In yeast, transcription factors mediate gene positioning at the nuclear periphery and interchromosomal clustering. These phenomena are regulated by several different strategies that lead to dynamic changes in the spatial arrangement of genes over different time scales.
Abstract
In budding yeast, targeting of active genes to the nuclear pore complex (NPC) and interchromosomal clustering is mediated by transcription factor (TF) binding sites in the gene promoters. For example, the binding sites for the TFs Put3, Ste12, and Gcn4 are necessary and sufficient to promote positioning at the nuclear periphery and interchromosomal clustering. However, in all three cases, gene positioning and interchromosomal clustering are regulated. Under uninducing conditions, local recruitment of the Rpd3(L) histone deacetylase by transcriptional repressors blocks Put3 DNA binding. This is a general function of yeast repressors: 16 of 21 repressors blocked Put3-mediated subnuclear positioning; 11 of these required Rpd3. In contrast, Ste12-mediated gene positioning is regulated independently of DNA binding by mitogen-activated protein kinase phosphorylation of the Dig2 inhibitor, and Gcn4-dependent targeting is up-regulated by increasing Gcn4 protein levels. These different regulatory strategies provide either qualitative switch-like control or quantitative control of gene positioning over different time scales.
Introduction
Within the nucleus of eukaryotic cells, genomes are spatially organized. Chromosomes loop, fold, and interact with subnuclear structures, occupying distinct “territories” (Cremer et al., 2006). Individual genes can assume different subnuclear positions, depending on their expression state (Pombo and Dillon, 2015). For example, in metazoan cells, developmentally regulated genes often move away from the nuclear lamina upon induction (Peric-Hupkes et al., 2010). Likewise, genes can reposition within chromosome territories and colocalize with RNA polymerase II foci called transcription factories (Brown et al., 2006; Xu and Cook, 2008; Schoenfelder et al., 2010). The spatial arrangement of the genome thus has the potential to create functionally distinct subdomains and contribute to transcriptional regulation.
The interaction of genes with stable nuclear structures can affect their regulation. Large transcriptionally repressed lamin-associated domains localize at the nuclear periphery and punctuate metazoan genomes (Guelen et al., 2008; Luperchio et al., 2014). In contrast, many active genes interact with nuclear pore proteins in yeast, flies, worms, and mammalian cells (Brickner and Walter, 2004; Casolari et al., 2004; Brown et al., 2008; Kalverda et al., 2010; Rohner et al., 2013). In budding yeast, these interactions occur at the nuclear pore complex (NPC), whereas in metazoan cells, the interactions occur both at the NPC and with soluble nuclear pore proteins in the nucleoplasm (Ahmed et al., 2010; Capelson et al., 2010; Kalverda et al., 2010). In both yeast and metazoan cells, interaction with nuclear pore proteins correlates with transcription (Brickner and Walter, 2004; Taddei et al., 2006; Brickner et al., 2007; Brown et al., 2008; Ahmed et al., 2010; Capelson et al., 2010; Kalverda et al., 2010; Liang and Hetzer, 2011). Thus, the interaction of genes with distinct compartments at the nuclear periphery can lead to opposite transcriptional outcomes.
Coregulated regions of the genome often cluster together (Pombo et al., 2000; Brown et al., 2006; Schoenfelder et al., 2010). For example, transcriptionally silent subtelomeric regions in yeast (Aparicio et al., 1991), tRNA genes (Thompson et al., 2003), and Klf1-regulated genes all cluster (Schoenfelder et al., 2010). The spatial proximity of coregulated genes through interchromosomal clustering may create distinct subnuclear environments that affect gene regulation. Alternatively, changes in chromatin state and expression can lead to the creation of subnuclear domains (Meister et al., 2011).
As a model for these phenomena, we have studied the spatial repositioning of inducible genes from the nucleoplasm to the NPC upon activation in budding yeast. Recruitment to the NPC is controlled by cis-acting transcription factor (TF) binding sites in gene promoters (Ahmed et al., 2010; Light et al., 2010; Brickner et al., 2012). These DNA elements function as DNA zip codes: they are necessary to recruit genes from the nucleoplasm to the nuclear periphery and promote stronger transcription, and they are sufficient to target ectopic sites to the NPC (Ahmed et al., 2010; Light et al., 2010; Brickner et al., 2012). Targeting to the nuclear periphery can also lead to interchromosomal clustering of genes that share zip codes (Brickner et al., 2012). For example, a DNA zip code called gene recruitment sequence I (GRS I) from the promoter of the INO1 gene (encoding inositol 1-phoshate synthase) interacts with the Put3 TF. Inserting GRS I beside the nucleoplasmic locus URA3 leads to targeting of URA3 to the nuclear periphery and clustering of URA3:GRS I with the endogenous INO1 gene (Ahmed et al., 2010; Brickner et al., 2012). Loss of Put3 disrupts both GRS I–mediated targeting and interchromosomal clustering. This suggests that some TFs can promote interaction with the NPC and clustering. (Consistent with this notion, interaction of Nup98 with genes in Drosophila is mediated by the MBD-R2 DNA binding protein [Pascual-Garcia et al., 2014]). If so, then genomes could encode their spatial organization through TF binding sites.
Yeast genes such as INO1, GAL1, HSP104, and TSA2 localize in the nucleoplasm before induction and relocalize to the nuclear periphery upon activation (Brickner and Walter, 2004; Casolari et al., 2004; Dieppois et al., 2006; Ahmed et al., 2010). Targeting of these genes requires DNA zip codes in their promoters (Ahmed et al., 2010). However, the molecular mechanisms that regulate zip code activity have not been described. Here we identify three different regulatory strategies used by yeast cells to provide dynamic control of gene positioning and interchromosomal clustering. We define the regulation of TFs representing different families: Put3 (a Zn+2-binuclear cluster TF), Ste12 (a helix-turn-helix TF), and Gcn4 (a basic leucine zipper TF). Targeting of the INO1 promoter to the nuclear periphery by Put3 is regulated through local recruitment of the Rpd3(L) histone deacetylase complex by transcriptional repressors. Many repressors are capable of blocking zip code function through Rpd3-dependent and -independent mechanisms. Ste12-mediated gene positioning is not affected by Rpd3 but is regulated downstream of DNA binding by MAPK phosphorylation of the inhibitor Dig2. Finally, Gcn4-mediated gene positioning is controlled by Gcn4 abundance. Each mechanism provides distinct advantages: repressor regulation leads to a slow switch, whereas MAPK signaling leads to a rapid switch. Changes in TF levels allow a quantitative increase in peripheral localization and interchromosomal clustering over an intermediate time scale.
Results
TF-dependent and stimulus-specific recruitment to the nuclear periphery
Our previous work suggests that targeting of inducible genes to the nuclear periphery is mediated by cis-acting TF binding sites that function as DNA zip codes (Ahmed et al., 2010). To test the generality of this model and to understand the regulation of gene positioning, we asked whether the inducible genes PRM1 (a cell surface transmembrane protein induced by mating pheromone) and HIS4 (a multifunctional histidine biosynthetic enzyme induced by histidine starvation) are also recruited to the nuclear periphery in a TF-dependent fashion. PRM1 is among a large set of genes previously shown to physically interact with the NPC in the presence of pheromone (Casolari et al., 2005), and HIS4 was identified as a putative NPC-interacting gene by in silico comparison of TF binding to promoters that interact with the NPC (Fig. S1; Casolari et al., 2004; Venters et al., 2011).
An array of 128 Lac repressor binding sites was integrated downstream of each of these genes as well as the nucleoplasmic gene URA3. Into these strains, GFP-tagged Lac repressor (GFP-LacI) and an mCherry ER/nuclear envelope marker were introduced (Egecioglu et al., 2014). Live cells were imaged by confocal microscopy, and the fraction of the cells in which the GFP-LacI focus was unresolvable from the nuclear envelope was scored under uninducing or inducing conditions (Fig. 1 A). For all three genes, the uninducing condition is the same (synthetic complete medium). However, the inducing conditions are distinct: INO1 (inositol starvation), PRM1 (mating pheromone stimulation), and HIS4 (amino acid starvation). In cells grown under uninducing conditions, URA3, INO1, and PRM1 colocalized with the nuclear periphery in ∼30% of the cells counted (Fig. 1 B), similar to the fraction of the nucleus that is unresolvable from the nuclear envelope by light microscopy (baseline, blue hatched line in Fig. 1 B and throughout; Brickner and Walter, 2004). In contrast, HIS4, which is modestly expressed in the presence of histidine (Mueller and Hinnebusch, 1986), colocalized with the nuclear envelope in ∼45% of cells, significantly above the baseline (Fig. 1, B and C). Under all inducing conditions, URA3 remained nucleoplasmic. In contrast, specifically under their respective inducing conditions, peripheral localization of INO1, PRM1, and HIS4 increased significantly to ∼60% of the cells (Fig. 1, B and C).
Figure 1.
TF-dependent and stimulus-specific recruitment of INO1, PRM1, and HIS4 to the nuclear periphery. (A) Confocal micrographs of cells having the Lac operator (LacO) array integrated at INO1, expressing GFP-LacI and an mCherry-ER marker (Egecioglu et al., 2014), scored as either peripheral (top) or nucleoplasmic (bottom). Bar, 1 µm. (B) Wild-type or mutant strains having the LacO array inserted at URA3, INO1, PRM1, or HIS4 were imaged by confocal microscopy under uninducing conditions (synthetic complete medium; gray bars), after inositol starvation overnight (black bars), after α-factor stimulation for 15–25 min (orange bars), or after histidine starvation for 45–75 min (cyan blue bars). (C) P-values (Fisher exact test) comparing the peripheral localization of the URA3 locus with INO1, PRM1, or HIS4 in the indicated conditions and strains from B. -ino, inositol starvation; +α-f, α-factor stimulation; -his, histidine starvation. (D) Peripheral localization of INO1, PRM1, and HIS4 in spt20Δ strain. *, P ≤ 0.05 (Fisher exact test) between SDC and inducing condition for specific strain. Mean and SEM from three of more biological replicates (30–60 cells per replicate).
We next defined the TFs required for the targeting of INO1, PRM1, and HIS4 to the nuclear periphery. INO1 targeting to the nuclear periphery requires one of two cis-acting DNA elements, GRS I and GRS II (Ahmed et al., 2010). The Put3 TF binds to the GRS I zip code and is necessary for GRS I–dependent positioning but is not required for GRS II–mediated positioning (Brickner et al., 2012). Because the TF Cbf1 binds near the GRS II upon INO1 induction (Shetty and Lopes, 2010), we tested whether Cbf1 is required for GRS II function. Indeed, whereas loss of Put3 alone (Ahmed et al., 2010; Brickner et al., 2012) or Cbf1 alone (Fig. S2) does not block targeting of INO1 to the nuclear periphery, strains lacking both Put3 and Cbf1 fail to target INO1 to the nuclear periphery (Fig. 1 B). This suggests that Cbf1 is required for GRS II–mediated gene positioning.
The positioning of PRM1 and HIS4 to the nuclear periphery requires the same TFs that regulate their expression. PRM1 transcription is controlled by cooperative binding of the Ste12 TF to three pheromone response elements (PREs; 5′-TGAAACA-3′) in its promoter (Hagen et al., 1991). Loss of Ste12 blocks PRM1 relocalization to the nuclear periphery in the presence of mating pheromone (Fig. 1 B). HIS4 expression requires binding of the Gcn4 TF to binding sites (5′-TGACTC-3′) in the promoter (Arndt and Fink, 1986), and loss of Gcn4 blocked positioning of HIS4 to the nuclear periphery (Fig. 1 B). Therefore, the subnuclear positioning of INO1, PRM1, and HIS4 requires TF binding to their promoters.
The SAGA histone acetyltransferase complex is required for recruitment of genes such as GAL1-10 and INO1 to the NPC (Rodríguez-Navarro et al., 2004; Luthra et al., 2007; Ahmed et al., 2010; Strambio-De-Castillia et al., 2010). SAGA is also required for the interaction of extrachromosomal circles with the NPC in yeast (Denoth-Lippuner et al., 2014). To test whether SAGA is required for recruitment of HIS4 and PRM1 to the nuclear periphery, we deleted SPT20 (which is required for the structural integrity of SAGA; Roberts and Winston, 1997). Loss of Spt20 blocked recruitment of both INO1 and HIS4 to the nuclear periphery (Fig. 1 D). However, PRM1 repositioning to the nuclear periphery was independent of Spt20 (Fig. 1 D). Thus, SAGA is necessary for recruitment of some, but not all, genes to the nuclear periphery.
TF binding sites function as DNA zip codes
We next asked whether the TFs that are required for peripheral localization mediate peripheral localization. Each binding site (BS) was inserted beside URA3 to test its sufficiency to promote peripheral localization (Ahmed et al., 2010). As we have shown previously, both GRS I and GRS II are sufficient to reposition URA3 to the nuclear periphery (Fig. 2 A). Put3 was required for GRS I–mediated gene positioning, Cbf1 was required for GRS II–mediated gene positioning, and SAGA was required for both (Fig. 2 A).
Figure 2.
TF BSs function as DNA zip codes. (A and B) Peripheral localization of the URA3 locus, ± indicated DNA BSs, grown under uninducing and inducing conditions. GRS I or GRS II (A), 3×PREs (B), or Gcn4 BS (B) were inserted at URA3 in wild-type (WT) and mutant strains. *, P ≤ 0.05 (Fisher exact test) comparing uninducing and inducing condition. Mean and SEM from three of more biological replicates (30–50 cells per replicate).
Insertion of three PREs (3×PRE) and the Gcn4 BS at URA3 was also sufficient to promote peripheral localization, and these zip codes required Ste12 and Gcn4, respectively (Fig. 2 B). Furthermore, the positioning to the nuclear periphery mediated by 3×PRE was SAGA independent, whereas the positioning mediated by the Gcn4 BS was SAGA dependent (Fig. 2 B).
Although repositioning of INO1, PRM1, and HIS4 to the nuclear periphery was mediated by TFs, the regulation of these three TF-dependent repositioning events was different. Under uninducing conditions, recruitment of INO1 and PRM1 to the nuclear periphery is near baseline levels. In contrast, HIS4 localization to the periphery under uninducing conditions is significantly higher than URA3 recruitment or that in the gcn4Δ strain. This suggests that, under uninducing conditions, HIS4 is targeted to the nuclear periphery by Gcn4 to an intermediate level and that it increases to a maximal level under inducing conditions. Also, whereas the zip codes from the INO1 promoter functioned constitutively at the ectopic site, 3×PRE and Gcn4BS were both regulated at the ectopic site. This suggested that the regulation of GRS I and GRS II function in the INO1 promoter is dependent on its local context and that the regulation of Ste12- and Gcn4-mediated positioning is independent of local context.
The Rpd3(L) histone deacetylase regulates targeting of INO1 to the nuclear periphery
To understand how GRS I and GRS II are regulated by their promoter context, we identified the cis- and trans-acting factors that block their function in the context of the INO1 promoter. Deletion of 100 bp of the INO1 promoter (Δ4; Fig. 3 A) led to constitutive localization to the nuclear periphery (Ahmed et al., 2010). This part of the promoter contains two inositol-sensitive upstream activating sequence (UASINO) elements as well as an upstream repressing sequence (URS) that regulate the transcription of INO1 (Fig. 3 A; Lopes et al., 1993; Nikoloff and Henry, 1994). Mutations that disrupt these UASINO elements prevent expression of INO1, and mutation of the URS element leads to constitutive expression of INO1 (Lopes et al., 1993; Bachhawat et al., 1995). Therefore, we hypothesized that the URS element might both repress transcription and prevent peripheral targeting in the presence of inositol. Indeed, mutation of the URS element led to constitutive targeting of INO1 to the nuclear periphery (Fig. 3 A). However, mutations that disrupted the UASINO elements also led to constitutive localization at the nuclear periphery (Fig. 3 A). Therefore, INO1 targeting to the nuclear periphery under uninducing conditions is blocked by a mechanism that requires both the URS element and the UASINO elements.
Figure 3.
Rpd3(L) histone deacetylase regulates INO1 gene recruitment to the nuclear periphery. (A and B) Peripheral localization of INO1. *, P ≤ 0.05 (Fisher exact test) between SDC and inducing condition. Mean and SEM from three of more biological replicates (30–50 cells per replicate). (A) Strains having the indicated cis-acting, trans-acting, and histone deacetylase mutants were scored for INO1 targeting to the nuclear periphery. A schematic of the INO1 promoter showing the 100-bp “segment 4,” GRS I, GRS II, UASINO, and URS elements is shown in C. WT, wild-type. (B) Peripheral localization of wild type, grsI, grsII, or grsI grsII mutant INO1 in the opi1Δ mutant strain grown under uninducing conditions. (C) Model for INO1 transcriptional regulation. Ino2/Ino4 binds the UASINO elements and recruits Opi1 to the promoter under uninducing conditions. Both Ume6 and Opi1 recruit Rpd3(L). (D) ChIP with anti–pan-acetyl antibody, quantified relative to ChIP with anti-histone H3 antibody from RPD3 or rpd3Δ strains. The recovery of the INO1 promoter from each IP, normalized to input, was quantified by real-time quantitative PCR (qPCR). NS, not significant. (E) ChIP of Gst-Put3 (Ahmed et al., 2010; Brickner et al., 2012) expressed in either RPD3 or rpd3Δ strains. The recovery of the INO1 promoter or the GAL1 coding sequence was quantified relative to input by qPCR. P-values determined by Student’s t test.
Transcriptional repression of INO1 is mediated by two mutually dependent repressors (Fig. 3 C). In the presence of inositol, the Ume6 repressor binds to the URS (Lopes et al., 1993; Kadosh and Struhl, 1997; Rundlett et al., 1998) and the Opi1 repressor interacts with the Ino2/Ino4 activator bound to the UASINO (Heyken et al., 2005). Neither mechanism is sufficient, because loss of either Ume6 or Opi1 leads to constitutive, high-level expression of INO1 (Fig. S3 B). Loss of Ume6, Opi1, Ino2, or Ino4 led to constitutive targeting of INO1 to the nuclear periphery (Fig. 3 A). Furthermore, a mutation in Ino2 (L118A) that disrupts binding of Opi1 (Heyken et al., 2005) had the same effect (Fig. 3 A). This is not related to derepression of INO1 transcription, because strains lacking Ino2 and Ino4 or the UASINO element show no expression of INO1 (Graves and Henry, 2000). Furthermore, a strain lacking Isw2, a chromatin-remodeling factor required for INO1 repression (Shetty and Lopes, 2010), showed normal, regulated peripheral targeting (Fig. 3 A). Therefore, this suggests that recruitment of Opi1 and Ume6 to the INO1 promoter blocks GRS I and GRS II function. Consistent with this idea, the peripheral targeting of INO1 in the opi1Δ mutant was lost when both GRS I and GRS II were mutated (Fig. 3 B).
Both Ume6 and Opi1 recruit the Sin3/Rpd3 histone deacetylase, which is essential for INO1 repression (Kadosh and Struhl, 1997; Wagner et al., 2001), so we tested whether this complex prevents repositioning of INO1 to the nuclear periphery under repressing conditions. Indeed, loss of Sin3 or Rpd3 or a catalytically inactive form of Rpd3 (His188A; Kadosh and Struhl, 1998) led to constitutive targeting of INO1 (Fig. 3 A). The peripheral targeting of INO1 in the rpd3Δ mutant was lost when GRS I and GRS II were mutated (Fig. 3 B). Therefore, local recruitment of Rpd3 deacetylase activity blocks GRS I and GRS II zip code activity.
Rpd3 is the catalytic subunit of two distinct complexes, Rpd3(L) and Rpd3(S), that have distinct protein components, interact with distinct genomic sites, and have distinct effects on gene expression (Keogh et al., 2005). Whereas Rpd3(L) is associated with promoters and functions as a corepressor, Rpd3(S) is recruited cotranscriptionally to gene bodies to prevent cryptic transcriptional initiation (Kadosh and Struhl, 1997; Carrozza et al., 2005). We tested mutants lacking complex-specific subunits. Loss of Rdp3(L) components Pho23 or Sap30 led to unregulated targeting of INO1 to the nuclear periphery, whereas loss of the Rpd3(S) component Rco1 did not (Fig. 3 A). Thus, Rpd3(L) blocks peripheral targeting of INO1.
To test whether Rpd3 affects local histone acetylation and Put3 binding to the GRS I, we performed chromatin immunoprecipitation (ChIP). Acetylation over the INO1 promoter increased under inositol starvation and was constitutively high in the absence of Rpd3 (Fig. 3 D). Likewise, unlike the wild-type strain, in which Put3 binds to the GRS I only under inducing conditions (Brickner et al., 2012), in the rpd3Δ mutant, Put3 binding to the INO1 promoter was constitutive (Fig. 3 E). This suggests that Rpd3 deacetylase activity regulates Put3 binding to GRS I in the INO1 promoter.
Rpd3(L) regulates interchromosomal clustering of INO1
Zip code–mediated targeting to the nuclear periphery also leads to interchromosomal clustering of genes that share the same zip codes (Brickner et al., 2012). For example, upon inositol starvation, the two alleles of INO1 reposition to the nuclear periphery and cluster (Brickner et al., 2012). This requires the zip codes, the TFs that bind to the zip codes, and nuclear pore proteins (Brickner et al., 2012). In haploid cells, this can be observed by comparing the position of two loci that are targeted to the nuclear periphery by the same zip code. For example, active INO1 clusters with both URA3:GRS I and another GRS I–containing gene, TSA2 (Brickner et al., 2012).
To test whether Rpd3(L) also regulates clustering of INO1 alleles in diploid cells, we measured the distribution of distances between the alleles of INO1 in a population of cells grown under both uninducing and inducing conditions (Fig. 4 A). Upon inositol starvation, the distribution of distances between alleles of INO1 shifts to significantly shorter distances (Brickner et al., 2012; Fig. 4 B; P = 2 × 10−4, Wilcoxon rank sum test). The fraction of cells in which the two alleles are ≤0.55 µm apart, an alternative metric for clustering (Brickner et al., 2012), was 24% in uninducing conditions and 49% under inositol starvation (Fig. 4 E; P = 6 × 10−5, Fisher exact test).
Figure 4.
Interchromosomal clustering of INO1 alleles is regulated by Rpd3(L). (A) Confocal micrographs of diploid cells having LacO arrays at each allele of INO1, expressing GFP-LacI and mCherry-ER marker. (B–D) Distances between INO1 alleles were measured and binned into 0.2-µm bins, and the distribution of distances within the population was compared. In B and C, the p-value is from a Wilcoxon rank sum test comparing the distributions. In panels C and D, the gray line (wild-type [WT] control) is the distribution from B under the inducing condition. (D, inset) P-values from Wilcoxon rank sum test for the indicated comparisons. (E) From the same measurements used to generate the distributions in B–D, the percentage of cells in which the two alleles are within 0.55 µm of each other. P-values from Fisher exact test.
In contrast to wild-type INO1, two alleles with mutated URS elements clustered constitutively, independent of inositol starvation (Fig. 4 C, P = 0.173). These alleles were ≤0.55 µm apart in 45.9% (uninduced) and 52.6% (induced) cells (Fig. 4 E, P = 0.31). Likewise, in the rpd3 H188A mutant strain, the alleles of INO1 were constitutively clustered, and this was dependent on GRS I and GRS II (Fig. 4, D and E). Thus, Rpd3(L) recruitment regulates both INO1 positioning and interchromosomal clustering.
Opi1 and Ume6 are sufficient to block GRS I and GRS II function
To test whether Opi1 and Ume6 are sufficient to regulate GRS I– and GRS II–mediated gene positioning, we inserted a LexA BS into the endogenous INO1 promoter 50 bp from the middle of GRS I and 395 bp from the middle of GRS II (Fig. 5 A). This experiment used the Ino2 L118A mutant strain, which bocks binding of Opi1, leading to constitutive targeting to the nuclear periphery (Fig. 3 A). LexA-Ume6 or LexA-Opi1 were expressed in these strains, as confirmed by immunoblotting against LexA (Fig. S4 A), and both proteins repressed INO1 transcription (Fig. S4 B). LexA-Ume6 and LexA-Opi1, but not LexA alone, blocked recruitment of INO1 to the nuclear periphery (Fig. 5 A). Therefore, Opi1 and Ume6 are sufficient to block GRS I and GRS II function in the context of the INO1 promoter.
Figure 5.
Opi1 and Ume6 are sufficient to block GRS I and GRS II zip code function, but not PRE zip code function. (A, top) Schematic of the INO1 promoter having a LexA BS in the ino2 L118A strain (which blocks the interaction of Ino2 with Opi1; indicated with an X). (bottom) Peripheral localization of ino2 L118A LexA BS-INO1 in strains expressing LexA, LexA-Opi1, or LexA-Ume6 grown under the uninducing condition. (B, top) Schematic of LacO128-LexA BS ± zip codes integrated at URA3. The spacing from the middle of the LexA BS to the middle of the zip codes was GRS I = 68 bp, GRS II = 58 bp, 3×PRE = 42 bp. (bottom) Peripheral localization of URA3-LexA BS ± the indicated zip codes in strains expressing LexA, LexA-Opi1, or LexA-Ume6. For A and B, the color indicates the p-value from the Fisher exact test, compared with the control. (C) Distribution of distances between INO1 and URA3-GRS I-LexA BS in strains expressing LexA or LexA-Opi1, grown under inositol starvation. (top) Mean distances and standard deviations. P-value from Wilcoxon rank sum test comparing the distributions. (D) Fraction of cells in which INO1 and URA3-GRS I-LexA BS were ≤0.55 µm apart (p-values from Fisher exact test).
We also reconstituted Opi1- and Ume6-mediated regulation of each zip code separately at the ectopic site by tethering LexA-Ume6 and LexA-Opi1 beside URA3:GRS I, URA3:GRS II, or URA3:3×PRE (Fig. 5 B). Tethering the repressors beside GRS I or GRS II blocked peripheral localization (Fig. 5 B). However, this effect was specific: neither repressor blocked Ste12-mediated targeting (Fig. 5 B). This suggests that Ste12 targeting must be regulated by a different mechanism and that repressors such as Ume6 and Opi1 can regulate peripheral targeting by some, but not all, TFs.
Finally, we asked whether tethered LexA-Opi1 was also sufficient to regulate clustering of INO1 with URA3:GRS I LexA BS. In cells expressing LexA alone, URA3:GRS I LexA BS clustered with INO1 under inositol starvation (Fig. 5, C and D). Expression of Lex-Opi1 disrupted this clustering (Fig. 5, C [P = 0.002] and D [P = 0.003]). Thus LexA-Opi1 was sufficient to regulate both peripheral localization and interchromosomal clustering mediated by GRS I.
General role for repressors in regulating zip code–mediated targeting to the nuclear periphery
We next asked whether the ability to repress zip code function is a general function of transcriptional repressors by creating LexA fusions to an additional 19 transcriptional repressors. These 24 fusion proteins were tested for expression (immunoblot), ability to repress INO1 transcription (by tethering to the LexA BS used in Fig. 5 A), and ability to block recruitment of URA3:GRS I LexA BS to the nuclear periphery (Fig. S4). Three repressor fusions (Leu3, Rpd3, and Pho23) failed all three of these tests and were excluded (Fig. S4 C). By measuring the peripheral localization of URA3:GRS I LexA BS in strains expressing the remaining 21 LexA-repressor fusions, we found that 16 blocked GRS I–mediated targeting to the nuclear periphery in a LexA BS–dependent manner (Fig. 6, A and B). There was no correlation between the ability to block GRS I function and repressor size, expression level, or ability to repress INO1 transcription (Fig. S4 C). Finally, 11 of the 16 repressors that blocked GRS I function required Rpd3; in the rpd3 H188A mutant, GRS I–mediated peripheral localization was restored. The remaining five repressors were Rpd3 independent. Therefore, many repressors are capable of regulating DNA zip codes, both through Rpd3 recruitment and by other mechanisms.
Figure 6.
A general role for repressors in regulating zip code–mediated targeting to the nuclear periphery. (A and B) Peripheral localization of URA3-GRS I-LexA BS (A) or URA3-GRS I (B) in strains expressing LexA alone (−) or LexA fused to the indicated full-length repressors (see www.yeastgenome.org for descriptions of all gene names). These LexA fusions were validated as described in Methods and in Fig. S3. LexA fusions were tested for their ability to block GRS I–mediated targeting in both RPD3 (A and B; light green or forest green) and rpd3 H188A (A; lavender or purple) strains. Repressors that resulted in a statistically significantly decrease in peripheral localization compared with LexA alone (P < 0.05; Fisher exact test) are indicated in light green (RPD3) and lavender (rpd3 H188A); repressors that did not affect GRS I–mediated peripheral localization are indicated in forest green (RPD3) and purple (rpd3 H188A) bars. Mean and SEM from three of more biological replicates (30–50 cells per replicate).
Regulation of Ste12-mediated gene positioning by MAPK signaling
Ste12 binds to the promoters of genes such as PRM1 constitutively (Zheng et al., 2010), but transcriptional activation (Roberts et al., 2000) and targeting to the nuclear periphery (Fig. 7 B) occur only in the presence of mating pheromone. The positioning mediated by 3×PRE is independent of context (Fig. 2), and tethering Opi1 and Ume6 beside 3×PRE failed to block targeting to the periphery (Fig. 3 B). Therefore, the regulation of Ste12 must be through a different mechanism.
Figure 7.
MAPK phosphorylation of Dig2 regulates Ste12-mediated peripheral targeting and interchromosomal clustering. (A) Schematic for regulation of Ste12-dependent transcriptional activation of PRM1 by Dig1 and Dig2. (B) Peripheral localization of the PRM1 gene in wild-type (WT) and mutant strains ± α-factor. Point mutations in Dig2 were introduced into the chromosomal DIG2 locus. (C, top) Schematic of URA3-LexA BS. (bottom) Peripheral localization of URA3-LexA BS in wild-type and dig2Δ strains expressing LexA or LexA-Ste12, grown under uninducing and inducing conditions. Mean and SEM from three of more biological replicates (30–50 cells per replicate); *, P ≤ 0.05 (Fisher exact test) between the uninducing and inducing condition. NS, not significant. (D) Distribution of distances in 0.2-µm bins between PRM1 and URA3:3×PRE in wild-type, dig1Δ, or dig2Δ MATa haploid strains ± α-factor. (top) Mean distances and standard deviations. (inset) P-values from Wilcoxon rank sum test for each comparison. (E) Fraction of cells in which PRM1 and URA3:3×PRE were ≤0.55 µm apart (p-values from Fisher exact test).
In the absence of mating pheromone, Ste12 binds to two inhibitors, Dig1 and Dig2, that independently inhibit Ste12 by different mechanisms (Olson et al., 2000). Stimulation of the MAPK pathway by pheromone leads to phosphorylation of Dig1 and Dig2, causing them to dissociate from Ste12 (Fig. 7 A). Therefore, we tested whether Dig1 and/or Dig2 blocked Ste12-mediated positioning of PRM1 at the nuclear periphery. Mutants lacking Dig1 showed normal, conditional targeting to the nuclear periphery (Fig. 7 B). However, mutants lacking Dig2 showed constitutive targeting of PRM1 to the periphery (Fig. 7 B). Targeting of PRM1 to the periphery in the dig2Δ mutant required Ste12 (Fig. 7 B). Therefore, although both Dig1 and Dig2 regulate Ste12-mediated transcription, Dig2 alone blocks Ste12-mediated gene positioning.
We next tested whether MAPK signaling relieves Dig2 repression to allow PRM1 targeting to the nuclear periphery. A phosphoproteomic study identified Ser34 of Dig2 as a pheromone-stimulated MAPK phosphorylation site (Gruhler et al., 2005). Ser34 was replaced with either an alanine (to block phosphorylation) or an aspartate (to mimic phosphorylation) in the chromosomal DIG2 gene. The Ser34Ala mutation blocked PRM1 targeting to the nuclear periphery (Fig. 7 B). Mimicking phosphorylation with the Ser34Asp mutation led to constitutive peripheral localization, like the dig2Δ mutant (Fig. 7 B). This suggests that MAPK phosphorylation of Dig2 Ser34 relieves inhibition of Ste12-mediated targeting to the nuclear periphery.
Ste12 is necessary and its BS is sufficient to control spatial positioning. To confirm that Ste12 is responsible for targeting, LexA-Ste12 was tethered to URA3:LexA BS (Fig. 7 C). Under uninducing conditions, URA3:LexA BS was nucleoplasmic (Fig. 7 C). However, in the presence of mating pheromone or in the dig2Δ mutant strain, LexA-Ste12 expression caused URA3:LexA BS localization at the nuclear periphery (Fig. 7 C). Thus, Ste12 is sufficient to induce peripheral positioning, and its regulation by Dig2 is independent of DNA binding.
To test whether Ste12-mediated targeting to the nuclear periphery leads to interchromosomal clustering, we created a MATa haploid yeast strain having LacO arrays at both PRM1 and URA3:3×PRE. In the absence of mating pheromone, the distances between PRM1 and URA3:3×PRE were broadly distributed, with 23% of the cells having distances ≤0.55 µm (Fig. 7 D). In the presence of mating pheromone, PRM1 and URA3:3×PRE clustered together, with a decrease in the mean distance (P = 0.0017; Fig. 7 D) and an increase to 43% of the cells with distances ≤0.55 µm (P = 0.0006; Fig. 7 E). Loss of Dig2 led to even higher levels of clustering in the absence of mating pheromone (Fig. 7, D and E). Therefore, Ste12-mediated positioning and interchromosomal clustering are regulated by MAPK signaling through Dig2 phosphorylation.
Increased peripheral gene positioning through regulated TF synthesis
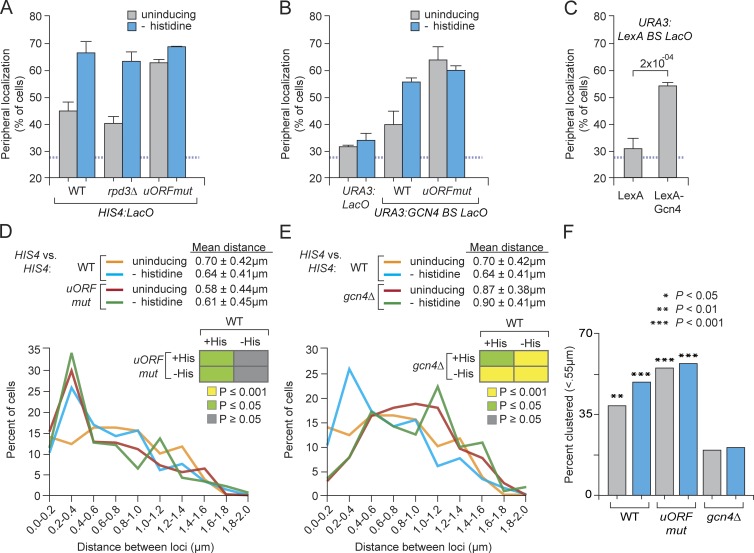
Mutant strains lacking Gcn4 fail to target both HIS4 (Fig. 1 B) and URA3:Gcn4 BS (Fig. 2 C) to the nuclear periphery. Therefore, we hypothesized that Gcn4 abundance, not DNA binding or activity, regulates Gcn4-mediated gene positioning. Several short upstream open reading frames (uORFs) in the 5′ end of the GCN4 mRNA compete with the GCN4 coding sequence for translation (Mueller and Hinnebusch, 1986; Hinnebusch, 2005) and, in the presence of amino acids, Gcn4 is poorly translated. Amino acid starvation leads to an accumulation of uncharged tRNAs, stimulating the Gcn2 kinase to phosphorylate eIF2α and leading to a global decrease in translation initiation rates (Cigan et al., 1993). This both reduces the global utilization of amino acids and, by decreasing translation initiation of the GCN4 uORFs, leads to increased translation of Gcn4 protein.
To test the idea that Gcn4-mediated gene positioning is controlled by Gcn4 protein concentrations, we mutated the initiation codon of the third and fourth uORF in the 5′ end of the GCN4 mRNA in the endogenous GCN4 gene, which leads to constitutive translation of Gcn4 at levels comparable to those observed during histidine starvation (Mueller and Hinnebusch, 1986). These uORF mutations led to constitutive, high-level localization of both HIS4 (∼65%; Fig. 8 A) and URA3:Gcn4 BS (∼60%; Fig. 8 B) at the nuclear periphery. LexA-Gcn4 is also sufficient to position URA3:LexA BS to the nuclear periphery, even under uninducing conditions (Fig. 8 C). These results suggest that the occupancy of Gcn4 on the DNA is regulated by the efficiency of Gcn4 translation and that the increase in occupancy leads to an increase in targeting to the nuclear periphery.
Figure 8.
Translational control of Gcn4-mediated peripheral targeting and interchromosomal clustering. (A–C) Peripheral localization of HIS4 (A), URA3 (B), URA3:GCN4 BS (B), and URA3:LexA BS (C) in the indicated strains grown under uninducing (gray bars) or inducing (cyan bars) conditions. Mutations in the uORFs were introduced into the chromosomal GCN4 locus (Methods). *, P ≤ 0.05 (Fisher exact test) between uninducing and inducing condition. WT, wild-type. (C) Peripheral localization of URA3-LexA BS in strains expressing LexA or LexA-Gcn4. Mean and SEM from three of more biological replicates (30–50 cells per replicate; p-value from Fisher exact test). (D and E) Distribution of distances, binned into 0.2-µm bins, between alleles of HIS4 in wild type, GCN4uORF (D), or gcn4Δ (E) homozygous diploid strains grown ± histidine. (insets) P-values from Wilcoxon rank sum test for each comparison. (F) Fraction of cells in which the two alleles of HIS4 were ≤0.55 µm apart (p-values from Fisher exact test compared with the gcn4Δ strain under uninducing conditions).
Gcn4-mediated targeting of HIS4 to the nuclear periphery also leads to interchromosomal clustering of HIS4 alleles in a diploid cell (Fig. 8 C). In the presence of histidine, the HIS4 alleles were partially clustered and, upon histidine starvation, this clustering increased (Fig. 8 C). Clustering requires Gcn4; in strains lacking Gcn4, clustering was completely lost under both uninducing and inducing conditions (Fig. 8 C). In a strain having the uORF mutations, HIS4 clustering was high and unregulated (Fig. 8 C). Thus, interchromosomal clustering of HIS4 is quantitatively controlled by Gcn4 protein levels.
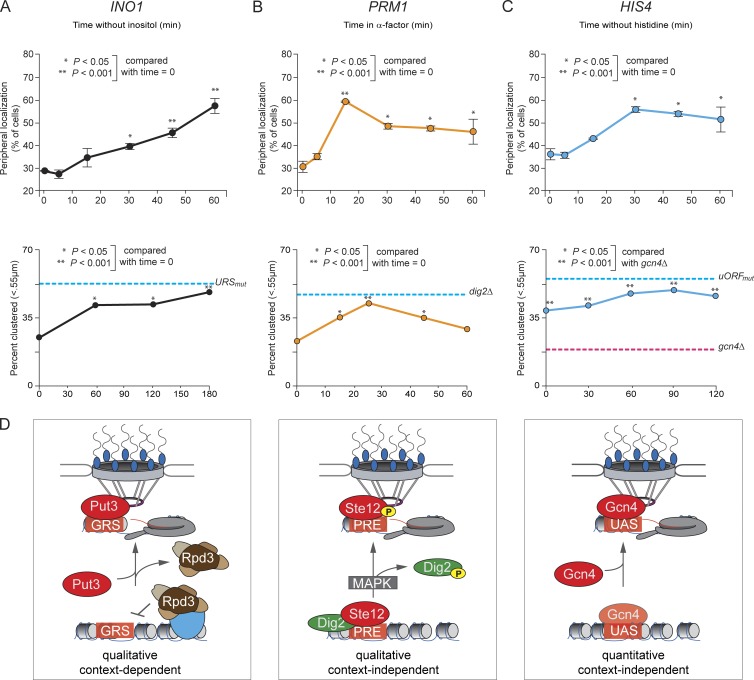
To compare the kinetics of spatial reorganization regulated by these three mechanisms, we analyzed the gene positioning and interchromosomal clustering of INO1, HIS4, or PRM1 after shifting cells from uninducing to inducing conditions (Fig. 9). In each case, interchromosomal clustering occurred more slowly than targeting to the nuclear periphery, consistent with the fact that interaction with the NPC is a prerequisite for clustering (Brickner et al., 2012). Dissociation of a histone deacetylase from the INO1 promoter led to peripheral localization within 1 h and interchromosomal clustering after 3 h (Fig. 9 A). MAPK phosphorylation of Dig2 led to relocalization of PRM1 to the nuclear periphery within 15 min and interchromosomal clustering within 30 min (Fig. 9 B). Relief of Gcn4 translational attenuation led to a statistically significant increase in peripheral localization within 30 min and interchromosomal clustering after 60 min (Fig. 9 C). Thus, these different mechanisms of regulation allow cells to change the spatial positioning of genes with respect to each other over different time scales.
Figure 9.
Different regulatory strategies lead to large-scale changes in nuclear organization over different time scales. (A–C) Time course after shifting to –inositol (A), +α-factor (B), or –histidine (C). (top) Peripheral localization of INO1 (A), PRM1 (B), and HIS4 (C). (bottom) Percentage of cells in which the two loci were ≤0.55 µm for INO1 versus INO1 in diploid cells (A), PRM1 versus URA3:3×PRE in haploid MATa cells (B), and HIS4 versus HIS4 in diploid cells. (D) Schematic for three distinct mechanisms of regulation of gene positioning. *, P ≤ 0.05; **, P ≤ 0.001 (Fisher exact test) between SDC and inducing condition.
Discussion
The work presented here argues that TFs play a critical role in controlling the spatial organization of the yeast genome and that their function can be regulated to allow this spatial organization to be dynamically altered. Put3, Cbf1, Ste12, and Gcn4 (and other TFs; unpublished data) mediate inducible repositioning of target genes from nucleoplasm to the nuclear periphery. Importantly, the BSs for these TFs function as DNA zip codes, encoding peripheral targeting and interchromosomal clustering (Brickner et al., 2012). These TFs represent four different families, suggesting that control of gene positioning is a common function of TFs. Not all TFs possess this activity; several TFs bind to the promoters of genes that interact with the NPC (Fig. S1). However, many of these BSs are not sufficient to confer targeting to the nuclear periphery (Fig. S1 and not depicted). Such TFs may be important transcriptional regulators of genes that are targeted to the NPC by different mechanisms. For example, although the Ino2/Ino4 BS is enriched among genes that interact with the NPC, these factors are neither necessary nor sufficient to promote peripheral localization. It will be important to understand what distinguishes the TFs that control spatial positioning from those that do not.
Our data suggest that TFs use multiple mechanisms to promote targeting to the nuclear periphery. Repositioning of INO1 and HIS4 or their respective zip codes to the nuclear periphery requires the SAGA complex, whereas repositioning of PRM1 or URA3:3×PRE does not. It remains unclear what role the SAGA complex plays in recruiting chromatin. Because the TFs that require SAGA are regulated at the level of TF occupancy, whereas Ste12 is regulated downstream of DNA binding, perhaps SAGA functions to promote TF binding. For example, the ability of Put3 to bind to GRS I and mediate recruitment to the nuclear periphery is blocked by a histone deacetylase. In the absence of SAGA-mediated acetylation, Put3, Cbf1, and Gcn4 may fail to bind to the DNA, preventing peripheral targeting. Alternatively, SAGA may be recruited by the TFs and serve a more direct role in recruiting chromatin to the nuclear periphery (Cabal et al., 2006; Luthra et al., 2007; Schneider et al., 2015).
Gene positioning is regulated by either genomic context or direct modulation of TF function (Fig. 9 D). In the context of the INO1 promoter, GRS I and GRS II zip code function is blocked by local recruitment of the conserved Rpd3(L) histone deacetylase, which regulates hundreds of genes in yeast (Bernstein et al., 2000; Yang and Seto, 2008). Under uninducing conditions, Rpd3(L) recruitment by Opi1 and Ume6 blocks Put3 binding to GRS I. The requirement for both Ume6 and Opi1 suggests that these repressors associate poorly with the INO1 promoter in the absence of one another. Upon induction, Opi1 dissociates from Ino2/Ino4 (Loewen et al., 2003; Brickner and Walter, 2004; Luthra et al., 2007; Schneider et al., 2015). This presumably leads to Ume6 dissociation as well, leading to loss of Rpd3(L) and increased histone acetylation, ultimately allowing binding of Put3. Consistent with this model, tethering either repressor to the INO1 promoter was sufficient to block peripheral localization (Fig. 5 A). Thus, transcriptional repressors can regulate gene positioning by blocking the interaction of the TFs that mediate targeting.
Regulation of GRS I–mediated targeting represents a novel assay for Rpd3 function. Although loss of Rpd3 affects the expression of hundreds of genes, in most cases the mechanism of repression has not been defined. Our data provide new insight into Rpd3 function. For example, despite the fact Rpd3 shows physical interactions with Whi5 and Tup1 and genetic interaction with Gal80, none of these factors requires Rpd3 catalytic activity to block GRS I targeting. Furthermore, of the 11 that are Rpd3 dependent, only three (Cup9, Oaf3, and Xbp1) been shown to interact with Rpd3. Our results suggest that the rest may also repress transcription and gene recruitment to the nuclear periphery through local recruitment of Rdp3(L).
Another GRS I target, TSA2, is induced by protein folding stresses and is not regulated by Ume6 or Opi1 (Ahmed et al., 2010; Brickner et al., 2012). This gene is induced and targeted to the nuclear periphery much more rapidly than INO1 (Ahmed et al., 2010), suggesting that Put3 binding can be can be regulated by other mechanisms. Consistent with this idea, we have found that many transcriptional repressors are capable of blocking Put3-dependent recruitment of an ectopic site to the nuclear periphery. These repressors regulate gene targeting by more than one mechanism: eleven repressors require Rpd3 deacetylase activity and five do not. Therefore, the same targeting mechanism can be regulated by different, context-specific regulatory strategies.
Ste12-dependent targeting is not dependent on its genomic context: when inserted at an ectopic site in the genome, 3×PRE mediated pheromone-responsive targeting to the nuclear periphery. Ste12 transcriptional activation is inhibited by Dig1 and Dig2. Previous work has suggested that Dig1 regulates the spatial arrangement of Ste12 target genes: in dig1Δ mutants, Ste12 forms foci in the nucleus, leading to increased intrachromosomal looping of Ste12 target genes (McCullagh et al., 2010). We found that Ste12-mediated targeting to the nuclear periphery and interchromosomal clustering was blocked by Dig2 but not Dig1. Blocking phosphorylation of serine 34 of Dig2 prevented Ste12-mediated targeting to the nuclear periphery in the presence of pheromone, and a phosphomimetic substitution for serine 34 led to unregulated peripheral targeting and interchromosomal clustering. Dig2 regulates Ste12 function, not DNA binding; although Ste12 binding to some sites in the genome is increased by pheromone signaling, binding to the PRM1 promoter is not (Zheng et al., 2010). Furthermore, tethering LexA-Ste12 to the URA3 locus resulted in pheromone- and Dig2-regulated targeting to the nuclear periphery. Therefore, Dig2 must block events downstream of DNA binding that lead to targeting to the nuclear periphery.
Unlike INO1 and PRM1, Gcn4 is capable of promoting significant peripheral targeting and interchromosomal clustering under uninducing conditions. When cells were starved for amino acids or when the translational regulation of Gcn4 was disrupted, targeting was much stronger. Thus, low-level production of Gcn4 leads to partial targeting to the nuclear periphery, and increased production of Gcn4 leads to increased targeting to the nuclear periphery, a quantitative, rather than a qualitative, change upon induction of its target genes (Fig. 9 D). Consistent with this idea, the number of Gcn4 BSs also impacts the efficiency of peripheral targeting. The peripheral localization of HIS4 (which has five Gcn4 BSs in its promoter) was consistently higher than that of URA3:Gcn4 BS.
Cells use many strategies to regulate TF function. Here we have defined three different ways that TF-mediated gene positioning is regulated in yeast, although it is likely that this list is incomplete. Why should cells use different regulatory schemes for different target genes? Because the rates of the three mechanisms we describe are very different, these regulatory strategies allow cells to dynamically alter the spatial reorganization of their genomes over very different time scales, which may have important adaptive value (Fig. 9).
Materials and Methods
Chemicals and media
All chemicals were purchased from Sigma-Aldrich, unless otherwise noted. Yeast media were obtained from Sunrise Science Products. Alpha factor was obtained from Zymo Research. Yeast and bacteria were grown with standard media as described previously (Maniatis et al., 1982). For experiments involving inositol starvation, cells were grown in SDC-inositol ± 100 µM myo-inositol. For experiments involving α factor, 100 µM (final concentration) was added to yeast suspended in 100 µl of SDC media.
Yeast strains
Yeast were transformed with plasmids described in Egecioglu et al. (2014). All yeast strains were derived from W303 (ade2-1 URA3-1 trp1-1 his3-11,15 leu2-3,112 can1-100) strains CRY1 (MATa) or CRY2 (MATα) and are listed in Table S1.
INO1 promoter mutants Δ4, grsIΔ, grsIIΔ, and grsIΔgrsIIΔ were created by transforming INO1promoterΔ strains with PCR of the INO1 promoter containing the desirable mutations and selecting on minimal medium without inositol. The following mutations were introduced at the endogenous loci by a different approach: UASmut INO1pro, URSmut INO1pro, ino2-L188A, rpd3-H188A, GCN4-uORF, dig2-S34A, and DIG2-S34D were created by integrating URA3 and SUP4-o ochre suppressor (Goodman et al., 1977) to replace the endogenous locus surrounding the mutation. Mutated versions of the endogenous loci were then integrated in place of the URA3-SUP4-o cassette by counterselection with 5-fluorooroitic acid (against URA3) or canavanine (against SUP4-o). For UASmut INO1pro, two UASINO elements at −178 (5′-CACATG-3′) to −172 and −243 (5′-CATGTG-3′) to −237 were mutated to (5′-CACTTC-3′) and (5′-GAAGTG-3′), respectively; for URSmut INO1pro, −260 (5′-TCGGCGGCT-3′) to −251 was mutated to (5′-GATTATTAG-3′); for GCN4, 5′ third uORF ATG was mutated to AGG and fourth uORF ATG was mutated to AUC; for rpd3-H188A, HIS188 5′-CAT-3′ was mutated to Ala 5′-GCT-3′; and for Dig2-S34A and DIG2-S34D, 5′-TCT-3′ was mutated to Ala 5′-GCT-3′ and 5′-GAT-3′, respectively.
Molecular biology
The plasmids p6LacO128 and p6LacO128-INO1 have been described (Brickner and Walter, 2004). The plasmids pmCh-ER03, pmCh-ER04, and pmCh-ER05 were derived from pAC08-mCh-L-TM (Meinema et al., 2011). The GAL1-10 promoter of pAC08-mCh-L-TM was replaced with the GPD1 promoter as a SacI–SpeI fragment to produce pGPD-mCh-ER16. The promoter, mCherry fusion, and 3′ UTR were then inserted into shuttle vectors pRS303 (HIS3), pRS304 (TRP1), and pRS305 (LEU2; Sikorski and Hieter, 1989) to generate pmCh-ER03, pmCh-ER04, and pmCh-ER05. These plasmids were digested with BstXI and integrated at HIS3, TRP1, or LEU2, respectively.
Plasmid pADH-LexA was derived from p414-ADH1 (Mumberg et al., 1995). LexA was inserted into p414-ADH1 as a NotI–PstI fragment. Repressors were PCR amplified from W303 genomic DNA and cloned into a pADH-LexA plasmid as either a BamHI C-terminal LexA fusion or a XhoI or PstI fragment creating an N-terminal LexA fusion (Fig. S3 C). Ste12 and Gcn4 were cloned into p414-ADH1 as XhoI and BamHI fragments, respectively.
The following plasmids were derived from p6LacO128: p6LacO128-GRS I, p6LacO128-GRS I-LexA, p6LacO128-GRS II-LexA, p6LacO128-3XPRE-LexA, p6LacO128-PRM1, p6LacO128-HIS4, and p6LacO128-GCN4. They were created as follows. Plasmid p6LacO128-GRS I: −266 to −366 of INO1 promoter was amplified and inserted into p6LacO128 as a SpeI–SacI fragment. For integration at URA3, p6LacO128-GRS I was linearized by digestion with StuI. Plasmid p6LacO128-GRS I-LexA: LexA BS was inserted into p6LacO128-GRS I as a SacI fragment with the sequence 5′-AAGGTTGGGAAGCCCTGCAAACTCATATACTGTATATATATACAGTATACAAGCT-3′. Plasmid p6LacO128-GRS II-LexA and p6LacO128-3×PRE-LexA were created by inserting the zip code along with LexA BS as BamHI fragments into the p6LacO128 plasmid. GRS II LexA fragment sequence is 5′-GATCCTTCCTACTGTTATTCTTCCCAGCAATCATTCACGCTTGCTACGTTGTATATGAAACGAGTAGTGATACTGTATATATATACAGTA-3′. 3×PRE LexA fragment sequence is 5′-GATCGAGTCCGGGTAATACATATGTTTCAATACTGTTTCAATACTGTTTCAGAAGTGCGTCACATATTATACTGTATATATATACAGTA-3′. Plasmid p6LacO128-PRM1: 1500 to 3067 of the PRM1 CDS and 3′ UTR was amplified and inserted as a BamHI–NotI fragment into p6LacO128. This plasmid was linearized by digestion with SnaBI to integrate at PRM1 locus. Plasmid p6LacO128-HIS4: 3171 to 3577 HIS4 CDS and 3′ UTR was amplified and inserted as a BamHI–SphI fragment into p6LacO128. p6LacO128-HIS4 was linearized by HindIII and integrated at HIS4. p6LacO128-GCN4: GCN4 BS 5′-CATGCACAGTGACTCACGTTTTTTT-3′ from the HIS4 promoter (228 to 253) was inserted into p6LacO128 as a HindIII fragment. Plasmid p6LacO128-GCN4 was linearized by StuI and integrated at URA3. To create the URA3:3×PRE LacO strain, the sequence 5′-TACATATGTTTCAATACTGTTTCAATACTGTTTCAGAAGTGCGTCACATATTAA-3′ was cloned into an integration cassette within the plasmid pZipKan using flanking StuI sites. It was then integrated directly into the backbone of the p6LacO128 as described previously (Egecioglu et al., 2014).
Microscopy
For all experiments, cells were maintained at OD600 <0.8. For inositol starvation experiments, strains were grown overnight in SDC-inositol in the presence or absence or 100 µM myo-inositol. For histidine starvation experiments, strains were grown overnight in SDC, harvested, resuspended in either SDC or SDC-His media, and incubated at 30°C for 45–75 min. For α-factor stimulation experiments, strains were grown overnight in SDC, and 100 µl of cells were transferred to a 1.5-ml Eppendorf tube with 100 µM final concentration of α-factor for 15–25 min. After treatment, cells were concentrated by brief centrifugation and imaged immediately. The images were captured as 0.34-µm-thick z-stacks (the yeast nucleus is ∼2 µm in diameter) with an SP5 II Line Scanning Confocal Microscope (Leica Biosystems) with 100× 1.44-NA (oil immersion) objective using Argon 488-nm and Diode Pumped Solid State 561-nm lasers in the Northwestern Biological Imaging Facility as described (Egecioglu et al., 2014). Cells were scored using LAS AF or LAS AF Lite software. For each individual cell, the z-stack with the brightest and most focused LacO dot was scored. The slice used was not necessarily the same for every cell and, for cells in which the dot was at the top or bottom of the nucleus, localization was not scored. If the center of the dot overlapped with the membrane, the cell was scored as peripheral.
Clustering analysis
Samples were visualized on a Spinning Disc Advanced Fluorescence confocal microscope (Leica Biosystems) in the Northwestern University Biological Imaging Facility. Cells were deposited on a double-cavity microscope slide (VWR) prepared with 4% low-melt agar. Slide covers were sealed with nail polish to prevent drying. Images of the cells consisted of 16 z-slices spaced 0.4 µm apart, with eight time points taken at 30-s intervals over 4 min. From the first time point of each image, the distance between the two loci was measured using ImageJ. When only a single locus was apparent in a cell but the entire cell was captured in the image, the additional time points were referenced to determine whether the spot subsequently later split into two loci. In those cases, a distance value of 0 µm was assigned to these tightly clustered loci. For images of the PRM1 versus URA3:3×PRE loci, only a single time point was taken in the interest of more rapid acquisition, and tightly clustered loci were excluded from our analysis. For each condition, >100 cells were measured. The distances were binned into 0.15- or 0.2-µm classes to generate distributions of distance used in mountain plots and heat plots. For heat plots, the mean fraction of cells and standard deviation for all bins was used. An R script was used to generate the grayscale heat map of the number of standard deviations from the mean for each bin, scaled as shown in Figs. 3, 5, and 6. To control for the effect of GFP dimerization on interchromosomal clustering, we tested a mutant monomeric form of GFP (A206K)-LacI (Zacharias et al., 2002; Mirkin et al., 2014). This mutation had no effect on the degree of interchromosomal clustering between two alleles of HIS4 (Fig. S5).
ChIP
ChIP experiments were performed as described in Egecioglu et al. (2014). For anti–pan-acetyl and anti-H3 ChIP, lysates were fixed for 15 min. For GST-Put3 ChIP, lysates were fixed for 1 h. 5 µl anti–pan-acetyl (sc-8649-R; Santa Cruz Biotechnology), 3 µl anti-H3 (ab180727; Abcam), 4 µl α-GST antibody (G7781; Sigma-Aldrich), and 8 µl α-rabbit IgG Dynabeads (11203D; Invitrogen) were incubated with 3.0 mg lysates. All immunoprecipitations were incubated overnight at 4°C.
Online supplemental material
Fig. S1 shows identifying TFs that mediate targeting to the nuclear periphery, related to Fig. 1. Fig. S2 shows that loss of Cbf1 does not block targeting of INO1 to the nuclear periphery. Fig. S3 shows trans- and cis-mutant effects on INO1 transcription, related to Fig. 2. Fig. S4 shows the phenotype of LexA-repressor fusions, related to Fig. 4. Fig. S5 shows the distribution of distances and fraction of cells between alleles of HIS4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201508068/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201508068.dv.
Supplementary Material
Acknowledgments
The authors thank members of the Brickner laboratory for helpful comments on the manuscript and M. Hogan for technical assistance.
This work was supported by National Institutes of Health grant GM080484. S. Ahmed was supported by a Rappaport Award for Research Excellence; V. Sood was supported by an American Heart Association predoctoral fellowship; R. Coukos was supported by a Northwestern Undergraduate Research Fellowship, a Program of Biological Sciences Summer Grant, and a Krieghbaum Award; and J.H. Brickner is the Soretta and Henry Shapiro Research Professor in Molecular Biology.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BS
- binding site
- ChIP
- chromatin immunoprecipitation
- GRS
- gene recruitment sequence
- NPC
- nuclear pore complex
- PRE
- pheromone response element
- TF
- transcription factor
- UASINO
- inositol-sensitive upstream activating sequence
- uORF
- upstream open reading frame
- URS
- upstream repressing sequence
References
- Ahmed S., Brickner D.G., Light W.H., Cajigas I., McDonough M., Froyshteter A.B., Volpe T., and Brickner J.H.. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12:111–118. 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O.M., Billington B.L., and Gottschling D.E.. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 66:1279–1287. 10.1016/0092-8674(91)90049-5 [DOI] [PubMed] [Google Scholar]
- Arndt K., and Fink G.R.. 1986. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc. Natl. Acad. Sci. USA. 83:8516–8520. 10.1073/pnas.83.22.8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhawat N., Ouyang Q., and Henry S.A.. 1995. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J. Biol. Chem. 270:25087–25095. 10.1074/jbc.270.42.25087 [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Tong J.K., and Schreiber S.L.. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA. 97:13708–13713. 10.1073/pnas.250477697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J.H., and Walter P.. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342 10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.C., Widom J., and Brickner J.H.. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Ahmed S., Meldi L., Thompson A., Light W., Young M., Hickman T.L., Chu F., Fabre E., and Brickner J.H.. 2012. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev. Cell. 22:1234–1246. 10.1016/j.devcel.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.R., Kennedy C.J., Delmar V.A., Forbes D.J., and Silver P.A.. 2008. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 22:627–639. 10.1101/gad.1632708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.M., Leach J., Reittie J.E., Atzberger A., Lee-Prudhoe J., Wood W.G., Higgs D.R., Iborra F.J., and Buckle V.J.. 2006. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 172:177–187. 10.1083/jcb.200507073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., and Nehrbass U.. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773. 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., and Hetzer M.W.. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 140:372–383. 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., and Workman J.L.. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 123:581–592. 10.1016/j.cell.2005.10.023 [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., and Silver P.A.. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439. 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Drubin D.A., Rando O.J., and Silver P.A.. 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 19:1188–1198. 10.1101/gad.1307205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A.M., Bushman J.L., Boal T.R., and Hinnebusch A.G.. 1993. A protein complex of translational regulators of GCN4 mRNA is the guanine nucleotide-exchange factor for translation initiation factor 2 in yeast. Proc. Natl. Acad. Sci. USA. 90:5350–5354. 10.1073/pnas.90.11.5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer M., Dietzel S., Müller S., Solovei I., and Fakan S.. 2006. Chromosome territories: a functional nuclear landscape. Curr. Opin. Cell Biol. 18:307–316. 10.1016/j.ceb.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Denoth-Lippuner A., Krzyzanowski M.K., Stober C., and Barral Y.. 2014. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. eLife. 3:3 10.7554/eLife.03790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G., Iglesias N., and Stutz F.. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 26:7858–7870. 10.1128/MCB.00870-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D.E., D’Urso A., Brickner D.G., Light W.H., and Brickner J.H.. 2014. Approaches to studying subnuclear organization and gene-nuclear pore interactions. Methods Cell Biol. 122:463–485. 10.1016/B978-0-12-417160-2.00021-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman H.M., Olson M.V., and Hall B.D.. 1977. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc. Natl. Acad. Sci. USA. 74:5453–5457. 10.1073/pnas.74.12.5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J.A., and Henry S.A.. 2000. Regulation of the yeast INO1 gene. The products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics. 154:1485–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhler A., Olsen J.V., Mohammed S., Mortensen P., Faergeman N.J., Mann M., and Jensen O.N.. 2005. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics. 4:310–327. 10.1074/mcp.M400219-MCP200 [DOI] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., and van Steensel B.. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 453:948–951. 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Hagen D.C., McCaffrey G., and Sprague G.F. Jr. 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11:2952–2961. 10.1128/MCB.11.6.2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyken W.T., Repenning A., Kumme J., and Schüller H.J.. 2005. Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol. Microbiol. 56:696–707. 10.1111/j.1365-2958.2004.04499.x [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407–450. 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- Kadosh D., and Struhl K.. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 89:365–371. 10.1016/S0092-8674(00)80217-2 [DOI] [PubMed] [Google Scholar]
- Kadosh D., and Struhl K.. 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12:797–805. 10.1101/gad.12.6.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B., Pickersgill H., Shloma V.V., and Fornerod M.. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 140:360–371. 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. . 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 123:593–605. 10.1016/j.cell.2005.10.025 [DOI] [PubMed] [Google Scholar]
- Liang Y., and Hetzer M.W.. 2011. Functional interactions between nucleoporins and chromatin. Curr. Opin. Cell Biol. 23:65–70. 10.1016/j.ceb.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W.H., Brickner D.G., Brand V.R., and Brickner J.H.. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell. 40:112–125. 10.1016/j.molcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Roy A., and Levine T.P.. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22:2025–2035. 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J.M., Schulze K.L., Yates J.W., Hirsch J.P., and Henry S.A.. 1993. The INO1 promoter of Saccharomyces cerevisiae includes an upstream repressor sequence (URS1) common to a diverse set of yeast genes. J. Bacteriol. 175:4235–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luperchio T.R., Wong X., and Reddy K.L.. 2014. Genome regulation at the peripheral zone: lamina associated domains in development and disease. Curr. Opin. Genet. Dev. 25:50–61. 10.1016/j.gde.2013.11.021 [DOI] [PubMed] [Google Scholar]
- Luthra R., Kerr S.C., Harreman M.T., Apponi L.H., Fasken M.B., Ramineni S., Chaurasia S., Valentini S.R., and Corbett A.H.. 2007. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 282:3042–3049. 10.1074/jbc.M608741200 [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., and Sambrook J.. 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 545 pp. [Google Scholar]
- McCullagh E., Seshan A., El-Samad H., and Madhani H.D.. 2010. Coordinate control of gene expression noise and interchromosomal interactions in a MAP kinase pathway. Nat. Cell Biol. 12:954–962. 10.1038/ncb2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema A.C., Laba J.K., Hapsari R.A., Otten R., Mulder F.A., Kralt A., van den Bogaart G., Lusk C.P., Poolman B., and Veenhoff L.M.. 2011. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 333:90–93. 10.1126/science.1205741 [DOI] [PubMed] [Google Scholar]
- Meister P., Mango S.E., and Gasser S.M.. 2011. Locking the genome: nuclear organization and cell fate. Curr. Opin. Genet. Dev. 21:167–174. 10.1016/j.gde.2011.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin E.V., Chang F.S., and Kleckner N.. 2014. Protein-mediated chromosome pairing of repetitive arrays. J. Mol. Biol. 426:550–557. 10.1016/j.jmb.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P.P., and Hinnebusch A.G.. 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 45:201–207. 10.1016/0092-8674(86)90384-3 [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller R., and Funk M.. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 156:119–122. 10.1016/0378-1119(95)00037-7 [DOI] [PubMed] [Google Scholar]
- Nikoloff D.M., and Henry S.A.. 1994. Functional characterization of the INO2 gene of Saccharomyces cerevisiae. A positive regulator of phospholipid biosynthesis. J. Biol. Chem. 269:7402–7411. [PubMed] [Google Scholar]
- Olson K.A., Nelson C., Tai G., Hung W., Yong C., Astell C., and Sadowski I.. 2000. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell. Biol. 20:4199–4209. 10.1128/MCB.20.12.4199-4209.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia P., Jeong J., and Capelson M.. 2014. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Reports. 9:433–442. 10.1016/j.celrep.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S.W., Solovei I., Brugman W., Gräf S., Flicek P., Kerkhoven R.M., van Lohuizen M., et al. . 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 38:603–613. 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A., and Dillon N.. 2015. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 16:245–257. 10.1038/nrm3965 [DOI] [PubMed] [Google Scholar]
- Pombo A., Jones E., Iborra F.J., Kimura H., Sugaya K., Cook P.R., and Jackson D.A.. 2000. Specialized transcription factories within mammalian nuclei. Crit. Rev. Eukaryot. Gene Expr. 10:21–29. 10.1615/CritRevEukarGeneExpr.v10.i1.40 [DOI] [PubMed] [Google Scholar]
- Roberts S.M., and Winston F.. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 147:451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C.J., Nelson B., Marton M.J., Stoughton R., Meyer M.R., Bennett H.A., He Y.D., Dai H., Walker W.L., Hughes T.R., et al. . 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 287:873–880. 10.1126/science.287.5454.873 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro S., Fischer T., Luo M.J., Antúnez O., Brettschneider S., Lechner J., Pérez-Ortín J.E., Reed R., and Hurt E.. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 116:75–86. 10.1016/S0092-8674(03)01025-0 [DOI] [PubMed] [Google Scholar]
- Rohner S., Kalck V., Wang X., Ikegami K., Lieb J.D., Gasser S.M., and Meister P.. 2013. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J. Cell Biol. 200:589–604. 10.1083/jcb.201207024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen A.A., Suka N., Turner B.M., and Grunstein M.. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 392:831–835. 10.1038/33952 [DOI] [PubMed] [Google Scholar]
- Schneider M., Hellerschmied D., Schubert T., Amlacher S., Vinayachandran V., Reja R., Pugh B.F., Clausen T., and Köhler A.. 2015. The nuclear pore-associated TREX-2 complex employs mediator to regulate gene expression. Cell. 162:1016–1028. 10.1016/j.cell.2015.07.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S., Sexton T., Chakalova L., Cope N.F., Horton A., Andrews S., Kurukuti S., Mitchell J.A., Umlauf D., Dimitrova D.S., et al. . 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42:53–61. 10.1038/ng.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A., and Lopes J.M.. 2010. Derepression of INO1 transcription requires cooperation between the Ino2p-Ino4p heterodimer and Cbf1p and recruitment of the ISW2 chromatin-remodeling complex. Eukaryot. Cell. 9:1845–1855. 10.1128/EC.00144-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., and Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., and Rout M.P.. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501. 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., and Gasser S.M.. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 441:774–778. 10.1038/nature04845 [DOI] [PubMed] [Google Scholar]
- Thompson M., Haeusler R.A., Good P.D., and Engelke D.R.. 2003. Nucleolar clustering of dispersed tRNA genes. Science. 302:1399–1401. 10.1126/science.1089814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters B.J., Wachi S., Mavrich T.N., Andersen B.E., Jena P., Sinnamon A.J., Jain P., Rolleri N.S., Jiang C., Hemeryck-Walsh C., and Pugh B.F.. 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 41:480–492. 10.1016/j.molcel.2011.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C., Dietz M., Wittmann J., Albrecht A., and Schüller H.J.. 2001. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 41:155–166. 10.1046/j.1365-2958.2001.02495.x [DOI] [PubMed] [Google Scholar]
- Xu M., and Cook P.R.. 2008. The role of specialized transcription factories in chromosome pairing. Biochim. Biophys. Acta. 1783:2155–2160. 10.1016/j.bbamcr.2008.07.013 [DOI] [PubMed] [Google Scholar]
- Yang X.J., and Seto E.. 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9:206–218. 10.1038/nrm2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., and Tsien R.Y.. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916. 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zheng W., Zhao H., Mancera E., Steinmetz L.M., and Snyder M.. 2010. Genetic analysis of variation in transcription factor binding in yeast. Nature. 464:1187–1191. 10.1038/nature08934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.