Abstract
Bacterial species comprise related genotypes that can display divergent phenotypes with important clinical implications. Staphylococcus epidermidis is a common cause of nosocomial infections and, critical to its pathogenesis, is its ability to adhere and form biofilms on surfaces, thereby moderating the effect of the host’s immune response and antibiotics. Commensal S. epidermidis populations are thought to differ from those associated with disease in factors involved in adhesion and biofilm accumulation. We quantified the differences in biofilm formation in 98 S. epidermidis isolates from various sources, and investigated population structure based on ribosomal multilocus typing (rMLST) and the presence/absence of genes involved in adhesion and biofilm formation. All isolates were able to adhere and form biofilms in in vitro growth assays and confocal microscopy allowed classification into 5 biofilm morphotypes based on their thickness, biovolume and roughness. Phylogenetic reconstruction grouped isolates into three separate clades, with the isolates in the main disease associated clade displaying diversity in morphotype. Of the biofilm morphology characteristics, only biofilm thickness had a significant association with clade distribution. The distribution of some known adhesion-associated genes (aap and sesE) among isolates showed a significant association with the species clonal frame. These data challenge the assumption that biofilm-associated genes, such as those on the ica operon, are genetic markers for less invasive S. epidermidis isolates, and suggest that phenotypic characteristics, such as adhesion and biofilm formation, are not fixed by clonal descent but are influenced by the presence of various genes that are mobile among lineages.
Introduction
Hospital acquired infections associated with implanted biomaterials such as intravenous catheters, joint prostheses, shunts and heart valves cause substantial morbidity and mortality [1]. Coagulase-negative staphylococci (CoNS), mainly Staphylococcus epidermidis, are currently the most frequent cause of such medical device-associated infections [2], responsible for an estimated 1.7 million infections in the US each year and 100,000 deaths [3, 4], at an estimated annual cost of $35–45 billion [5]. A major concern of biomedical device-related infections is their chronic persistence due to biofilm formations, and failure to respond to antibiotics, often necessitating the removal of the device associated with the infection.
A major challenge to reduce the burden of nosocomial infection is to identify if certain S. epidermidis lineages have a greater pathogenic potential than others. DNA sequencing techniques, including multi-locus sequence typing (MLST) and whole genome sequencing have been instructive in showing that commensal and clinical S. epidermidis comprise of highly diverse assemblages of related strains [2, 6]. However, the factors involved in generating and maintaining this genetic structure, and how they relate to pathogenicity-associated phenotypes, are poorly understood. For example, closely related isolates can display different phenotypes, and among the most characterized differences between isolates from the skin and infected sites is a greater propensity for biofilm formation among clinical isolates [7]. Such differences may allow them to make the transition from the commensal skin environment to implant-associated infections. The formation of complex adherent bacterial biofilms involves four steps: (i) primary attachment; (ii) accumulation; (iii) maturation; and (iv) detachment. The initial adhesion to an implant surface involves cell-wall-anchored (CWA) proteins/adhesins, such as the microbial surface components recognising adhesive matrix molecules (MSCRAMMs) [8], that are responsible for binding the bacteria directly to inert surfaces or to the host’s extracellular proteins coating the surface. S. epidermidis adhesins to fibrinogen (Fbe/SdrG), fibronectin (Embp), collagen (SdrF), vitronectin (AtlE, Aae) and elastin (EbpS) have all been identified in S. epidermidis [9–12]. Three independent mechanisms of biofilm accumulation have been identified. The first is mediated by the polysaccharide intercellular adhesin (PIA), synthesised by icaADBC encoded proteins [13, 14]. The other two, mediated by proteinaceous factors independent of the icaADBC locus, involve the accumulation associated protein (Aap) [15], and the extracellular matrix-binding protein (Embp) [16]. However, little is known about the extent to which variations in biofilm phenotypes are the result of widespread acquisition of known adhesins and other proteinaceous factors, or clonal expansion of specific lineages.
Here, we characterize variation in 117 S. epidermidis isolates by genotyping using Multi Locus Sequence Typing (rMLST) [17], an approach which indexes variation of the 53 genes encoding the bacterial ribosome protein subunits (rps and rpl genes). This has been compared to phenotypic variation in biofilm formation as quantified by confocal microscopy. Using these data, our aim was to investigate (i) the level of variation in biofilm phenotype in S. epidermidis isolates; (ii) the correlation between genotypic groupings (clade, clonal complex and sequence type), known biofilm associated genes and biofilm phenotype classification, and (iii) the extent to which the phenotype varies among closely related S. epidermidis isolates and to what extent this is linked to isolate source (commensal and clinical).
Materials and Methods
Bacterial isolates
S. epidermidis isolates (n = 117) were collected from clinical and commensal sites (Table 1). Clinical isolates were obtained from ENDO-Klinik Hamburg, and University Hospital Hamburg-Eppendorf, Germany [18, 19]; and the Laboratory of Molecular Genetics of the Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Portugal [6, 20]. The commensal isolates consisted of human nasal mucosa and skin surface isolates collected from healthy volunteers at Swansea University, and animal isolates from the Institute of Farm Animal Genetics, Friedrich-Loeffler-Institut, Germany [21]. All isolates were streaked from frozen stocks onto Columbia agar supplemented with 5% horse blood (E&O Laboratories Ltd, Bonnybridge, UK), and incubated overnight at 37°C; and then used in the subsequent experiments.
Table 1. List of the S. epidermidis isolates used in the study.
| Isolate source | No. isolates | Reference | |
|---|---|---|---|
| Clinical | Prosthetic joint infections | 38 | 18 |
| Catheter infections | 26 | 19 | |
| Other infections | 6 | 6 | |
| Commensal | Nasal mucosa | 20 | This study |
| Skin surface | 20 | This study | |
| Animal | 7 | 21 | |
Biofilm morphotype analysis
Characteristics of the biofilm phenotype of the S. epidermidis isolates were determined using confocal laser scanning microscopy (CLSM) and image analysis, as in previous studies [22–25]. Briefly, each isolate was cultured from a 3 h pre-culture into an 8-well chamber slide (ibidi GmbH, Planegg-Martinsried, Germany) to a starting OD600 of 0.05, and incubated stationary at 37°C for 20–24 h. Non-adherent bacteria were then removed by washing the wells with phosphate buffered saline and the adherent bacteria stained with SYTO9 (Life Technologies Ltd, Paisley, UK), before being visualised with a Zeiss LSM 510-META Confocal laser scanning microscope and Zen software (Carl Zeiss, Jena, Germany). Four image stacks were taken randomly from three independent samples per isolate (12 stacks in total). The image stacks were then analysed using the COMSTAT software [23]. A fixed threshold value and connected volume filtration was used for all image stacks; and the biofilm parameters average biofilm thickness (a measure of the spatial size of the biofilm in μm), biovolume (the volume (μm3) of the biomass per μm2 of substratum area; μm3 per μm2) and roughness coefficient (Ra, correlates with biofilm heterogeneity and measured in μm) were determined for each image stack [23]. Regression analysis was used to estimate the relationship between biovolume, thickness and Ra, and Fisher’s exact test was used to examine the significance in the association between biovolume, thickness and Ra measurements, with the level of significance set at p ≤ 0.05. An agglomerative hierarchical cluster analysis (AHC) was used to examine the relationship between biovolume, thickness, Ra for the biofilms formed by the S. epidermidis isolates. The AHC was an unweighted pair-group average agglomeration with Euclidean distance for dissimilarity. From the COMSTAT measurements, the AHC dendrogram and CLSM images, it was possible to group the isolates into 5 biofilm structure patterns as defined in Table 2.
Table 2. Definitions of the biofilm structure patterns described in this study.
| Biofilm structure pattern | Thickness (μm) | Biovolume (μm3 per μm2) | Ra (μm) |
|---|---|---|---|
| dense thick uneven | > 10 | > 10 | 0.04–0.20 |
| dense thick smooth | > 10 | > 10 | < 0.1 |
| patchy thin smooth | < 10 | < 10 | < 0.1 |
| patchy thin rough | < 10 | < 10 | < 0.2 |
| patchy thin uneven | < 10 | < 10 | 0.04–0.20 |
DNA extraction and sequencing
For DNA extraction, the 117 S. epidermidis isolates were grown in 2 ml peptone yeast (PY) broth overnight at 37°C, harvested by centrifugation, and chromosomal DNA extracted using a Qiagen QiAmp DNA mini kit (Qiagen, Crawley, UK) following the manufacturer’s instructions using 1 μg/mL lysostaphin (Sigma-Aldrich, Gillingham, UK) to facilitate cell lysis. DNA was quantified using a Nanodrop spectrophotometer, and the Quant-iT DNA Assay Kit (Life Technologies, Paisley, UK) before sequencing. High-throughput genome sequencing was performed using a HiSeq 2500 machine (Illumina, San Diego, CA, USA) and the 100bp short read paired-end data was assembled using the de novo assembly algorithm within Velvet software [26] (version 1.2.08). Resulting data was then archived in the Staphylococcal Bacterial Isolate Genome Sequence database (BIGSdb) [27]. This software uses a gene-by-gene approach for genome alignment and comparison supported by the BLAST algorithm, for identification of allelic orthologs, and file generation for investigating phylogenetic networks, and locus information for the investigation of gene function. Isolate information, including genome sequence files are publically available (Dryad, doi: 10.5061/dryad.82jq4 for the 14 previously published isolates and doi: 10.5061/dryad.br0h8 for the 103 other isolates).
Genotype analysis
The S. epidermidis RP62A reference genome [28] was used as a basis for defining locus designations, and reference sequences for each of these were stored in BIGSdb (http://zoo-talisker.zoo.ox.ac.uk/perl/bigsdb/bigsdb.pl?db=staphylococcus). Isolate genomes, stored as concatenated contiguous sequence files—including gaps for missing nucleotides (or entire genes), were aligned on a gene-by-gene basis using MUSCLE software [29], and the BLAST algorithm was used to identify gene orthologs at all loci in the reference genome. These were defined as reciprocal best hits for a sequence with ≥70% nucleotide identity and a 50% difference in alignment length.
The rMLST approach was used to investigate the genetic relationship between the 117 isolates [17]. Orthologs for the 53 genes encoding the bacterial ribosome protein subunits (rps and rpl genes) were defined in all isolates by comparison to the annotated genome of S. epidermidis RP62A [17]. Reciprocal best hits identifying 11,168 rps and rpl alleles were identified using BLAST as described above. CLONALFRAME, a model-based approach to determining microevolution in bacteria, was used to estimate the genealogies for these alignments [30]. This program differentiates mutation and recombination events on each branch of the tree based on the density of polymorphisms. Clusters of polymorphisms were likely to have arisen from recombination, and scattered polymorphisms were considered likely to have arisen from mutation. The program was run with 50,000 burn-in iterations, followed by 50,000 sampling iterations. The consensus tree represents combined data from three independent runs with 75% consensus required for inference of relatedness. Recombination events were defined as sequences with a length of >50 bp with a probability of recombination ≥75% over the length, reaching 95% in at least one site.
Presence of biofilm associated genes
The presence of known biofilm-associated genes was investigated by BLAST comparison to a reference genome [27, 31, 32]. First, a reference gene list was assembled from the publicly available S. epidermidis strain RP62A genome [28] (GenBank: NC_002976), including genes that have been shown to be associated with S. epidermidis adhesion and biofilm formation. Adhesion and biofilm-associated genes including aae, aap, atlE, bhp, embp, ebpS, fbe/sdrG, icaADBC, sdrF, sesC, sesE, sesG, sesH and sesI [9–16, 33, 34], were considered as being present when a BLAST match with a >70% nucleotide sequence identity on ≥50% of sequence length was recorded. Where present, genes where mapped onto the CLONALFRAME tree to examine the significance of association between the clonal frame substructure and the presence of a specific gene. Data were analysed statistically by Fisher’s exact test and Chi-square, with the level of significance set at p ≤ 0.05.
Results
Clonal structure based on rMLST genes
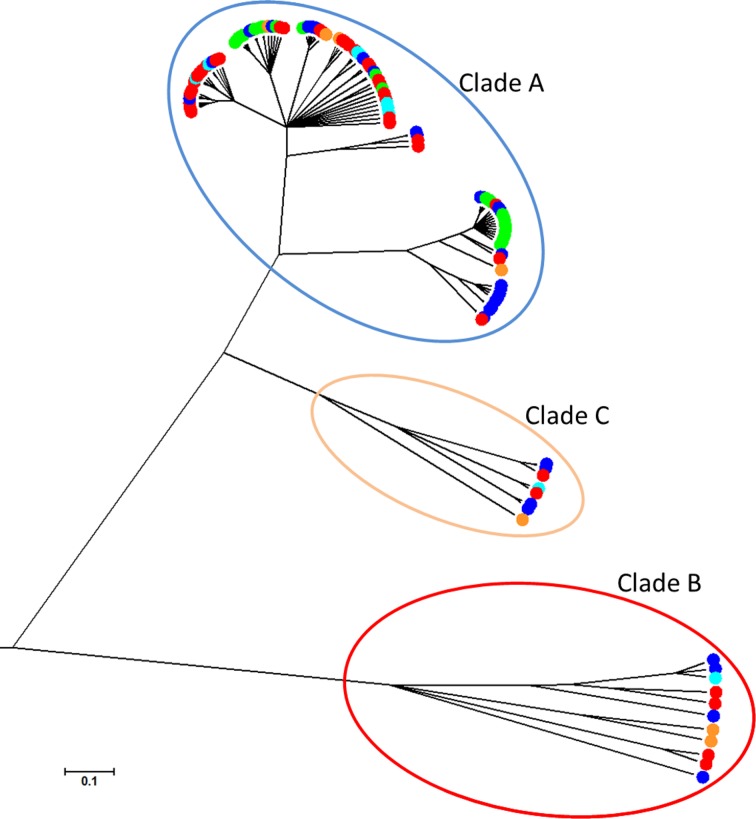
The rMLST locus genealogy, estimated using CLONALFRAME, showed that the S. epidermidis isolates clustered into 3 clades (Fig 1), as previously described [32]. Commensal and pathogenic isolates were distributed in multiple sequence clusters (p ≤ 0.05). Clade A comprised of 98 isolates that clustered into three groups, and contained isolates belonging to 7-locus MLST clonal complexes 2, 5, 89 and 291 [6]; whilst the isolates in Clades B (11 isolates) and C (8 isolates) did not belong to any previously described clonal complexes. There was a significant association between clonal complex and sub-clade sequence-cluster distribution (p ≤ 0.001, S1 Table). For example CC291 corresponded to a single lineage; however, some clonal complexes were represented by isolates in separate sequence clusters including CC2, CC5 and CC8. There was a significant association between clonal complex and isolate source (p = 0.001), with 54% of infection isolates belonging to CC2 compared to only 9% from healthy carriage and none in the animal isolates. CC5 and CC89 were more represented among isolates from healthy carriage constituting 31% and 28% of the isolates from this source respectively.
Fig 1. Population structure of 117 S. epidermidis isolates constructed from 53 rMLST genes and implemented in CLONALFRAME, with the 3 clades highlighted.
The scale (0.1) represents the number of substitutions per site. Isolates are coloured according to source: PJI (blue); catheter (green); undefined infection (orange); healthy nasal and skin (red); animal (turquoise).
Biofilm morphotype analysis
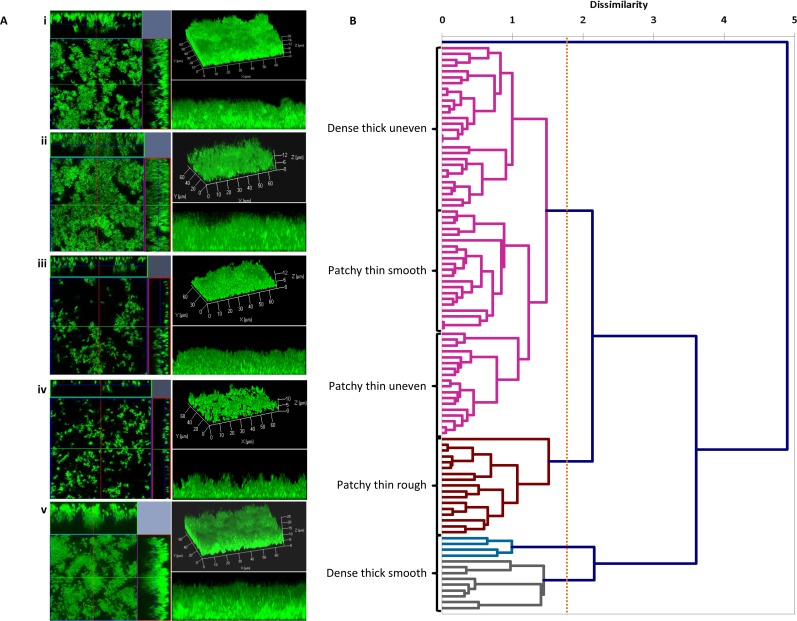
Among the 98 S. epidermidis isolates in Clade A, confocal microscopy (CLSM) imaging revealed that all isolates were adherent and formed biofilms in vitro, with microscale variability in their biofilm structure patterns (Fig 2A). Isolates adhered and formed patchy to multi-layered biofilms, with varying thicknesses from 3.42 μm (thin) to 30.97 μm (thick). Biofilm formation was consistent among three independent replicate assays (standard deviation values +/- 4 μm). Analysis of the Z-stacks using the Zen software orthogonal view and 3D imaging functions (Carl Zeiss, Jena, Germany) revealed that isolates either adhered and formed thick dense biofilms with an uneven topography (Fig 2Ai and 2Av) or patchy biofilms with different sized gaps between the bacterial aggregates (Fig 2Aii–2Aiv). COMSTAT software was then used to quantify the average biovolume (μm3 per μm2), average thickness (μm) and roughness coefficient (Ra; μm) of the CLSM Z-stacks from each isolate (S2 Table). These measurements support the visual observations of variation in biofilm morphotypes. T the largest biofilm biovolume and thickness measurements (>10 μm3 per μm2 and >10 μm2 respectively) were seen in isolates that formed thick dense biofilms with either a smooth or uneven topography (Fig 2Ai and 2Av). These werecompared to the thinner patchier biofilms (<10 μm3 per μm2 and <10 μm2 respectively; Fig 2Aii–2Aiv). Roughness coefficient (Ra) values were more varied falling into two groups, Ra <0.1 μm, smooth and homogenous, or Ra >0.1 μm, rough and heterogeneous. Regression analysis confirmed a correlation between biovolume and thickness but not with roughness coefficient (S1 Fig). AHC dendrogram is a multivariate cluster analysis using the biovolume, thickness and Ra measurements from each isolate, and clustered isolates together based on structure metrics (Fig 2B). Five biofilm morphotypes were identified based on the AHC dendogram. Isolates were classified as: dense thick uneven (29%), patchy thin smooth (21%), patchy thin uneven (18%), patchy thin rough (18%), and dense thick smooth (13%).
Fig 2. Confocal laser scanning microscopy (CLSM) biofilm images from different isolates and agglomerative hierarchical clustering (AHC) dendrogram labelled with biofilm morphotypes.
(A) Example of CLSM images comprising orthogonal view of Z-stacks (left panel), 3D image of the Z-stacks (top right panel), and 3D cross-section of Z-stacks (bottom right panel). Scale of in μm. (B) Each set of images corresponds to a biofilm structure group defined according to CLSM measurements (average thickness and roughness coefficient) (i) dense thick uneven; (ii) patchy thin smooth; (iii) patchy thin uneven; (iv) patchy thin rough; and (v) dense thick smooth. The AHC dendrogram is a multivariate cluster analysis examining the relationship between the biovolume, thickness and Ra measurements for the biofilms formed by each S. epidermidis isolate. The x-axis represents the dissimilarity score of merged clusters.
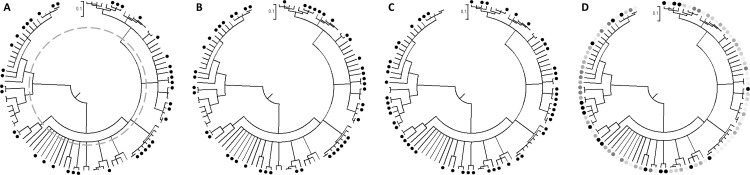
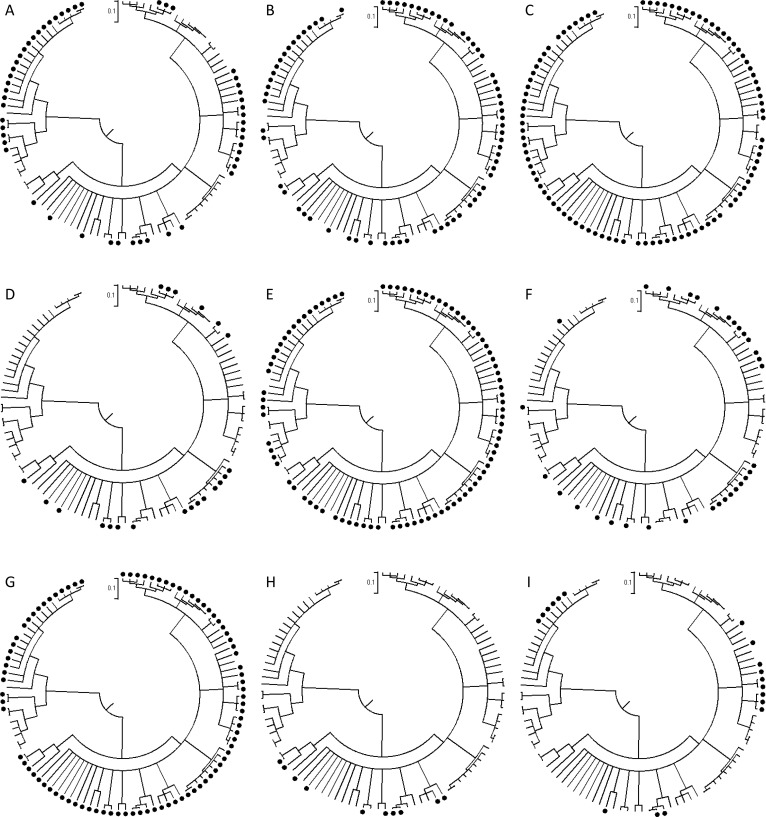
Biofilm morphotype, biovolume, thickness and roughness coefficient, were mapped onto the Clade A rMLST CLONALFRAME tree (Fig 3). All 19 multi-ST lineages contained isolates with one or more biofilm morphotype. Isolates producing biovolumes >10 μm3 per μm2 were present in 12/19 lineages, with the percentage of high biovolume morphotypes per cluster ranging from 22% to 100%. Isolates producing biofilm thickness >10 μm were present in 13/19 clusters and the proportion of thick biofilms ranged from 22% to 100%. Isolates producing rough biofilms (Ra>0.1) were present in 14/19 clusters and the proportion of rough morphotypes per cluster ranged from 40% to 100%.
Fig 3.
rMLST CLONALFRAME genealogies of clade A, showing COMSTAT results: A) Biovolume: black circle >10 μm3 per μm2; B) Thickness, black circle >10 μm; C) Roughness coefficient (Ra), black circle >0.10 μm; and D) Morphotypes, patchy thin rough (black circle), dense thick smooth (dark grey circle), patchy thin uneven (grey circle), patchy thin smooth (light grey circle), dense thick uneven (pale grey circle). The scale bar (0.1) indicates the genetic distance in coalescent units and represents the number of substitutions per site. Dashed line in image A at 0.3 coalescent units was used to characterise 19 lineages.
For each morphotype metric, chi-square tests for homogeneity were carried out to test the null hypothesis that populations (lineages) are homogeneous in their biofilm phenotypes. In the case of biovolume, roughness coefficient or morphotype (p = 0.22, 0.72 and 0.23 respectively), there was no evidence to reject the null hypothesis. In the case of biofilm thickness (p = 0.05) there was a significant difference in the incidence of thick biofilms in certain lineages, with thick biofilms being more common among CC2 isolates, accounting for 20% of the 98 isolates.
Variation in the presence of known biofilm-associated genes
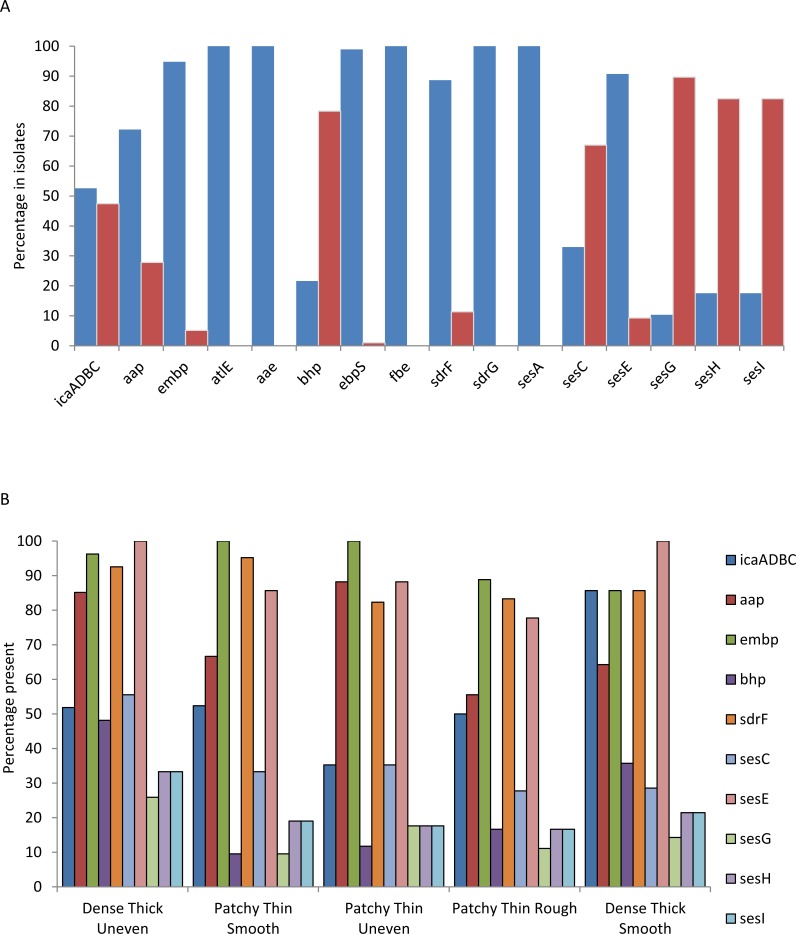
The 98 genomes from clade A were analysed for the presence of genes previously described as being involved in adhesion [9–16, 33]. bhp, sdrF, sesC, sesE, sesG, sesH and sesI genes were present in 20%, 89%, 32%, 91%, 10%, 17% and 17% of the isolates respectively, whilst atlE, aae, ebpS, and fbe/sdrG were present in all the isolates (Fig 4A and S2 Table). The loci icaADBC, aap and embp known to be involved in biofilm formation [13–16] were present in 52%, 71% and 95% of the isolates respectively. The gene mecA was also identified in 56% of the isolates, with 82% of which were from a pathogenic source (results not shown). The presence of these genes was examined in relation to the 5 biofilm morphotypes (Fig 4B). The icaADBC cluster was most common in isolates that produced the dense thick rough morphotype (64%) and least common in those producing the patchy thick smooth (33%), whilst 93% of the dense thick rough morphotypes contained the aap gene compared to only 62% in the patchy thin rough morphotype. embp was found in 100% of the patchy thick smooth morphotypes versus 96% in the patchy thin rough, patchy thin smooth and dense thick smooth, and only in 86% of the dense thick rough morphotypes. The bhp and sesE genes were more associated with the patchy thick smooth biofilm morphotype (38% and 100%, respectively) than with the thinner biofilm morphotypes (19% and 81%, respectively); whilst the sesG, sesH, and sesI genes were present in higher numbers (14%, 43% and 43%, respectively) in the dense thick rough morphotype isolates compared to 8% and 4% respectively in the patchy thin rough morphotype isolates. Despite these differences there was no significant difference in the distribution of genes within the 5 biofilm patterns (p = 0.406).
Fig 4.
Graphs showing A) the presence and absence of the previously described adhesion and biofilm associated genes (9–16, 33) in clade A isolates, present (blue), absent (red); and B) percentage genes present in the isolates based on the 5 biofilm morphotypes.
The distribution of icaADBC, aap, embp, bhp, sdrF, sesC, sesE, sesG, sesH and sesI in clade A were mapped onto the rMLST CLONALFRAME tree (Fig 5). Distributions of the different genes on the tree varied depending on the gene, and correspond to the presence and absence data in Fig 4. Statistical analysis of the distribution of genes on the different sub-clades using Fisher’s Exact test and Chi-square, suggested a strong clade association for icaADBC, aap, bhp, sdrF, sesC, sesE, sesG, sesH, and sesI (p ≤ 0.05; S1 Table) but not for embp (p = 0.55). The allele numbers for each locus were analysed. With the exception of aap and bhp which showed sub-clade specific allelic variation, identical alleles were present in (2–14) sub-clades. This is consistent with the action of multiple recombination at these loci.
Fig 5.
rMLST CLONALFRAME genealogies of Clade A showing the presence of specific genes in the isolates: (A) icaADBC, (B) aap, (C) embp, (D) bhp, (E) sdrF, (F) sesC, (G) sesE, (H) sesG, (I) sesH/ sesI. atlE, aae, ebpS and fbe/sdrG were present in all isolates. The scale bar (0.1) indicates the genetic distance in coalescent units.
Discussion
S. epidermidis infections associated with biomedical implants have become much more common with the increased use of biomaterials in medicine. In recent years, some studies have shown that commensal S. epidermidis populations differ from disease-causing strains in the frequency of the carriage of virulence factors such as those involved in adhesion and biofilm formation [18, 35–38]. The genealogical reconstruction of S. epidermidis genotypes from commensal and pathogenic sources showed evidence of clusters of related isolates consistent with clade and clonal complex designations previously described using whole genome sequencing and MLST [6, 32]. This provided the basis for testing if variation in adhesion and biofilm formation could mirror the genetic structure and distinguish virulent bacterial strains as particular lineages.
The combination of fine-scale phenotypic information with genotype information can provide insight into the evolution of S. epidermidis. In some bacteria for instance those that occupy multiple niches, genetic structuring of the population can reflect phenotypic differences. For example, in Campylobacter jejuni, different clonal complexes can dominate in different animal hosts [39], and in Helicobacter pylori population structure of the bacteria reflects the country of origin of the human from which it was isolated [40], whilst in S. aureus different clonal complexes are specifically associated with epidemic clones from specific countries [41, 42]. This niche segregation can be used to explain genetic structure in a population, according to an evolutionary model where isolates become separated in different niches and their genomes progressively diversify over time, accompanied by the accumulation of amino acid differences and concomitant phenotypic diversification.
Under this evolutionary model, S. epidermidis sequence clusters should display similar phenotypes. The biofilm phenotype is complex, incorporating live and dead bacteria [43]. Biofilms can be considered a two-step process involving primary attachment of bacterial cells to a surface via adhesins, followed by accumulation of the attached bacteria into a multi-layered biofilm via intercellular interactions [44]. When conditions are such that the ability to adhere and form a biofilm confers a strong selective advantage, for example on an implant, lineages that trace ancestry to strong biofilm formers will increase as a proportion of the population [45]. Under these circumstances, a phylogenetic tree will show clusters of biofilm forming lineages that have expanded clonally. However, while there was some evidence of sequence clusters of isolates that form thick biofilms (p = 0.05), statistical analyses of the CLONALFRAME tree showed random clustering of other biofilm variables and morphotypes in relation to clade distribution. There are a number of possible reasons for this. First, the confocal microscopy-derived biofilm variables contain less phenotypic information than is required for discrimination at this level. An explanation for this is that the biovolume, thickness and roughness variables mask other more informative biofilm-associated variables. Second, it may also be that the evolutionary model of linked genotypic and phenotypic diversification is too simplistic, particularly when applied to bacteria, such as S. epidermidis, that can undergo transition between multiple niches.
Some of the genes involved in biofilm formation have been well studied in S. epidermidis, but little is known about variation in gene presence among genetically diverse populations [33]. Biofilm-associated genes including atlE, aae, ebpS, and fbe/sdrG were found in all isolates in this study, and icaADBC, aap, embp, bhp, sdrF, sesC, sesE, sesG, sesH and sesI showed variation between the isolates and clades. The presence of aap or sesE showed a significant association with biofilm. The accumulation-associated protein (Aap) encoded by aap is known to be involved in biofilm formation, whilst an actual function for SesE, a cell wall surface anchor protein like Aap, has yet to be elucidated [10, 28]. However, the results in this study suggest the sesE gene product may have some involvement in biofilm accumulation.
The role of other biofilm associated genes in biofilm morphotype variation may be more complex. For example, PIA, encoded by the ica genes, is the most common molecule associated with biofilm formation in S. epidermidis [13, 14] and it has been suggested that the absence of the ica operon is more common in commensal S. epidermidis strains [46, 47]. However, the results presented here, show that the PIA-independent aap gene, that also mediates primary adhesion and/or biofilm accumulation [15, 48], was more frequently found in both pathogenic and commensal isolates than the ica operon, which is consistent with other studies [28, 49] and a complex interaction among biofilm associated genes that has been previously described and which may involve: (i) switching from PIA production to Aap mechanisms resulting in differences in biofilm substructures [50]; (ii) a role for Aap in anchoring PIA to the cell surface [51]; (iii) phase variation and the ‘switching on and off’ of adhesion and biofilm formation genes or other mechanisms of transcriptional regulation [52–54]. All of these factors make establishing a direct link between biofilm genes and phenotype variation more difficult.
The presence of genes associated with adhesion and biofilm formation in divergent rMLST sequence clusters (Fig 4) is evidence for horizontal gene transfer. Genome variation in S. epidermidis is generally thought to be generated more often by homologous recombination than by point mutation [55] compared to S. aureus [32, 56]. In addition, as in S. aureus, much of the genetic variation between S. epidermidis strains is associated with accessory genome variation in genomic islands, phage elements, and integrated plasmids [28, 46]. This can have major implications for resistance and virulence development in S. epidermidis [28]. Thus the mobility of adhesion and biofilm associated genes, including those in the ica operon [55], may be important in the increased abundance of phenotypes that aid adaptation to novel environments, and the transition from the commensal environment to infection.
Conclusion
This study highlights the genetic diversity of S. epidermidis isolates from commensal and clinical samples and demonstrates the widespread capacity of these bacteria to adhere and form multilayer biofilms in in vitro assays. Biofilm phenotypes were highly variable and could be classified into various morphotypes. However, these did not correlate well with the clonal frame of the S. epidermidis isolates. Furthermore, while there was some correlation between genotypic groupings (clade, clonal complex and sequence type) and known biofilm-associated genes, there was also evidence for the widespread acquisition and mobility of these genes between lineages. Taken together, these findings are consistent with a multifactorial cell-cell adhesion processes resulting in different biofilm structures, involving complex adaptive mechanisms potentially involving multiple determinants of biofilm formation.
Supporting Information
Regression analysis graphs: A) biovolume versus thickness; B) biovolume versus roughness coefficient (Ra); and C) thickness versus roughness coefficient (Ra). The results show a correlation between biovolume and thickness but not with roughness coefficient.
(TIF)
Level of significance was set at p ≤ 0.05; statistically significant samples are in bold.
(DOCX)
(DOCX)
Acknowledgments
This study was funded by grants from the Medical Research Council (MRC), and the Wellcome Trust. We thank the High-Throughput genomics group at the Wellcome Trust for Centre for Human Genetics (funded by the Wellcome Trust grant reference 090532/Z/09/Z) for the generation of sequences, and Dr Noel McCarthy (Department of Zoology, University of Oxford, Oxford, UK) for assisting with statistical analyses.
Data Availability
All relevant data are within the paper and its Supporting Information files. Geneome sequence files available at Dryad doi:10.5061/dryad.82jq4 and doi:10.5061/dryad.br0h8.
Funding Statement
This work was supported by Medical Research Council grant numbers MR/M501608/1 and MR/L015080/1 awarded to SKS, and G0801929 awarded to LGH and DM (www.mrc.ac.uk); Wellcome Trust grant number 088786/C/09/Z awarded to SKS, and grant number 090532/Z/09/Z awarded to RB and KAJ (www.wellcome.ac.uk). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mack D, Rohde H, Harris LG, Davies AP, Horstkotte MA, Knobloch JK. Biofilm formation in medical device-related infection. Int J Artif Organs. 2006;29:343–59. [DOI] [PubMed] [Google Scholar]
- 2.Piette A, Verschraegen G. Role of coagulase-negative staphylococci in human disease. Veterinary Microbiology. 2009;134(1–2):45–54. doi: 10.1016/j.vetmic.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Klevens RM, Edwards JR, Richards CL Jr., Horan TC, Gaynes RP, Pollock DA, et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007;122(2):160–6. Epub 2007/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29(11):996–1011. Epub 2008/10/25. 10.1086/591861 [DOI] [PubMed] [Google Scholar]
- 5.Scott II RD. The direct medical costs of healthcare-associated infections in US hospitals and the benefits of prevention 2009.
- 6.Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189(6):2540–52. Epub 2007/01/16. doi: JB.01484-06 [pii] 10.1128/JB.01484-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mack D, Horstkotte M, Rohde H, Knobloch JKM. Coagulase-negative staphylococci In: Pace JL RM, Finch RG, editor. Biofilms, infection, and antimicrobial therapy: CRC Press, Boca Raton; 2006. p. 109–53. [Google Scholar]
- 8.Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12(1):49–62. Epub 2013/12/18. 10.1038/nrmicro3161 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arrecubieta C, Lee M- H, Macey A, Foster TJ, Lowy FD. SdrF, a Staphylococcus epidermidis Surface Protein, Binds Type I Collagen. J Biol Chem. 2007;282(26):18767–76. 10.1074/jbc.M610940200 [DOI] [PubMed] [Google Scholar]
- 10.Bowden MG, Visai L, Longshaw CM, Holland KT, Speziale P, Hook M. Is the GehD Lipase from Staphylococcus epidermidis a Collagen Binding Adhesin? J Biol Chem. 2002;277(45):43017–23. 10.1074/jbc.M207921200 [DOI] [PubMed] [Google Scholar]
- 11.Hartford O, O'Brien L, Schofield K, Wells J, Foster TJ. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology. 2001;147(9):2545–52. [DOI] [PubMed] [Google Scholar]
- 12.Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekotter A, Peters G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149(10):2769–78. [DOI] [PubMed] [Google Scholar]
- 13.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20(5):1083–91. 10.1111/j.1365-2958.1996.tb02548.x [DOI] [PubMed] [Google Scholar]
- 14.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178(1):175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55(6):1883–95. [DOI] [PubMed] [Google Scholar]
- 16.Christner M, Franke GC, Schommer NN, Wend t, Wegert K, Pehle P, et al. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 2010;75(1):187–207. 10.1111/j.1365-2958.2009.06981.x [DOI] [PubMed] [Google Scholar]
- 17.Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM, et al. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology. 2012;158(Pt 4):1005–15. Epub 2012/01/28. 10.1099/mic.0.055459-0 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28(9):1711–20. [DOI] [PubMed] [Google Scholar]
- 19.Rohde H, Kalitzky M, Kroger N, Scherpe S, Horstkotte MA, Knobloch JK- M, et al. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J Clin Microbiol. 2004;42(12):5614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolo J, de Lencastre H, Miragaia M. Strategies of adaptation of Staphylococcus epidermidis to hospital and community: amplification and diversification of SCCmec. J Antimicrob Chemother. 2012;67(6):1333–41. Epub 2012/03/17. 10.1093/jac/dks068 [pii]. . [DOI] [PubMed] [Google Scholar]
- 21.Weiss S, Kadlec K, Fessler AT, Schwarz S. Identification and characterization of methicillin-resistant Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus pettenkoferi from a small animal clinic. Veterinary Microbiology. 2013;167(3–4):680–5. 10.1016/j.vetmic.2013.07.036 . [DOI] [PubMed] [Google Scholar]
- 22.Lawrence JR, Korber DR, Hoyle BD, Costerton JW, Caldwell DE. Optical sectioning of microbial biofilms. J Bacteriol. 1991;173(20):6558–67. Epub 1991/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146 (Pt 10):2395–407. Epub 2000/10/06. . [DOI] [PubMed] [Google Scholar]
- 24.Murga R, Stewart PS, Daly D. Quantitative analysis of biofilm thickness variability. Biotechnol Bioeng. 1995;45(6):503–10. Epub 1995/03/20. 10.1002/bit.260450607 . [DOI] [PubMed] [Google Scholar]
- 25.Beyenal H, Donovan C, Lewandowski Z, Harkin G. Three-dimensional biofilm structure quantification. J Microbiol Methods. 2004;59(3):395–413. [DOI] [PubMed] [Google Scholar]
- 26.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–9. Epub 2008/03/20. 10.1101/gr.074492.107 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595 Epub 2010/12/15. 10.1186/1471-2105-11-595 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill SR, Fouts DE, Archer GL, Mongodin EF, DeBoy RT, Ravel J, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187(7):2426–38. 10.1128/jb.187.7.2426-2438.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. Epub 2004/03/23. 10.1093/nar/gkh340 32/5/1792 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175(3):1251–66. Epub 2006/12/08. doi: genetics.106.063305 [pii] 10.1534/genetics.106.063305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheppard SK, Jolley KA, Maiden MCJ. A Gene-By-Gene Approach to Bacterial Population Genomics: Whole Genome MLST of Campylobacter. Genes. 2012;3(2):261–77. 10.3390/genes3020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meric G, Miragaia M, de Been M, Yahara K, Pascoe B, Mageiros L, et al. Ecological Overlap and Horizontal Gene Transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol Evol. 2015;7(5):1313–28. 10.1093/Gbe/Evv066 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151(Pt 5):1453–64. 10.1099/mic.0.27534-0 . [DOI] [PubMed] [Google Scholar]
- 34.Nilsson M, Frykberg L, Flock JI, Pei L, Lindberg M, Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infection and Immunity. 1998;66(6):2666–73. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galdbart J- O, Morvan A, Desplaces N, el Solh N. Phenotypic and genomic variation among Staphylococcus epidermidis strains infecting joint prostheses. J Clin Microbiol. 1999;37(5):1306–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klingenberg C, Rønnestad A, Anderson AS, Abrahamsen TG, Zorman J, Villaruz A, et al. Persistent strains of coagulase-negative staphylococci in a neonatal intensive care unit: virulence factors and invasiveness. Clin Microbiol Infect. 2007;13(11):1100–11. [DOI] [PubMed] [Google Scholar]
- 37.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48(6):1429–49. 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- 38.Schommer NN, Christner M, Hentschke M, Ruckdeschel K, Aepfelbacher M, Rohde H. Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infect Immun. 2011;79(6):2267–76. Epub 2011/03/16. 10.1128/IAI.01142-10 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheppard SK, Colles FM, McCarthy ND, Strachan NJ, Ogden ID, Forbes KJ, et al. Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol Ecol. 2011;20(16):3484–90. Epub 2011/07/19. 10.1111/j.1365-294X.2011.05179.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–5. Epub 2003/03/08. 10.1126/science.1080857 299/5612/1582 [pii]. . [DOI] [PubMed] [Google Scholar]
- 41.Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infection, Genetics and Evolution. 2008;8(6):747–63. doi: 10.1016/j.meegid.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Gomes AR, Vinga S, Zavolan M, de Lencastre H. Analysis of the Genetic Variability of Virulence-Related Loci in Epidemic Clones of Methicillin-Resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 2005;49(1):366–79. 10.1128/AAC.49.1.366-379.2005 PMC538922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the Natural environment to infectious diseases. Nat Rev Micro. 2004;2(2):95–108. [DOI] [PubMed] [Google Scholar]
- 44.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–45. [DOI] [PubMed] [Google Scholar]
- 45.Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–74. Epub 2010/01/23. 10.1126/science.1182395 327/5964/469 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y-Q, Ren S-X, Li H-L, Wang Y-X, Fu G, Yang J, et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Molecular Microbiology. 2003;49(6):1577–93. [DOI] [PubMed] [Google Scholar]
- 47.Chokr A, Leterme D, Watier D, Jabbouri S. Neither the presence of ica locus, nor in vitro-biofilm formation ability is a crucial parameter for some Staphylococcus epidermidis strains to maintain an infection in a guinea pig tissue cage model. Microb Pathog. 2007;42(2–3):94–7. Epub 2006/11/07. doi: S0882-4010(06)00123-9 [pii] 10.1016/j.micpath.2006.09.001 . [DOI] [PubMed] [Google Scholar]
- 48.Conlon BP, Geoghegan JA, Waters EM, McCarthy H, Rowe SE, Davies JR, et al. Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J Bacteriol. 2014;196(24):4268–75. Epub 2014/10/01. 10.1128/JB.01946-14 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Los R, Sawicki R, Juda M, Stankevic M, Rybojad P, Sawicki M, et al. A comparative analysis of phenotypic and genotypic methods for the determination of the biofilm-forming abilities of Staphylococcus epidermidis. FEMS Microbiol Lett. 2010;310(2):97–103. Epub 2010/08/21. 10.1111/j.1574-6968.2010.02050.x [pii]. . [DOI] [PubMed] [Google Scholar]
- 50.Hennig S, Nyunt Wai S, Ziebuhr W. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int J Med Microbiol. 2007;297(2):117–22. [DOI] [PubMed] [Google Scholar]
- 51.von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet InfectDis. 2002;2(11):677–85. [DOI] [PubMed] [Google Scholar]
- 52.Ziebuhr WK V; Rachid S; Lossner I; Gotz F; Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–56. [DOI] [PubMed] [Google Scholar]
- 53.Handke LD, Conlon KM, Slater SR, Elbaruni S, Fitzpatrick F, Humphreys H, et al. Genetic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J Med Microbiol. 2004;53(5):367–74. [DOI] [PubMed] [Google Scholar]
- 54.Christensen GB LM; Simpson WA. Phenotypic variation of Staphylococcus epidermidis slime production in vitro and in vivo. Infect Immun. 1987;55:2870–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol. 2005;43(9):4751–7. Epub 2005/09/08. doi: 43/9/4751 [pii] 10.1128/JCM.43.9.4751-4757.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, et al. How clonal Is Staphylococcus aureus? J Bacteriol. 2003;185(11):3307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Regression analysis graphs: A) biovolume versus thickness; B) biovolume versus roughness coefficient (Ra); and C) thickness versus roughness coefficient (Ra). The results show a correlation between biovolume and thickness but not with roughness coefficient.
(TIF)
Level of significance was set at p ≤ 0.05; statistically significant samples are in bold.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Geneome sequence files available at Dryad doi:10.5061/dryad.82jq4 and doi:10.5061/dryad.br0h8.