Abstract
The 2 most abundant human pancreatic islet cell types are insulin-producing β-cells and glucagon-producing α-cells. Defined cis-regulatory elements from rodent Insulin genes have permitted genetic labeling of human islet β-cells, enabling lineage tracing and generation of human β-cell lines, but analogous elements for genetically labeling human α-cells with high specificity do not yet exist. To identify genetic elements that specifically direct reporter expression to human α-cells, we investigated noncoding sequences adjacent to the human GLUCAGON and ARX genes, which are expressed in islet α-cells. Elements with high evolutionary conservation were cloned into lentiviral vectors to direct fluorescent reporter expression in primary human islets. Based on the specificity of reporter expression for α- and β-cells, we found that rat glucagon promoter was not specific for human α-cells but that addition of human GLUCAGON untranslated region sequences substantially enhanced specificity of labeling in both cultured and transplanted islets to a degree not previously reported, to our knowledge. Specific transgene expression from these cis-regulatory sequences in human α-cells should enable targeted genetic modification and lineage tracing.
Islet β-cells are defective or destroyed in diabetes mellitus. Replacement of β-cells from other cell types is one approach for diabetes therapy and a current focus of intensive investigations worldwide (1). In mice with extensive β-cell loss or targeted genetic modification (2–4), α-cells are capable of conversion into progeny resembling β-cells, an interpretation substantiated by lineage tracing. Whether α-to-β-cell conversion is possible in human tissue remains unclear, primarily because methods for indelible genetic labeling of human α-cells do not yet exist, to our knowledge. Thus, assessment of human α-cell lineage conversion has been limited to identifying coexpression of glucagon and other islet hormones (5). By contrast, genetic elements specific for human β-cells have enabled lineage tracing of β-cell dedifferentiation during in vitro culture or during pancreatic development, including characterization of ARX-dependent β-cell conversion to α-like cells during in vitro culture (6–8). Thus, identifying genetic tools to label human α-cells with high specificity could enable measurement of cell conversion and accelerate regenerative approaches to diabetes therapy.
Human α-cell biology would also benefit from generation of immortalized cell lines with appropriately regulated glucagon secretion. The use of genetic elements that specify gene expression in human β-cells has led recently to development of the first human β-cell lines (9, 10). Using rat Ins1 promoter (RIP) elements, lentiviral expression of SV40, Simian Virus 40, large T-antigen and human telomerase reverse transcriptase (11) led to successful transformation and immortalization of human fetal pancreatic cells with regulated insulin secretion (9, 12). Thus, identification of DNA elements sufficient to direct transgene expression to human α-cells could similarly lead to development of new human α-cell lines.
Here, we report identification of DNA sequences that direct expression of a reporter gene in primary human α-cells. We focused on candidate regulatory elements at the GLUCAGON and ARX loci, as expression of these genes is restricted to α-cells in human islets. The genetic α-cell labeling methods described here could enable long-term lineage tracing, cell purification, or generation of immortalized human α-cell lines.
Materials and Methods
Human tissue
All studies were conducted in accordance with Stanford University Institutional Review Board guidelines. Human islet tissue from adult donors with body mass index less than 30 and without a clinical history of diabetes mellitus was obtained through the Integrated Islet Distribution Program.
Enhancer cloning and lentivirus production
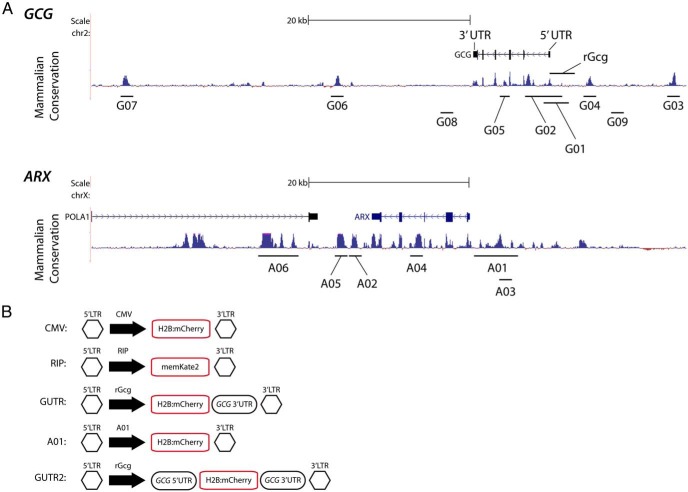
Candidate conserved DNA elements were identified by assessing mammalian conservation (PhastCons and PhyloP algorithms) (13, 14) at the human GLUCAGON and ARX loci in the UCSC Genome Browser. Sequences larger than 300 bp were considered. Primer sequences used to amplify each region are provided in Supplemental Table 1, and further details regarding candidate regions are presented in Supplemental Table 2. Candidate regions were amplified by PCR using Accuprime Pfx DNA polymerase (Life Technologies) from bacterial artificial chromosomes encompassing the GLUCAGON (RP11–664H19, CHORI) and ARX (RP11–56L21, CHORI) loci. PCR products were purified and cloned into pCDH lentiviral plasmids (System Biosciences) with the Histone H2B:mCherry cDNA (20972; Addgene) and a pTA minimal promoter (Clontech). To generate the pCDH-RIP-memKate2 plasmid, the 361-bp sequence upstream of the rat Ins1 transcription start site was cloned into pCDH to direct expression of palmitoylated mKate2 (memKate2; Evrogen) sequence, which directs expression of mKate2 to the cell membrane. pTA promoter sequences were omitted for constructs expected to have promoter activity, including CMV, RIP, rat Glucagon promoter (rGcg), and A01 regions, and the rGcg derivatives Glucagon promoter untranslated region (GUTR) and GUTR2. Construct generation was confirmed by restriction digestion and Sanger sequencing. The rGcg sequence (15) was a gift from Dr P. Herrera (University of Geneva).
Lentiviruses were produced by transient transfection of 293T cells with pCDH lentiviral backbone vectors and pMD2.G (12259; Addgene) and psPAX2 (12260; Addgene) packaging constructs. Turbofect reagents were used for transfection (Life Technologies). Supernatants were collected and purified using PEG-it (System Biosciences). Concentrated lentivirus was stored at −80°C for transduction of primary human cells.
All constructs generated in this manuscript are available upon request. Identifying sequence characteristics (size, position, 5′, and 3′ primer sequences) are included in Supplemental Tables 1 and 2.
Islet culture
Upon receipt, islets were cultured in ultralow-attachment plates and infected with lentivirus overnight as intact islets. Approximately 250 islets were tested from each donor for each candidate DNA element for in vitro labeling experiments. Multiplicities of infection on the order of 105-106 viral particles per islet were used. Islets were cultured in RPMI 1640 (100-mg/dL glucose), 10% fetal bovine serum (HyClone, GE Life Sciences), and 1% penicillin-streptomycin. The next day, islets were washed with culture media, embedded in Matrigel (CB-40230; Fisher Scientific), and cultured for 4 days. Matrigel was used to maintain islet architecture and enhance islet survival over the culture period (16). Fluorescence was evaluated by epifluorescence microscopy after 48 hours to confirm islet labeling by CMV-driven expression constructs. After 4 days, islets were released from Matrigel by digestion with Dispase (Life Technologies), washed with PBS, embedded in collagen gel (EMD Millipore), and fixed in 4% paraformaldehyde overnight at 4°C. Samples were embedded in optimal cutting temperature (OCT) compound and cryosectioned for histological analysis.
Transplantation in the renal capsular space
All animal experiments were performed in accordance with guidelines from the Stanford University Institutional Animal Care and Use Committee. For transplantation experiments, islets were processed as for in vitro culture, but 1000 islets were tested per candidate DNA element. Islets were resuspended in cold Matrigel and transferred into the left renal capsular space of host animals using a glass microcapillary tube. Transplant recipients were 2- to 4-month-old male NOD scid IL2Rγnull mice (stock number 005557; The Jackson Laboratory) and were anesthetized using ketamine/xylazine. Appropriate depth of anesthesia was confirmed by lack of toe-pinch response. After 2 weeks, kidneys with grafts were removed, fixed in 4% paraformaldehyde, and processed for cryosectioning and immunohistology.
Immunostaining and cell counting
Islet sections were immunostained as previously described (17). Primary antibodies used were guinea pig polyclonal antiglucagon (1:5000, M182; Clontech), goat polyclonal anti-PDX1 (1:250, AF2419; R&D Systems), goat polyclonal anti-somatostatin (1:500, sc-7189; Santa Cruz Biotechnologies, Inc), goat polyclonal anti-ghrelin (1:200, sc-10368; Santa Cruz Biotechnologies, Inc), and mouse monoclonal anti-mCherry 1C51 (1:500, ab125096; Abcam). PDX1 staining was used to identify a cell population enriched for β- and δ-cells and to facilitate unambiguous identification of nuclear mCherry+ cells. Contaminating ductal epithelial cells would also be PDX1+. Percentages in some experiments add to greater than 100% because of occasional coexpression of GCG and PDX1, as reported by others (6). Samples were imaged using an AxioM1 microscope with ×20 and ×40 objectives.
Cells were counted using the ImageJ Cell Counter plugin (18). Total numbers of H2B:mCherry-labeled cells were calculated for at least 5 images (×20 magnification) from each islet group, and mCherry+ cells were subsequently scored for expression of either GCG or PDX1. Data were exported to Microsoft Excel and GraphPad Prism for analysis and presentation.
Results and Discussion
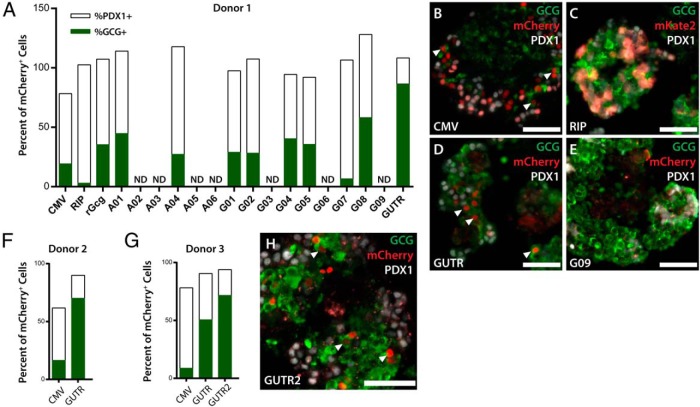
To assess the specificity of transgene expression from cis-regulatory DNA sequences in human α-cells, we used a lentiviral strategy to infect intact primary human islets. Each lentivirus encoded a fluorescent reporter and a minimal promoter fused to a candidate cis-regulatory sequence (Figure 1, A and B). Lentiviral transduction of cultured islets using a CMV reporter labeled approximately 30%, of all α-cells, ranging from 11% to 34% (Supplemental Table 3). Lentiviruses were used to transduce primary human islets from adult tissue donors with each candidate construct; then the frequency of GCG+ cells and PDX1+ cells as a percentage of total H2B:mCherry+ cells was calculated after 4 days of in vitro culture. In this setting, 99.6% of reporter expression from the RIP colocalized in PDX1+ cells (Figure 2, A and C), indicating that specific expression of transgenes was feasible in islet cells using lentiviral delivery of regulatory elements, as previously reported (7). By contrast, the nonspecific CMV promoter directed H2B:mCherry reporter expression to a population in which 19% of H2B:mCherry+ cells were GCG+ and 59% were PDX1+ (Figure 2, A and B). Previously, a 1.6-kb rGcg element was used to label mouse islet α-cells with high specificity in vivo (15). This element is 58% conserved between rat and human, is not highly conserved in mammals, and in humans aligns directly upstream of the transcription start site (positions −1541 to +206) (Figure 1A). However, this element only achieved 35% H2B:mCherry+ GCG+ colabeling in primary human islets, whereas 72% of H2B:mCherry+ cells were PDX1+ (Figure 2A). Thus, although transgene expression from the rGcg element was readily detected in primary human islets, this element only marginally enhanced the specificity of α-cell transgene expression compared with a CMV promoter. These results indicating divergent regulation in rodents and humans are consistent with data from previous studies (19). Therefore, we sought additional cis-regulatory elements to enhance the specificity of human α-cell genetic labeling.
Figure 1.
Identification of candidate regulatory sequences at the GCG and ARX loci. A, Identification of evolutionarily conserved regions near the human GLUCAGON and ARX loci. B, Design of lentiviral constructs for assessing labeling of human α-cells. CMV, CMV promoter; G, GCG; A, ARX. Detailed information regarding candidate DNA sequences is included in Supplemental Tables 1 and 2.
Figure 2.
Labeling of α-cells in vitro. A, Initial testing of labeling specificity of 17 DNA elements by lentiviral transduction of primary human islets. Percent of reporter+ cells expressing PDX1 is shown in white, percent expressing GCG is shown in green. Elements that caused expression in 10 or fewer cells were excluded from analysis (ND). B–E, Representative images from immunofluorescence analysis demonstrating labeling of islet cells by lentiviral transduction. F–H, Repeated in vitro labeling experiments with GUTR construct and initial testing of GUTR2 construct, which includes both 5′ and 3′ UTR sequences from the human GCG gene. Arrowheads label GCG+ mCherry+ cells. CMV, CMV promoter; G, GCG; A, ARX; ND, not detected. Detailed cell counting data in Supplemental Table 4. Scale bars, 100 μm.
We identified evolutionarily conserved regions within a 50-kb region, including approximately 20-kb 5′ and 20-kb 3′ to the human GLUCAGON and ARX loci, and assessed their capacity to direct reporter expression specifically to α-cells (Figure 1A). In addition, we analyzed candidate human GLUCAGON regulatory elements identified in previous studies (19–21). We also assessed human GLUCAGON untranslated regions (UTRs) based on findings in many systems that gene expression can be modulated through interactions with mRNA UTRs (22–25). For example, previous work suggests that 3′ and 5′ UTRs in rodent Glucagon mRNA may regulate expression levels or specificity in cell lines (26–28). Here, we tested whether flanking H2B:mCherry reporter sequences with human GLUCAGON 3′ and 5′ UTRs could increase reporter specificity.
We tested a total of 19 candidate regulatory regions, including control CMV and RIPs (Figure 1A), by cloning each candidate into a lentiviral vector with a minimal promoter and a reporter transgene. After transduction of primary islets from an index human tissue donor with reporter lentiviruses, we found that fusing the human GLUCAGON 3′ UTR to a reporter construct expressed from the rGcg promoter (GUTR) substantially enhanced specificity for α-cells (19% with CMV, 35% with rGcg, 87% with GUTR) (Figure 2, A–E). Only a subset of constructs including putative regulatory elements at the ARX locus directed any expression in islet cells, potentially related to specific roles for ARX in other organs including the central nervous system or to possible repressive effects on gene expression by specific regulatory elements. Despite variability in baseline α-cell labeling frequency, the GUTR construct consistently enhanced reporter specificity for α-cells in 2 additional islet donors (Figure 2, D, F, and G). Based on the improved specificity with the GCG 3′ UTR, we reasoned that incorporation of additional elements contributing to post-transcriptional regulation might further enhance reporter specificity for α-cells. We found that the combination of the GCG 5′ and 3′ UTRs flanking H2B:mCherry and expressed from the rGcg element (GUTR2) increased specific labeling of GCG+ cells compared with the GUTR construct (Figure 2, G and H). Thus, inclusion of UTR elements enhanced the specificity of in vitro α-cell labeling in cultured human islets.
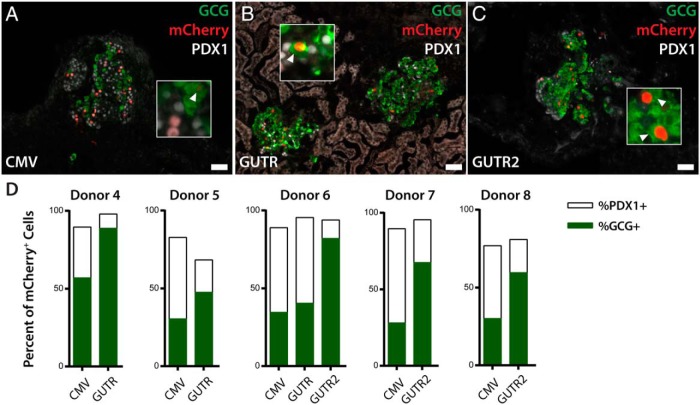
Dedifferentiation of islet cells in vitro (6, 8, 29) could complicate interpretation of lineage labeling. For example, human β-cells begin to express α-cell markers including ARX during in vitro culture (6). To address this concern, we used the lentiviral constructs with the highest specific in vitro labeling of GCG+ cells for studies of islets after transplantation. After lentiviral transduction, primary human islets were transplanted into the renal subcapsular space of immunodeficient mice, and the specificity of GCG+ cell labeling was quantified after 2 weeks. Cells that coexpressed PDX1 and GLUCAGON proteins were rare, present at a frequency of 1.7 ± 0.4% (mean ± SEM). Human ARX conserved upstream sequences labeled GCG+ cells in transplants with higher specificity than during in vitro experiments, although we observed substantial donor-to-donor variability in specificity (Supplemental Figure 1). This variability could derive from differences in islet preparation or purity, integration sites of reporters, or donor genetic characteristics. Similar to in vitro labeling experiments, we observed that the GUTR and GUTR2 constructs labeled GCG+ cells with greater specificity than CMV promoter controls (Figure 3, A–D). These constructs reduced the frequency of labeled somatostatin+ δ-cells by greater than 70% relative to CMV controls, from 4%–6% to 1%–1.8% (Supplemental Figure 2). Ghrelin+ cells were rare in these studies and were not labeled by CMV or GUTR-based constructs (data not shown). Overall, labeling specificities with GUTR and GUTR2 in transplanted islets were higher than in donor-matched islets infected and cultured in vitro (Figure 3, A–D). Thus, the elements described here are capable of specifically labeling human α-cells both in vitro and in transplanted islets.
Figure 3.
Labeling of α-cells in transplanted islets. A–C, Immunofluorescence analysis of lentiviral labeling of islets transplanted into the renal subcapsular space for 14 days. Insets highlight mCherry+ cells that also express GCG. D, Quantification of islet α- and β-cell labeling frequencies in transplanted islets from multiple donors. Percent of H2B:mCherry+ cells expressing PDX1 is shown in white, percent expressing GCG is shown in green. Arrowheads label GCG+ mCherry+ cells. Abbreviations as for Figure 1. Scale bars, 100 μm.
The experiments presented here have identified effects of human GLUCAGON 5′ and 3′ UTR elements on reporter specificity. These results suggest a role for posttranscriptional mechanisms in enhancing the specificity of hormone expression in human islet cell subsets. This is consistent with previous findings from studies of the rat Glucagon 5′ UTR, which suggested UTR sequences enhanced expression of Glucagon in islet, but not nonislet, cell lines (26, 27). The need for post-transcriptional mechanisms to control protein expression in islet cells is also implied by findings that single islet cells simultaneously express mRNAs encoding multiple hormones, including Insulin and Glucagon transcripts (30, 31). Precedence for restricted translation of specific transcripts in specific cell types through UTR-based regulation exists. For example, high-level Pax3 expression is maintained in muscle stem cells through incorporation of an alternative 3′ UTR form in progenitors compared with differentiating cells (23), and 5′ UTR-based translational regulation influences Hox gene expression in anterior-posterior patterning (32).
In summary, we have identified DNA sequences sufficient to direct reporter gene expression specifically in human α-cells in vitro and in transplanted islets. In the best cases, we achieved genetic labeling of GCG+ cells approaching 90% specificity. In contrast to previous studies of human GLUCAGON gene regulation (19–21, 26–28), which relied on experimental assessment in rodent cell lines and tissues, we assessed function of candidate elements in primary human islets. Divergence between our results and those of rodent-based studies highlights the importance of conducting work of this type in human tissue. We detected variability in labeling of α-cells in islets from different human donors. Thus, for interpreting future experiments involving these lentiviral reporters, controls to establish the basal specificity of α-cell labeling will be important for interpreting outcomes. The genetic elements described here should allow specific genetic labeling of human α-cells. Adaptation of the technical advances described here to deliver lineage tracing constructs or cDNA sequences for targeted genetic modification might enable assessment of cell conversion and accelerate establishment of human α-cell lines.
Additional material
Supplementary data supplied by authors.
Acknowledgments
We thank members of the Kim Lab for helpful discussion and advice, particularly C. Downie for technical assistance. We also thank Dr P. Herrera (University of Geneva) for the gift of the rat Glucagon promoter construct.
This work was supported by the Howard Hughes Medical Institute (HHMI) Medical Fellows program, by the Stanford Medical Scientist Training Program, and by the United States National Institutes of Health (NIH) Grant F30DK102301 (to P.T.P.). Work in the Kim lab was supported by a Targeted Resource Generation Project from the NIH Beta Cell Biology Consortium, HHMI, JDRF, the Helmsley Charitable Trust, and the HL Snyder Medical Foundation.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CMV
- cytomegalovirus
- rGcg
- rat glucagon promoter
- GUTR
- Glucagon promoter plus untranslated region
- RIP
- rat Ins1 promoter
- UTR
- untranslated region.
References
- 1. Puri S, Folias AE, Hebrok M. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell. 2015;16(1):18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collombat P, Xu X, Ravassard P, et al. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into α and subsequently β cells. Cell. 2009;138(3):449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thorel F, Népote V, Avril I, et al. Conversion of adult pancreatic α-cells to β-cells after extreme β-cell loss. Nature. 2010;464(7292):1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang YP, Thorel F, Boyer DF, Herrera PL, Wright CV. Context-specific α-to-β-cell reprogramming by forced Pdx1 expression. Genes Dev. 2011;25(16):1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bramswig NC, Everett LJ, Schug J, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest. 2013;123(3):1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spijker HS, Ravelli RB, Mommaas-Kienhuis AM, et al. Conversion of mature human β-cells into glucagon-producing α-cells. Diabetes. 2013;62(7):2471–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scharfmann R, Xiao X, Heimberg H, Mallet J, Ravassard P. β Cells within single human islets originate from multiple progenitors. PLoS One. 2008;3(10):e3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bar Y, Russ HA, Knoller S, Ouziel-Yahalom L, Efrat S. HES-1 is involved in adaptation of adult human β-cells to proliferation in vitro. Diabetes. 2008;57(9):2413–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ravassard P, Hazhouz Y, Pechberty S, et al. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121(9):3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scharfmann R, Pechberty S, Hazhouz Y, et al. Development of a conditionally immortalized human pancreatic β cell line. J. Clin. Invest. 2014;124(5):2087–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ravassard P, Bricout-Neveu E, Hazhouz Y, et al. A new strategy to generate functional insulin-producing cell lines by somatic gene transfer into pancreatic progenitors. PLoS One. 2009;4(3):e4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersson LE, Valtat B, Bagge A, et al. Characterization of stimulus-secretion coupling in the human pancreatic EndoC-βH1 β cell line. PLoS One. 2015;10(3):e0120879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Dev Camb Engl. 2000;127(11):2317–2322. [DOI] [PubMed] [Google Scholar]
- 16. Beattie GM, Montgomery AM, Lopez AD, et al. A novel approach to increase human islet cell mass while preserving β-cell function. Diabetes. 2002;51(12):3435–3439. [DOI] [PubMed] [Google Scholar]
- 17. Pauerstein PT, Sugiyama T, Stanley SE, et al. Dissecting human gene functions regulating islet development with targeted gene transduction. Diabetes. 2015;64(8):3037–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nian M, Drucker DJ, Irwin D. Divergent regulation of human and rat proglucagon gene promoters in vivo. Am J Physiol. 1999;277(4):G829–G837. [DOI] [PubMed] [Google Scholar]
- 20. Nian M, Gu J, Irwin DM, Drucker DJ. Human glucagon gene promoter sequences regulating tissue-specific versus nutrient-regulated gene expression. Am J Physiol Regul Integr Comp Physiol. 2002;282(1):R173–R183. [DOI] [PubMed] [Google Scholar]
- 21. Zhou L, Nian M, Gu J, Irwin DM. Intron 1 sequences are required for pancreatic expression of the human proglucagon gene. Am J Physiol Regul Integr Comp Physiol. 2006;290(3):R634–R641. [DOI] [PubMed] [Google Scholar]
- 22. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boutet SC, Cheung TH, Quach NL, et al. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10(3):327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–126. [DOI] [PubMed] [Google Scholar]
- 25. Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular “hotspot” for pathology? Nat Med. 2000;6(6):637–641. [DOI] [PubMed] [Google Scholar]
- 26. Drucker DJ, Philippe J, Jepeal L, Habener JF. Glucagon gene 5′-flanking sequences promote islet cell-specific gene transcription. J Biol Chem. 1987;262(32):15659–15665. [PubMed] [Google Scholar]
- 27. Philippe J, Drucker DJ, Knepel W, Jepeal L, Misulovin Z, Habener JF. α-Cell-specific expression of the glucagon gene is conferred to the glucagon promoter element by the interactions of DNA-binding proteins. Mol Cell Biol. 1988;8(11):4877–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee YC, Drucker DJ. Glucagon gene 3′-flanking sequences direct formation of proglucagon messenger RNA 3′-ends in islet and nonislet cells lines. Mol Endocrinol. 1990;4(6):800–806. [DOI] [PubMed] [Google Scholar]
- 29. Negi S, Jetha A, Aikin R, Hasilo C, Sladek R, Paraskevas S. Analysis of β-cell gene expression reveals inflammatory signaling and evidence of dedifferentiation following human islet isolation and culture. PLoS One. 2012;7(1):e30415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiang MK, Melton DA. Single-cell transcript analysis of pancreas development. Dev Cell. 2003;4(3):383–393. [DOI] [PubMed] [Google Scholar]
- 31. Katsuta H, Akashi T, Katsuta R, et al. Single pancreatic β cells co-express multiple islet hormone genes in mice. Diabetologia. 2010;53(1):128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517(7532):33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.