Abstract
We report here NMR and ITC studies of the binding of ionizable guests (carboxylate acids) to a deep-cavity cavitand. These studies reveal that the shortest guests favored 1:1 complex formation, but the longer the alkyl chain the more the 2:1 host-guest capsule is favored. For intermediate-sized guests, the equilibrium between these two states is controlled by pH; at low values the capsule containing the carboxylic acid guest is favored, whereas as the pH is raised deprotonation of the guest favors the 1:1 complex. Interestingly, for one host-guest pair the energy required to de-cap the 2:1 capsular complex and form the 1:1 complex is sufficient to shift the pKa of the guest by ~ 3–4 orders of magnitude (4.1–5.4 kcal mol−1). The two largest guests examined form stable 2:1 capsules, with in both cases the guest adopting a relatively high energy J-shaped motif. Furthermore, these 2:1 complexes are sufficiently stable that at high pH guest deprotonation occurs without de-capping of the capsule.
Graphical Abstract
Introduction
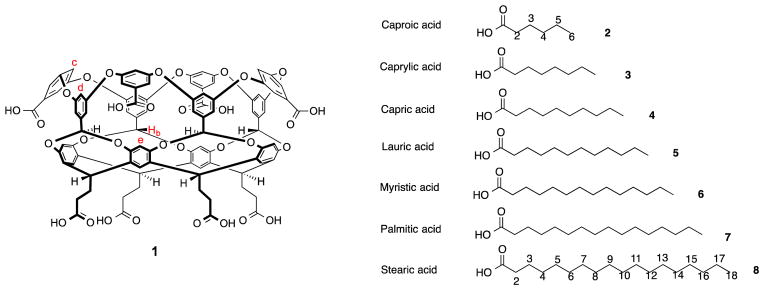
With the long term goal of examining the hydrophobic effect at the molecular scale, we detail here the binding interactions and, in selected cases, the preferred binding motif of fatty acids within deep-cavity cavitand 1 (Figure 1) and its corresponding dimeric nano-capsule.1 Specifically, using a combination of NMR and Isothermal Titration Calorimetry (ITC) we examine the complexes formed by 1 and seven fatty acids (2–8) as a function of host-guest ratio, temperature, and pH. The details revealed by these analytical techniques provide useful insight into the structural and thermodynamic nature of these complexes.
Figure 1.
Chemical structures of Cavitand 1 and guests 2–8.
Saturated fatty acids play key roles in biology, including: as components of membranes, as protein structure stabilizers, in energy storage/cytosolic transport, in signaling processes, and in gene regulation.2 The insolubility of long-chain fatty acids means that transporters – fatty acid binding proteins (FABPs) – are required for distribution of this key class of molecules between extra- and intracellular membranes.3 FadL is an exemplar of this class of protein.4 Key to its transport properties is its flattened, 14-stranded β-barrel tertiary structure forming a large hydrophobic pocket for guest binding. In the obtained crystal structures of FABPs such as FadL, the chain of the guest fatty acid has been seen to adopt at least two binding motifs: extended (‘linear’) and J– or U–shaped, whilst the carboxylate group is found near the portal of the pocket forming salt-bridges and hydrogen bonds with nearby residues. There are however many questions relating to this guest packing motif in the solution phase, and how this plays a role in the thermodynamics of fatty acid binding and release.5
Unsurprisingly, supramolecular chemistry has shown considerable interest in this remarkably straightforward, yet complicated, class of guest. Both with an eye on sensor development and understanding the fundamentals of how flexible alkyl chains can be recognized, a wide variety of investigations into the complexation of fatty acids and long-chain alkanes have been reported. By way of example, cyclodextrins,6 and various calixarene7 and resorcinarene8 derivatives have proven to be particularly popular.
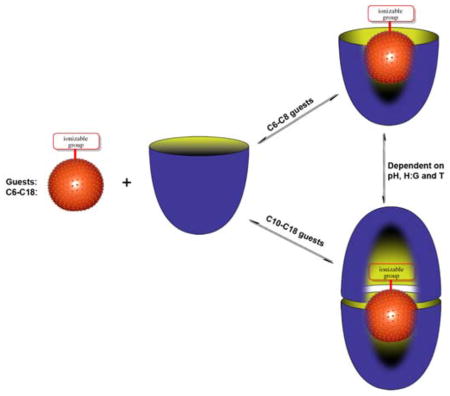
Building on the above thoughts, we report here the NMR and ITC analysis of complexes formed between deep-cavity cavitand 1 and saturated fatty acids 2–8. We have previously shown that cavitand 1 is predisposed to assemble into supramolecular nano-capsules when in the presence of relatively hydrophobic guests.1d, 9 However, in the presence of anionic guests such as small inorganic anions and small carboxylates the host forms 1:1 complexes instead.10 Only approximately ten non-hydrogen atoms can fit within the binding pocket of 1. Consequently, we anticipated that as the chain length is increased in the guest series 2–8, so the complexes formed with 1 would transition from 1:1 to 2:1 host-guest complex, and reveal details about guest packing motifs and the need for carboxylate protonation for formation of the 2:1 capsular complex. This notion and a summary of our observations are presented in Figure 2.
Figure 2.
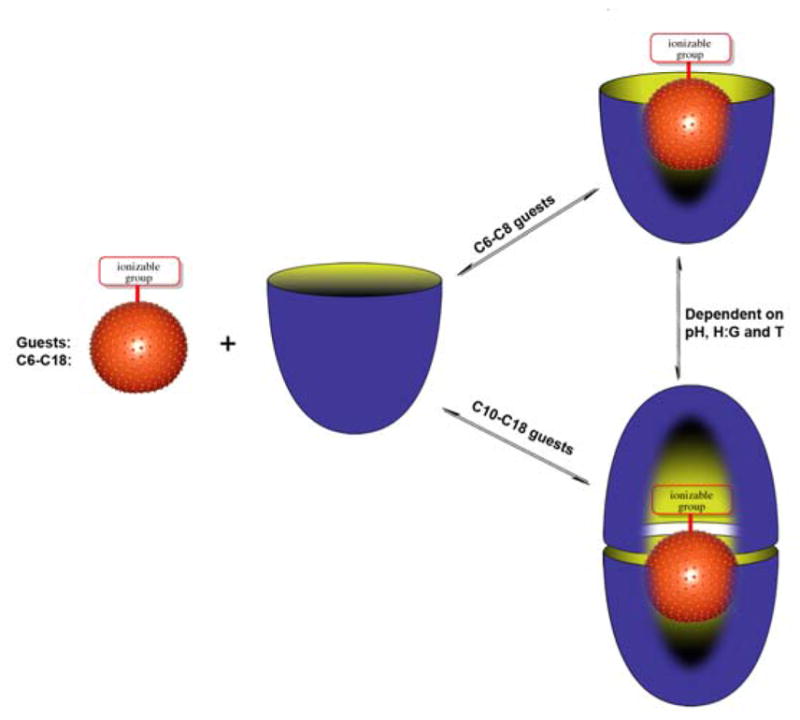
Schematic of the formation of 1:1 complexes or 2:1 assemblies during the binding of C6–C18 saturated fatty acids 2–8 with cavitand 1.
Results and Discussion
Complex with guest 2
We began with previously reported guest 2 (studied in sodium tetraborate buffer).10d As expected, guest 2 formed a 1:1 complex (Figures 3, S1, and S2). The 1H NMR spectra – which showed little change as a function of buffer at basic pH – confirmed a fast exchange rate close to the NMR time-scale. Peak assignment was based on the fact that as these kinds of amphiphilic guests bind with their carboxylates at the portal of the pocket, the nearer a group is to the terminal methyl the deeper it is bound in the truncated cone-shaped pocket. Consequently, Δδ values and signal width were seen to increase from C2–C6. The ITC data for this complex (Table 1) revealed binding to be enthalpically driven and essentially entropically neutral.
Figure 3.
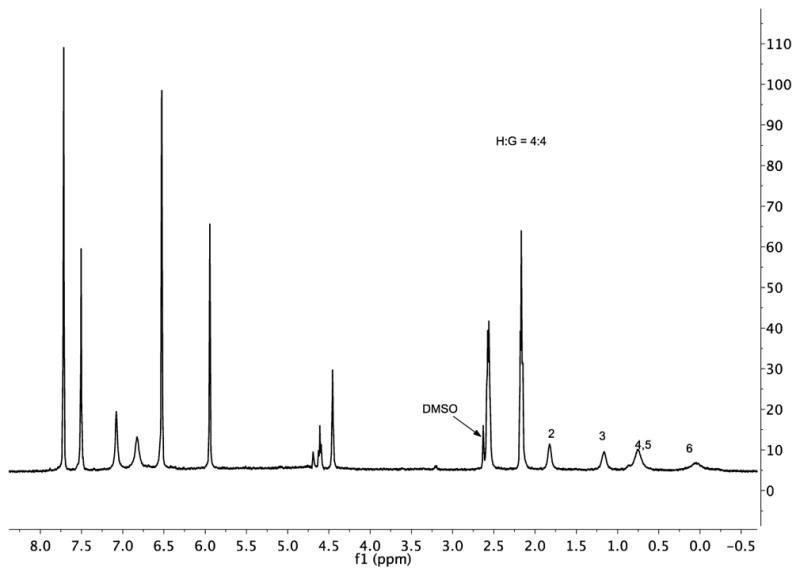
1H NMR spectrum (pre-saturation water suppression) of the 1:1 complex formed between 1 and 2 (both 1 mM) in 10 mM borate buffer, pD 9.73. Bound guest signals are marked. The residual DMSO-d6 signal arose from the necessity to dissolve the guest in a small volume of the solvent to aid mixing.
Table 1.
Thermodynamic dataa for binding of the saturated fatty acids 2–5 to host 1.
| Guest | Ka (M−1) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | −TΔS (kcal mol−1) |
|---|---|---|---|---|
| 2 | 6.75 x 103 | −5.22 | −5.13 | −0.097 |
| 3 | 8.07 x 104 | −6.70 | −6.09 | −0.602 |
| 4 | 3.25 x 105 | −7.52 | −6.52 | −1.000 |
| 5 | 4.19 x 105 | −8.08 | −8.82 | 0.762 |
All determinations at 298 K except guest 5 (313 K). Data is an average of three titrations with experimental errors between runs of < 5%. Sodium phosphate buffer concentration was 50 mM (pH = 11.5).
Complex with guest 3
As reported previously10d this guest formed a 1:1 complex with the host (Figure S4). The 1H NMR spectra confirmed an exchange rate faster, but close to the NMR time scale. Relative to guest 2, the complexation of 3 was stronger, more exothermic, and weakly entropically favorable rather that unfavorable (Table 1 and Figure S5).
Complex with guest 4
The complex between 1 and capric acid 4 was previously noted parenthetically and not studied in detail.11 The binding properties of this guest are more complex that those of guests 2 and 3. 1H NMR revealed that at pD = 9.73 and a host-guest ratio of 4:1, free host and both its 1:1 and 2:1 complexes were formed (Figure S6). This was most evident from the host Hd-signals (see assignments in Figure 1) and two highfield, bound-methyl signals. As anticipated, as the amount of guest was increased, so the amount of 1:1 complex increased. However, at the end of the titration (1:1 ratio) there was still evidence of 2:1 complex.
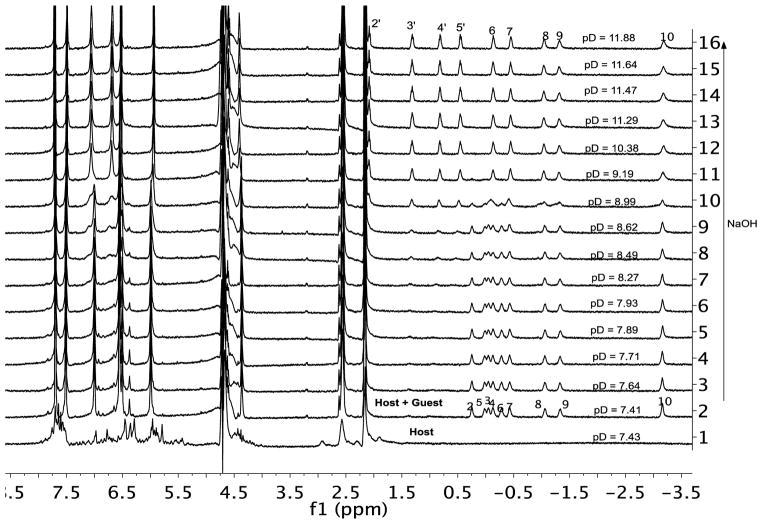
To investigate this further, a 2:1 host-guest mixture was used in a pH titration 1H NMR experiment (Figure 4). At the beginning of the titration (pD = 7.43) the host was extensively aggregated. However aggregation was greatly reduced in the presence of guest 4, and at pD = 7.93 COSY (Figure S7) and diffusion NMR (D0 = 1.40 × 10−6 cm2 s−1)12 revealed essentially pure capsular complex. COSY NMR also revealed that the guest adopted an extended motif within the capsule, with the terminal methyl group in the base of one pocket and the carboxylic acid in the base of the other.
Figure 4.
1H NMR spectra of capric acid 4 encapsulated in host 1 (H:G = 2:1) as a function of pD in 50 mM phosphate buffer. The lower most spectrum corresponding to 1 alone in 50 mM phosphate buffer. pH was controlled with small amounts of NaOH.
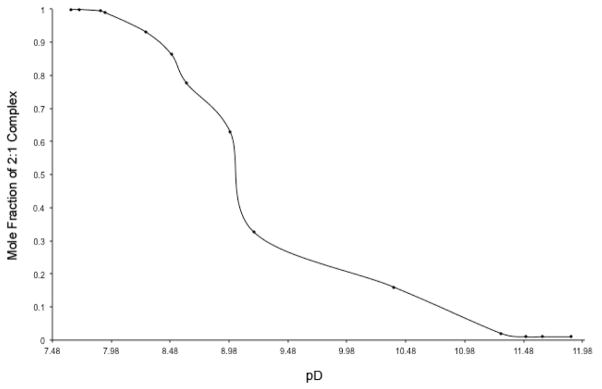
As the pD was increased to 11.88 there was an increase in the anisotropic peak distribution in the guest binding region, and diffusion NMR revealed this new species to be the corresponding 1:1 complex (D0 = 1.79 × 10−6 cm2 s−1). Thus the capsular complex underwent de-capping at the pD was increased. There are two possible sources of this de-capping: changes to either the protonation state of the host or the guest. A graph of the mole-fraction of the 2:1 complex as a function of pD is shown in Figure 5. We attribute the steep inflection point to the singular deprotonation of the protonated guest (rather than any multiple-step deprotonation of the host which would give a more gradual change). Assuming that the system behaves as a two-state model in which the 2:1 capsular complex contains the protonated guest is the acid, and the 1:1 complex involves the deprotonated, the Henderson-Hasselbalch equation reveals the pKa of the encapsulated guest to be ~8.95. This is some three to four orders of magnitude less acidic than a free carboxylic acid (4.1–5.4 kcal mol−1 of stabilization), but is consistent with the pKa values of carboxylic acids found in the hydrophobic environment of, for example, enzymes.13 The high pKa value is also consistent with data from solvatochromic probes bound within the capsule formed by 1 which reveal no water of encapsulation when a guest is present.14 In the dry, inner environment of the capsule guest 4 resists deprotonation because this would result in de-capping, and hence generation of exposed hydrophobic surfaces of the free host, the rim of the host in the complex, and the C2–C4 methylenes of the bound guest.
Figure 5.
Plot of mole fraction of the 2:1 complex using the guest H2 protons signals.
As with guests 2 and 3, the ITC experiments with 4 were carried out at concentration one order of magnitude lower than the NMR studies, and under these conditions 1 and 4 formed exclusively a 1:1 complex (Figure S9). This complex proved to be half an order of magnitude stronger than 3 and almost two orders stronger than 2 (Table 1). Relative to 3, the binding of 4 was more exothermic and the favorable entropy change larger.
Complex with guest 5
At a 4:1 ratio of host 1 and guest 5 the 1H NMR (Figure S10) revealed a sharp terminal methyl peak of the bound guest indicative of a 2:1 complex. Diffusion NMR however indicated that this combination of host and guest formed a mixture of 1:1 and 2:1 complex (D0 = 1.51 × 10−6 cm2 s−1). As more guest was added, the terminal methyl peak broadened considerably and, as suspected, diffusion NMR pointed towards an increase in the mole ratio of the 1:1 complex (D0 = 1.70 × 10−6 cm2 s−1). Because of peak overlap in the 4:1 ratio case, and peak broadening in the 1:1 ratio case, it was not possible to obtain information from COSY 1H NMR. However, a VT NMR experiment of a 4:1 mixture of 1 and 5 (Figure S11) supported this dynamic exchange between 1:1 and 2:1 complex. Thus as the temperature was increased from 25 °C to 85 °C, the bound guest signal anisotropy increase significantly to reveal all of the eleven bound guest signals. Overall, these results demonstrate that 1 and 5 form mixtures of 1:1 and 2:1 complexes that exchange close to the NMR timescale.
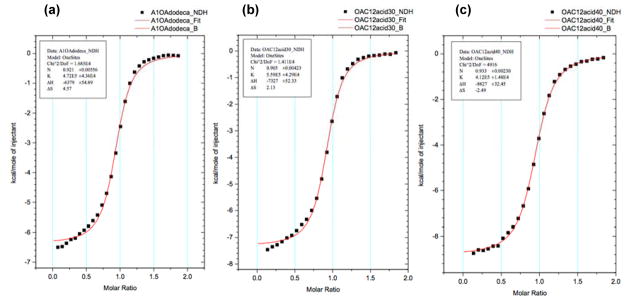
Similarly, VT ITC provided further evidence of the tendency of host 1 and guest 5 to form mixtures of complexes (Figure 6). As expected, at 25 °C the data fitted neither a 1:1 nor a 2:1 binding model suggesting that, as observed by NMR, a mixture of complexes was present. However, increasing the temperature to 40 °C (and disfavoring the 2:1 complex) led to an excellent fit to a 1:1 model. At this temperature the binding constant was determined to be Ka = 4.19 x 10−5 M−1, i.e., slightly higher than the binding of 2,3 and 4 at 25 °C.
Figure 6.
Variable temperature ITC data for the complexes formed between 1 and 5. A 1.5 mM solution of 5 was titrated into 0.15 mM solution of 1. Both host and guest were in 10 mM phosphate buffer, pH 11.3. Temperatures are: a) 25 °C; b) 30 °C and; c) 40 °C (See also Figures S12–S14).
Molecular models of guest 5 inside 1 reveal that the guest is too long to be bound in the pocket of the host and that even with maximal compression several of the methylene groups nearest the carboxylate group must necessarily be exposed to the aqueous environment. This is not an idea situation. Capping the 1:1 complex would minimize the solvation of the hydrocarbon surfaces but force desolvation of the carboxylate. Consequently, the two complexes are roughly isoenergetic, but increases in pD, T or equivalents of guest pushes the equilibrium towards the 1:1 species.
Complex with guest 6
The properties of the myristic acid 6 complex were found to be similar to those of lauric acid 5, but that overall it formed a more stable capsular complex and showed slower exchange between the free and bound states. At a host-guest ratio of 4:1 where the capsular complex predominated (D0 = 1.43 × 10−6 cm2 s−1), only the signals from what were presumed to be the C9–C14 methylenes were observed by 1H NMR and COSY (Figure S15 and S16). The other methylenes of the encapsulated guest presumably were evident in the broad signals at, for example, −0.75 ppm. Considering that within the capsule formed by 1 smaller guests adopt extended binding motifs whilst larger guest adopt J-shaped motifs (vide infra), we believe that this guest lies at the transition between these two motifs and that the limited NMR signals arises because the exchange between these two states is on the NMR-timescale. In contrast, at a 1:1 host-guest ratio where the mixture was composed principally of 1:1 complex (D0 = 1.64 × 10−6 cm2 s−1) all thirteen signals could be observed, and COSY 1H NMR showed (Figure S17) that the signals from the H-atoms on C14 had much larger Δδ values than those of C2 and C3 methylene groups suggesting the typical carboxylate-at-portal binding motif.
ITC demonstrated that guest 6 had a greater tendency to form capsular complexes than 5. Thus, at 25 °C the ITC thermogram was distinctly non-monotonic, tending towards a distinct two-step assembly event (Figure S18). Even at 60 °C the fit to the 1:1 binding model was poor (Figure S19) confirming that 6 formed a more stable capsule.
Complex with guest 7
1H NMR showed that at a host guest ratio of 4:1 palmitic acid 7 formed a 2:1 capsular complex (Figure S20), and this was confirmed by diffusion NMR (D0 = 1.35 × 10−6 cm2 s−1). COSY 1H NMR and the calculated Δδ values (Figure S21 and S22) demonstrated that rather than adopting an extended packing motif as its small homologues do, guest 7 adopts a J-shape conformation within the capsule with the terminal methyl in one ‘pole’ of the capsule, a bend in the alkyl chain at the other, and the carboxylic acid at the equator. Thus, the signals from the H-atoms on the terminal C16, and mid-chain C7 and C8 methylenes undergo larger shifts (are more deeply bound) than those of the C11–C13 and C2–C5 methylenes. Hence, not only is this guest too long to form a stable 1:1 complex, but it is too long to fit within the capsule in an extended form and must necessarily adopt a higher energy J-motif.8a Variable temperature NMR revealed that at this host-guest ratio the capsule began to break down at 50 °C (Figure S23).
At the 1:1 host-guest ratio, signal broadening (Figure S20) suggested the formation of some 1:1 complex, and diffusion NMR (D0 = 1.59 × 10−6 cm2 s−1) confirmed a mixture of both 1:1 and 2:1 complex. Molecular models and the relative small shift and coincidence of the C3–C10 methylenes suggest that in the 1:1 complex this part of the guest is not within the pocket of the host but is solvent exposed and tightly packed.
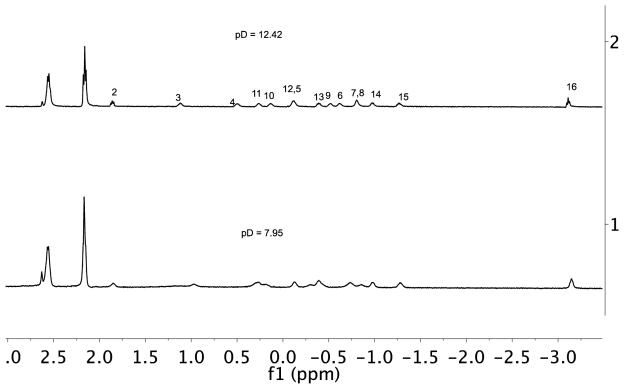
We also examined the effect of pD on the stability of the capsule formed by 7 (Figure 7 and S24). Diffusion NMR demonstrated that for a 2:1 mixture of host and guest the mixture was mostly 2:1 capsular complex across the pD range of 7.95 to 12.42. Nevertheless, over this pD range subtle changes to the signals of the guest-binding region did occur (Figure 7). One possibility for these changes is a protonation state change to the host. However, a more likely possibility is that at pD 7.95 the encapsulated guest is protonated, but that at higher pD the carboxylic acid group is deprotonated. The guest is however so long that de-capping would expose considerable amounts of hydrocarbon surface, and consequently even the carboxylate binds in the capsule. This deprotonation is likely aided by the preferred packing motif of the guest; the J-shaped motif ensures the carboxylate is located at the equatorial region of the capsule where it can either be partially hydrated, or even may protrude out of the capsule to be solvated by external water.
Figure 7.
Guest binding region of 1H NMR spectra of palmitic acid 7 encapsulated in host 1 (H:G = 2:1) at pD = 7.95 (lower spectrum) and at pD = 12.42 (upper) in 50 mM phosphate buffer.
Complex with guest 8
As expected, 1H NMR revealed (Figure S25) that guest 8 demonstrated the greatest tendency to form a capsular complex. This was confirmed by diffusion NMR experiments showing that at 4:1 and 1:1 host-guest ratios the diffusion constants varied from 1.26 × 10−6 cm2 s−1 to 1.33 × 10−6 cm2 s−1. COSY NMR (Figures S26) allowed the calculation of the Δδ values (Figure S27) for the guest signals and revealed that this guest adopts a J-shaped binding motif similar to 7.
Conclusions
Our studies of the binding of carboxylate acids to host 1 reveal several interesting phenomena. First, as the chain length of the guest is systematically lengthened, so more of its chain must necessarily be exposed to water in a 1:1 complex. Consequently, longer guests have an increased tendency to form 2:1 capsular complexes rather than the open 1:1 species. Second, for intermediate-sized guests the equilibrium between the 1:1 and 2:1 states can be controlled by pH. At low pH the capsule is favored. However as the pH is raised, so the conflict between the solvation of hydrophobic surfaces on host and guest, and the solvation of the latent carboxylate of the guest, favors the latter and the 1:1 complex. Interestingly, the energy required to de-cap the capsular complex formed between 1 and 4 is sufficient to shift the pKa of the guest by three to four orders of magnitude (4.1–5.4 kcal mol−1). Third, the two largest guests examined form stable 2:1 capsules, but in both cases the guest is longer than the interior of the capsule and it must necessarily adopt a higher energy J-shaped motif. Finally, in these stable 2:1 complexes there are subtle changes to the bound guest region of their NMR spectra as a function of pH. The precise cause of these changes is unclear, but the most likely candidate is deprotonation of the bound carboxylic aid to form the corresponding encapsulated carboxylate. Further studies of these types of complex are ongoing.
Supplementary Material
Acknowledgments
The authors would like to thank the National Science Foundation (CHE 1244024) and National Institutes of Health (GM098141) for generous support of this work.
References
- 1.(a) Gibb CLD, Gibb BC. J Am Chem Soc. 2004;126:11408–11409. doi: 10.1021/ja0475611. [DOI] [PubMed] [Google Scholar]; (b) Liu S, Gibb BC. Chem Commun. 2008:3709–3716. doi: 10.1039/b805446k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liu S, Whisenhunt-Ioup SE, Gibb CLD, Gibb BC. Supramolecular Chemistry. 2011;24:480–485. doi: 10.1080/10610278.2010.550290. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Jordan JH, Gibb BC. Chem Soc Rev. 2015;44(2):547–85. doi: 10.1039/c4cs00191e. [DOI] [PubMed] [Google Scholar]
- 2.(a) Watson RR, editor. Fatty Acids in Health Promotion and Disease Causation. AOCS Press; 2009. p. 857. [Google Scholar]; (b) Begley TP, editor. Wiley Encyclopedia of Chemical Biology; Volume 2; Metabolism: Fatty Acid Desaturases, Chemistry of to Metabolic Diseases: Biological Mechanisms. John Wiley & Sons, Inc; 2009. p. 785. [Google Scholar]; (c) Mostofsky DI, Yehuda S, Salem N Jr, editors. Fatty Acids: Physiological and Behavioral Functions. Humana; 2001. p. 435. [Google Scholar]; (d) Michal G, Schomburg D. Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology. 2. John Wiley and Sons; New York: 2012. [Google Scholar]; (e) Choi JK. The Journal of Lipid Research. 2002;43(7):1000–1010. doi: 10.1194/jlr.m200041-jlr200. [DOI] [PubMed] [Google Scholar]
- 3.(a) Furuhashi M, Hotamisligil GS. Nature Reviews Drug Discovery. 2008;7(6):489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Esteves A, editor. Fatty Acid-Binding Proteins. Transworld Research Network; 2009. p. 87. [Google Scholar]
- 4.van den Berg B, Black PN, Clemons WMJ, Rapoport TA. Science. 2004;304:1506–1509. doi: 10.1126/science.1097524. [DOI] [PubMed] [Google Scholar]
- 5.Matsuoka S, Sugiyama S, Matsuoka D, Hirose M, Lethu S, Ano H, Hara T, Ichihara O, Kimura SR, Murakami S, Ishida H, Mizohata E, Inoue T, Murata M. Angew Chem Int Ed Engl. 2015;54(5):1508–11. doi: 10.1002/anie.201409830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rekharsky MV, Inoue Y. Chem Rev. 1998;98:1875–1917. doi: 10.1021/cr970015o. [DOI] [PubMed] [Google Scholar]
- 7.(a) Hou JL, Ajami D, Rebek J., Jr J Am Chem Soc. 2008;130:7810–7811. doi: 10.1021/ja802288k. [DOI] [PubMed] [Google Scholar]; (b) Ajami D, Rebek J., Jr Nature Chemistry. 2009;1:87–90. doi: 10.1038/nchem.111. [DOI] [PubMed] [Google Scholar]; (c) Asadi A, Ajami D, Rebek J., Jr J Am Chem Soc. 2011;133(28):10682–4. doi: 10.1021/ja203602u. [DOI] [PubMed] [Google Scholar]; (d) Gavette JV, Zhang KD, Ajami D, Rebek J., Jr Org Biomol Chem. 2014;12(34):6561–3. doi: 10.1039/c4ob01032a. [DOI] [PubMed] [Google Scholar]; (e) Zhang KD, Ajami D, Gavette JV, Rebek J., Jr J Am Chem Soc. 2014;136(14):5264–6. doi: 10.1021/ja501685z. [DOI] [PubMed] [Google Scholar]; (f) Shorthill BJ, Avetta CT, Glass TE. J Am Chem Soc. 2004;126:12732–12733. doi: 10.1021/ja047639d. [DOI] [PubMed] [Google Scholar]; (g) Avetta CT, Shorthill BJ, Ren C, Glass TE. J Org Chem. 2012;77(2):851–7. doi: 10.1021/jo201791a. [DOI] [PubMed] [Google Scholar]
- 8.(a) Liu S, Russell DH, Zinnel NF, Gibb BC. J Am Chem Soc. 2013;135(11):4314–4324. doi: 10.1021/ja310741q. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gibb CLD, Gibb BC. Chem Commun. 2007:1635–1637. doi: 10.1039/b618731e. [DOI] [PubMed] [Google Scholar]
- 9.Laughrey Z, Gibb BC. Chem Soc Rev. 2011;40(1):363–86. doi: 10.1039/c0cs00030b. [DOI] [PubMed] [Google Scholar]
- 10.(a) Carnagie R, Gibb CLD, Gibb BC. Angew Chem Int Ed. 2014;53(43):11498–11500. doi: 10.1002/anie.201405796. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gibb CL, Gibb BC. J Am Chem Soc. 2011;133:7344–7347. doi: 10.1021/ja202308n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gibb CLD, Gibb BC. J Comp Aided Molec Design. 2014;28(4):319–325. doi: 10.1007/s10822-013-9690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Sun H, Gibb CLD, Gibb BC. Supramolecular Chemistry. 2008;20(1–2):141–147. [Google Scholar]
- 11.Liu S, Gan H, Hermann AT, Rick SW, Gibb BC. Nat Chem. 2010;2(10):847–852. doi: 10.1038/nchem.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibb CLD, Gibb BC. Tetrahedron. 2009;65(35):7240–7248. doi: 10.1016/j.tet.2009.01.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petsko GA, Ringe D. Protein Structure and Function. New Science Press; Sunderland, MA: 2004. [Google Scholar]
- 14.Porel M, Jayaraj N, Kaanumalle LS, Maddipatla MVSN, Parthasarathy A, Ramamurthy V. Langmuir. 2009;25:3473–3481. doi: 10.1021/la804194w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.