Abstract
PE_PGRS represent a large family of proteins typical of pathogenic mycobacteria whose members are characterized by an N-terminal PE domain followed by a large Gly-Ala repeat-rich C-terminal domain. Despite the abundance of PE_PGRS-coding genes in the Mycobacterium tuberculosis (Mtb) genome their role and function in the biology and pathogenesis still remains elusive. In this study, we generated and characterized an Mtb H37Rv mutant (MtbΔ33) in which the structural gene of PE_PGRS33, a prototypical member of the protein family, was inactivated. We showed that this mutant entered macrophages with an efficiency up to ten times lower than parental or complemented strains, while its efficiency in infecting pneumocytes remained unaffected. Interestingly, the lack of PE_PGRS33 did not affect the intracellular growth of this mutant in macrophages. Using a series of functional deletion mutants of the PE_PGRS33 gene to complement the MtbΔ33 strain, we demonstrated that the PGRS domain is required to mediate cell entry into macrophages, with the key domain encompassing position 140–260 amino acids of PE_PGRS33. PE_PGRS33-mediated entry into macrophages was abolished in TLR2-deficient mice, as well as following treatment with wortmannin or an antibody against the complement receptor 3 (CR3), indicating that PE_PGRS33-mediated entry of Mtb in macrophages occurs through interaction with TLR2.
Introduction
Tuberculosis (TB) is still one of the most relevant public health issues worldwide and primarily in developing countries where it poses a tremendous health, economic and social burden [1]. Decisive prophylactic and therapeutic tools are still missing, in part because many aspects of TB pathogenesis and of Mycobacterium tuberculosis (Mtb) biology remain elusive [2,3]. In the last two decades, a new understanding of this ancient disease has been gained following the deciphering of the Mtb genome [4], which uncovered unexpected and surprising features of the tubercle bacilli [5,6]. The disclosure that almost 10% of the Mtb genome coding capacity is devoted to the mycobacterial-specific PE and PPE protein families raised much interest on their role in Mtb biology [7–10].
All PE proteins are characterized by a highly conserved N-terminal domain of ≅ 100 amino acids with the presence of a proline-glutamic acid (PE) motif at position 8–9 [4]. Out of the 99 PE genes found in Mtb H37Rv [4], 63 were annotated as PE_PGRS though some of these were pseudogenes or lacked some of the typical PE_PGRS features, so that only 51 PE_PGRS potentially functional proteins are expressed [8]. PE_PGRSs proteins share the same molecular architecture, characterized by the presence, beyond a PE domain, of i) a typical PGRS domain varying in sequence and size containing a variable number of GGA-GGN repeats; ii) a highly conserved putative transmembrane domain linking the PE and PGRS domains with a GRPLI motif around position 115; iii) a unique C-terminal domain which is usually less than 30 amino acids long, but that in some cases (such as in PE_PGRS30) can be as large as 300 amino acids [7,11,12]. PE_PGRS-encoding genes are found only in few mycobacterial species (M. marinum, M. ulcerans and members of the Mtb complex), are scattered throughout the genome, and are differently regulated. The transcriptional regulation of some of them appears to be finely tuned depending on the environmental signals encountered during the complex steps of the infectious process, while others (as that encoding PE_PGRS33) are constitutively expressed [7,8,13,14]. The paucity of experimental data on PE_PGRSs has so far hampered a sufficient understanding of their role in TB pathogenesis.
PE_PGRS33 (Rv1818c), which can be considered a model for the family, is a 498 amino acids protein whose PE domain mediates its translocation on the mycobacterial outer membrane [15], and was used to deliver antigens on the mycobacterial surface [16–18]. Its surface localization makes it available for interaction with host components, as suggested by studies carried out with the avirulent vaccine strain Mycobacterium bovis BCG [19]. Interestingly, PE_PGRS33 was previously shown to trigger macrophage cell death by inducing secretion of pro-inflammatory cytokines [20,21] and activation of pro-apoptotic or pro-necrotic signals involving mitochondria [22–24]. Basu et al. [23] showed that PE_PGRS33 was capable of inducing TLR2-dependent apoptosis in macrophages. All the studies aimed at investigating the pro-inflammatory role of PE_PGRS33 were carried out by ectopically expressing PE_PGRS33 in the avirulent species M. smegmatis [20,21], by directly using the purified recombinant protein obtained in Escherichia coli [22,23] or by expressing the mycobacterial protein directly in host cells using an eukaryotic expression plasmids [24].
In this study, we generated an Mtb Rv1818c null mutant, which was complemented with a series of Rv1818c genes manipulated to functionally dissect PE_PGRS33 structure. The resulting strains were used to investigate the role of PE_PGRS33 in Mtb cell entry gaining insights in the involvement of this protein in TB pathogenesis.
Material and Methods
Bacterial Strains
Mtb was grown at 37°C in Middlebrook 7H9 or 7H10 (Difco Becton-Dickinson), supplemented with 0.2% glycerol (Sigma-Aldrich), ADC 10% (Becton-Dickinson), and 0.05% v/v Tween 80 (Sigma-Aldrich). The PE_PGRS33 mutant was generated in Mtb H37Rv by allelic exchange using the recombineering system [25]. Briefly, we constructed a pJSC-derivative vector, in which a region upstream Rv1818c (-563 to -1 bp) and a region internal to its coding sequence (561 to 1004 bp) were cloned at the flanks of an hygromycin cassette. The resulting recombination substrate was digested, purified and introduced by electroporation in Mtb H37Rv competent cells containing pJV53, carrying the recombinases and conferring kanamycin resistance. Transformants were first selected on 7H11/OADC-Tween 80 plates containing hygromycin (50 μg ml-1) at 37°C for 3–4 weeks and selected colonies analyzed by PCR to demonstrate Rv1818c deletion (S1 Fig). One mutant with the correct deletion was sub-cultured in drug-free media for about 10 generations to allow the loss of pJV53 and then plated on solid medium containing hygromycin at 37°C. Single colonies were picked and analyzed for loss of the kanamycin resistance to isolates a mutant without pJV53. The Rv1818c null mutant was then complemented using the integrative pMV306-derivative pAL79 [17] containing a copy of Rv1818c gene fused with the sequence encoding the HA epitope under the control of its own putative promoter (S2 Fig) [16].
Generation of PE_PGRS33 Functional Deletion Mutants for the PGRS Domain
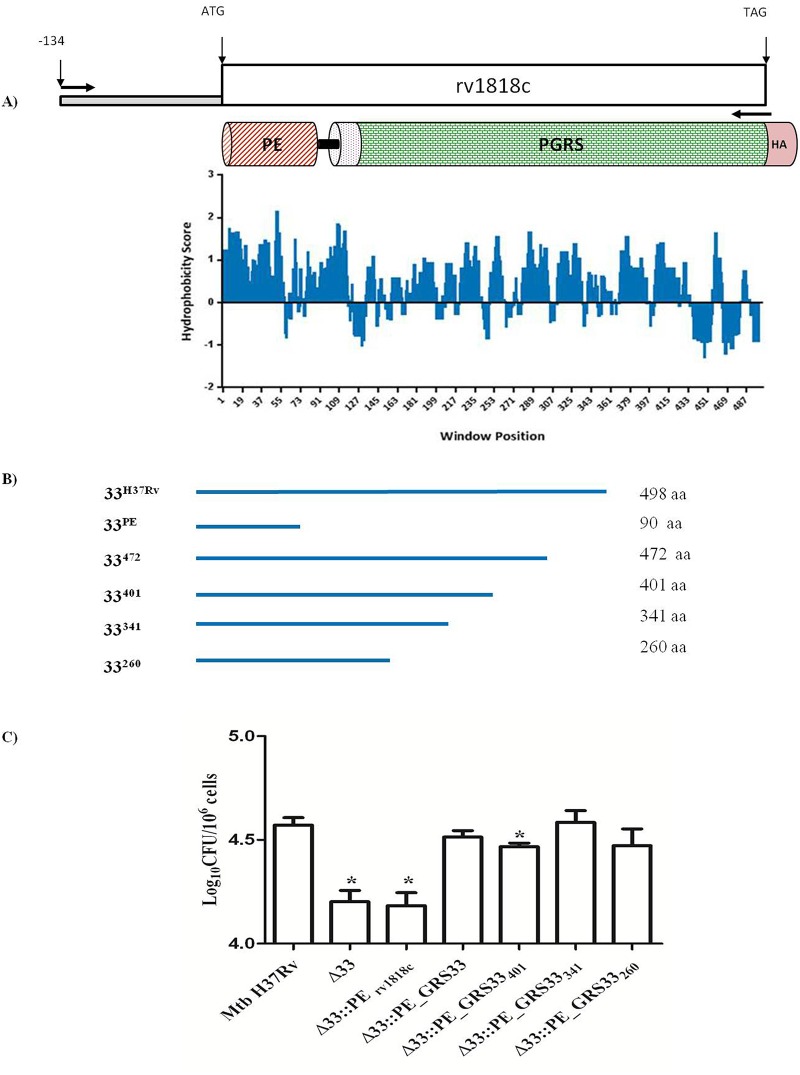
The Mtb H37Rv PE_PGRS33 hydrophobicity profile was obtained by in silico analysis using the Kyte-Doolittle scale (CLC Main Workbench 6.9). Based on the hydrophobic/hydrophilic characteristics at the C-terminal PGRS domain, four Rv1818c fragments encoding the first 472, 401, 341 and 260 amino acids, lacking variable size of the PGRS domain, were amplified from the Mtb H37Rv genome (using primers indicated in S1 Table) and cloned under the control of the Rv1818c native promoter into the integrative plasmid pMV306 upstream and in frame with the HA epitope sequence.
Cell Line Culture
The J774 cell line (murine macrophage) were grown in RPMI 1640 medium containing 10% fetal calf serum (FCS) supplemented with glutamine (2 mM), streptomycin (100 μg/ml), penicillin (100 U/ml), and sodium pyruvate (1 mM). Cells were kept in a humidified atmosphere containing 5% CO2 at 37°C. The cells were diluted to 1.2 x106 per ml in 48 and 24-well plates with 2% FCS. Human THP-1 monocytic cells were grown in RPMI 1640 supplemented with glutamine (2 mM) and 10% FCS. Cells were treated with 20 nM PMA (Sigma-Aldrich, St. Louis, MO) for 24 h to induce their differentiation into macrophage-like cells, then washed three times with PBS and maintained in 2% FCS.
The human lung adenocarcinoma epithelial cell line, A549 (ATCC, Rockville, MD), were used as model of human type II alveolar epithelial cells. Cells were grown in complete medium consisting of RPMI 1640 supplemented with 10% FCS, 2 mM L-glutamine and 5 μg/ml of gentamicin, and split when a confluent cell monolayer was attained [26].
Primary Murine Peritoneal Macrophages (pMM0)
C57BL/6 mice were purchased from Harlan (Italy) and maintained in pathogen-free micro-isolator cages. The animals were housed in a temperature-controlled environment with 12 h light/dark cycles, and received food and water ad libitum. All animal experiments were authorized by the Ethical Committee of the Università Cattolica del Sacro Cuore (n°T21/2011) and performed in compliance with the legislative decree of the Italian Government 27 January 1992, n. 116 and the Health Minister memorandum 14 May 2001, n. 6. All manipulations were performed under isoflurane anesthesia, and all efforts were made to minimize suffering. Nine-fourteen weeks-old female mice were euthanized by inhalation of isoflurane (Baxter, Warsaw, Poland). Inflammatory peritoneal cells, elicited with 1 ml of aged 3% thioglycollate (Difco, Detroit, MI), injected intraperitoneal 4–5 days earlier, were washed out with phosphate buffered saline (PBS) and collected into centrifuge tubes kept on ice. After washing once, peritoneal cells were re-suspended in the RPMI/FBS medium and plated at 1.2×106 cells/ml in 48-well tissue culture plates. Following overnight incubation, non-adherent cells were removed by washing, whereas adherent macrophages were used for experiments. Similar procedures were used to isolate pMM0 from TLR2-/- mice.
Mycobacterial Infection and Bacterial Counts In Vitro
Cells were infected with mycobacteria at different multiplicity of infection (MOI) and incubated for 4 hours post-infection at 37°C in a 5% CO2 atmosphere. Cells were then washed three times with PBS to remove extracellular bacteria and then incubated with complete RPMI 2% of FCS without antibiotics. To assess the ability of the Mtb strains to survive intracellularly, macrophages were infected at different MOIs (1:10, 1:1 or 10:1) and infection in A549 cells was carried at MOI of 5:1. Cells were washed with PBS three times to remove extracellular bacteria and then incubated in fresh medium. At different time points, cells were lysed in 0.1% of Triton X-100 and intracellular bacteria determined by CFU counting by serially diluting lysates in PBS containing Tween80 (0.05%) and plating on 7H11/OADC agar plates [13]. Colony counting was then performed in triplicate. In some cases, after infection the supernatant, containing not internalized bacteria was collected and plated on 7H11/OADC agar plates.
pMM0 were treated before infection with wortmannin (Calbiochem, San Diego, CA) at the concentration of 100 nM and 300 nM and with mAb anti-CR3 at final concentration of 10 μg/ml (clone M1/70 BD, Biosciences) for 30 minutes at 37°C and then infected with Mtb strains at MOI 1:10 and intracellular bacteria counted as above. Statistical analysis to assess differences between the MtbΔ33 mutants strains and parental and complemented strains was performed using the Student’s t-test and ANOVA with GraphPad Prism software version 5.0 (GraphPad software, CA, USA).
Results
Generation of an Rv1818c Mutant and Its Complementation
To investigate the role of PE_PGRS33 in Mtb pathogenesis, inactivation of the Rv1818c gene by recombineering [25] was carried out on the Mtb H37Rv strain as shown in S1 Fig, to generate the MtbΔ33 mutant strain. The lack of an antibody specific for PE_PGRS33 prevented the demonstration of the gene inactivation by immunoblot [27]. The mutant strain was complemented by transformation with an integrative plasmid containing Rv1818c fused with the sequence encoding the HA epitope, expressed from its own promoter [16,17] (S2 Fig). Expression of the PE_PGRS33HA in the MtbΔ33 complemented strain (MtbΔ33::PE_PGRS33HA) was demonstrated by immunoblotting using an anti-HA antibody (S2 Fig). Conversely, from what previously observed with the BCG mc21525 mutant, where inactivation of the Rv1818c was obtained by transposon mutagenesis [19], no significant differences in the growth features of the Mtb H37Rv, MtbΔ33 and MtbΔ33::PE_PGRS33HA in solid media and liquid culture were observed (data not shown). In any case, the lack of a phenotype in axenic culture for the MtbΔ33 mutant provided a reliable model to investigate the role of PE_PGRS33 in Mtb cell entry.
PE_PGRS33 Is Required for Mtb Cell Entry into Macrophages
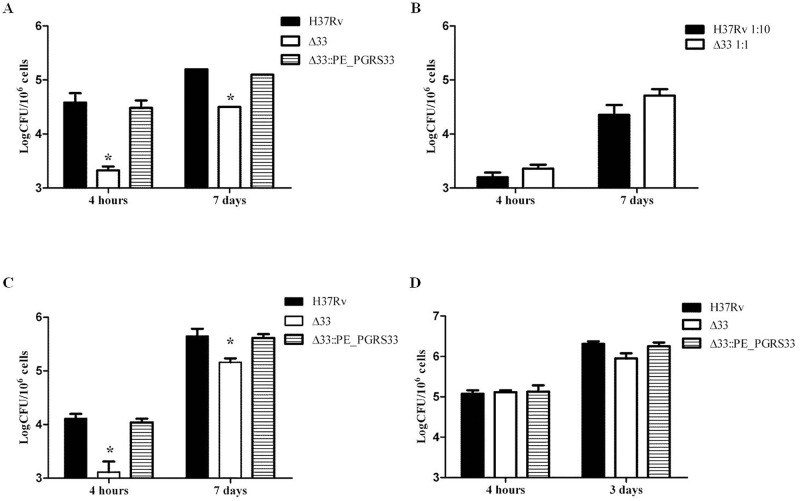
Seminal studies showed that PE_PGRS33 is exposed on the surface and that lack of PE_PGRS33 in a BCG transposon insertion mutant prevents proper bacilli entry into host cells [19]. To assess whether this was also true for virulent Mtb, J774 murine macrophages were infected with H37Rv, the MtbΔ33 mutant and the MtbΔ33::PE_PGRS33HA complemented strain. The mutant strain was strongly impaired in its ability to enter macrophages by a reduction of 1.3 Log CFU of intracellular bacteria after 4h of infection compared to wild type strain, (p<0.01), although the intracellular replication rate of the mutant that entered macrophages was similar to that of the parental or complemented strains (Fig 1A). Interestingly, when the infection was repeated increasing the multiplicity of infection (MOI) of the MtbΔ33 mutant, keeping the number of the parental Mtb strain constant, at 4h post-infection a similar number of intracellular bacteria for the two strains was found and their number increased after 7 days of infection (Fig 1B).
Fig 1. MtbΔ33 is impaired in its ability to enter macrophages.
Murine J774 macrophages (A and B), human monocyte-derived macrophages THP-1 (C) and human type II pneumocytes A549 (D) were infected with Mtb H37Rv, MtbΔ33 and MtbΔ33::PE_PGRS33 strains at a MOI 1:10 (A-C) or 5:1 (D) and following incubation cells were washed and at the different time points intracellular bacteria were determined by CFU counting. In the experiment shown in panel B, the Mtb H37Rv was infected at MOI 1:10, while the MtbΔ33 at MOI 1:1. Results of one representative experiments from at least three assays are shown (*p<0.,01). CFUs were expressed as mean ± SD and were analysed by two-way ANOVA followed by Bonferroni posttest.
The role of PE_PGRS33 in Mtb entry was also confirmed in human monocyte-derived macrophages (THP-1), with MtbΔ33 impaired in its ability to enter these cells (Fig 1C; - 0.99 Log CFU in the MtbΔ33 compared to the Mtb H37Rv parental strain; p<0.01), yet invading organisms were still capable of intracellular replication as shown by intracellular CFUs measured at day 7 (Fig 1A–1C). Taken together these results indicate that PE_PGRS33 significantly contributes to the ability of Mtb to enter macrophages, but is dispensable for Mtb intracellular replication.
Interestingly, when the same experiments were performed in type II human pneumocytes (A549) following usual experimental settings [26], no differences in the ability to infect these cells were observed among the three strains (Fig 1D), suggesting that PE_PGRS33 is required for Mtb entry in macrophages but not in epithelial cells.
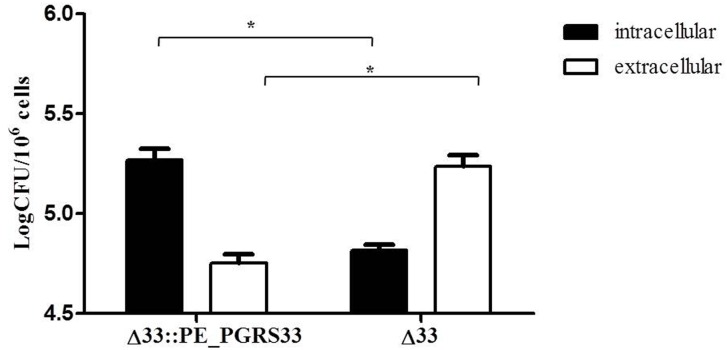
To further investigate the role of PE_PGRS33 in Mtb cell entry, murine peritoneal macrophages (pMM0) from C57Bl/6 mice were infected with Mtb H37Rv, MtbΔ33 and MtbΔ33::PE_PGRS33HA and the number of intracellular and extracellular bacilli was assessed by CFU counting. As shown in Fig 2, while most of the MtbΔ33::PE_PGRS33HA bacilli were detected intracellularly, the MtbΔ33 mutant was primarily detected in the supernatant and an inverse correlation among the two Mtb strains was observed, clearly indicating that viable MtbΔ33 mutant organisms are indeed impaired in their ability to enter these macrophages.
Fig 2. Deletion of PE_PGRS33 gene is associated with a defect in bacterial entry in pMM0.
pMM0 were infected with Mtb H37Rv, MtbΔ33 and MtbΔ33::PE_PGRS33 strains at MOI 1:1 to measure the number of intracellular vs extracellular bacteria at 4h post infection. Results of one representative experiments from at least three assays are shown (*p<0.05). CFUs were expressed as mean ± SD and were analysed by two-way ANOVA followed by Bonferroni posttest.
PE_PGRS33-Mediated Mtb Cell Entrance in Macrophages Occurs through Interaction with TLR2
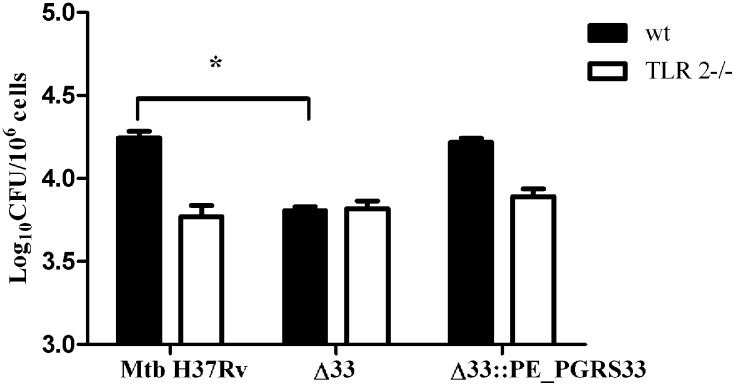
A number of mycobacterial components are known to interact with TLR2, such as lipoarabinomannan (LAM), lipomannan (LM), phosphatidil-myo-inositol mannoside (PIM), the 19-kDa lipoprotein [28]. The immunomodulatory properties of the mycobacterial surface protein PE_PGRS33 were found to be dependent upon the ability of the protein to bind and trigger TLR2 [22,23]. Interestingly, interactions of microbial ligands with TLR2 were shown to mediate entry of certain bacteria into macrophages [29]. To establish whether PE_PGRS33-mediated entrance of Mtb in macrophages was dependent on TLR2, pMM0 isolated from wild type and TLR2-deficient mice were infected with H37Rv, MtbΔ33 and the complemented strain. As shown in Fig 3, no differences were observed between MtbΔ33 and the parental or complemented strains in their ability to enter in TLR2-/- pMM0. These results demonstrate that PE_PGRS33 mediates entrance of Mtb in macrophages in a TLR2-dependent mechanism.
Fig 3. PE_PGRS33-mediated Mtb cell entry in macrophages occurs through interaction with TLR2.
pMM0 isolated from C57Bl/6 and syngeneic TLR2-/- mice were infected with Mtb H37Rv, MtbΔ33 and MtbΔ33::PE_PGRS33HA and intracellular CFUs were determined at 4h post-infection. Results of one representative experiments of at least three assays are shown (**p<0,.01). CFUs were expressed as mean ± SD and were analysed by two-way ANOVA followed by Bonferroni posttest.
PE_PGRS33 Activates the TLR2-Dependent Pro-Adhesive Pathway in Macrophages
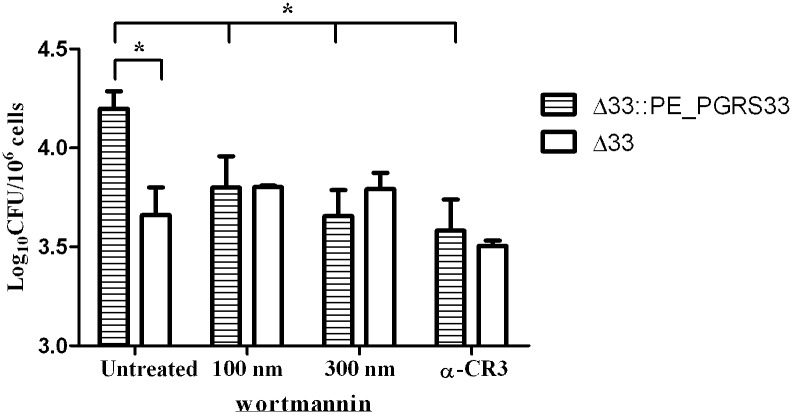
Interaction of bacterial surface components with TLR2 has been shown to trigger the pro-adhesive pathway by enhancing the avidity of the CR3 receptor through the activity of phosphatidylinositol 3-kinase (PI3K) [30,31]. To investigate whether this was also the case in the PE_PGRS33-mediated entrance of Mtb, pMM0 were previously treated with wortmannin, an inhibitor of PI3K [31]. As shown in Fig 4, pMM0 treated with wortmannin were impaired in their ability to phagocytize MtbΔ33::PE_PGRS33HA, but not MtbΔ33, compared to untreated pMM0 indicating that inhibition of PI3K abolishes the PE_PGRS33-mediated entrance in macrophages. Equally, incubation of pMM0 with an antibody directed against CR3 inhibited entrance of MtbΔ33::PE_PGRS33HA, but not of MtbΔ33 into macrophages (Fig 4) providing further evidences that the activation of TLR2 by PE_PGRS33 triggers the pro-adhesive pathway which enhances CR3 avidity for mycobacteria. The PE_PGRS33-mediated entrance of Mtb in macrophages was observed in opsonized and non-opsonized bacilli (data not shown) suggesting that this phenomena is not restricted for non-opsonic entrance [32]. Further characterization of the MtbΔ33 mutant, including in vivo studies, will provide insights on the role of PE_PGRS33 in the early and late steps of the TB infectious process.
Fig 4. PE_PGRS33 activates the TLR2-dependent pro-adhesive pathway in macrophages.
pMM0 isolated from C57Bl/6 mice were incubated with wortmannin or with an anti-CR3 antibody (M1/70), or left untreated, and then infected (MOI 1:10) with MtbΔ33 and the complemented strain MtbΔ33::PE_PGRS33HA and intracellular CFUs were determined at 4h post-infection. Results of one representative experiments of at least three assays are shown (*p<0,.001). CFUs were expressed as mean ± SD and were analysed by two-way ANOVA followed by Bonferroni posttest.
The PGRS Domain Encompassing Position 140–260 of the Protein Is Responsible for PE_PGRS33-Mediated Mtb Entry into Macrophages
In an attempt to identify the functional region(s) of PE_PGRS33 implicated in the entry of Mtb into macrophages, we analyzed the protein hydrophobicity pattern (Fig 5A), which revealed the presence of numerous hydrophobic regions intercalated by shorter hydrophilic segments, primarily located in the PGRS domain and likely available on Mtb surface for interaction with TLR2 [16,17,23]. Based on these observations, four PE_PGRS33 gene fragments corresponding to the sequences encoding the first 472, 401, 341 and 260 amino acid residues of PE_PGRS33 (PE_PGRS33472, PE_PGRS33401, PE_PGRS33341, PE_PGRS33260) were cloned in an integrative plasmid under the transcriptional control of the native promoter. These sequences were inserted in frame with the HA epitope sequence to demonstrate protein expression of the functional deletion mutants by immunoblot (data not shown). These four plasmids and the plasmid encoding the functional mutant lacking the entire PGRS domain (PE1818c) [17] were used to complement the MtbΔ33 strain (Fig 5B). pMM0 cells were infected at a MOI of 1:10 with Mtb H37Rv, MtbΔ33, MtbΔ33::PE_PGRS33 and the panel of complemented strains and bacterial entry was measured by counting intracellular mycobacteria at 4 hours post-infection. As expected [17,23,33], complementation of the MtbΔ33 mutant with the plasmid expressing the PE1818c domain only did not restore the wild type phenotype (Fig 5C). Conversely, complementation of the mutant strain with the four functional deletion mutants for the PGRS region of PE_PGRS33 restored the wild type phenotype, with the only exception of the MtbΔ33::PE_PGRS33401 strain. Taken together these results suggest that the protein domain of PE_PGRS33 that contributes to Mtb entry in macrophages resides within the first 260 amino acids of the protein.
Fig 5. PE_PGRS33 encoding the first 260 amino acids is capable to mediate the entrance phenotype into macrophages.
Schematic of PE_PGRS33 fragments (A-C). Truncated forms of Rv1818c were cloned downstream the native promoter PE_PGRS33 and in frame with the HA epitope sequence into the integrative plasmid pMV306. A) Rv1818c gene and its native putative promoter located 134 nucleotides upstream the start codon. Schematic overview showing PE_PGRS33 domains: PE domain (red stripped), transmembrane region (magenta dotted), PGRS domain (green netting), HA epitope (rose). The hydrophobicity profile of PE_PGRS33 was obtained using CLC Main Workbench 6.9 (Kyte-Doolittle scale, window size 9). B) Stylized representation of full-length PE_PGRS33, PE domain and the 472, 401, 341 and 260 amino acid in length fragments (blue lines). Cloning and protein information are reported beside. C) pMM0 isolated from C57BL/6 mice were infected at a MOI of 1:10 with Mtb H37Rv, MtbΔ33, MtbΔ33::PE_PGRS33HA and complemented strains expressing the 4 PE_PGRS33 functional deletion mutants. Intracellular CFUs were enumerated at 4h post-infection. Results of one representative experiment of at least three assays are shown. * p<0.05 for Mtb H37Rv vs MtbΔ33::PE_PGRS33 401 and *** p<0.001 compared with for Mtb H37Rv vs MtbΔ33 and MtbΔ33::PE Rv1818c. CFUs were expressed as mean ± SD and were analysed by one-way ANOVA followed by Dunnett’s multiple comparison test.
Discussion
The appearance and expansion of PE_PGRS genes in certain mycobacteria has been linked to the acquisition of new pathogenetic properties that made these organisms capable of causing disease in mammals [4,9]. Despite the great interest raised by these findings, studies aimed at addressing the role of PE_PGRS proteins in Mtb biology and TB pathogenesis are still limited [34]. In a seminal work, the role of PE_PGRS33 was partially characterized in BCG, using a strain were the PE_PGRS33 was inactivated by random mutagenesis [19]. In this work, we extended these findings and further dissected the role of PE_PGRS33 in the virulent strain Mtb. The results obtained demonstrate that PE_PGRS33 is required for efficient Mtb cell entry in macrophages, but not in type II pneumocytes, while is dispensable for bacilli intracellular replication. PE_PGRS33-mediated entrance into macrophages is dependent upon the TLR2 receptor and this interaction occurs through the PGRS domain of the protein, specifically involving the first 260 amino acids of PE_PGRS33. These results indicate that PE_PGRS33 plays a crucial role during Mtb infection and provide experimental evidences that shed some light on the functional role of this protein in TB pathogenesis.
Lack of PE_PGRS33 was associated with an impaired ability of the MtbΔ33 mutant strain to enter macrophages, probably because of a defect in bacterial attachment to host cells. This defect was not observed during infection of A549 pneumocytes, where Mtb entry is mediated by molecules different from those involved during macrophages entry [35], thereby suggesting a specific interaction of PE_PGRS33 with macrophage surface components. The reduction in intracellular CFUs measured for the MtbΔ33 mutant was robust and similar to the reduction observed when the most important macrophage receptors involved in Mtb phagocytosis were blocked by specific antibodies [36]. Since Mtb enters macrophages by receptor-mediated phagocytosis [37] these results raised the possibility that PE_PGRS33 may directly interact with a macrophage receptor.
Several major host-receptors are involved in phagocytosis of Mtb, such as complement receptors (CR1, CR3 and CR4), the mannose receptor (MR) and the Fcγ receptors [37]. Fcγ is known to play a role in the presence of specific antibodies and entry via this receptor is expected to generate a vigorous host response [36]. Lipoarabinomannans, mannans and mannoproteins present on the Mtb surface are accessible to the MR and contribute to the adhesion of the bacilli to macrophages [38]. The MR also mediates uptake of mycobacteria other than Mtb [39], which do not express any PE_PGRS protein and therefore, in our view, it is unlikely that PE_PGRS33-mediated phagocytosis occurs through interaction with MR.
In previous studies [15] we showed that overexpression of PE_PGRS33 impacts on cell morphology and modifies the surface of bacilli. It may be hypothesized that lack of PE_PGRS33 may alter the proper conformation and/or the composition of mycobacterial surface constituents, such as for instance LAM or other non-proteinaceous components involved in Mtb cell entry and activation of pro-inflammatory cytokines secretion through CD14 and TLR2 [40–43]. In this scenario, the effects observed following overexpression of PE_PGRS33, or in its absence, might be considered simple epiphenomena. However, we did not observe any major difference in the cell morphology of the MtbΔ33 compared with the wild type and complemented strains. It remains to be determined the causes of the differences observed between the mutants of PE_PGRS33 obtained in BCG [19] and Mtb, although immunoblot analysis with an anti-PGRS antibody of the total cell fractions of BCG and Mtb showed differences in the protein profile, suggesting that PE_PGRS expression and eventually localization in these two mycobacteria might be different [27].
The results obtained with the MtbΔ33 complemented with the wild type gene PE_PGRS33 (MtbΔ33::PE_PGRS33HA) and the MtbΔ33::PE1818c, indicate that the PE domain of PE_PGRS33 does not contain the information sufficient to mediate PE_PGRS33-mediated entry in macrophages, in line with previous findings [19,23,33]. In an attempt to identify the domain, within the PGRS region, responsible for the observed phenotype a panel of functional deletion mutants expressing PE_PGRS33 protein missing fragments of increasing size of the C-terminal domain were generated (Fig 5). Interestingly, complementation of the wild type phenotype was obtained even with the mutant expressing the first 260 amino acids of the protein, which contains only a relatively small part of the PGRS domain. These results suggest that the PGRS domain residing between position 140 and 260 of the protein contains the information sufficient to interact with TLR2. The finding that the mutant MtbΔ33::PE_PGRS33401 could only partially complement the phenotype, suggests that folding of the downstream domain of the PGRS is critical for the proper exposure of the first 260 amino acids of PE_PGRS33. Sequencing of the PE_PGRS33 gene in Mtb clinical isolates indicated that most of the genetic polymorphisms resides in the PGRS region, but mainly downstream of the first 260 amino acids [44–46], somehow supporting the key role of this domain. Dissecting the impact on function of these naturally occurring PE_PGRS alleles will be important to understand the role of the PGRS domain, from a pathogenetic and evolutionary perspective.
Using purified recombinant PE_PGRS33 protein obtained in E. coli, Basu et al. showed that PE_PGRS33 interacts directly with TLR2 ectopically expressed in RAW 264.7 or HEK293 cells; that triggering of TNF-α was dependent on the PGRS domain and that variations in the polymorphic repeats of the PGRS domain differentially modulated TNF-α secretion [23]. Recent data obtained in our laboratory, in the model of ectopic expression in M. smegmatis, demonstrated that PE_PGRS33-dependent TNF-α secretion in macrophages is dependent on TLR2 [33], thereby suggesting that PE_PGRS33 may indeed interact with TLR2 on the macrophage surface. Remarkably, the defect in macrophage entry observed for the MtbΔ33 could not be observed when pMM0 from TLR2-deficient mice were used, suggesting that PE_PGRS33-mediated entry of Mtb into macrophages is likely dependent on a direct interaction of PE_PGRS33 with TLR2. Unfortunately, the lack of any information on the structure of PE_PGRS33, or of any other PE_PGRS protein, prevents to hypothesize the mechanism of interaction with TLR2 [47].
The involvement of TLR2 in bacterial cell entry in macrophages has been proposed for several pathogens. Recognition through the CD14 of the Porphyromonas gingivalis fimbriae activates a TLR2-dependent signalling pathway promoting binding to and internalization by macrophages [48,49]. Interaction of these fimbriae with the CD14/TLR2 complex was shown to induce a PI3K-mediated inside-out signalling that activates the ligand-binding capacity of the CR3 and entry through CR3 was known to promote bacterial survival and virulence [31,48,50,51]. Similarly, it has been shown that Bacillus anthracis spores contain an hair-like nap formed by a glycoprotein that, by interacting with the CD14/TLR2 present on macrophages surface, activates the PI3K inside-out signalling pathway that enhances spore internalization by increasing the avidity of the Mac-1 integrin [52]. The relevance of this inside-out signalling, involving CD14, TLR2, PI3K and cytohesin and leading to the conversion of the low avidity CR3 into an active receptor, has been demonstrated also for mycobacteria [53,54]. The results of this study, indicating that treatment of pMM0 with a PI3K inhibitor wortmannin or with the anti-CR3 antibody abolishes the PE_PGRS33-mediated entrance in macrophages (Fig 5), suggest that interaction of PE_PGRS33 with TLR2 activates the pro-adhesive pathway. While the mechanism of PE_PGRS33-mediated entrance needs to be better defined, it can be hypothesized that interaction of PE_PGRS33 with the TLR2 on macrophages in the early steps of infection activates an inside-out signalling which contributes to Mtb entry through the CR3 receptor.
This hypothesis is consistent with the TLR2-dependent pro-inflammatory properties of PE_PGRS33 that have been demonstrated by several studies [20,22,23,33]. In fact, it has been shown that P. gingivalis fimbriae induced two distinct TLR2 pathways mediating pro-inflammatory and pro-adhesive effects [30] and, more recently, that a fine tuning of the TLR2-CR3 crosstalk mediates inflammation and phagocytosis during Francisella tularensis infection [55]. In this scenario, PE_PGRS33 may be considered one of the Mtb proteins that plays a relevant role in the host-pathogen interplay, by fine tuning fundamental biological processes that can impact the outcomes of infection.
The relevance of PE_PGRS33 for Mtb pathogenesis can be inferred from the results obtained following genomic analysis of smooth tubercle bacilli [56]. The gene encoding PE_PGRS33 was found only in the MTBC genome but not in the smooth tubercle bacilli genome, and PE_PGRS33 was not found in M. marinum, despite the large number of PE_PGRS genes present in the genome of this mycobacterium [56]. The authors concluded that the PE_PGRS33 structural gene was inserted in MTBC next to the Rv1817 gene [56] and may thus have contributed to the acquistion of the specific pathogenic properties that made MTBC a highly successfull global pathogen.
In a previous study [11], we showed that the PGRS domain of PE_PGRS30 is involved in the arrest of phagosome acidification and in Mtb growth inside macrophages, while it is not involved in the entry of the bacteria inside macrophages. In this work, we demonstrate that the PGRS domain of PE_PGRS33 is involved in the bacterial entry in macrophages, but not in their intracellular replication. The reason why two very similar domains can be involved in so different cellular functions is still totally unknown and suggests that, despite the high similarity and redundancy of the GGA-GGN repeats, PE_PGRS proteins can be involved in a wide range of functions in Mtb pathophysiology.
Supporting Information
(PDF)
(PDF)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funded by EU (FP7/2007–2013) under grant agreement n°201762 awarded to GD and RM and by MIUR PRIN (project number 2008 Y8RZTF) awarded to GD and RM.
References
- 1.World Health Organization (2004) Global Tuberculosis Control: Surveillance, Planning, Financing. [Google Scholar]
- 2.Dorhoi A, Reece ST, Kaufmann SH (2011) For better or for worse: the immune response against Mtb balances pathology and protection. Immunol Rev 240: 235–251. 10.1111/j.1600-065X.2010.00994.x [DOI] [PubMed] [Google Scholar]
- 3.Delogu G, Manganelli R, Brennan MJ (2014) Critical research concepts in TB vaccine development. Clin Microbiol Infect 20 Suppl 5: 59–65. 10.1111/1469-0691.12460 [DOI] [PubMed] [Google Scholar]
- 4.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. (1998) Deciphering the biology of Mtb from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 5.Chao MC, Rubin EJ (2010) Letting sleeping dos lie: does dormancy play a role in TB? Annu Rev Microbiol 64: 293–311. 10.1146/annurev.micro.112408.134043 [DOI] [PubMed] [Google Scholar]
- 6.Gengenbacher M, Kaufmann SH (2012) Mtb: success through dormancy. FEMS Microbiol Rev 36: 514–532. 10.1111/j.1574-6976.2012.00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan MJ, Delogu G (2002) The PE multigene family: a 'molecular mantra' for mycobacteria. Trends Microbiol 10: 246–249. [DOI] [PubMed] [Google Scholar]
- 8.Delogu G, Cole ST, Brosch R (2008) The PE and PPE Protein Families of Mtb In: Kaufmann SH, Rubin E, editors. Handbook of Tuberculosis. Weinheim: Wiley-VCH Verlag GmbH%Co. KGaA; pp. 131–150. [Google Scholar]
- 9.Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM (2006) Evolution and expansion of the Mtb PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol 6: 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson SL (2011) Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol 2011: 497203 10.1155/2011/497203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iantomasi R, Sali M, Cascioferro A, Palucci I, Zumbo A, Soldini S, et al. (2012) PE_PGRS30 is required for the full virulence of Mtb. Cell Microbiol 14: 356–367. 10.1111/j.1462-5822.2011.01721.x [DOI] [PubMed] [Google Scholar]
- 12.De Maio F, Maulucci G, Minerva M, Anoosheh S, Palucci I, Iantomasi R, et al. (2014) Impact of protein domains on PE_PGRS30 polar localization in Mycobacteria. PLoS ONE 9: e112482 10.1371/journal.pone.0112482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delogu G, Sanguinetti M, Pusceddu C, Bua A, Brennan MJ, Zanetti S, et al. (2006) PE_PGRS proteins are differentially expressed by Mtb in host tissues. Microbes Infect 8: 2061–2067. [DOI] [PubMed] [Google Scholar]
- 14.Dheenadhayalan V, Delogu G, Sanguinetti M, Fadda G, Brennan MJ (2006) Variable expression patterns of Mtb PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J Bacteriol 188: 3721–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delogu G, Pusceddu C, Bua A, Fadda G, Brennan MJ, Zanetti S (2004) Rv1818c-encoded PE_PGRS protein of Mtb is surface exposed and influences bacterial cell structure. Mol Microbiol 52: 725–733. [DOI] [PubMed] [Google Scholar]
- 16.Cascioferro A, Delogu G, Colone M, Sali M, Stringaro A, Arancia G, et al. (2007) PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Mol Microbiol 66: 1536–1547. [DOI] [PubMed] [Google Scholar]
- 17.Cascioferro A, Daleke MH, Ventura M, Dona V, Delogu G, Palu G, et al. (2011) Functional dissection of the PE domain responsible for translocation of PE_PGRS33 across the mycobacterial cell wall. PLoS ONE 6: e27713 10.1371/journal.pone.0027713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sali M, Di SG, Cascioferro A, Zumbo A, Nicolo C, Dona V, et al. (2010) Surface expression of MPT64 as a fusion with the PE domain of PE_PGRS33 enhances Mycobacterium bovis BCG protective activity against Mtb in mice. Infect Immun 78: 5202–5213. 10.1128/IAI.00267-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan MJ, Delogu G, Chen Y, Bardarov S, Kriakov J, Alavi M, et al. (2001) Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect Immun 69: 7326–7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dheenadhayalan V, Delogu G, Brennan MJ (2006) Expression of the PE_PGRS 33 protein in Mycobacterium smegmatis triggers necrosis in macrophages and enhanced mycobacterial survival. Microbes Infect 8: 262–272. [DOI] [PubMed] [Google Scholar]
- 21.Singh PP, Parra M, Cadieux N, Brennan MJ (2008) A comparative study of host response to three Mtb PE_PGRS proteins. Microbiology 154: 3469–3479. 10.1099/mic.0.2008/019968-0 [DOI] [PubMed] [Google Scholar]
- 22.Balaji KN, Goyal G, Narayana Y, Srinivas M, Chaturvedi R, Mohammad S (2007) Apoptosis triggered by Rv1818c, a PE family gene from Mtb is regulated by mitochondrial intermediates in T cells. Microbes Infect 9: 271–281. [DOI] [PubMed] [Google Scholar]
- 23.Basu S, Pathak SK, Banerjee A, Pathak S, Bhattacharyya A, Yang Z, et al. (2007) Execution of macrophage apoptosis by PE_PGRS33 of Mtb is mediated by Toll-like receptor 2-dependent release of tumor necrosis factor-alpha. J Biol Chem 282: 1039–1050. [DOI] [PubMed] [Google Scholar]
- 24.Cadieux N, Parra M, Cohen H, Maric D, Morris SL, Brennan MJ (2011) Induction of cell death after localization to the host cell mitochondria by the Mtb PE_PGRS33 protein. Microbiology 157: 793–804. 10.1099/mic.0.041996-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Kessel JC, Hatfull GF (2007) Recombineering in Mtb. Nat Methods 4: 147–152. [DOI] [PubMed] [Google Scholar]
- 26.Greco E, Santucci MB, Sali M, De Angelis FR, Papi M, De SM, et al. (2010) Natural lysophospholipids reduce Mtb-induced cytotoxicity and induce anti-mycobacterial activity by a phagolysosome maturation-dependent mechanism in A549 type II alveolar epithelial cells. Immunology 129: 125–132. 10.1111/j.1365-2567.2009.03145.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jimenez C, et al. (2009) PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol 73: 329–340. 10.1111/j.1365-2958.2009.06783.x [DOI] [PubMed] [Google Scholar]
- 28.Quesniaux V, Fremond C, Jacobs M, Parida S, Nicolle D, Yeremeev V, et al. (2004) Toll-like receptor pathways in the immune responses to mycobacteria. Microbes Infect 6: 946–959. [DOI] [PubMed] [Google Scholar]
- 29.Hajishengallis G, Lambris JD (2011) Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol 11: 187–200. 10.1038/nri2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajishengallis G, Wang M, Liang S (2009) Induction of distinct TLR2-mediated proinflammatory and proadhesive signaling pathways in response to Porphyromonas gingivalis fimbriae. J Immunol 182: 6690–6696. 10.4049/jimmunol.0900524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harokopakis E, Albzreh MH, Martin MH, Hajishengallis G (2006) TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J Immunol 176: 7645–7656. [DOI] [PubMed] [Google Scholar]
- 32.Klink M, Brzezinska M, Szulc I, Brzostek A, Kielbik M, Sulowska Z, et al. (2013) Cholesterol oxidase is indispensable in the pathogenesis of Mtb. PLoS ONE 8: e73333 10.1371/journal.pone.0073333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zumbo A, Palucci I, Cascioferro A, Sali M, Ventura M, D'Alfonso P, et al. (2013) Functional dissection of protein domains involved in the immunomodulatory properties of PE_PGRS33 of Mtb. Pathog Dis 69: 232–239. 10.1111/2049-632X.12096 [DOI] [PubMed] [Google Scholar]
- 34.Fishbein S, van WN, Warren RM, Sampson SL (2015) Phylogeny to function: PE/PPE protein evolution and impact on Mtb pathogenicity. Mol Microbiol 96: 901–916. 10.1111/mmi.12981 [DOI] [PubMed] [Google Scholar]
- 35.Pethe K, Alonso S, Biet F, Delogu G, Brennan MJ, Locht C, et al. (2001) The heparin-binding haemagglutinin of Mtb is required for extrapulmonary dissemination. Nature 412: 190–194. [DOI] [PubMed] [Google Scholar]
- 36.Schlesinger LS, Bellinger-Kawahara CG, Payne NR, Horwitz MA (1990) Phagocytosis of Mtb is mediated by human monocyte complement receptors and complement component C3. J Immunol 144: 2771–2780. [PubMed] [Google Scholar]
- 37.Fenton MJ, Riley LW, Schlesinger LS (2008) Receptor-mediated recognition of Mtb by host cells In: Cole ST, editors. TBs and the Tubercle Bacillus. Washington,D.C.: ASM Press; pp. 405–426. [Google Scholar]
- 38.Schlesinger LS, Hull SR, Kaufman TM (1994) Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mtb to human macrophages. J Immunol 152: 4070–4079. [PubMed] [Google Scholar]
- 39.Astarie-Dequeker C, N'Diaye EN, Le C, V, Rittig MG, Prandi J, Maridonneau-Parini I (1999) The mannose receptor mediates uptake of pathogenic and nonpathogenic mycobacteria and bypasses bactericidal responses in human macrophages. Infect Immun 67: 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan ED, Morris KR, Belisle JT, Hill P, Remigio LK, Brennan PJ, et al. (2001) Induction of inducible nitric oxide synthase-NO* by lipoarabinomannan of Mtb is mediated by MEK1-ERK, MKK7-JNK, and NF-kappaB signaling pathways. Infect Immun 69: 2001–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan J, Fan XD, Hunter SW, Brennan PJ, Bloom BR (1991) Lipoarabinomannan, a possible virulence factor involved in persistence of Mtb within macrophages. Infect Immun 59: 1755–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Means TK, Lien E, Yoshimura A, Wang S, Golenbock DT, Fenton MJ (1999) The CD14 ligands lipoarabinomannan and lipopolysaccharide differ in their requirement for Toll-like receptors. J Immunol 163: 6748–6755. [PubMed] [Google Scholar]
- 43.Zhang Y, Doerfler M, Lee TC, Guillemin B, Rom WN (1993) Mechanisms of stimulation of interleukin-1 beta and tumor necrosis factor-alpha by Mtb components. J Clin Invest 91: 2076–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talarico S, Cave MD, Marrs CF, Foxman B, Zhang L, Yang Z (2005) Variation of the Mtb PE_PGRS 33 gene among clinical isolates. J Clin Microbiol 43: 4954–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talarico S, Zhang L, Marrs CF, Foxman B, Cave MD, Brennan MJ, et al. (2008) Mtb PE_PGRS16 and PE_PGRS26 genetic polymorphism among clinical isolates. Tuberculosis (Edinb) 88: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Huang Y, Zhang A, Zhu C, Yang Z, Xu H (2011) DNA polymorphism of Mtb PE_PGRS33 gene among clinical isolates of pediatric TB patients and its associations with clinical presentation. Tuberculosis (Edinb) 91: 287–292. [DOI] [PubMed] [Google Scholar]
- 47.Borrello S, Nicol X, Delogu G, Pandolfi F, Ria F (2011) TLR2: a crossroads between infections and autoimmunity? Int J Immunopathol Pharmacol 24: 549–556. [DOI] [PubMed] [Google Scholar]
- 48.Hajishengallis G, Wang M, Harokopakis E, Triantafilou M, Triantafilou K (2006) Porphyromonas gingivalis fimbriae proactively modulate beta2 integrin adhesive activity and promote binding to and internalization by macrophages. Infect Immun 74: 5658–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harokopakis E, Hajishengallis G (2005) Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. Eur J Immunol 35: 1201–1210. [DOI] [PubMed] [Google Scholar]
- 50.Hajishengallis G, Shakhatreh MA, Wang M, Liang S (2007) Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol 179: 2359–2367. [DOI] [PubMed] [Google Scholar]
- 51.Wang JY, Lee LN, Lai HC, Hsu HL, Liaw YS, Hsueh PR, et al. (2007) Prediction of the TB reinfection proportion from the local incidence. J Infect Dis 196: 281–288. [DOI] [PubMed] [Google Scholar]
- 52.Oliva C, Turnbough CL Jr., Kearney JF (2009) CD14-Mac-1 interactions in Bacillus anthracis spore internalization by macrophages. Proc Natl Acad Sci U S A 106: 13957–13962. 10.1073/pnas.0902392106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lasunskaia EB, Campos MN, de Andrade MR, Damatta RA, Kipnis TL, Einicker-Lamas Met al.(2006) Mycobacteria directly induce cytoskeletal rearrangements for macrophage spreading and polarization through TLR2-dependent PI3K signaling. J Leukoc Biol 80: 1480–1490. [DOI] [PubMed] [Google Scholar]
- 54.Sendide K, Reiner NE, Lee JS, Bourgoin S, Talal A, Hmama Z (2005) Cross-talk between CD14 and complement receptor 3 promotes phagocytosis of mycobacteria: regulation by phosphatidylinositol 3-kinase and cytohesin-1. J Immunol 174: 4210–4219. [DOI] [PubMed] [Google Scholar]
- 55.Dai S, Rajaram MV, Curry HM, Leander R, Schlesinger LS (2013) Fine tuning inflammation at the front door: macrophage complement receptor 3-mediates phagocytosis and immune suppression for Francisella tularensis. PLoS Pathog 9: e1003114 10.1371/journal.ppat.1003114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, et al. (2013) Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mtb. Nat Genet 45: 172–179. 10.1038/ng.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.