Biochemical, cytological, and lipidomic approaches show that lipid droplets are dispensable as membrane sources for autophagy, but are required for ER homeostasis by buffering fatty acid synthesis and ER stress and maintaining phospholipid composition to allow autophagy regulation and autophagosome biogenesis.
Abstract
Lipid droplets (LDs) are conserved organelles for intracellular neutral lipid storage. Recent studies suggest that LDs function as direct lipid sources for autophagy, a central catabolic process in homeostasis and stress response. Here, we demonstrate that LDs are dispensable as a membrane source for autophagy, but fulfill critical functions for endoplasmic reticulum (ER) homeostasis linked to autophagy regulation. In the absence of LDs, yeast cells display alterations in their phospholipid composition and fail to buffer de novo fatty acid (FA) synthesis causing chronic stress and morphologic changes in the ER. These defects compromise regulation of autophagy, including formation of multiple aberrant Atg8 puncta and drastically impaired autophagosome biogenesis, leading to severe defects in nutrient stress survival. Importantly, metabolically corrected phospholipid composition and improved FA resistance of LD-deficient cells cure autophagy and cell survival. Together, our findings provide novel insight into the complex interrelation between LD-mediated lipid homeostasis and the regulation of autophagy potentially relevant for neurodegenerative and metabolic diseases.
Introduction
Macroautophagy (hereafter autophagy) is a highly conserved homeostasis and stress response mechanism characterized by de novo formation of autophagosomes (APs), double-membrane structures that deliver cargo to vacuoles/lysosomes for degradation (Kraft and Martens, 2012; Lamb et al., 2013). Through hierarchical assembly and function, a multicomponent autophagy machinery drives membrane rearrangements, which nucleate, expand, and close nascent APs (Suzuki et al., 2007; Feng et al., 2014). Several membrane sources for AP biogenesis have been identified, including ER (Axe et al., 2008; Hayashi-Nishino et al., 2009), ER exit sites (ERES)/ER–Golgi intermediate compartment (Ge et al., 2013; Graef et al., 2013; Suzuki et al., 2013), Golgi apparatus (Young et al., 2006; Mari et al., 2010; Nair et al., 2011), endosomes (Longatti et al., 2012), mitochondria (Hailey et al., 2010), and plasma membrane (Ravikumar et al., 2010), but their relative contribution and underlying regulatory mechanisms remain unclear. Recent studies suggest that lipid droplets (LDs) function as a critical lipid source for AP biogenesis (Dupont et al., 2014; Li et al., 2015; Shpilka et al., 2015). LDs are conserved organelles originating from ER membranes that are comprised of a neutral lipid core formed by triacylglycerols (TGs) and sterol esters (SEs) and a surrounding monolayer of phospholipids (PLs; Kohlwein, 2010; Walther and Farese, 2012; Koch et al., 2014; Wilfling et al., 2014). Number and size of LDs vary substantially between different cell types and dynamically adapt to cellular needs. On one hand, LDs store excess fatty acids (FA) and lipids as carbon sources and thereby buffer potential cytotoxic effects (Garbarino et al., 2009; Petschnigg et al., 2009). On the other hand, they provide precursors for energy conversion, PL biosynthesis, and signaling molecules by lipolysis or selective turnover by autophagy (Singh et al., 2009; Henry et al., 2012; van Zutphen et al., 2014; Wang et al., 2014). A variety of metabolic and neurodegenerative diseases are associated with conditions of FA/lipid stress and commonly show defects in autophagy (Hotamisligil, 2010; Yang et al., 2010; Harris and Rubinsztein, 2011; Nixon, 2013; Quan et al., 2013). Hence, knowledge of the mechanisms connecting the function of LDs and autophagy is crucial for the understanding of underlying pathogeneses.
To dissect the functional role of LDs for autophagy, we took advantage of the facile yeast system and analyzed cells lacking the ability to form LDs by biochemical, cytological, and lipidomic approaches. Our study demonstrates that LDs are dispensable as membrane source for autophagy, but they are required for ER homeostasis by buffering de novo FA synthesis and ER stress and maintaining PL composition to allow intact autophagy regulation and AP biogenesis.
Results and discussion
LD deficiency conditionally blocks autophagy
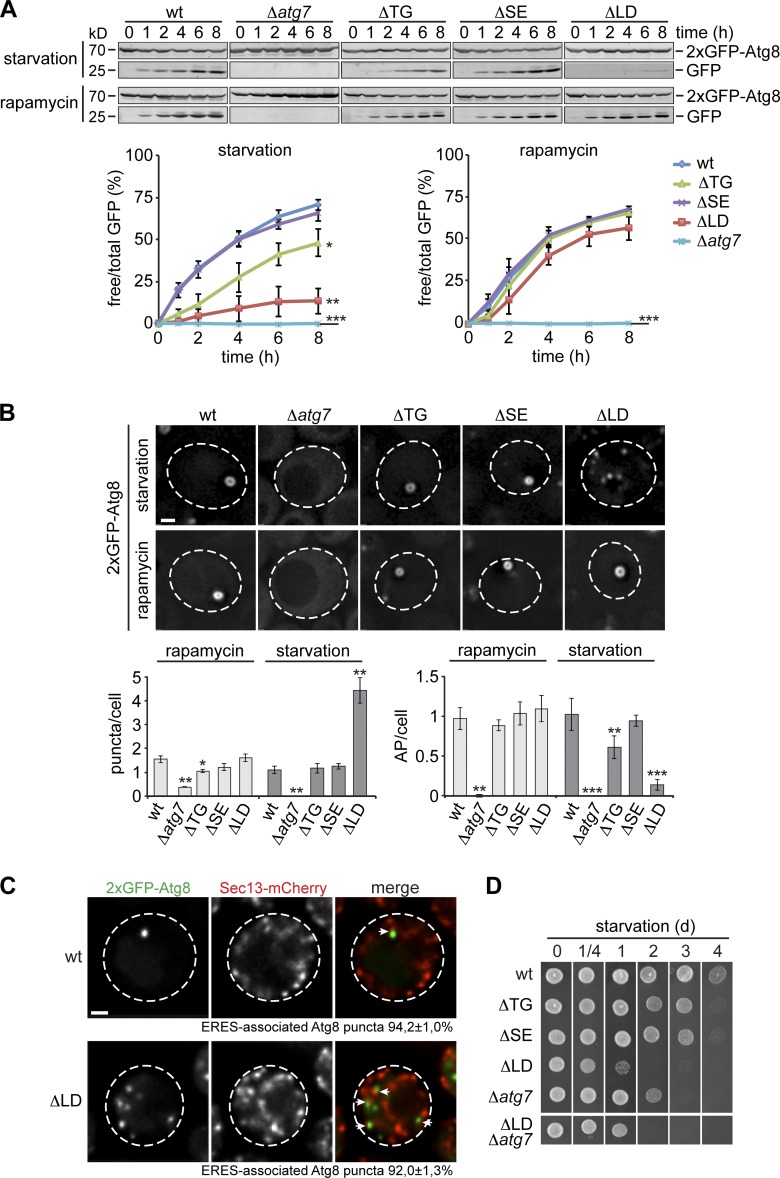
To investigate the functional relationship between LDs and autophagy, we analyzed yeast strains carrying gene deletions in DGA1 and LRO1 (Δdga1Δlro1; ΔTG) or in ARE1 and ARE2 (Δare1Δare2; ΔSE) required for synthesis of TGs or SEs, respectively, and a strain deleted for all four genes (Δare1Δare2Δdga1Δlro1; ΔLD), completely devoid of LDs, and compared them with a wild-type (wt) and an autophagy-deficient Δatg7 strain (Yang et al., 1996; Tanida et al., 1999; Oelkers et al., 2000, 2002; Sandager et al., 2002; Sorger and Daum, 2002). Cells were cultured in synthetic medium, which requires cells to synthesize FAs de novo, to avoid any influence of external FA on cellular lipid homeostasis. First, we induced autophagy by shifting wt, Δatg7, ΔTG, ΔSE, and ΔLD cells expressing a genomically integrated 2xGFP-ATG8 reporter to nitrogen starvation (starvation) and monitored autophagy flux using the GFP-Atg8 assay (Shintani and Klionsky, 2004). While autophagy was completely blocked in Δatg7 cells, we observed similar or partially reduced autophagy flux in ΔSE or ΔTG cells, respectively, compared with wt cells (Fig. 1 A, starvation). Interestingly, LD-deficient cells showed almost completely impaired autophagy flux, indicating that the presence of LDs is required for autophagy in line with recent studies (Fig. 1 A, starvation; Li et al., 2015; Shpilka et al., 2015). However, when we triggered autophagy by inhibiting target of rapamycin complex 1 (TORC1) pharmacologically by rapamycin treatment, ΔTG, ΔSE, and ΔLD cells induced wt-like autophagy flux (Fig. 1 A, rapamycin). We obtained similar results, when we analyzed wt, ΔLD, and Δatg7 cells expressing a plasmid-encoded cytosolic Rosella (cytRosella; pHluorin-mCherry), which reports on autophagy-mediated turnover of cytosol (Rosado et al., 2008). LD-deficient cells were defective in the autophagy-dependent transfer of cytRosella to the vacuole during starvation, but not after rapamycin treatment (Fig. S1 A). Collectively, these data demonstrate that the autophagy machinery is functionally intact in ΔLD cells, but conditionally depend on the presence of LDs during starvation.
Figure 1.
LD deficiency conditionally impairs autophagy. (A) Autophagy flux of wt, Δatg7, ΔTG, ΔSE, and ΔLD cells expressing 2xGFP-ATG8 during starvation or rapamycin treatment. Data are means ± SD (n = 4). (B) Cells were treated as in A and imaged by fluorescence microscopy after 1 h. Mean number of Atg8 puncta (left panel) and APs (right panel) per cell are shown as mean ± SD (≥150 cells; n = 3). (C) wt and ΔLD cells coexpressing 2xGFP-ATG8 and genomically tagged SEC13-mCherry were shifted to starvation for 1 h before imaging. Arrows indicate Atg8 puncta associated with ERES (Sec13-mCherry). Data are means ± SD (≥150 cells; n = 3). Dashed lines indicate cell boundaries. (D) Survival of indicated strains during starvation. Bars, 1 µm. t test in A and B: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
These conditional effects are not a consequence of differential TORC1 regulation because all tested strains showed the same shift in the TORC1-dependent phosphorylation pattern of endogenous Atg13, indicating Atg1 kinase complex activation and autophagy induction, in response to starvation or rapamycin treatment in Western blot analysis (Fig. S1 B; Kamada et al., 2000).
To explore the basis for the conditional defect in autophagy flux during starvation and rapamycin treatment in LD-deficient cells, we monitored autophagy by fluorescence microscopy. After 1 h of autophagy induction, we detected on average one punctum and one ring-like AP marked by 2xGFP-Atg8 per wt cell under both conditions (Fig. 1 B). ΔTG, ΔSE, and ΔLD cells displayed wt levels of Atg8 puncta and APs after 1 h of rapamycin treatment, confirming unaffected autophagy flux (Fig. 1 B, rapamycin). In contrast, during starvation, we observed a slight reduction in the number of APs in ΔTG cells compared with wt and ΔSE cells, paralleling the effects on autophagy flux in these cells (Fig. 1 B, starvation). Significantly, in the absence of LDs, AP biogenesis was severely defective during starvation: ΔLD cells showed a dramatic reduction of APs and an increased number of Atg8 puncta (Fig. 1 B, starvation). Interestingly, we detected quantitative spatial association of these Atg8 puncta with ERES, specifically marked by Sec13-mCherry, in ΔLD cells consistent with initiation of biogenesis, but compromised expansion and maturation of APs during starvation-induced autophagy (Fig. 1 C; Graef et al., 2013; Suzuki et al., 2013).
To test for physiological consequences of defects in LD biogenesis and autophagy, we monitored survival of wt, Δatg7, ΔTG, ΔSE, ΔLD, and Δatg7ΔLD cells during starvation. ΔLD cells were highly sensitive to starvation, similar to Δatg7 or Δatg7ΔLD cells linking LD biogenesis to autophagy regulation and nutrient stress survival (Fig. 1 D; Tsukada and Ohsumi, 1993).
In summary, our data demonstrate that LDs are required for biogenesis of APs, autophagy flux, and cell survival during starvation. However, LD-deficient cells possess a functional autophagy machinery capable of mediating a full, pharmacologically induced autophagy response questioning a general role of LDs as membrane source for AP biogenesis and suggesting that LDs function in autophagy regulation.
FA synthesis impairs autophagy in LD-deficient cells during starvation
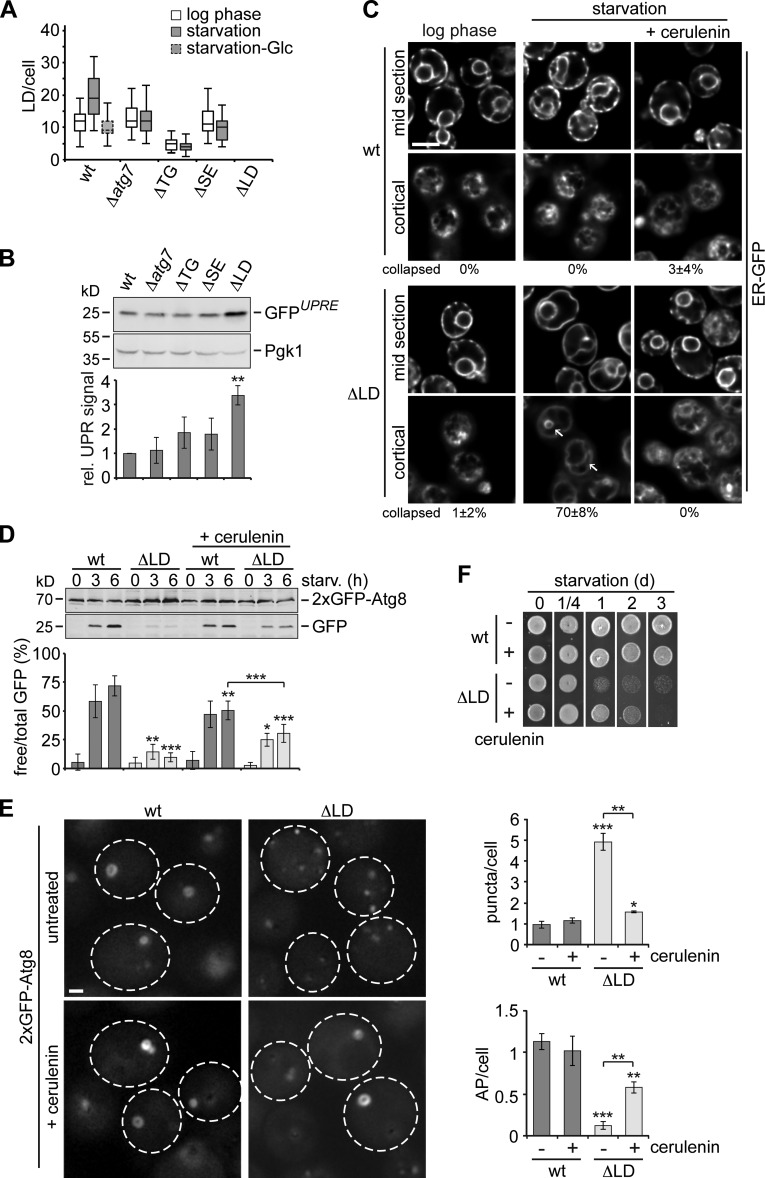
wt cells contained a significant number of LDs during log phase and induced LD biogenesis during starvation in a glucose-, TG and SE synthesis–, and autophagy-dependent manner (Figs. 2 A and S1 C), the latter being consistent with a previous study linking the autophagy machinery to LD biogenesis (Shibata et al., 2010). Because LDs originate from the ER, we hypothesized that LD deficiency might interfere with ER homeostasis. To test this notion, we first examined unfolded protein response (UPR) signaling as a measure for ER stress. We found almost fourfold-induced UPR signaling, specifically in ΔLD cells relative to wt cells using the synthetic reporter 4xUPRE-GFP, consistent with previous studies (Fig. 2 B; Jonikas et al., 2009; Petschnigg et al., 2009; Olzmann and Kopito, 2011). However, UPR signaling did not interfere with autophagy regulation. wt and ΔLD cells were unaffected in autophagy flux or cell survival during starvation in the absence of the unfolded protein sensor IRE1 or its downstream target HAC1 and upon expression of a constitutively active allele of IRE1, ire1C (Fig. S1, D–F; Patil and Walter, 2001; Papa et al., 2003; Walter and Ron, 2011). Next, we monitored ER morphology as a sensitive readout for ER homeostasis by fluorescence microscopy. Despite chronic UPR signaling, we could not detect differences in the morphology of the ER labeled by ER-targeted GFP in the absence of LDs in log phase (Fig. 2 C). However, the interconnected network of tubular and sheet-like cortical ER collapsed into a simplified network of dilated and continuous tubules in LD-deficient cells after 1 h of starvation (Fig. 2 C, starvation, arrows), reminiscent of altered ER structure induced by excess of external saturated FA (Pineau et al., 2009). Since cells rely on internal FA under our tested condition, we reasoned that de novo FA synthesis exceeding the storage capacity of LD-deficient cells might cause alterations in ER morphology. Indeed, upon chemical inhibition of de novo FA synthesis by FA synthase–specific inhibitor cerulenin during starvation (Vance et al., 1972), ΔLD cells maintained a wt-like ER morphology (Fig. 2 C). Thus, LDs are critical for maintenance of ER homeostasis by buffering de novo FA synthesis. Of note, we did not observe structural changes in the ER when we treated wt and ΔLD cells with rapamycin (Fig. S1 G), indicating that impaired ER homeostasis might be casually linked to defective autophagy.
Figure 2.
Inhibition of de novo FA synthesis improves autophagy, ER morphology, and cell survival of LD-deficient cells during starvation. (A) Number of BODIPY493/503-stained LDs in wt, Δatg7, ΔTG, ΔSE, and ΔLD cells during log phase and starvation ± glucose (Glu) after 6 h (150 cells per strain) analyzed by fluorescence microscopy as shown in Fig. S1 C. (B) wt, Δatg7, ΔTG, ΔSE, and ΔLD cells expressing 4xUPRE-GFP grown to log phase were analyzed by whole-cell extraction and Western blot analysis using α-GFP and α-Pgk1 antibodies. GFP signals were normalized to Pgk1 and expressed relative (rel.) to wt cells in log phase (set as one). Data are means ± SD (n = 4). (C) wt and ΔLD cells expressing GFP-HDEL (ER-targeted GFP) were imaged in log phase or after 1 h of starvation ± cerulenin (10 µg/ml). Images show single cortical and mid sections of the same Z-stack. Quantifications of cells with a collapsed ER network of dilated tubules (arrows) as seen for ΔLD cells during starvation are mean ± SD (≥150 cells; n = 3). Bar, 5 µm. (D) Autophagy flux of wt and ΔLD cells expressing 2xGFP-ATG8 during starvation ± cerulenin (10 µg/ml). Data are means ± SD (n = 5). (E) Cells were treated as described in D and imaged and analyzed by fluorescence microscopy as described in Fig. 1 B. Data are means ± SD (≥150 cells; n = 3). Dashed lines indicate cell boundaries. Bar, 1 µm. (F) Survival of indicated strains treated as in D during starvation. t test in B, D, and E: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test whether unbuffered FA synthesis causes defects in autophagy, we compared the autophagy response of wt and ΔLD cells in the presence or absence of cerulenin during starvation. Inhibition of FA synthesis mildly reduced autophagy flux in wt cells (Fig. 2 D). However, we observed a moderate, but statistically significant, improvement in autophagy flux in treated compared with untreated LD-deficient cells and relative to untreated (15% of wt) and treated wt cells (48% of wt) after 6 h of starvation (Fig. 2 D). Importantly, cytological analysis revealed clearly improved AP biogenesis in LD-deficient cells upon inhibition of FA synthesis: we found a wt-like number of Atg8 puncta and significantly increased formation of APs in ΔLD cells, demonstrating that unbuffered FA synthesis is linked to both aberrations (Fig. 2 E). A block in FA synthesis also extended survival of LD-deficient cells in an autophagy-dependent manner up to 2 d during starvation (Figs. 2 F and S1 H). Together, these data indicate that LDs fulfill crucial functions as buffers of de novo FA synthesis to maintain ER homeostasis, intact autophagy, and nutrient stress resistance.
Altered PL composition is linked to autophagy defects in LD-deficient cells
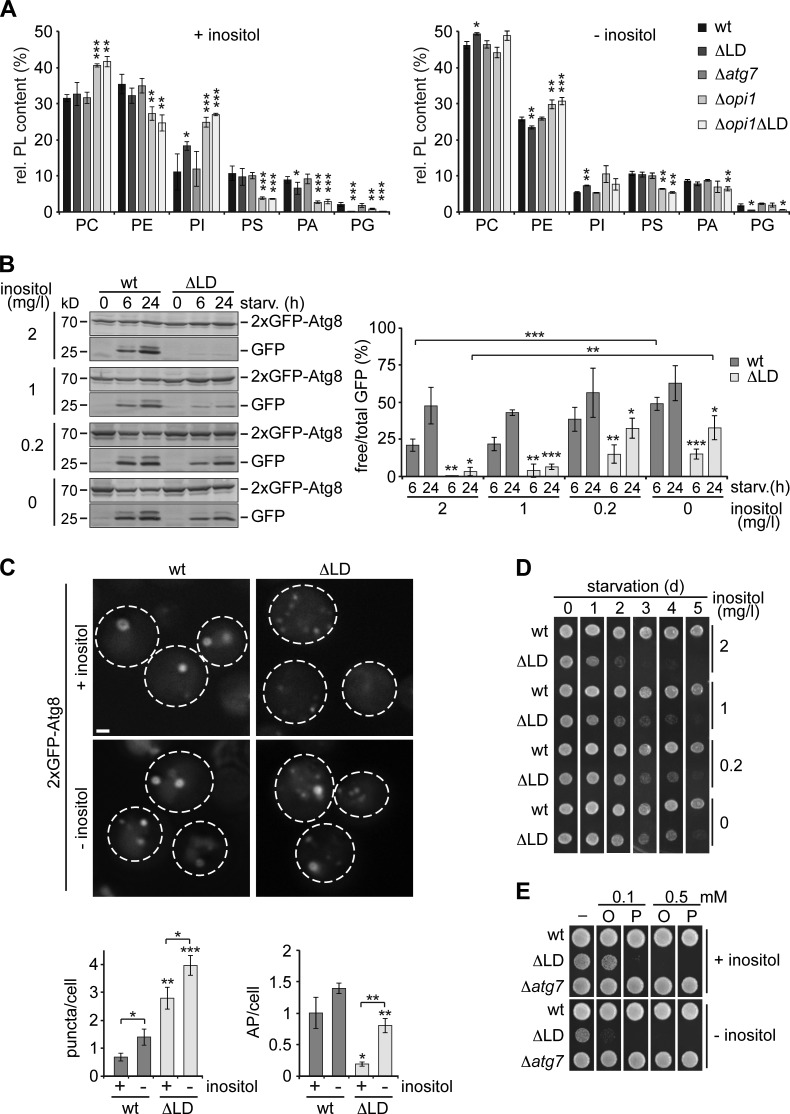
Partial improvement of autophagy upon inhibition of FA synthesis suggested that additional physiological changes in the absence of LDs might interfere with autophagy regulation. Therefore, we turned to analysis of the membrane composition of major PL in whole wt, ΔLD, and Δatg7 cells during log phase by mass spectrometry. Interestingly, while a block in autophagy in Δatg7 cells did not alter the PL composition, we observed a statistically significant increase in the relative phosphatidylinositol (PI) content and a decrease in phosphatidic acid (PA) and phosphatidylglycerol (PG) content in LD-deficient cells compared to wt cells (Fig. 3 A, +inositol). PA, PI, and PI derivatives are signaling lipids, which regulate cellular processes, including PL synthesis and autophagy, and their relative levels are interdependently linked by availability of inositol (Henry et al., 2012; Dall’Armi et al., 2013). Hence, to test for a possible regulatory link between PL composition and autophagy, we first analyzed the PL composition of wt, ΔLD, and Δatg7 cells grown to log phase in inositol-free media. We observed a general decrease in PI content in all strains under these conditions, as expected, and moreover, a reduced or nonsignificant difference in PI or PA content, respectively, in LD-deficient cells compared to wt and Δatg7 cells in the absence of inositol (Fig. 3 A, −inositol). Thus, we identified metabolic conditions that allowed us to alleviate the differences in PL composition caused by LD deficiency. Noteworthy, growing cells in the absence of inositol or treating cells with rapamycin led to a similar reshaping of the PL composition in wt and ΔLD cells (Figs. 3 A and S2 A).
Figure 3.
Restoring altered PL composition in LD-deficient cells improves autophagy and cell survival during starvation. (A) wt, ΔLD, Δatg7, Δopi1, and Δopi1ΔLD cells were grown to log phase ± inositol (2 mg/l), and PLs were analyzed by mass spectrometry as described in the Materials and methods. Relative (rel.) distribution of PC, PE, PI, PS, PA, and PG is shown as mean ± SD (n ≥ 3). (B) Autophagy flux of wt and ΔLD cells expressing 2xGFP-ATG8 during starvation (starv.) in the presence of indicated inositol concentrations. Data are means ± SD (n = 3). (C) wt and ΔLD cells expressing 2xGFP-ATG8 were grown to log phase and shifted to starvation ± inositol (2 mg/l). Cells were imaged and analyzed as described in Fig. 1 B. Dashed lines indicate cell boundaries. Bar, 1 µm. (D) Survival of indicated strains treated as in C during starvation. (E) Growth of wt, ΔLD, or Δatg7 strains on plates containing oleate (O) or palmitoleate (P) at indicated concentrations ± inositol (2 mg/l). t test in A, B, and C: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
When we monitored autophagy flux in wt and ΔLD cells, we found increasing autophagy flux in wt cells and, strikingly, significantly improving autophagy flux in LD-deficient cells relative to wt cells in response to starvation medium containing decreasing inositol concentrations (Fig. 3 B). Importantly, cytological analysis revealed that whereas aberrant Atg8 puncta formation occurred in an inositol-independent manner, AP biogenesis was significantly restored in ΔLD cells in the absence of inositol (Fig. 3 C). Lowering inositol concentrations also prolonged survival of LD-deficient cells in an autophagy-dependent manner during starvation (Figs. 3 D and S2 B). Hence, these data indicate that cells are more permissive for autophagy in the absence of inositol and that LD-deficient cells are specifically improved in AP biogenesis, autophagy flux, and cell survival when differences in PL composition are alleviated.
Since de novo FA synthesis interfered with autophagy in LD-deficient cells, we tested whether inositol levels modulated autophagy through changes in FA resistance. ΔLD cells are sensitive to high concentrations of external FA oleate or palmitoleate and display mild inositol auxotrophy (Fig. 3 E; Sandager et al., 2002; Petschnigg et al., 2009; Gaspar et al., 2011). Interestingly, absence of inositol further increased the sensitivity of ΔLD cells to external FA, suggesting that external inositol is linked to FA buffering and that the positive effects on autophagy in the absence of inositol are distinct from FA resistance (Fig. 3 E). These data raise the possibility that excess FA are buffered by increased PI synthesis and might provide a rationale for the mild inositol auxotrophy of LD-deficient cells. Consistently, LD-deficient cells showed elevated Ino1 levels compared to wt cells, indicating elevated inositol synthesis even in the presence of external inositol (Fig. S1 I). Furthermore, all strains showed an increase in PI levels after 6 h of starvation, a condition of induced FA synthesis and LD biogenesis (Fig. S2 C).
In summary, our data support a model in which LD-deficient cells use external inositol for increased PI synthesis to buffer excess FA synthesis and improve FA resistance. As a consequence, however, these changes in PL composition interfere with autophagy regulation and nutrient stress resistance in LD-deficient cells.
Improving FA resistance in the absence of inositol cures autophagy in ΔLD cells during starvation
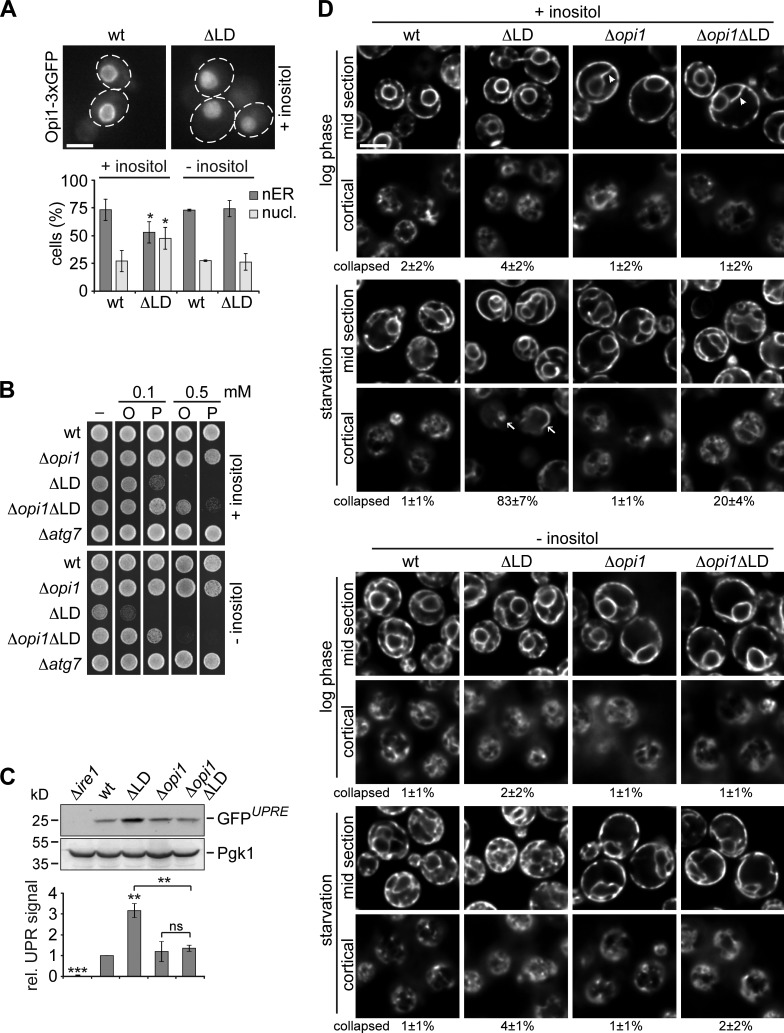
So far, we identified de novo FA synthesis and, likely, changed PL composition as factors that compromise the ability of LD-deficient cells to regulate autophagy. In addition, ΔLD cells suffer from chronic ER stress and starvation-induced ER deformation. We hypothesized that artificially increasing FA buffer capacity of LD-deficient cells could improve ER homeostasis and autophagy regulation. Opi1 is a central transcriptional inhibitor of PL biosynthesis genes and its deletion leads to expansion of ER membranes capable of alleviating unfolded protein stress (Carman and Henry, 2007; Schuck et al., 2009). The activity of Opi1 is regulated by differential localization to nuclear lumen or ER membranes dependent on PA levels (Loewen et al., 2004; Hofbauer et al., 2014). Consistent with decreased PA levels in ΔLD cells, 50% of LD-deficient cells showed a diffuse nuclear signal for genomically modified OPI1-3xGFP compared to <25% in the wt cell population when grown in media containing inositol (Fig. 4 A). Interestingly, Opi1 localized to the nuclear ER membrane in LD-deficient cells indistinguishable from wt cells in the absence of inositol, conditions that restore PA levels (Fig. 4 A). Next, we tested whether uncoupled PL biosynthesis caused by OPI1 deletion could expand the ER and improve FA resistance in LD-deficient cells. Independent of LDs, both Δopi1 and Δopi1ΔLD cells showed expanded ER characterized by peripheral ER extensions in the cytosol in log phase (Fig. 4 D, arrowheads; Schuck et al., 2009). Strikingly, consistent with our hypothesis, deletion of OPI1 in LD-deficient cells improved resistance to external FA (Fig. 4 B), reduced chronic UPR signaling to wt levels in log phase (Fig. 4 C), and partially suppressed the collapse of cortical ER into dilated tubules during starvation (Fig. 4 D), indicating improved ER homeostasis in Δopi1ΔLD cells compared to ΔLD cells.
Figure 4.
Deletion of OPI1 buffers FA and improves ER stress and morphology in LD-deficient cells during starvation. (A) Localization of Opi1-3xGFP in wt and ΔLD cells in log phase ± inositol (2 mg/l) analyzed by fluorescence microscopy. Single focal planes of representative cells during growth in +inositol media are shown. Quantifications of cells with nuclear ER (nER) or diffuse nuclear (nucl.) Opi1 localization (≥100 cells; n = 3). (B) Growth of indicated strains on plates containing oleate (O) or palmitoleate (P) at indicated concentrations ± inositol (2 mg/l). (C) wt, Δire1, ΔLD, Δopi1, and Δopi1ΔLD cells expressing 4xUPRE-GFP in log phase. Cells were analyzed as described in Fig. 2 B. (D) wt, ΔLD, Δopi1, and Δopi1ΔLD cells expressing GFP-HDEL (ER-GFP) were grown to log phase and starved ± inositol (2 mg/l) for 1 h and analyzed as in Fig. 2 C. Arrowheads and arrows mark ER extensions or dilated tubules, respectively. Bar, 5 µm. t test in A and C: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
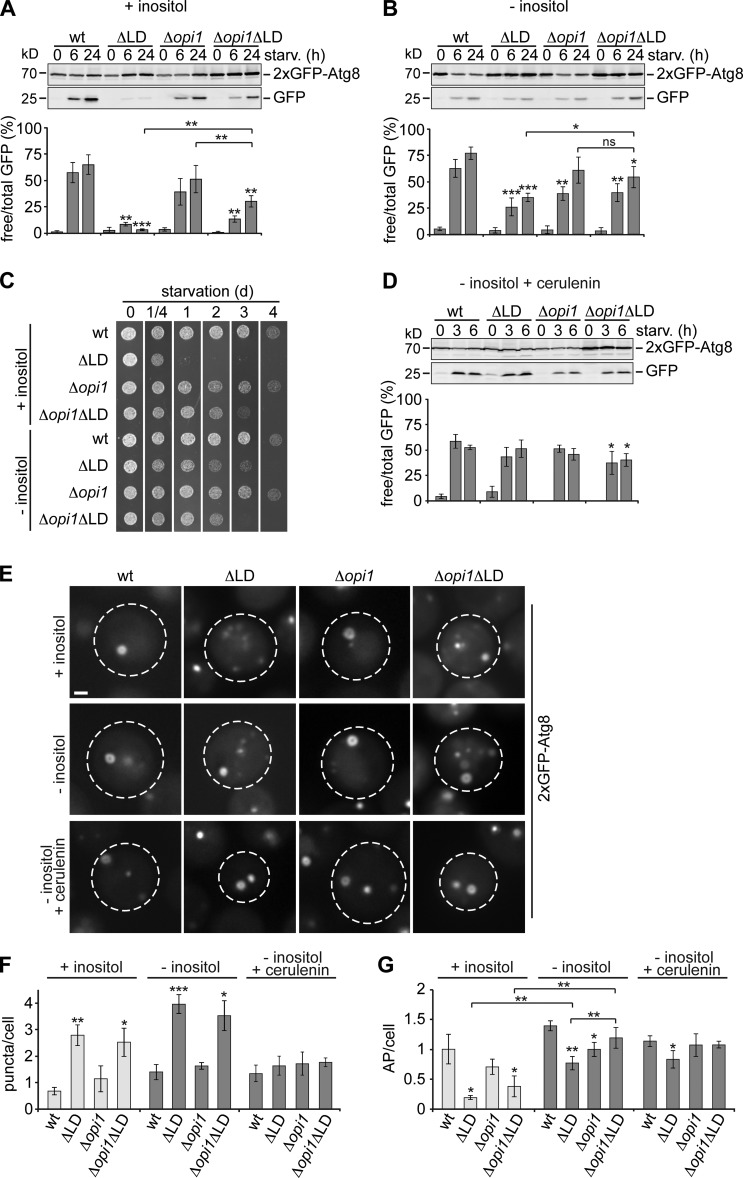
To test whether improved ER homeostasis in the absence of Opi1 affected autophagy in LD-deficient cells, we monitored autophagy flux, AP biogenesis, and cell survival in wt, ΔLD, Δopi1, and Δopi1ΔLD cells during starvation. Indeed, we observed moderate, but significant, improvement in autophagy flux (Fig. 5 A), AP biogenesis (Fig. 5, E and G, +inositol), and cell survival in Δopi1ΔLD cells compared to ΔLD cells (Fig. 5 C, +inositol). Interestingly, ΔLD and Δopi1ΔLD cells showed the same intermediate autophagy flux when we starved cells in the presence of cerulenin (Fig. S2 D), placing OPI1 deletion and cerulenin treatment functionally in the same pathway and indicating that deletion of OPI1 improves autophagy mainly by FA buffering in LD-deficient cells. We reasoned that, in addition to FA resistance, other alterations in LD-deficient cells have to be corrected to allow for intact autophagy. Thus, we analyzed the PL composition of Δopi1 and Δopi1ΔLD cells and found significant changes compared with wt cells, including a strong increase in PI and a decrease in PA content (Fig. 3 A, +inositol). These data raise the possibility that altered PI and PA levels interfere with autophagy regulation also in Δopi1ΔLD cells. The fact that PI and PA levels in both Δopi1 and Δopi1ΔLD cells were restored to wt-like levels in the absence of inositol (Fig. 3 A, −inositol) enabled us to test for their effects on FA resistance, ER morphology, and autophagy. Both, ΔLD and Δopi1ΔLD cells showed an increased sensitivity toward external FA in the absence of inositol, but FA resistance still increased upon deletion of OPI1 in LD-deficient cells (Fig. 4 B). Interestingly, starvation in inositol-free media fully prevented the collapse of cortical ER in ΔLD and Δopi1ΔLD cells despite increased FA sensitivity linking inositol metabolism, potentially through PL composition, to ER morphology (Fig. 4 D, −inositol). We tested whether concomitant improvement of FA resistance and PL composition is sufficient to restore autophagy in LD-deficient cells. Strikingly, consistent with this notion, the absence of inositol and deletion of OPI1 resulted in an additive improvement in LD-deficient cells restoring autophagy flux and AP biogenesis in Δopi1ΔLD cells to identical levels as in Δopi1 cells and thus allowed for a LD-independent autophagy response during starvation (Fig. 5, B, E, and G, −inositol). In line with the conclusion that OPI1 deletion and cerulenin treatment function in the same pathway, ΔLD cells displayed wt-like autophagy flux when we starved cells in the presence of cerulenin in inositol-free media (Fig. 5 D). Interestingly, cerulenin treatment also prevented the formation of aberrant Atg8 puncta observed in ΔLD and Δopi1ΔLD cells during starvation in the absence of inositol with minor effects on cell survival (Fig. 5, E and F; and Fig. S2 E), indicating that puncta formation is highly sensitive to excess FA even when FA resistance is improved in Δopi1ΔLD cells.
Figure 5.
Increased FA resistance and restored PL composition cures AP biogenesis and autophagy flux in LD-deficient cells. (A and B) Autophagy flux in wt, ΔLD, Δopi1, and Δopi1ΔLD cells expressing 2xGFP-ATG8 during starvation (starv.) in the presence (A) or absence (B) of inositol (2 mg/l). Data are mean ± SD (n = 4). (C) Survival of indicated strains during starvation ± inositol (2 mg/l). (D) Autophagy flux in wt, ΔLD, Δopi1, and Δopi1ΔLD cells grown in inositol-free media and starved in the presence of cerulenin (10 µg/ml). Data are mean ± SD (n = 4). (E–G) wt, ΔLD, Δopi1, and Δopi1ΔLD cells expressing 2xGFP-ATG8 were treated as described in A, B, and D and imaged and analyzed as described in Fig. 1 B. Bar, 1 µm. Data are means ± SD (150 cells/condition, n = 3). t test in A, B, D, F, and G: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In summary, these findings demonstrate that AP biogenesis and autophagy flux can be fully cured in the absence of LDs and thus provide unequivocal evidence that LDs are dispensable as a membrane source for autophagy. Our data support a model in which LDs fulfill two fundamental functions required for maintaining ER homeostasis, intact autophagy regulation, and nutrient stress resistance—buffering of excess FA and maintenance of PL composition (Fig. S2 F). Interestingly, we provided evidence suggesting that both functions might be linked in that, when LD storage capacity is exceeded, FAs are buffered by increased PI levels improving FA resistance of cells. Since we analyzed LD-deficient cells in the absence of external FA, our study revealed a remarkable inability of cells to adjust the level of de novo FA synthesis to maintain growth without simultaneously compromising autophagy regulation and nutrient stress resistance. Our observations indicate that the regulatory circuits controlling FA synthesis evolved inherently depending on the FA buffer capacity of LDs to maintain cellular function (Tehlivets et al., 2007).
Defects in autophagy and chronic ER stress are common features of neurodegenerative diseases, obesity, and diabetes (Ozcan et al., 2004; Nixon, 2013; Hetz and Mollereau, 2014). Analysis of hepatic ER derived from obese mice previously revealed alterations in PL composition, specifically in the phosphatidylcholine (PC)/phosphatidylethanolamine (PE) ratio (PI was not analyzed), causing chronic ER stress (Fu et al., 2011). Intriguingly, these hepatocytes are also compromised in AP biogenesis upon starvation (Yang et al., 2010). Hence, these and our data raise the exciting possibility that alterations in PL composition upon exceeded LD storage capacity might be a general underlying principle for defects in autophagy regulation. Additional work will be necessary to unravel how changes in PA, PI, or PI derivatives previously shown to affect autophagy, might interfere with the autophagy machinery during AP formation (Petiot et al., 2000; Moreau et al., 2012; Li et al., 2014; Vicinanza et al., 2015). However, our work sheds light on principle metabolic and genetic alterations that in combination fully compensate for LD deficiency. Hence, these pathways might represent suitable targets for therapeutic intervention in the aforementioned pathologies.
One of the central questions in the autophagy field is where the membranes for AP formation are coming from. Our data demonstrate unequivocally that LDs are not required as a membrane source. However, LDs may contribute FA or lipids to AP biogenesis in the presence of abundant, nontoxic external FA (Dupont et al., 2014; Shpilka et al., 2015). A major challenge remains to determine how cells regulate the contribution of multiple membrane sources in response to diverse metabolic conditions.
Materials and methods
Strains and media
All strains used in this study are derivatives of w303 and listed in Table S1. Gene deletions were generated by replacing complete ORFs by indicated marker cassettes using PCR-based targeted homologous recombination as previously described (Longtine et al., 1998). Double mutants ΔTAG (Δdga1::TRP1 Δlro1::HIS3) and ΔSE (Δare1::TRP1 Δare2::HIS3) were generated by crossing of haploid strains carrying single deletions and sporulation. The quadruple mutant ΔLD (Δdga1::TRP1 Δlro1::HIS3 Δare1::TRP1 Δare2::HIS3) was obtained by crossing of the two double mutants ΔTAG and ΔSE followed by sporulation, tetrade dissection, and analysis.
Strains were grown in synthetic complete medium (0.7% [wt/vol] yeast nitrogen base [BD] or 0.7% [wt/vol] yeast nitrogen base without inositol [Formedium] complemented with indicated concentrations of inositol [Sigma-Aldrich]; 2% [wt/vol] α-d-glucose [Sigma-Aldrich]) at 30°C. For starvation experiments, cells were grown to early log phase and shifted to SD-N media (0.17% [wt/vol] yeast nitrogen base without aa and ammonium sulfate [BD] or 0.17% [wt/vol] yeast nitrogen base without aa, ammonium sulfate, and inositol [Formedium] complemented with indicated concentrations of inositol; 2% [wt/vol] α-d-glucose [Sigma-Aldrich]). Rapamycin (400 ng/ml), tunicamycin (1 µg/ml; both Calbiochem), or cerulenin (10 µg/ml; Sigma-Aldrich) in DMSO were added to the media when indicated.
For growth analyses, 106 cells were spotted on YPD plates (1% [wt/vol] yeast extract [Serva], 2% [wt/vol] peptone [Merck], and 2% [wt/vol] α-d-glucose [Sigma-Aldrich]) and grown for 2 d at 30°C. For test of FA resistance, 106 cells were spotted on synthetic complete medium plates containing 0.6% (vol/vol) ethanol/tyloxapol (5:1 vol/vol) and indicated concentrations of oleate or palmitoleate (both Sigma-Aldrich) and grown for 2 d at 30°C as described previously (Garbarino et al., 2009).
Plasmids pRS306-prATG8-2xyEGFP-ATG8, pRS305-DsRed-HDEL, and pRS305-yEGFP-HDEL, were described previously (gift from J. Nunnari, University of California, Davis, Davis, CA; Graef et al., 2013; Lackner et al., 2013). pRS315-pHluorin-mCherry was constructed according to Rosado et al. (2008) by integrating the ADH1 promoter (–717 to –1 bp; primer forward: 5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGCGAATTGATCCTTTTGTTGTTTCCGGG-3′; primer reverse: 5′-GTTCTTCTCCTTTACTCATTGTATATGAGATAGTTGATTGTATG-3′) fused to pHluorin amplified from pRS315-pHluorin-ATG8 (pSW12; gift from D. Teis, Medical University of Innsbruck, Innsbruck, Austria; Müller et al., 2015; primer forward: 5′-CATACAATCAACTATCTCATATACAATGAGTAAAGGAGAAGAAC-3′; primer reverse: 5′-TAAACCAGCACCGTCACCTTTGTATAGTTCATCCATGCCATGTGTAATC-3′) and mCherry amplified from pFA6a-mCherry (gift from J. Nunnari; primer forward: 5′-GATTACACATGGCATGGATGAACTATACAAAGGTGACGGTGCTGGTTTA-3′; primer reverse: 5′-CAAGCTCGGAATTAACCCTCACTAAAGGGAACAAAAGCTGGCAAGCTAAACAGATC-3′) by “gap repair” in vivo homologous recombination (Oldenburg et al., 1997) into BamHI- and HindIII-linearized pRS315. The 4xUPRE-GFP reporter was genomically integrated as described previously using primers oMJ007 and oMJ014 to amplify from pKT007 a PCR product containing 4xUPRE repeats driving GFP followed by the URA3 marker, flanked by sequences homologous to the URA3 locus (gift from J. Weissman, University of California, San Francisco, San Francisco, CA; Jonikas et al., 2009).
Whole-cell extraction, western blot analysis, and quantification
Cells corresponding to 0.2 OD600 units were collected and lysed by alkaline whole-cell extraction (0.255 M NaOH and 1% [vol/vol] β-mercaptoethanol). Protein extracts were analyzed by SDS-PAGE and immunoblotting (α-Atg13, rabbit polyclonal, gift from D. Klionsky, University of Michigan, Ann Arbor, MI; α-GFP, mouse monoclonal; Covance; α-HA, mouse monoclonal; Covance; and α-Pgk1, mouse monoclonal; Abcam) and visualized using corresponding secondary goat antibodies conjugated to IRDye (800CW; Li-COR Biosciences). Quantifications were performed using the Odyssey Infrared Imaging System (Li-COR Biosciences).
Fluorescence microscopy
Cells were viewed in 96-well microplates with a glass bottom (Greiner bio-one) containing indicated growth or starvation media at RT with an inverted microscope (Ti-E; Nikon) using a Plan Apochromat IR 60× 1.27 NA objective (Nikon), Spectra X LED light source (Lumencor), and acquisition software NIS elements AR (Nikon). 3D light microscopy data were collected using the triggered Z-Piezo system (Nikon) and orca flash 4.0 camera (Hamamatsu). 3D data were processed using NIS-elements AR; deconvolution of imaging data was performed with 3D deconvolution 3D-Fast in NIS-elements AR (Figs. 2 E, 3 C, 4 A, and 5 A) or Huygens Professional 15.10 (Scientific Volume Imaging; Figs. 1, B and C; Fig. 2 C; Fig. 4 D; and Fig. S1, A, C, and G). Photoshop (Adobe) software was used to make linear adjustments in brightness.
Mass spectrometric lipid analysis
Mass spectrometric analysis was performed essentially as described with some optimization for the analysis of phospholipids from whole yeast cell (Connerth et al., 2012). Lipids were extracted from whole yeast cells in the presence of internal standards of major PLs (PC 17:0–14:1, PE 17:0–14:1, PI 17:0–14:1, phosphatidylserine [PS] 17:0–14:1, PG 17:0–14:1, PA 17:0–14:1, all from Avanti Polar Lipids). Extraction was performed according to Bligh and Dyer (1959), with modifications. In brief, 0.5 OD600-unit cells were resuspended in 250 µl water and mixed with 1 ml of chloroform/methanol/25% HCl (40:80:0.6 [vol/vol]) with lipid standards in a glass vial. 300-µl glass beads were added to the vial, and cells were treated by rigorous mixing for 30 min. After adding 250 µl of chloroform and 250 µl of water, the sample was mixed again for 1 min, and phase separation was induced by centrifugation (800× g, 2 min). The lower chloroform phase was carefully transferred to a second vial and was mixed with 400 µl water (washing step). After vortexing for 30 s, phase separation was induced by centrifugation, and the lower chloroform phase was carefully transferred to a clean glass vial. The upper water phase after the first phase separation was mixed with 300 µl of chloroform, and extraction was repeated. After phase separation, lower chloroform phase was mixed with the upper water phase after the first washing step. After vortexing for 30 s, phase separation was induced by centrifugation, and the lower chloroform phase was carefully transferred to the glass vial with the chloroform phase from the first extraction. The solvent was evaporated by a gentle stream of argon at 37°C. Lipids were dissolved in 10 mM ammonium acetate in methanol and analyzed on a QTRAP 6500 triple quadrupole mass spectrometer (SCIEX) equipped with a nano-infusion splay device (TriVersa NanoMate; Advion) under the following settings: (QT 6500) Curtain gas, 20; Collision gas, medium; Interface heater temperature, 90°C; Entrance potential, 10; mode: high mass; step size, 0.1 D; setting time, 0 ms; scan rate, 200 D/s; pause 5 ms; CEM, 2,300; sync, LC sync; scan mode, profile (NanoMate) sample infusion volume, 12 µl; volume of air to aspirate after sample, 1 µl; air gap before chip, enabled; aspiration delay, 0 s; prepiercing, with mandrel; spray sensing, enabled; temperature, 12°C; gas pressure, 0.4 psi; ionization voltage, 1.15 kV; polarity, positive; vent headspace, enabled; prewetting, 1×; volume after delivery, 0.5 µl; contact closure delay, 1 s; volume timing delay, 0 s; aspiration depth, 1 mm; prepiercing depth, 9 mm; and output contact closure, Rel 1–2.5-s duration. The quadrupoles Q1 and Q3 were operated at unit resolution. PC analysis was performed in positive ion mode by scanning for precursors of mass to charge ratio 184 at a collision energy (CE) of 50 eV. PE, PI, PS, PG, PA, and CDP-DAG measurements were performed in positive ion mode by scanning for neutral losses of 141, 277, 185, 189, 115, and 403 D at CEs of 25, 30, 20, 30, 25, and 40 eV, respectively. Mass spectra were processed by the LipidView Software Version 1.2 (SCIEX) for identification and quantification of lipids. Lipid amounts (picomoles) were corrected for response differences between internal standards and endogenous lipids.
Statistical analysis
Error bars represent the SD as indicated in the figure legends. Data were processed in Excel (Microsoft). Statistical analysis of differences between the two groups was performed using a two-tailed, unpaired t test; *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Only statistically significant comparisons are indicated in the figures.
Online supplemental material
Fig. S1 provides further evidence for the conditional defect of LD-deficient cells in autophagy flux, data showing unaltered dephosphorylation of Atg13, primary data for Fig. 2 A, evidence that UPR signaling does not affect autophagy flux or starvation survival, unchanged ER morphology during rapamycin treatement in wt and LD-deficient cells, and evidence showing an autophagy-dependent increase in survival of LD-deficient cells upon inhibition of FA synthesis during starvation. Fig. S2 provides additional analysis of PL composition, starvation survival, autophagy flux analysis in dependence of inositol and cerulenin treatment, and a model for the role of LDs for ER homeostasis and autophagy regulation. Table S1 provides a list of all Saccharomyces cerevisiae strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201508102/DC1.
Supplementary Material
Acknowledgments
We would like to thank members of the Graef Laboratory, Professors Jodi Nunnari, Thomas Langer, Adam Antebi, and Linda Partridge for discussion, and Professors Jonathan Weissman, David Teis, and Daniel Klionsky for providing materials. We would like to thank the FACS & Imaging Core Facility at the Max Planck Institute for Biology of Ageing for support in data processing.
This work was supported by the Max Planck Society.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AP
- autophagosome
- ERES
- ER exit site
- FA
- fatty acid
- LD
- lipid droplet
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PL
- phospholipid
- PS
- phosphatidylserine
- SE
- sterol ester
- TG
- triacylglycerol
- TORC1
- target of rapamycin complex 1
- UPR
- unfolded protein response
- wt
- wild type
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E.G., and Dyer W.J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Carman G.M., and Henry S.A.. 2007. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282:37293–37297. 10.1074/jbc.R700038200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., and Langer T.. 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 338:815–818. 10.1126/science.1225625 [DOI] [PubMed] [Google Scholar]
- Dall’Armi C., Devereaux K.A., and Di Paolo G.. 2013. The role of lipids in the control of autophagy. Curr. Biol. 23:R33–R45. 10.1016/j.cub.2012.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N., Chauhan S., Arko-Mensah J., Castillo E.F., Masedunskas A., Weigert R., Robenek H., Proikas-Cezanne T., and Deretic V.. 2014. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr. Biol. 24:609–620. 10.1016/j.cub.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., and Klionsky D.J.. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., Lin X., Watkins S.M., Ivanov A.R., and Hotamisligil G.S.. 2011. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 473:528–531. 10.1038/nature09968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino J., Padamsee M., Wilcox L., Oelkers P.M., D’Ambrosio D., Ruggles K.V., Ramsey N., Jabado O., Turkish A., and Sturley S.L.. 2009. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J. Biol. Chem. 284:30994–31005. 10.1074/jbc.M109.050443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar M.L., Hofbauer H.F., Kohlwein S.D., and Henry S.A.. 2011. Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J. Biol. Chem. 286:1696–1708. 10.1074/jbc.M110.172296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Melville D., Zhang M., and Schekman R.. 2013. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2:e00947 10.7554/eLife.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M., Friedman J.R., Graham C., Babu M., and Nunnari J.. 2013. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 24:2918–2931. 10.1091/mbc.E13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., and Lippincott-Schwartz J.. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–667. 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H., and Rubinsztein D.C.. 2011. Control of autophagy as a therapy for neurodegenerative disease. Nat. Rev. Neurol. 8:108–117. 10.1038/nrneurol.2011.200 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., and Yamamoto A.. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437. 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Henry S.A., Kohlwein S.D., and Carman G.M.. 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 190:317–349. 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., and Mollereau B.. 2014. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 15:233–249. 10.1038/nrn3689 [DOI] [PubMed] [Google Scholar]
- Hofbauer H.F., Schopf F.H., Schleifer H., Knittelfelder O.L., Pieber B., Rechberger G.N., Wolinski H., Gaspar M.L., Kappe C.O., Stadlmann J., et al. 2014. Regulation of gene expression through a transcriptional repressor that senses acyl-chain length in membrane phospholipids. Dev. Cell. 29:729–739. 10.1016/j.devcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G.S. 2010. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 140:900–917. 10.1016/j.cell.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J.S., and Schuldiner M.. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 323:1693–1697. 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., and Ohsumi Y.. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507–1513. 10.1083/jcb.150.6.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B., Schmidt C., and Daum G.. 2014. Storage lipids of yeasts: a survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol. Rev. 38:892–915. 10.1111/1574-6976.12069 [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D. 2010. Triacylglycerol homeostasis: insights from yeast. J. Biol. Chem. 285:15663–15667. 10.1074/jbc.R110.118356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., and Martens S.. 2012. Mechanisms and regulation of autophagosome formation. Curr. Opin. Cell Biol. 24:496–501. 10.1016/j.ceb.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Lackner L.L., Ping H., Graef M., Murley A., and Nunnari J.. 2013. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA. 110:E458–E467. 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Li D., Song J.Z., Li H., Shan M.H., Liang Y., Zhu J., and Xie Z.. 2015. Storage lipid synthesis is necessary for autophagy induced by nitrogen starvation. FEBS Lett. 589:269–276. 10.1016/j.febslet.2014.11.050 [DOI] [PubMed] [Google Scholar]
- Li Y., Li S., Qin X., Hou W., Dong H., Yao L., and Xiong L.. 2014. The pleiotropic roles of sphingolipid signaling in autophagy. Cell Death Dis. 5:e1245 10.1038/cddis.2014.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Gaspar M.L., Jesch S.A., Delon C., Ktistakis N.T., Henry S.A., and Levine T.P.. 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 304:1644–1647. 10.1126/science.1096083 [DOI] [PubMed] [Google Scholar]
- Longatti A., Lamb C.A., Razi M., Yoshimura S., Barr F.A., and Tooze S.A.. 2012. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J. Cell Biol. 197:659–675. 10.1083/jcb.201111079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J., and Reggiori F.. 2010. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190:1005–1022. 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K., Ravikumar B., Puri C., and Rubinsztein D.C.. 2012. Arf6 promotes autophagosome formation via effects on phosphatidylinositol 4,5-bisphosphate and phospholipase D. J. Cell Biol. 196:483–496. 10.1083/jcb.201110114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Schmidt O., Angelova M., Faserl K., Weys S., Kremser L., Pfaffenwimmer T., Dalik T., Kraft C., Trajanoski Z., et al. 2015. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. eLife. 4:e07736 10.7554/eLife.07736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., Yen W.L., Griffith J., Nag S., Wang K., Moss T., et al. 2011. SNARE proteins are required for macroautophagy. Cell. 146:290–302. 10.1016/j.cell.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R.A. 2013. The role of autophagy in neurodegenerative disease. Nat. Med. 19:983–997. 10.1038/nm.3232 [DOI] [PubMed] [Google Scholar]
- Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J.T., and Sturley S.L.. 2000. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 275:15609–15612. 10.1074/jbc.C000144200 [DOI] [PubMed] [Google Scholar]
- Oelkers P., Cromley D., Padamsee M., Billheimer J.T., and Sturley S.L.. 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277:8877–8881. 10.1074/jbc.M111646200 [DOI] [PubMed] [Google Scholar]
- Oldenburg K.R., Vo K.T., Michaelis S., and Paddon C.. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451–452. 10.1093/nar/25.2.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J.A., and Kopito R.R.. 2011. Lipid droplet formation is dispensable for endoplasmic reticulum-associated degradation. J. Biol. Chem. 286:27872–27874. 10.1074/jbc.C111.266452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U., Cao Q., Yilmaz E., Lee A.H., Iwakoshi N.N., Ozdelen E., Tuncman G., Görgün C., Glimcher L.H., and Hotamisligil G.S.. 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 306:457–461. 10.1126/science.1103160 [DOI] [PubMed] [Google Scholar]
- Papa F.R., Zhang C., Shokat K., and Walter P.. 2003. Bypassing a kinase activity with an ATP-competitive drug. Science. 302:1533–1537. 10.1126/science.1090031 [DOI] [PubMed] [Google Scholar]
- Patil C., and Walter P.. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349–355. 10.1016/S0955-0674(00)00219-2 [DOI] [PubMed] [Google Scholar]
- Petiot A., Ogier-Denis E., Blommaart E.F., Meijer A.J., and Codogno P.. 2000. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275:992–998. 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C.F., Natter K., and Kohlwein S.D.. 2009. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284:30981–30993. 10.1074/jbc.M109.024752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau L., Colas J., Dupont S., Beney L., Fleurat-Lessard P., Berjeaud J.M., Bergès T., and Ferreira T.. 2009. Lipid-induced ER stress: synergistic effects of sterols and saturated fatty acids. Traffic. 10:673–690. 10.1111/j.1600-0854.2009.00903.x [DOI] [PubMed] [Google Scholar]
- Quan W., Jung H.S., and Lee M.S.. 2013. Role of autophagy in the progression from obesity to diabetes and in the control of energy balance. Arch. Pharm. Res. 36:223–229. 10.1007/s12272-013-0024-7 [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Moreau K., Jahreiss L., Puri C., and Rubinsztein D.C.. 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12:747–757. 10.1038/ncb2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado C.J., Mijaljica D., Hatzinisiriou I., Prescott M., and Devenish R.J.. 2008. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy. 4:205–213. 10.4161/auto.5331 [DOI] [PubMed] [Google Scholar]
- Sandager L., Gustavsson M.H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., and Stymne S.. 2002. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277:6478–6482. 10.1074/jbc.M109109200 [DOI] [PubMed] [Google Scholar]
- Schuck S., Prinz W.A., Thorn K.S., Voss C., and Walter P.. 2009. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187:525–536. 10.1083/jcb.200907074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M., Yoshimura K., Tamura H., Ueno T., Nishimura T., Inoue T., Sasaki M., Koike M., Arai H., Kominami E., and Uchiyama Y.. 2010. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem. Biophys. Res. Commun. 393:274–279. 10.1016/j.bbrc.2010.01.121 [DOI] [PubMed] [Google Scholar]
- Shintani T., and Klionsky D.J.. 2004. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279:29889–29894. 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T., Welter E., Borovsky N., Amar N., Mari M., Reggiori F., and Elazar Z.. 2015. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 34:2117–2131. 10.15252/embj.201490315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., and Czaja M.J.. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D., and Daum G.. 2002. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J. Bacteriol. 184:519–524. 10.1128/JB.184.2.519-524.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., and Ohsumi Y.. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218. 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Suzuki K., Akioka M., Kondo-Kakuta C., Yamamoto H., and Ohsumi Y.. 2013. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 126:2534–2544. 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., and Kominami E.. 1999. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 10:1367–1379. 10.1091/mbc.10.5.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehlivets O., Scheuringer K., and Kohlwein S.D.. 2007. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta. 1771:255–270. 10.1016/j.bbalip.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Tsukada M., and Ohsumi Y.. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169–174. 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- Vance D., Goldberg I., Mitsuhashi O., and Bloch K.. 1972. Inhibition of fatty acid synthetases by the antibiotic cerulenin. Biochem. Biophys. Res. Commun. 48:649–656. 10.1016/0006-291X(72)90397-X [DOI] [PubMed] [Google Scholar]
- van Zutphen T., Todde V., de Boer R., Kreim M., Hofbauer H.F., Wolinski H., Veenhuis M., van der Klei I.J., and Kohlwein S.D.. 2014. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 25:290–301. 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza M., Korolchuk V.I., Ashkenazi A., Puri C., Menzies F.M., Clarke J.H., and Rubinsztein D.C.. 2015. PI(5)P regulates autophagosome biogenesis. Mol. Cell. 57:219–234. 10.1016/j.molcel.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., and Ron D.. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Walther T.C., and Farese R.V. Jr. 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81:687–714. 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Miao Y.H., and Chang Y.S.. 2014. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206:357–366. 10.1083/jcb.201404115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Haas J.T., Walther T.C., and Farese R.V. Jr. 2014. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 29:39–45. 10.1016/j.ceb.2014.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bard M., Bruner D.A., Gleeson A., Deckelbaum R.J., Aljinovic G., Pohl T.M., Rothstein R., and Sturley S.L.. 1996. Sterol esterification in yeast: a two-gene process. Science. 272:1353–1356. 10.1126/science.272.5266.1353 [DOI] [PubMed] [Google Scholar]
- Yang L., Li P., Fu S., Calay E.S., and Hotamisligil G.S.. 2010. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 11:467–478. 10.1016/j.cmet.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.R., Chan E.Y., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., and Tooze S.A.. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900. 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.