Abstract
In fetal growth restriction (FGR), fetal growth is limited by reduced nutrient and oxygen supply. Insulin-like growth factor I (IGF-I) is a key regulator of fetal growth and IGF binding protein -1(IGFBP-1) is the principal regulator of fetal IGF-I bioavailability. Phosphorylation enhances IGFBP-1's affinity for IGF-I. Hypoxia induces IGFBP-1 hyperphosphorylation, markedly decreasing IGF-I bioavailability. We recently reported that fetal liver IGFBP-1 hyperphosphorylation is associated with inhibition of the mechanistic target of rapamycin (mTOR) in a nonhuman primate model of FGR. Here, we test the hypothesis that IGFBP-1 hyperphosphorylation in response to hypoxia is mediated by mTOR inhibition. We inhibited mTOR either by rapamycin or small interfering RNA (siRNA) targeting raptor (mTOR complex [mTORC]1) and/or rictor (mTORC2) in HepG2 cells cultured under hypoxia (1% O2) or basal (20% O2) conditions. Conversely, we activated mTORC1 or mTORC1+mTORC2 by silencing endogenous mTOR inhibitors (tuberous sclerosis complex 2/DEP-domain-containing and mTOR-interacting protein). Immunoblot analysis demonstrated that both hypoxia and inhibition of mTORC1 and/or mTORC2 induced similar degrees of IGFBP-1 phosphorylation at Ser101/119/169 and reduced IGF-I receptor autophosphorylation. Activation of mTORC1+mTORC2 or mTORC1 alone prevented IGFBP-1 hyperphosphorylation in response to hypoxia. Multiple reaction monitoring-mass spectrometry showed that rapamycin and/or hypoxia increased phosphorylation also at Ser98 and at a novel site Ser174. In silico structural analysis indicated that Ser174 was in close proximity to the IGF-binding site. Together, we demonstrate that signaling through the mTORC1 or mTORC2 pathway is sufficient to induce IGFBP-1 hyperphosphorylation in response to hypoxia. This study provides novel understanding of the cellular mechanism that controls fetal IGFBP-1 phosphorylation in hypoxia, and we propose that mTOR inhibition constitutes a mechanistic link between hypoxia, reduced IGF-I bioavailability and FGR.
Fetal growth restriction (FGR) is associated with increased risk of perinatal hypoxia, stillbirth and neonatal morbidity. In addition, FGR infants are susceptible to developing cardiovascular disease, obesity, and diabetes in childhood and as adults (1). The most common cause of FGR is uteroplacental insufficiency, which is often associated with fetal hypoxia in utero (2). Elegant studies in the chicken embryo have demonstrated that hypoxia per se is sufficient to cause FGR (3); however, the molecular mechanisms linking hypoxia to restricted fetal growth are not well understood. Earlier studies provide evidence to suggest that hypoxia influences fetal growth via the IGF signaling system (4). Fetal liver is the major source of IGF binding protein-1(IGFBP-1), the major IGF-binding protein in fetal life (5). Uteroplacental insufficiency is associated with increased fetal hepatic IGFBP-1 mRNA and protein expression and markedly elevated circulating IGFBP-1 in umbilical cord blood (5). In addition, IGFBP-1 levels in cord blood are inversely correlated with birth weight and fetal cord pO2 levels (6).
IGFBP-1 sequesters IGF-I and regulates the bioavailability of free IGF-I in the fetal circulation (7). The IGFBP-1 gene has a consensus sequence for the hypoxia-response element that binds hypoxia-inducible factor-1 and causes significant induction in IGFBP-1 expression in fetal liver (5). In zebrafish, hypoxia induces IGFBP-1 mRNA and protein expression, resulting in FGR (8). Increased expression of IGFBP-1 is considered a marker of nutritional deprivation and hypoxia that cause FGR (6, 8, 9–15). Using HepG2 cells, we have previously demonstrated that hypoxia causes IGFBP-1 hyperphosphorylation that markedly decreases IGF-I bioavailability and inhibits IGF-I-stimulated cell growth (16). These data are consistent with the model that increased IGFBP-1 phosphorylation due to hypoxia sequesters IGF-I, which inhibits IGF-I-mediated fetal growth, thereby contributing to FGR.
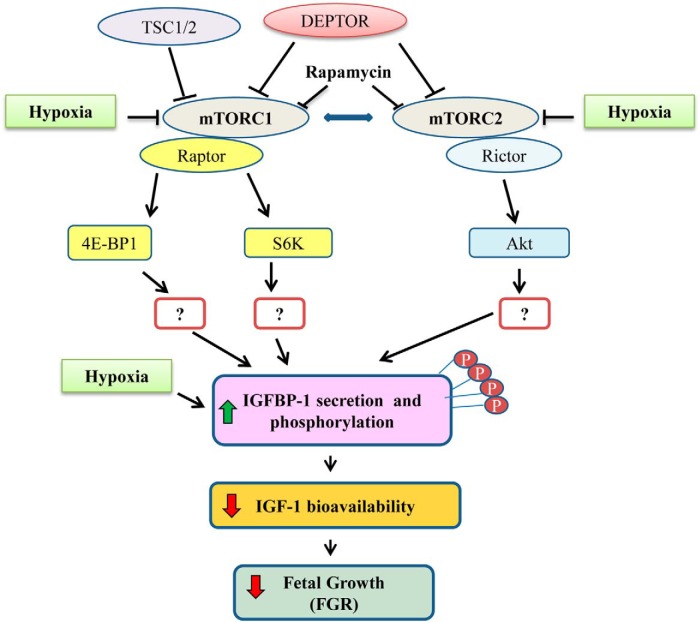
Mechanistic target of rapamycin (mTOR) is a conserved serine/threonine kinase that controls cell growth and metabolism, which is primarily mediated by effects on protein translation (17). As shown in Figure 1, mTOR exists in 2 complexes, mTOR complex (mTORC)1 and mTORC2, with the protein raptor associated to mTORC1 (18, 19) and rictor associated to mTORC2 (20). mTORC1 phosphorylates ribosomal protein S6 kinase beta-1 (21) and eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1) (22), resulting in increased protein translation (23). mTORC2 phosphorylates Akt, a serine/threonine kinase also known as protein kinase B (24), protein kinase Cα (25), and serum and glucocorticoid-regulated kinase 1 (26) and regulates cell survival and metabolism (27). It is well established that mTORC1 signaling is inhibited by hypoxia (28, 29) and decreased amino acid availability (30, 31). We recently demonstrated a marked inhibition of mTOR signaling together with IGFBP-1 hyperphosphorylation in fetal liver from a baboon model of FGR (32). However, the molecular mechanisms linking hypoxia to increased IGFBP-1 phosphorylation are unknown.
Figure 1.
Functionally important mTOR-related proteins linking mTOR to the regulation of IGFBP-1 secretion and phosphorylation. Schematic diagram representing a proposed model connecting mTOR signaling to IGFBP-1 secretion and phosphorylation. Key target proteins for silencing and functional readouts for mTORC1 and mTORC2 activity are presented.
Using HepG2 cells as a model for fetal hepatocytes (16, 32–35), we tested the hypothesis that IGFBP-1 hyperphosphorylation in response to hypoxia is mediated by mTOR inhibition (Figure 1). We applied pharmacological and RNA interference (RNAi) approaches to inhibit and/or activate mTORC1/mTORC2 in HepG2 cells cultured under normoxic and hypoxic conditions. Using cell media, we performed immunoblot analysis (custom IGFBP-1 phosphosite antibodies) and applied multiple reaction monitoring (MRM)/selective reaction monitoring-mass spectrometry (36, 37) as an alternative approach to establish mTOR signaling as a mechanistic link between hypoxia and IGFBP-1 hyperphosphorylation. The functional effects of IGFBP-1 phosphorylation on IGF-I bioactivity were assessed using our IGF-I receptor (IGF-1R) autophosphorylation assay (32).
Materials and Methods
Cell culture
Human hepatoma HepG2 cells (ATCC) demonstrate biotransformation characteristics and the gene expression similar to primary human and baboon fetal hepatocytes (32–35) and are therefore appropriate to conduct this mechanistic study. HepG2 cells were cultured in DMEM/F-12 supplemented with 10% fetal bovine serum (FBS) (Invitrogen Corp) at 37°C in atmospheric air 20% O2 with 5% CO2 (at 75% confluence) for 24 hours as described previously (16, 38). Before treatments, the media were changed to 2% FBS for 12 hours. Mouse embryo fibroblast P6 cells that overexpress human IGF-1R (a kind gift from Dr R. Baserga, Thomas Jefferson University, Philadelphia, PA) were used to perform in vitro IGF-1R autophosphorylation assay. P6 cells were cultured in DMEM with sodium pyruvate and treatments were performed in FBS-free conditions as described (32, 39).
Rapamycin treatment
As reported in our previous study (32), HepG2 cells were incubated in 100nM rapamycin for 24 hours (32). Control media contained 2% FBS. After treatments, the cell media/lysate were collected and stored at −80°C.
Hypoxia
HepG2 cells were cultured in hypoxia or normoxia (16, 38). Briefly, cells were placed in an incubator either with atmospheric air and 5% CO2 (normoxia) or a hypoxia chamber (Billups-Rothenburg), which was flushed with a 1% O2, 5% CO2, balanced N2 gas mixture (BOC Canada Ltd) for 5 minutes to ensure saturation. The cells in the sealed chamber were placed in a tissue-culture incubator at 37°C on an orbital shaker. Cell media and cell lysate were collected after 24 hours of exposure.
Cell viability assay
We tested the effect of rapamycin and hypoxia treatments on cell viability using the trypan blue exclusion assay to ensure treatments did not cause increased cell death. After treatments, cells were trypsinized and resuspended 1:1 with 0.4% trypan blue. Cells were counted using the Countess Automated Cell Counter (Life Technologies), and viability was determined as a measure of live/total cells.
RNAi-mediated silencing
Silencing raptor and/or rictor, DEP-domain-containing and mTOR-interacting protein (DEPTOR) and tuberous sclerosis complex 2 (TSC2) in HepG2 cells (∼1.5 × 105 cells per well in 12-well plate) was achieved using transfection with 100nM small interfering RNA (siRNA) (Sigma-Aldrich) and Dharmafect transfection reagent 4 (Thermo Scientific) as described (32). Based on the time course for maximal silencing (data not shown), immediately after transfection, cells were cultured for 48 hours in normoxia (20% O2), followed by an additional 24 hours either in normoxia or in hypoxia (1% O2). The efficiency of target silencing was determined using Western blot analysis.
SDS-PAGE and Western blotting
Equal amounts of HepG2 cell lysate protein (20–50 μg) were used to determine phosphorylation and total expression of 4E-BP1 at Thr70, Akt at Ser473, IGF-1R at Tyr1135, as well as expression of siRNA targets. For analysis, the band intensity for phosphorylated proteins (4E-BP1, Akt, and IGF-1R) were normalized to respective total protein which were normalized to β-actin and compared with respective control as in our previous studies (16, 32, 39). IGFBP-1 secretion (8 μL) and phosphorylation (40 μL) in cell media were normalized via plating equal number of cells and loading equal volume of cell media, because there are no valid loading controls available for secreted proteins. The sources, dilution, and species of all primary antibodies are listed in Supplemental Table 1. Secondary antibodies were peroxidase-labeled goat-antimouse or goat-antirabbit antibodies 1:10000 (Bio-Rad Laboratories, Inc). Band intensities were determined using densitometry and Image Lab (Beta 3) software (Bio-Rad). 2-Dimensional (2-D) immunoblot analysis was performed with HepG2 cell media (∼150 μL) using polyclonal IGFBP-1 antibody as described (16, 32, 39–41).
Immunoprecipitation and immunodepletion of IGFBP-1
Equal volume of pooled triplicate HepG2 cell media (biological replicates) from various treatments were buffer exchanged against PBS (Centricon filters; Pall). The samples were incubated overnight at 4°C with highly specific and well-established monoclonal antihuman IGFBP-1 monoclonal antibody 6303 (100 ng) together with Protein A Sepharose (50 μL, 50% slurry) to immunoprecipitate (IP) the IGFBP-1 immune complex as we described previously (41) and were used for multiple reaction monitoring-mass spectrometry (MRM-MS) assessments. Furthermore, for immunodepletion, supernatants obtained after IP as described above were used as IGFBP-1 depleted samples (negative controls) for IGF-1R autophosphorylation assay.
IGF-1R activation assay
Using P6 cells, we determined IGF-I mediated-functional effects on IGF-1R stimulation as described (32, 39). In brief, aliquots of HepG2 cell media containing equal concentrations of total IGFBP-1 were incubated with recombinant human IGF-I for 2 hours at room temperature. The IGFBP-1/IGF-I complexes were used to activate IGF-1R in P6 cells for 10 minutes. Subsequently, P6 cells were lysed and tested on immunoblots for IGF-1R autophosphorylation using phospho-IGF-1Rβ (Tyr1135) antibody. Furthermore, in order to confirm that the effects on IGF-1R were specific to IGFBP-1 in HepG2 cell secretion, equal aliquots of the cell media from control or rapamycin-treated samples were subjected to IGFBP-1 immunodepletion as described above, and then the supernatant (depleted of IGFBP-1) was tested for IGF-I-induced IGF-1R autophosphorylation in tandem with the experimentally treated samples.
MRM-mass spectrometry
IGFBP-1 (IP) samples obtained as described above (3 pooled biological replicates of each treatment) were resuspended in endoproteinase Asp-N digestion buffer (50mM sodium phosphate; pH 8.0) and digested with Asp-N (Roche Diagnostics) overnight at 37°C. After Asp-N digestion, half of each sample was subjected to a subsequent digestion with sequencing grade trypsin (Roche Diagnostics) over night at 37°C. The digest was analyzed by positive electrospray ionization liquid chromatography-mass spectrometry (LC-MS/MS) on a triple quadruple mass spectrometer (4000 QTRAP AB Sciex) with Q3 used as a linear ion trap. The in silico protease digest patterns and the corresponding MRM-MS transitions were compiled with the Skyline software (Supplemental Table 1) (42). Transitions that were larger than the precursor ion were selected on the basis of the Skyline predictions and the specific b/y ions that allow unambiguous identification of the selected phosphorylated IGFBP-1 serine residues were included. Relative change in IGFBP-1 phosphorylation status was determined by the total peak height of combined transitions. For MRM absolute quantitation, peptides sequences unique to the targeted IGFBP-1 phosphorylation sites were selected. An internal IGFBP-1-specific synthetic peptide (NH2-ALPGEQQPLHALTR-COOH) was used as standard to normalize all IGFBP-1 phosphorylation data to IGFBP-1 total protein abundance.
In silico analysis of IGFBP-1 Ser174 phosphorylation
Structural data was obtained to determine the location of the newly identified IGFBP-1 phosphorylated residue Ser174 relative to IGF binding to IGFBP-1. The structure of the IGFBP-1 C terminus was created using the SWISS-MODEL server (http://swissmodel.expasy.org/) (43). Molecular modeling of the IGFBP C terminus and IGF-IB complex was based on the available crystal structure (2DSQ). The C terminus of IGFBP-1 was selected from the available PDB 2DSQ:G crystal structure (44). Next, the IGFBP-1 C terminus model was protonated and optimized by energy minimization using MMFF94 × force field model in Molecular Operating Environment software (v.2011.10) (Chemical Computing Group). The molecular surface (ActiveLP) areas of IGF-I were calculated using the Molecular Operating Environment software (42–44).
Data presentation and statistics
Statistics were performed using GraphPad Prism 5 (GraphPad Software, Inc). For each protein quantified, the mean band intensity of the control sample was assigned an arbitrary value of 100. All individual densitometry values were expressed relative to this mean. To compare means, Student's t test and one-way ANOVA and Dunnet's multiple comparison test were used and the results were expressed as mean ± SEM. Significance was accepted at P < .05.
Results
Rapamycin and/or hypoxia inhibit mTOR signaling
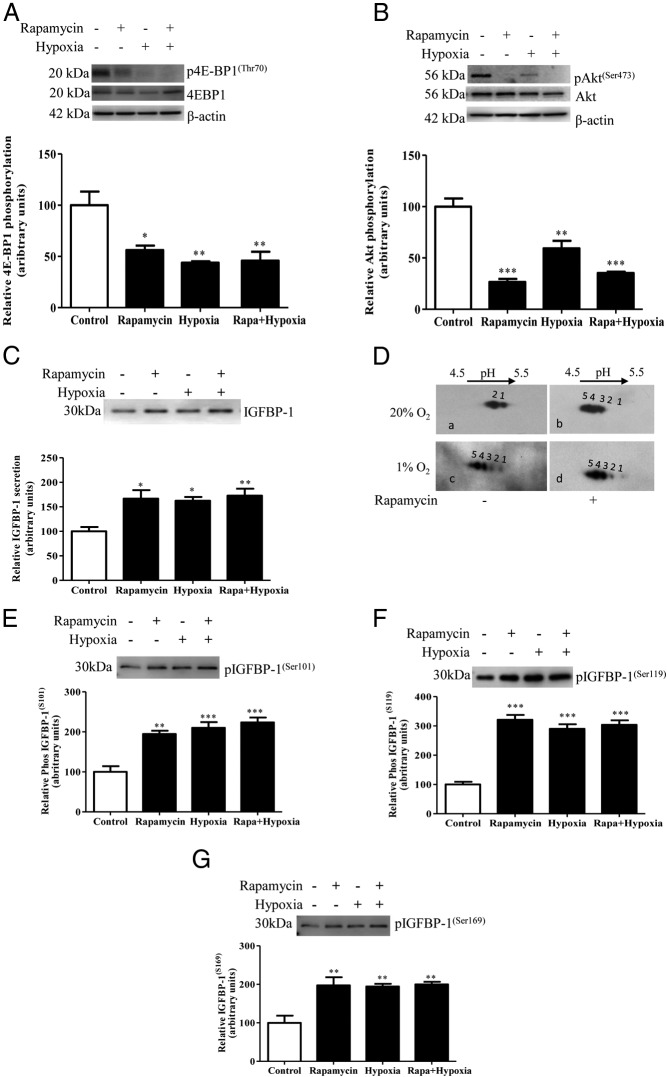
Phosphorylation of mTORC1 target, 4E-BP1 (Thr70) and mTORC2 target, Akt (Ser473) in HepG2 cell lysate was decreased in response to rapamycin treatment, in agreement with our previous report (32). Furthermore, phosphorylation of 4E-BP1 (Thr70) and Akt (Ser473) was also markedly inhibited by hypoxia, and rapamycin combined with hypoxia. These data demonstrate that hypoxia and rapamycin inhibit mTORC1 and mTORC2 signaling to a similar extent (Figure 2, A and B).
Figure 2.
The effect of hypoxia and/or rapamycin treatment on IGFBP-1 secretion and phosphorylation. Representative Western blottings of the functional readouts of (A) mTORC1, 4E-BP1 (Thr70) and (B) mTORC2, Akt (Ser473) phosphorylation. Phosphorylation of both mTORC1 and C2 was markedly inhibited by rapamycin, hypoxia, and rapamycin+hypoxia. C, A representative Western blotting of IGFBP-1 secreted from rapamycin, hypoxia, or rapamycin+hypoxia-treated cells. Rapamycin or hypoxia treatment significantly increased IGFBP-1 secretion. Combined rapamycin+hypoxia did not increase IGFBP-1 secretion more than either treatment alone. D, A representative 2-D Western blotting of IGFBP-1 phosphoisoforms in cell media from rapamycin, hypoxia, or rapamycin+hypoxia-treated cells. Rapamycin (top right) and hypoxia (bottom left) induced IGFBP-1 phosphorylation compared with normoxia (top left). In the presence of rapamycin, hypoxia did not cause any additional increases in phosphorylation (bottom right) compared with either treatment alone. E–G, Representative Western blottings of phosphorylated IGFBP-1 at Ser101, Ser119, and Ser169 in cell media of rapamycin, hypoxia, or rapamycin+hypoxia-treated cells. Rapamycin or hypoxia treatment significantly increased IGFBP-1 phosphorylation at all 3 serine sites. Combined rapamycin+hypoxia did not cause any additional increases in phosphorylation. Values are displayed as mean ± SEM; *, P < .05; **, P = .001-.05; ***, P < .0001 vs control; one-way ANOVA; Dunnet's multiple comparison test.
mTOR inhibition and hypoxia increase IGFBP-1 secretion and phosphorylation
Next, we examined the effects of hypoxia and rapamycin on IGFBP-1 secretion/phosphorylation using HepG2 cell media. A cell viability assay ensured treatments were not increasing cell death (data not shown). We demonstrated that IGFBP-1 secretion was significantly increased to a similar degree by rapamycin, hypoxia, or rapamycin and hypoxia combined (67%, 62%, and 73%, respectively; P = .011) as compared with the control (Figure 2C).
To determine IGFBP-1 phosphorylation, we resolved IGFBP-1 phosphoisoforms by qualitative 2-D immunoblotting (Figure 2D), where more spots shifting to the left (positive) represent increased phosphorylation. Multiple spots were resolved; however, the most prominent spots with hypoxic treatment were (#4–5) towards acidic pH consistent with our previous report showing one highly phosphorylated form (16). 2-D immunoblotting was restricted to visual assessment of spot profiles within the same set of experimental treatments. Most notably, hypoxia and/or rapamycin or the 2 combined caused similar increases in the intensity and number of spots concomitantly, with a shift towards the left compared with control. Rapamycin caused a pronounced hyperphosphorylation of IGFBP-1 (Figure 2Db) and hypoxia had a similar effect (Figure 2Dc). Furthermore, in the presence of rapamycin, hypoxia did not cause any additional phosphorylation of IGFBP-1 (Figure 2Dd). These results support the hypothesis that IGFBP-1 hyperphosphorylation in hypoxia is mediated by mTOR inhibition.
Using phospho-site-specific antibodies, we then demonstrated a pronounced increase in IGFBP-1 phosphorylation at the 3 specific sites in response to rapamycin (Ser101 [95%, P = .0004], Ser119 [221%, P = .0001], and Ser169 [98%, P = .0036]) (Figure 2, E–G). Moreover, hypoxia alone or hypoxia combined with rapamycin induced IGFBP-1 phosphorylation to a similar degree as rapamycin alone indicating that hypoxia did not increase IGFBP-1 phosphorylation further (Figure 2, E–G). Because we previously have shown that predominant increase in IGF-I bioactivity in HepG2 cells occurs due to concerted changes in sites and degree of phosphorylation and not in response to total IGFBP-1 secretion (16), for consistency and accurate representation, data here were assessed using equal volume of cell media without normalizing phosphorylation to total IGFBP-1 as in our previous reports (16, 32). Overall, these findings are consistent with the hypothesis that induction of IGFBP-1 phosphorylation in response to hypoxia is mediated via inhibition of mTOR signaling.
Increase in IGFBP-1 phosphorylation inhibits IGF-I bioactivity
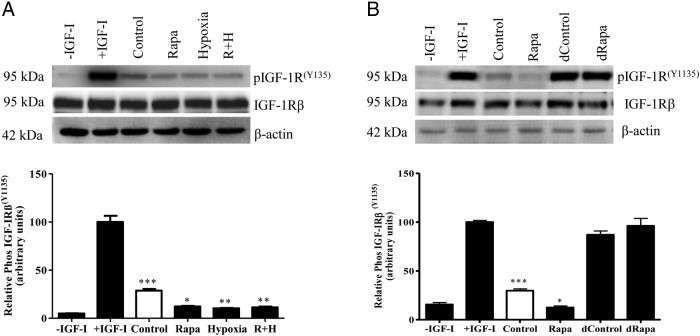
To determine effects on IGF-I bioactivity in response to rapamycin, hypoxia, or rapamycin+hypoxia treatments, IGF-1Rβ autophosphorylation assay was performed using P6 cells (32, 39). P6 cells incubated with IGF-I alone (+IGF-I, positive control) demonstrated a marked increase (20-fold) in IGF-1R phosphorylation (100%, P = .0001) compared with cells without IGF-I (−IGF-I, negative control) (Figure 3A, lane 2). This demonstrated the ability of IGF-I to significantly stimulate IGF-1R autophosphorylation. When P6 cells were incubated with HepG2 cell media+IGF-I (untreated, control) (Figure 3A, lane 3), a 72% (P = .0001) reduction in IGF-1R autophosphorylation was observed compared with P6 cells incubated with IGF-I alone. This suggested that basal levels of IGFBP-1 secreted by HepG2 cells (control) can sequester bioavailable IGF-I and subsequently reduce IGF-1R signaling. A further reduction in IGF-1R autophosphorylation was observed when P6 cells were incubated with HepG2 cell media from rapamycin or hypoxia, respectively (Figure 3A, lanes 4 and 5) (88%, P = .01, 89%, P = .004), compared with the positive control (+IGF-I). The effects of rapamycin+hypoxia combined were not different (89%, P = .002) (Figure 3A, lane 6) than the 2 separate treatments. Because equal concentration of total IGFBP-1 from HepG2 cell media were used to activate IGF-1R in P6 cells, these data suggest that inhibition of IGF-1R autophosphorylation was caused mainly by increased phosphorylation of IGFBP-1 and not total IGFBP-1. Collectively, these data provide evidence for a key role of mTOR inhibition in linking hypoxia to IGFBP-1 hyperphosphorylation and reduced IGF-I bioavailability in HepG2 cells.
Figure 3.
The effect of IGFBP-1 hyperphosphorylation on IGF-1Rβ autophosphorylation. A representative Western blotting of IGF-1R autophosphorylation in P6 cells. A, Equal concentrations of total IGFBP-1 from HepG2 cell media from rapamycin, hypoxia, or rapamycin+hypoxia were used to induce P6 cell IGF-1R autophosphorylation in presence of IGF-I. After that equal concentration of P6 cell lysate protein was used for immunoblot analysis to detect IGF-1Rβ (Tyr1135) autophosphorylation. Increased IGFBP-1 phosphorylation due to rapamycin, hypoxia, or combined rapamycin+hypoxia (R+H) resulted in significantly decreased IGF-1R activation compared with control. B, To validate that IGFBP-1 in HepG2 cell media was the source of IGF-1R inhibition, equal aliquots of cell media from control or rapamycin-treated cells were subjected to IGFBP-1 immunodepletion and used to then treat P6 cells in the presence of IGF-I. Immunodepleted cell media from control or rapamycin-treated HepG2 cells resulted in IGF-1Rβ autophosphorylation in P6 cells similar to the positive control (+IGF-I only), whereas the nondepleted control or rapamycin-treated samples retained significant inhibition of IGF-1Rβ activation (P < .0001). The percentage of inhibition of IGF-1R activation was considered significant at *, P < .05; **, P = .001-.05; ***, P < .0001 vs control; one-way ANOVA; Dunnet's multiple comparison test.
Furthermore, to demonstrate that the key reactive component in HepG2 cell media which effects IGF-1R phosphorylation is IGFBP-1, we performed similar treatments using HepG2 cell media immunodepleted of IGFBP-1 from treated cells. Using immunodepleted samples (control or rapamycin) (Figure 3B, lanes 5 and 6), IGF-1R autophosphorylation levels were not significantly different from those observed with, IGF-I alone. In contrast, nondepleted control (untreated) and rapamycin-treated HepG2 cell media (without depletion) (Figure 3B, lanes 3 and 4) resulted in the expected decrease (71% and 88%, respectively) in IGF-1R phosphorylation, validating our findings that effects on IGF-I bioactivity are specific to IGFBP-1.
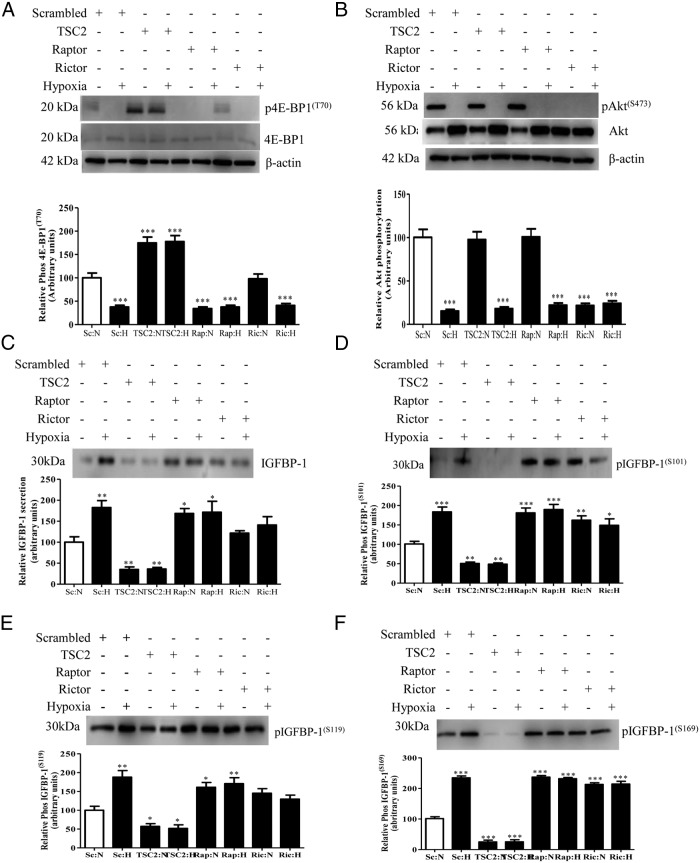
mTOR inhibition by raptor or rictor silencing induces IGFBP-1 secretion and phosphorylation in normoxia
We further tested our hypothesis using an RNAi strategy to silence raptor or rictor individually in presence and absence of hypoxia. We first confirmed that our RNAi approach efficiently silenced the mTORC1 and mTORC2 signaling pathways in normoxic conditions (Supplemental Figure 1, A and B). As shown in Supplemental Figure 1, C–F, raptor, rictor individually, or raptor+rictor silencing combined increased IGFBP-1 secretion and phosphorylation to similar levels. For example, raptor silencing resulted in increase in IGFBP-1 phosphorylation at Ser101 (61%, P = .0002), Ser119 (90%, P = .0001), and Ser169 (129%, P = .0001) compared with the control (scramble, 100%). Furthermore, rictor or raptor+rictor silencing also significantly induced IGFBP-1 phosphorylation to similar levels as raptor separately, consistent with our previous findings (32). These data indicate that both mTORC1 and mTORC2 regulate IGFBP-1 secretion and phosphorylation in normoxia.
Inhibition of mTORC1 and mTORC2 combined prevents additional increase in IGFBP-1 secretion and phosphorylation in response to hypoxia
Next, we silenced both raptor and rictor in the same experiment to inhibit mTORC1+mTORC2 signaling in presence of hypoxia. Silencing efficiency of raptor or rictor, remained similar in hypoxia (Supplemental Figure 2, B and C) as we reported in normoxia. As shown in Figure 4, A and B, in normoxia, raptor+rictor silencing caused a pronounced decrease in mTORC1 and mTORC2 activity as shown by reduction in phosphorylation of 4E-BP1 (Thr70) and Akt (Ser473). Hypoxia alone markedly inhibited the activity of mTORC1 and mTORC2 to similar levels. Additionally, raptor+rictor silencing inhibited mTORC1 and mTORC2 activity, respectively, to a similar extent in hypoxia (Figure 4, A and B). Hypoxia induced IGFBP-1 secretion and phosphorylation (Figure 4, C–F) in raptor+rictor-silenced cells to a level similar to that of hypoxia, which shows that hypoxia did not stimulate further total or phosphorylated IGFBP-1 than in normoxic raptor+rictor-silenced cells (Figure 4, C–F). Together, combined mTORC1+mTORC2 inhibition in hypoxia provided evidence that IGFBP-1 secretion/phosphorylation in response to hypoxia are mediated by inhibition of the mTORC1 and/or mTORC2 signaling pathway.
Figure 4.
The effect of mTORC1+mTORC2 inhibition and/or activation with and without hypoxia (H) on IGFBP-1 secretion and phosphorylation. HepG2 cells were treated with DEPTOR siRNA or raptor+rictor siRNA, and then cells were additionally cultured in normoxia (N) or in H. Western blottings were performed on cell lysates or cell media from cells transfected with scramble (N), scramble (H), DEPTOR (N), DEPTOR (H), raptor+rictor (N), and rictor+rictor (H). A representative Western blotting of (A) 4E-BP1 phosphorylation (Thr70); H alone significantly inhibited 4E-BP1 phosphorylation. When treated with DEPTOR siRNA, 4E-BP1 phosphorylation was significantly increased regardless of hypoxic status, suggesting that DEPTOR knock down prevented mTORC1 inhibition due to H alone. When treated with raptor+rictor siRNA, 4E-BP1 phosphorylation was significantly inhibited regardless of hypoxic status. B, Akt phosphorylation (Ser473). H alone significantly inhibited Akt phosphorylation (Ser473). With DEPTOR siRNA, Akt phosphorylation (Ser473) was significantly increased regardless of hypoxic status, suggesting that DEPTOR knock down prevented mTORC2 inhibition due to H alone. Using raptor+rictor siRNA, Akt phosphorylation was significantly inhibited regardless of hypoxic status. A representative Western blotting of (C) IGFBP-1 secretion and (D–F) phosphorylated IGFBP-1 at Ser101, Ser119, and Ser169 from cell media. H alone significantly increased both IGFBP- 1 secretion and phosphorylation. When treated with DEPTOR siRNA, IGFBP-1 secretion and phosphorylation was not significantly different from controls regardless of hypoxic status, suggesting that constitutively activated mTOR signaling prevented the induction of IGFBP-1 secretion and phosphorylation due to H. Conversely, when treated with combined raptor+rictor siRNA, IGFBP-1 secretion and phosphorylation was significantly increased to levels similar to H alone, suggesting that H exerts it effects on both IGFBP-1 secretion and phosphorylation via mTORC1+C2 signaling. These data indicate that inhibition of mTOR during H is responsible for the regulation of IGFBP-1 phosphorylation. Values are displayed as mean ± SEM; *, P < .05; **, P = .001-.05; ***, P < .0001 vs control; one-way ANOVA; Dunnet's multiple comparison test.
Activation of mTORC1 and mTORC2 by DEPTOR silencing reduces IGFBP-1 secretion and phosphorylation in response to hypoxia
To further confirm that mTORC1 and/or mTORC2 mediate the changes in IGFBP-1 secretion and phosphorylation in response to hypoxia, we determined the effect of hypoxia on IGFBP-1 secretion and phosphorylation in cells in which mTORC1 and mTORC2 were activated. To activate mTOR, we silenced DEPTOR, an endogenous mTORC1 and mTORC2 inhibitor. We validated that DEPTOR silencing reduced DEPTOR protein expression in normoxia (52%, P = .018) (Supplemental Figure 2A). Phosphorylation of 4E-BP1 (Thr70) and Akt (Ser473) were significantly increased 83% (P = .0001) and 49% (P = .01), respectively, after DEPTOR silencing, confirming activation of mTORC1 and mTORC2 (Figure 4, A and B) in normoxia. Furthermore, mTORC1 and mTORC2 activation by DEPTOR silencing significantly reduced IGFBP-1 secretion (70%, P = .034) (Figure 4C) and phosphorylation at Ser101 (53%, P = .031), Ser119 (53%, P = .012), and Ser169 (60%, P = .0032) (Figure 4, D–F) under normoxic condition. Collectively, these data show that activation of mTOR signaling under normoxia decreases IGFBP-1 secretion and phosphorylation. As indicated earlier, hypoxia alone inhibited both mTORC1 and mTORC2 to similar levels (Figure 4, A and B). Importantly, DEPTOR silencing led to activation of mTORC1 and mTORC2 and completely prevented hypoxia-induced IGFBP-1 secretion and phosphorylation (Figure 4, C–F). Together, these data provide conclusive evidence that IGFBP-1 secretion and phosphorylation in response to hypoxia are mediated by inhibition of mTORC1 and/or mTORC2 signaling pathway.
Activation of mTORC1 signaling decreases IGFBP-1 secretion and phosphorylation in normoxia
Next, in order to prove that mTORC1 signaling is involved in mediating the effects of hypoxia on IGFBP-1, we silenced the endogenous mTORC1 inhibitor TSC2 to specifically activate mTORC1 in HepG2 cells. We validated silencing efficiency of TSC2, and as shown in Supplemental Figure 3A, total TSC2 protein expression was reduced (47%, P < .004). Silencing of TSC2 led to significant increase in mTORC1 signaling as determined by phosphorylation of 4E-BP1 at (Thr70) 75% (P = .0001) (Figure 5A), whereas mTORC2 signaling was not changed to a significant level using phosphorylation of Akt (Ser473) as a readout for mTORC2 activity (Figure 5B). Therefore, these data confirmed that TSC2 silencing results in increased mTORC1 activity without significantly effecting mTORC2 in HepG2 cells under normoxia. We then determined changes in IGFBP-1 secretion/phosphorylation as a result of mTORC1 activation in normoxia. TSC2 silencing resulted in reduction in IGFBP-1 secretion (65%, P = .005) (Figure 5C) and IGFBP-1 phosphorylation at Ser101 (50%, P = .0016), Ser119 (42%, P = .039), and Ser169 (77%, P = .0008) (Figure 5, D–F). Together, these data provide evidence that mTORC1 signaling is an important regulator of IGFBP-1 secretion and phosphorylation in normoxia.
Figure 5.
The effect of mTORC1 activation and individual mTORC1 and mTORC2 inhibition with/without hypoxia on IGFBP-1 secretion/phosphorylation. HepG2 cells were treated with TSC2, raptor, or rictor siRNA, and then cells were additionally cultured in normoxia or in hypoxia. Western blottings were performed on cell lysates or cell media from cells transfected with scramble (normoxia), scramble (hypoxia), TSC2 (normoxia), TSC2 (hypoxia), raptor (normoxia), raptor (hypoxia), rictor (normoxia), and rictor (hypoxia) siRNA. A, A representative Western blotting of 4E-BP1 phosphorylation (Thr70). Hypoxia alone significantly inhibited 4E-BP1 (Thr70) phosphorylation. When treated with TSC2 siRNA, 4E- BP1 phosphorylation was significantly increased from control levels regardless of hypoxic status. Raptor siRNA significantly inhibited 4E-BP1 phosphorylation regardless of hypoxic status. Rictor siRNA did not affect 4E-BP1 phosphorylation, but 4E-BP1 phosphorylation was significantly reduced in rictor siRNA+hypoxia treatment. B, A representative Western blotting of Akt phosphorylation (Ser473). Hypoxia alone significantly inhibited Akt phosphorylation (Ser473). TSC2 siRNA did not affect Akt phosphorylation, although Akt phosphorylation was significantly reduced as a result of hypoxia during TSC2 siRNA. Similarly, raptor siRNA alone did not affect Akt phosphorylation, but raptor siRNA+hypoxia significantly reduced Akt phosphorylation. Rictor siRNA significantly reduced Akt phosphorylation regardless of hypoxic status. Representative Western blottings of (C) IGFBP-1 secretion and (D–F) phosphorylated IGFBP-1 at Ser101, Ser119, and Ser169 from cell media. Hypoxia alone significantly increased IGFBP-1 secretion and phosphorylation. When treated with TSC2 siRNA, IGFBP-1 secretion and phosphorylation was significantly reduced from control levels regardless of hypoxic status, suggesting that constitutively activated mTORC1 signaling prevented the induction of IGFBP-1 secretion and phosphorylation due to hypoxia. Conversely, when treated with individual raptor or rictor siRNA, IGFBP-1 secretion and phosphorylation was significantly increased to levels similar to hypoxia alone, suggesting that hypoxia exerts it effects on IGFBP-1 secretion and phosphorylation via mTORC1 or combined mTORC1+C2 inhibition. Values are displayed as mean ± SEM; *, P < .05; **, P = .001-.05; ***, P < .0001 vs control; one-way ANOVA; Dunnet's multiple comparison test.
mTORC1 activation prevents the induction of IGFBP-1 secretion/phosphorylation in response to hypoxia
To further explore the role of mTORC1 in mediating the effect of hypoxia on IGFBP-1, we studied the effects of hypoxia in cells with mTORC1 activation by TSC2 silencing. To validate these findings, in parallel we silenced raptor or rictor individually to inhibit mTORC1 and mTORC2 separately in cells cultured in hypoxia or normoxia. As shown in Supplemental Figure 3, A–C, protein expression of TSC2, raptor, and rictor were all decreased after corresponding siRNA silencing and the silencing efficiency was similar in hypoxic cells.
As shown earlier (Figure 5, A and B), hypoxia inhibited mTORC1 and mTORC2 activity. However, TSC2 silencing prevented hypoxia induced mTORC1 inhibition as evident by similar levels of 4E-BP1 (Thr70) phosphorylation between normoxia and hypoxia in TSC2-silenced cells (Figure 5A). Furthermore, TSC2 silencing also reduced IGFBP-1 secretion (Figure 5C) and phosphorylation at all 3 sites in hypoxia (Ser101 [52%, P = .0023], Ser119 [48%, P = .013], and Ser169 [76%, P = .0001]) (Figure 5, D–F). These effects were comparable with as detected in normoxia indicated as above with a decrease in secretion and phosphorylation to the same extent (Ser101, 50%; Ser119, 42%; and Ser169, 77%). These data show that TSC2 silencing prevented the increase in IGFBP-1 secretion and phosphorylation in response to hypoxia and that mTORC1 signaling is involved in the hypoxia mediated IGFBP-1 secretion and phosphorylation. mTORC2 signaling, on the other hand, was not affected by TSC2 or raptor siRNA but was significantly inhibited due to hypoxia (88%, P = .0005) or rictor siRNA (78%, P = .0001) (Figure 5B). Together, these data demonstrate that mTORC1 signaling specifically regulates IGFBP-1 secretion/phosphorylation in response to hypoxia.
Rictor silencing on the other hand significantly increased IGFBP-1 phosphorylation at Ser101 and Ser169, whereas an apparent increase in IGFBP-1 secretion and phosphorylation at Ser119 was not significant. Importantly, hypoxia did not increase IGFBP-1 secretion and phosphorylation further in rictor-silenced cells. These data suggest that inhibition of either mTORC1 or mTORC2 are able to drive IGFBP-1 secretion and phosphorylation in hypoxia.
MRM-MS based validation of immunoblot data and discovery of mTOR/hypoxia-responsive IGFBP-1 phosphorylation at Ser174
We finally used MRM-MS as an independent approach to validate Western immunoblot data as well as to aid the discovery of novel/unique IGFBP-1 phosphorylation site linked to mTOR in hypoxia. Targeted MRM-MS strategy allowed for the relative quantification of IGFBP-1 phosphorylation within the hypoxic state and all efforts were assigned for scanning only sequence-specific fragment ions for reliable and technically reproducible detection. IGFBP-1 isolated (IP) from cell media samples pooled from HepG2 cell treatments performed in triplicate (biological replicates) were subjected to MRM-MS analysis and select transitions (MRM-MS) were used to validate phosphorylation at each of the IGFBP-1 phosphorylated residues (Supplemental Figure 4, A–H, and Supplemental Table 2). Each sample was subsequently run in technical replicate. For analysis, all our data were normalized to an internal IGFBP-1-specific peptide. The data is represented as total transition peak intensity (phosphorylated IGFBP-1 peptides) relative to an internal IGFBP-1 standard peptide and compared with untreated (control).
As opposed to pSer101 or pSer169 being detected singly using Western blot analysis, the digests from IGFBP-1 in MRM-MS analysis were able to identify both single and dual phosphorylated peptides, Ser98+Ser101 (Supplemental Figure 4, A–C) and Ser169+Ser174 (Supplemental Figure 4, E–G), compared with an IGFBP-1 internal peptide (Supplemental Figure 4H). Although phosphorylation of IGFBP-1 at Ser98 has been identified previously (16, 39, 40; see also references 59 and 60 bellow), Ser174 phosphorylation has not yet been reported. To better reflect corresponding changes detected by Western blotting, we presented MRM-MS results for Ser101 and Ser169 so that they also include the 2 doubly phosphorylated peptides, Ser98+Ser101 (Figure 6A and Supplemental Figure 4, A–C) and Ser169+Ser174 (Figure 6A and Supplemental Figure 4, E–G). MRM-MS data confirmed that rapamycin increased IGFBP-1 phosphorylation at all sites observed.
Figure 6.
MRM-MS analysis for assessment of IGFBP-1 phosphorylation sites and validation of immunoblot data. A, MRM transitions used to detect relative rapamycin-induced phosphorylation of IGFBP-1 at Ser98, Ser101, Ser119, Ser169, and Ser174 (see Supplemental Table 1 for transition list). B–D, Relative IGFBP-1 phosphorylation induced by rapamycin, hypoxia, or rapamycin+hypoxia-treated HepG2 cells. Distinct phosphorylation sites were detected in IGFBP-1 (IP) samples using MRM-MS. Shown is Ser98+Ser101 value displayed as combined Ser101+dual phosphorylated, Ser98+Ser101; Ser169+Ser174 value displayed as combined Ser169+dual phosphorylated Ser169+Ser174 values. E, Relative IGFBP-1 phosphorylation individually at Ser169, Ser174, and dual phosphorylated Ser169+Ser 174 after DEPTOR siRNA and DEPTOR siRNA+hypoxia treatments. Data are represented as total transition peak intensity relative to untreated control samples (set to a value of 1) and normalized to a nonphosphorylated internal peptide within the IGFBP-1 protein (standard).
We similarly examined IGFBP-1 phosphorylation from HepG2 cell media after hypoxia and combined rapamycin+hypoxia treatments. MRM-MS analysis run in technical replicate detected increased phosphorylation of IGFBP-1 where the relative levels of IGFBP-1 phosphorylation detected in response to rapamycin, hypoxia, and rapamycin+hypoxia were increased to a similar extent at all 3 serine residues (Ser101, Ser119, and Ser169) (Figure 6, B–D, and Supplemental Table 3), corroborating Western blotting findings. Notably, using MRM-MS, we were able to detect dual phosphorylation at Ser98+Ser101 and Ser169+Ser174 and Ser119 singly (Figure 6, B–D). Although Ser169 phosphorylation was increased in hypoxia as we have shown using Western blot analysis, MRM-MS analysis allowed us to distinguish between the sole pSer169 and the combined tandem phosphorylation of Ser169 together with Ser174. Due to lack of pSer174-specific antibody, we were unable to determine whether the total phosphorylation signal on Ser169 detected by Western blotting is cumulative of the 2 phosphorylation events on Ser169/Ser174 peptide. Therefore, with MRM-MS, we have distinguished between the sole pSer169 (which did not appear to be changed during hypoxia in this study) and the combined tandem phosphorylation of Ser169 together with Ser174, which was stimulated.
Considering limitations with Western blot analysis, targeted MRM-MS normalized signals with a standard peptide within IGFBP-1 (internal peptide) yielded highly precise, reliable, and reproducible data. Overall, targeted MRM-MS corroborated Western blotting findings and confirmed that induction of IGFBP-1 phosphorylation due to hypoxia is mediated via inhibition of mTOR signaling. Importantly, MRM-MS analysis was also instrumental in discovery of a novel phosphorylation site Ser174, which is hypoxia/mTOR responsive.
MRM-MS analysis of DEPTOR silencing confirmed reduction in IGFBP-1 phosphorylation at novel site Ser174
Next, using MRM-MS, we validated whether activation of mTORC1+C2 by DEPTOR silencing prevents hypoxia-induced IGFBP-1 phosphorylation. For MRM-MS analysis, pooled cell media samples from HepG2 cell treatments performed in triplicate (biological replicates) into a single sample were used. Each sample was then run in technical replicate to corroborate Western immunoblot findings. MRM-MS analysis of media from cells treated with DEPTOR siRNA demonstrated a marked decrease in IGFBP-1 phosphorylation at all residues to almost undetectable levels. Select transitions were considered specifically for Ser174 for which immunoblot data was not available (due to lack of pSer174 antibody). Because pSer174 was detected singly as well as with pSer169, as shown in Figure 6E and Supplemental Table 4, MRM-MS data is represented individually for evaluation of IGFBP-1 phosphorylation at Ser169, Ser174, as well as the doubly phosphorylated Ser169+Ser174 peptide. There was a marked reduction in IGFBP-1 phosphorylation, most importantly at Ser169+Ser174 residues, which remained the same in hypoxia. Thereby using MRM-MS as a targeted approach with normalization using an internal peptide for absolute measurement of IGFBP-1 phosphorylation, we confirmed a mechanistic role for mTOR signaling in hypoxia-induced IGFBP-1 phosphorylation. Notably these data also highlight the significance of Ser174 phosphorylation singly and/or combined with Ser169 in being an mTOR-sensitive IGFBP-1 phosphorylation site that may be potentially involved in IGF-I binding.
IGFBP-1 structural analysis indicates the newly identified phosphorylation site Ser174 near IGF-I-binding site
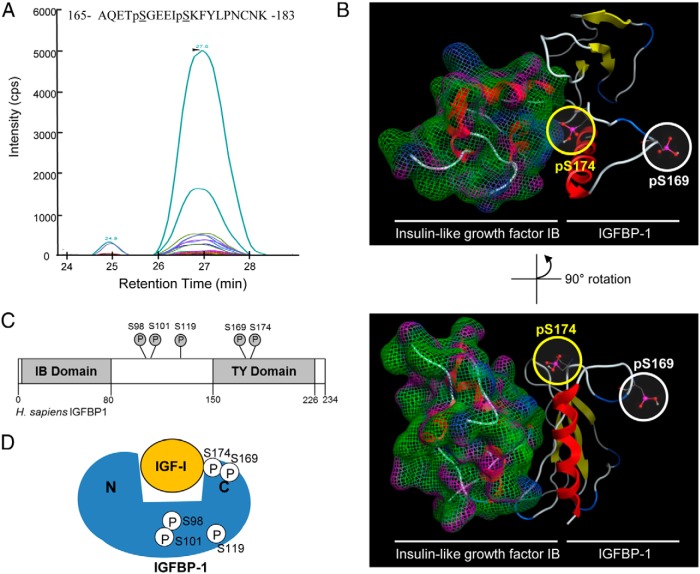
Our aim using structural modeling strategy was to demonstrate the proximity of Ser174 residue to the IGF-I-binding site on IGFBP-1 to determine its potential relevance to IGF-I binding. Synthetic peptides containing either single Ser174 phosphorylation, or double Ser169+Ser174 phosphorylation, were created in order to validate transitions selected to detect and monitor phosphorylation at novel site Ser174. We performed MRM-MS analysis using known transitions that were unique to the Ser174 phosphorylated IGFBP-1 peptide as are shown in Figure 7A and Supplemental Figure 5, A and B.
Figure 7.
Discovery of IGFBP-1 (Ser174) phosphorylation. MRM transitions used for the discovery and detection of dual phosphorylation at Ser169 and Ser174. Colored traces represent the detection and specificity of each transition ion generated specifically from the dual phosphorylated IGFBP-1 at Ser169 and Ser174 (see Supplemental Table 2 for transition list). B, Molecular modeling of the IGFBP C terminus and IGF-IB complex, based on the available crystal structure (2DSQ). Both the phosphorylated Ser174 and Ser169 residues are highlighted. The C terminus of IGFBP-1 was modeled from the available PDB 2DSQ:G crystal structure. C, Schematic of IGFBP-1 and phosphorylation sites (Ser98, Ser101, Ser119, Ser169, and Ser174) in proximity to the IGFBP-1-IGF-binding (IB) and TY domains. Phosphorylation sites Ser98, Ser101, and Ser119 are found within the interdomain region, whereas both Ser169 and Ser174 are within the TY domain. D, Proximity of the studied phosphorylation sites within the IGFBP-1 protein, relative to each other and to the IGF-I-binding site.
To explore a possible function for the Ser174 site, we mapped the proximity for pSer174 binding with IGF-I using molecular modeling where IGFBP-1 protein was modeled (in silico) in complex with IGF-I based on the known crystal structures (PDB entry 2DSQ:G). Shown in Figure 7B are data using a structural modeling illustrating the relative proximity of Ser174 and Ser169 with IGF-I in the thyroglobulin type-I (TY) domain of IGFBP-1. Ser174 in the IGFBP-1-IGF-I complex model was found to be near regions of IGFBP-1 involved in IGF-I binding, providing support for a close interaction of Ser174 with the structured regions of IGFBP-1 involved in IGF-I binding. Although we speculate that phosphorylation at residue Ser174 could affect IGF-I affinity (Figure 7, C and D), further structure-functional data are required to obtain conclusive experimental proof to assign a functional role to this novel residue using site directed mutagenesis (39).
Discussion
We provide novel mechanistic evidence that mTOR inhibition mediates IGFBP-1 hyperphosphorylation in response to hypoxia, which increases the affinity of IGFBP-1 to bind IGF-I resulting in decreased biological action of IGF-I (Figure 1). Importantly, using immunobloting and MRM-MS analysis, our data suggest that both mTORC1 and mTORC2 signaling regulate IGFBP-1 secretion/phosphorylation in a coordinated manner and that inhibition of either complex is sufficient to cause hypoxia induced IGFBP-1 hyperphosphorylation. Additionally, using MRM-MS, we identified a novel IGFBP-1 phosphorylation site (Ser174), which was mTOR/hypoxia-sensitive and is near the structural region containing the IGF-I-binding site. These findings support the main tenets of our overall hypothesis and are consistent with the possibility that increased IGFBP-1 phosphorylation in response to hypoxia is mediated by mTOR inhibition to regulate the IGF-I actions in restriction of fetal growth.
Elevated IGFBP-1 has been proposed to be an important mechanism restricting fetal growth in both human FGR and in animal models of chronic intrauterine hypoxia (8, 12–14). Hypoxia up-regulates IGFBP-1 mRNA and protein expression in vitro (5, 16), and in vivo (8). It is known that mTORC1 promotes the expression of hypoxia-inducible factor-1α, which activates the transcription of several genes responsive to hypoxia (45), including IGFBP-1 (5). Hypoxia-inducible factors provide a molecular framework for the hypoxic control of differentiation in many cell types (46–47). Our previous data demonstrate that hypoxia also results in IGFBP-1 hyperphosphorylation, which markedly increases its binding affinity to IGF-I and inhibits IGF-I-stimulated cell growth (16). We recently provided evidence that IGFBP-1 is hyperphosphorylated in human FGR and established an association between IGFBP-1 hyperphosphorylation and mTOR inhibition in baboon FGR in vivo (32). However, if mTOR inhibition is mechanistically linked to IGFBP-1 hyperphosphorylation in response to hypoxia remained unknown.
It is recognized that mTORC1 signaling is inhibited by hypoxia (28, 29). With respect to mTORC2, previous studies have generated mixed results, reporting both increased and/or decreased mTORC2 activity in response to hypoxia (48). Here, we investigated the mTOR-mediated mechanistic link between hypoxia and IGFBP-1 phosphorylation using HepG2 cells as a model for human fetal hepatocytes (32–35). We performed our study using 1% O2 (hypoxia) and 20% O2 (normoxia), which were based on animal models of hypoxia in vivo (49–53) and our previous in vitro studies (16, 38). Using systematic mTOR inhibition/activation strategies, we demonstrated that inhibition of either mTORC1 or mTORC2 was sufficient to induce IGFBP-1 secretion/phosphorylation to the same extent as with hypoxia alone. Importantly, hypoxia did not induce IGFBP-1 secretion/phosphorylation further in cells where mTORC1 or mTORC2 signaling was inhibited. This suggests that inhibition of either mTORC is able to induce IGFBP-1 secretion/phosphorylation in response to hypoxia. Furthermore, additional support for the involvement on mTOR signaling in linking hypoxia to IGFBP-1 was generated by DEPTOR silencing, which activated mTORC1+mTORC2 (54–56) and reduced IGFBP-1 secretion/phosphorylation as well as prevented increase in IGFBP-1 in response to hypoxia. These findings are highly consistent with the hypothesis that both mTORC1 and mTORC2 are involved cooperatively in the regulation of total and phosphorylated IGFBP-1.
We targeted TSC2, which is a negative regulator of mTORC1 (57, 58) and, by silencing TSC2, which specifically activates mTORC1; we confirmed that mTORC1 activation decreases IGFBP-1 secretion/phosphorylation. Furthermore, TSC2-mediated mTORC1 activation prevented the hypoxia-induced increases in IGFBP-1 secretion/phosphorylation, providing direct mechanistic evidence for an important role for mTORC1 linking hypoxia to total and phosphorylated IGFBP-1. Together, these data suggest that induction of total and phosphorylated IGFBP-1 can be mediated by signaling events which affect either mTORC, representing a wide array of possible mechanisms by which IGF-I bioavailability and fetal growth, can be regulated. Importantly, application of MRM-MS corroborated our immunoblot data, providing additional evidence for a mechanistic link between hypoxia, mTOR inhibition, and IGFBP-1 phosphorylation.
Our earlier data have shown that regulation of IGF-I bioavailability in hypoxia predominantly occurs due to an increase in the abundance of IGFBP-1 phosphoisoforms (site and degree) rather than increases in total IGFBP-1 levels or even total phosphorylated IGFBP-1 (16). These data provide support for the model that hypoxia in FGR inhibits mTOR, resulting in increased IGFBP-1 phosphorylation, which elicits functional effects on IGF-I signaling in FGR. It is known that IGFBP-1 can be phosphorylated at Ser95, Ser98, Ser101, and Ser119 in the midlinker region and Ser169 in the C-terminal domain (Figure 7C) (15, 16, 39, 40, 41, 59, 60). Using LC-MS we have earlier shown IGFBP-1 hyperphosphorylation at Ser98/101/119 and Ser169 in FGR (41) and in response to hypoxia in vitro (16). We also previously confirmed the functional role of these 4 residues using mutagenesis (9). In the current study, using highly sensitive MRM-MS, we have now identified a novel residue Ser174, which was hyperphosphorylated in a doubly phosphorylated peptide with Ser169 in a rapamycin and hypoxia-sensitive manner. This suggests that mTOR inhibition affects phosphorylation of more than one residue at the C terminus of IGFBP-1. Phosphorylation at Ser174, singly or in combination has, to the best of our knowledge, not been reported previously.
Using structural modeling, we demonstrated earlier that Ser169 is localized near IGF-binding site (41). The N-terminal region of IGFBP-1 has been implicated in IGF binding (61). In silico structural analysis in the current study demonstrated a close proximity between the pSer174 residue and the IGF-I-binding site. Although structure-functional proof is currently lacking, we speculate that increased phosphorylation at Ser169 together with Ser174 at the C-terminal end may cooperatively function to increase the binding affinity of IGFBP-1 to IGF-I through much stronger interactions. This effect, in tandem with other IGFBP-1 phosphorylation sites in the linker region could potentially prolong binding of IGF-I to IGFBP-1 and enhance the function of IGFBP-1 in reducing IGF-I bioavailability. These findings suggest that decreased IGF-I bioavailability in response to hypoxia in utero mediated by mTOR inhibition may involve a previously unknown synergistic effect of IGFBP-1 hyperphosphorylation at Ser174 combined with Ser169 (C-terminal), or possibly other residues in the mid linker region.
Previously, an in vivo study using chick embryos demonstrated that both hypoxia and nutrient deprivation were associated with FGR (3); however, prenatal hypoxia and undernutrition may have differential effects on fetal development (62) possibly involving diverse regulatory mechanisms. Recently, we showed that amino acid response pathway plays a key role in modulating IGFBP-1 phosphorylation in response to leucine deprivation (63). Whether IGFBP-1 phosphorylation at newly discovered Ser174 along with Ser169 is mTOR/hypoxia responsive or is involved in amino acid deprivation-induced mechanisms is unknown. It is nonetheless an intriguing possibility that synergistic interactions of multiple IGFBP-1 residues, provide a unique and more sophisticated mechanism for regulation of IGF-I bioavailability under diverse cellular environments (64).
Together, using 2 independent strategies, this study has demonstrated for the first time that signaling through either the mTORC1 or mTORC2 pathway is sufficient to induce effects on IGF-I activity in response to hypoxia. We have also identified a new major phosphorylation site Ser174 in IGFBP-1, which is regulated by hypoxia and mTOR, and additional studies are required to elucidate its functional role in binding IGF-I. These data have demonstrated a novel mechanistic link between hypoxia and IGFBP-1 hyperphosphorylation mediated by inhibition of mTOR, which we propose contributes to reduced IGF-I-mediated fetal growth in response to hypoxia.
Additional material
Supplementary data supplied by authors.
Acknowledgments
S.S.-C.L. holds a Canada Research Chair in Functional Genomics and Cellular Proteomics.
This work was supported by the National Institute of Health Grant HD 078313 (to M.B.G. and T.J.) and in part by Natural Science and Engineering Council of Canada (NSERC) (Discovery grant), the Lawson Health Research Institute, the Children's Health Research Institute, University of Western Ontario, Canada (M.B.G.) and the Canadian Cancer Society (S.S.-C.L.). I.D. received Children's Health Research Institute and Department of Pediatrics (UWO) Graduate Student Scholarships. K.K.B. is the recipient of an NSERC Banting fellowship.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- 2-D
- 2-dimensional
- Akt
- a serine/threonine kinase also known as protein kinase B
- DEPTOR
- DEP-domain-containing and mTOR-interacting protein
- 4E-BP1
- eukaryotic translation initiation factor 4E binding protein 1
- FBS
- fetal bovine serum
- FGR
- fetal growth restriction
- IGF-1R
- IGF-I receptor
- IGF
- binding protein-1(IGFBP-1)
- IP
- immunoprecipitate
- MRM
- multiple reaction monitoring
- MS
- mass spectrometry
- mTOR
- mechanistic target of rapamycin
- mTORC
- mTOR complex
- RNAi
- RNA interference
- siRNA
- small interfering RNA
- TSC2
- tuberous sclerosis complex 2
- TY
- thyroglobulin type-I.
References
- 1. Eleftheriades M, Creatsas G, Nicolaides K. Fetal growth restriction and postnatal development. Ann NY Acad Sci. 2006;1092:319–330. [DOI] [PubMed] [Google Scholar]
- 2. Kinzler WL, Vintzileos AM. Fetal growth restriction: a modern approach. Curr Opin Obstet Gynecol. 2008;20:125–131. [DOI] [PubMed] [Google Scholar]
- 3. Ruijtenbeek K, Kessels LC, De Mey JG, Blanco CE. Chronic moderate hypoxia and protein malnutrition both induce growth retardation, but have distinct effects on arterial endothelium-dependent reactivity in the chicken embryo. Pediatr Res. 2003;53:573–579. [DOI] [PubMed] [Google Scholar]
- 4. Randhawa R, Cohen P. The role of the insulin-like growth factor system in prenatal growth. Mol Genet Metab. 2005;86:84–90. [DOI] [PubMed] [Google Scholar]
- 5. Tazuke SI, Mazure NM, Sugawara J, et al. Hypoxia stimulates insulin-like growth factor binding protein 1 (IGFBP-1) gene expression in HepG2 cells: a possible model for IGFBP-1 expression in fetal hypoxia. Proc Natl Acad Sci USA. 1998;95:10188–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verhaeghe J, Van Herck E, Billen J, Moerman P, Van Assche FA, Giudice LC. Regulation of insulin-like growth factor-I and insulin-like growth factor binding protein-1 concentrations in preterm fetuses. Am J Obstet Gynecol. 2003;188:485–491. [DOI] [PubMed] [Google Scholar]
- 7. Lee PD, Giudice LC, Conover CA, Powell DR. Insulin-like growth factor binding protein-1: recent findings and new directions. Proc Soc Exp Biol Med. 1997;216:319–357. [DOI] [PubMed] [Google Scholar]
- 8. Kajimura S, Aida K, Duan C. Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc Natl Acad Sci USA. 2005;102:1240–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Langford K, Blum W, Nicolaides K, Jones J, McGregor A, Miell J. The pathophysiology of the insulin-like growth factor axis in fetal growth failure: a basis for programming by undernutrition? Eur J Clin Invest. 1994;24:851–856. [DOI] [PubMed] [Google Scholar]
- 10. Chard T. Insulin-like growth factors and their binding proteins in normal and abnormal human fetal growth. Growth Regul. 1994;4:91–100. [PubMed] [Google Scholar]
- 11. El Khattabi I, Remacle C, Reusens B. The regulation of IGFs and IGFBPs by prolactin in primary culture of fetal rat hepatocytes is influenced by maternal malnutrition. Am J Physiol Endocrinol Metab. 2006;291:E835–E842. [DOI] [PubMed] [Google Scholar]
- 12. Giudice LC, Martina NA, Crystal RA, Tazuke S, Druzin M. Insulin-like growth factor binding protein-1 at the maternal-fetal interface and insulin-like growth factor-I, insulin-like growth factor-II, and insulin-like growth factor binding protein-1 in the circulation of women with severe preeclampsia. Am J Obstet Gynecol. 1997;176:751–757; discussion 757–758. [DOI] [PubMed] [Google Scholar]
- 13. Grobman WA, Kazer RR. Serum insulin, insulin-like growth factor-I, and insulin-like growth factor binding protein-1 in women who develop preeclampsia. Obstet Gynecol. 2001;97:521–526. [DOI] [PubMed] [Google Scholar]
- 14. Ingec M, Gursoy HG, Yildiz L, Kumtepe Y, Kadanali S. Serum levels of insulin, IGF-I, and IGFBP-1 in pre-eclampsia and eclampsia. Int J Gynaecol Obstet. 2004;84:214–219. [DOI] [PubMed] [Google Scholar]
- 15. Jones JI, Busby WH, Jr, Wright G, Smith CE, Kimack NM, Clemmons DR. Identification of the sites of phosphorylation in insulin-like growth factor binding protein-1. Regulation of its affinity by phosphorylation of serine 101. J Biol Chem. 1993;268:1125–1131. [PubMed] [Google Scholar]
- 16. Seferovic MD, Ali R, Kamei H, et al. Hypoxia and leucine deprivation induce human insulin-like growth factor binding protein-1 hyperphosphorylation and increase its biological activity. Endocrinology. 2009;150:220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hall MN. mTOR-what does it do? Transplant Proc. 2008;40:S5–S8. [DOI] [PubMed] [Google Scholar]
- 18. Hara K, Maruki Y, Long X, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. [DOI] [PubMed] [Google Scholar]
- 19. Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. [DOI] [PubMed] [Google Scholar]
- 20. Jacinto E, Facchinetti V, Liu D, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. [DOI] [PubMed] [Google Scholar]
- 21. Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. [DOI] [PubMed] [Google Scholar]
- 22. Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. [DOI] [PubMed] [Google Scholar]
- 23. Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sarbassov DD, Ali SM, Sengupta S, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. [DOI] [PubMed] [Google Scholar]
- 25. Oh WJ, Wu CC, Kim SJ, et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci. 2009;34:620–627. [DOI] [PubMed] [Google Scholar]
- 27. Dada S, Demartines N, Dormond O. mTORC2 regulates PGE2-mediated endothelial cell survival and migration. Biochem Biophys Res Commun. 2008;372:875–879. [DOI] [PubMed] [Google Scholar]
- 28. Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol Cell. 2010;40:509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. [DOI] [PubMed] [Google Scholar]
- 30. Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. [DOI] [PubMed] [Google Scholar]
- 31. Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. [DOI] [PubMed] [Google Scholar]
- 32. Abu Shehab M, Damerill I, Shen T, et al. Liver mTOR controls IGF-I bioavailability by regulation of protein kinase CK2 and IGFBP-1 phosphorylation in fetal growth restriction. Endocrinology. 2014;155:1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maruyama M, Matsunaga T, Harada E, Ohmori S. Comparison of basal gene expression and induction of CYP3As in HepG2 and human fetal liver cells. Biol Pharm Bull. 2007;30:2091–2097. [DOI] [PubMed] [Google Scholar]
- 34. Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31:1035–1042. [DOI] [PubMed] [Google Scholar]
- 35. Kelly JH, Darlington GJ. Modulation of the liver specific phenotype in the human hepatoblastoma line Hep G2. In Vitro Cell Dev Biol. 1989;25:217–222. [DOI] [PubMed] [Google Scholar]
- 36. Liu H, Galka M, Mori E, et al. A method for systematic mapping of protein lysine methylation identifies functions for HP1β in DNA damage response. Mol Cell. 2013;50:723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD. Multiple reaction monitoring to identify sites of protein phosphorylation with high sensitivity. Mol Cell Proteomics. 2005;4:1134–1144. [DOI] [PubMed] [Google Scholar]
- 38. Seferovic MD, Chen S, Pinto DM, Gupta MB. Altered liver secretion of vascular regulatory proteins in hypoxic pregnancies stimulate angiogenesis in vitro. J Proteome Res. 2011;10:1495–1504. [DOI] [PubMed] [Google Scholar]
- 39. Abu Shehab M, Iosef C, Wildgruber R, Sardana G, Gupta MB. Phosphorylation of IGFBP-1 at discrete sites elicits variable effects on IGF-I receptor autophosphorylation. Endocrinology. 2013;154:1130–1143. [DOI] [PubMed] [Google Scholar]
- 40. Nissum M, Abu Shehab M, Sukop U, et al. Functional and complementary phosphorylation state attributes of human insulin-like growth factor-binding protein-1 (IGFBP-1) isoforms resolved by free flow electrophoresis. Mol Cell Proteomics. 2009;8:1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abu Shehab M, Khosravi J, Han VK, Shilton BH, Gupta MB. Site specific IGFBP-1 hyper-phosphorylation in fetal growth restriction: clinical and functional relevance. J Proteome Res. 2010;9:1873–1881. [DOI] [PubMed] [Google Scholar]
- 42. MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. [DOI] [PubMed] [Google Scholar]
- 44. Sitar T, Popowicz GM, Siwanowicz I, Huber R, Holak TA. Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proc Natl Acad Sci USA. 2006;103:13028–13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Düvel K, Yecies JL, Menon S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mohyeldin A, Garzón-Muvdi T, Quiñones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–161. [DOI] [PubMed] [Google Scholar]
- 47. Csete M. Oxygen in the cultivation of stem cells. Ann NY Acad Sci. 2005;1049:1–8. [DOI] [PubMed] [Google Scholar]
- 48. Li W., Petrimpol M., Molle K. D., Hall M. N., Battegay E. J., Humar R. Hypoxia-induced endothelial proliferation requires both mTORC1 and mTORC2. Circ Res. 2007;100:79–87. [DOI] [PubMed] [Google Scholar]
- 49. Tchirikov M, Eisermann K, Rybakowski C, Schröder HJ. Doppler ultrasound evaluation of ductus venosus blood flow during acute hypoxemia in fetal lambs. Ultrasound Obstet Gynecol. 1998;11:426–431. [DOI] [PubMed] [Google Scholar]
- 50. Semenza G. L. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–78. [DOI] [PubMed] [Google Scholar]
- 51. Wu D, Yotnda P. (2011) Induction and testing of hypoxia in cell culture. J Vis Exp. 2011; 12 (54): pii: 2899. doi: 10.3791/2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ivanovic Z. Hypoxia or in situ normoxia: The stem cell paradigm. J Cell Physiol. 2009;219:271–5. [DOI] [PubMed] [Google Scholar]
- 53. Mottet D, Dumont V, Deccache Y, et al. Regulation of hypoxia-inducible factor-1α protein level during hypoxic conditions by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3β pathway in HepG2 cells. J Biol Chem. 2003;278:31277–31285. [DOI] [PubMed] [Google Scholar]
- 54. Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang D, Li L, Liu H, et al. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ. 2013;20:235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kazi AA, Hong-Brown L, Lang SM, Lang CH. Deptor knockdown enhances mTOR activity and protein synthesis in myocytes and ameliorates disuse muscle atrophy. Mol Med. 2011;17:925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14–3-3 shuttling. Genes Dev. 2008;22:239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dolcini L, Sala A, Campagnoli M, et al. Identification of the amniotic fluid insulin-like growth factor binding protein-1 phosphorylation sites and propensity to proteolysis of the isoforms. FEBS J. 2009;276:6033–6046. [DOI] [PubMed] [Google Scholar]
- 60. Temporini C, Dolcini L, Abee A, et al. Development of an integrated chromatographic system for on-line digestion and characterization of phosphorylated proteins. J Chromatogr A. 2008;1183:65–75. [DOI] [PubMed] [Google Scholar]
- 61. Brinkman A, Kortleve DJ, Schuller AG, Zwarthoff EC, Drop SL. Site-directed mutagenesis of the N-terminal region of IGF binding protein 1; analysis of IGF binding capability. FEBS Lett. 1991;291:264–268. [DOI] [PubMed] [Google Scholar]
- 62. Camm EJ, Hansell JA, Kane AD, et al. Partial contributions of developmental hypoxia and undernutrition to prenatal alterations in somatic growth and cardiovascular structure and function. Am J Obstet Gynecol. 2010;203;495.e24–495.e34. [DOI] [PubMed] [Google Scholar]
- 63. Malkani N, Jansson T, Gupta MB. IGFBP-1 hyperphosphorylation in response to leucine deprivation is mediated by the AAR pathway. Mol Cell Endocrinol. 2015;412:182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gupta MB. The role and regulation of IGFBP-1 phosphorylation in fetal growth restriction. J Cell Commun Signal. 2015;9:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.