Abstract
The kinetochore-associated kinase Mps1 controls the spindle assembly checkpoint, but the regulation of its kinetochore recruitment and activity is unclear. In this issue, Isokane et al. (2016. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201408089) show that interaction with and phosphorylation of its substrate, ARHGEF17, regulates Mps1 kinetochore retention, suggesting an autoregulated, timer-like mechanism.
To achieve mitotic fidelity, an elaborate mechanism called the spindle assembly checkpoint (SAC) has evolved to ensure proper chromosome–spindle attachment and alignment before anaphase. Through the action of many proteins found on centromeres and kinetochores, the SAC inhibits the anaphase promoting complex/cyclosome (APC/C) to prevent mitotic exit. The SAC must be highly tunable to rapidly respond to even a single misaligned chromosome, yet still allow mitosis to proceed in a timely manner once any alignment defects have been corrected. Mps1, a dual specificity kinase, is a core SAC protein that regulates kinetochore recruitment of other SAC proteins, including the protein kinases Bub1 and BubR1 and the APC/C inhibitor Mad2 (Maciejowski et al., 2010). Previous studies have implicated Mps1 dimerization and autoactivation (Hewitt et al., 2010; Jelluma et al., 2010), as well as the kinetochore component Ndc80/Hec1 and the Aurora B protein kinase, in targeting Mps1 to kinetochores (Saurin et al., 2011; Nijenhuis et al., 2013). The regulation of this targeting is less well understood, as are the mechanisms that ensure Mps1 is removed from kinetochores in a timely fashion. In this issue, Isokane et al. add ARHGEF17 to the list of Mps1 targeting machinery and propose that it mediates a timing mechanism that limits the duration of Mps1 activity at kinetochores.
Isokane et al. (2016) became interested in ARHGEF17 after they uncovered this gene in the MitoCheck genome-wide RNAi screen for mitotic regulators (Neumann et al., 2010), but MitoCheck lacked the temporal resolution to determine the mitotic function of ARHGEF17. Using high-resolution confocal live-cell imaging, Isokane et al. (2016) showed that ARHGEF17-depleted HeLa cells exhibited accelerated mitoses with chromosome congression, biorientation, and segregation defects and performed phenotypic rescue using a mouse ARHGEF17 transgene. Consistent with a SAC function, Isokane et al. (2016) found that mitosis in ARHGEF17-depleted cells was not arrested in response to the microtubule poison nocodazole and that kinetochore localization of several core SAC proteins was deficient, including Bub1, BubR1, and Mad2 (Hewitt et al., 2010; Jelluma et al., 2010; Maciejowski et al., 2010). These functions of ARHGEF17 are conferred by its central Rho GEF domain (Rümenapp et al., 2002), which Isokane et al. (2016) found to be both necessary and sufficient for SAC activity. However, the SAC functions of ARHGEF17 are independent of its GEF activity, as the inactive mutant ARHGEF17Y1216A (Zheng, 2001; Rümenapp et al., 2002) fully rescued the mitotic phenotypes.
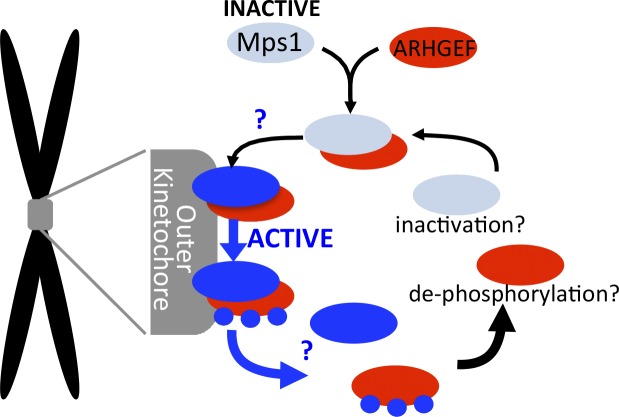
Because ARHGEF17 depletion phenocopied Mps1 inhibition, Isokane et al. (2016) explored a connection between the two and found that Mps1 interacts with the ARHGEF17 central domain and phosphorylates ARHGEF17 at three sites. Fluorescence cross-correlation spectroscopy (Wachsmuth et al., 2015) suggested that ARHGEF17 preferentially binds to inactive Mps1 in the cytoplasm, forming a complex required for localization of Mps1 and phosphorylation of its substrate, KNL1, at kinetochores (Yamagishi et al., 2012). ARHGEF17 localized to kinetochores independently of Mps1, but its only function in the SAC appears to be delivering Mps1 to kinetochores, as a kinetochore-tethered Mps1–CENP-B fusion bypassed the requirement for ARHGEF17 in the SAC. Interestingly, retention of the Mps1–ARHGEF17 complex at kinetochores is negatively regulated by Mps1 itself. The Mps1 inhibitor reversine increased recovery times for both Mps1 and ARHGEF17 in fluorescence recovery after photobleaching assays, indicating that Mps1 activity promotes their release from kinetochores. Based on these observations, Isokane et al. (2016) suggest an exciting model in which ARHGEF17 binding acts as a timer for retention of Mps1 at kinetochores: inactive Mps1 must form a complex with ARHGEF17 to bind to unattached/unaligned kinetochores, but once activated Mps1 limits its own retention at kinetochores by breaking apart the Mps1–ARHGEF17 complex (Fig. 1). Presumably, an individual molecule of Mps1 is retained at kinetochores for the time it takes it to phosphorylate the three sites on ARHGEF17 identified by Isokane et al. (2016).
Figure 1.
Proposed timer model for Mps1 retention at kinetochores. ARHGEF17 (red) binds inactive Mps1 (light blue) in the cytoplasm, and the complex binds to the outer kinetochore (gray) where Mps1 becomes activated (blue). Mps1 phosphorylates ARHGEF17 (blue circles), causing dissociation of both from kinetochores. It is still unknown where and how Mps1 is activated upon ARHGEF17 binding or where ARHGEF17 dissociates from Mps1 (blue question marks), and if and how Mps1 is inactivated or ARHGEF17 is dephosphorylated upon returning to the cytoplasm.
This model will be very intriguing to the SAC field because it suggests a mechanism by which kinetochores achieve the proper amount of Mps1 at individual kinetochores to properly respond to chromosome alignment defects in a timely fashion. However, data presented in this study will have to be put into the context of other studies showing requirements for Ndc80/Hec1 and Aurora B in recruiting Mps1 to the kinetochore (Martin-Lluesma et al., 2002; Saurin et al., 2011; Nijenhuis et al., 2013; Zhu et al., 2013). Although Isokane et al. (2016) show that ARHGEF17 depletion has no effect on Ndc80 or Aurora B and demonstrate that its effect on Mps1 targeting is direct, how ARHGEF17 cooperates with Ndc80 and Aurora B remains to be determined. Perhaps, like Mps1, ARHGEF17 binds to Ndc80 and/or is regulated by Aurora B or perhaps Mps1 and ARHGEF17 cooperate by binding to different kinetochore components. The proposed timer model raises several exciting questions. The authors speculate that ARHGEF17 binding might activate Mps1, but this is not yet tested. It will also be important to determine the fates of Mps1 and ARHGEF17 once released from kinetochores. Does Mps1 remain active to support functions in the cytoplasm, or is it inactivated to participate in further cycles of kinetochore targeting and SAC activity? Is ARHGEF17 dephosphorylated to participate in further cycles and, if so, what is the phosphatase? The exciting study by Isokane et al. (2016) is just the first step toward addressing how and where Mps1 activity is regulated to in turn ensure timely activation and silencing of the SAC.
Acknowledgments
The authors declare no competing financial interests.
References
- Hewitt L., Tighe A., Santaguida S., White A.M., Jones C.D., Musacchio A., Green S., and Taylor S.S.. 2010. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1–C-Mad2 core complex. J. Cell Biol. 190:25–34. 10.1083/jcb.201002133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isokane M., Walter T., Mahen R., Nijmeijer B., Hériché J.-K., Miura K., Maffini S., Ivanov M.P., Kitajima T.S., Peters J.-M., and Ellenberg J.. 2016. ARHGEF17 is an essential spindle assembly checkpoint factor that targets Mps1 to kinetochores. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201408089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N., Dansen T.B., Sliedrecht T., Kwiatkowski N.P., and Kops G.J.P.L.. 2010. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J. Cell Biol. 191:281–290. 10.1083/jcb.201003038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., George K.A., Terret M.-E., Zhang C., Shokat K.M., and Jallepalli P.V.. 2010. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 190:89–100. 10.1083/jcb.201001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V.M., and Nigg E.A.. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270. 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- Neumann B., Walter T., Hériché J.-K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., et al. . 2010. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 464:721–727. 10.1038/nature08869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijenhuis W., von Castelmur E., Littler D., De Marco V., Tromer E., Vleugel M., van Osch M.H.J., Snel B., Perrakis A., and Kops G.J.P.L.. 2013. A TPR domain–containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 201:217–231. 10.1083/jcb.201210033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rümenapp U., Freichel-Blomquist A., Wittinghofer B., Jakobs K.H., and Wieland T.. 2002. A mammalian Rho-specific guanine-nucleotide exchange factor (p164-RhoGEF) without a pleckstrin homology domain. Biochem. J. 366:721–728. 10.1042/bj20020654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.T., van der Waal M.S., Medema R.H., Lens S.M.A., and Kops G.J.P.L.. 2011. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2:316 10.1038/ncomms1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth M., Conrad C., Bulkescher J., Koch B., Mahen R., Isokane M., Pepperkok R., and Ellenberg J.. 2015. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 33:384–389. 10.1038/nbt.3146 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.-H., Tanno Y., and Watanabe Y.. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752. 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Zheng Y. 2001. Dbl family guanine nucleotide exchange factors. Trends Biochem. Sci. 26:724–732. 10.1016/S0968-0004(01)01973-9 [DOI] [PubMed] [Google Scholar]
- Zhu T., Dou Z., Qin B., Jin C., Wang X., Xu L., Wang Z., Zhu L., Liu F., Gao X., et al. . 2013. Phosphorylation of microtubule-binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore. J. Biol. Chem. 288:36149–36159. 10.1074/jbc.M113.507970 [DOI] [PMC free article] [PubMed] [Google Scholar]