Abstract
The global prevalence of metabolic disorders is an immediate threat to human health. Genetic features, environmental aspects and lifestyle changes are the major risk factors determining metabolic dysfunction in the body. Autophagy is a housekeeping stress-induced lysosomal degradation pathway, which recycles macromolecules and metabolites for new protein synthesis and energy production and regulates cellular homeostasis by clearance of damaged protein or organelles. Recently, a dramatically increasing number of literatures has shown that defects of the autophagic machinery is associated with dysfunction of multiple metabolic tissues including pancreatic β cells, liver, adipose tissue and muscle, and is implicated in metabolic disorders such as obesity and insulin resistance. Here in this review, we summarize the representative works on these topics and discuss the versatile roles of autophagy in the regulation of cellular metabolism and its possible implication in metabolic diseases.
Keywords: autophagy, selective autophagy, metabolism, metabolic disease, obesity, diabetes
Introduction
Literally defined as cellular “self-eating,” autophagy is an evolutionarily conserved catabolic process that targets long-lived cytoplasmic proteins or damaged organelles for lysosomal degradation (De Duve and Wattiaux, 1966). Autophagy occurs constitutively at a low level, which is important for intracellular quality control through constitutive turnover of cytoplasmic component (Kuma and Mizushima, 2010). The autophagic pathway can be potently induced by stress, such as deprivation of nutrients or growth factors, hypoxia, oxidative stress, pathogen infection and physical exercise. The resulting degraded small- and macro-molecules can be released and recycled back to the cytosol for new biosynthesis processes and energy production to maintain cellular function and survival under unfavorable conditions (Levine and Klionsky, 2004; Kroemer et al., 2010; Rabinowitz and White, 2010). Different from the ubiquitin-proteasome system (UPS), which recognizes and degrades primarily short-lived proteins, autophagy is more than a self-destruction pathway and plays key roles in a broader spectrum of physiological processes including differentiation, development, cellular metabolism and energy homeostasis (Qu et al., 2007; Cecconi and Levine, 2008).
Three major types of autophagy have been characterized, which differ in the manner of cargo transportation to the lysosome for degradation (Mizushima and Komatsu, 2011): Macroautophagy is the main autophagy pathway (herein referred to as autophagy). It is characterized by the formation of a double-membrane vesicle, known as autophagosome, surrounding the targeted cargos. Autophagosome completion is followed by its fusion with the lysosome (Levine and Kroemer, 2008; Kim and Lee, 2014) (Fig. 1). Chaperonemediated autophagy is a mechanism for degradation of specific proteins that carry a recognition sequence (KFERQ). This motif is recognized by the Hsc70 chaperones, which transport these proteins into the lysosome via the lysosomal membrane receptor LAMP 2A (lysosome-associated membrane protein type 2A) (Kaushik and Cuervo, 2012). Microautophagy refers to the sequestration of cargo elements in proximity to the lysosomal membrane and subsequent invagination into the lysosomal lumen (Santambrogio and Cuervo, 2011).
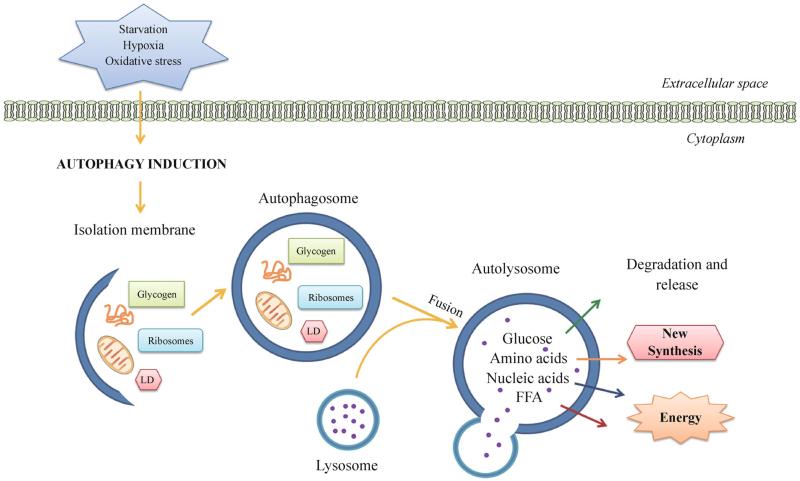
Figure 1.
Autophagy in organelle turnover and nutrient recycling. Autophagy is initiated by the formation of an isolation membrane (or phagophore) under stress conditions, such as nutrient deprivation, hypoxia and oxidative stress. The isolation membrane enwraps bulk cytosol or specific cargos (misfolded proteins, mitochondria, glycogen, ribosomes, lipid droplets, etc.), and elongates and forms a double-membrane autophagosome. It then fuses with the lysosome into an autolysosome, where the resident hydrolytic enzymes digest cargos and various resulting metabolites and macromolecules (including amino acids, glucose, nucleic acids and free fatty acids) are released back to the cytosol as new building blocks or energy sources. LD, lipid droplets; FFA, free fatty acids.
Regulation of the autophagy machinery
Formation of an autophagosome is the first step of the autophagy process. Induced by starvation or other stress, the construction of the autophagosome is orchestrated by the Atg (autophagy-related) proteins (Fig. 2). These proteins have been first discovered in yeast, and are found highly conserved in higher eukaryotes, ranging from drosophila to mammals (Mizushima et al., 2011). Autophagosome nucleation and elongation is initiated by the serine-threonine protein kinase ULK1 complex (the Atg1 homolog), which consists of several autophagy proteins including ULK1, Atg13, FIP200 and Atg101 (Hara et al., 2008; Mizushima, 2010). Under nutrient-rich conditions, ULK1 is suppressed by mTORC1, a major negative regulator of autophagy. Upon autophagy induction, mTORC1 is inhibited; meanwhile, AMPK (AMP-activated protein kinase) can be activated. Either condition, dephosphorylation of ULK1 by mTORC1 inhibition or ULK1 phosphorylation by AMPK, activates ULK1 (Jung et al., 2010; Shang and Wang, 2011). Once activated, the ULK1 complex is able to upregulate the class III phosphatidylinositol (PtdIns) 3-kinase (PtdIns3K) complex, comprising Beclin 1/Atg6, Atg14, Vps15, Vps34 and Ambra1. Induction of the kinase complex generates the lipid phosphatidylinositol 3-phospate (PI3P),which recruits more PI3P binding proteins essential for vesicle formation, including WIPI and DFCP1 (Proikas-Cezanne et al., 2004; Axe et al., 2008; Jung et al., 2009; Burman and Ktistakis, 2010).
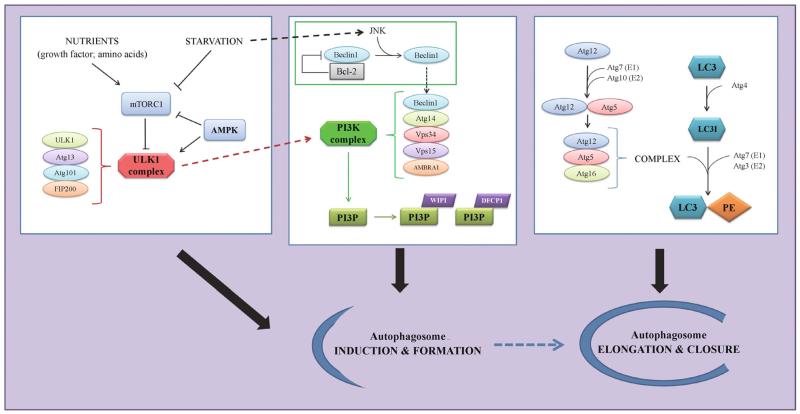
Figure 2.
Regulation of autophagosome formation. Under nutrient-rich conditions, active mTORC1 inhibits the ULK1 kinase complex; whereas starvation activates AMPK, suppresses mTORC1 and induces ULK1, which upregulates another kinase complex, the PtdIns3K complex. Recruitment of Beclin 1 to the PtdIns3K complex is essential for the kinase activity, and is negatively regulated by sequestration of Beclin 1 by Bcl-2 and positively regulated by release of Beclin 1 upon JNK-mediated phosphorylation of Bcl-2. The PtdIns3K complex generates PI3P, which recruits additional PI3P-binding proteins (including WIPI and DFCP1) to promote autophagosome formation. Elongation of autophagosome membrane requires two ubiquitin-like conjugation systems, Atg12-Atg5-Atg16 and LC3-PE; the former acts as an E3 ligase to assist the conjugation of LC3 to PE. All the above molecules and pathways cooperate on the induction and formation of autophagosomes under stress conditions.
In addition, two ubiquitin-like conjugation systems, Atg12-Atg5-Atg16 and Atg8/LC3-PE (phosphatidylethanolamine), are required for membrane elongation (Hanada et al., 2007; Suzuki et al., 2007; Geng and Klionsky, 2008). The former complex serves as an ubiquitin E3-like ligase for the latter LC3-PE system. In brief, Atg12 is conjugated to Atg5 by an E1-like activating enzyme Atg7 and an E2-like conjugating enzyme Atg10. Atg16 then interacts with the Atg12-Atg5 conjugates to facilitate LC3-PE formation. Similarly, in the LC3-PE system, cytosolic LC3 is processed by the Atg4 protease, and conjugated to PE by Atg7 (E1), Atg3 (an E2-like enzyme) and the Atg12-Atg5-Atg16 complex. PE-conjugated LC3 is then incorporated into autophagosomal membrane, and thus can be used as a marker protein for autophagosomes.
In addition to mTORC1 inhibition and AMPK activation, the PtdIns3K activity is also regulated by direct sequestration of Beclin 1 by the anti-apoptosis and anti-autophagy protein Bcl-2. When nutrients are abundant, Beclin 1 forms a complex with ER-associated Bcl-2 protein, which inhibits the recruitment and function of Beclin 1 in the PtdIns3K complex; in response to starvation, Beclin 1 is released from the inhibitory binding of Bcl-2 for functioning in autophago-some formation (Pattingre et al., 2005; Matsunaga et al., 2009; Zhong et al., 2009). Disassociation of Beclin 1-Bcl-2 binding is mediated through phosphorylation of Bcl-2 on three sites (T69, S70 and S84) by JNKs (c-Jun N-terminal kinases) upon starvation (Wei et al., 2008), and further investigation is needed to determine whether other upstream kinases are involved in autophagy induction under other stress conditions.
Recent works have also revealed the roles of many metabolic factors in the transcriptional regulation of autophagy, including PPARα (peroxisome proliferator-activated receptor-α), CREB (cAMP response element binding protein), TFEB (basic helix-loop-helix transcription factor EB), FoxO1/3 (Forkhead box O1 and O3) and FXR (farnesoid X receptor) (Zhao et al., 2007; Mammucari et al., 2007; Sengupta et al., 2009; Settembre et al., 2011; Warr et al., 2013; Seok et al., 2014; Lee et al., 2014). Among them, PPARα, CREB, TFEB and FoxO3 are positive regulators of autophagy genes under fasting or stress conditions, and FXR is a nuclear receptor that senses and is activated by the nutrient-rich state and represses expression of autophagy genes. However, transcriptional regulation of the autophagy machinery in many organs is still unclear, and the roles of numerous other transcription factors and co-factors in autophagy are waiting to be discovered.
Selective autophagy in metabolism
Although autophagy is usually considered a non-selective process for bulk degradation of cytoplasmic components, several types of selective autophagy have been described in the past decade (Bjørkøy et al., 2005; Pankiv et al., 2007; Geisler et al., 2010). Ubiquitination is found to be the key signal for the cargo proteins to be recognized by the receptors of selective autophagy, such as the LC3 binding protein p62/SQSTM1. This protein contains a ubiquitin binding domain (UBA) that specifically interacts with ubiquitinated proteins and drives their sequestration into autophagosomes via LC3 binding (Johansen and Lamark, 2011; Shaid et al., 2013). This is the main mechanism of clearing toxic aggregated-prone proteins in cells (Ravikumar et al., 2002; Jeong et al., 2009; Knaevelsrud and Simonsen, 2010). In addition to p62, several other receptors of selective autophagy, including NBR1, Alfy, NDP52 and optineurin (Simonsen et al., 2004; Kirkin et al., 2009; Boyle and Randow, 2013; Korac et al., 2013), have been identified. Specific targeting of different cargo proteins, such as mitochondria or pathogenic proteins, has also been described (Levine and Kroemer, 2008; Zheng et al., 2009; Ogawa et al., 2011; Wong et al., 2012).
Autophagy is essential for the maintenance of cellular metabolism during nutrient deprivation. Autophagic degradation of molecules and organelles generates an important reservoir of metabolites that can be reused for de novo synthesis or energy production. In cells, glycogen, protein, ribosomes and lipid droplets (LDs) are the major nutrient storage of carbohydrates, amino acids, nucleic acids and fatty acids, respectively. In response to starvation, the autophagy machinery gets access to these storages for lysosomal degradation, and the resulting molecules serve as building blocks or fuels for ATP production through aerobic or anaerobic processes (Rabinowitz and White, 2010; Ryter et al., 2013; Kim and Lee, 2014) (Fig. 1).
Lipophagy and glycophagy
As a mechanism to mobilize lipid contents in LDs, autophagy is involved in lipid metabolism, termed as “lipophagy.” In addition to cytosolic lipases, autophagic components associate with LDs and autophagy can mediate LD breakdown by hydrolysis within the lysosomal lumen in various cell types, including hepatocytes, macrophages and hypothalamic neurons (Singh et al., 2009a; Ouimet et al., 2011). Upon nutrient deprivation in hepatocytes and mouse liver, activated lipophagy of triglyceride and cholesterol contents in LDs generates free fatty acids (FFAs) for mitochondrial β oxidation; on the other hand, inhibition of autophagy increases triglyceride storage in LDs, results in lipid accumulation and consequently impairs cellular energy homeostasis due to reduced FFA availability, which becomes a risk factor for insulin resistance, obesity and diabetes. Furthermore, autophagy also plays a role in cholesterol efflux from macrophage foam cells, a key component responsible for atherosclerotic lesions. This specific autophagy-dependent cholesterol mobilization from LDs seems to be mediated by the ATP binding cassette (ABC) transporter ABCA1 (Ouimet et al., 2011). A following study demonstrated that this process also involves the Wip1 phosphatase (Le Guezennec et al., 2012). Through an indirect deactivating effect on the upstream mTOR pathway, Wip1 deficiency increases autophagy-mediated cholesterol efflux, and results in attenuated early foam cell formation and a delay of atherosclerotic plaque accumulation. Thus, these findings support the hypothesis that impairment of macrophage autophagy promotes accumulation of atherosclerotic lipids.
In the hypothalamus, which controls many metabolic features in the central nervous system including appetite and energy expenditure, autophagy is essential for neuronal function and thus exerts opposite effects on metabolic regulation in functionally antagonizing neuronal cell types, such as appetite-promoting AgRP (agouti-related peptide) neurons versus anti-obesity POMC (pro-opiomelanocortin) neurons. Lipid mobilization by lipophagy in AgRP neurons promotes AgRP expression and increases food intake and bodyweight (Kaushik et al., 2011); whereas in POMC neurons, autophagy activity prevents animals from glucose intolerance and adiposity induced by high-fat diet (HFD) (Coupéet al., 2012; Kaushik et al., 2012; Quan et al., 2012a).
In addition to LDs, glycogen can also be degraded by selective autophagy (termed as “glycophagy”), through the Starch binding domain-containing protein 1 (Stbd1) (Jiang et al., 2010; Jiang et al., 2011). Stbd1 Interacts with the Atg8/LC3 homolog GABARAP and may function as a receptor linking glycogen to the autophagic machinery. Stbd1 has been shown to express in skeletal muscle, liver and spleen, suggesting that glycophagy may occur in multiple tissues, although the in vivo importance of glycophagy in these tissues still needs to be further investigated.
Mitophagy
Aerobic respiration is dependent on mitochondria. Dysfunction or depolarization of mitochondria causes cell damage via excessive release of reactive oxygen species (ROS). Thus, removal of the defective mitochondria by selective autophagy (defined as “mitophagy”) plays an essential role in the organelle quality control and the prevention of cells from oxidative damages. The PINK1-Parkin pathway is the major mechanism that mediates depolarization-induced mitophagy (Campello et al., 2014). PINK1 is a serine-threonine kinase that is recruited to mitochondrial outer membrane upon loss of mitochondrial membrane potential, for example, after treatment of the uncoupling agent CCCP. PINK1 phosphorylates and activates the E3 ubiquitin ligase Parkin; in addition, recent findings from three independent groups showed that PINK1-mediated phosphorylation of ubiquitin at Ser65 is also required to recruit and activate Parkin responsible for mitochondrial ubiquitination (Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014). Ubiquitinated mitochondria are subsequently recognized by the ubiquitin binding adaptor protein p62 in the autophagy machinery and degraded by selective autophagy (Narendra et al., 2008; Geisler et al., 2010).
Ribophagy
Ribosomes are another example of intracellular organelles degraded by selective autophagy, referred to as “ribophagy.” Although ribosomes can be eliminated through non-selective autophagic degradation of bulk cytoplasm, the ribosomal proteins showed a higher turnover kinetic compared to cytoplasmic proteins after a prolonged period of starvation, suggesting that starvation-induced ribophagy is a selective process (Kraft et al., 2008). Ribophagy targets both large and small ribosomal subunits for degradation, and the mechanism is most clearly studied in yeast. In contrast to other types of selective autophagy, where the ubiquitin signaling promotes cargo recognition and degradation by the autophagy pathway, ubiquitination of the large subunit 60S ribosomes by the Ltn1 E3 ubiquitin ligase prevents their degradation by ribophagy in yeast (Ossareh-Nazari et al., 2014). Meanwhile, the Ubp3ubiquitin protease and its activator Bre5 are required for ribophagy of the 60S subunit (Kraft et al., 2008). It still needs to be investigated what specific cargo receptors mediate this process.
Regarding the metabolic roles of ribophagy, since the ribosomes represent a huge portion of cellular mass, they may serve as an important source of metabolites for the cell during nutrient limitation. Furthermore, degradation of ribosomes upon environmental challenges may be essential to adjust the level of protein translation in order to preserve energy and nutrient for cell homeostasis (Cebollero et al., 2012). However, further studies are needed to test this hypothesis in higher organisms, especially in mammals.
Metabolic functions of autophagy in tissues
Metabolic disorders, such as type 2 diabetes and its major risk factor obesity, affect the population worldwide and are the combined effects of several factors such as genetics, environment and life style. The most recent estimation reveals that in 2013 382 million people have diabetes, and this number is expected to increase within years (Guariguata et al., 2014). In these circumstances, it is important to understand how stress-induced autophagy exerts its diverse impacts in the pathogenesis of metabolic disorders. Here, we will discuss the specific metabolic roles of autophagy in different tissues as follows (Fig. 3).
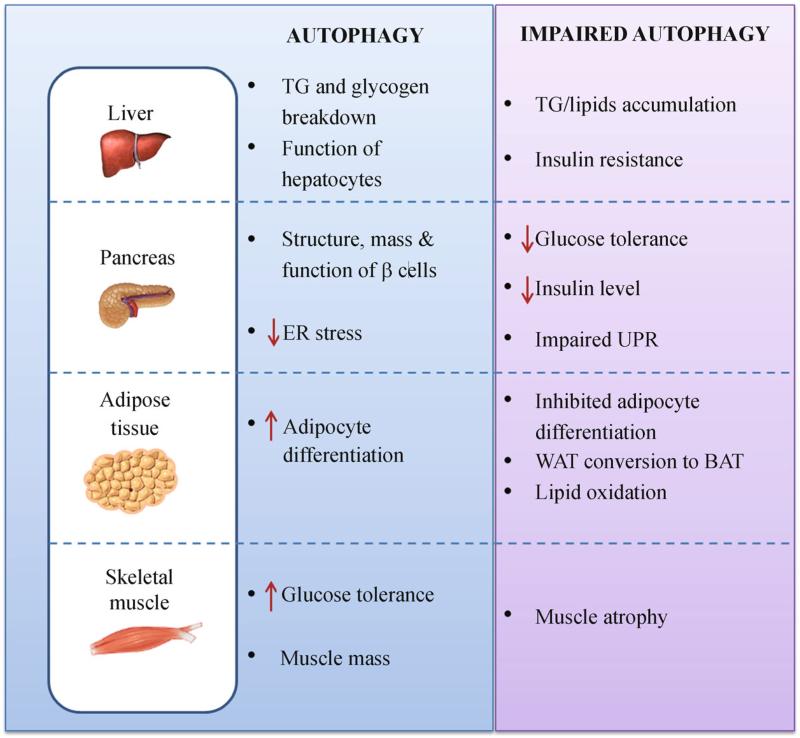
Figure 3.
Functions of autophagy in multiple metabolic tissues. The roles of autophagy in selective tissues are summarized, based on the results from both in vitro and in vivo studies. Impaired autophagy imposes opposite effects in specific organs, leading to diverse metabolic abnormalities and susceptibility to diseases. TG, triglycerides; UPR, unfolded protein response; WAT, white adipose tissue; BAT, brown adipose tissue.
Pancreas
The high blood glucose level in diabetes is a result of impaired insulin-producing β cells that are unable to compensate for peripheral insulin insensitivity (Kahn et al., 2006); thus, pancreatic β cell failure is a key point in the progression of diabetes. Autophagy has been found to be important for the maintenance of the structure, cell mass and function of β cells. For example, β cell-specific deletion of Atg7 leads to defective basal autophagy, morphological abnormalities of islets and decreased insulin secretion, which are symptoms typical of both type 1 and type 2 diabetes (Ebato et al., 2008; Jung et al., 2008). In response to stress such as HFD treatment, these mice develop more severe diabetic phenotypes such as hyperglycemia and impaired glucose tolerance. Similarly, β cell-ablation of Atg7 in obese mice (ob/ob background) also results in decreased function and mass of β cells and glucose intolerance, possibly due to a combination of mitochondrial dysfunction, ER stress, accumulation of misfolded proteins and altered unfolded protein response (UPR) (Jung and Lee, 2010; Quan et al., 2012b). On the other hand, an elevation of autophagy activity in islets hasbeen observed in response to HFD treatment, indicating that upregulated autophagy may be an adaptive response of β cells to combat diet-induced insulin insufficiency and insulin resistance (Ebato et al., 2008).
Adipose tissue
In the adipose tissue, autophagy plays an important role in the regulation of adipocyte development and differentiation. Genetic ablation of Atg7 or Atg5 specifically in adipose tissue causes abnormal lipid accumulation and adipose tissue composition (Baerga et al., 2009; Singh et al., 2009b; Zhang et al., 2009; Zhang et al., 2013). In brief, inhibition of autophagy leads to a decrease of adipocyte differentiation into the white adipose tissue (WAT). Concurrent with a mass loss of WAT, the WAT assumes a phenotype similar to the brown adipose tissue (BAT), characterized by more mitochondria and a higher rate of lipid β oxidation, due to a failure in mitochondrial removal by mitophagy (Singh et al., 2009b; Goldman et al., 2010). Autophagy repression is also accompanied by upregulation of the compensatory UPS system, leading to increased degradation of PPARγ2, a transcriptional factor required for adipogenesis (Zhang et al., 2013; Lee et al., 2014). Accordingly, inhibition of autophagy in adipose tissue results in a lean and obesity-resistant phenotype. These studies highlighted the tissue specificity and functional diversity of autophagy in metabolic regulation, and revealed an interesting link connecting autophagy and obesity rooting from tissue development.
Muscle
Basal autophagy in skeletal muscle plays a role in supporting muscle mass and morphology, and stimulated autophagy by exercise has systematic beneficial effects on glucose homeostasis and metabolic profile. There is strong in vivo evidence that suppression of basal autophagy is associated with accumulation of toxic aggregated proteins and muscle atrophy, based on studies on skeletal muscle-specific deletion of Atg7 by Cre recombinase driven by the Mlc1f (myosin light chain 1 fast) promoter, or deletion of Atg5 by Cre driven by the HAS (human skeletal actin) promoter (Raben et al., 2008; Masiero et al., 2009). Interestingly, a recent study using the same skeletal muscle-specific Atg7 knockout (KO) mice (Atg7 flox/flox; Mlc1f-Cre) showed that they are protected from HFD-induced obesity and insulin resistance. This study demonstrated that these mice lose not only muscle mass, but are also subject to a reduction in fat mass, mitochondrial dysfunction and an induction of Fgf21 (fibroblast growth factor 21), a myokine that leads to increased fat usage, higher β oxidation and consequent browning of WAT (Kim et al., 2013). However, it is not yet clear whether the increased expression of Fgf21 is caused by a compensatory response due to developmental or structural defects of skeletal muscle; it will be interesting to study whether Atg7 ablation under the control of an inducible promoter leads to the same phenotypes.
In addition to starvation as the most known physiological inducer of autophagy, a number of studies have shown that physical exercise potently stimulates autophagy in skeletal muscle, by disrupting the inhibitory Beclin 1-Bcl-2 complex (Grumati et al., 2011; He et al., 2012; Lira et al., 2013). Of note, using two different mouse models deficient in exercise-induced autophagy (BCL2 AAA knock-in mice and beclin1+/− KO mice), researchers have demonstrated that these mice fail to show muscle adaptation, endurance improvement and protection from HFD-induced diabetic symptoms after chronic exercise training. These lines of evidences strongly suggest that autophagy activation in muscle may have a systematic metabolic effect and may underlie the benefits of exercise against metabolic diseases. Further studies are required to sort out the mechanisms of autophagy induction during exercise.
Liver
Liver is one of the most metabolically active organs, controlling both carbohydrate and lipid metabolism; therefore, the importance of hepatic autophagy have been broadly explored. The first line of evidence came from a conditional Atg7 KO mouse model, in which the loss of Atg7 in hepatocyte causes accumulation of swollen and deformed mitochondria and an increased number of ubiquitinated protein aggregates and lipids droplets (Komatsu et al., 2005). Another study using poly (I:C)-inducible Atg7 KO mice suggest that these pathogenic effects in the liver may be due to impaired turnover of p62 (Komatsu et al., 2010). The researchers found that p62 competes with the stress-responsive transcription factor Nrf2 in binding Keap1, an ubiquitin ligase of Nrf2, leading to stabilization and persistent activation of Nrf2 and overproduction of its target antioxidant proteins (Komatsu et al., 2010; Ichimura et al., 2013; Kageyama et al., 2014). Other studies based on liver-specific ablation of autophagy genes demonstrated an increase in hepatic lipid content (Jaber et al., 2012; Settembre et al., 2013), which may be caused by defects in lipophagy regulated by the transcription factor TFEB (Singh et al., 2009a; Settembre et al., 2013). In summary, it becomes evident that hepatic autophagy plays a crucial role in the regulation of lipid metabolism, nutritional status and energy balance in the whole body.
Systematic autophagy in metabolic diseases
There has been increasing studies suggesting that the global autophagy activity is important in the maintenance of whole body metabolism. The potential relationship between systematic autophagy and obesity-associated type 2 diabetes arises from several concepts as described below.
Recent studies on a number of different genetic models of global autophagy deficiency have demonstrated the diverse roles of autophagy in systemic regulation of metabolism. The first line of evidence came from a study on the p62 global KO mouse model, which develops severe insulin resistance together with increased body fat. The authors revealed that p62 is able to inhibit adipogenesis by blocking the ERK activity (Rodriguez et al., 2006). Since p62 is a fundamental modulator/receptor of selective autophagy, it would be interesting to investigate how loss of p62 influences the degradation of specific cargos involved in adipocyte differentiation and insulin resistance in mice. Additional genetic models include heterozygous KO mouse models of Atg7 or Beclin 2, as homozygous deletion of either gene causes embryonic lethality (He et al., 2013; Lim et al., 2014). Specifically, Atg7+/− heterozygous KO mice do not show metabolic abnormalities until crossed with the ob/ob mice or under the treatment of HFD. Atg7+/− ob/ob mice, or HFD-treated Atg7+/− mice, have exacerbated insulin resistance, elevated lipid storage and inflammation. Application of imatinib or trehalose, both with autophagy-inducing activities, rescues the diabetic phenotypes in these mice (Lim et al., 2014). However, it is not clear whether the effects of either drug are directly through autophagy upregulation or through autophagy-independent functions. In addition, monoallelic loss of Beclin 2, a newly identified member in the beclin family, also leads to metabolic dysregulation (including elevated food intake, increased bodyweight and insulin resistance). Different from the models mentioned above that are specifically impaired in autophagy, the metabolic abnormalities seen in beclin2+/− KO mice are at least partially due to increased levels of a brain GPCR (G protein-coupled receptor) cannabinoid 1 receptor, which directly upregulates food intake and energy storage upon activation. Strong experimental evidence has shown that Beclin 2 regulates both autophagy via binding to components of the PtdIns3K complex, and agonist-induced lysosomal degradation of GPCRs via binding to an adaptor protein GASP1 (GPCR-associated sorting protein 1). Because the genetic locus of beclin 2 is also linked to obesity and diabetes related traits in multiple ethnic groups in human, Beclin 2 likely represents an important genetic determinant of human obesity and diabetes. Future investigations are needed to evaluate how defects in Beclin 2-regulated autophagy contribute to the pathogenesis of metabolic diseases.
Interestingly, recent studies have shown that autophagy is suppressed in genetic or dietary-induced mouse models of obesity in various tissues, including liver, skeletal muscle and cardiac muscle (Liu et al., 2009; Yang et al., 2010; He et al., 2012). This is likely caused by constantly high levels of circulating insulin (an autophagy-inhibitory hormone) due to insulin resistance and hyperinsulinemia induced by HFD (Liu et al., 2009). These studies also revealed that the expression of several key autophagy genes, including Vps34, Atg12, Atg8, Atg5, Atg7 and Beclin 1, is downregulated in hepatic cells from HFD-treated mice or ob/ob mice, which may involve FoxO1-mediated gene transcription and thus provides another layer of regulatory mechanism of autophagy inhibition during hyperinsulinemia. On the other hand, adenoviral restoration of Atg7 in ob/ob mice increases autophagy activity and protects the animals from insulin resistance (Yang L. et al., 2010).
Accordingly, these findings revealed a promising direction to pursue the therapeutic potential of autophagy induction in metabolic diseases. Studies have shown that the FDA-approved anti-diabetic drug metformin can induce autophagy activity (Kalender et al., 2010; Jiang et al., 2014). In addition, several autophagy-inducing compounds, such as rapamycin, trehalose or imatinib, have been tested on animal models of diabetes and have shown beneficial effects on insulin sensitivity and β cells protection (Newgard et al., 2009; Zhou et al., 2009; Gonzalez et al., 2011; Elzinga et al., 2013; Marselli et al., 2013;). However, future investigations will be necessary to analyze whether the beneficial effects of these drugs are directly through autophagy.
Conclusions
In the past decade, there has been increasing evidence on the important roles of autophagy in metabolic homeostasis. Despite the numerous evidences that autophagy exerts a protective role in obesity and insulin resistance in general, there are many answered questions for further study. For example, tissue-specific roles of autophagy at different developmental or aging stages are not completely understood, and transcriptional regulation of autophagy in many metabolic tissues under various stress conditions is largely unclear. On the other hand, autophagy itself is regulated by the levels of metabolic hormones and nutrients; thus, the reciprocal relationship between autophagy and metabolic disorders seems more complicated and needs to be better elucidated. In addition, more studies are required to evaluate the therapeutic potential and safety of autophagy modulation and to discover new pharmacological inducers of autophagy in the prevention of metabolic diseases. It will also be interesting to explore the autophagy-mediated beneficial mechanisms of existing clinically used drugs.
Given the critical functions of autophagy in the prevention of many metabolic-related disorders other than obesity and diabetes, including cancer, cardiovascular diseases, bone disorders and aging (Levine and Kroemer, 2008), the understanding of the mechanisms underlying the protective roles of autophagy in metabolism will help develop or improve therapies of a variety of widespread human diseases. Various cell-based and animal models generated in recent studies have provided useful genetic tools for the future research in these areas.
Acknowledgements
This work was supported by the NIH Pathway to Independence Award R00 DK094980 to C.H.
Footnotes
Compliance with ethics guidelines
Altea Rocchi and Congcong He declare that they have no conflict of interest. This manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee.
References
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidy-linositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5(8):1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle KB, Randow F. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol. 2013;16(3):339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010;584(7):1302–1312. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Campello S, Strappazzon F, Cecconi F. Mitochondrial dismissal in mammals, from protein degradation to mitophagy. Biochim Biophys Acta. 2014;1837(4):451–460. doi: 10.1016/j.bbabio.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Cebollero E, Reggiori F, Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol. 20122012:182834. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15(3):344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupé B, Ishii Y, Dietrich MO, Komatsu M, Horvath TL, Bouret SG. Loss of autophagy in pro-opiomelanocortin neurons perturbs axon growth and causes metabolic dysregulation. Cell Metab. 2012;15(2):247–255. doi: 10.1016/j.cmet.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28(1):435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, Kawamori R, Fujitani Y, Watada H. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8(4):325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Elzinga BM, Nyhan MJ, Crowley LC, Donovan TR, Cahill MR, McKenna SL. Induction of autophagy by Imatinib sequesters Bcr-Abl in autophagosomes and down-regulates Bcr-Abl protein. Am J Hematol. 2013;88(6):455–462. doi: 10.1002/ajh.23428. [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9(9):859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S, Zhang Y, Jin S. Autophagy and adipogenesis: implications in obesity and type II diabetes. Autophagy. 2010;6(1):179–181. doi: 10.4161/auto.6.1.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez CD, Lee MS, Marchetti P, Pietropaolo M, Towns R, Vaccaro MI, Watada H, Wiley JW. The emerging role of autophagy in the pathophysiology of diabetes mellitus. Autophagy. 2011;7(1):2–11. doi: 10.4161/auto.7.1.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Coletto L, Schiavinato A, Castagnaro S, Bertaggia E, Sandri M, Bonaldo P. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy. 2011;7(12):1415–1423. doi: 10.4161/auto.7.12.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract. 2014;103(2):137–149. doi: 10.1016/j.diabres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, Levine B. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Wei Y, Sun K, Li B, Dong X, Zou Z, Liu Y, Kinch LN, Khan S, Sinha S, Xavier RJ, Grishin NV, Xiao G, Eskelinen EL, Scherer PE, Whistler JL, Levine B. Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell. 2013;154(5):1085–1099. doi: 10.1016/j.cell.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51(5):618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, Zong WX. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci USA. 2012;109(6):2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, Yamamoto A, Krainc D. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137(1):60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, Wells CD, Skurat AV, Roach PJ. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem. 2010;285(45):34960–34971. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. 2011;413(3):420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Huang W, Wang J, Xu Z, He J, Lin X, Zhou Z, Zhang J. Metformin plays a dual role in MIN6 pancreatic β cell function through AMPK-dependent autophagy. Int J Biol Sci. 2014;10(3):268–277. doi: 10.7150/ijbs.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, Jeong YT, Han MS, Lee MK, Kim KW, Shin J, Lee MS. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8(4):318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Jung HS, Lee MS. Role of autophagy in diabetes and mitochondria. Ann N Y Acad Sci. 2010;1201(1):79–83. doi: 10.1111/j.1749-6632.2010.05614.x. [DOI] [PubMed] [Google Scholar]
- Kageyama S, Sou YS, Uemura T, Kametaka S, Saito T, Ishimura R, Kouno T, Bedford L, Mayer RJ, Lee MS, Yamamoto M, Waguri S, Tanaka K, Komatsu M. Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J Biol Chem. 2014;289(36):24944–24955. doi: 10.1074/jbc.M114.580357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444(7121):840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- Kalender A, Selvaraj A, Kim SY, Gulati P, Brûlé S, Viollet B, Kemp BE, Bardeesy N, Dennis P, Schlager JJ, Marette A, Kozma SC, Thomas G. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010;11(5):390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205(2):143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Arias E, Kwon H, Lopez NM, Athonvarangkul D, Sahu S, Schwartz GJ, Pessin JE, Singh R. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. 2012;13(3):258–265. doi: 10.1038/embor.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22(8):407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, Singh R. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14(2):173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460(1):127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim H, Hur KY, Kim HK, Ko T, Han J, Kim HL, Kim J, Back SH, Komatsu M, Chen H, Chan DC, Konishi M, Itoh N, Choi CS, Lee MS. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19(1):83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- Kim KH, Lee MS. Autophagy—a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10(6):322–337. doi: 10.1038/nrendo.2014.35. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Øvervatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 2010;584(12):2635–2645. doi: 10.1016/j.febslet.2010.04.041. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J, Dikic I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013;126(Pt 2):580–592. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21(7):683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Le Guezennec X, Brichkina A, Huang YF, Kostromina E, Han W, Bulavin DV. Wip1-dependent regulation of autophagy, obesity, and atherosclerosis. Cell Metab. 2012;16(1):68–80. doi: 10.1016/j.cmet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516(7529):112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YM, Lim H, Hur KY, Quan W, Lee HY, Cheon H, Ryu D, Koo SH, Kim HL, Kim J, Komatsu M, Lee MS. Systemic autophagy insufficiency compromises adaptation to metabolic stressand facilitates progression from obesity to diabetes. Nat Commun. 2014;5:4934. doi: 10.1038/ncomms5934. [DOI] [PubMed] [Google Scholar]
- Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, Breen DS, Hoehn KL, Yan Z. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J. 2013;27:4184–4193. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284(45):31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Marselli L, Bugliani M, Suleiman M, Olimpico F, Masini M, Petrini M, Boggi U, Filipponi F, Syed F, Marchetti P. β-Cell inflammation in human type 2 diabetes and the role of autophagy. Diabetes Obes Metab. 2013;15(Suppl 3):130–136. doi: 10.1111/dom.12152. [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27(1):107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS, Jr, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, Piao Z, Ashida H, Yoshida M, Kakuta S, Koyama T, Goto Y, Nagatake T, Nagai S, Kiyono H, Kawalec M, Reichhart JM, Sasakawa C. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 2011;9(5):376–389. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Niño CA, Bengtson MH, Lee JW, Joazeiro CA, Dargemont C. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J Cell Biol. 2014;204(6):909–917. doi: 10.1083/jcb.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13(6):655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23(58):9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128(5):931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Quan W, Kim HK, Moon EY, Kim SS, Choi CS, Komatsu M, Jeong YT, Lee MK, Kim KW, Kim MS, Lee MS. Role of hypothalamic proopiomelanocortin neuron autophagy in the control of appetite and leptin response. Endocrinology. 2012a;153(4):1817–1826. doi: 10.1210/en.2011-1882. [DOI] [PubMed] [Google Scholar]
- Quan W, Lim YM, Lee MS. Role of autophagy in diabetes and endoplasmic reticulum stress of pancreatic β-cells. Exp Mol Med. 2012b;44(2):81–88. doi: 10.3858/emm.2012.44.2.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17(24):3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330(6009):1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11(9):1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Durán A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3(3):211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Ryter SW, Cloonan SM, Choi AMK. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol Cells. 2013;36(1):7–16. doi: 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santambrogio L, Cuervo AM. Chasing the elusive mammalian microautophagy. Autophagy. 2011;7(6):652–654. doi: 10.4161/auto.7.6.15287. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284(41):28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516(7529):108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination andselective autophagy. Cell Death Differ. 2013;20(1):21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang L, Wang X. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy. 2011;7(8):924–926. doi: 10.4161/auto.7.8.15860. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117(Pt 18):4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009a;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009b;119(11):3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegué E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494(7437):323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Bejarano E, Rakshit M, Lee K, Hanson HH, Zaarur N, Phillips GR, Sherman MY, Cuervo AM. Molecular determinants of selective clearance of protein inclusions by autophagy. Nat Commun. 2012;3:1240. doi: 10.1038/ncomms2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11(6):467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, He Y, Okutsu M, Ong LC, Jin Y, Zheng L, Chow P, Yu S, Zhang M, Yan Z. Autophagy is involved in adipogenic differentiation by repressesing proteasome-dependent PPARγ2 degradation. Am J Physiol Endocrinol Metab. 2013;305(4):E530–E539. doi: 10.1152/ajpendo.00640.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA. 2009;106(47):19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183(9):5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Sebhat IK, Zhang BB. AMPK activators—potential therapeutics for metabolic and other diseases. Acta Physiol (Oxf) 2009;196(1):175–190. doi: 10.1111/j.1748-1716.2009.01967.x. [DOI] [PubMed] [Google Scholar]