Analysis of luminal lysosomal pH in combination with heterologous expression of lysosomal-associated proteins indicates that peripheral lysosomes are more alkaline than juxtanuclear ones and that depletion of Rab7 and its effector, RILP, are associated with and can account for the reduced acidification.
Abstract
We examined the luminal pH of individual lysosomes using quantitative ratiometric fluorescence microscopy and report an unappreciated heterogeneity: peripheral lysosomes are less acidic than juxtanuclear ones despite their comparable buffering capacity. An increased passive (leak) permeability to protons, together with reduced vacuolar H+–adenosine triphosphatase (V-ATPase) activity, accounts for the reduced acidifying ability of peripheral lysosomes. The altered composition of peripheral lysosomes is due, at least in part, to more limited access to material exported by the biosynthetic pathway. The balance between Rab7 and Arl8b determines the subcellular localization of lysosomes; more peripheral lysosomes have reduced Rab7 density. This in turn results in decreased recruitment of Rab-interacting lysosomal protein (RILP), an effector that regulates the recruitment and stability of the V1G1 component of the lysosomal V-ATPase. Deliberate margination of lysosomes is associated with reduced acidification and impaired proteolytic activity. The heterogeneity in lysosomal pH may be an indication of a broader functional versatility.
Introduction
Lysosomes, the terminal organelles of the endocytic pathway, are characterized by a highly acidic lumen that is rich in hydrolytic enzymes. Lysosome functions are diverse and include digestion of macromolecules taken up by endocytosis or macropinocytosis (Saftig and Klumperman, 2009), degradation of organelles sequestered by autophagy (Shen and Mizushima, 2014), and elimination of pathogens engulfed by phagocytosis (Saftig and Klumperman, 2009). Lysosomes also regulate metal ion homeostasis (Shawki et al., 2012) and can sense nutrient availability, thus controlling autophagy, energy metabolism, and organelle biogenesis (Settembre et al., 2011; Roczniak-Ferguson et al., 2012). Finally, lysosomes are integral to antigen processing, degrading antigenic proteins to peptides that are loaded onto major histocompatibility complex class II molecules for presentation to T cells (Trombetta et al., 2003; Furuta et al., 2013).
Like other compartments of the endocytic pathway, lysosomes generate and maintain an acidic lumen by means of the vacuolar H+-ATPase (V-ATPase). The acidic lysosomal lumen is well suited for the activity of hydrolases (de Duve and Wattiaux, 1966; Ng et al., 2012), many of which have pH optima between 4.5 and 5.5 (Mellman et al., 1986). The protonmotive force generated by the V-ATPase also drives the coupled transport of ions and small molecules (Hinton et al., 2009; Scott and Gruenberg, 2011), including amino acids by members of the SLC36 family (Thwaites and Anderson, 2011) and chloride by the ClC-7 antiporter (Scott and Gruenberg, 2011). In addition, luminal acidification is required for efficient cargo sorting along recycling and degradative pathways; accordingly, dissipation of the transmembrane pH gradient using weak bases, ionophores, or V-ATPase inhibitors causes mistargeting of multiple ligands and proteases (Gonzalez-Noriega et al., 1980; Basu et al., 1981; Tycko et al., 1983; Schwartz et al., 1984; Brown et al., 1986; Johnson et al., 1993; Presley et al., 1993, 1997; Chapman and Munro, 1994; Reaves and Banting, 1994; van Weert et al., 1995). Alkalinizing agents also alter membrane traffic because budding of carrier vesicles from endosomes is dependent on functional V-ATPases (Clague et al., 1994; van Weert et al., 1995; Aniento et al., 1996). Luminal acidification is seemingly required for the recruitment of Arf1 and β-COP (Aniento et al., 1996) as well as Arf6 and ARNO (Hurtado-Lorenzo et al., 2006) to endosomal membranes. Lastly, formation of intraluminal vesicles is similarly dependent on an acidic endosomal lumen (Falguières et al., 2008).
Although lysosomes are generally conceived as a uniform compartment, there is evidence of both structural (Baccino et al., 1971; Goldstone and Koenig, 1974; Pertoft et al., 1978; de Duve, 1983; Luzio et al., 2007; Saftig and Klumperman, 2009; Huotari and Helenius, 2011) and functional heterogeneity (Nilsson et al., 1997; Terman et al., 2006; Kurz et al., 2008; Lima et al., 2012), even within individual cells. Neither the source nor the consequences of this heterogeneity are known. We reasoned that a detailed analysis of lysosomal pH would provide insight into lysosomal heterogeneity. The luminal pH of a large number of individual lysosomes can be measured accurately by noninvasive means in intact cells, yielding robust data that can be correlated with parameters such as subcellular location. Using this approach, in combination with heterologous expression of lysosomal-associated proteins, we found that peripheral lysosomes are more alkaline than juxtanuclear ones and that depletion of Rab7 and its effector, Rab-interacting lysosomal protein (RILP), is associated with and can account for the reduced acidification.
Results
Lysosomal pH is heterogeneous
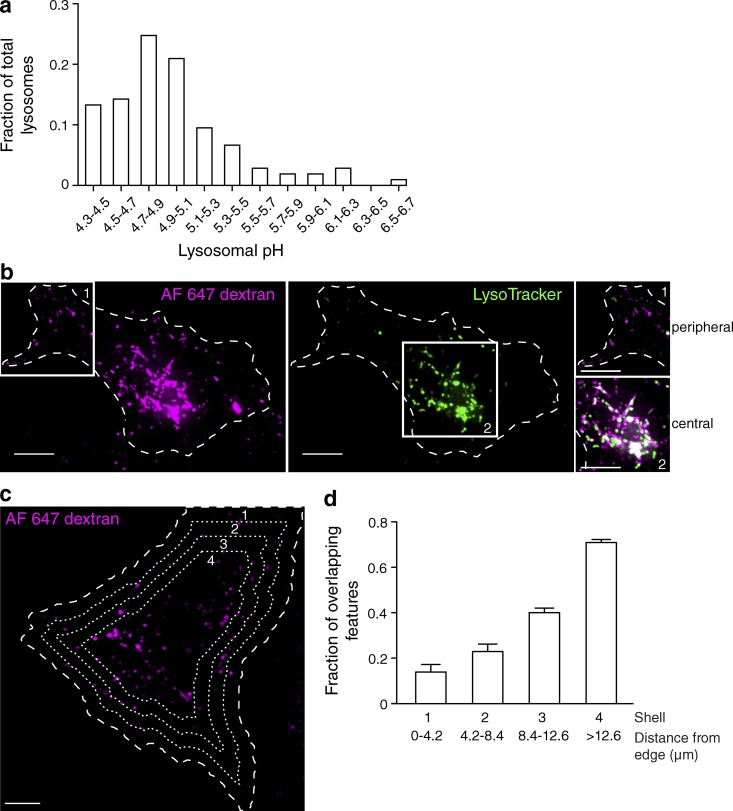
We assessed lysosomal heterogeneity within the cell by measuring the pH of individual lysosomes using ratiometric fluorescence microscopy. The lysosomes of HeLa cells were loaded with two fluorescently tagged probes: the pH-sensitive Oregon green 488–dextran and the pH-insensitive tetramethylrhodamine-dextran. Oregon green 488 has a pKa of 4.7, which is well suited to measure the acidic pH of the lysosome lumen. The emission of the two dyes was separately determined in individual lysosomes, and the fluorescence ratio was converted to absolute pH using the internal calibration procedure described in the Conversion of fluorescence ratio to pH section in Materials and methods. A 6-h pulse with the dextrans was followed by an overnight (12–16 h) chase to ensure that the probes had fully traversed the endocytic pathway and were confined to lysosomes. Accordingly, the fluorescent dextrans showed a high degree of overlap with the conventional lysosomal marker, lysosomal associated membrane protein 1 (LAMP1; Fig. S1 a). Near the cell center, 90 ± 1% of the dextran-containing compartments colocalized with LAMP1-RFP, whereas the overlap was 82 ± 2% at the cell periphery. LAMP1, in turn, colocalized extensively with saposin C, a protein thought to be restricted to lysosomes, where it facilitates glycolipid catabolism (Fig. S1 b). Having validated that the dextran is located predominantly in lysosomes, we proceeded to measure the pH of this compartment. As illustrated in Fig. 1 a, ratiometric fluorescence microscopy using Oregon green 488 revealed an unexpectedly broad distribution: although the largest fraction of lysosomes had a luminal pH (pHl) between 4.7 and 4.9 (mean pHl was 4.76 ± 0.03), consistent with earlier determinations (Ohkuma and Poole, 1978), a sizable subpopulation was more alkaline, reaching pH values ≥6.
Figure 1.
Lysosomal pH is heterogeneous. (a) The lysosomes of HeLa cells were loaded with two fluorescently tagged dextrans: the pH-sensitive Oregon green 488–dextran and the pH-insensitive tetramethylrhodamine-dextran. Individual lysosomal pH was measured by ratiometric fluorescence microscopy, and fluorescence values were converted to pH as described in the Conversion of fluorescence ratio to pH section in Materials and methods. The fraction of total lysosomes as a function of pH was plotted (n = 105 individual lysosomes from three independent experiments). (b) The lysosomes of HeLa cells were loaded with Alexa Fluor 647–dextran (magenta) and then loaded with the acidotropic dye LysoTracker (green) and imaged by confocal microscopy. The cell of interest is outlined with a dashed line. White boxes delineate two regions, labeled 1 and 2, which were used to superimpose the two channels in the right-hand insets. Here and elsewhere, white indicates overlap of the signals. (c) Cells expressing soluble EGFP were loaded with Alexa Fluor 647–dextran and LysoTracker. The outline of the cells was defined from the pattern of EGFP fluorescence (not depicted) using Volocity software and was degraded inward by 4.2 µm every cycle (three times) to create four concentric shells (dashed lines), which were used for the calculations shown in d. (d) The fraction of overlapping features (dextran-positive lysosomes containing LysoTracker) for each shell was determined and plotted as a function of distance from the edge of the cell. Error bars represent SEM (n = 10 cells from three independent experiments). Bars, 10 µm.
Peripheral lysosomes are more alkaline than central lysosomes
The heterogeneity of the lysosomal pH was validated using an alternative approach: lysosomes were loaded with dextran conjugated to Alexa Fluor 647, a pH-insensitive far red–emitting fluorophore, and chased overnight as above. The cells were then incubated with LysoTracker, a fluorescent weak base that accumulates in highly acidic organelles. In agreement with the ratiometric determinations, we found that most, but not all, lysosomes accumulated LysoTracker. We also noted that a greater proportion of the most acidic (LysoTracker positive) lysosomes resided in the perinuclear region, whereas less LysoTracker accumulated in peripheral lysosomes (Fig. 1 b). A similar pattern was observed in C2C12 murine myoblasts and in human microvascular endothelial cells (Fig. S2, a and b). However, this feature was not universal: primary dendritic cells, which have long tubular lysosomes, and CHO cells displayed more homogeneous lysosomal pH (Fig. S2, c and d).
We used HeLa cells hereafter to analyze the source of the heterogeneity in lysosomal pH. To quantitatively assess whether luminal pH is correlated with subcellular localization, we analyzed the LysoTracker content of individual lysosomes as a function of their distance from the edge of the cell (Fig. 1, c and d). The boundary of the cells was determined by outlining the fluorescence of cytosolic EGFP. This boundary was degraded inward by 4.2-µm increments to create four concentric shells, as illustrated in Fig. 1 c. The degree of overlap between dextran-containing and LysoTracker-positive lysosomes was quantified in each shell and is plotted as a function of distance from the edge of the cell in Fig. 1 d. The fraction of overlapping features increased progressively from the cell periphery to the cell center, confirming that central lysosomes are more likely to accumulate LysoTracker. The pH gradient correlated with the distance of the lysosomes from the microtubule-organizing center (MTOC) rather than to their closeness to the plasma membrane. This was established by measuring the overlap between dextran and LysoTracker as a function of their distance from pericentrin, a component of the MTOC (Fig. S3, a and c), or from wheat germ agglutinin, added externally as a plasmalemmal label (Fig. S3, b and d). These observations not only confirm that the lysosomal pH is heterogeneous, but indicate that in HeLa cells, the peripheral lysosomes (those further away from the MTOC) tend to be more alkaline than those in the perinuclear region.
Margination of lysosomes elevates their luminal pH
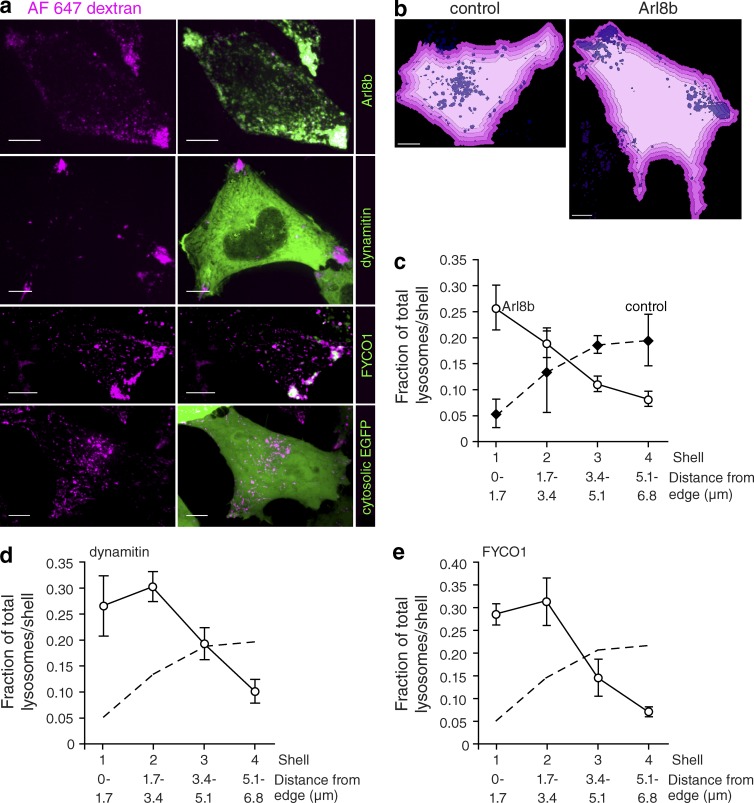
The preceding experiments indicated that the luminal pH of lysosomes was correlated with their position within the cell. To determine whether these events are causally related, we sought to manipulate lysosome localization. Lysosomes move bidirectionally along microtubules, propelled by dynein and kinesins (Matteoni and Kreis, 1987; Jordens et al., 2001). Arl8b, a small GTPase found on the lysosomal surface (Hofmann and Munro, 2006), binds the centrifugal motor kinesin-1 through its effector SKIP (SifA and kinesin-interacting protein; Rosa-Ferreira and Munro, 2011). FYCO1 (FYVE and coiled-coil domain–containing 1) and RILP both associate with Rab7, another lysosomal GTPase (Bucci et al., 2000). Like SKIP, FYCO1 binds kinesin-1 (Pankiv et al., 2010), whereas RILP associates with the centripetal motor dynein (Jordens et al., 2001). We took advantage of the reported ability of these proteins, when expressed ectopically, to modify lysosomal positioning. As reported (Bagshaw et al., 2006), we found that expression of Arl8b or FYCO1 results in accumulation of lysosomes at the cell periphery (Fig. 2 a). We were similarly able to displace lysosomes peripherally by inhibiting dynein. The activity of dynein requires the protein complex dynactin, which can be disrupted by suprastoichiometric expression of one of its components, dynamitin (Burkhardt et al., 1997). As shown in Fig. 2 a, expression of dynamitin unmasked the unopposed activity of kinesin, displacing the lysosomes peripherally. The redistribution of lysosomes was quantified for cells expressing Arl8b, dynamitin, and FYCO1 using the concentric shell analysis described above (Fig. 2 b). As summarized in Fig. 2 (c–e), in control cells, the largest fraction of lysosomes was found in the innermost shells, whereas the opposite was true for cells expressing Arl8b, dynamitin, or FYCO1. Furthermore, expression of Arl8b or dynamitin caused an overall reduction in the accumulation of LysoTracker, suggesting alkalinization of the lysosomal pH (Fig. S4, a–e).
Figure 2.
Peripheral lysosome phenotype is magnified by expression of proteins that cause lysosomal redistribution. (a) The lysosomes of HeLa cells were loaded with Alexa Fluor 647–dextran (magenta) and transfected with Arl8b-mCherry, dynamitin-EGFP, FYCO1-mCherry, or cytosolic EGFP cDNAs (all shown in green). (b) Using Volocity software, the boundary of control or Arl8b-expressing cells was determined as in Fig. 1 and was degraded inward by 1.7 µm every cycle (four times) to create four shells (illustrated by shades of pink). Lysosomes are shown in purple. (c) The fraction of total lysosomes per shell in control cells and Arl8b-expressing cells was determined and plotted as a function of distance from the edge of the cell. n = 4 cells from four independent experiments. (d and e) Using a similar analysis, the fraction of total lysosomes per shell in dynamitin-expressing cells (d) and FYCO1-expressing cells (e) was determined and plotted as a function of distance from the edge of the cell. n = 6–16 cells from at least three independent experiments. The dashed lines indicate the fraction of control lysosomes, reproduced from c for comparison. Error bars represent SEM. Bars, 10 µm.
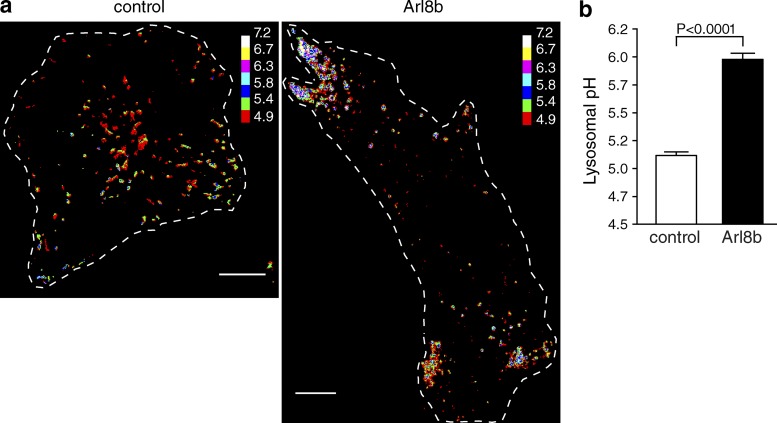
To determine whether positioning within the cell could affect lysosome acidification, we quantified luminal pH in cells where lysosomes were displaced to the periphery by Arl8b expression. The lysosomes of HeLa cells were initially loaded with the ratiometric pH-sensitive fluorescein-dextran (Ohkuma and Poole, 1978) and subsequently transfected with Arl8b during the chase period. For these experiments, fluorescein was used instead of Oregon green 488 because its higher pKa (6.4) more accurately reports the more alkaline pH detected in peripheral lysosomes. As before, lysosomal pH was determined by ratiometric fluorescence microscopy; a pseudocolor scale was used to indicate pH. As illustrated in Fig. 3 a, peripheral lysosomes tend to be less acidic than perinuclear ones. More importantly, when margination of lysosomes is imposed by expression of Arl8b, a much larger fraction of the lysosomes is less acidic. The mean pH of control lysosomes was 5.12 ± 0.03, whereas that of Arl8b-expressing lysosomes was 5.98 ± 0.05 (Fig. 3 b). Thus, the data are consistent with the notion that a more peripheral location is associated with and may cause elevation of the lysosomal pH.
Figure 3.
Lysosomes displaced to the cell periphery by expression of Arl8b are more alkaline. The lysosomes of HeLa cells were loaded with fluorescein-dextran, a ratiometric pH-sensitive dye. Cells were then either mock transfected or transfected with Arl8b-mCherry cDNA. Lysosomal pH was measured by ratiometric fluorescence microscopy as described in the Ratiometric fluorescence microscopy section in Materials and methods. (a) Representative images of control and Arl8b-expressing cells with fluorescein fluorescence ratios converted to pH (see calibration bar). Cells of interest are outlined with dashed lines. Bars, 10 µm. (b) Lysosomal pH was determined in control and Arl8b-expressing cells. Error bars represent SEM (n = 30 and 39 cells from at least nine independent experiments for control and Arl8b transfectants, respectively).
Mechanism of peripheral lysosome alkalinization
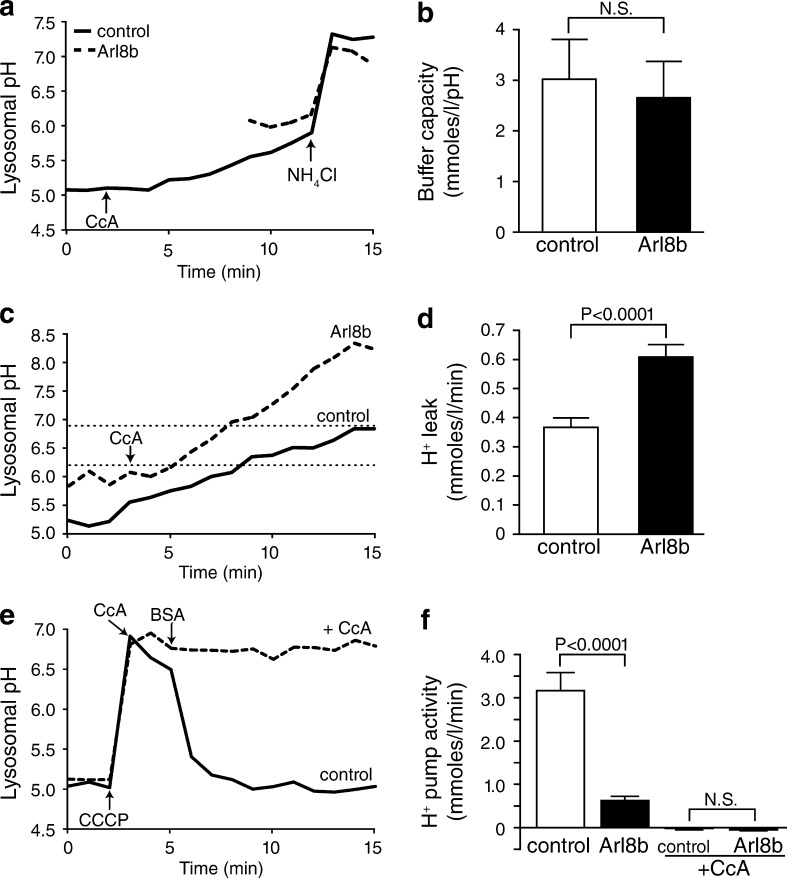
We proceeded to investigate the possible mechanisms underlying the differential pH of juxtanuclear versus more peripheral lysosomes. We initially measured lysosomal buffering capacity, pulsing untransfected or Arl8b-expressing cells with membrane-permeant weak electrolytes (Loiselle and Casey, 2010). Because buffering power often varies as a function of pH, we ensured that the measurements were performed under comparable conditions. To this end, the control cells were pretreated with the V-ATPase inhibitor concanamycin A (CcA), and their pH was allowed to drift upwards until it reached values comparable to those measured in the peripheral lysosomes of Arl8b-expressing cells (Fig. 4 a). At that stage, NH4Cl was added, and the resulting change in lysosomal pH was quantified. As illustrated in Fig. 4 b, the intrinsic buffering capacity of (predominantly perinuclear) lysosomes of control cells and (largely peripheral) lysosomes of Arl8b transfectants—calculated as Δ[NH4+]l/ΔpHl—was indistinguishable.
Figure 4.
Buffering capacity is indistinguishable in central and peripheral lysosomes, whereas peripheral lysosomes have greater H+ leak and lower H+ pump activity. The lysosomes of HeLa cells were loaded with fluorescein-dextran. Cells were then mock transfected or transfected with Arl8b-mCherry cDNA. Lysosomal pH was measured by ratiometric fluorescence microscopy. (a) Lysosomal buffering capacity was assessed using the NH4Cl pulse technique, as described in the Buffer capacity section in Materials and methods. Control lysosomes were treated with 2 µM of the V-ATPase inhibitor CcA when indicated by the arrow. 3.75 mM NH4Cl was added to both samples where indicated by the arrow. (b) Quantification of lysosomal buffer capacity in control and Arl8b-expressing cells. n = 6–9 cells for each condition from at least six independent experiments. (c) Lysosomal H+ leak was assessed by measuring the rate of alkalinization in control and Arl8b-expressing cells after addition of 2 µM CcA (indicated by arrow). H+ leak was calculated in the pH 6.2–6.9 interval (indicated by dotted lines). (d) Quantification of H+ leak in control and Arl8b-expressing cells. n = 21–27 cells for each condition from at least six independent experiments. (e and f) Lysosomal H+ pump activity was measured in control and Arl8b-expressing cells in the presence or absence of 2 µM CcA as indicated. (e) 30 µM protonophore CCCP was added to untransfected cells either alone or together with CcA, where indicated by the arrow, to dissipate the transmembrane ΔpH. CCCP was removed by addition of 5 mg/ml of defatted BSA when indicated. (f) Quantification of H+ pump activity determined from the rate of reacidification recorded upon removal of CCCP. n = 6–9 cells for each condition from at least three independent experiments. Error bars represent SEM.
The rate of passive H+ (equivalent) permeability, i.e., the H+ “leak,” was measured next. The leak was estimated from the rate change of the lysosomal pH (ΔpHl/t) after inhibition of the V-ATPase (Fig. 4 c). The rate of change was measured between pH 6.2 and 6.9 in all cases to ensure that the driving forces and buffering power were comparable. The rate of H+ leakage in the peripheral lysosomes of Arl8b-transfected cells was considerably larger than in lysosomes of control cells (0.61 ± 0.04 mmol/l/min vs. 0.37 ± 0.03 mmol/l/min, respectively; Fig. 4 d).
The activity of the lysosomal H+ pump was compared next. The pumping rate decreases as the lumen acidifies, becoming increasingly difficult to detect. To facilitate its measurement, we initially dissipated the lysosomal transmembrane pH gradient using carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a conductive protonophore. This maneuver also ensured that measurements in peripheral and juxtanuclear lysosomes would be performed under thermodynamically equivalent conditions. The activity of the V-ATPase was assessed from the rate of reacidification, which was triggered by removing the protonophore by washing the cells with medium containing defatted albumin. Scavenging CCCP in this manner initiated a rapid acidification that was attributable to the activity of the V-ATPase because it was fully prevented by addition of CcA (Fig. 4 e). The H+ pump activity, calculated as the slope of the reacidification measured during the initial 90 s after removal of CCCP multiplied by the buffering power, was 3.17 ± 0.42 mmol/l/min in control cells (Fig. 4 f). H+ pump activity in Arl8b-expressing cells was 0.63 ± 0.10 mmol/l/min, markedly less than in control cells (Fig. 4 f). Together, these data indicate that the less acidic pH of peripheral lysosomes is attributable to a combination of reduced H+ pumping and increased rates of H+ leakage.
Interaction of lysosomes with the plasma membrane and with endosomes
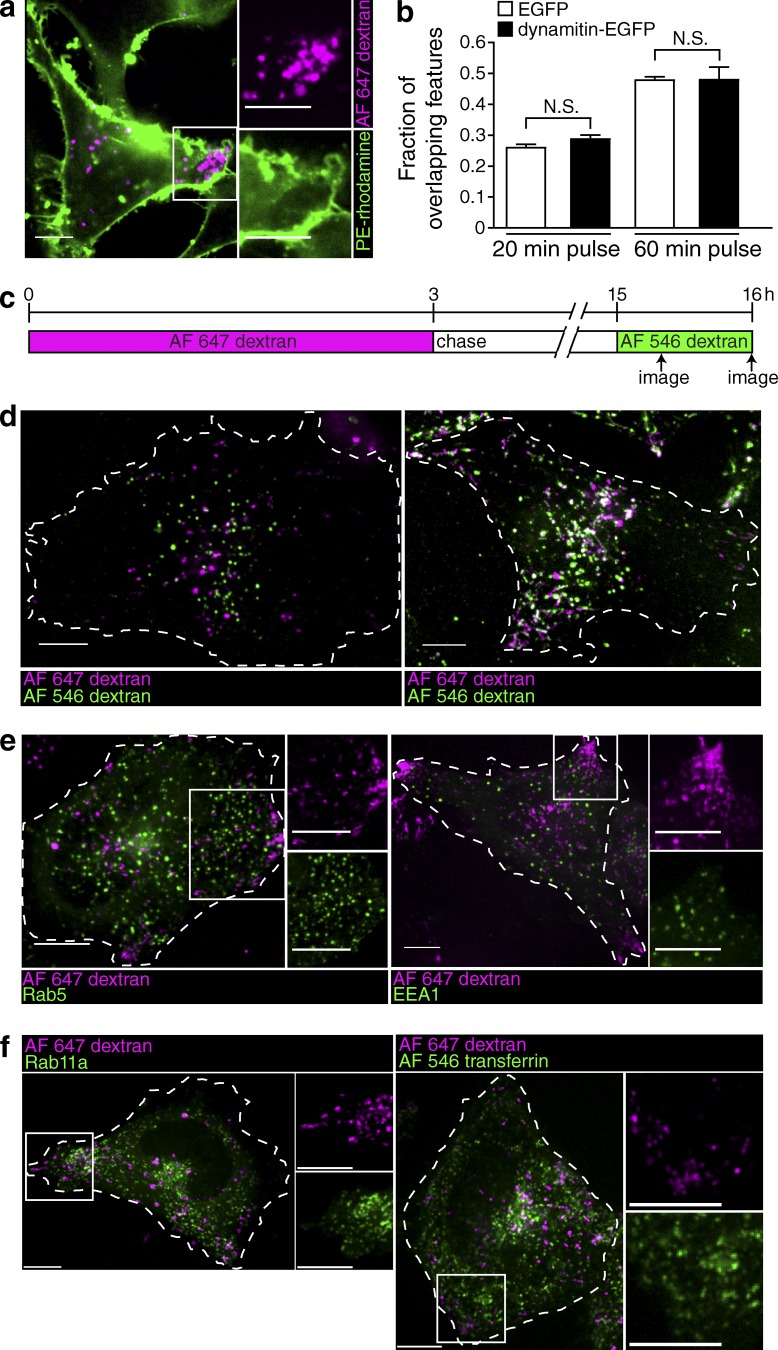
Under some conditions, lysosomes can fuse with the plasma membrane. We reasoned that, by virtue of their location, peripheral lysosomes may be prone to interact transiently with the plasma membrane, which could explain their increased leakage and may result in loss or inactivation of V-ATPases. This possibility was tested by analyzing the ability of plasmalemmal components to access peripheral lysosomes in Arl8b-transfected cells. The plasma membrane was labeled with phosphatidylethanolamine (PE)-rhodamine, whereas lysosomes were identified by preloading them with Alexa Fluor 647–dextran as above. Confocal microscopy revealed no evidence of continuity or preferential vesicular traffic between the two compartments when the fate of PE-rhodamine was monitored for incubation periods of up to 15 min (Fig. 5 a).
Figure 5.
Peripheral lysosomes do not exhibit enhanced interaction with the plasma membrane, nor do they colocalize with early or recycling endocytic markers. (a) The lysosomes of HeLa cells were loaded with Alexa Fluor 647–dextran (magenta) and transfected with dynamitin-EGFP cDNA (not depicted). The plasma membrane was stained with the fluorescent marker PE-rhodamine (green), and the cells were imaged by confocal microscopy in the 5–10-min interval after membrane labeling. The white box indicates the region magnified in the right-hand insets. (b) The fraction of overlapping features (Alexa Fluor 647–dextran-positive lysosomes containing Alexa Fluor 546–dextran) at 20 and 60 min was determined in lysosomes of both control (EGFP transfected) and dynamitin-EGFP–transfected cells. Error bars represent SEM (n = 6 cells for each condition from three independent experiments). (c) Diagrammatic presentation of the protocol used: lysosomes were loaded with Alexa Fluor 647–dextran for 3 h, chased for 12 h, and then loaded with Alexa Fluor 546–dextran for 20 or 60 min. Cells were imaged by confocal microscopy when indicated by the arrows. (d) Representative images of control cells imaged 20 min (left) or 60 min (right) after addition of Alexa Fluor 546–dextran. Cells of interest are outlined with dashed lines. (e) Lysosomes were loaded with Alexa Fluor 647–dextran (magenta) and transfected with Rab5-EGFP (green) or EEA1-EGFP (green). Cells were imaged by confocal microscopy. White boxes indicate the regions magnified in the right-hand insets. (f) Lysosomes were loaded with Alexa Fluor 647–dextran (magenta) and transfected with Rab11a (green) or loaded for 20 min with Alexa Fluor 546–transferrin (green). Cells were imaged by confocal microscopy. White boxes indicate the regions magnified in the right-hand insets. Bars, 10 µm.
Rather than interacting with the plasma membrane, it is conceivable that peripheral lysosomes fuse more readily with earlier compartments of the endocytic pathway, which are known to be less acidic. To assess this possibility, lysosomes were loaded with Alexa Fluor 647–dextran for 3 h and chased for 12 h, and the cells were then allowed to internalize Alexa Fluor 546–dextran for 20 or 60 min (Fig. 5 c). The rate of delivery of endocytosed material to lysosomes was compared in dynamitin-EGFP–transfected cells and in cells transfected with EGFP alone. Dynamitin-EGFP was used in this instance to displace lysosomes centrifugally to enable the detection of the red- and far red–labeled dextrans (for reasons that are unclear, EGFP-tagged constructs of Arl8b were found to be much less effective than Arl8b-mCherry). Representative control cells imaged 20 min (left) or 60 min (right) after addition of Alexa Fluor 546–dextran are shown in Fig. 5 d. Quantification of the overlap of the two dextran markers revealed no significant difference in the rate of endosome to lysosome fusion between control (EGFP transfected) and dynamitin-EGFP–expressing cells (Fig. 5 b).
The preceding data suggest that the altered pH homeostasis of peripheral lysosomes cannot be attributed to interaction with the plasma membrane or to accelerated fusion with earlier components of the endocytic pathway. In agreement with this conclusion, the peripheral lysosomes of HeLa cells were found to be devoid of Rab5 and early endosome antigen 1 (EEA1), markers of early/sorting endosomes (Fig. 5 e), and of Rab11a and transferrin, markers of recycling endosomes (Fig. 5 f).
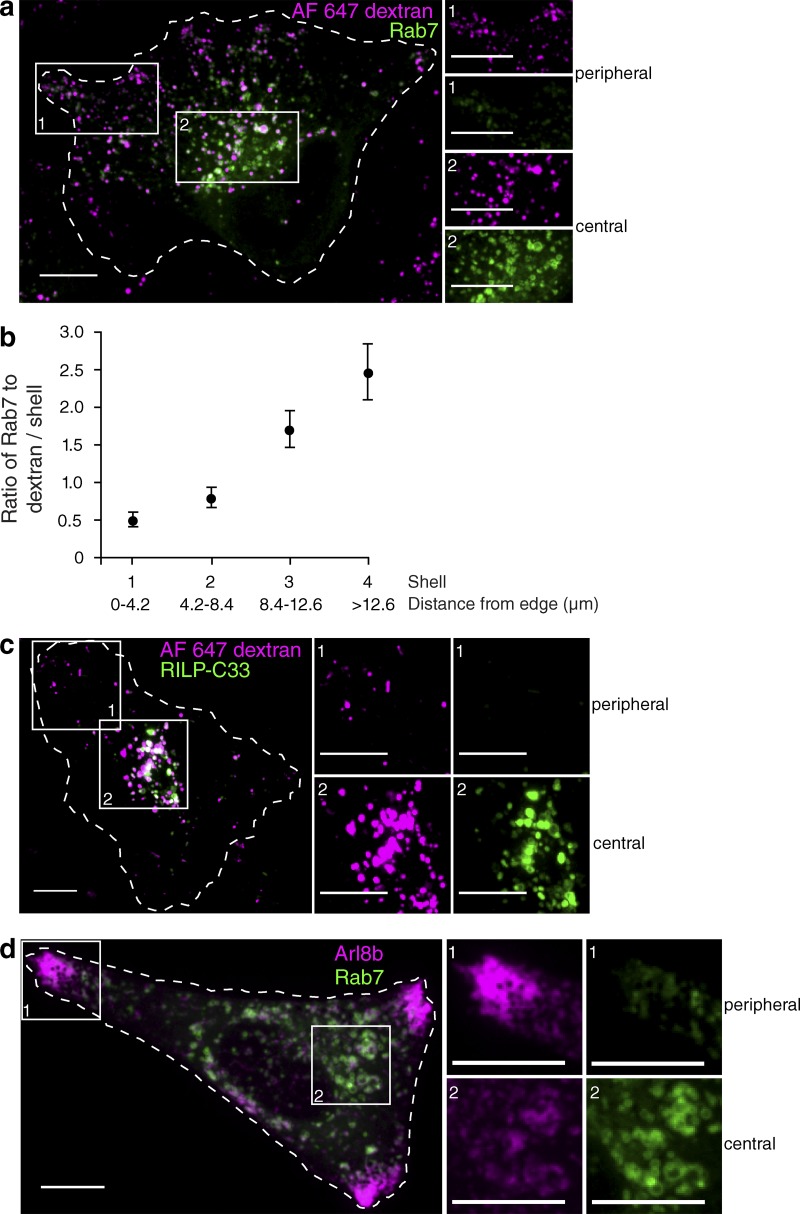
Peripheral lysosomes have decreased levels of Rab7
The activity of the V-ATPase, which we found to be reduced in peripheral lysosomes, was recently described to be regulated by RILP, an effector of GTP-bound Rab7 (De Luca et al., 2014). It was therefore conceivable that the density of active Rab7 differs between peripheral and juxtanuclear lysosomes. It is noteworthy that RILP also serves to recruit dynein to Rab7-positive organelles; a reduced association of RILP with lysosomes could therefore also account for their more peripheral distribution. We therefore compared the relative density of Rab7 on the membrane of juxtanuclear versus peripheral lysosomes. In cells transfected with wild-type EGFP-Rab7 but otherwise untreated, Rab7 was more abundant in central lysosomes (Fig. 6 a). Quantitation of the ratio of fluorescence of Rab7 to that of the dextran used to label the lysosomes, followed by segmentation of the cells in concentric shells, as in Fig. 1, revealed that the density of Rab7 increased progressively from the outermost to the innermost lysosomes (Fig. 6 b). We used RILP-C33 to ensure that the uneven distribution of ectopically expressed EGFP-Rab7 was a reliable indication of the behavior of the endogenous Rab7. Although heterologous expression of full-length RILP causes lysosomal clustering due to excessive recruitment of dynein, the C-terminal fragment (RILP-C33) does not have this effect (Cantalupo et al., 2001). As shown in Fig. 6 c, peripheral lysosomes bound less RILP-C33 than more central ones, recapitulating the observations made with EGFP-Rab7. Importantly, because RILP-C33, like full-length RILP, associates only with GTP-bound Rab7, these data indicate that it is the active form of Rab7 that is unevenly distributed among lysosomes. Moreover, because RILP modulates the activity of the V-ATPase, the observed depletion of this protein in peripheral lysosomes can explain their reduced acidification. This possibility was tested by silencing RILP by means of siRNA. Using a pool of four oligonucleotides, RILP was knocked down by 90 ± 3%, as determined by quantitative PCR. Under these conditions, pHl increased to 6.3 ± 0.1 compared with 5.1 ± 0.1 in control cells treated with nontargeting siRNA.
Figure 6.
Peripheral lysosomes have decreased levels of Rab7 and RILP-C33. (a) Lysosomes of HeLa cells were loaded with Alexa Fluor 647–dextran (magenta) and transfected with EGFP-Rab7 (green). Cells were imaged by confocal microscopy. (b) The boundary of cells was determined and degraded inward by 4.2 µm every cycle to create four concentric shells as in Fig. 1. The ratio of Rab7 to dextran fluorescence intensity per shell was determined and plotted as a function of distance from the edge of the cell. Error bars represent SEM (n = 13 cells). (c) Lysosomes were loaded with Alexa Fluor 647–dextran (magenta) and transfected with EGFP-RILP-C33 (green). Cells were imaged by confocal microscopy. (d) HeLa cells were transfected with Arl8b-mCherry (magenta) and EGFP-Rab7 (green) and imaged by confocal microscopy. (a, c, and d) The numbered white boxes indicate the regions magnified in the right-hand insets. Bars, 10 µm.
Collectively, our data suggest that depletion of active Rab7 is associated with, and in fact may lead to, a more peripheral localization of lysosomes within the cell. Indeed, it is possible that the balance between active Rab7 and Arl8b dictates the positioning of the lysosomes within the cell and also their luminal pH. The experiment illustrated in Fig. 6 d supports this contention. When cells were cotransfected with both Rab7 and Arl8b, the more centrally located organelles had considerably higher levels of Rab7 than of Arl8b, whereas the reverse was true for the marginated lysosomes.
Delayed delivery of proteins from the secretory pathway to peripheral lysosomes
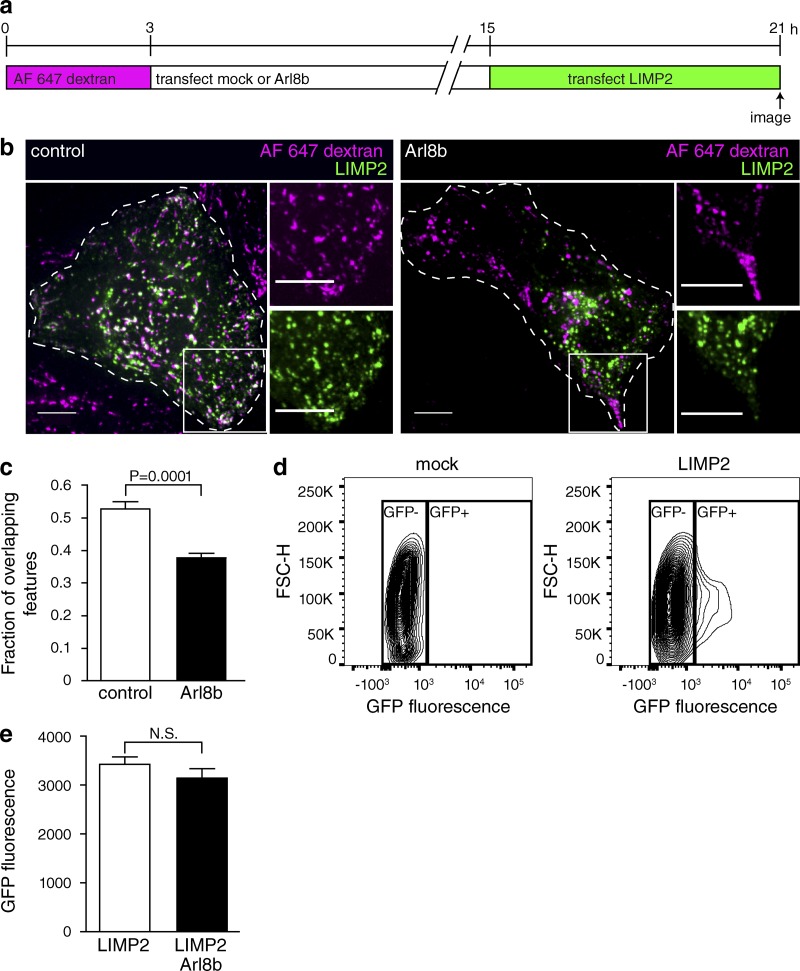
Although reduced RILP content can explain the lower V-ATPase activity of peripheral lysosomes, their elevated H+ leakage remained unaccounted for. In this regard, it is most likely that components other than Rab7 and RILP are also altered in these lysosomes. The composition of lysosomes is dictated by a balance between acquisition of endocytic components and components originating from the secretory pathway. Because delivery of endosomal constituents was not visibly abnormal (Fig. 5), we analyzed the ability of lysosomes located in different regions to import secretory material. The molecular nature of the components of the H+ leak pathway is unknown, but they are likely to be transmembrane proteins. We therefore used lysosomal integral membrane protein 2 (LIMP2), an abundant prototypical lysosomal protein that spans the membrane twice, to compare the ability of central and peripheral lysosomes to acquire transmembrane proteins from the Golgi complex.
Lysosomes were loaded with Alexa Fluor 647–dextran and either mock transfected or transfected with Arl8b-mCherry to induce margination of the lysosomes. 12 h later, the cells were transfected with LIMP2-GFP and imaged by confocal microscopy 6 h later, the minimum time required for the synthesis of detectable amounts of GFP (Fig. 7 a). As illustrated in Fig. 7 b, newly synthesized LIMP2-GFP was rapidly delivered to preformed lysosomes in cells not expressing Arl8b (left), but delivery was delayed in cells transfected with Arl8b (right), an effect that was particularly noticeable in the most peripheral lysosomes. The overlap between dextran and LIMP2-GFP was significantly higher in control cells than in Arl8b-expressing cells (Fig. 7 c). Importantly, the total expression of LIMP2-GFP, quantified by flow cytometry, was not altered by transfection of Arl8b (Fig. 7, d and e). These data are consistent with the notion that peripheral lysosomes are less accessible to de novo synthesized material emerging from the Golgi and hence subject to gradual changes in composition that may account for their increased H+ permeability.
Figure 7.
Delayed delivery of newly synthesized components from Golgi to peripheral lysosomes. (a) Diagrammatic presentation of the protocol used: lysosomes were loaded with Alexa Fluor 647–dextran for 3 h and then mock transfected or transfected with Arl8b-mCherry. 12 h later, cells were transfected with the lysosomal membrane protein LIMP2-GFP and imaged by confocal microscopy 6 h later. (b) Representative images of control and Arl8b-expressing cells treated as in panel a. Cells of interest are outlined with dashed lines. The white boxes indicate the regions magnified in the right-hand insets. Bars, 10 µm. (c) The fraction of overlapping features (dextran-positive lysosomes containing LIMP2-GFP) was determined for both control and Arl8b-expressing cells. n = 10–13 cells for each condition from three independent experiments. (d) The expression of LIMP2-GFP was assessed by flow cytometry. The gating of mock-transfected or LIMP2-GFP–transfected cells is shown. (e) Quantification of GFP fluorescence in cells expressing LIMP2 or LIMP2 and Arl8b, gated as illustrated in d. Error bars represent SEM. n = 3 independent experiments.
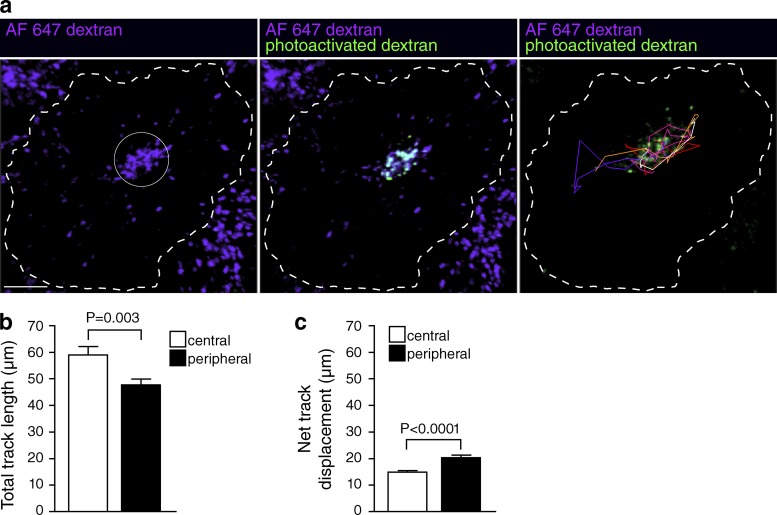
Because the properties of peripheral and juxtanuclear lysosomes differ, we sought to determine the rate at which these two populations exchange with each other. To this end, we generated a photoactivatable dextran (CMNB-caged carboxyfluorescein-dextran) and, using a focused beam, generated localized subpopulations of fluorescent lysosomes either near the cell center or at the periphery (Fig. 8 a). The fluorescent features were then tracked for periods of up to 120 min. Total track length was assessed by acquiring images every 1 min to more accurately assess overall (nondirectional) lysosome mobility; gradual photobleaching limited these measurements to 30 min. Central and peripheral lysosomes exhibited total track lengths of 59 ± 3 µm and 48 ± 2 µm, respectively (Fig. 8 b). Net displacement from the starting position, measured over 120 min and acquiring images every 4 min, was 15 ± 1 µm and 20 ± 1 µm for central and peripheral lysosomes, respectively. Because the distance from the MTOC (pericentrin) to the farthest edge of the cells averaged 30.5 ± 1.7 µm, it is clear that exchange between the two pools of lysosomes is at best slow, requiring many hours.
Figure 8.
Assessment of lysosomal mobility by photoactivation. (a) Lysosomes were loaded with Alexa Fluor 647–dextran (purple) and CMNB-caged carboxyfluorescein-dextran. A ROI (indicated by solid white circle) was selectively illuminated using a 405-nm laser to photoactivate the CMNB-caged carboxyfluorescein-dextran (green in middle and right panels). The resulting fluorescence was imaged using a 488-nm laser every 1 min for 30 min or every 4 min for 2 h. Photoactivated lysosomes were tracked using Imaris software (right). (b) Lysosomes were tracked in cells where CMNB-caged carboxyfluorescein-dextran was photoactivated in either the cell center or the cell periphery. The total track length over 30 min was quantified. Error bars represent SEM (n = 104–204 lysosomes from three to five cells from five independent experiments). (c) The net displacement over 2 h was quantified. Error bars represent SEM (n = 137–285 lysosomes from three to seven cells from two independent experiments).
Functional differences between juxtanuclear and peripheral lysosomes
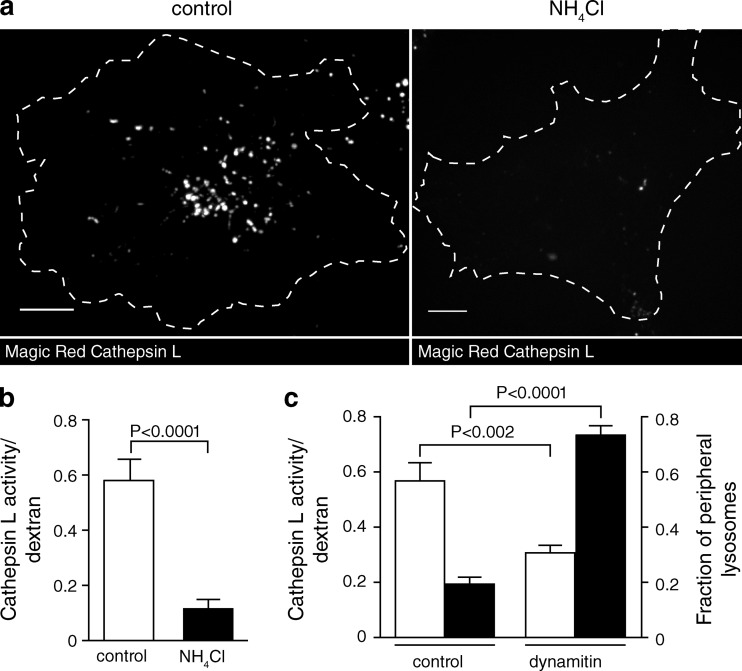
Does the more alkaline pH of peripheral lysosomes have physiological consequences? Several of the degradative enzymes present in lysosomes display acidic pH optima; it is therefore reasonable to assume that such enzymes might have lower degradative capacity in peripheral lysosomes. We tested this hypothesis using a fluorogenic assay of cathepsin L activity (Magic Red cathepsin L). Cathepsin L is pH sensitive, its activity being maximal between pH 4.5 and 5.5 (Mason et al., 1985). We first verified the pH responsiveness of the reaction by comparing the fluorescence generated by untreated versus NH4Cl-treated lysosomes. The data were normalized to the lysosomal content of each cell, determined by preloading with labeled dextran, as above. As anticipated based on the reported pH optimum of the enzyme, cathepsin L activity was much greater in the untreated (acidic) lysosomes than in those alkalinized by NH4+ (Fig. 9, a and b).
Figure 9.
Peripheral lysosomes exhibit lower cathepsin L activity than central lysosomes. (a) Lysosomes were loaded with Alexa Fluor 647–dextran (not depicted) and incubated with Magic Red cathepsin L substrate (shown in gray/white). On the right, lysosomes were alkalinized with NH4Cl before Magic Red addition. Cells were imaged by confocal microscopy. Bars, 10 µm. (b) Cathepsin L activity was quantified in untreated and NH4Cl-treated cells by normalizing Magic Red fluorescence to Alexa Fluor 647–dextran fluorescence. n = 15–17 cells for each condition from three independent experiments. (c) Lysosomes were loaded with Alexa Fluor 647–dextran followed by transfection with EGFP or dynamitin-EGFP. Finally, the cells were incubated with Magic Red cathepsin L substrate and imaged by confocal microscopy. Cathepsin L activity (left y axis, unshaded bars) was quantified by normalizing Magic Red fluorescence to Alexa Fluor 647–dextran fluorescence. n = 20–26 cells for each condition from three independent experiments. The fraction of peripheral lysosomes (right y axis, shaded bars) was determined using Volocity software. The boundary of the cells was determined and was degraded inward by 9.1 µm to create two shells. The fraction of total lysosomes in the outermost shell for control cells and dynamitin-EGFP–expressing cells was determined and plotted. n = 13 from three independent experiments. Error bars represent SEM.
We next quantified cathepsin L activity in control (EGFP transfected) and dynamitin-EGFP–expressing cells. As discussed in the third Results section, dynamitin-EGFP was used to marginate the lysosomes in these experiments to enable the detection of the fluorescence of Magic Red. As shown by the shaded bars in Fig. 9 c, dynamitin-EGFP effectively displaced a large fraction of the lysosomes to the cell periphery. More importantly, lysosome margination was accompanied by a highly significant decrease in the overall activity of cathepsin L (Fig. 9 c, unshaded bars).
Discussion
While in resting cells in culture, the density of lysosomes is greatest near the MTOC, a considerable fraction is more peripheral. The subcellular distribution of lysosomes varies among cell types and can be affected by several parameters, including their metabolic state (Korolchuk et al., 2011). Lysosome positioning is thought to reflect the dynamic balance between centripetal and centrifugal microtubule motors. These are linked to the lysosomal membrane via adapters, which are in turn associated with active GTPases. Arl8b recruits kinesin-1 via its adapter, SKIP, fostering displacement toward the (peripheral) plus ends of microtubules. Rab7, in contrast, has dual effects: like Arl8b, it promotes centrifugal motion by associating with kinesins via FYCO1, but it is also able to bind RILP, which recruits the dynein complex, a minus end–directed motor. The net effect of Rab7 appears to be centripetal, inasmuch as lysosomes concentrate around the MTOC in cells transfected with constitutively active Rab7 and disperse peripherally when dominant-negative Rab7(T22N) is expressed (Bucci et al., 2000). One would therefore expect Rab7-depleted lysosomes to be more peripheral, as documented in Fig. 6.
Not only is the subcellular distribution of lysosomes heterogeneous, but so are their contents and, as our data indicate, their luminal pH. The relationship between lysosomal positioning, motility, and pH had been studied before, but only in the context of the cytosolic (as opposed to luminal) pH. Heuser and colleagues first reported that acidification of the cytosol caused lysosomes to move to the periphery of macrophages, to the basolateral region of epithelial cells, and into the processes of neurons (Heuser, 1989; Parton et al., 1991); cytosolic alkalinization had the converse effect. The dependence on cytosolic pH was also proposed to account for the effects of nutrients on lysosomal positioning: addition of amino acids was found to cause cytosolic acidification, which could account for the accompanying centripetal displacement of the lysosomes (Korolchuk et al., 2011). These effects are most simply explained by differential pH sensitivity of the microtubule-associated centripetal and centrifugal motors and/or of the molecules that link them to the lysosomes. Of note, all of these components are exposed to the cytosol, sensing the prevailing pH, but cannot directly perceive the luminal pH. Hence, we believe that the more acidic pH of the juxtanuclear lysosomes is not the cause, but a consequence of their subcellular location. Accordingly, deliberate margination of lysosomes by expression of Arl8b or dynamitin (Fig. 3 and Fig. S4) was associated with elevated luminal pH. Furthermore, accumulation of the acidotropic dye LysoTracker was reported to be deficient in cells expressing Rab7(T22N) (Bucci et al., 2000).
Two separate effects contribute to the more alkaline pH of peripheral lysosomes: decreased V-ATPase activity and increased H+ leakage. We were unable to compare the number of V-ATPases in lysosomes located in different regions of the cell because available antibodies were not sufficiently sensitive for quantitative immunostaining. However, differences in the rate of pumping need not result from altered density of pumps and could instead reflect reduced activity per pump. Indeed, the elevated pH of peripheral lysosomes can be attributed to a reduced pumping rate caused by the inability of RILP to associate with the V1G1 subunit of the V-ATPase (De Luca et al., 2014). RILP regulates recruitment to the lysosome of V1G1 (also known as ATP6V1G1), which is essential for the correct assembly and operation of the V-ATPase (De Luca et al., 2014). In good accordance, our data indicate that lysosomal pH was significantly increased in cells depleted of RILP. Because they are deficient in Rab7, peripheral lysosomes are expected to contain less RILP, which was validated using RILP-C33, a surrogate marker.
The source of the increased H+ leakage is unclear because the pathways capable of passively translocating H+ or its equivalents across the lysosomal membrane have not been characterized (Haines, 2001). It has been proposed that the transient receptor potential cation channel, mucolipin subfamily 1 (TRP-ML1), can serve as a H+ channel in lysosomes (Soyombo et al., 2006). Accordingly, lysosomes of TRP-ML1–deficient cells are overacidified. It is noteworthy, however, that such cells process lipids abnormally and that in cells that accumulate lipids as a result of lysosomal storage disorders, like Niemann-Pick disease, lysosomes accumulate in the perinuclear region (Ko et al., 2001; Lebrand et al., 2002). Thus, the overacidification may be a consequence of the altered subcellular localization of the lysosomes. Regardless of the precise identity of the pathways responsible for H+ leakage, it is conceivable that the membrane composition of central versus peripheral lysosomes differs substantially. Our data indicate that delivery of newly synthesized proteins proceeds at different rates for central versus peripheral lysosomes, which will inevitably alter their composition.
The biological significance of lysosomal heterogeneity has not been explored, but several possibilities can be contemplated. More peripheral lysosomes may be poised to serve in membrane repair, in which case reduced luminal acidification, and consequently reduced protease and lipase activities, may be advantageous. Alternatively, heterogeneous lysosomal positioning may be an indicator and possibly a determinant of their metabolic state. Korolchuk et al. (2011) reported that margination of lysosomes upon expression of Arl8b increased the activity of mTORC1 (mammalian target of rapamycin complex 1). The change in mTORC1 activity may have resulted from the associated change in luminal pH. However, our results (Fig. S5, a and b) and those of others (Zoncu et al., 2011; Settembre et al., 2012) suggest that luminal alkalinization depresses the activity of mTORC1, an effect opposite to that reported by Korolchuk et al. (2011). Clearly, the relationship between lysosomal localization, pH, and mTORC1 activity requires further definition.
Lastly, it is conceivable that the differential positioning of lysosomes is a consequence and perhaps an indicator of their development, aging, or disease. Hirota et al. (2004) described the existence of a postlysosomal compartment endowed with membrane proteins characteristic of lysosomes, e.g., LIMP2, yet devoid of cathepsins; like the peripheral lysosomes of HeLa cells, such postlysosomes were less acidic than bona fide lysosomes containing cathepsins. The prevalence of such postlysosomes and their pH regulatory features remains to be investigated in detail. Lysosomal aging has not been rigorously studied, but like other exhausted organelles, old lysosomes likely become damaged (leaky) and are cleared by autophagy. This is indeed the fate of lysosomes that are purposefully damaged (Hung et al., 2013; Maejima et al., 2013). Intentional damage of lysosomes is associated with impaired acidification (Hung et al., 2013), resembling the phenotype of the peripheral lysosomes. As found for other organelles, membrane leakiness may signal the initiation of lysosomal autophagy.
In summary, our findings are consistent with the notion that lysosome function is not homogeneous. It can be modulated by changes in its enzymatic composition resulting from altered connectivity to the biosynthetic pathway and/or to recycling of selected components to earlier compartments of the endocytic pathway. However, the functional responsiveness of lysosomes may also be changed acutely by altering their pH, a level of regulation that could be exerted more rapidly by the GTPases. The manner whereby the balance between Rab7 and Arl8b is maintained remains to be defined, but competition for finite binding sites or reciprocal feedback inhibition may cause polarization of lysosomes within the cell.
Materials and methods
Reagents
Oregon green 488–dextran, tetramethylrhodamine-dextran, Alexa Fluor 647–dextran, Alexa Fluor 546–dextran, LysoTracker red DND-99, CMNB-caged carboxyfluorescein, SE (5-carboxyfluorescein-bis-[5-carboxymethoxy-2-nitrobenzyl]ether and β-alanine-carboxamide succinimidyl ester), amino-dextran, Alexa Fluor 546–transferrin, Oregon green 488–labeled wheat germ agglutinin, nigericin, 18-mm round glass coverslips, and Slide-A-Lyzer 10K Dialysis Cassettes were from Thermo Fisher Scientific. Fluorescein-dextran and monensin were from Sigma-Aldrich. All dextrans were 10,000 mol wt. FuGene 6 transfection reagent was from Promega. HiPerFect was from QIAGEN. Arl8b-mCherry cDNA was a gift from J. Kay (State University of New York at Buffalo, Buffalo, NY). Arl8b was subcloned from Arl8b-GFP (Kaniuk et al., 2011) into pmCherry-N1 (Takara Bio Inc.). The generation of plasmids used for the expression of p50 dynamitin–EGFP (Harrison et al., 2003), FYCO1-mCherry (Pankiv et al., 2010), Rab5-EGFP (Scott et al., 2002), EEA1-EGFP (Gillooly et al., 2000), EGFP-Rab11a (Choudhury et al., 2002), EGFP-Rab7 (Bucci et al., 2000), EGFP-RILP-C33 (Colucci et al., 2005), LAMP1-RFP (Sherer et al., 2003), and P3X-FLAG-EGFP-pericentrin-myc (provided by V. Menella, Hospital for Sick Children, Toronto, Canada; Kim and Rhee, 2014) is described in the references provided. pEGFP-N1 was from Takara Bio Inc. LIMP2-GFP cDNA was a gift from D. Neculai (Hospital for Sick Children, Toronto, Canada). Full-length hLIMP2 was amplified from a mammalian gene collection cDNA template (AT8-G3) using forward and reverse primers (5′-CTCGAGCTCAAGCTTATGGGCCGATGCTGCTTCTACAC-3′ and 5′-GATCCCGGGCCCGCGGTACCGGTTCGAATGAGGGGTGC-3′, respectively) and cloned into pEGFP-N1 using the In-Fusion CG Dry-Down PCR Cloning kit (Takara Bio Inc.). CcA was from Abcam. PE-rhodamine was from Avanti Polar Lipids, Inc. Magic Red cathepsin L detection kit was from ImmunoChemistry Technologies, LLC. Mouse anti–human LAMP1 antibody (H4A3) was from the Developmental Studies Hybridoma Bank. Rabbit anti–saposin C antibody (H-81) was from Santa Cruz Biotechnology, Inc. Rabbit antiphospho-p70 S6 kinase antibody was from Cell Signaling Technology. HRP-conjugated anti–mouse and anti–rabbit secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc. ON-TARGETplus human RILP siRNA SMARTpool was from GE Healthcare. TaqMan Gene Expression Assays (RILP FAM and CDKN1A VIC) were from Applied Biosystems. Excitation/emission filters were from Semrock, Inc. Dichroics were from Chroma Technology Corp.
Tissue culture
EGFP, Arl8b-mCherry, p50 dynamitin–EGFP, FYCO1-mCherry, Rab5-EGFP, EEA1-EGFP, EGFP-Rab11a, EGFP-Rab7, EGFP-RILP-C33, LAMP1-RFP, LIMP2-GFP, and P3X-FLAG-EGFP-pericentrin-myc constructs were expressed by transient transfection of HeLa cells using FuGene 6 transfection reagent according to the manufacturer’s protocol. Cells were grown at 37°C in an air/CO2 (19:1) environment in DMEM supplemented with 5% (vol/vol) FBS.
RILP knockdown and quantitative PCR
HeLa cells were plated sparsely in 6-well tissue culture plates and grown overnight. The next day, cells were transfected with four pooled siRNA oligonucleotides against human RILP (SMARTpool; L-008787-01-0005) using HiPerFect transfection reagent according to the manufacturer’s protocol. As a control, a nontargeting SMARTpool siRNA oligonucleotide set (D-001810-10) was used. After 48 h, cells were washed and transfected again. After a further 24 h, RNA was isolated using the GeneJET RNA Purification kit (Thermo Fisher Scientific). Equal amounts of RNA were loaded as template for the generation of cDNA using the SuperScript VILO cDNA Synthesis kit (Thermo Fisher Scientific). Quantitative PCR was performed in a 96-well plate using the TaqMan System (Thermo Fisher Scientific) on a Step One Plus Real-Time PCR thermal cycler with Step One software (v2.2.2; Applied Biosystems). The Taqman gene expression assays for the reference gene (CDKN1; Hs00355782_m1) and target gene (RILP; Hs01018707_g1) were duplexed in triplicate. Target gene expression was determined by relative quantification (ΔΔCt method) to the CDKN1 reference gene and the control sample.
Lysosomal markers
For pH measurements, lysosomes of HeLa cells were loaded with 0.5 mg/ml dextran (Oregon green 488– and tetramethylrhodamine- or fluorescein-conjugated dextran) for 6 h at 37°C (5% CO2 balance air), transfected as noted in figure legends, and chased overnight. For all other experiments, lysosomes were loaded with 0.1 mg/ml dextran for 3 h at 37°C (5% CO2 balance air), transfected as indicated in figure legends, and chased overnight. Cells were imaged the next day or loaded with 200 nM LysoTracker for 5 min, washed, and imaged immediately.
Immunofluorescence
Lysosomes of HeLa cells were loaded by pulsing cells with 0.1 mg/ml Alexa Fluor 647–dextran for 3 h, and then cells were chased for 1 h and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. Cells were permeabilized with 0.1% saponin for 15 min at room temperature and blocked for 1 h with 1% BSA in PBS, 10% goat serum, 0.3 M glycine, and 0.1% saponin. Samples were incubated with primary and secondary antibodies for 1 h at room temperature in 1% BSA.
Transferrin loading
Lysosomes of HeLa cells were loaded by pulsing cells with 0.1 mg/ml Alexa Fluor 647–dextran for 3 h at 37°C (5% CO2 balance air) and chasing them overnight. The next day, cells were pulsed with 0.05 mg/ml Alexa Fluor 546–transferrin for 20 min, washed, and imaged immediately by confocal microscopy.
Ratiometric fluorescence microscopy
HeLa cells grown on glass coverslips were mounted in a Chamlide magnetic chamber and overlaid with Hepes-buffered HBSS or isotonic Na+ buffer (140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose, and 20 mM Hepes, pH 7.4). The chamber was placed in a Leiden microincubator maintained at 37°C on the stage of an inverted microscope (DM IRB; Leica Biosystems) equipped with a 40×/1.25 NA oil objective (Leica Biosystems), a lamp (X-Cite 120; EXFO Life Sciences Group), and filter wheels (Sutter Instrument) that control excitation and emission filters. For experiments using Oregon green 488– and tetramethylrhodamine-dextran, excitation wavelengths were alternated between 485 ± 10 nm and 543 ± 11 nm, with emitted light filtered alternately through a 505-nm dichroic and a 535 ± 20–nm emission filter or a 495/565-nm dichroic and a 580 ± 30–nm emission filter, respectively. For experiments using fluorescein-dextran, excitation wavelengths of 485 ± 10 nm and 438 ± 12 nm were selected, and emitted light was filtered through a 505-nm dichroic and a 535 ± 20–nm emission filter. Light was captured by an electron-multiplied charge-coupled device camera (Cascade II; Photometrics). The filter wheel and camera were under the control of MetaFluor software (Molecular Devices). Images were acquired at 60-s intervals.
Conversion of fluorescence ratio to pH
To convert fluorescence values to pH, cells were sequentially bathed in isotonic K+ solutions (143 mM KCl, 5 mM glucose, 1 mM MgCl2, 1 mM CaCl2, and 20 mM Hepes) buffered to a pH ranging from 4.5 to 7.5 and containing 10 µM nigericin and 5 µM monensin. For calibration, data were acquired 5 min after the addition of each solution to ensure equilibration of pH across compartments (Thomas et al., 1979). A calibration curve relating themean fluorescence ratio (corrected by subtracting the background fluorescence at each wavelength) at each pH to the medium pH value was fitted by least squares and used to transform fluorescence ratio measurements to intracellular pH.
Spinning disk confocal microscopy
Confocal images were acquired using a spinning disk system (WaveFX; Quorum Technologies Inc.). The instrument consists of a microscope (Axiovert 200M; ZEISS), scanning unit (CSU10; Yokogawa Electric Corporation), electron-multiplied charge-coupled device (C9100-13; Hamamatsu Photonics), five-line (405, 443, 491, 561, and 655 nm) laser module (Spectral Applied Research), and filter wheel (MAC5000; Ludl) and is operated by Volocity software version 4.3.2 or 6.2.1 (PerkinElmer). Images were acquired using a 63×/1.4 NA oil objective (ZEISS) coupled to an additional 1.5 magnifying lens and the appropriate emission filter. Cells were maintained at 37°C using an environmental chamber (Live Cell Instruments).
Generation of photoactivatable dextran
The succinimidyl ester of CMNB-caged carboxyfluorescein was incubated with amino-dextran in borate buffer (50 mM borate, pH 8.5) for 1 h at 24°C and dialyzed overnight using a dialysis cassette (Slide-A-Lyzer 10K) in PBS at 4°C to generate CMNB-caged carboxyfluorescein-dextran.
Point-scanning confocal microscopy for photoactivation
Photoactivation experiments were conducted on a confocal microscope (A1R; Nikon) using a 60×/1.4 NA oil objective (Nikon). Using NIS Elements software (Nikon), a region of interest was selectively illuminated using a 405-nm laser, and the resulting fluorescence was imaged using a 488-nm laser with a 1.2-AU pinhole and 2× line averaging every 1 min for 30 min or every 4 min for 2 h. Focus was maintained using MCL Piezo Drive and Nikon Perfect Focus. Cells were maintained at 37°C using a temperature/CO2 environmental control chamber (Tokai Hit). Lysosome tracking analysis was conducted using Imaris 8.1.2 software (Bitplane).
Image processing and quantification
Images acquired using Volocity or MetaFluor were exported to ImageJ 1.48 (National Institutes of Health) for contrast enhancement, analysis, and quantification, unless otherwise noted. Analysis was performed on optical slices acquired 0.4–0.8 µm from the ventral surface of the cell.
For pH measurements, fluorescence values were obtained using the freehand tool in ImageJ to select regions of interest (ROIs) delimiting individual lysosomes. In cases where individual lysosomes could not be resolved because they formed dense clusters, e.g., in Arl8b-expressing cells, ROIs were drawn to encompass the entire cluster. The fluorescence intensity ratio in each ROI was corrected by subtracting the background fluorescence at each wavelength and converted to pH by curve fitting as described in the Conversion of fluorescence ratio to pH section.
For pseudocolor display, a thresholding algorithm in ImageJ was used to select lysosomes, and a calibration curve was applied to convert fluorescence ratio to colors corresponding to the indicated pH.
Overlap analysis was conducted using Volocity 6.3.1. In brief, a thresholding algorithm was used to identify populations of individual lysosomes (marked with dextran, LAMP1, saposin C, LysoTracker, and/or LIMP2). Using the compartmentalize function, features of diameters >0.4 µm sharing significant overlap were identified and scored as a fraction of the total population.
Lysosome position analysis
The boundary of the cells was determined by outlining the fluorescence of cytosolic EGFP or dynamitin-EGFP using Volocity software and was degraded inward to create multiple shells of the indicated thickness. Background-corrected lysosome intensity for the total cell or for each shell was determined using the following equation: background-corrected lysosome intensity = sum intensitylysosomes − (sum intensitybackground/areabackground × arealysosomes).
Distance measurements from pericentrin or plasma membrane
To assess distances from the MTOC, lysosomes of HeLa cells were loaded with 0.1 mg/ml Alexa Fluor 647–dextran for 3 h at 37°C, transfected with P3X-FLAG-EGFP-pericentrin-myc, and chased overnight. The next day, cells were loaded with 200 nM LysoTracker red for 5 min, washed, and imaged immediately.
To assess distance to the membrane, lysosomes were loaded with 0.1 mg/ml Alexa Fluor 647–dextran as in the Lysosomal markers section. The next day, cells were loaded with 200 nM LysoTracker red for 5 min, washed, and then labeled with 10 µg/ml Oregon green 488–wheat germ agglutinin at 4°C, washed, and imaged immediately.
Optical slices were used to reconstruct sagittal (x vs. z) views near the middle of the cell, and the distances between the MTOC (pericentrin) or the plasma membrane (wheat germ agglutinin) and dextran-positive lysosomes were measured using the measure distance function in Volocity. Lysosomes were scored manually as being positive or negative for LysoTracker, and the LysoTracker-positive fraction is reported.
Buffer capacity
Lysosomal pH was measured by ratiometric fluorescence microscopy as described in a previous section. The lysosomes were loaded with fluorescein-dextran and washed, and the cells were then transiently transfected with Arl8b-mCherry cDNA or mock transfected. The next day, cells were bathed in isotonic Na+ buffer. 2 µM CcA was added to mock-transfected cells, and the pH was allowed to drift upward so that buffering capacity in control and Arl8b-expressing cells would be determined at comparable initial pH values. Fluorescence images were acquired as described in the Ratiometric fluorescence microscopy section. The bathing solution was switched to Na+ buffer containing 3.75 mM NH4Cl, and the resulting pH change was measured immediately. At the end of each experiment, a standard calibration was performed as described in the Conversion of fluorescence ratio to pH section, and fluorescence values were converted to pH. The intralysosomal NH4+ concentration ([NH4+]l) was calculated using the Henderson-Hasselbalch equation, and the intrinsic buffer capacity (in millimoles/liter/pH) was calculated as Δ([NH4+]l/ΔpHl).
H+ leak
Lysosomes were loaded with fluorescein-dextran and washed, and the cells were then transiently transfected with Arl8b-mCherry cDNA or mock transfected. The next day, cells were bathed in isotonic Na+ buffer, and images were acquired for pH determination. The bathing solution was then switched to Na+ buffer containing 2 µM CcA, and pH was measured for 20 min. At the end of each experiment, a standard calibration was performed, and fluorescence values were converted to pH. Rates of H+ leak (in millimoles/liter/minute) were determined by multiplying the slope (ΔpH/Δt) of the line between pH 6.2 and 6.9 (fitted by the least squares method) by the buffer capacity determined as in the previous paragraph in the same pH range.
H+ pump activity
The lysosomes were loaded with fluorescein-dextran and washed, and the cells were transiently transfected with Arl8b-mCherry cDNA or mock transfected. The next day, cells were bathed in isotonic Na+ buffer, and images were acquired for pH determination. The bathing solution was switched to Na+ buffer containing 30 µM CCCP for 3 min to dissipate the existing pH gradient, and the ionophore was then rapidly washed away with Na+ buffer containing 5 mg/ml fatty acid–free BSA. Where indicated, 2 µM CcA was present during the incubations with CCCP and BSA. At the end of each experiment, a standard calibration was performed, and fluorescence values were converted to pH. Rates of pH change during the initial 90 s after the addition of BSA were determined as the slope (ΔpH/Δt) of the line fitted by least squares. H+ pump activity (in millimoles/liter/minute) was determined by multiplying ΔpH/Δt by the buffer capacity determined in the same pH range.
PE-rhodamine loading
The lysosomes were loaded with 0.1 mg/ml Alexa Fluor 647–dextran, and the cells were then transfected with dynamitin-EGFP cDNA. The next day, cells were incubated with the fluorescent marker PE-rhodamine (dissolved in 1:1 [vol/vol] chloroform/methanol and then dried under constant N2 flow and reconstituted in 0.1% BSA) for 15 min at 10°C to label the plasma membrane. Cells were washed with HBSS at 10°C and immediately imaged by confocal microscopy.
Flow cytometry
HeLa cells were mock transfected or transfected with Arl8b-mCherry. 12 h later, cells were again mock transfected or transfected with the lysosomal membrane protein LIMP2-GFP. 6 h later, the cells were lifted from the plates by incubation with 0.05% trypsin, pelleted by centrifugation for 5 min at 500 g, and resuspended in PBS. Cells were passed through a cell strainer, run through a flow cytometer (LSRII; BD), and analyzed using FlowJo software (Tree Star).
Cathepsin L activity assay
The lysosomes were loaded with 0.1 mg/ml Alexa Fluor 647–dextran for 3 h and washed, and the cells were then transfected with EGFP or dynamitin-EGFP cDNAs. The next day, cells were incubated with Magic Red cathepsin L substrate for 15 min and then washed. The fluorescence generated by hydrolysis of the Magic Red substrate was imaged by confocal microscopy. Where specified, lysosomes were alkalinized with NH4Cl before Magic Red addition; in these instances, NH4Cl remained present for the duration of the experiment.
Cathepsin L activity was quantified by normalizing Magic Red fluorescence to Alexa Fluor 647–dextran fluorescence, thereby standardizing the measurements per lysosomal volume.
Immunoblotting
HeLa cells were incubated at 37°C for 1 h in DMEM supplemented with 5% FBS alone (control), DMEM supplemented with 5% FBS containing 1 µM CcA and 10 mM NH4Cl, or 2 µM torin1. Cell lysates were harvested in Laemmli buffer containing 2% (vol/vol) 2-mercaptoethanol plus protease and phosphatase inhibitors. Samples were boiled for 5 min and then resolved on 12% acrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes by electrophoresis, blocked by incubation for 1 h at 20°C in PBS containing 0.1% (vol/vol) Tween 20 and 10% (vol/vol) nonfat dry milk (PBST-M), and then incubated at 4°C for 16 h in PBST-M containing rabbit antiphosphorylated p70 S6 kinase (1:1,000) and mouse anti-GAPDH antibodies (1:20,000). Blots were incubated for 1 h with PBST-M containing donkey anti–rabbit and donkey anti–mouse IgG conjugated to horseradish peroxidase (1:3,000). Blots were visualized using ECL Prime Western Blot Detection reagent (GE Healthcare) and an Odyssey FC imaging system (LI-COR Biosciences) and quantified using ImageJ.
Statistical analysis
Data are presented as mean ± SEM of the number of determinations indicated. Statistical significance of data was determined using an unpaired t test (PRISM 4; GraphPad Software), with P < 0.05 considered significant.
Online supplemental material
Fig. S1 shows the distribution of LAMP1 and saposin C in lysosomes. Fig. S2 examines lysosomal pH heterogeneity in several cell types. Fig. S3 shows the localization of lysosomes as a function of their distance from either the MTOC or the plasma membrane. Fig. S4 shows that lysosomes displaced to the cell periphery by expression of Arl8b or dynamitin accumulate reduced levels of LysoTracker. Fig. S5 shows that alkalinizing lysosomes decreases mTORC1 activity. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507112/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Amra Saric and Johnathan Canton for help and advice.
D.E. Johnson is supported by a postdoctoral fellowship provided by the Restracomp from the Hospital for Sick Children. This work was supported by a foundation grant from the Canadian Institutes of Health Research (FDN-143202) to S. Grinstein.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CcA
- concanamycin A
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- EEA1
- early endosome antigen 1
- FYCO1
- FYVE and coiled-coil domain–containing 1
- LAMP1
- lysosomal associated membrane protein 1
- LIMP2
- lysosomal integral membrane protein 2
- MTOC
- microtubule-organizing center
- PE
- phosphatidylethanolamine
- RILP
- Rab-interacting lysosomal protein
- ROI
- region of interest
- SKIP
- SifA and kinesin-interacting protein
- V-ATPase
- vacuolar H+-ATPase
References
- Aniento F., Gu F., Parton R.G., and Gruenberg J.. 1996. An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J. Cell Biol. 133:29–41. 10.1083/jcb.133.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccino F.M., Rita G.A., and Zuretti M.F.. 1971. Studies on the structure-bound sedimentabolity of some rat liver lysosome hydrolases. Biochem. J. 122:363–371. 10.1042/bj1220363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw R.D., Callahan J.W., and Mahuran D.J.. 2006. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem. Biophys. Res. Commun. 344:1186–1191. 10.1016/j.bbrc.2006.03.221 [DOI] [PubMed] [Google Scholar]
- Basu S.K., Goldstein J.L., Anderson R.G., and Brown M.S.. 1981. Monensin interrupts the recycling of low density lipoprotein receptors in human fibroblasts. Cell. 24:493–502. 10.1016/0092-8674(81)90340-8 [DOI] [PubMed] [Google Scholar]
- Brown W.J., Goodhouse J., and Farquhar M.G.. 1986. Mannose-6-phosphate receptors for lysosomal enzymes cycle between the Golgi complex and endosomes. J. Cell Biol. 103:1235–1247. 10.1083/jcb.103.4.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., and van Deurs B.. 2000. Rab7: A key to lysosome biogenesis. Mol. Biol. Cell. 11:467–480. 10.1091/mbc.11.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J.K., Echeverri C.J., Nilsson T., and Vallee R.B.. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139:469–484. 10.1083/jcb.139.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo G., Alifano P., Roberti V., Bruni C.B., and Bucci C.. 2001. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20:683–693. 10.1093/emboj/20.4.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R.E., and Munro S.. 1994. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO J. 13:2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A., Dominguez M., Puri V., Sharma D.K., Narita K., Wheatley C.L., Marks D.L., and Pagano R.E.. 2002. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J. Clin. Invest. 109:1541–1550. 10.1172/JCI0215420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague M.J., Urbé S., Aniento F., and Gruenberg J.. 1994. Vacuolar ATPase activity is required for endosomal carrier vesicle formation. J. Biol. Chem. 269:21–24. [PubMed] [Google Scholar]
- Colucci A.M.R., Campana M.C., Bellopede M., and Bucci C.. 2005. The Rab-interacting lysosomal protein, a Rab7 and Rab34 effector, is capable of self-interaction. Biochem. Biophys. Res. Commun. 334:128–133. 10.1016/j.bbrc.2005.06.067 [DOI] [PubMed] [Google Scholar]
- de Duve C. 1983. Lysosomes revisited. Eur. J. Biochem. 137:391–397. 10.1111/j.1432-1033.1983.tb07841.x [DOI] [PubMed] [Google Scholar]
- de Duve C., and Wattiaux R.. 1966. Functions of lysosomes. Annu. Rev. Physiol. 28:435–492. 10.1146/annurev.ph.28.030166.002251 [DOI] [PubMed] [Google Scholar]
- De Luca M., Cogli L., Progida C., Nisi V., Pascolutti R., Sigismund S., Di Fiore P.P., and Bucci C.. 2014. RILP regulates vacuolar ATPase through interaction with the V1G1 subunit. J. Cell Sci. 127:2697–2708. 10.1242/jcs.142604 [DOI] [PubMed] [Google Scholar]
- Falguières T., Luyet P.-P., Bissig C., Scott C.C., Velluz M.-C., and Gruenberg J.. 2008. In vitro budding of intralumenal vesicles into late endosomes is regulated by Alix and Tsg101. Mol. Biol. Cell. 19:4942–4955. 10.1091/mbc.E08-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K., Walseng E., and Roche P.A.. 2013. Internalizing MHC class II–peptide complexes are ubiquitinated in early endosomes and targeted for lysosomal degradation. Proc. Natl. Acad. Sci. USA. 110:20188–20193. 10.1073/pnas.1312994110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.-M., Parton R.G., and Stenmark H.. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588. 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone A., and Koenig H.. 1974. Synthesis and turnover of lysosomal glycoproteins. Relation to the molecular heterogeneity of the lysosomal enzymes. FEBS Lett. 39:176–181. 10.1016/0014-5793(74)80045-1 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Noriega A., Grubb J.H., Talkad V., and Sly W.S.. 1980. Chloroquine inhibits lysosomal enzyme pinocytosis and enhances lysosomal enzyme secretion by impairing receptor recycling. J. Cell Biol. 85:839–852. 10.1083/jcb.85.3.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines T.H. 2001. Do sterols reduce proton and sodium leaks through lipid bilayers? Prog. Lipid Res. 40:299–324. 10.1016/S0163-7827(01)00009-1 [DOI] [PubMed] [Google Scholar]
- Harrison R.E., Bucci C., Vieira O.V., Schroer T.A., and Grinstein S.. 2003. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: Role of Rab7 and RILP. Mol. Cell. Biol. 23:6494–6506. 10.1128/MCB.23.18.6494-6506.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J. 1989. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J. Cell Biol. 108:855–864. 10.1083/jcb.108.3.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton A., Bond S., and Forgac M.. 2009. V-ATPase functions in normal and disease processes. Pflugers Arch. 457:589–598. 10.1007/s00424-007-0382-4 [DOI] [PubMed] [Google Scholar]
- Hirota Y., Masuyama N., Kuronita T., Fujita H., Himeno M., and Tanaka Y.. 2004. Analysis of post-lysosomal compartments. Biochem. Biophys. Res. Commun. 314:306–312. 10.1016/j.bbrc.2003.12.092 [DOI] [PubMed] [Google Scholar]
- Hofmann I., and Munro S.. 2006. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119:1494–1503. 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- Hung Y.H., Chen L.M., Yang J.Y., and Yang W.Y.. 2013. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat. Commun. 4:2111 10.1038/ncomms3111 [DOI] [PubMed] [Google Scholar]
- Huotari J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G.-H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D.A., et al. 2006. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8:124–136. 10.1038/ncb1348 [DOI] [PubMed] [Google Scholar]
- Johnson L.S., Dunn K.W., Pytowski B., and McGraw T.E.. 1993. Endosome acidification and receptor trafficking: Bafilomycin A1 slows receptor externalization by a mechanism involving the receptor’s internalization motif. Mol. Biol. Cell. 4:1251–1266. 10.1091/mbc.4.12.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J.. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11:1680–1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Kaniuk N.A., Canadien V., Bagshaw R.D., Bakowski M., Braun V., Landekic M., Mitra S., Huang J., Heo W.D., Meyer T., et al. 2011. Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cell. Microbiol. 13:1812–1823. 10.1111/j.1462-5822.2011.01663.x [DOI] [PubMed] [Google Scholar]
- Kim S., and Rhee K.. 2014. Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One. 9:e87016 10.1371/journal.pone.0087016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D.C., Gordon M.D., Jin J.Y., and Scott M.P.. 2001. Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol. Biol. Cell. 12:601–614. 10.1091/mbc.12.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T., Terman A., Gustafsson B., and Brunk U.T.. 2008. Lysosomes in iron metabolism, ageing and apoptosis. Histochem. Cell Biol. 129:389–406. 10.1007/s00418-008-0394-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrand C., Corti M., Goodson H., Cosson P., Cavalli V., Mayran N., Fauré J., and Gruenberg J.. 2002. Late endosome motility depends on lipids via the small GTPase Rab7. EMBO J. 21:1289–1300. 10.1093/emboj/21.6.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima P.D., Croy B.A., Degaki K.Y., Tayade C., and Yamada A.T.. 2012. Heterogeneity in composition of mouse uterine natural killer cell granules. J. Leukoc. Biol. 92:195–204. 10.1189/jlb.0312136 [DOI] [PubMed] [Google Scholar]
- Loiselle F.B., and Casey J.R.. 2010. Measurement of intracellular pH. Methods Mol. Biol. 637:311–331. 10.1007/978-1-60761-700-6_17 [DOI] [PubMed] [Google Scholar]
- Luzio J.P., Pryor P.R., and Bright N.A.. 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8:622–632. 10.1038/nrm2217 [DOI] [PubMed] [Google Scholar]
- Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., and Yoshimori T.. 2013. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32:2336–2347. 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.W., Green G.D., and Barrett A.J.. 1985. Human liver cathepsin L. Biochem. J. 226:233–241. 10.1042/bj2260233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteoni R., and Kreis T.E.. 1987. Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Cell Biol. 105:1253–1265. 10.1083/jcb.105.3.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., Fuchs R., and Helenius A.. 1986. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55:663–700. 10.1146/annurev.bi.55.070186.003311 [DOI] [PubMed] [Google Scholar]
- Ng E.L., Gan B.Q., Ng F., and Tang B.L.. 2012. Rab GTPases regulating receptor trafficking at the late endosome-lysosome membranes. Cell Biochem. Funct. 30:515–523. 10.1002/cbf.2827 [DOI] [PubMed] [Google Scholar]
- Nilsson E., Ghassemifar R., and Brunk U.T.. 1997. Lysosomal heterogeneity between and within cells with respect to resistance against oxidative stress. Histochem. J. 29:857–865. 10.1023/A:1026441907803 [DOI] [PubMed] [Google Scholar]
- Ohkuma S., and Poole B.. 1978. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. USA. 75:3327–3331. 10.1073/pnas.75.7.3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Alemu E.A., Brech A., Bruun J.-A., Lamark T., Øvervatn A., Bjørkøy G., and Johansen T.. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end–directed vesicle transport. J. Cell Biol. 188:253–269. 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Dotti C.G., Bacallao R., Kurtz I., Simons K., and Prydz K.. 1991. pH-induced microtubule-dependent redistribution of late endosomes in neuronal and epithelial cells. J. Cell Biol. 113:261–274. 10.1083/jcb.113.2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertoft H., Wärmegård B., and Höök M.. 1978. Heterogeneity of lysosomes originating from rat liver parenchymal cells. Metabolic relationship of subpopulations separated by density-gradient centrifugation. Biochem. J. 174:309–317. 10.1042/bj1740309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J.F., Mayor S., Dunn K.W., Johnson L.S., McGraw T.E., and Maxfield F.R.. 1993. The End2 mutation in CHO cells slows the exit of transferrin receptors from the recycling compartment but bulk membrane recycling is unaffected. J. Cell Biol. 122:1231–1241. 10.1083/jcb.122.6.1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J.F., Mayor S., McGraw T.E., Dunn K.W., and Maxfield F.R.. 1997. Bafilomycin A1 treatment retards transferrin receptor recycling more than bulk membrane recycling. J. Biol. Chem. 272:13929–13936. 10.1074/jbc.272.21.13929 [DOI] [PubMed] [Google Scholar]
- Reaves B., and Banting G.. 1994. Vacuolar ATPase inactivation blocks recycling to the trans-Golgi network from the plasma membrane. FEBS Lett. 345:61–66. 10.1016/0014-5793(94)00437-4 [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., and Ferguson S.M.. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira C., and Munro S.. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21:1171–1178. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P., and Klumperman J.. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623–635. 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Schwartz A.L., Bolognesi A., and Fridovich S.E.. 1984. Recycling of the asialoglycoprotein receptor and the effect of lysosomotropic amines in hepatoma cells. J. Cell Biol. 98:732–738. 10.1083/jcb.98.2.732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.C., and Gruenberg J.. 2011. Ion flux and the function of endosomes and lysosomes: pH is just the start. The flux of ions across endosomal membranes influences endosome function not only through regulation of the luminal pH. BioEssays. 33:103–110. 10.1002/bies.201000108 [DOI] [PubMed] [Google Scholar]
- Scott C.C., Cuellar-Mata P., Matsuo T., Davidson H.W., and Grinstein S.. 2002. Role of 3-phosphoinositides in the maturation of Salmonella-containing vacuoles within host cells. J. Biol. Chem. 277:12770–12776. 10.1074/jbc.M110399200 [DOI] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawki A., Knight P.B., Maliken B.D., Niespodzany E.J., and Mackenzie B.. 2012. H+-coupled divalent metal-ion transporter-1: Functional properties, physiological roles and therapeutics. Curr. Top. Membr. 70:169–214. 10.1016/B978-0-12-394316-3.00005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.M., and Mizushima N.. 2014. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem. Sci. 39:61–71. 10.1016/j.tibs.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Sherer N.M., Lehmann M.J., Jimenez-Soto L.F., Ingmundson A., Horner S.M., Cicchetti G., Allen P.G., Pypaert M., Cunningham J.M., and Mothes W.. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 4:785–801. 10.1034/j.1600-0854.2003.00135.x [DOI] [PubMed] [Google Scholar]
- Soyombo A.A., Tjon-Kon-Sang S., Rbaibi Y., Bashllari E., Bisceglia J., Muallem S., and Kiselyov K.. 2006. TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J. Biol. Chem. 281:7294–7301. 10.1074/jbc.M508211200 [DOI] [PubMed] [Google Scholar]
- Terman A., Kurz T., Gustafsson B., and Brunk U.T.. 2006. Lysosomal labilization. IUBMB Life. 58:531–539. 10.1080/15216540600904885 [DOI] [PubMed] [Google Scholar]
- Thomas J.A., Buchsbaum R.N., Zimniak A., and Racker E.. 1979. Intracellular pH measurements in Ehrlich ascites tumor cells utilizing spectroscopic probes generated in situ. Biochemistry. 18:2210–2218. 10.1021/bi00578a012 [DOI] [PubMed] [Google Scholar]
- Thwaites D.T., and Anderson C.M.H.. 2011. The SLC36 family of proton-coupled amino acid transporters and their potential role in drug transport. Br. J. Pharmacol. 164:1802–1816. 10.1111/j.1476-5381.2011.01438.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta E.S., Ebersold M., Garrett W., Pypaert M., and Mellman I.. 2003. Activation of lysosomal function during dendritic cell maturation. Science. 299:1400–1403. 10.1126/science.1080106 [DOI] [PubMed] [Google Scholar]
- Tycko B., Keith C.H., and Maxfield F.R.. 1983. Rapid acidification of endocytic vesicles containing asialoglycoprotein in cells of a human hepatoma line. J. Cell Biol. 97:1762–1776. 10.1083/jcb.97.6.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weert A.W., Dunn K.W., Geuze H.J., Maxfield F.R., and Stoorvogel W.. 1995. Transport from late endosomes to lysosomes, but not sorting of integral membrane proteins in endosomes, depends on the vacuolar proton pump. J. Cell Biol. 130:821–834. 10.1083/jcb.130.4.821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D.M.. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science. 334:678–683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.