Abstract
NOTCH plays a role in regulating stem cell function and fate decision. It is involved in tooth development and injury repair. Information regarding NOTCH expression in human dental root apical papilla (AP) and its residing stem cells (SCAP) is limited. Here we investigated the expression of NOTCH3, its ligand JAG1, and mesenchymal stem cell markers CD146 and STRO-1 in the AP or in the primary cultures of SCAP isolated from AP. Our in situ immunostaining showed that in the AP NOTCH3 and CD146 were co-expressed and associated with blood vessels having NOTCH3 located more peripherally. In cultured SCAP, NOTCH3 and JAG1 were co-expressed. Flow cytometry analysis showed that 7%, 16% and 98% of the isolated SCAP were positive for NOTCH3, STRO-1 and CD146, respectively with a rare 1.5% subpopulation of SCAP co-expressing all three markers. The expression level of NOTCH3 reduced when SCAP underwent osteogenic differentiation. Our findings are the first step towards defining the regulatory role of NOTCH3 in SCAP fate decision.
Keywords: Apical papilla, CD146, JAG1, NOTCH3, Osteogenesis, SCAP, Stemness, STRO-1
Introduction
Since the discovery of stem cells from apical papilla (SCAP), new clinical treatment concepts have emerged. A clinical regenerative protocol has been proposed based on some research studies and many clinical case reports.1, 2, 3, 4, 5 The theory behind this protocol is that SCAP in the apical papilla may be induced to regenerate damaged or lost pulp tissue in the canal space.6, 7, 8 This possibility is further supported by the capacity of SCAP to regenerate pulp-dentin-like tissues in vivo in animal models.6, 9 However, despite such clinical endeavors to practice regenerative treatments, the understanding of the biology of SCAP is still limited.
NOTCH signaling pathway plays a key role in the development and morphogenesis of different organs and tissues in various species. It promotes or suppresses cell proliferation, initiates or inhibits cell differentiation and determines the fate of different types of stem cells.10 There are four NOTCH receptors (NOTCH1–4), which are activated by direct contact with their membrane-bound ligands (JAGGED (JAG) 1, JAG2, Delta-like (DLL) 1, DLL3 and DLL4) on neighboring cells. Upon activation of NOTCH receptors, enzymatic activities are triggered resulting in the cleavage of NOTCH intracellular domain (NICD), which is then translocated into the nucleus to activate the transcription of target genes, such as HEY1 and HEY2.10, 11 NOTCH signaling pathway is also believed to be involved and play an important role in the process of tooth development and pulp regeneration and repair after injury.12, 13, 14 NOTCH3 and its ligand JAG1 are upregulated during tooth development in the vicinity of blood vessels and in the subodontoblast layer, but not in odontoblasts. Similar expression pattern was found in pulp tissue undergoing repair and regeneration after pulp exposure and capping.12, 15, 16 Interestingly, NOTCH3 is expressed in the cervical loop (stem cell niche) of continuously erupting teeth (mouse incisors and vole molars), suggesting its possible role in maintaining the undifferentiated state of cells within that niche.13
Little is known regarding the expression of NOTCH3 and its ligand JAG1 in apical papilla (AP) and SCAP. The aim of this study was to investigate whether and where NOTCH3 is expressed in AP and its expression along with JAG1 in cultured SCAP, as well as its co-expression with mesenchymal stem cell markers CD146 and STRO-1.
Material and methods
Sample collection
This study was approved by the Human Research Ethics Committee of United Arab Emirates University (#11/10) and Boston University Medical Institutional Review Board (#H-28882). Freshly extracted, intact human teeth were obtained from 10 to 24 years old consented healthy patients (n = 5). The teeth were caries-free and had incompletely formed root apices. The root apical papillae were microdissected from extracted teeth and SCAP were isolated and cultured as described below based on a previous report.6
Immunohistochemical staining of apical papillae
Apical papillae were obtained as mentioned above and processed for cryosectioning; 8 μm-thick sections were fixed with cold acetone at −20 °C for 15 min, washed in PBS, treated with 1.5% hydrogen peroxide for 30 min and blocked with 2.5% normal horse serum (Vectastain Elite ABC kit; Vector laboratories) for 1 h. Tissue sections were then incubated with mouse monoclonal anti-NOTCH3 antibodies (dilution 1:100, Abcam, USA) for 1 h at room temperature, followed by washing and incubation with biotinylated anti-mouse immunoglobulin G (secondary antibody) for another 1 h. After washing, avidin-peroxidase complex was added and incubated for 30 min, followed by washing and the addition of peroxidase substrate solution for 5 min. Sections were counterstained with hematoxylin solution (Sigma, USA). Negative control slides were prepared in parallel without adding the primary antibody.
For immunofluorescence staining, frozen sections of AP were fixed with cold acetone at −20 °C for 15 min, washed and blocked for 1 h. Sections were then incubated with primary antibodies: NOTCH3 (dilution 1:100, Abcam, USA) and CD146 (dilution 1:50, Invitrogen, USA) for 1 h at room temperature, followed by washing and incubation with the appropriate fluorophore-conjugated secondary antibodies for another 1 h.
Isolation and culture of SCAP
The papillae were minced and digested in a physiological solution containing 3 mg/mL collagenase type I (GIBCO/Invitrogen) and 4 mg/mL dispase (GIBCO/Invitrogen) for 45 min at 37 °C. Isolated cells were then plated in culture dishes containing alpha-modification of Eagle's medium (GIBCO/Invitrogen) supplemented with 10% fetal bovine serum (GIBCO/Invitrogen), 2 mM l-glutamine (GIBCO/Invitrogen), 100 U/mL penicillin and 100 mg/mL streptomycin (GIBCO/Invitrogen) and incubated in a humidified incubator (Thermo Scientific) at 37 °C in 5% CO2. Once the cells reached ∼80% confluence, they were trypsinized and passaged.
Immunocytochemical and immunofluorescence staining of SCAP
For immunocytochemistry, isolated SCAP of passage 2 were seeded on sterile coverslips. At 80% confluence, cells were fixed using 4% paraformaldehyde (PFA) for 30 min, washed with phosphate-buffered saline (PBS) and permeabilized with 0.1% Triton-X100 (Sigma, USA). To inhibit endogenous peroxidase activity, cells on coverslips were incubated in 1% hydrogen peroxide in PBS for 35 min. Nonspecific binding was blocked by incubating cells in 1% bovine serum albumin (BSA) containing 0.5% Tween-20 in PBS for 45 min. Cells were then incubated with goat polyclonal anti-NOTCH3 antibody (dilution 1:25, clone M-20, Santa Cruz Biotechnology Inc., USA) overnight at 4 °C. Cells on coverslips were washed with PBS and then incubated with biotinylated donkey-anti-goat immunoglobulin G (dilution 1:500, Jackson ImmunoResearch Laboratories Inc., USA) for 1 h and then were incubated in extravidin/peroxidase conjugate (dilution 1:1000, Sigma, USA) for 1 h. The antigen–antibody binding sites were revealed by incubating tissue sections with 3,3'-diaminobenzidine tetrahydrochloride (DAB, Sigma, USA). Negative control slides were prepared in parallel without adding the primary antibody.
For immunofluorescence staining, SCAP were grown on chamber slides and they were fixed, when reaching 80% confluence, in 4% PFA for 30 min, permeabilized, blocked and incubated with primary antibodies; NOTCH3 (dilution 1:100, Abcam, USA) and JAG1 (dilution 1:100, Santa Cruz Biotechnology, USA) for 1 h. The samples were subsequently incubated with the corresponding secondary antibodies for another 1 h followed by counterstaining with DAPI (Invitrogen, USA). The samples were then observed and images recorded under a fluorescence microscope.
Flow cytometry of SCAP
SCAP at passage 3 were grown in a T25 mm flask to reach 80% confluence. They were harvested, washed with PBS, and resuspended in PBS containing 2% fetal bovine serum. Conjugated mouse IgG1κ anti-human monoclonal antibodies specific for CD146-APC, STRO-1-FITC and NOTCH3-PE or their corresponding isotype controls were added according to the recommended manufacturer concentrations (Biolegend, USA) for 30 min at 4 °C. Cells were washed and resuspended in PBS. Flow cytometry was performed on FACSCalibur cytometer (BD Biosciences), and data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR).
Osteogenic differentiation of SCAP
SCAP were grown to 80% confluence and incubated in osteogenic differentiation media containing 10 nM dexamethasone, 10 mM β-glycerophosphate, 50 μg/mL ascorbate phosphate, 10 nM 1,25-dihydroxyvitamin D3, and 10% FBS (Gibco/Invitrogen USA) for 14 days. SCAP were processed and stained for NOTCH3 for flow cytometry analysis as mentioned above.
Results
Immunohistochemical analysis of the apical papilla
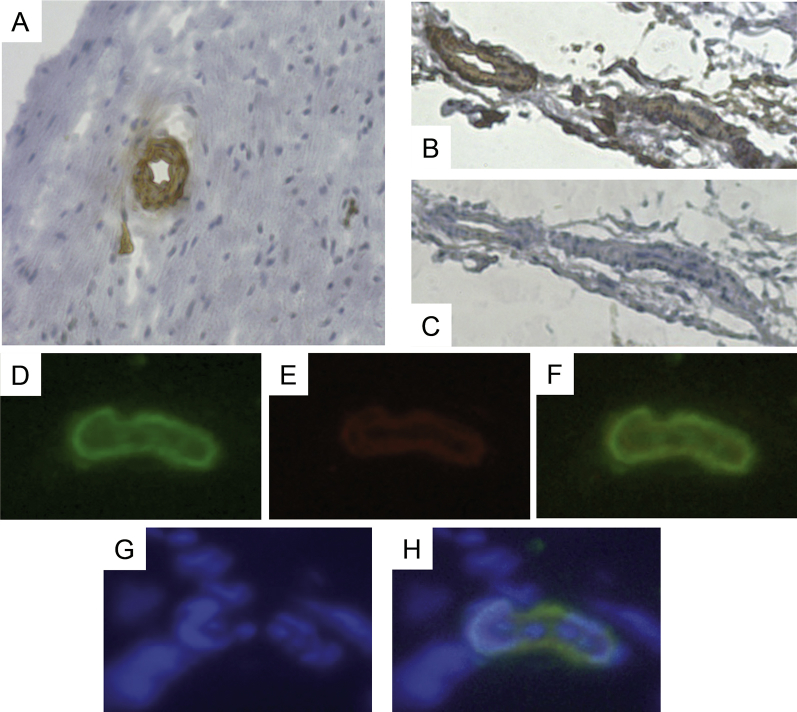
The immunohistochemical staining showed that NOTCH3 expression is associated with blood vessels (Fig. 1). Immunoflorescence staining for NOTCH3 and CD146 confirmed the results of the immunohistochemical staining, and showed that cells in and around the wall of blood vessels were stained positive for NOTCH3 and CD146 with NOTCH3 located at the outer layer of the vascular vessel wall.
Fig. 1.
Detection of NOTCH3 and CD146 in human apical papilla. (A) Immunohistochemical staining of the apical papilla, showing NOTCH3 staining is associated with blood vessels. (B) Staining for NOTCH3, (C) control. (D–H) Immunoflourescence staining of the apical papilla showing blood vessels stained for D) NOTCH3 (green), (E) CD146 (red), (F) co-localization of NOTCH3 and CD146, (G) DAPI (blue), (H) co-localization of NOTCH3, CD146 and DAPI. Data are representative of independent experiments of samples from 2 donors each performed in triplicate.
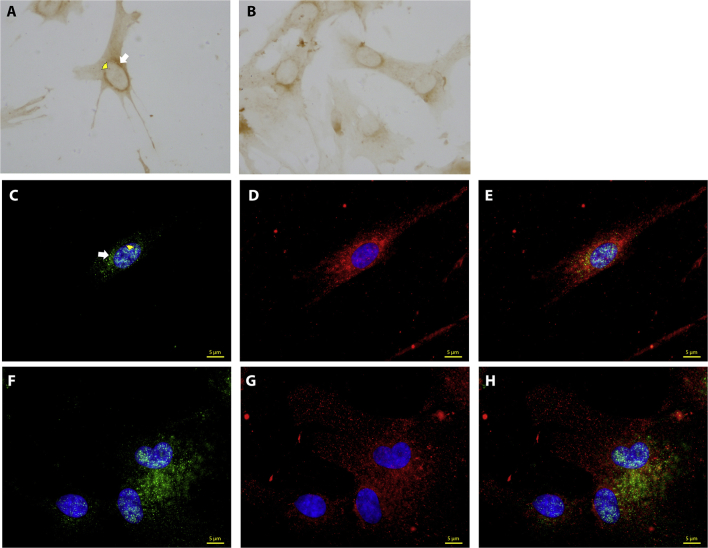
Detection of NOTCH3 in cultured SCAP
Immunocytochemical staining revealed the localization of NOTCH3 in the nucleus and the cytoplasm of SCAP especially in the perinuclear region (Fig. 2A,B). Immunofluorescence double-staining showed that NOTCH3 and its ligand, JAG1 were co-localized in SCAP; NOTCH3 was detected in the perinuclear area of the cytoplasm and also in the nucleus, while JAG1 is concentrated in the perinuclear cytoplasm (Fig. 2C,D).
Fig. 2.
Expression of NOTCH3 and JAG1 in SCAP at passage 3. (A, B) Immunocytochemical staining of SCAP, showing positive staining for NOTCH3 in the perinuclear region (indicated by the white arrow in A) and in the nucleus (yellow arrow head in A). (C–H) Immunoflourescence staining of SCAP showing positive staining for NOTCH3 (green) and JAG1 (red). Note that NOTCH3 is located in the perinuclear region and in the nucleus while JAG1 only in the cytplasm and perinuclear region. In (C–E), isolated single cells showing both NOTCH3 and JAG1 in the same cell. Similar finding is in (F–H) when cells are in contact with each other. Data are representative of independent experiments of samples from 3 donors each performed in triplicate.
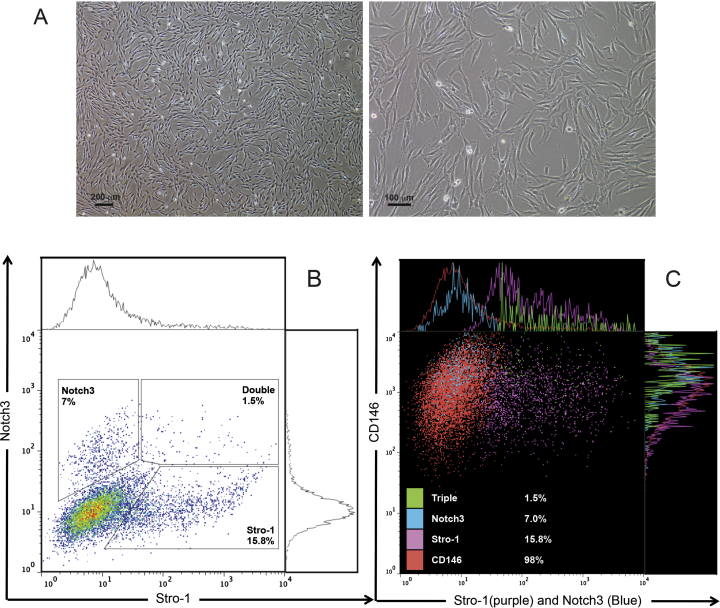
Quantification and characterization of NOTCH3-expressing subpopulations of SCAP
Flow cytometry analysis showed that 7%, 16% and 98% of the isolated SCAP were positive for NOTCH3, STRO-1 and CD146, respectively. All the 7% NOTCH3+ SCAP expressed CD146, and only 1.5% of the total SCAP population co-expressed NOTCH3, STRO-1 and CD146 (Fig. 3).
Fig. 3.
Flow cytometry analysis of SCAP at passage 3. (A) Representative images of SCAP of ∼80% confluence before subjected to flow cytometry. (B) SCAP double staining with NOTCH3 and STRO-1: 7% is expressing NOTCH3, 15.8% expressing STRO-1 and 1.5% expressing both STRO-1 and NOTCH3. (C) Flow cytometry dot plots showing triple- staining of SCAP for CD146 (red), STRO-1 (purple) and NOTCH3 (blue). Note the triple positive population of cells are shown in green. Data are representative of 2 experiments performed in triplicate.
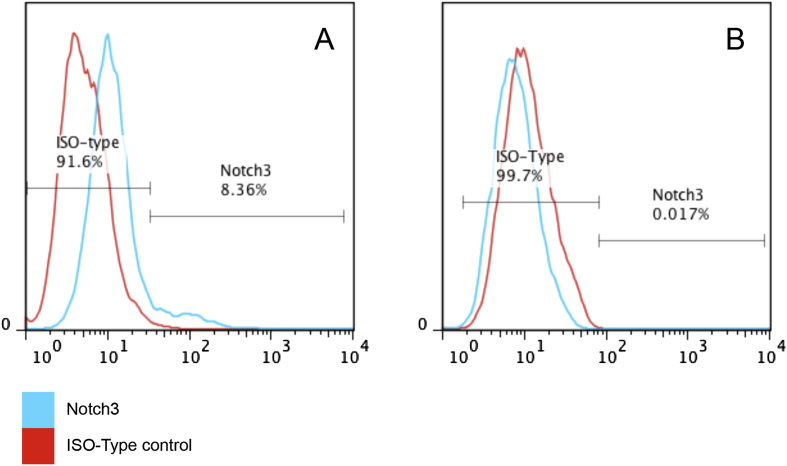
SCAP lose NOTCH3 expression after differentiation
To examine the relationship between SCAP differentiation and NOTCH3 expression, flow cytometry analysis was performed for SCAP grown in the regular medium or osteogenic differentiation medium. The flow cytometry results showed that NOTCH3 expression was detected on SCAP when cultured in the regular medium while the expression was absent when cultured in the osteogenic differentiation medium (Fig. 4).
Fig. 4.
Flow cytometry analysis of SCAP at passage 3 underwent osteogenesis. (A) Control SCAP expressing NOTCH3; (B) SCAP after osteogenic differentiation for 14 days losing NOTCH3 expression. Representative data from 2 experiments in triplicate.
Discussion
To understand the possible role of NOTCH signaling pathway in SCAP we undertook the first step by examining the expression of NOTCH3 and its ligand JAG1. We found that a subpopulation of SCAP is expressing NOTCH3 (Fig. 2, Fig. 3) similar to recent findings reported by Sun et al.17 These results suggest a possible role of NOTCH3 in regulating SCAP properties and behaviors. Mitsiadis et al found that during development, NOTCH3 is expressed in the subodontoblast layer but not in odontoblast.12 In another study, they found it expressed in the stem cell niche of cervical loop of mouse incisor, but this expression decreases and disappears as the cells leave the niche and move coronally and start to differentiate into ameloblasts.13 They suggest that NOTCH3 could be responsible for maintaining the cells in their undifferentiated state, and that once the cells differentiate they lose NOTCH3 expression. Our findings shown in Fig. 4 that SCAP lose NOTCH3 expression after osteo-induction suggests that NOTCH3 is associated with less differentiated SCAP. This indicates that NOTCH3 may play a role in maintaining SCAP in their undifferentiated state, as is suggested in the recent studies of SCAP and DPSCs by other investigators.17, 18 However, further experiments are needed to confirm this possibility and to explore the association of NOTCH3 with SCAP stemness and differentiation status.
STRO-1 and CD146 are considered as early markers of many types mesenchymal stem cells (MSCs) and it was found that 82% and 96% of the dental pulp derived colony forming cells were represented in the STRO-1+ and CD146+ subpopulations, respectively.19 In addition, it was found that the STRO-1+ subpopulation demonstrated higher expression of embryonic and MSCs makers and had a better odontogenic potential than the STRO-1- subpopulation.20, 21 To investigate whether NOTCH3+ SCAP share the expression of STRO-1 and CD146, our flow cytometry analysis showed that 7%, 16% and 98% of the isolated SCAP were positive for NOTCH3, STRO-1 and CD146 respectively. The latter two markers were examined by Sonoyama et al and the respective percentage is similar to our findings.6 Our flow cytometry using triple staining is the first to demonstrate the presence of these subpopulations of SCAP. While our data indicate that almost all isolated SCAP expressed CD146, there are 4 distinct different subpopulations: 7% NOTCH3+STRO1-, ∼16% NOTCH3-STRO-1+, ∼75% NOTCH3-STRO-1- and a rare population of NOTCH3+STRO-1+ (Fig. 3). Further investigation is warranted to explore the characteristics of these subpopulations such as NOTCH3+ SCAP, especially in the context of regenerative potential, as other MSCs are known to be heterogeneous containing different subpopulations of varying regenerative potential.19, 20, 22
As mentioned earlier, studies have indicated that during tooth development and after pulp injury, NOTCH3 is upregulated and expressed on the wall of blood vessels.15, 16 Lovschall et al found a similar expression pattern and that NOTCH3 was co-expressed with RGS5, which is one of the markers for peicytes.15 These findings suggest that those NOTCH3-expressing cells likely reside in the perivascular niche and may be of pericyte origin. Our immunohistochemical staining indicates that NOTCH3 expression in apical papillae is exclusively associated with blood vessels. The doubled staining (NOTCH3 and CD146) further showed that both are restricted and co-localized in and around the vascular walls with NOTCH3 located more in the periphery (Fig. 1). The reason why we chose CD146 in addition to NOTCH3 was that 1) all the 7% NOTCH+ SCAP are expressing CD146, and 2) CD146 is one of the pericyte markers.19, 23 This expression pattern of NOTCH3 in SCAP and their localization on the outer layer of the vascular wall are in favor of the possibility that NOTCH3+ SCAP is of pericyte origin in apical papillae.
In NOTCH signaling, its receptors are activated by binding to one of its ligands on neighboring cells and this require direct physical contact between two cells.10 We attempted to identify other subpopulations of SCAP that provide the ligands to NOTCH+ SCAP. To our surprise, we found that NOTCH3 and JAG1 were co-localized on the same cells in culture (Fig. 2C–H). To confirm this finding, we repeated this immunohistochemical localization with antibodies from different sources, and the results were consistent (Data not shown). As mentioned earlier, NOTCH activation will result in the cleavage of NICD and its translocation to the nucleus.10, 11 Our data showed that NOTCH3 was also localized in the nucleus (Fig. 2C,F) and the anti-NOTCH3 antibodies we used were bound to the intracellular domain. This suggests that the receptor might have been already activated as it was translocated into the nucleus. This NOTCH3 activation could be a result of binding to a ligand (JAG1 or other ligands such as Delta-1) on adjacent cells; or via a different mechanism from the classical NOTCH activation mechanism, possibly in a cell-autonomous NOTCH activation mode, in which both NOTCH receptor and its ligand are present on the same cell. The later possibility is supported by the expression of both NOTCH3 and JAG1 in the same cells, NOTCH3 localization in the nucleus, and this expression pattern is consistent when cells are isolated (Fig. 2C–E) or in contact with other cells (Fig. 2F–H). This mode of activation has been reported in Drosophila and in vascular smooth muscle cells in pulmonary arteries.11, 24
Conclusion
Our findings set the basis for further studies to explore NOTCH3 signaling in SCAP and its role in controlling SCAP behavior and faith decision. A better understanding of SCAP biology may help develop clinical strategies to advance regenerative endodontics.
Conflicts of interest
All authors have none to declare.
Acknowledgments
This work was supported in part by a grant from the Emirates Foundation – United Arab Emirates UAE University/NRF Grant and a grant from the National Institutes of Health RO1 DE019156 (G.T.-J.H.). The authors deny any conflicts of interest.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Mohamed Jamal, Email: mjamal@bu.edu.
Sami M. Chogle, Email: schogle@bu.edu.
Sherif M. Karam, Email: skaram@uaeu.ac.ae.
George T.-J. Huang, Email: gtjhuang@uthsc.edu.
References
- 1.Windley W., Teixeira F., Levin L., Sigurdsson A., Trope M. Disinfection of immature teeth with a triple antibiotic paste. J Endod. 2005;31:439–443. doi: 10.1097/01.don.0000148143.80283.ea. [DOI] [PubMed] [Google Scholar]
- 2.Shimizu E., Ricucci D., Albert J., Alobaid A.S., Gibbs J.L., Huang G.T., Lin. L.M. Clinical, radiographic, and histological observation of a human immature permanent tooth with chronic apical abscess after revitalization treatment. J Endod. 2013;39:1078–1083. doi: 10.1016/j.joen.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 3.Iwaya S.I., Ikawa M., Kubota M. Revascularization of an immature permanent tooth with apical periodontitis and sinus tract. Dent traumatol. 2001;17:185–187. doi: 10.1034/j.1600-9657.2001.017004185.x. [DOI] [PubMed] [Google Scholar]
- 4.Banchs F., Trope M. Revascularization of immature permanent teeth with apical periodontitis: new treatment protocol? J Endod. 2004;30:196–200. doi: 10.1097/00004770-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Chueh L.H., Huang G. Immature teeth with periradicular periodontitis or abscess undergoing apexogenesis: a paradigm shift. J Endod. 2006;32:1205–1213. doi: 10.1016/j.joen.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Sonoyama W., Liu Y., Fang D. Mesenchymal stem cell-mediated functional tooth regeneration in swine. PloS one. 2006;1:e79. doi: 10.1371/journal.pone.0000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonoyama W., Liu Y., Yamaza T. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. J Endod. 2008;34:166–171. doi: 10.1016/j.joen.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang G.T., Sonoyama W., Liu Y., Liu H., Wang S., Shi S. The hidden treasure in apical papilla: the potential role in pulp/dentin regeneration and bioroot engineering. J Endod. 2008;34:645–651. doi: 10.1016/j.joen.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang G.T.J., Yamaza T., Shea L.D. Stem/progenitor cell–mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng Part A. 2009;16:605–615. doi: 10.1089/ten.tea.2009.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopan R., Ilagan M.X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh S., Paez-Cortez J.R., Boppidi K. Activation dynamics and signaling properties of Notch3 receptor in the developing pulmonary artery. J biol Chem. 2011;286:22678–22687. doi: 10.1074/jbc.M111.241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitsiadis T.A., Lardelli M., Lendahl U., Thesleff I. Expression of Notch 1, 2 and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J cell Biol. 1995;130:407–418. doi: 10.1083/jcb.130.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitsiadis T.A., Hirsinger E., Lendahl U., Goridis C. Delta-notch signaling in odontogenesis: correlation with cytodifferentiation and evidence for feedback regulation. Dev Biol. 1998;204:420–431. doi: 10.1006/dbio.1998.9092. [DOI] [PubMed] [Google Scholar]
- 14.Mitsiadis T.A., Regaudiat L., Gridley T. Role of the Notch signalling pathway in tooth morphogenesis. Archives oral Biol. 2005;50:137–140. doi: 10.1016/j.archoralbio.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Lovschall H., Mitsiadis T.A., Poulsen K., Jensen K.H., Kjeldsen A.L. Coexpression of Notch3 and Rgs5 in the pericyte-vascular smooth muscle cell axis in response to pulp injury. Int J Dev Biol. 2007;51:715–721. doi: 10.1387/ijdb.072393hl. [DOI] [PubMed] [Google Scholar]
- 16.Lovschall H., Tummers M., Thesleff I., Füchtbauer E.-M., Poulsen K. Activation of the Notch signaling pathway in response to pulp capping of rat molars. Eur J oral Sci. 2005;113:312–317. doi: 10.1111/j.1600-0722.2005.00221.x. [DOI] [PubMed] [Google Scholar]
- 17.Sun F., Wan M., Xu X. Crosstalk between miR-34a and Notch Signaling Promotes Differentiation in Apical Papilla Stem Cells (SCAPs) J Dent Res. 2014;93:589–595. doi: 10.1177/0022034514531146. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C., Chang J., Sonoyama W., Shi S., Wang C.Y. Inhibition of human dental pulp stem cell differentiation by notch signaling. J Dent Res. 2008;87:250–255. doi: 10.1177/154405910808700312. [DOI] [PubMed] [Google Scholar]
- 19.Shi S., Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res. 2003;18:696–704. doi: 10.1359/jbmr.2003.18.4.696. [DOI] [PubMed] [Google Scholar]
- 20.Yang X., van den Dolder J., Walboomers X.F. The odontogenic potential of STRO-1 sorted rat dental pulp stem cells in vitro. J tissue Eng Regen Med. 2007;1:66–73. doi: 10.1002/term.16. [DOI] [PubMed] [Google Scholar]
- 21.Bakopoulou A., Leyhausen G., Volk J., Koidis P., Geurtsen W. Comparative characterization of STRO-1(neg)/CD146(pos) and STRO-1(pos)/CD146(pos) apical papilla stem cells enriched with flow cytometry. Archives oral Biol. 2013;58:1556–1568. doi: 10.1016/j.archoralbio.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Gronthos S., Mankani M., Brahim J., Robey P.G., Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crisan M., Yap S., Casteilla L. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Coumailleau F., Furthauer M., Knoblich J.A., Gonzalez-Gaitan M. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 2009;458:1051–1055. doi: 10.1038/nature07854. [DOI] [PubMed] [Google Scholar]