Increased stiffness in the tumor microenvironment generates forces on cells that promote cancer development. A signaling circuit is identified through which cells sense and respond to these changes. Patients whose tumors express these genes exhibit a better overall survival prognosis.
Abstract
Breast tumor progression is accompanied by changes in the surrounding extracellular matrix (ECM) that increase stiffness of the microenvironment. Mammary epithelial cells engage regulatory pathways that permit dynamic responses to mechanical cues from the ECM. Here, we identify a SLIT2/ROBO1 signaling circuit as a key regulatory mechanism by which cells sense and respond to ECM stiffness to preserve tensional homeostasis. We observed that Robo1 ablation in the developing mammary gland compromised actin stress fiber assembly and inhibited cell contractility to perturb tissue morphogenesis, whereas SLIT2 treatment stimulated Rac and increased focal adhesion kinase activity to enhance cell tension by maintaining cell shape and matrix adhesion. Further investigation revealed that a stiff ECM increased Robo1 levels by down-regulating miR-203. Consistently, patients whose tumor expressed a low miR-203/high Robo1 expression pattern exhibited a better overall survival prognosis. These studies show that cells subjected to stiffened environments up-regulate Robo1 as a protective mechanism that maintains cell shape and facilitates ECM adherence.
Introduction
Cancerous tissues are typically stiffer than their normal counterparts because of increased deposition of collagen and other extracellular components. Extracellular density can also affect normal tissues. For example, mammographic density, defined by the ratio of stromal collagen to adipose tissue, may be a contributing risk factor for breast carcinoma (Boyd et al., 2007). ECM deposition and cross-linking is associated with matrix stiffness that in turn impacts cellular behavior by altering signaling pathways. Cells encountering stiff matrices respond by generating tension between the ECM and actin cytoskeleton; in contrast, cells in compliant tissue environments contract and remodel the ECM. Cells respond to perturbations in the extracellular mechanical microenvironment via integrins and other ECM receptors and funnel this information principally through cell matrix adhesions (CMAs). These large, dynamic assemblages of proteins function as “adhesomes,” acting through interactions with the cytoskeleton to translate changes in ECM mechanics into cellular responses (Roca-Cusachs et al., 2012; Case and Waterman, 2015). Studies have documented how integrins and other collagen receptors mediate behavioral responses of cells to increased collagen deposition and matrix stiffness (Wozniak et al., 2003; Paszek et al., 2005; Zhang et al., 2013), but much less is known about how other extracellular factors function in mechanotransduction.
SLITs are large, multidomain proteins that are part of the basement membrane, interacting with numerous ECM constituents, including heparin sulfate proteoglycans and collagen-XV/XVIII (Ballard and Hinck, 2012). They signal through ROBO receptors, which belong to the immunoglobulin superfamily and contain no catalytic intracellular domains. Instead, ROBOs share several conserved regions that interact with adaptor proteins, such as Nck/Dock, and other signaling proteins, such as Rho GTPases (Ballard and Hinck, 2012). These GTPases govern the contractile activity of cells in compliant environments and are also activated in response to extracellular stiffness. It is well established that Rho enhances contractility and builds cellular tension by regulating the actin cytoskeleton in response to ECM density and stiffness (Wozniak et al., 2003; Paszek et al., 2005; Ponik et al., 2013). Recent evidence suggests that Rac may similarly function in enhancing intracellular tension by promoting the capture and assembly of MIIA minifilaments into maturing focal adhesions (FAs; Pasapera et al., 2015). In addition, Rac activation can perpetuate a FAK–Cas–Rac feedback loop that generates and enhances intracellular tension (Bae et al., 2014). These studies suggest a thus far underappreciated role for Rac in organizing the response of cells to external forces downstream of CMAs.
miRNAs exert profound effects on tissue morphology and function by controlling suites of genes during normal development and tumorigenesis. An example is miR-203, which has been shown to both suppress and enhance tumor progression through gene regulation (Ding et al., 2013; Benaich et al., 2014). Many bona-fide miR-203 targets have been identified, but none are located near the miR-203 locus, suggesting that transcriptional regulation of miR-203 is uncoupled from the protein coding genes it regulates (Saini et al., 2011; Sonkoly et al., 2012). Currently, other than allelic deletion occurring in glioblastomas (Dontula et al., 2013), there is little information about regulatory mechanisms leading to the initial suppression of miR-203 that occurs in different types of epithelial tumors. One possibility is that miR-203 is regulated by global changes in tissue architecture arising during transformation. Indeed, ECM stiffness, which itself is regulated by tumor cells, has recently been shown to modulate miRNA expression, resulting in accelerated tumorigenesis (Mouw et al., 2014). Understudied, however, are the potential mechanisms used by cells to maintain tensional homeostasis and resist transformation in changing ECM environments. Here, we explore such a signaling circuit in which changes in ECM density and stiffness down-regulate miR-203 expression, resulting in increased Robo1 that serves a protective function by activating Rac and FAK to maintain cell shape and enhance CMAs.
Results
SLIT2/ROBO1 regulates actin organization in tissue and epithelial contraction in compliant matrices
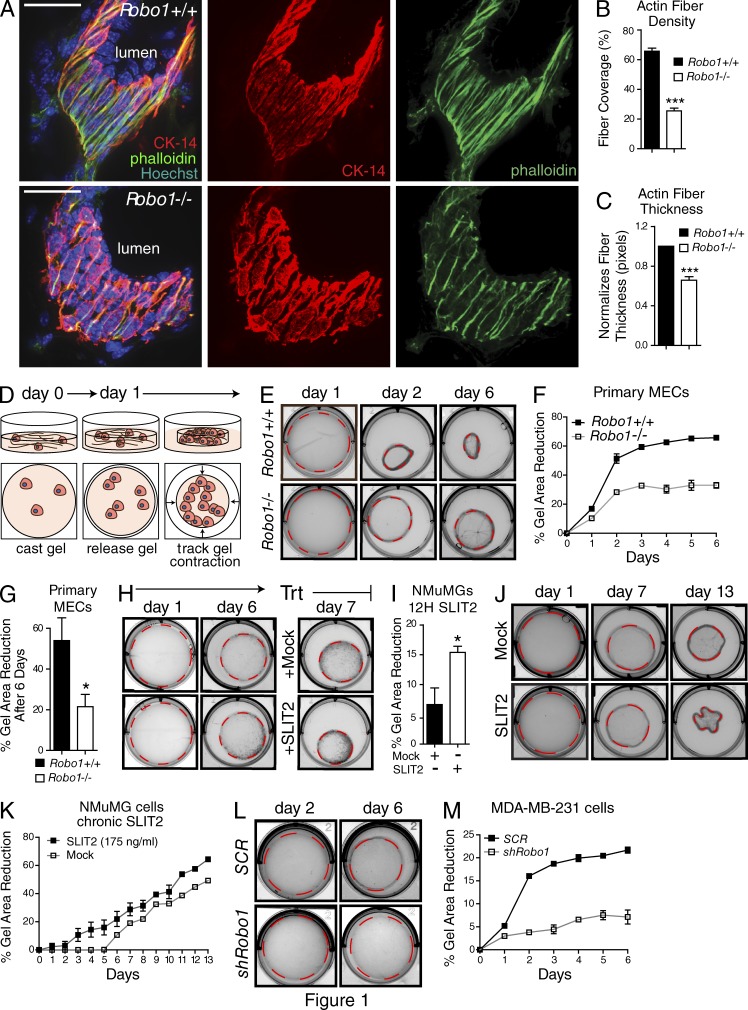
Breast cells sense physical changes in their microenvironment through CMAs and respond by modulating their cytoskeleton to mechanotransduce these extracellular alterations (DuFort et al., 2011; Schedin and Keely, 2011). One candidate pathway for regulating this behavior is the SLIT/ROBO family, originally identified through its role in axon guidance. During breast (mammary gland) development, ROBO1 expression is initially restricted to the outer myoepithelial layer of this bilayered tissue, but it is eventually expressed in the mature gland by a subset of inner luminal cells (Strickland et al., 2006; Marlow et al., 2008). In exploring ROBO1 function in the developing gland, we observed a marked morphologic change in Robo1−/− myoepithelial cells (Fig. 1, A–C). Cytokeratin-14 immunostaining of grazing tissue sections revealed that Robo1−/− myoepithelial cells are not as tightly packed as Robo1+/+ cells. They also contain actin fibers that are significantly narrower than the broadly distributed actin fibers of Robo1+/+ tissue. This suggested that loss of Robo1 impacts the ability of these outer layer cells to organize their cytoskeleton.
Figure 1.
SLIT2/ROBO1 regulates actin organization in the mammary gland and epithelial contraction of compliant matrices. (A) Grazing sections of Robo1+/+ and Robo1−/− murine mammary glands stained with Cytokeratin-14 (CK-14; red), phalloidin (green), and Hoechst (blue). Bars, 30 µm. (B and C) Quantification of fiber density (B) and fiber thickness (C) in grazing sections (n = 3 pairs of animals). (D) Cartoon depicting cell contraction in LD, 3D collagen gels. (E, H, J, and L) Representative micrographs of collagen gels containing (E) primary MECs, (H and J) NMuMG cells, and (L) MDA–MB-231 cells. Dashed outline represents measured area. (F, G, I, K, and M) Quantification of the percentage reduction in collagen gel area for gels embedded with Robo1+/+ and Robo1−/− MECs (F and G), NMuMG cells treated with SLIT2 (I and K), and MDA–MB-231 cells infected with Robo1 or scrambled (SCR) shRNA lentivirus (M) (n = 3 experiments). Asterisks denote significance by t test: *, P < 0.05; ***, P < 0.001.
To explore the behavior of Robo1+/+ and Robo1−/− cells, we cultured primary mammary epithelial cells (MECs) in low-density (LD) gels. This generates a compliant 3D ECM that recapitulates the stiffness of normal mammary gland (Paszek et al., 2005) and is capable of being remodeled by resident MECs through cytoskeletal contraction of the collagen fibers (Fig. 1 D; Gehler et al., 2009). In this assay, cell contractility and tension are approximated by percentage reduction in gel size. We examined the effects of losing Robo1 by culturing MECs, which express SLIT2 (Strickland et al., 2006) from mature virgin mice (Fig. 1, E–G). After 2 days in culture, gels containing Robo1+/+ cells were 50% the area of gels containing Robo1−/− cells, which remained smaller over the 6-d time course, showing that Robo1−/− MECs have a diminished capacity for contraction.
We also examined the effect of SLIT2/ROBO1 signaling on gel contraction using a nontransformed murine line (NMuMG) that is derived from normal glandular tissue and expresses ROBO1 similar to luminal cells harvested from a mature gland (Fig. S1 A; Strickland et al., 2006). After 6 d in LD collagen, the gel size was reduced by ∼30% (Fig. 1, H and I). At this time, cultures were treated with SLIT2 (500 ng/ml), resulting in an additional twofold decrease in gel size. In contrast, NMuMG cells cultured in high-density (HD) collagen gels, which approximate the stiffness of the tumor microenvironment (Paszek et al., 2005), showed no reduction in gel size because the rigid ECM formed by high collagen concentration resists the cellular force exerted on it (Fig. S2, A and B; Wozniak et al., 2003; Wozniak and Keely, 2005). Next, we titrated SLIT2 to a level that does not significantly inhibit cell proliferation (Fig. S3, A and B) and performed the assay with this level of chronic SLIT2 (175 ng/ml; Fig. 1, J and K). Under this condition, gels contracted within 3 days, as opposed to 6 days for Mock treatment, and SLIT2-treated gels contracted significantly more each day. We also examined whether transformed cells display a similar behavior by knocking down Robo1 in the basal-like cancer cell line, MDA–MB-231 that expresses Robo1 despite being transformed, unlike other lines such as BT20 or SKBR3 (Fig. S3 C; Marlow et al., 2008; Chang et al., 2012). Again, we observed a threefold decrease in gel contraction when Robo1 expression is reduced (Fig. 1, L and M). Together, these experiments show that SLIT2/ROBO1 signaling enhances the ability of the cell’s cytoskeleton to pull on collagen fibers, resulting in smaller gels in compliant environments.
SLIT/ROBO1 activates Rac signaling to modulate contractility
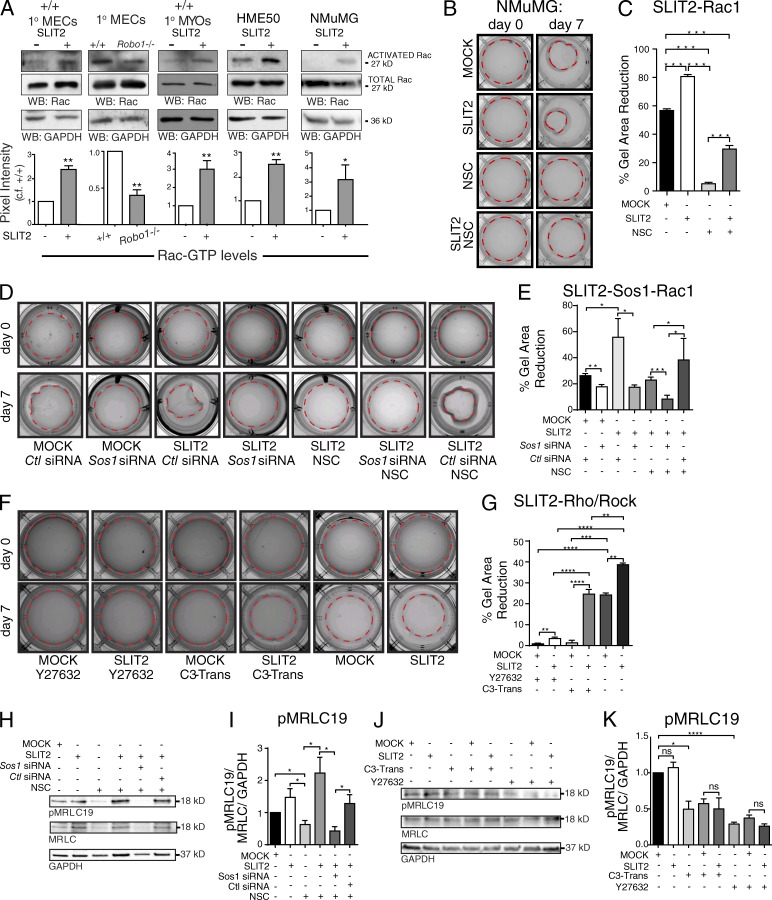
Cells use the Rho family of small GTPases to orchestrate the cytoskeletal changes that accompany ECM remodeling. Depending on cellular context, SLIT/ROBO signaling is capable of both suppressing and activating these small GTPases (Ballard and Hinck, 2012). To assess the response of Rho and Rac to SLIT2 in mammary tissue and breast cells, we performed pull-down assays using GST fusion proteins that bind activated Rac/Cdc42 or Rho. We prepared lysates from cultured MECs, harvested from developing (5 wk) virgin glands and treated with Mock or SLIT2 (175 ng/ml). It was difficult to reproducibly detect activated Rho in MEC lysates (Fig. S4 A). In contrast, we found an ∼2.5-fold increase in Rac activity upon SLIT2 treatment of wild-type MECs and a concordant ∼0.66-fold decrease in activated Rac in Robo1−/− MECs (Fig. 2 A). Because we observed a striking change in the morphology of the myoepithelial layer in Robo1−/− mammary gland tissue (Fig. 1 A), we further tested the specificity of Rac activation in isolated myoepithelial cells (Macias et al., 2011). There was an approximately threefold increase in Rac activation in the Robo1-expressing myoepithelial fraction upon SLIT2 treatment (Fig. 2 A) and no change in the luminal fraction (Fig. S4 B). Because ROBO1 is expressed in basal cells during development and luminal cells in the mature gland, we also probed lysates generated from basal-like human breast epithelial cell line, HME50, and from the luminal NMuMG line, both of which express ROBO1 (Fig. S1 A). Again, there was a significant increase in Rac activity with SLIT2 treatment (Fig. 2 A).
Figure 2.
SLIT2/ROBO1 signaling promotes Rac-mediated cell contraction of compliant matrices. (A, top) Representative blots for GTP-Rac, total Rac, and GAPDH. (bottom) Quantification of Rac activation (n = 3 experiments). (B, D, and F) Images of 3D LD gels with embedded NMuMG cells subjected to indicated treatments: Mock, SLIT2, NSC23766 (NSC), Sos1 or control siRNA, Y27632 (ROCK Inhibitor), or C3-Transferase (Rho inhibitor, C3-Trans). Dashed outline of gels represent measured areas. (C, E, and G) Quantification of the percentage reduction in collagen gel area for treated gels (n = 3 experiments). (H and J) Representative blots of pMRLC19, MRLC, and GAPDH from NMuMG cells treated with Mock or SLIT2 and indicated siRNA and inhibitors. (I and K) Quantification of pMRLC normalized to total MRLC and GAPDH (n = 3 experiments). Asterisks denote significance by t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. ns, not significant.
Next, we investigated whether Rac activation is responsible for SLIT2/ROBO1-induced remodeling of collagen fibers and subsequent reduction in the size of compliant gels. We took advantage of a widely used Rac inhibitor (NSC23766) that targets two Rac-specific guanine nucleotide exchange factors (GEFs), TrioN and Tiam1. In contrast, a different GEF, Son of Sevenless 1 (Sos1), is responsible for activating Rac downstream of SLIT2/ROBO1 signaling (Yang and Bashaw, 2006). We cultured NMuMG cells in LD collagen gels chronically treated with Mock, SLIT2 (175 ng/ml), and/or NSC2376 (Fig. 2, B and C). Similar to before (Fig. 1, H and I), we observed an ∼20% increase in gel contraction upon SLIT2 treatment. In contrast, treatment with NSC23766 dramatically inhibited contraction. Cotreatment of gels with NSC23766 + SLIT2, however, partially rescued the effect of the inhibitor, leading to an ∼20% increase in gel contraction, similar to the original increase observed with SLIT2. These data show that Rac plays an important role in cell-mediated collagen remodeling, which occurred under Mock but not inhibitor conditions and resulted in gel contraction. SLIT2 rescued the ability of cells to contract gels by activating Rac through a different GEF because this enhancement is observed in the presence of NSC23766 that inhibits TrioN and Tiam1, but not Sos1. We repeated the experiment in the presence of control (Ctl) or Sos1 siRNA (Fig. 2, D and E). In the absence of NSC23766, we again observed significantly increased gel contraction with SLIT2 + Ctl siRNA treatment. This increase in contraction was significantly reduced by Sos1 siRNA (Fig. S5, F and G), to the level observed in Mock + Sos1 siRNA (Fig. 2, D and E). In the presence of NSC23766, SLIT2, both alone and with Ctl siRNA, elicited gel contraction that was significantly greater than the contraction observed with SLIT2 + Sos1 siRNA. Together, these data demonstrate that SLIT2/ROBO1 signaling activates Rac through Sos1, resulting in contraction of compliant collagen gels.
Although our data suggest that Rac is the major target of SLIT2 in MECs (Fig. 2 A), SLIT can also activate Rho (Fig. S4 A). To investigate whether Rho/ROCK is downstream of SLIT2/ROBO1 signaling in NMuMG cells in complaint environments, we performed contraction assays in the presence of either a Rho inhibitor, exoenzyme C3 Transferase, or ROCK inhibitor, Y27632 (Fig. 2, F and G). With Mock treatment there was very little contraction (<3%). In contrast, SLIT2 treatment resulted in significantly increased contraction in the presence of the inhibitors. With ROCK inhibition, this increase was very small (<5%), indicating that ROCK mediates contraction downstream of SLIT2/ROBO1/Rac signaling, consistent with its key role mediating contraction in compliant environments (Wozniak et al., 2003). In contrast, with Rho inhibition, SLIT2 treatment resulted in a significantly greater degree of gel contraction (>25%), consistent with cells activating Rac to mediate gel contraction under this condition. Because SLIT2 treatment alone resulted in more gel contraction than achieved with the SLIT2 + Rho inhibitor, the data do not rule out a role for Rho downstream of SLIT2/ROBO1 signaling. Although SLIT2 may activate both Rac and Rho to achieve maximal contraction, our data support a subsidiary role for Rho activation.
To remodel and reduce the size of the 3D collagen gels, cells apply tension to the collagen fibers. A major contributor to this dynamic cellular behavior is nonmuscle myosin II (NMII) that cross-links and contracts actin. NMII activity is enhanced by phosphorylation, especially on serine 19 of the myosin regulatory light chain (pMRLC19; Aguilar-Cuenca et al., 2014). To evaluate if SLIT2/ROBO1 signaling increases pMRLC19 through Rac, NMuMG cells were treated with SLIT2 along with NSC23766, Sos1, and Ctl siRNA (Fig. 2, H and I). Compared with Mock, SLIT2 resulted in a trending but not significant increase in pMRLC19. Treating with NSC23766, however, significantly reduced pMRLC19 levels, which could then be significantly increased by SLIT2 treatment in a Sos1-dependent manner. Thus, SLIT2/ROBO1 signaling through Sos1 activated Rac and increased pMRLC19 when TrioN and Tiam1 were inhibited.
We also examined pMRLC19 levels in the presence of Rho (C3 Transferase) and ROCK (Y27632) inhibitors because ROCK directly phosphorylates MRLCS19 in response to Rho, while also indirectly increasing pMRLC19 by inhibiting myosin phosphatase (Vicente-Manzanares et al., 2009). We found that inhibiting either Rho or ROCK significantly diminished the level of pMRLC19, and this reduction was not mitigated by SLIT2 treatment (Fig. 2, J and K). Collectively, our studies suggest that although SLIT2/ROBO1 signaling may contribute to gel contraction by leading to MRLC activation, MRLC is not a primary target of SLIT2. Instead, additional targets downstream of ROBO1/Rac signaling are likely activated to achieve the enhanced gel contraction observed upon SLIT2 treatment.
SLIT2/ROBO1 activation of Rac increases FAK in CMAs
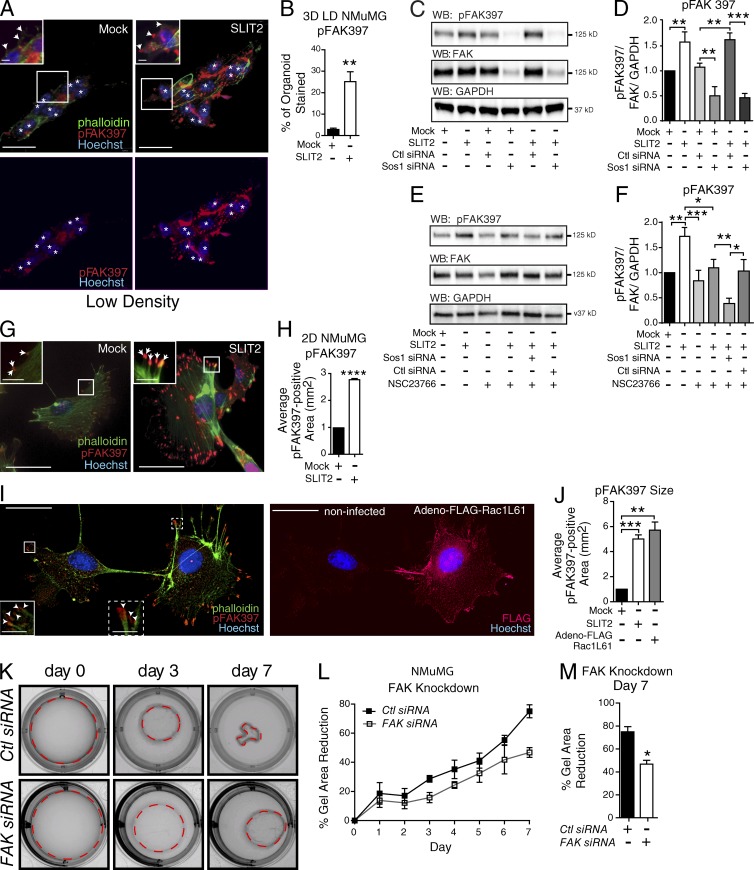
Cells sense ECM changes through mechanosensitive receptors, assembling CMAs that conduct and translate these changes into cells (Roca-Cusachs et al., 2012; Case and Waterman, 2015). Our studies in compliant gels demonstrate that SLIT2/ROBO1 signaling enhances the ability of cells to pull on the ECM, possibility by increased signaling through CMAs. FAK is a major constituent of CMAs, functioning as a key mechanosensory protein. During CMA maturation, FAK activates by autophosphorylation on tyrosine 397 and subsequently phosphorylates and activates other proteins. We examined the phosphorylation status of FAK in NMuMG cells cultured in LD collagen with and without SLIT2 (Fig. 3, A and B). After 3 d, cells under Mock conditions were organized into nascent tubules with a diffuse low level of pFAK397 staining. In contrast, SLIT2 treatment resulted in irregularly shaped aggregates that contained a 20% increase in pFAK397 concentrated in large puncta. To confirm FAK activation by SLIT2, we treated NMuMG cells with SLIT2 (175 ng) and assayed pFAK397 by Western blot, observing a significant increase that was dependent on Sos1 (Fig. 3, C and D). In the presence of the Rac GEF inhibitor, NSC23766, there was a trending but not significant increase in pFAK397 upon SLIT2 treatment (Fig. 3, E and F). Knockdown of Sos1 led to a significant decrease in baseline FAK397 phosphorylation in the absence or presence of NSC23766 (Fig. 3, C–F), suggesting that Rac activation through all three GEFs, but specifically Sos1 downstream of SLIT2/ROBO1 signaling, plays an important role in generating pFAK397.
Figure 3.
SLIT2/ROBO1/Rac signaling increases pFAK397 in CMAs. (A) Representative images of NMuMG cells in LD 3D collagen gels after Mock or SLIT2 treatment and stained for pFAK397 (red), phalloidin (green), and Hoechst (blue). White asterisks indicate nuclei. Arrowheads point to CMAs. (B) Mean percentage of the organoid (3+ nuclei) stained for pFAK397 (n = 12+ organoids). (C and E) Representative blots of pFAK397, total FAK, and GAPDH from NMuMG cells treated with Mock or SLIT2 and indicated siRNA and Rac Inhibitor, NSC. (D and F) Quantification of pFAK normalized to total FAK and GAPDH (n = 3 experiments). (G) NMuMG cells treated with Mock or SLIT2 and immunostained for pFAK397 (red), phalloidin (green), and Hoechst (blue). Arrows point to CMAs. (H) Quantification of pFAK397-labeled puncta (n = 3 experiments). (I, left) Representative images of NMuMG cells stained for pFAK397 (red), phalloidin (green), and Hoechst (blue); on right, FLAG (red). (inset left) Magnified images of boxed regions with arrowheads pointing to CMAs. (J) Quantification of pFAK397-labeled puncta (n = 3 experiments). (K) Representative images of 3D LD gels embedded with NMuMG cells transfected with control or FAK siRNA. (L and M) Quantification of K showing percentage reduction in collagen gel area (n = 3 experiments). Bars, 30 µm. Black asterisks denote significance by t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To better resolve whether these puncta represent FAs, we turned to single-cell immunocytochemical analysis using NMuMG cells because CMAs are difficult to detect in spindle-shaped HME50 cells. We plated NMuMG cells for 12 h, immunostained with pFAK397 and phalloidin, and then measured the size of peripheral pFAK397-positive puncta, located at the tips of labeled actin fibers. We found a threefold increase in the size of pFAK397-labeled puncta in SLIT2 treated cells (Fig. 3, G and H). Next, we investigated the consequences of activating Rac by infecting NMuMG cells with a constitutively active form and staining for pFAK397, FLAG, and phalloidin after 24 h (Adeno–FLAG–Rac L61; Fig. 3, I and J). Analysis revealed that cells expressing activated Rac had significantly larger FAs, similar in size to those observed in SLIT2-treated cells. To test whether gel contraction requires FAK activation, we reduced the expression of FAK ∼75% using siRNA (Fig. S5, A and B) and observed a significant decrease in the ability of cells to remodel the ECM and reduce gel size (Fig. 3, K–M). Together, these data suggest that recruitment of activated FAK to FAs is enhanced by SLIT2/ROBO1-activating Rac through Sos1 and that together this facilitates the contractile behavior of cells in compliant ECM environments.
ROBO1 is present in CMAs and its loss disrupts these structures
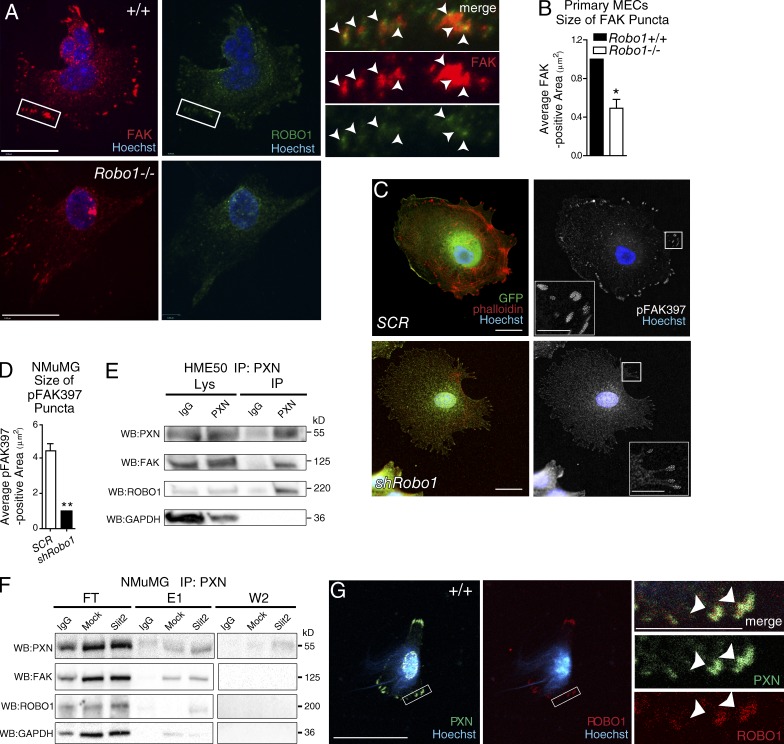
Our data suggest that SLIT2/ROBO1 is involved in CMA signaling. We next asked whether ROBO1 is detected in these structures by performing immunocytochemistry on Robo1+/+ and Robo1−/− MECs plated on fibronectin, using an antibody directed against the ROBO1 cytoplasmic domain. We observed ROBO1 distributed throughout the cell in a punctate pattern and observed colabeling of ROBO1 with FAK (∼40%) in peripheral puncta (Fig. 4 A). In Robo1−/− cells there was little or no ROBO1 staining, and FAK was much less visible, particularly at the edges of cells where quantification revealed reduced accumulation in peripheral puncta (Fig. 4, A and B).
Figure 4.
ROBO1 colocalizes and complexes with CMA components. (A) Representative images of MECs from wild-type (+/+) and Robo1−/− mice stained for FAK (red), ROBO1 (green), and Hoechst (blue). Magnified boxed are the regions of +/+ cells; arrowheads point to colocalization. Bars, 10 µm. (B) Quantification of FAK-labeled FAs in MECs (n = 10+ cells). (C) Representative images of NMuMG cells infected with Robo1 or scrambled (SCR) shRNA lentivirus and stained for GFP (green), phalloidin (red), and Hoechst (blue), and in next panel, pFAK397 (white). Inset: Magnified versions of boxed regions with dashed lines encircling FAs. (D) Quantification of the size of pFAK397-labeled puncta (n = 3 experiments). (E and F) Representative blots from column coimmunoprecipitation on HME50 (E) and NMuMG (F) using anti-PXN bait. Samples of flowthrough (FT), first and second wash (W1/W2), and first and second elutions (E1/E2) were blotted for PXN, FAK, ROBO1, and GAPDH. (G) Representative images of +/+ MECs stained for PXN (green), ROBO1 (red), and Hoechst (blue). Magnified boxed; arrowheads point to colocalization. Bars, 30 µm, unless labeled otherwise. Asterisks denote significance by t test: *, P < 0.05; **, P < 0.01.
To further explore ROBO1’s functional role in CMAs, we infected NMuMG cells with bicistronic Robo1 shRNA-GFP (shRobo1) or scramble shRNA-GFP (SCR; Fig. S5, C and D), treated with SLIT2 and stained for pFAK397 and phalloidin (Fig. 4, C and D). Analyzing CMAs in GFP-positive knockdown cells, we observed reduced accumulation of pFAK397 in puncta in cells expressing Robo1 shRNA. To determine if ROBO1 complexes with CMA proteins, we performed column coimmunoprecipitation using a paxillin (PXN) antibody covalently linked to beads (Fig. 4, E and F). Extracts from HME50 and NMuMG cells were passed over the column, followed by extensive washing, and finally elution of bound proteins. As expected, PXN was eluted after washing. In addition, we found FAK and ROBO1 in the eluted fractions, evidence that ROBO1 may be one of a large number of proteins that can be found associated with CMAs (Geiger and Zaidel-Bar, 2012). We also examined PXN and ROBO1 by immunocytochemistry on wild-type cells and observed their colocalization in puncta at the cell periphery (Fig. 4 G). Together, our data suggest that ROBO1 is both present in CMAs and plays a role in their maturation.
ECM stiffness up-regulates ROBO1
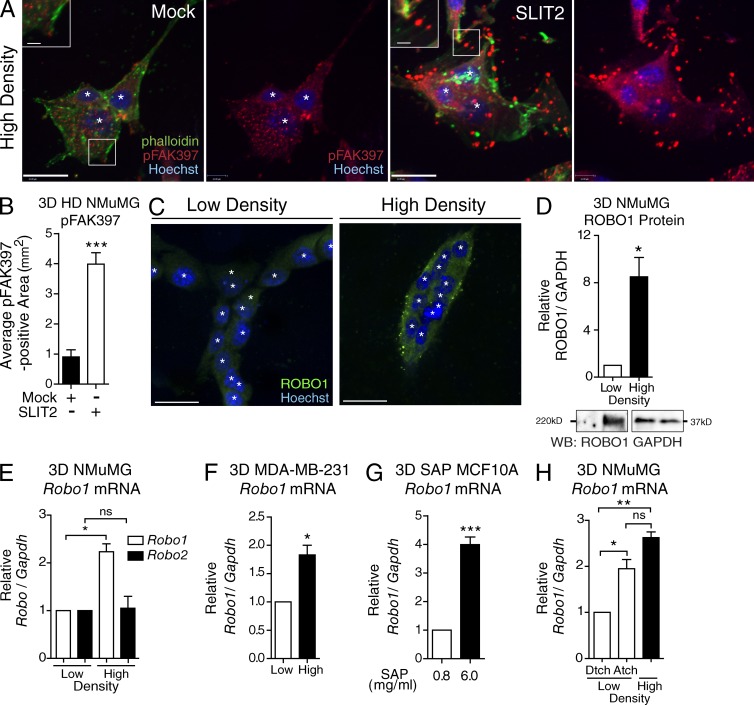
To investigate how SLIT2/ROBO1 signaling responds to extracellular stiffness in a 3D environment, we cultured NMuMG cells in gels tuned to a defined stiffness (∼2,000 Pa) by controlling collagen concentration (Paszek et al., 2005). To examine CMAs, we subjected aggregated NMuMG cells to Mock or SLIT2 and stained with pFAK397 and phalloidin (Fig. 5, A and B). As previously observed (Fig. 3, F and G), SLIT2 resulted in robust recruitment of pFAK397 to CMAs detected as puncta at the end of labeled actin fibers. Next, we allowed cells to mature for 7 d in HD 3D culture before staining for ROBO1. In contrast with LD conditions where cells generate structures resembling a mammary tubule (Fig. 5 C), cells in HD gels appeared as clusters with dramatically up-regulated ROBO1 (Fig. 5 C), an eightfold increase revealed by Western blot (Fig. 5 D). To determine whether Robo2 also exhibited such a dramatic response to extracellular collagen density, we extracted RNA from LD and HD gels and performed quantitative RT-PCR. There was no change in Robo2 expression, but Robo1 was significantly increased (Fig. 5 E), consistent with its up-regulation in microarray studies that compared LD/HD conditions (Provenzano et al., 2009). We also examined whether Robo1 is up-regulated in transformed cells under HD conditions by examining MDA–MB-231 cells. Again, we observed a significant up-regulation of Robo1 expression in HD (Fig. 5 F).
Figure 5.
ROBO1 up-regulation and pFAK397 FA enrichment occurs in high-stiffness 3D collagen systems. (A) Representative images of NMuMG aggregates in HD 3D collagen gels treated with Mock or SLIT2 and stained for pFAK397 (red), phalloidin (green), and Hoechst (blue). (B) Area of pFAK397 puncta located at the end of phalloidin-labeled fibers in acini (3+ nuclei; n = 15+ acini). (C) Representative images of NMuMG acini in LD and HD 3D culture and stained for ROBO1 (green) and Hoechst (blue). (D–F) Analysis of ROBO1 protein (D) and mRNA (E and F) of mammary cells cultured in LD and HD collagen matrices for 7 d. (D, top) Quantification of ROBO1 protein normalized to GAPDH from NMuMG cells (n = 3 experiments). (bottom) Representative blots for ROBO1 and loading control GAPDH. (E and F) Expression of Robo mRNA in NMuMG (E) or MDA–MB-231 (F) cells from 7 d, 3D cultures (n = 3 experiments). (G) MCF10A acini cultured in 3D SAP gels were analyzed for Robo1 (n = 3 experiments). (H) Robo1 expression in NMuMG cells cultured in either LD detached (Dtch), LD attached (Atch), or HD (n = 3 experiments). For mRNA analysis, expression was normalized to Gapdh of total mRNA. Bars, 30 µm. White asterisks denote individual nuclei. Black asterisks denote significance by t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant.
In these experiments, collagen concentration was increased to boost gel rigidity. Collagen, however, is a ligand for integrin and may stimulate integrin signaling. Consequently, the observed increase in Robo1 expression may be a response to enhanced collagen-integrin signaling rather than ECM stiffness. To investigate, we increased ECM stiffness using synthetic self-assembling peptides (SAPs) to construct 3D extracellular microenvironments with tunable mechanical properties. When exposed to physiologic salt solutions, SAPs form fibrils that generate gels of defined stiffness depending on peptide concentration (Miroshnikova et al., 2011). We performed these assays using a human nonmalignant cell line, MCF10A, that undergoes multicellular epithelial morphogenesis in compliant ECM environments (Mroue and Bissell, 2013). Cells were cultured in Matrigel to form acini, transferred into SAP gels calibrated to low (600 Pa) or high (2,000 Pa) stiffness, and then harvested for quantitative RT-PCR analysis. In cells cultured in rigid SAP gels, we observed a fourfold increase in Robo1 (Fig. 5 G). We also performed assays on NMuMG cells using LD gels cultured under attached (higher-stiffness) or detached (after 12 h, lower-stiffness) conditions (Wozniak and Keely, 2005; Fig. 5 H). In the high-stiffness condition, we observed an approximately twofold increase in Robo1 expression that is similar to the increase observed in HD 3D collagen gels but only one-half that observed in SAP gels. Previous studies have shown that even short periods of gel attachment allow cells to generate isometric tension because the matrix is restrained (Lee et al., 2003). Thus, the relative up-regulation of Robo1 in the detached/attached experiment is likely greater than what the data show because the detached gels were attached for 12 h while polymerizing. Collectively, these two studies, in which ligand concentration was held constant even as gel rigidity was increased, demonstrate that Robo1 is up-regulated by ECM stiffness.
miR-203 regulates Robo1 in an ECM stiffness-dependent manner
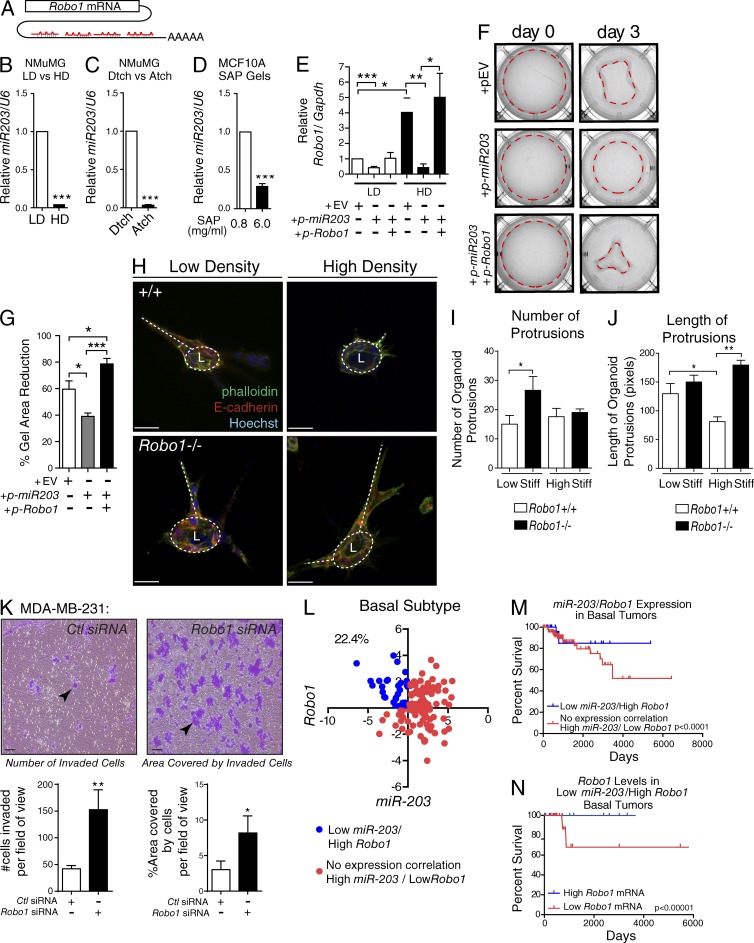
miRNAs control gene expression, thereby regulating many cellular processes, including the interaction of cells with the ECM. ECM stiffness, in turn, regulates the expression of many miRNAs (Mouw et al., 2014), one of which is miR-203 that regulates Robo1 by targeting binding sites in its 3′ UTR (Fig. 6 A). To determine whether extracellular stiffness governs miR-203 expression in our assays, we examined its expression in NMuMG cells cultured in HD and LD collagen, in detached and attached LD gels, and in SAP gels (Fig. 6, B–D). In all cases, miR-203 was significantly down-regulated by high stiffness, correlating with Robo1 up-regulation (Fig. 5, C and D).
Figure 6.
ECM stiffness inhibits miR-203 expression and elevates Robo1 expression to regulate mammary cell contraction and organoid morphology and is associated with improved survival. (A) Cartoon of four miR-203 binding sites on Robo1’s 3′UTR. (B–D) MiR-203 expression analyzed from NMuMG cells cultured in LD or HD (B), LD detached (Dtch) or attached (Atch; C), or 3D SAP gels (D; n = 3 experiments). (E) Robo1 expression analyzed from NMuMG cells transfected with an empty vector (EV), a vector overexpressing premiR-203, or a vector overexpressing Robo1 without a 3′UTR then cultured in LD or HD gels for 3 d (n = 3 experiments). (F) Representative images of LD gels from E, with dashed line encircling the gel area. (G) Quantification of the percent reduction in the gel area (n = 3 experiments). (H) Representative z-stack projections of MECs cultured in LD and HD collagen gels and stained for E-cadherin (red), phalloidin (green), and Hoechst (blue). Circular dashed lines mark lumens (L), and base of protrusions are traced by dashed lines. (I–J) Quantification of number (I) and length of protrusions (J) per organoid (n = 3 experiments). (K) Invasion assay using MDA–MB-231 cells. Arrowheads point to clusters of migrated cells (L) Scatterplot expression analysis of basal subtype breast tumors with low miR-203 + high Robo1 expressing samples indicated as blue dots and all other samples indicated as red dots. Percentage represents the proportion of samples: (low miR-203 + high Robo1 expressing tumors)/(total basal tumor; 22.4%; n = 142). (M) Survival curve analysis of indicated populations from K (n = 142). (N) Survival curve analysis of bottom and top 25% Robo1 expressing basal tumor samples with Low miR-203 + High Robo1 expression (n = 30). Bars: (H) 30 µm; (K) 100 µm. Asterisks denote significance using t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Survival curve p-values determined using Renyi analysis, with weighted values of P = 0 and q = 1 optimized for long-term survival. Stiff, stiffness.
This reduction in miR-203 expression is expected to relieve its repression of Robo1 and may be responsible for the increased Robo1 expression in high-stiffness conditions (Fig. 5, C–H). To investigate, we overexpressed miR-203 in NMuMG cells, embedded the cells in LD and HD gels, harvested them after 3 d, and evaluated Robo1 levels by quantitative RT-PCR (Fig. 6 E). Overexpression of miR-203 resulted in a significant decrease in Robo1 expression, regardless of collagen density. This decrease was not observed when Robo1 expression was reestablished in both HD and LD conditions by cotransfecting miR-203 with a Robo1 plasmid containing no miR-203 binding sites (Fig. 6 E). Because HD gels do not contract, we probed the function of manipulating miR-203 expression by overexpressing it in NMuMG cells and culturing them in LD gels (Fig. 6, F and G). As previously observed (Fig. 1, H–K), NMuMG cells pulled on the ECM and reduced gel size, but this ability was significantly inhibited by miR-203 overexpression. We also cotransfected miR-203 with Robo1 plasmid and observed successful rescue of the cells’ ability to contract the gels (Fig. 6, F and G). These data are consistent with a model in which increasing extracellular stiffness inhibits miR-203 expression, resulting in Robo1 up-regulation and an increased ability of cells to remodel the ECM in compliant environments.
Next, we cultured Robo1+/+ and Robo1−/− primary MECs in LD and HD gels. These cells aggregated and proliferated to form bilayered acini-containing lumens (Macias et al., 2011). We found that in LD gels, both Robo1+/+ and Robo1−/− colonies had multicellular protrusions of similar length, but there were significantly more protrusions when acini were generated using Robo1−/− cells (Fig. 6, H–J). We also observed these cellular extensions in HD gels, but under this condition there was not an increase in the number of extensions emanating from the Robo1−/− acini. Instead, we observed a significant increase in the length of the Robo1−/− extensions. This is in contrast with Robo1+/+ acini that have significantly shorter protrusions in the HD condition. To further investigate, we performed invasion assays on MDA–MB-231 cells and found that knockdown of Robo1 results in a threefold increase in invasiveness, suggesting that ROBO1 is involved in restricting the behavior of cells by maintaining cell shape and positional contact with the ECM (Fig. 6 K and Fig. S5 E).
Breast tumors have the ability to remodel their ECM to create a stiffened tumor microenvironment, leading us to interrogate publicly available clinical datasets for tumors displaying down-regulated miR-203 and corresponding up-regulated Robo1 expression. We focused on basal tumors (n = 142) because SLIT/ROBO signaling is most frequently altered in this tumor subtype (Cancer Genome Atlas, 2012). We found that the percentage of basal tumors with a low miR-203/high Robo1 expression pattern is 22.4% (Fig. 6 L). To evaluate clinical significance, we analyzed whether this inverse expression pattern improved the long-term survival of patients compared with those who do not have this signature. We found a significant increase in long-term survival for patients with basal tumors characterized by low miR-203/high Robo1 (Fig. 6 M). Next, we stratified these low miR-203/high Robo1 expressing basal tumors into lower and upper quartiles of Robo1 expression, and they were analyzed for survivability. Again, we saw a significant increase in long-term survival for patients with basal tumors expressing higher levels of Robo1 compared with those expressing lower levels (Fig. 6 N). Although these data do not take into consideration tumor stiffness, combined, our findings suggest that increasing Robo1 occurs when the ECM stiffness down-regulates miR-203. This may function as a protective, homeostatic mechanism that maintains cell morphology in response to protumorigenic events, thereby significantly increasing the long-term survival of patients.
Discussion
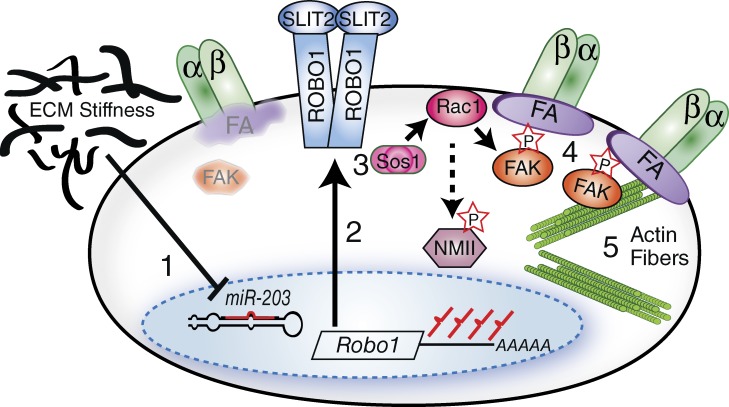
At each stage of tumor development—initiation, promotion, and progression—changes in the microenvironment, specifically the molecular and physical characteristics of the ECM, influence cell behavior. Although much research focuses on the deleterious consequences to cells of these ECM changes, there has been less attention paid to the self-protective measures cells take in response to these changes. Here, we present our studies identifying an miRNA-based response to increased extracellular density that generates an antimetastasis barrier by up-regulating the tumor suppressor, Robo1, which, in turn, enhances CMAs. Previous studies have shown that SLIT2/ROBO1 signaling stabilizes cell–cell contacts by recruiting β-catenin to the cell membrane (Prasad et al., 2008; Tseng et al., 2010; Macias et al., 2011). Here, our studies show that SLIT2/ROBO1 signaling also functions to enhance interactions with the ECM by boosting the size of FAs through Rac and FAK signaling (Fig. 7).
Figure 7.
Cartoon model of SLIT2/ROBO1 signaling circuit. (1) Increased ECM stiffness down-regulates miR-203 derepressing Robo1. (2) ROBO1 expression is increased. (3) SLIT2/ROBO1 signals through Sos1/Rac to activate FAK; Rac also contributes to NMII activation. (4) This results in FA maturation and (5) enhanced cell contractility.
ROBO1 is a well-characterized axon and cell guidance receptor (Ballard and Hinck, 2012). Studies performed primarily in Drosophila melanogaster and some tumor cell lines show that it remodels the cytoskeleton via the direct and indirect interaction of its cytoplasmic domain with GTPase-activating proteins and GEFs and the abelson tyrosine and p21-activated kinases (PAKs; Ballard and Hinck, 2012). ROBO1, like ROBO2, contains four conserved intracellular domains (CC0–CC3), whereas ROBO3 contains three domains, and ROBO4, which is expressed primarily in the vasculature, contains only two (CCO and CC2). Studies on ROBO4 have identified a PXN interaction motif that is located between the CCO and CC2 domains and directly interacts with PXN to inhibit Rac (Jones et al., 2009). This region, however, is only poorly conserved in ROBO1. Consequently, even though we found that ROBO1 coimmunoprecipitates with PXN from breast cell lysates, it may be that this interaction is indirect rather than direct, consistent with ROBO1 being identified in only one of three replicate runs in a recent proteomic analysis of isolated FAs (Kuo et al., 2011). Structure/function studies in Drosophila have shown that the CC3/CC4 domains bind to the adaptor protein Dock/Nck, which recruits both the Rac GEF, Sos, and PAK (Fan et al., 2003; Yang and Bashaw, 2006). This stimulates a series of events with Sos activating Rac, which in turn activates PAK. Currently, there is controversy concerning whether Rac promotes or inhibits cellular contractility via PAKs or other mechanisms. There is evidence that Rac activation of PAK results in the phosphorylation of the NMII light chain and enhanced contraction through the regulation of the actin cytoskeleton and FAs (Kiosses et al., 1999; Brzeska et al., 2004; Szczepanowska et al., 2006). Rac has also been shown to promote the PKC-dependent phosphorylation and recruitment of the NMII heavy chain to FAs (Pasapera et al., 2015). Altogether, these data are consistent with our observations that SLIT2/ROBO1 signaling results in increased Rac and FAK activation and larger FAs. A more detailed analysis of the events occurring subsequent to ROBO1 activation will be required to fully understand the precise sequence of events that enable ROBO1 to enhance contractility in compliant environments while increasing cellular stability in stiff ones.
The cells of tissues are constantly monitoring their environment and responding to maintain external appearances. The comparatively stable outward appearance of adult organisms belies the continuous internal work of cells required to maintain homeostasis. One such kind of homeostasis is tensional homeostasis, whereby cells maintain defined levels of tension in relation to their surroundings, despite the application of mechanical stresses. Integrins are well-known mechanoresponsive receptors that generate signals to the cytoskeleton, which in turn responds by creating reciprocal intracellular contractility. We conclude from our studies that ROBO1 is a mechanoresponsive receptor that similarly functions to coordinate a cell’s response to extracellular stiffness by regulating the actin cytoskeleton. Dysregulation of tensional homeostasis occurs in many tumor microenvironments and certainly contributes to the progression of breast cancer by driving the disorganization of the tissue (Pickup et al., 2014). SLIT/ROBO signaling is altered in many types of cancers, including breast, in which 40.7% of basal, 12.3% of luminal A, and 26.3% of luminal B type tumors display aberrant expression (Cancer Genome Atlas, 2012). Although the expression of Slits is usually silenced in tumors, the Robo1 expression is variable, with some tumors silencing the gene (Dallol et al., 2002; Mitra et al., 2012) and others up-regulating it (Chang et al., 2012). This likely explains the variability of Robo1’s description in the literature as tumor suppressor in some circumstances and oncogene in others (Harburg and Hinck, 2011). In the breast, we observed that Robo1 is up-regulated in response to increased ECM stiffness, which is associated with tumor formation. Eventually, however, only 20% of tumors contain a low miR-203/high Robo1 gene signature, leading us to speculate that ROBO1 mediates a bimodal response. It is initially up-regulated by cells to act as a homeostatic mechanism that maintains intracellular tension and tissue integrity in response to protumorigenic events; this up-regulation is associated with improved breast cancer prognosis (Fig. 6; Chang et al., 2012). Eventually, however, Robo1 expression is frequently silenced by the tumor, or its function coopted to promote promigratory behaviors that facilitate metastasis. Nevertheless, as a “first” responder to transforming signals, with low miR-203/high Robo1 expression patterns signifying a better survival prognosis, the SLIT2/ROBO1 pathway represents a potential therapeutic target for early stage breast cancer treatment.
Materials and methods
Mouse strains
Robo1 mice were generated by inserting the LacZ gene between exons 3 and 4 using standard recombination techniques and genotyped using a PCR screen with the following primers: 5′-TGGCACGAAGGTATATGTGC-3′ forward primer for both wild-type and mutant alleles, 5′-GAAGGACTGGTGGTTTTGAG-3′ reverse primer for wild-type alleles, and 5′-CCTCCGCAAACTCCTATTTC-3′ reverse primer for mutant alleles (Long et al., 2004; Strickland et al., 2006). This study conformed to guidelines set by the University of California, Santa Cruz Animal Care Committee.
2D cell culture
MDA–MB-231 cells were obtained from American Type Culture Collection and cultured in growth medium (DMEM; Gibco) supplemented with 10% heat-inactivated FBS (Atlanta Biologicals) and 1× penicillin streptomycin (Gibco; Marlow et al., 2008). HME50 were cultured in DMEM F12 Nutrient Mixture (Gibco) supplemented with 1× mammary epithelial growth supplement (Gibco) and 1× antibiotic-antimycotic (Gibco). NMuMG epithelial cells were obtained from Clonetics and cultured in growth medium supplemented with 10 µg/ml bovine insulin (Sigma-Aldrich). Primary MECs were harvested from tissue using combined collagenase type III (Worthington) and dispase class II digestion for 12 h, collected by 40-µm sieve filtration, and cultured in standard tissue culture conditions (Macias et al., 2011). For FA immunofluorescence studies, cover-slips were acid washed for 12 h, autoclaved, and coated with human fibronectin (0.5 µg/cm2) for 45 min at 37°C. The slips were thoroughly rinsed with Dulbecco’s PBS (Gibco) and dried in a sterile environment for 1 h before seeding cells (Schober et al., 2007). For experiments using pharmacologic inhibitors, all treatments were 3 h at the following concentrations: SLIT2 (500 ng/ml), NSC23766 (10 µM), C3-exoenzyme (1.0 µg/ml), and Y27632 (10 µM).
3D collagen culture
NMuMG, MDA–MB-231, and primary murine MECs were cultured in rat tail collagen type I (BD; Corning) gels with concentrations optimized for compliancy and stiffness for each cell type by titrating each lot of collagen according to the manufacturer’s recommendations (Provenzano et al., 2010). Compliant, LD conditions were determined by greater than 30% contraction of the gels; and stiff, HD conditions were determined with the lowest concentration of collagen required for <5% contraction of the gel. One day after the gels were cast in tissue culture, attached gels were left attached to the dish and remained at the bottom of media-filled wells. The detached gels were detached from the sides and bottom of the wells and allowed to float to the top of media-filled wells. Gels were fed with fresh media changed every 2–3 d. Gels were cultured for 3–14 d. For contractility studies, gels were imaged every 24 h using Bio-Rad ChemiDoc MP Imager and recorded with ImageLab software (Bio-Rad). Area of the gels was measured and analyzed using ImageJ (National Institutes of Health) and GraphPad Prism (GraphPad Software).
RNA extraction and quantitative RT-PCR
Total RNA was harvested from cells embedded in collagen gels using TRIzol reagent (Invitrogen) and phase separated according to the manufacturer’s protocols with an additional overnight RNA precipitation step in ethanol (Macias et al., 2011). The RNA was further purified with TURBO DNase (Ambion) treatment. Total RNA quality was analyzed by agarose gel electrophoresis and quantified with an ND-1000 spectrophotometer (NanoDrop). cDNA libraries were prepared from 1 µg of total RNA using an iScript cDNA synthesis kit (Bio-Rad). miR-203 and U6 syntheses were prepared from 10 ng of total RNA using Tetro Reverse transcription (Bioline) and Taqman MicroRNA Assays (Life Technologies). Quantitative RT-PCR was performed in triplicate using LightCycler 480 SYBR Green I Master (Roche) and quantified using Bio-Rad CFX Connect Real-Time System and CFX Manager software (Bio-Rad). Results were normalized to GAPDH or U6. Primers used for quantitative PCR were as follows: Robo1-F: 5′-TTATGGTGATGTGGACCTTAGTA-3′, Robo1-R: 5′-GGTTGTATGGGATGGTTGGAG-3′, GAPDH: 5′-F-CATGGCCTTCCGTGTTCCTA-3′, and GAPDH-R: 5′-CCTGCTTCACCACCTTCTTGAT-3′, as previously described (Harburg et al., 2014).
Western blotting
Protein lysates from adherent cells were prepared by direct lysing into 1× sample buffer and analyzed by Western blot. Lysates from 3D gel systems were made with 12-h incubation of diced gels in 2× RIPA buffer supplemented with 2× protease and phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich) at 4°C. Sample buffer (4×) was added to the lysates, and protein analysis was done by Western blot. For phosphor blots, cells were treated 3 h with SLIT2 and the different inhibitors followed by harvest. Phosphospecific primary antibodies pFAK397 (SCBT), ROBO1 (Abcam), and pMRLC19 (Cell Signaling) were prepared in 4% BSA. Antibodies targeting MRLC (SCBT) and FAK (SCBT) were prepared in 5% milk. All proteins were detecting using clarity ECL or FEMTO chemiluminescent substrate (Bio-Rad) on a Bio-Rad ChemiDoc MP Imager and quantified using ImageLab or ImageJ software.
Coimmunoprecipitation
PXN antibody (SCBT) was purified using an Antibody Clean-Up kit (Pierce) and covalently conjugated to agarose beads in the Co-Immunoprecipitation kit (Pierce). Coimmunoprecipitation studies were conducted according to manufacturer protocols. Lysates, flow through, washes, and elutions were run on SDS-PAGE gels and analyzed via Western blot.
Invasion assay
MDA–MB-231 cells were plated for 24 h and transfected with Ctl or Sos1 siRNA using RNAiMAX (Invitrogen) for 24 h. Then, the cells were serum starved for 12 h. Cells were detached using EDTA, and each well was seeded with 200,000 cells in DMEM into the upper well. FBS (10%) was used as the chemoattractant in the bottom of each well. Invasion was allowed to occur for 4 h, then the top part of the upper wells were cleaned with cotton swabs. To visualize invaded cells, they were fixed in methanol and stained with 0.1% crystal violet. A mean of 10 images were taken per well using a 10× objective and quantified using ImageJ.
Immunofluorescence antibodies and imaging
Antibodies and markers used were as follows: pFAK397 (Invitrogen), PXN (SCBT), PXN (Thermo Fisher Scientific), FAK (BD Biosciences), ROBO1 (Abcam), E-Cad (R&D Systems), Alexa Fluor–546 phalloidin (Thermo Fisher Scientific) for actin fibers (Invitrogen), and Hoechst for nuclei (Invitrogen). The cells (2D) or gels (3D) were fixed in 4% paraformaldehyde in PBS for 15 min, washed in PBS for 10 min, and washed in 0.15 M glycine in PBS for 10 min on a shaker. Then they were permeabilized in 0.02% Triton in PBS for 10 min with a subsequent wash in PBS for another 10 min. The sample was then blocked using 10% serum (to secondary host animal) in PBS and incubated with primary antibody diluted in 5% serum in a humidified chamber for 12 h at 4°C or 1 h at RT. After three washes in PBS (2D) or five washes in PBS (3D) for 15 min each, the samples were incubated with secondary antibody and a nuclear stain diluted in 5% serum for 45 min in a humidified chamber at RT. Control samples were also incubated with secondary (without primary) to ensure specificity of antibodies. Samples and controls were washed three times in PBS for 15 min each. Gels (3D) had an additional wash in water for 10 min, then all samples were mounted onto slides or cover-slipped with Fluoromount-G (SouthernBiotech). Fluorescence images of 3D collagen gels were captured using PerkinElmer Volocity Spinning Disk Confocal Microscope. FAs in 2D culture were imaged on a Digital Widefield Microscope (BZ-9000; Keyence Biorevo).
Image analysis
Assessment of actin fiber thickness was calculated in ImageJ and determined per cell. Each cell of a grazing section was identified by a nuclei in the plane of the cross section, and a straight line was drawn through the cell and perpendicular to the actin fibers. Along this line the intensity of the phalloidin stain was measured and is represented by peaks, where the width of the peaks represent the width of the fibers. The distance between the base of the peaks represents the relative distance between each fiber. Each N represents two age-matched littermates and the averaged measurements of at least 25 cells per animal. Measurements of matrix adhesions were also determined using ImageJ. Before analysis, the scale of the images was set using the calibrate function. Then, the adhesions were identified by zooming into peripheral regions of cells where clear puncta were found at the tips of phalloidin-labeled fibers. The phalloidin channel was reduced to barely visible for the puncta to be prominently displayed. Then, using the freehand tool in ImageJ, we circled the puncta and measured the area.
Rho/Rac activity assays
Primary MECs and fractionated myoepithelial cells or HME50 and NMuMG cells were cultured as described in the 2D cell culture section and treated with SLIT2 at 70% confluency for 30 min before lysate preparation. Pull-downs from primary MECs required 36 glands per pull-down and from myoepithelial cells 80 glands per pull-down. Lysates were assayed for active GTPase levels by using the RhoA/Rac/Cdc42 Activation Assay Combo kit (Cell Biolabs) according to the manufacturer’s protocols.
Plasmids, transfections, and infections
Expression of recombinant proteins was conducted with either Lipofectamine 2000 (Thermo Fisher Scientific), RNAi-max (Thermo Fisher Scientific), or Polyethylenimine, Linear (MW 25,000; Polyscience)-mediated transfection with 3:1 transfectant-DNA ratio. Plasmids used for Robo1 expression were based on the pSecTagB backbone (p-Robo1). Plasmids used for premiR203 overexpression were based on the pTarget backbone (p-miR203, p-EV, pRobo1-si: gift from S. Lakka, University of Illinois, Chicago, IL). Production of lentiviral particles for scrambled and Robo1 knockdown experiments involved combination transfection of psPAX2, pMD2.G, and pLVTHM-scrambled-GFP (SCR) or pLVTHM-shRobo1-GFP (shRobo1) into HEK293T cells. Filtered (0.45 µm) viral particles were then diluted in media to infect target mammary lines (MDA–MB-231 and NMuMG). Constitutively active Rac production in NMuMG cells was accomplished by infection with adenovirus encoding Rac L61 mutant (Ad-FLAG-Rac L61; Cell Biolabs) according to the manufacturer’s recommendations.
Bioinformatics and statistics
To assess expression from publicly available breast tumor data, a TCGA-BRCA (breast cancer) data matrix was uploaded into the UCSC Cancer Browser Xena analysis tool. Data for basal subtype tumors were identified according to their PAM50 signature and further segregated into low and high mir-203/Robo1 populations based on expression level. Survival curve (Kaplan–Meier) plots were constructed using GraphPad Prism. The p-values for the curves were determined using the survdiff function with weighted values of P = 0 and q = 1 within R. All other statistical analyses (t test, two-way analysis of variance) were conducted on GraphPad Prism. The p-values of <0.05 were considered significant. Graph columns represent the mean, and error bars represent the SEM.
Online supplemental material
Fig. S1 shows mammary cells expressing ROBO1. Fig. S2 shows HD collagen gels resist contractile forces from NMuMG cells even in the presence of SLIT2. Fig. S3 shows SLIT2 inhibits mammary cell proliferation. Fig. S4 shows SLIT2/ROBO1 modestly, but irreproducibly, activates Rho in MECs and does not activate Rac in luminal MECs. Fig. S5 shows the knockdown of Robo1, FAK, or Sos1 via lentivirus or siRNA. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507054/DC1.
Supplementary Material
Acknowledgments
We thank Dr. S. Lakka for pmir-203 and pRobo1-si constructs; Dr. B. Abrams for support of the University of California Santa Cruz Life Sciences Microscopy Center; and Dr. Jing Zhu for bioinformatics support. We thank Santa Cruz Biotechnology for the gift of siRNA, FAK/Ctl, and antibodies: PXN, GAPDH, HSP70, pFAK397, MRLC, and Rho.
This work was supported by the California Institute for Regenerative Medicine (FA1-00617-1 and CL1-00506-1.2; facilities); National Institutes of Health (GM-098897 to L. Hinck, GM098897-02S1 to O. Cazares, and CA142833 and CA114462 to P.J. Keely); National Science Foundation (DGE 0809125 predoctoral fellowship to L.T.-N. Le); Department of Defense Breast Cancer Research Program BC062562 (W81XWH-07-1-0538 to J.K. Mouw) and BC122990 (W81XWH-13-1-0216 to V.M. Weaver); and University of California Office of the President UC Leads fellowship to J. Ramos.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CMA
- cell matrix adhesion
- Ctl
- control
- FA
- focal adhesion
- GEF
- guanine nucleotide exchange factor
- HD
- high-density
- LD
- low-density
- MEC
- mammary epithelial cell
- MRLC
- Myosin Regulatory Light Chain
- NMII
- nonmuscle myosin II
- NMuMG
- normal murine mammary gland
- PAK
- P21-activated kinase
- PXN
- paxillin
- SAP
- self-assembling peptide
References
- Aguilar-Cuenca R., Juanes-García A., and Vicente-Manzanares M.. 2014. Myosin II in mechanotransduction: master and commander of cell migration, morphogenesis, and cancer. Cell. Mol. Life Sci. 71:479–492. 10.1007/s00018-013-1439-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y.H., Mui K.L., Hsu B.Y., Liu S.L., Cretu A., Razinia Z., Xu T., Puré E., and Assoian R.K.. 2014. A FAK-Cas-Rac-lamellipodin signaling module transduces extracellular matrix stiffness into mechanosensitive cell cycling. Sci. Signal. 7:ra57 10.1126/scisignal.2004838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard M.S., and Hinck L.. 2012. A roundabout way to cancer. Adv. Cancer Res. 114:187–235. 10.1016/B978-0-12-386503-8.00005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaich N., Woodhouse S., Goldie S.J., Mishra A., Quist S.R., and Watt F.M.. 2014. Rewiring of an epithelial differentiation factor, miR-203, to inhibit human squamous cell carcinoma metastasis. Cell Reports. 9:104–117. 10.1016/j.celrep.2014.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd N.F., Guo H., Martin L.J., Sun L., Stone J., Fishell E., Jong R.A., Hislop G., Chiarelli A., Minkin S., and Yaffe M.J.. 2007. Mammographic density and the risk and detection of breast cancer. N. Engl. J. Med. 356:227–236. 10.1056/NEJMoa062790 [DOI] [PubMed] [Google Scholar]
- Brzeska H., Szczepanowska J., Matsumura F., and Korn E.D.. 2004. Rac-induced increase of phosphorylation of myosin regulatory light chain in HeLa cells. Cell Motil. Cytoskeleton. 58:186–199. 10.1002/cm.20009 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N., Cancer Genome Atlas Network 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490:61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case L.B., and Waterman C.M.. 2015. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17:955–963. 10.1038/ncb3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P.H., Hwang-Verslues W.W., Chang Y.C., Chen C.C., Hsiao M., Jeng Y.M., Chang K.J., Lee E.Y., Shew J.Y., and Lee W.H.. 2012. Activation of Robo1 signaling of breast cancer cells by Slit2 from stromal fibroblast restrains tumorigenesis via blocking PI3K/Akt/β-catenin pathway. Cancer Res. 72:4652–4661. 10.1158/0008-5472.CAN-12-0877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallol A., Forgacs E., Martinez A., Sekido Y., Walker R., Kishida T., Rabbitts P., Maher E.R., Minna J.D., and Latif F.. 2002. Tumour specific promoter region methylation of the human homologue of the Drosophila Roundabout gene DUTT1 (ROBO1) in human cancers. Oncogene. 21:3020–3028. 10.1038/sj.onc.1205421 [DOI] [PubMed] [Google Scholar]
- Ding X., Park S.I., McCauley L.K., and Wang C.Y.. 2013. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 288:10241–10253. 10.1074/jbc.M112.443655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontula R., Dinasarapu A., Chetty C., Pannuru P., Herbert E., Ozer H., and Lakka S.S.. 2013. MicroRNA 203 Modulates Glioma Cell Migration via Robo1/ERK/MMP-9 Signaling. Genes Cancer. 4:285–296. 10.1177/1947601913500141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuFort C.C., Paszek M.J., and Weaver V.M.. 2011. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12:308–319. 10.1038/nrm3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Labrador J.P., Hing H., and Bashaw G.J.. 2003. Slit stimulation recruits Dock and Pak to the roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron. 40:113–127. 10.1016/S0896-6273(03)00591-9 [DOI] [PubMed] [Google Scholar]
- Gehler S., Baldassarre M., Lad Y., Leight J.L., Wozniak M.A., Riching K.M., Eliceiri K.W., Weaver V.M., Calderwood D.A., and Keely P.J.. 2009. Filamin A-beta1 integrin complex tunes epithelial cell response to matrix tension. Mol. Biol. Cell. 20:3224–3238. 10.1091/mbc.E08-12-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., and Zaidel-Bar R.. 2012. Opening the floodgates: proteomics and the integrin adhesome. Curr. Opin. Cell Biol. 24:562–568. 10.1016/j.ceb.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Harburg G.C., and Hinck L.. 2011. Navigating breast cancer: axon guidance molecules as breast cancer tumor suppressors and oncogenes. J. Mammary Gland Biol. Neoplasia. 16:257–270. 10.1007/s10911-011-9225-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harburg G.C., Compton J., Liu W., Iwai N., Zada S., Marlow R., Strickland P., Zeng Y.A., and Hinck L.. 2014. SLIT/ROBO2 signaling promotes mammary stem cell senescence by inhibiting Wnt signaling. Stem Cell Rep. 3:385–393. 10.1016/j.stemcr.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.A., Nishiya N., London N.R., Zhu W., Sorensen L.K., Chan A.C., Lim C.J., Chen H., Zhang Q., Schultz P.G., et al. . 2009. Slit2-Robo4 signalling promotes vascular stability by blocking Arf6 activity. Nat. Cell Biol. 11:1325–1331. 10.1038/ncb1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiosses W.B., Daniels R.H., Otey C., Bokoch G.M., and Schwartz M.A.. 1999. A role for p21-activated kinase in endothelial cell migration. J. Cell Biol. 147:831–844. 10.1083/jcb.147.4.831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J.C., Han X., Hsiao C.T., Yates J.R. III, and Waterman C.M.. 2011. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13:383–393. 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.J., Ho C.H., and Grinnell F.. 2003. LPA-stimulated fibroblast contraction of floating collagen matrices does not require Rho kinase activity or retraction of fibroblast extensions. Exp. Cell Res. 289:86–94. 10.1016/S0014-4827(03)00254-4 [DOI] [PubMed] [Google Scholar]
- Long H., Sabatier C., Ma L., Plump A., Yuan W., Ornitz D.M., Tamada A., Murakami F., Goodman C.S., and Tessier-Lavigne M.. 2004. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron. 42:213–223. 10.1016/S0896-6273(04)00179-5 [DOI] [PubMed] [Google Scholar]
- Macias H., Moran A., Samara Y., Moreno M., Compton J.E., Harburg G., Strickland P., and Hinck L.. 2011. SLIT/ROBO1 signaling suppresses mammary branching morphogenesis by limiting basal cell number. Dev. Cell. 20:827–840. 10.1016/j.devcel.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow R., Strickland P., Lee J.S., Wu X., Pebenito M., Binnewies M., Le E.K., Moran A., Macias H., Cardiff R.D., et al. . 2008. SLITs suppress tumor growth in vivo by silencing Sdf1/Cxcr4 within breast epithelium. Cancer Res. 68:7819–7827. 10.1158/0008-5472.CAN-08-1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnikova Y.A., Jorgens D.M., Spirio L., Auer M., Sarang-Sieminski A.L., and Weaver V.M.. 2011. Engineering strategies to recapitulate epithelial morphogenesis within synthetic three-dimensional extracellular matrix with tunable mechanical properties. Phys. Biol. 8:026013 10.1088/1478-3975/8/2/026013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S., Mazumder-Indra D., Mondal R.K., Basu P.S., Roy A., Roychoudhury S., and Panda C.K.. 2012. Inactivation of SLIT2-ROBO1/2 pathway in premalignant lesions of uterine cervix: clinical and prognostic significances. PLoS One. 7:e38342 10.1371/journal.pone.0038342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw J.K., Yui Y., Damiano L., Bainer R.O., Lakins J.N., Acerbi I., Ou G., Wijekoon A.C., Levental K.R., Gilbert P.M., et al. . 2014. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat. Med. 20:360–367. 10.1038/nm.3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroue R., and Bissell M.J.. 2013. Three-dimensional cultures of mouse mammary epithelial cells. Methods Mol. Biol. 945:221–250. 10.1007/978-1-62703-125-7_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapera A.M., Plotnikov S.V., Fischer R.S., Case L.B., Egelhoff T.T., and Waterman C.M.. 2015. Rac1-dependent phosphorylation and focal adhesion recruitment of myosin IIA regulates migration and mechanosensing. Curr. Biol. 25:175–186. 10.1016/j.cub.2014.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek M.J., Zahir N., Johnson K.R., Lakins J.N., Rozenberg G.I., Gefen A., Reinhart-King C.A., Margulies S.S., Dembo M., Boettiger D., et al. . 2005. Tensional homeostasis and the malignant phenotype. Cancer Cell. 8:241–254. 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Pickup M.W., Mouw J.K., and Weaver V.M.. 2014. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 15:1243–1253. 10.15252/embr.201439246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponik S.M., Trier S.M., Wozniak M.A., Eliceiri K.W., and Keely P.J.. 2013. RhoA is down-regulated at cell-cell contacts via p190RhoGAP-B in response to tensional homeostasis. Mol. Biol. Cell. 24:1688–1699: S1–S3. 10.1091/mbc.E12-05-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A., Paruchuri V., Preet A., Latif F., and Ganju R.K.. 2008. Slit-2 induces a tumor-suppressive effect by regulating beta-catenin in breast cancer cells. J. Biol. Chem. 283:26624–26633. 10.1074/jbc.M800679200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano P.P., Inman D.R., Eliceiri K.W., and Keely P.J.. 2009. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 28:4326–4343. 10.1038/onc.2009.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano P.P., Eliceiri K.W., Inman D.R., and Keely P.J.. 2010. Engineering three-dimensional collagen matrices to provide contact guidance during 3D cell migration. Curr Protoc Cell Biol. Chapter 10:Unit 10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., Iskratsch T., and Sheetz M.P.. 2012. Finding the weakest link: exploring integrin-mediated mechanical molecular pathways. J. Cell Sci. 125:3025–3038. 10.1242/jcs.095794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S., Majid S., Yamamura S., Tabatabai L., Suh S.O., Shahryari V., Chen Y., Deng G., Tanaka Y., and Dahiya R.. 2011. Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis. Clin. Cancer Res. 17:5287–5298. 10.1158/1078-0432.CCR-10-2619 [DOI] [PubMed] [Google Scholar]
- Schedin P., and Keely P.J.. 2011. Mammary gland ECM remodeling, stiffness, and mechanosignaling in normal development and tumor progression. Cold Spring Harb. Perspect. Biol. 3:a003228 10.1101/cshperspect.a003228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M., Raghavan S., Nikolova M., Polak L., Pasolli H.A., Beggs H.E., Reichardt L.F., and Fuchs E.. 2007. Focal adhesion kinase modulates tension signaling to control actin and focal adhesion dynamics. J. Cell Biol. 176:667–680. 10.1083/jcb.200608010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E., Lovén J., Xu N., Meisgen F., Wei T., Brodin P., Jaks V., Kasper M., Shimokawa T., Harada M., et al. . 2012. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis. 1:e3 10.1038/oncsis.2012.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland P., Shin G.C., Plump A., Tessier-Lavigne M., and Hinck L.. 2006. Slit2 and netrin 1 act synergistically as adhesive cues to generate tubular bi-layers during ductal morphogenesis. Development. 133:823–832. 10.1242/dev.02261 [DOI] [PubMed] [Google Scholar]
- Szczepanowska J., Korn E.D., and Brzeska H.. 2006. Activation of myosin in HeLa cells causes redistribution of focal adhesions and F-actin from cell center to cell periphery. Cell Motil. Cytoskeleton. 63:356–374. 10.1002/cm.20125 [DOI] [PubMed] [Google Scholar]
- Tseng R.C., Lee S.H., Hsu H.S., Chen B.H., Tsai W.C., Tzao C., and Wang Y.C.. 2010. SLIT2 attenuation during lung cancer progression deregulates beta-catenin and E-cadherin and associates with poor prognosis. Cancer Res. 70:543–551. 10.1158/0008-5472.CAN-09-2084 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S., and Horwitz A.R.. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790. 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak M.A., and Keely P.J.. 2005. Use of three-dimensional collagen gels to study mechanotransduction in T47D breast epithelial cells. Biol. Proced. Online. 7:144–161. 10.1251/bpo112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak M.A., Desai R., Solski P.A., Der C.J., and Keely P.J.. 2003. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 163:583–595. 10.1083/jcb.200305010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., and Bashaw G.J.. 2006. Son of sevenless directly links the Robo receptor to rac activation to control axon repulsion at the midline. Neuron. 52:595–607. 10.1016/j.neuron.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Zhang K., Corsa C.A., Ponik S.M., Prior J.L., Piwnica-Worms D., Eliceiri K.W., Keely P.J., and Longmore G.D.. 2013. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat. Cell Biol. 15:677–687. 10.1038/ncb2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.