Abstract
Background:
Andrographis paniculata Nees is a medicinal plant with multiple pharmacological properties. It has been used over many centuries as a household remedy. A. paniculata products sold on the markets are in processed forms so it is difficult to authenticate. Therefore buying the herbal products poses a high-risk of acquiring counterfeited, substituted and/or adulterated products. Due to these issues, a reliable method to authenticate products is needed.
Materials and Methods:
High resolution melting analysis coupled with DNA barcoding (Bar-HRM) was applied to detect adulteration in commercial herbal products. The rbcL barcode was selected to use in primers design for HRM analysis to produce standard melting profile of A. paniculata species. DNA of the tested commercial products was isolated and their melting profiles were then generated and compared with the standard A. paniculata.
Results:
The melting profiles of the rbcL amplicons of the three closely related herbal species (A. paniculata, Acanthus ebracteatus and Rhinacanthus nasutus) are clearly separated so that they can be distinguished by the developed method. The method was then used to authenticate commercial herbal products. HRM curves of all 10 samples tested are similar to A. paniculata which indicated that all tested products were contained the correct species as labeled.
Conclusion:
The method described in this study has been proved to be useful in aiding identification and/or authenticating A. paniculata. This Bar-HRM analysis has allowed us easily to determine the A. paniculata species in herbal products on the markets even they are in processed forms.
SUMMARY
We propose the use of DNA barcoding combined with High Resolution Melting analysis for authenticating of Andrographis paniculata products.
The developed method can be used regardless of the type of the DNA template (fresh or dried tissue, leaf, and stem).
rbcL region was chosen for the analysis and work well with our samples
We can easily determine the A. paniculata species in herbal products tested.
Abbreviations used: bp: Base pair, Tm: Melting temperature
Keywords: Andrographis paniculata, authentication, DNA barcoding, herbal product, high resolution melting, species identification
INTRODUCTION
Andrographis paniculata (Burm.f.) Wall.ex Nees which belongs to the Acanthaceae (common name Andrographis) family is a medicinally important annual herb essentially distributed in tropical Asia. It is locally named as “Fah Talai Joan” in Thailand. The leaves and underground stem are used in the production of traditional medicine and have been used over many centuries as a household remedy in Asia. A. paniculata has now become popular also in Scandinavia for colds and influenza treatments and begins to become available in the United States as well. Diterpenoids and flavonoids are the main active constituents of A. paniculata.[1,2] Although Andrographis is frequently used for preventing or treating common cold and flu, it is also used for treatment of other mild to severe medical conditions (i.e. sore throat, diarrhoea, stomach pain, diabetes, hepatitis, and malaria, etc.). It has multiple pharmacological properties such as anti-malarial, anti-inflammatory, anti-oxidant, anti-hepatitic, anti-hyperglycemic, anthelmintic, antibacterial, antipyretic, and anti-cancer activity.[3] Recent pharmacological and clinical studies suggest the potential of A. paniculata for beneficial effects in killer diseases such as cancer[4,5] and HIV infections.[6,7] Since many disease conditions were commonly treated with A. paniculata in traditional Thai medical system, the plant was included in Thai national list of essential medicine.
During the past decade, traditional or indigenous health system has gained importance in the field of medicine. In most of the developing countries, a large number of people depend on traditional remedies, which in turn are dependent on the medicinal plant. Thai people spend several million baht a year on unproven herbal products that promise everything from curbing hot flashes to fighting off stomach ache and sore throat soothing. As it seems it is not only Thai people who spend so much on the herbal products or what we call alternative drugs, Western people such as Americans, Canadians, and Europeans also found to rely on the herbal products as supplements. Americans spend over 5 billion US $ a year; these tremendous numbers are corresponding with the World Health Organization estimation which is that 70–80% of the developed-world populations have used alternative medicines.[8]
Herbal products on the market are often perceived to be safe as fewer side effects could be observed when compared with synthetic drugs. However, there is an alarm for consumers to beware. Adulterated, counterfeit, and substitute products pose serious safety issues for the consumers from money loss to severe allergy or other medical conditions. Traditionally, biological species are authenticated according to their morphological features which are still the main basis of taxonomy.[9] Traditional taxonomy studies usually require the expertise of an experienced professional taxonomist. However, in some cases where the morphological character needed for identification of a specimen is lacking, it hinders even the specialists to recognise a species correctly. Especially in herbal products, the authentication becomes complicated because the original identifying characteristics are absent as these products are mainly sold in processed forms such as capsules and tablets, or are dried parts. Therefore, herbal products are often perceived to be safe due to their natural origin, however, adulterated, counterfeit, substituted of economically vulnerable materials with low-quality products pose serious safety threats to consumers and could prove fatal.[8,10] Common problems with raw drug trade are included not having readily distinguishable keys for materials that exhibit similarity in morphological features, materials sharing similar vernacular names in local languages, and the substitution or admixtures of economically valuable materials with inexpensive ones.[11,12,13] Medicinal plant materials may be substituted accidentally by herbs from closely related species or adulterated intentionally by materials from unrelated plants. In addition, lack of cultivation was observed in most of developing countries so that herbal materials are predominantly collected from the wild, by local farmers or collectors who often rely only on their experience in identifying the species being collected at the same time taxonomists are rarely availed for authentication. Frequently, admixtures could also be deliberate due increase profit from the adulteration.[12] It is undeniable that quality and effectiveness of the drug are directly linked to the quality of the raw materials in which the consequences of species admixtures can range from reducing the efficacy of the herbal products to lowering the trade value[14] not to mention misidentified materials could lead to the admixtures of toxic or otherwise unsuitable species and increasing the risk of accidental poisonings.[15,16,17] Therefore, appropriate measures should be taken if a reliable method for species identification and authentication of medicinal plant products existed which is not only critical for the enforcement of regulations, and to avoid adverse health and economic outcomes, but they are also important to reduce the negative environmental effects associated with purchasing protected species marketed as unprotected.
DNA barcoding referred as a short DNA sequences provides a way to confirm the identification of a variety of plant species including medicinal plants. A considerable amount of literature has been published and these studies showed a potential of DNA barcodes effectively distinguish medicinal plants, as well as identifying herbal medicinal materials and establishing a level of quality assurance.[18,19] Recently, DNA tests in Canadian research[20] show that many herbal products sold on the markets were found to be contaminated or substituted with alternative plant species that are not listed on the labels as they are replaced entirely by powdered rice, wheat and soybean. One-third of the herbal supplement products tested were nothing same as their labels. Noticing they found that Echinacea supplements, common products for Americans to prevent and treat colds, were contaminated with ground up bitter weed, Parthenium hysterophorus, an invasive plant found in India and Australia which can caused rashes, nausea and flatulence. Moreover, they found two bottles of supplements which labeled as St. John's wort contained none of the herb. Following a number of smaller studies performed in recent years suggests herbal products on the markets are not what they promised to be.[21,22]
Although, the DNA barcoding approach has been proved to be efficient and informative for both species identification and herbal products authentication, it is relatively costly and time consume which it is not suitable for a developing country like Thailand. Not to mention that the DNA barcoding method can be quite challenging in generating barcodes and analyzing the data to determine discrimination power. Thus, if the method is more rapid, simple, and economic, the DNA barcoding technique might have been far more persuasive for the developing countries like Thailand where the majority of people are using alternative medicines. Thus, the establishment of a novel effective and relatively cheap method for the routine medicinal plant materials would be beneficial for consumers and traders. High resolution melting (HRM) analysis coupled with DNA barcoding has a great potential to be applied for species identification. The principle of HRM is to distinguish the differences of DNA fragments using the thermodynamic properties of DNA sequences. HRM coupled with DNA barcoding, single-nucleotide polymorphism marker and microsatellite have been applied in aiding the taxonomical identification and the detection of adulteration in food and agriculture products.[23,24,25,26] However, the application of this approach as a potential use for authentication of herbal products has never been exerted, although the hybrid barcoding method showed good results in other applications as mentioned earlier. Here, we propose the use of availed DNA data from online database combined with HRM temperature to advance species identification and authenticating of A. paniculata products sold on the market in Thailand.
MATERIALS AND METHODS
Reference materials
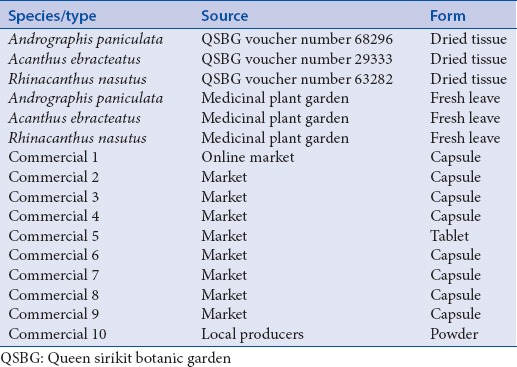
Specimens of authentic species including A. paniculata and related species were provided by Queen Sirikit Botanic Garden (QSBG). All plant material (dried samples) obtained from the QSBG came with the voucher number. Fresh plant material was collected and identified by an expert at the Medicinal Plant Garden, Faculty of Pharmacy, Chiang Mai University. Herbal products were purchased from markets (Chiang Mai, Thailand). All plants and herbal products were listed in Table 1.
Table 1.
The values of Tm (°C) gaining for HRM analysis using rbcL markers of Andrographis paniculata and related species

DNA isolation
DNA isolation from dried samples and herbal products was performed using 0.1 g of starting material in the form of fine powder by employing the Qiagen DN easy plant mini kit according to manufacturer's instructions. DNA concentration was estimated by standard spectrophotometric methods at 260 nm and 280 nm UV lengths using a BioDrop™ µLITE. DNA integrity was tested by gel electrophoresis in a 0.8% agarose gel. Samples were then diluted to a 20 ng/µL concentration. We used individually isolated triplicate samples. The extracted DNA solution was stored at −20°C until further use.
DNA barcodes data and Oligonucleotides primers
The sequences of A. paniculata and relevant species were extracted from GenBank (at the end of December 2013) using the keyword “The name of locus” and “the name of species” in the annotations. Generally, sequences obtaining from the public database include GenBank do not pose following the access to vouchers and/or low-quality often observed. Therefore, all sequences were subjected to processing and low-quality sequences were cut out. After processing and multiple alignments, primers design for HRM analysis was then performed using the SeqMan program. The primer pairs expected to generate a polymerase chain reaction (PCR) product not exceeding 200 bp that cover the variation site to enable discrimination of these different species.
Polymerase chain reaction amplification for high resolution melting analysis
To obtain the characteristic melting temperature (Tm) that was capable to distinguish the A. paniculata and other related species, DNA amplification using real-time PCR was performed using Eco™ Real-Time PCR system (illumina®, San Diego, California, USA). The reaction mixture for real-time PCR and HRM analysis was carried out in 10 µl of total volume contained 5 µl of 2x THUNDERBIRD® SYBR qPCR Mix, 0.2 µM forward primer, 0.2 µM reverse primer and 1 µl of 25 ng DNA. The primers rbcL_F; 5’-GGTACATGGACAACTGTGTGGA-3’ and rbcL_R; 5’-ACAGAACCTTCTTCAAAAAGGTCTA-3’ were synthesized by Pacific Science (Thailand). SYBR fluorescence dye was used to monitor the accumulation of amplified product during PCR and HRM process to derive Tm value.
PCR protocol was conducted in 48-well plate Helixis using an initial denaturing step at 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 57°C for 30 s and 72°C for 20. The fluorescent data were acquired at the end of each extension step during PCR cycles. Before HRM, the products were denatured at 95°C for 15 s and then annealed at 50°C for 15 s to randomly form DNA duplexes. For HRM experiments, fluorescence data were collected every 0.1°C. The Eco™ software (version 4.0.7.0) was used to analysis the Tm. The negative derivative of fluorescence (F) over temperature (T) (dF/dt) curve primarily displaying the Tm, the normalized raw curve depicting the decreasing fluorescence versus increasing temperature. To generate normalized melt curves and difference melt curves, pre- and post-melt normalization regions are set to define the temperature boundaries of the normalized and difference plot that were mainly used A. paniculata was set as a baseline species.
Authenticating Andrographis paniculata products
The developed method was used further to analyze the product labeled as A. paniculata (Fah Talai Joan) which were sold on the Thai markets using the DNA template extracted from the QSBG dried sample as a reference.
RESULTS AND DISCUSSION
DNA barcodes data and primer pair
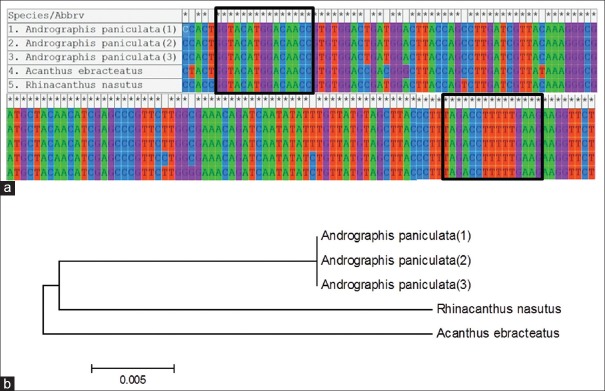
In order to develop a new method combining the DNA barcoding and HRM analysis for discrimination of A. paniculata and other related species and also its further use for the authenticating the species products on the market, availed DNA barcodes data on the online databases were collected. From the alignment of the extracted data [Figure 1a], the primer pair named rbcL_F and rbcL_R was chose for HRM analysis as products expected from this primers contained nucleotides variation of the different species which would allow their discrimination. The sequences of the species tested were used to construct phylogenetic tree [Figure 1b].
Figure 1.
Comparative alignment of rbcL consensus DNA barcodes of Andrographis paniculata, Acanthus ebracteatus and Rhinacanthus nasutus and phylogenetic tree using MEGA5. (a) Common DNA barcodes in three Acanthaceae were generated based on DNA sequences obtained from available DNA database sequences information from NCBI for rbcL region. The boxes denote the region were the primer pair was designed. (b) Phylogenetic NJ tree based on DNA barcodes derived from DNA database sequences information from NCBI for rbcL region (Accession numbersof each sample, Andrographis paniculata; JF949965.2, JQ922118.1 and JQ230990.1, Acanthus ebracteatus; AY289682.1, Rhinacanthus nasutus; GQ436493.1)
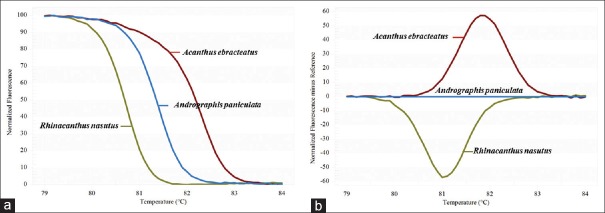
A real-time PCR protocol was applied for the identification and quantitative determination of A. paniculata in commercial herbal products. Table 1 depicts the results of the SYBR assay for the discrimination of the A. paniculata and related species. Herbal samples from different species could be distinguished using HRM analysis and the designed primers rbcL. The melting profiles of the rbcL amplicons of the three closely related herbal species (A. paniculata, Acanthus ebracteatus and Rhinacanthus nasutus) are illustrated in Figure 2a and b. A distinct melting curve was generated for each herbal species presenting one inflection point. Analysis of the normalised HRM curves with the barcode marker rbcL [Figure 2a and b] revealed that the three species could easily be distinguished. Assigning species A. paniculata as a genotype we were able by subtracting the area (difference graph) from the rest of the produced melting curves by the other species, to estimate the confidence value of similarity between the three species used [Figure 2b]. GCPs were calculated and a cut off value of 90% was used to assign a genotype for each barcode region [Table 2]. Furthermore, a closer examination of the A. paniculata HRM difference curve, with the mean of A. ebracteatus and R. nastus with A. paniculata curve as the baseline, revealed part of the curve sitting outside the 90% confidence interval (CI) curve, suggesting that the A. ebracteatus and R. nastus HRM curves are indeed different [Figure 2b]. The HRM analysis with the designed primer pairs proved to be a powerful tool in the identification of more or less closely related Acanthaceae species. The reproducible individual melting curves were achieved from different species with triplicate. Similar melting curves were achieved from the same species regardless of the type of the DNA template (fresh or dried tissue, leaf, and stem) (data not shown). Thus, the method can be applicable also for the processed herbal products that still contain tissue material with intact DNA.
Figure 2.
Barcoding of three species tested using high resolution melting analysis with the designed rbcL chloroplast marker. (a) Difference graph of the three species using Andrographis paniculata as reference. (b) Melting curves of the rbcL amplicons from the three species using Andrographis paniculata as reference. Color code box with the species used
Table 2.
Commercial Andrographis paniculata or Fah Talai Joan products and related species used in this study
Detection of Andrographis paniculata in herbal products
After the confirmation that each tested species (three Acanthaceae species of which A. paniculata was used as references) can be identified by HRM analysis we applied the same approach for the identification of the species used in herbal products [Table 2]. The DNA extracted from all products tested yielded a specific amplification product with the rbcL primers. The normalized HRM curves for the amplicons, from the species, studied and 10 commercial “Fah Talai Joan” herbal products, based on HRM analysis with barcode marker rbcL are shown in Figure 3. The products tested produced a unique melting plot that was easily to spot. Thus, all commercial samples could be successfully assigned to the species. All 10 products contained the A. paniculata or “Fah Talai Joan” that was promised or labeled as can be seen from their normalized HRM curves [Figure 3]. Furthermore, closer examination of the HRM difference curve, with all the tested samples with A. paniculata curve as the baseline, revealed part of the curve sitting outside the 90% CI curve, only for A. ebracteatus and R. nasutus suggesting that the A. ebracteatus and R. nasutus HRM curves are indeed different and all the other samples tested are similar to A. paniculata. Thus, DNA barcode coupled with HRM analysis methodology has allowed us easily to determine the A. paniculata species in herbal products on the markets even they are in processed forms.
Figure 3.
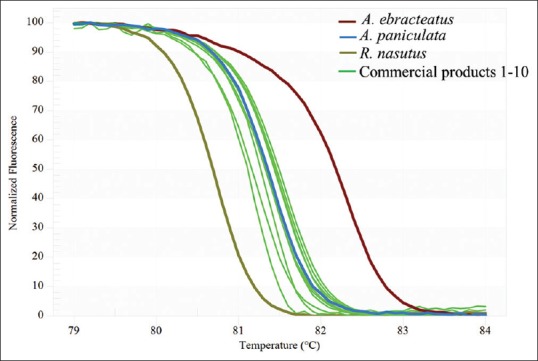
High resolution melting analysis using the designed rbcL primers analysis of 10 commercial products labeled as Andrographis paniculata. (a) Normalized curves of the three species Andrographis paniculata, Acanthus ebracteatus, Rhinacanthus nasutus and 10 commercial products. (b) Melting curves of the rbcL amplicons from the three species and 10 commercial products. Color code box with the samples used
This is partly indicated that the regulations of quality control in the herbal industry are seem to be appropriate and rigorous in Thailand, although we could not conclude this for other herbal products. In Thailand, A. paniculata is one among popular species used by a vast number of people. Luckily, misidentification has not been an issue for A. paniculata species but several medicinal plants of Thailand are currently facing the issue. The method developed in this study would be useful for authentication of others products too. Both traders and consumers would ultimately gain trust and confidence as there is a reliable way to authenticate the products. The developed method could be improved further if it is used along with a more species-specific primer and more species tested, which has been demonstrated for resolving individuals contributing trace amounts of DNA to highly complex mixtures.
CONCLUSION
Bar-HRM analysis was proven to be a fast and accurate technique for authentication testing of herbal products. Here, we describe the development of a Bar-HRM method for adulteration testing of A. paniculata commercial products, which commonly used as a household remedy by Thai people. Various forms of the A. paniculata such as fine powder, capsule and tea bag were observed on the markets. These processed forms the A. paniculata making it almost impossible to identify the products. The method developed here was proven to be effective and accurate detecting A. paniculata species. The DNA extracted from all products tested yielded a specific amplification product with the rbcL primers. The normalized HRM curves for the amplicons, from the three species (A. paniculata, A. ebracteatus and R. nasutus) and 10 commercial “A. paniculata” herbal products, based on HRM analysis with barcode marker rbcL were easily to spot, and all commercial samples could be successfully assigned to the A. paniculata species that were promised or labeled. The developed method could be easily used for rapid and low-cost authentication testing in commercial products without a doubt.
Financial support and sponsorship
This work was financially supported by the new researcher grant from the Thailand Research Fund (grant number TRG5780027).
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We thank Mr. Kittiphong Kertsawang (The Botanical Garden Organization, Ministry of Natural Resources and Environment, Thailand) and the late Mr. James F. Maxwell (Curator of CMUB herbarium, Chiang Mai University) for providing, identifying and collecting the samples.
REFERENCES
- 1.Tang W, Eisenbrand G. Berlin: Springer Verlag; 1992. Chinese Drugs of Plant Origin, Chemistry, Pharmacology and Use in Traditional and Modern Medicine; pp. 97–103. [Google Scholar]
- 2.Saxena S, Jain DC, Bhakuni R, Sharma RP. Chemistry and pharmacology of Andrographis species. Indian Drugs. 1998;35:458–67. [Google Scholar]
- 3.Jayakumar T, Hsieh CY, Lee JJ, Sheu JR. Experimental and clinical pharmacology of Andrographis paniculata and its major bioactive phytoconstituent andrographolide. Evid Based Complement Alternat Med 2013. 2013 doi: 10.1155/2013/846740. 846740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao F, He EQ, Wang L, Liu K. Anti-tumor activities of andrographolide, a diterpene from Andrographis paniculata, by inducing apoptosis and inhibiting VEGF level. J Asian Nat Prod Res. 2008;10:467–73. doi: 10.1080/10286020801948334. [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Wu D, Luo K, Wu S, Wu P. Andrographolide enhances 5-fluorouracil-induced apoptosis via caspase-8-dependent mitochondrial pathway involving p53 participation in hepatocellular carcinoma (SMMC-7721) cells. Cancer Lett. 2009;276:180–8. doi: 10.1016/j.canlet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Calabrese C, Berman SH, Babish JG, Ma X, Shinto L, Dorr M, et al. A phase I trial of andrographolide in HIV positive patients and normal volunteers. Phytother Res. 2000;14:333–8. doi: 10.1002/1099-1573(200008)14:5<333::aid-ptr584>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 7.Reddy VL, Reddy SM, Ravikanth V, Krishnaiah P, Goud TV, Rao TP, et al. A new bis-andrographolide ether from Andrographis paniculata Nees and evaluation of anti-HIV activity. Nat Prod Res. 2005;19:223–30. doi: 10.1080/14786410410001709197. [DOI] [PubMed] [Google Scholar]
- 8.Who.int. World Health Organization. [Last updated on 2003 May; Last cited on 2013 Oct 30]. Available from: http://www.who.int/mediacentre/factsheets/2003/fs134/en .
- 9.Wendy A. Texas: American Botanical Council; 2006. The Identification of Medicinal Plants: A Handbook of the Morphology of Botanicals in Commerce. [Google Scholar]
- 10.Mine Y, Young D. Regulation of natural health products in Canada. Food Sci Technol. 2009;15:459–68. [Google Scholar]
- 11.Khatoon S, Rai V, Rawat AK, Mehrotra S. Comparative pharmacognostic studies of three Phyllanthus species. J Ethnopharmacol. 2006;104:79–86. doi: 10.1016/j.jep.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 12.Mitra SK, Kannan R. A note on unintentional adulterations in ayurvedic herbs. Ethnobotanical Lealf. 2007;11:11–5. [Google Scholar]
- 13.Sunita G. New Delhi: Periodical Experts Book Agency; 1992. Substitute and Adulterant Plants. [Google Scholar]
- 14.Wieniawski W. Risk assessment as an element of drug control. WHO Drug Inf. 2011;15:7–11. [Google Scholar]
- 15.Song J, Yao H, Li Y, Li X, Lin Y, Liu C, et al. Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique. J Ethnopharmacol. 2009;124:434–9. doi: 10.1016/j.jep.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 16.Barthelson RA, Sundareshan P, Galbraith DW, Woosley RL. Development of a comprehensive detection method for medicinal and toxic plant species. Am J Bot. 2006;93:566–74. doi: 10.3732/ajb.93.4.566. [DOI] [PubMed] [Google Scholar]
- 17.Ize-Ludlow D, Ragone S, Bruck IS, Bernstein JN, Duchowny M, Peña BM. Neurotoxicities in infants seen with the consumption of star anise tea. Pediatrics. 2004;114:e653–6. doi: 10.1542/peds.2004-0058. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Cao H, But PP, Shaw PC. Identification of herbal medicinal materials using DNA barcodes. J Syst Evol. 2011;49:271–83. [Google Scholar]
- 19.Techen N, Parveen I, Pan Z, Khan IA. DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol. 2014;25:103–10. doi: 10.1016/j.copbio.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Newmaster SG, Grguric M, Shanmughanandhan D, Ramalingam S, Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11:222. doi: 10.1186/1741-7015-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Baker DA, Stevenson DW, Little DP. DNA barcode identification of black cohosh herbal dietary supplements. J AOAC Int. 2012;95:1023–34. doi: 10.5740/jaoacint.11-261. [DOI] [PubMed] [Google Scholar]
- 22.Wallace LJ, Boilard SM, Eagle SH, Spall JL, Shokralla S, Hajibabaei M. DNA barcodes for everyday life: Routine authentication of natural health products. Food Res. 2012;49:446–52. [Google Scholar]
- 23.Ganopoulos I, Madesis P, Darzentas N, Argiriou A, Tsaftaris A. Barcode high resolution melting (Bar-HRM) analysis for detection and quantification of PDO “Fava Santorinis” (Lathyrus clymenum) adulterants. Food Chem. 2012;133:505–12. doi: 10.1016/j.foodchem.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Jaakola L, Suokas M, Häggman H. Novel approaches based on DNA barcoding and high-304 resolution melting of amplicons for authenticity analyses of berry species. Food Chem. 2010;123:494–1305. [Google Scholar]
- 25.Madesis P, Ganopoulos I, Argiriou A, Tsaftaris A. The application of Bar-HRM (barcode DNA-high resolution melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control. 2012;25:576–82. [Google Scholar]
- 26.Faria MA, Magalhães A, Nunes ME, Oliveira MB. High resolution melting of trnL amplicons in fruit juices authentication. Food Control. 2013;33:136–41. [Google Scholar]