results with similar analyses of the red1Δ and mmi1Δ transcriptome profiles. We found significant overlap between Erh1-regulated genes and Red1-regulated genes. Specifically, 246 of 602 transcripts (41%) that were upregulated in erh1Δ were also upregulated in red1Δ (p< 4.14×10−67) (Figure 2C, right), whereas 43 of 356 transcripts (12%) that were downregulated in erh1Δ were also downregulated in red1Δ (p< 5.54×10−29) (Figure 2D, right). Interestingly, the overlap between the set of Erh1-regulated genes and Mmi1-regulated genes was even more pronounced: 370 of 602 transcripts (62%) that were upregulated in erh1Δ were also upregulated in mmi1Δ (p< 1.86×10−200) (Figure 2C, left), whereas 116 of 356 transcripts (33%) that were downregulated in erh1Δ were also downregulated in mmi1Δ (p< 4.89×10−100) (Figure 2D, left). These results indicate that in vegetative cells, Erh1 regulates the expression of target transcripts that are shared with Red1 and Mmi1, but also that Erh1 function is more closely related to Mmi1 than to Red1.
Erh1 colocalizes with the Mei2-meiRNA dot and Is required for Its formation
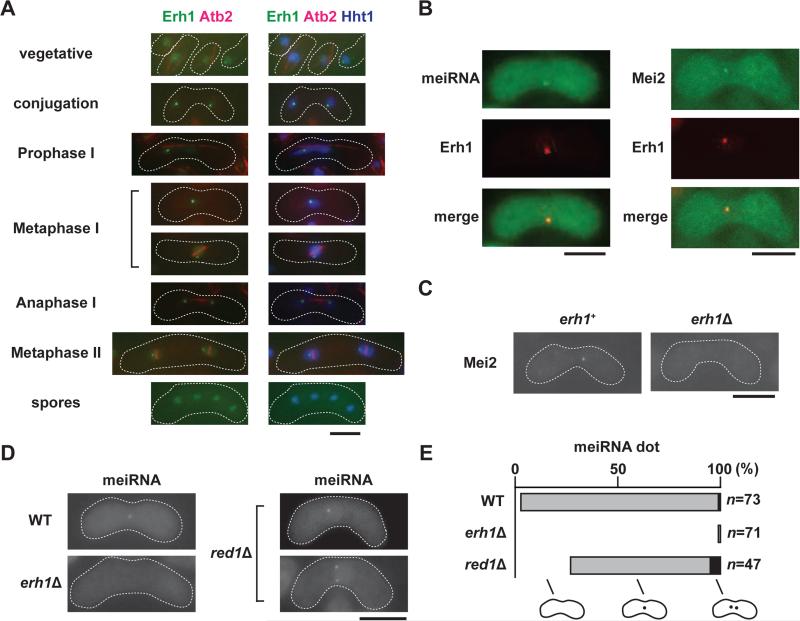
We next determined the localization of Erh1 during meiosis. In vegetative cells, Erh1 was evenly distributed in the nucleoplasm and formed nuclear foci (Figures 1A and 3A). Upon meiotic induction, Erh1 converged into a single distinct dot (Figure 3A). In asci, the Erh1 signal coincided with histone H3, but Erh1 foci in spores were not as obvious as in mitotically growing cells (Figure 3A).
Figure 3. Erh1 localization and function in meiotic cells.
(A) Localization of Erh1 during meiosis. Meiosis was induced in homothallic cells expressing Erh1-GFP, mCherry-Atb2 and Hht1-CFP, and the localization of these fluorescent proteins was determined. (B) Erh1 colocalizes with Mei2 and the meiRNA dot. Homothallic strains expressing Erh1-mCherry (Erh1) with either meiRNA, which was visualized using the MS2-loop and MS2-GFP system, or Mei2-GFP were spotted onto minimal (PMG) plates and then incubated at 26°C for 1-2 days to induce meiosis prior to imaging. (C) The Mei2 focus disappears in erh1Δ. The localization of Mei2-GFP in meiotic WT and erh1Δ cells is shown. (D) Erh1 is essential for meiRNA dot formation. The meiRNA dot in meiotic WT, erh1Δ, and red1Δ cells was visualized by the MS2-loop and MS2-GFP system. (E) The percentages of cells with 0 (white), 1 (grey) or 2 (black) meiRNA dots are shown in the bar graph. “n” indicates the number of cells examined for each strain. Representative images are shown, and the white dashed lines indicate the cell shape. Bars, 5 μm. See also Figure S3.
The localization patterns we observed for Erh1 were reminiscent of Mmi1, which colocalizes with Mei2-meiRNA, a ribonucleoprotein essential for meiosis (Yamamoto, 1996b), during early meiosis (Ding et al., 2012; Harigaya et al., 2006). As shown in Figure 3B, mCherry-tagged Erh1 (Erh1-mCherry) also colocalized with GFP-tagged Mei2 (Mei2-GFP) and with meiRNA, which was visualized using the MS2-GFP system (Shichino et al., 2014), indicating that Erh1 colocalizes with Mmi1 and meiRNA in meiotic cells.
Mmi1 dots converge during early meiosis in mei4Δ, but this convergence does not occur in mei2Δ or in sme2Δ (sme2+ encodes meiRNA) (Harigaya et al., 2006; Sugiyama and Sugioka-Sugiyama, 2011). Similarly, we observed a single Erh1 focus in mei4Δ, and two Erh1 dots in mei3Δ and sme2Δ (Figure S3A). Since Mei2 cannot be active in mei3Δ (Yamamoto, 1996b), these observations suggest that the formation of the Erh1 single dot requires both Mei2 and meiRNA. We next conversely asked whether Mei2 dot formation requires Erh1. The lack of a Mei2 dot in erh1Δ (Figure 3C) indicates that Erh1 facilitates proper Mei2 localization. In addition to Mei2, the meiRNA dot was rarely observed in erh1Δ: only 1% (1 out of 75 cells examined) of erh1Δ cells had a meiRNA dot, in contrast to 93% (71 out of 73 cells examined) of WT cells (Figures 3D and 3E). In contrast, we found an increased number of red1Δ cells either without a meiRNA dot (27.7% vs 2.7%) or with two meiRNA dots (10.6% vs 1.3%), and a large fraction of red1Δ cells (61.7%) had a single meiRNA dot (Figures 3D and 3E). Moreover, we noticed that haploid red1Δ cells often had a single dot (Figure S3B). These results indicate that unlike Red1, Erh1 and Mmi1 are essential for meiRNA dot formation.
Erh1 and Mmi1 form a complex called EMC
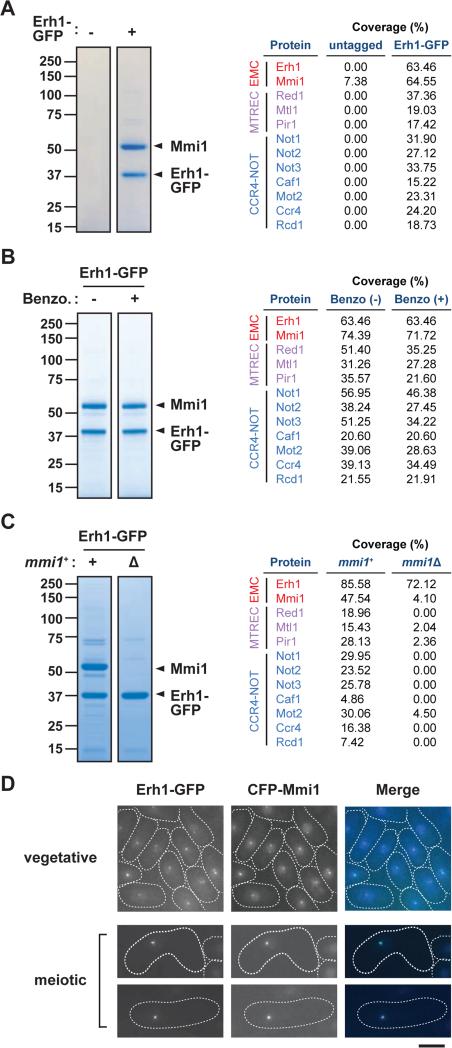
Our findings together with previous studies (Harigaya et al., 2006; Shichino et al., 2014) strongly suggest that the role of Erh1 closely reflects that of Mmi1. To investigate whether Erh1 interacts with Mmi1, we purified Erh1-GFP and found two distinct protein bands of approximately 55 and 39 kDa specifically in the Erh1-GFP purified fraction (Figure 4A). The intensity of the bands indicated that the proteins co-purified in stoichiometric amounts. Mass-spectrometry analyses revealed that the 39 kDa protein was Erh1-GFP, consistent with Western blot results (Figure S4A), and that the 55 kDa protein was Mmi1 (Figure 4A and Table S3). Because Mmi1 associates with both target mRNAs and chromatin (Hiriart et al., 2012; Tashiro et al., 2013), we included a Benzonase treatment step to remove all forms of DNA and RNA during the Erh1-GFP purification. As shown in Figure 4B, Benzonase treatment did not disrupt the interaction between Erh1 and Mmi1, and mass-spectrometry analyses indicated that Erh1 maintained its association with Mmi1 (Table S4). These data indicate that in vegetative cells, Erh1 stoichiometrically and directly associates with Mmi1 to form EMC.
Figure 4. Erh1 forms a complex with the YTH domain containing protein Mmi1.
(A) Erh1 purification. Extracts from the parental (untagged) or Erh1-GFP strains were purified on anti-GFP agarose. Erh1-binding proteins were visualized on an SDS-PAGE gel by CBB staining (left). The total peptide coverage (%) of the identified proteins is shown (right). (B) Erh1 purification with or without Benzonase treatment. The extract from the Erh1-GFP strain was purified on anti-GFP agarose with (+) or without (−) Benzonase treatment. The purified fractions were analyzed on an SDS-PAGE gel followed by CBB staining (left). The total peptide coverage (%) of the identified proteins is shown (right). (C) Erh1 purification from mmi1Δ cells. Extracts from WT (+) or mmi1Δ (Δ) strains expressing Erh1-GFP were subjected to affinity-purification using anti-GFP agarose. Purified proteins were resolved by SDS-PAGE and visualized by CBB staining (left). The total peptide coverage (%) of the identified proteins is shown (right). (D) Erh1 colocalizes with Mmi1. Representative images of vegetative or meiotic cells expressing Erh1-GFP and CFP-Mmi1 are shown. Bar, 5μm. See also Figure S4 and Tables S3-S5.
EMC interacts with two distinct protein complexes, MTREC and CCR4-NOT
In addition to Mmi1, other proteins were co-purified with Erh1-GFP including MTREC components Red1 and Mtl1 as well as Pir1/Iss10 (Figure 4A and Table S3). The interaction between EMC and MTREC is consistent with previous reports of Mmi1 co-purification with Red1 and Mtl1 (Egan et al., 2014; Lee et al., 2013; Zhou et al., 2015). Interestingly, all of the conserved subunits of the CCR4-NOT complex (Miller and Reese, 2012) were also co-purified. Caf4 and Caf16, also annotated as CCR4-NOT subunits, were not found in the Erh1 purified fraction, and are apparently missing from CCR4-NOT in higher eukaryotes. Purification of Ccr4-GFP from S. pombe revealed that EMC, MTREC and Pab2 were co-purified with CCR4-NOT (Figure S4B), further confirming the interaction between EMC and CCR4-NOT. Moreover, the association of Erh1 with MTREC and CCR4-NOT was retained after Benzonase treatment, whereas the recovery of RNA-binding proteins such as Vgl1 and Pabp was substantially reduced (Figure 4B and Table S4), indicating that EMC directly engages two distinct protein complexes, MTREC and CCR4-NOT.
We next asked whether the interaction with Mmi1 is required for Erh1 to interact with its partner proteins. To address this, we purified Erh1 from mmi1Δ. Although the steady-state level of Erh1-GFP was not affected in mmi1Δ (Figure S4C), staining of the fractions revealed that several bands were missing (Figure 4C). Mass-spectrometry analysis revealed that in the absence of Mmi1, MTREC and CCR4-NOT were not recovered with Erh1 (Figure 4C and Table S5), indicating that the integrity of EMC is essential for its interactions with MTREC and CCR4-NOT.
The localization of Erh1 and Mmi1 is mutually dependent
We found Erh1 colocalized with Mmi1 in both vegetative and meiotic cells (Figure 4D), further supporting the notion that Erh1 and Mmi1 form a stable complex. In addition, we observed that Erh1 foci were lost in mmi1Δ, and Mmi1 foci were not evident in erh1Δ (Figures S4D and S4E). Since the steady-state level of Erh1 or Mmi1 did not change upon deletion of mmi1+ or erh1+, respectively (Figures S4C and S4F), these results indicate that the assembly of EMC is a prerequisite for its proper localization.
Unlike the loss of Mmi1, the loss of Red1 did not greatly change the level of Erh1-GFP or alter its localization (Figures S4G and S4H). Moreover, erh1Δ affected neither the Red1 protein level nor Red1 localization (Figures S4I and S4J). These results indicate that although Erh1 and Red1 colocalize and interact with each other, they are present in distinct functional modules, EMC and MTREC, respectively, which have both separate and overlapping functions. Indeed, MTREC affects several loci that are not regulated by Mmi1 (Lee et al., 2013; Yamanaka et al., 2013)
Erh1 directs the assembly of heterochromatin islands
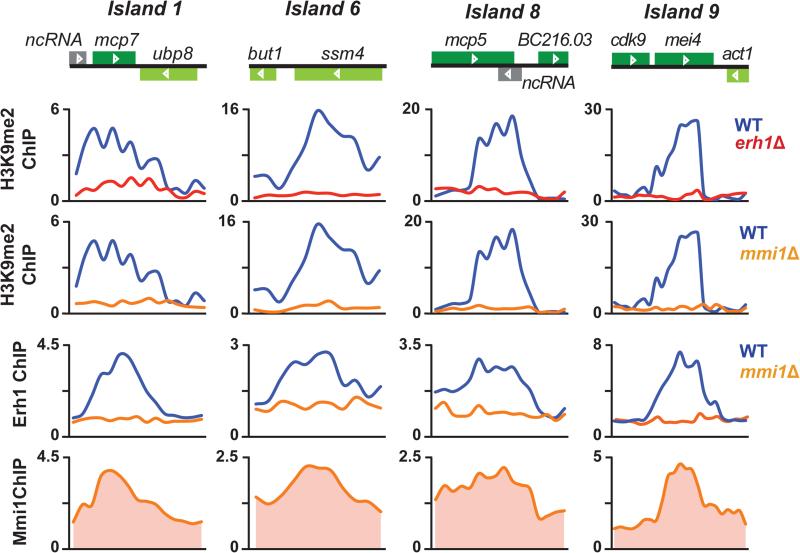
In addition to constitutive heterochromatin assembled at centromeres, telomeres and the mating-type (mat) locus, discrete blocks of heterochromatin islands are found throughout the S. pombe genome (Cam et al., 2005; Zofall et al., 2012). Many heterochromatin islands are formed at meiotic genes, and are assembled in a Red1-dependent manner (Egan et al., 2014; Lee et al., 2013; Tashiro et al., 2013; Zofall et al., 2012). Since both Erh1 and Red1 play a role in meiotic mRNA decay, we predicted that Erh1 is also required for the assembly of heterochromatin islands. As expected, we found that dimethylated lysine 9 of histone H3 (H3K9me2), which is a hallmark of heterochromatin, was lost in erh1Δ and mmi1Δ, and that both Erh1 and Mmi1 localized to Red1-dependent heterochromatin islands (Figure 5). ChIP followed by quantitative PCR (ChIP-qPCR) analyses confirmed that at two heterochromatin islands, mei4 and ssm4, H3K9me2 levels were greatly reduced in erh1Δ and mmi1Δ, whereas H3 occupancy levels were not affected (Figure S5A). This result indicates that H3K9me2, but not H3, was reduced at these loci in erh1Δ and mmi1Δ. In contrast to these heterochromatin islands, constitutive heterochromatin domains were unaffected by the deletion of either erh1+ or mmi1+, and neither Erh1 nor Mmi1 appeared to localize to these regions (Figure S5B). These results demonstrate that EMC is essential for Red1-dependent assembly of heterochromatin islands.
Figure 5. EMC is required for the assembly of heterochromatin islands.
ChIP-chip of H3K9me2 at Red1-dependent (1, 6, 8 and 9) islands in WT and erh1Δ, and in WT and mmi1Δ is shown in the top panels. ChIP-chip of Erh1-GFP in WT and mmi1Δ, and of CFP-Mmi1 in WT is shown in the bottom panels. See also Figure S5.
EMC associates with several loci in a heterochromatin-independent manner
In addition to heterochromatin islands, EMC also localized to several loci with no detectable H3K9me2 such as at non-coding RNAs (ncRNAs) located upstream of byr2+ and tgp1+ (Figures S5C and S5D). We found that Erh1 localization to two ncRNA loci, SPNCRNA.1197 and SPNCRNA.1459, was dependent on Mmi1, but not Red1 or Ccr4 (Figure S5E). EMC also associated with rpl3002+ and genes encoding other ncRNAs such as snoRNAs and meiRNA (Figure S5F). Since most of these genes were upregulated in erh1Δ, mmi1Δ (Table S1), and other RNA elimination factor mutants (Lee et al., 2013), it is likely that EMC and MTREC associate with these genes to target their transcripts for degradation or processing.
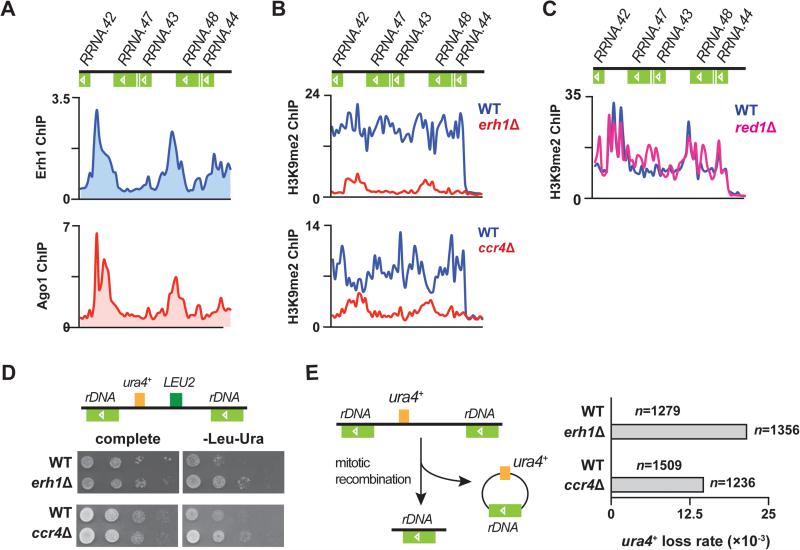
Erh1 and CCR4-NOT, but not MTREC, are essential for rDNA integrity
Erh1 also associated with rDNA repeats (Figure 6A, top). The binding profile of Erh1 at rDNA strikingly resembles that of the RNAi component Ago1 (Figure 6A, bottom), as we described previously (Cam et al., 2005). Given that RNAi is essential for heterochromatin formation and silencing at rDNA repeats (Cam et al., 2005), we hypothesized that Erh1 is also involved in the assembly of rDNA heterochromatin. As expected, we observed a significant decrease in H3K9me2 at rDNA clusters in erh1Δ (Figure 6B, top). Considering that we identified MTREC and CCR4-NOT as Erh1-binding partners, we wondered whether Erh1 cooperates with MTREC and/or CCR4-NOT to facilitate heterochromatin assembly at rDNA. We found that H3K9me2 levels at rDNA repeats were considerably decreased in ccr4Δ (Figure 6B, bottom), but were not affected in red1Δ (Figure 6C). These results indicate that Erh1 and CCR4-NOT are involved in the assembly of rDNA heterochromatin, whereas MTREC is dispensable.
Figure 6. Erh1 and Ccr4 regulate heterochromatin formation at rDNA repeats to protect rDNA integrity.
(A) ChIP-chip of Erh1-GFP (top) and myc-Ago1 (bottom) at rDNA repeats in WT. (B) ChIP-chip of H3K9me2 at rDNA repeats in WT and erh1Δ (top), and in WT and ccr4Δ (bottom). (C) ChIP-chip of H3K9me2 at rDNA repeats in WT and red1Δ. (D) LEU2 and ura4+ markers inserted at rDNA repeats (Ylp2.4pUCura4+-7) are de-repressed in erh1Δ and ccr4Δ. A schematic representation of Ylp2.4pUCura4+-7 is shown (top). WT and erh1Δ (middle) or WT and ccr4Δ (bottom) carrying Ylp2.4pUCura4+-7 were spotted onto complete medium or minimal medium lacking leucine and uracil (-Leu-Ura) and incubated at 30°C for 2 days. (E) Increased frequency of mitotic recombination at tandem rDNA repeats in erh1Δ and ccr4Δ. A schematic representation of rDNA::ura4+ loss is shown (left). WT, erh1Δ and ccr4Δ carrying Ylp2.4pUCura4+-7 were first plated on medium lacking leucine, transferred to complete medium, and then replica plated to medium lacking uracil (-Ura). Mitotic recombination at rDNA loci was indicated by the lack of growth on -Ura plates, and was confirmed by genomic PCR. “n” indicates the number of cells counted for each strain. See also Figure S6.
To further confirm heterochromatin loss at rDNA repeats, we first examined whether rDNA silencing is impaired in erh1Δ and ccr4Δ. As expected, ura4+ and LEU2 markers embedded within rDNA repeats were de-repressed in erh1Δ and ccr4Δ (Figure 6D). We next explored whether heterochromatin reduction at rDNA clusters in erh1Δ and ccr4Δ causes increased mitotic recombination at these loci. To assess mitotic recombination, we utilized the rDNA::ura4+ marker, which is lost by mitotic recombination when heterochromatin assembly is compromised (Figure 6E, left) (Cam et al., 2005). We observed a marked increase in mitotic recombination at rDNA repeats in erh1Δ (WT: 2.36×10−3; erh1Δ: 2.14×10−2) and in ccr4Δ (WT: 2.65×10−3; ccr4Δ: 1.46×10−2), as determined by the loss of rDNA::ura4+ (Figures 6E, right, S6A and S6B). These results clearly demonstrate that Erh1 and Ccr4 are required to maintain rDNA integrity, and indicate that Erh1, CCR4-NOT and RNAi machinery cooperate to establish and/or maintain heterochromatin at rDNA.
Subtelomeric heterochromatin is partially affected in ccr4Δ
We next investigated whether Ccr4 affects the formation of constitutive heterochromatin. We found that H3K9me2 levels at centromeres and the mat locus were unaffected in ccr4Δ (Figure S6C). Interestingly, whereas heterochromatin at regions containing dh-like elements (within tlh1+ and near SPAC212.06c) was maintained, we observed considerable decrease in H3K9me2 at subtelomeric domains containing genes upregulated during sexual differentiation and genes encoding transmembrane proteins (Cam et al., 2005; Yamanaka et al., 2013) in ccr4Δ (Figures S6D). Consistent with this H3K9me2 reduction, RNA-Seq analysis revealed many upregulated subtelomeric genes in ccr4Δ (Figure S6E). Taken together, these data indicate that Ccr4 is required to establish and/or maintain heterochromatin and to repress genes embedded within a specific region at subtelomeres.
Ccr4 plays only a minor role in heterochromatin island assembly, meiotic mRNA decay and meiRNA dot formation
Mmi1 facilitates the assembly of facultative heterochromatin islands via RNAi-independent pathways at many meiotic genes (Hiriart et al., 2012; Tashiro et al., 2013; Zofall et al., 2012). Our finding that Erh1 is essential for the formation of meiotic heterochromatin islands (Figure 5) prompted us to explore whether CCR4-NOT also participates in this process. We found that ccr4Δ showed only a slight reduction in H3K9me2 at meiotic islands, with the exception of the mei4+ locus (Figure S7A, island 1, 6, 8 and 9), and caused no change at non-meiotic islands (Figure S7A, island 14 and 15). The minor contribution of CCR4-NOT to forming islands other than at the mei4+ locus is also supported by a recent report (Cotobal et al., 2015). These observations are in contrast to the complete loss of H3K9me at meiotic islands observed in cells lacking Mmi1, Erh1 or MTREC (Figure 5) (Hiriart et al., 2012; Tashiro et al., 2013; Zofall et al., 2012), and indicates that of the two EMC-associated complexes, MTREC is largely responsible for meiotic heterochromatin islands, whereas CCR4-NOT plays only a minor role.
Although ccr4Δ had a minor effect on heterochromatin islands, there was a possibility that, like Erh1, CCR4-NOT mediates degradation of meiotic mRNAs in vegetative cells. We performed RNA-Seq and found that transcripts that accumulated in erh1Δ were not significantly increased in ccr4Δ (Figures S7B and S7C), and the increase in the Mmi1-regulon (Chen et al., 2011), with the exception of sme2, was less than 2 fold in two independent ccr4Δ RNA-Seq datasets (Figures S7C). We also examined whether ccr4Δ could rescue the sporulation defect of sme2Δ, which can be suppressed by mutations that impair meiotic mRNA degradation (Chalamcharla et al., 2015; Harigaya et al., 2006; Sugiyama et al., 2013; Yamashita et al., 2013). However, we observed almost no sme2Δ suppression by ccr4Δ (Figure S7D), further supporting our conclusion that Ccr4 is dispensable for meiotic mRNA decay in vegetative cells. Moreover, meiRNA dot formation, which depends on Erh1 (Figure 3) and Mmi1 (Shichino et al., 2014), was not considerably affected in the absence of Ccr4 (Figure S7E). We conclude that CCR4-NOT is largely dispensable for heterochromatin island assembly, meiotic mRNA degradation and meiRNA dot formation.
Loss of Ccr4 affects RNAi-dependent facultative heterochromatin at developmental genes and retrotransposons
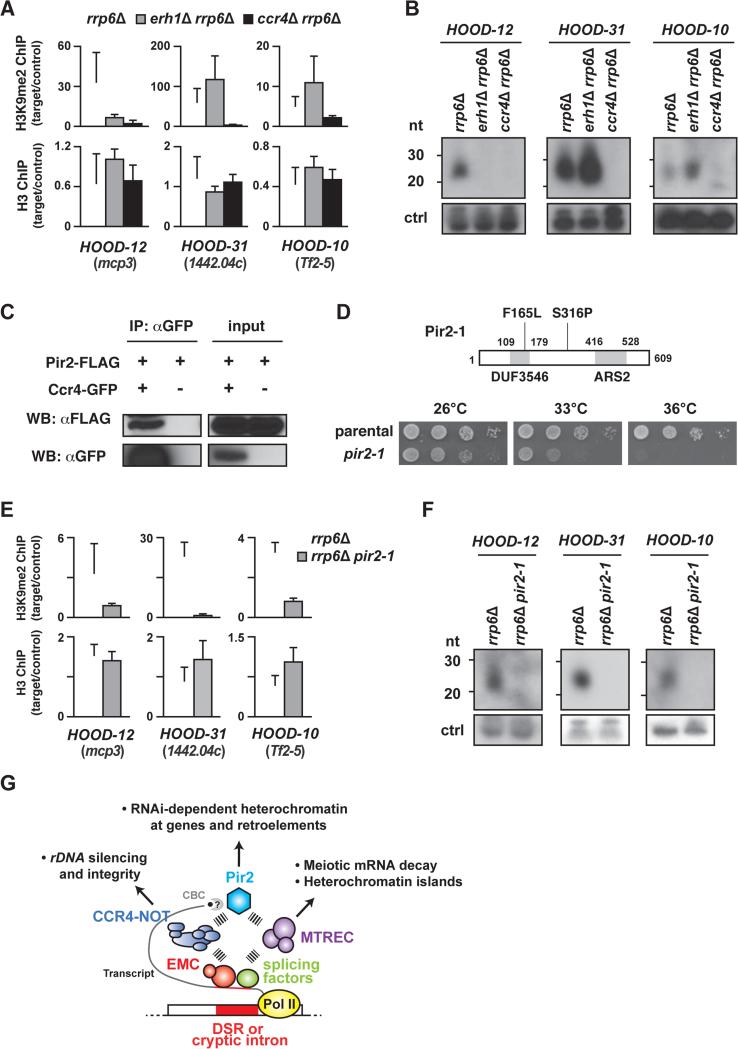
HOODs, another class of facultative heterochromatin detected under specific growth conditions or upon deletion of the nuclear exosome subunit Rrp6 (Marasovic et al., 2013; Shah et al., 2014; Yamanaka et al., 2013), coat developmentally and environmentally regulated genes and retrotransposons. HOOD assembly depends on RNAi, and Mmi1 is also required for the formation of some HOODs (Shah et al., 2014; Yamanaka et al., 2013), raising the possibility that Erh1 and CCR4-NOT are also required for HOOD assembly. To test this possibility, we performed H3K9me2 ChIP analyses in rrp6Δ, erh1Δ rrp6Δ and ccr4Δ rrp6Δ. We found that H3K9me2 at the mcp3+ locus (HOOD-12), which is Mmi1-dependent, was abolished in erh1Δ rrp6Δ, whereas H3K9me2 at SPCC1442.04c (HOOD-31) and the Tf2-5 retrotransposon (HOOD-10), both of which are Mmi1-independent, was slightly increased (Figures 7A and S7F). Consistent with this, small RNAs derived from HOOD-12 were lost whereas small RNAs from HOOD-31 and -10 were increased in erh1Δ rrp6Δ (Figure 7B). Such increases at Mmi1-independent HOODs were also observed in mmi1Δ (Yamanaka et al., 2013), indicating that Erh1 and Mmi1 have similar effects on HOOD assembly. Surprisingly, ccr4Δ resulted in the loss of H3K9me2 at both EMC-dependent and -independent HOODs (Figures 7A and S7F). Moreover, small RNAs from these HOODs were not detected in ccr4Δ rrp6Δ (Figure 7B). These results contradict a recent paper reporting that a ccr4 mutation did not decrease H3K9me2 at HOODs (Cotobal et al., 2015), and indicate that Ccr4 is indeed indispensable for RNAi-dependent H3K9me2 and small RNA generation at HOODs.
Figure 7. Ccr4 interacts with Pir2/ARS2 to promote small RNA production and H3K9 methylation at HOODs.
(A) ChIP-qPCR of H3K9me2 and H3 at Mmi1-dependent (HOOD-12) and Mmi1-independent HOODs (HOOD-31 and -10) in rrp6Δ (white bars), erh1Δ rrp6Δ (grey bars) and ccr4Δ rrp6Δ (black bars). The fold enrichment values of the target loci relative to the control locus vps33 are shown as the mean+SD (n=3). (B) Northern blot analysis of small RNA generation at three HOODs. Non-specific bands (~90 nt) served as a loading control (ctrl). (C) Co-immunoprecipitation of Pir2-FLAG with Ccr4-GFP. Cell lysates from the indicated strains were subjected to immunoprecipitation using anti-GFP antibody followed by Western blotting. (D) Schematic showing the pir2-1 allele, which encodes Pir2 protein containing Phe165 to Leu (F165L) and Ser316 to Pro (S316P) substitutions. Two conserved domains, DUF3546 and ARS2, are shown as grey boxes (top). Serial dilutions of parental wild-type (parental) and pir2-1 cultures were spotted onto rich plates and then incubated at the indicated temperatures (bottom). (E) ChIP-qPCR of H3K9me2 and H3 at Mmi1-dependent (HOOD-12) and Mmi1-independent HOODs (HOOD-31 and -10) in rrp6Δ (white bars) and rrp6Δ pir2-1 (grey bars). Fold enrichment of H3K9me2 and H3 at three target loci relative to the control locus (vps33) are shown as the mean+SD from three biological replicates. (F) Northern blot analysis of small RNAs generation at three HOODs. Non-specific bands (~90 nt) served as a loading control (ctrl). (G) Model showing EMC, CCR4-NOT, MTREC and Pir2 involvement in RNA-based regulation of gene expression and heterochromatin assembly. Transcripts containing a DSR are recognized by EMC, which in turn recruits MTREC and/or CCR4-NOT. MTREC is predominantly responsible for meiotic mRNA decay and formation of heterochromatin islands. On other hand, CCR4-NOT facilitates heterochromatin assembly at rDNA repeats and interacts with Pir2 (which also associates with Cap binding complex: CBC) and Pab2 (not depicted) to promote HOOD assembly at genes and retrotransposons. Pir2 is also known to associate with MTREC, implicated in formation of certain HOODs. In addition to the DSR, cryptic introns and splicing factors, which were previously implicated in the assembly of HOODs, likely also engage Pir2, CCR4-NOT and MTREC to promote mRNA decay and generate small RNAs mapping to HOODs. See also Figure S7.
Ccr4 cooperates with Pir2/ARS2 to form facultative heterochromatin
To investigate whether CCR4-NOT is linked to factors involved in RNAi, we performed a detailed examination of results from biochemical purifications including mass spectrometry analyses of various RNA elimination factors. These analyses led to an unexpected observation that all seven subunits of CCR4-NOT co-purified with an evolutionarily conserved protein, Pir2/Ars2, which also associates with MTREC and the Cap-binding complex (CBC) (Egan et al., 2014; Lee et al., 2013; Zhou et al., 2015). We confirmed that Pir2-FLAG co-immunoprecipitates with Ccr4-GFP (Figure 7C). Intriguingly, ARS2 is involved in RNAi in Drosophila and Arabidopsis (Sabin et al., 2009; Yang et al., 2006), which suggested a potential biological significance for the CCR4-NOT-Pir2 interaction.
We isolated a temperature-sensitive loss of function pir2 allele (pir2-1), since pir2+ is essential for cell viability (Kim et al., 2010). The pir2-1 mutant contained two amino acid substitutions: F165L in the DUF3546 domain, whose function is unknown, and S316P between the DUF3546 and ARS2 domains (Figure 7D, top). The pir2-1 strain grew slower than WT at 26°C and 33°C, and did not grow at 36°C (Figure 7D, bottom). The pir2-1 mutant showed a reduction in H3K9me2 and silencing at centromeres (G.T., J.D. and S.I.S.G., unpublished data). Remarkably, we also found that Pir2 affects RNAi-mediated heterochromatin assembly at both EMC-dependent and -independent HOODs, in a manner similar to CCR4-NOT. H3K9me2 levels at both EMC-dependent and -independent HOODs were reduced in rrp6Δ pir2-1 (Figure 7E). Moreover, our Northern blot analysis showed that small RNA generation at these HOODs was also severely impaired (Figure 7F). These results clearly show that CCR4-NOT works with Pir2/ARS2 for the assembly of facultative heterochromatin at retrotransposons as well as at developmentally regulated genes.
DISCUSSION
Here we identify Mmi1 as a binding partner of Erh1, and demonstrate that EMC cooperates with conserved RNA processing factors to promote meiotic mRNA degradation and facultative heterochromatin assembly. In addition to engaging MTREC for mRNA decay and assembly of heterochromatin islands at meiotic genes, EMC also interacts with CCR4-NOT to promote the formation of RNAi-dependent heterochromatin domains at developmental genes, retrotransposons and rDNA. However, unlike EMC and MTREC, CCR4-NOT plays only a minor role in silencing meiotic genes and forming heterochromatin islands in vegetative cells. Thus, it appears that EMC connects with distinct factors to perform different functions. Our results provide insights into the functions of Erh1 and its associated factors in the regulation of gene expression and the maintenance of genome stability.
EMC, a complex involved in meiotic mRNA decay
Although functionally connected, EMC and MTREC comprise distinct complexes based on the following observations: (1) Erh1 purification identified Mmi1 as a stoichiometric binding partner, but recovered a relatively low amount of MTREC subunits; (2) the localization of Erh1 and Red1 is not mutually dependent; (3) Erh1, but not Red1, is required for Mei2-meiRNA dot formation and for maintenance of rDNA integrity; and (4) previous purification studies did not indicate that Red1 interacts with Erh1 (Egan et al., 2014; Lee et al., 2013; Zhou et al., 2015). Therefore, we propose that EMC, like MTREC and the nuclear exosome, is another complex essential for meiotic mRNA degradation, and that these complexes cooperate to degrade meiotic mRNAs.
Mmi1 is essential for viability and temperature-sensitive mmi1 mutants cannot be used to study meiosis because high temperatures inhibit meiosis (Sabatinos and Forsburg, 2010). Thus, the fact that the erh1Δ strain is viable and does sporulate makes it a particularly useful tool to study EMC functions in meiosis such as meiRNA formation and homologous chromosome pairing (Ding et al., 2012; Shichino et al., 2014). Moreover, these results indicate that Mmi1 likely has additional functions independent of Erh1, either in meiotic mRNA elimination or other biological processes.
The possible function of Erh1 in EMC
The proteins of the ERH family are small (~12 kDa) and have reported roles in different kinds of nuclear transactions, including transcriptional repression, transcriptional elongation, mRNA splicing and translation (Weng and Luo, 2013). These observations suggest that ERH modulates the function(s) of its binding partners, rather than suggesting ERH has multiple activities. Given that mammalian ERH forms homodimers in solution (Arai et al., 2005; Wan et al., 2005), it is possible that Erh1 mediates Mmi1 dimerization, thereby allowing Mmi1 to exert its RNA-binding activity in vivo. Indeed, dimerization is known to be a prerequisite for the function of STAR, which is a subfamily of KH domain-containing RNA-binding proteins (Feracci et al., 2014), and hnRNP C forms tetramers to measure RNA length (McCloskey et al., 2012).
Another not mutually exclusive possibility is that the Mmi1 nuclear foci, which depend on Erh1, are required for Mmi1 function. Although we previously suggested that the formation of Red1 foci may correlate with mRNA degradation activity (Sugiyama and Sugioka-Sugiyama, 2011), it remains unclear whether the assembly of nuclear bodies by RNA elimination factors is required for mRNA degradation, and whether they are the place where RNA degradation occurs. Further analyses, such as the isolation of mutants that affect the localization of RNA elimination factors, may reveal the biological significance of nuclear body formation.
Erh1, CCR4-NOT and Pir2/ARS2: additional factors involved in H3K9 methylation
Our analyses also revealed that Erh1 is required for heterochromatin formation at meiotic genes. Moreover, defects in CCR4-NOT affect heterochromatin distribution at rDNA, HOODs and subtelomeres. At subtelomeric regions, heterochromatin consists of two distinct domains: dh-like sequences and the adjacent regions that contain meiosis-upregulated genes and transmembrane protein genes (Cam et al., 2005; Yamanaka et al., 2013). Impaired H3K9me2 in ccr4Δ is largely restricted to the latter region. Since RNA elimination factors (Sugiyama and Sugioka-Sugiyama, 2011; Yamanaka et al., 2013) and Ccr4 (this study) control the expression of genes embedded within these regions at subtelomeres, it is conceivable that CCR4-NOT directly participates in this process through the assembly of heterochromatin domains coating these loci. These heterochromatin domains located adjacent to blocks of telomeric heterochromatin resemble HOODs that also require CCR4-NOT and RNA elimination factors for small RNA production and H3K9 methylation.
Independently of our study, CCR4-NOT function in H3K9me was recently reported (Cotobal et al., 2015). Although both studies show the role of CCR4-NOT in H3K9me, there are profound differences in our conclusions. First, we demonstrate that CCR4-NOT is required for rDNA heterochromatin and rDNA integrity, a finding that was not reported by Cotobal et al., (Cotobal et al., 2015). Second, we show that CCR4-NOT is a critical player for H3K9 methylation and small RNA generation at HOODs, whereas Cotobal and colleagues reported that ccr4 and caf1 mutations do not affect HOODs (Cotobal et al., 2015). Third, we find that ccr4Δ has only a minor impact on Red1-dependent heterochromatin islands, with the exception of island 9 (the mei4+ locus). Although these results are in agreement with the data recently reported by Cotobal et al., (2015), the authors claimed that CCR4-NOT is essential for Mmi1-dependent islands (Cotobal et al., 2015). Based on our current and previous results (Lee et al., 2013; Yamanaka et al., 2013; Zofall et al., 2012), we propose that: (1) EMC and MTREC are the major players in heterochromatin island assembly, and (2) CCR4-NOT facilitates heterochromatin assembly at rDNA repeats, subtelomeres, and HOODs (Figure 7G).
We find that Ccr4 and Pir2/ARS2, both of which interact with each other, play an essential role in HOOD assembly, and that Pab2 also co-purifies with Ccr4. As HOOD assembly depends on RNAi, Pab2, cryptic introns and splicing factors (Lee et al., 2013; Yamanaka et al., 2013), it is plausible that CCR4-NOT and Pir2 are recruited by splicing factors (and EMC at certain loci) and cooperate with RNAi to assemble HOODs. Supporting this idea, it has been shown that: (1) ARS2 interacts with Dicer in Drosophila and Arabidopsis and is involved in RNAi (Sabin et al., 2009; Yang et al., 2006); (2) C. elegans ntl-9, a subunit of CCR4-NOT, is required to generate siRNAs from its endogenous targets (Fischer et al., 2013); and (3) human ARS2 co-purifies with the spliceosome (Rappsilber et al., 2002). Alternatively, Pir2 may facilitate HOOD assembly through non-canonical transcription termination. This idea is based on the following studies: (1) ARS2 has been shown to promote transcription termination (Andersen et al., 2013; Gruber et al., 2012; Hallais et al., 2013); and (2) Dhp1/Xrn2, a transcription termination factor, plays an essential role in HOOD assembly (Chalamcharla et al., 2015).
Perspective
Our findings are likely to have important biological implications. Heterochromatin formation at HOODs is controlled by growth conditions and developmental signals (Yamanaka et al., 2013). Since the functions of CCR4-NOT can be modulated by environmental and developmental cues (Miller and Reese, 2012), this machinery may facilitate cellular adaptation to different growth conditions. Furthermore, inappropriate expression of germline/meiosis-specific genes in somatic cells can induce tumor formation. In Drosophila, mutations in lethal (3) malignant brain tumor [l(3)mbt] are associated with brain tumors and are accompanied by ectopic expression of germline genes including piwi, aub and nos. Mutation of piwi, aub or nos is sufficient to suppress the brain tumor caused by l(3)mbt (Janic et al., 2010), indicating that Piwi, Aub or Nos plays a direct role in tumorigenesis. In recent years, much interest is focused on the identification of tumor-specific antigens, which can provide potential targets for immunotherapy. The continuing search for human tumor-specific antigens has identified more than 40 genes that are normally expressed in gametes. Called C/T (cancer/testis) antigens, their abnormal expression in somatic cells is suspected to induce genomic instability (Simpson et al., 2005; Whitehurst, 2014). Therefore, it has become clinically relevant to uncover the mechanism(s) by which aberrant expression of germline-specific genes is suppressed in somatic cells. It would be of great biological and clinical interest to test whether conserved RNA processing factors contribute to suppression of C/T antigen expression in somatic cells of higher eukaryotes.
EXPERIMENTAL PROCEDURES
Yeast strain construction and cell culture were performed using standard methods (Moreno et al., 1991). The pir2-1 mutant was constructed using an error-prone PCR-based method as described previously (Lee et al., 2013; Sugiyama et al., 2013). Strains and primers used in this study are listed in Tables S6 and S7, respectively. Dilution analyses, mating efficiency assays, and rDNA::ura4+ loss assays were performed as described previously (Cam et al., 2005; Sugiyama and Sugioka-Sugiyama, 2011). Briefly, for the rDNA::ura4+ loss assay, strains carrying LEU2 and ura4+ reporter genes inserted at tandem rDNA repeats were plated on medium lacking leucine, propagated in non-selective liquid YEA and plated on YEA medium plates followed by replica plating onto medium lacking uracil (-Ura). Cells that grew on YEA but not on -Ura were counted as ura4+ loss clones. Loss of ura4+ was confirmed by PCR using Terra™ PCR Direct Polymerase Mix (TaKaRa Bio). A DeltaVision Elite (Applied Precision, GE Healthcare) was used for differential interference contrast and fluorescence microscopy. The raw images were captured and analyzed using softWoRx (Applied Precision, GE Healthcare). RNA sequencing, Northern blotting, Western blotting, ChIP, ChIP-chip, protein purification and mass spectrometry analyses were performed as described previously (Cam et al., 2005; Lee et al., 2013; Sugiyama et al., 2007). Detailed procedures and associated references are available in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Erh1 stoichiometrically interacts with Mmi1 to form a complex called EMC
EMC engages MTREC and CCR4-NOT to assemble facultative heterochromatin
Ccr4 cooperates with Pir2/ARS2 to trigger RNAi-mediated facultative heterochromatin
Erh1 and CCR4-NOT promote rDNA silencing and the maintenance of rDNA integrity
ACKNOWLEDGEMENT
We thank J. Cooper, A. Kelly, K. Sawin, K. Tatebayashi, Y. Watanabe, M. Yamamoto, A. Yamashita and the National Bio Resource Project (NBRP) of the MEXT, Japan for S. pombe strains and materials; H. P. Cam, H. D. Folco, S. Yamanaka and M. Zofall for technical advice; J. Barrowman for her valuable help in editing the manuscript; and members of the Grewal lab for helpful discussions. T.S. thanks R. Sugioka-Sugiyama for her support. This research was supported by the intramural Research Program of the National Institutes of Health and by the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
T.S. and S.I.S.G. designed the experiments; T.S., G.T., V.R.C., Z.M., V.B., J.D. and M.Z. performed the experiments; T.S., G.T., Z.M., M.Z. and S.I.S.G. analyzed the data; T.S. and S.I.S.G wrote the manuscript.
ACCESSION NUMBER
All ChIP-chip and RNA-sequencing datasets can be accessed at the NCBI GEO under the accession code GSE76114.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, 7 Figures and 7 Tables and can be found with this article online.
REFERENCES
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 2013;20:1367–1376. doi: 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai R, Kukimoto-Niino M, Uda-Tochio H, Morita S, Uchikubo-Kamo T, Akasaka R, Etou Y, Hayashizaki Y, Kigawa T, Terada T, et al. Crystal structure of an enhancer of rudimentary homolog (ERH) at 2.1 Angstroms resolution. Protein Sci. 2005;14:1888–1893. doi: 10.1110/ps.051484505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ard R, Tong P, Allshire RC. Long non-coding RNA-mediated transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nat. Commun. 2014;5:5576. doi: 10.1038/ncomms6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Chalamcharla VR, Folco HD, Dhakshnamoorthy J, Grewal SI. Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc. Natl. Acad. Sci. USA. 2015;112:15548–15555. doi: 10.1073/pnas.1522127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Futcher B, Leatherwood J. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PloS One. 2011;6:e26804. doi: 10.1371/journal.pone.0026804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotobal C, Rodriguez-Lopez M, Duncan C, Hasan A, Yamashita A, Yamamoto M, Bahler J, Mata J. Role of Ccr4-Not complex in heterochromatin formation at meiotic genes and subtelomeres in fission yeast. Epigenet. Chromatin. 2015;8:28. doi: 10.1186/s13072-015-0018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, Hiraoka Y. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science. 2012;336:732–736. doi: 10.1126/science.1219518. [DOI] [PubMed] [Google Scholar]
- Egan ED, Braun CR, Gygi SP, Moazed D. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA. 2014;20:867–881. doi: 10.1261/rna.044479.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feracci M, Foot J, Dominguez C. Structural investigations of the RNA-binding properties of STAR proteins. Biochem. Soc. Trans. 2014;42:1141–1146. doi: 10.1042/BST20140081. [DOI] [PubMed] [Google Scholar]
- Fischer SE, Pan Q, Breen PC, Qi Y, Shi Z, Zhang C, Ruvkun G. Multiple small RNA pathways regulate the silencing of repeated and foreign genes in C. elegans. Genes Dev. 2013;27:2678–2695. doi: 10.1101/gad.233254.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, Thompson CB. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol. Cell. 2012;45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallais M, Pontvianne F, Andersen PR, Clerici M, Lener D, Benbahouche Nel H, Gostan T, Vandermoere F, Robert MC, Cusack S, et al. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 2013;20:1358–1366. doi: 10.1038/nsmb.2720. [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Harigaya Y, Yamashita A, Tanaka K, Yamamoto M. [Selective elimination of meiosis-specific mRNA operates during the mitotic cell cycle]. Tanpakushitsu Kakusan Koso. 2007;52:1–10. [PubMed] [Google Scholar]
- Hiriart E, Vavasseur A, Touat-Todeschini L, Yamashita A, Gilquin B, Lambert E, Perot J, Shichino Y, Nazaret N, Boyault C, et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012;31:2296–2308. doi: 10.1038/emboj.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzanowski MK, Kozlowska E, Kozlowski P. Identification and functional analysis of the erh1+ gene encoding enhancer of rudimentary homolog from the fission yeast Schizosaccharomyces pombe. PloS One. 2012;7:e49059. doi: 10.1371/journal.pone.0049059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, Balachandran V, Dhakshnamoorthy J, Taneja N, Yamanaka S, Zhou M, et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013;155:1061–1074. doi: 10.1016/j.cell.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasovic M, Zocco M, Halic M. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol. Cell. 2013;52:173–183. doi: 10.1016/j.molcel.2013.08.046. [DOI] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335:1643–1646. doi: 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- Miller JE, Reese JC. Ccr4-Not complex: the control freak of eukaryotic cells. Criti. Rev. Biochem. Mol. Biol. 2012;47:315–333. doi: 10.3109/10409238.2012.667214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 2010;470:759–795. doi: 10.1016/S0076-6879(10)70032-X. [DOI] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Wittmann S, Kilchert C, Vasiljeva L. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014;28:231–244. doi: 10.1101/gad.230177.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichino Y, Yamashita A, Yamamoto M. Meiotic long non-coding meiRNA accumulates as a dot at its genetic locus facilitated by Mmi1 and plays as a decoy to lure Mmi1. Open Biol. 2014;4:140022. doi: 10.1098/rsob.140022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J. Biol. Chem. 2010;285:27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Sugioka-Sugiyama R. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 2011;30:1027–1039. doi: 10.1038/emboj.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Wanatabe N, Kitahata E, Tani T, Sugioka-Sugiyama R. Red5 and three nuclear pore components are essential for efficient suppression of specific mRNAs during vegetative growth of fission yeast. Nucleic Acids Res. 2013;41:6674–6686. doi: 10.1093/nar/gkt363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S, Asano T, Kanoh J, Ishikawa F. Transcription-induced chromatin association of RNA surveillance factors mediates facultative heterochromatin formation in fission yeast. Genes Cells. 2013;18:327–339. doi: 10.1111/gtc.12038. [DOI] [PubMed] [Google Scholar]
- Wan C, Tempel W, Liu ZJ, Wang BC, Rose RB. Structure of the conserved transcriptional repressor enhancer of rudimentary homolog. Biochemistry. 2005;44:5017–5023. doi: 10.1021/bi047785w. [DOI] [PubMed] [Google Scholar]
- Weng MT, Luo J. The enigmatic ERH protein: its role in cell cycle, RNA splicing and cancer. Protein Cell. 2013;4:807–812. doi: 10.1007/s13238-013-3056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annu. Rev. Pharmacol. Toxicol. 2014;54:251–272. doi: 10.1146/annurev-pharmtox-011112-140326. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Wojcik E, Murphy AM, Fares H, Dang-Vu K, Tsubota SI. Enhancer of rudimentaryp1, e(r)p1, a highly conserved enhancer of the rudimentary gene. Genetics. 1994;138:1163–1170. doi: 10.1093/genetics/138.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem. Sci. 1996a;21:18–22. [PubMed] [Google Scholar]
- Yamamoto M. Regulation of meiosis in fission yeast. Cell Struct. Funct. 1996b;21:431–436. doi: 10.1247/csf.21.431. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Mehta S, Reyes-Turcu FE, Zhuang F, Fuchs RT, Rong Y, Robb GB, Grewal SI. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–560. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010;29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Shichino Y, Tanaka H, Hiriart E, Touat-Todeschini L, Vavasseur A, Ding DQ, Hiraoka Y, Verdel A, Yamamoto M. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol. 2012;2:120014. doi: 10.1098/rsob.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Takayama T, Iwata R, Yamamoto M. A novel factor Iss10 regulates Mmi1-mediated selective elimination of meiotic transcripts. Nucleic Acids Res. 2013;41:9680–9687. doi: 10.1093/nar/gkt763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu J, Schermann G, Ohle C, Bendrin K, Sugioka-Sugiyama R, Sugiyama T, Fischer T. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat. Commun. 2015;6:7050. doi: 10.1038/ncomms8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.