Abstract
Background
Human herpesvirus 6 (HHV-6) has a unique ability to integrate into chromosomal telomeres. Vertical transmission via germ cell integration results in offspring with inherited chromosomally integrated (ci)HHV-6 in all nucleated cells, affecting ~1% of the population.
Objectives
Inherited ciHHV-6 may be a direct or indirect mediator of human disease, but efficient identification of affected individuals is a fundamental roadblock to larger studies exploring the clinical importance of this condition.
Study design
A group testing strategy was designed to efficiently identify individuals with inherited ciHHV-6. DNA was extracted from 2496 cellular samples from hematopoietic cell transplant (HCT) donor–recipient pairs. Pools of 12 samples were screened for HHV-6 DNA with quantitative (q)PCR. Individual samples from high positive pools were tested with qPCR, and high positive individual samples were tested for inherited ciHHV-6 using droplet digital (dd)PCR to determine HHV-6 DNA copies/cellular genome.
Results
Thirty-one pools had high positive HHV-6 DNA detection with >103 HHV-6 DNA copies/μg. Each pool had one sample with >104 copies/μg HHV-6 DNA. Inherited ciHHV-6 was confirmed by ddPCR in every high positive sample (>103 HHV-6 DNA copies/μg), yielding a prevalence of 1.5% in HCT recipients and 0.96% in donors. We performed 580 qPCR tests to screen 2496 samples for inherited ciHHV-6, a 77% reduction in testing.
Conclusions
Inherited ciHHV-6 can be efficiently identified by specimen pooling coupled with modern molecular techniques. This algorithm can be used to facilitate cost-effective identification of patients with inherited ciHHV-6, thereby removing a major hurdle for large-scale study of its clinical impact.
Keywords: Herpes, PCR, HHV-6, Chromosomally integrated, Diagnostic, PCR
1. Background
Human herpesvirus 6 (HHV-6) species B infects >95% of people during childhood and causes exanthema subitum (roseola) [1]. HHV-6B frequently reactivates in immunocompromised patients and causes significant morbidity [2]. The epidemiology of HHV-6A infection is poorly understood, and this virus does not have clear disease associations. Like other herpesviruses, HHV-6A and HHV-6B establish life-long latency after primary infection. However, they have a unique ability to establish latency by integrating into chromosomal telomeres [3,4]. If viral integration occurs in a germ-line cell, vertical transmission of chromosomally integrated HHV-6 (ciHHV-6) via Mendelian inheritance results in offspring with the entire HHV-6 genome in every nucleated cell of their body. This condition of inherited ciHHV-6 will be passed on to 50% of children from affected individuals. Population-based studies demonstrate that inherited ciHHV-6 is found in ~1% of the population (~70 million people) [4].
In vitro and in vivo evidence suggest that inherited ciHHV-6 may play a role in complications ranging from encephalitis to heart disease due to direct (e.g. pathogenic viral reactivation) or indirect (e.g. telomeric instability and deletions) mechanisms [5–12]. However, the clinical impact of inherited ciHHV-6 remains poorly understood due to the low prevalence of this condition, lack of routine screening, and the diagnostic challenge of identifying affected patients [13]. If a patient is suspected to harbor inherited ciHHV-6, confirmation requires specialized methods such as fluorescence in situ hybridization (FISH) or droplet digital polymerase chain reaction (ddPCR) assays [4,14]. As a result, studies have been limited to case reports and case series, aside from one recent study that required extensive resources to test ~20,000 patients [15].
2. Objectives
To facilitate larger studies of the clinical significance of inherited ciHHV-6 and whether routine screening of all or select patient populations is warranted, efficient testing strategies are needed. An early approach to resourceful identification of low prevalence conditions was developed by Robert Dorfman in 1943 using group testing to identify military recruits with syphilis [16], a strategy that is particularly well-suited for targeting inherited ciHHV-6. We have coupled group testing with modern molecular techniques to design an algorithm for rapid and cost-effective identification of patients with inherited ciHHV-6.
3. Study design
3.1. Specimens
Specimens for this study were obtained from the Fred Hutchinson Cancer Research Center Research Cell Bank, which routinely collects peripheral blood mononuclear cells (PBMCs) leftover from human leukocyte antigen (HLA) typing of donors and recipients for hematopoietic cell transplantation (HCT). Most of these samples have been infected with Epstein–Barr virus (EBV) to create immortalized beta lymphoblastoid cell lines (LCLs) stored at −80 °C. Only complete donor–recipient pairs from 1992–2014 were used for this study in an effort to establish an informative cohort for future study. All samples were collected prior to HCT, when HHV-6 DNA detection due to viral reactivation is rare [17] and most likely due to inherited ciHHV-6.
LCLs were grown at 37 °C to a concentration of 20 million cells. The cells were pelleted and lysed in white cell lysis buffer. Supernatants were isolated in 100% Isopropanol after centrifugation (3000 rpm, 15 min, ambient). The DNA was precipitated and resuspended in 70% ethanol followed by centrifugation (3000 rpm, 5 min, ambient). After being decanted and air-dried, DNA was hydrated with DNA Hydration Solution (10 mM Tris, 1 mM EDTA).
Approximately 5 μg of DNA/sample were aliquoted at concentrations of ~200 ηg/μL. Given that 1 μg of cellular DNA represents ~1.5 × 105 cells, each μL of a sample contained ~30,000 cells. Cell lines with inherited ciHHV-6 contain the HHV-6 genome in every cell, so each μL of sample from an affected cell line will contain ~30,000 copies of the HHV-6 genome.
3.2. Laboratory assays
HHV-6 DNA targets were amplified and detected using a real-time quantitative fluorescent probe polymerase chain reaction (qPCR) assay as previously described [18]. Briefly, each 30 μl real-time PCR reaction contained 15 μl of 2x QuantiTect multiplex PCR master mix (Qiagen, CA), 830 nM primer (5R(A): GTT AGG ATA TAC CGA TGT GCG TGA T; 5R(B): TAC AGA TAC GGA GGC AAT AGA TTT G), 100 μM probe (5R(P): FAM-TCC GAA ACA ACT GTC TGA CTG GCA AAA-TAMRA), and 0.03 units of uracil-DNA-/glycosylase and EXO internal control. The thermocycling conditions were 50 °C for 2 min and 95 °C for 15 min followed by 45 cycles of 94 °C for 1 min and 60 °C for 1 min. A conserved region of the U67/68 gene was amplified to distinguish between species HHV-6A and HHV-6B [19]. The primer and probe sequences that target HHV-6 U67/68 are as follows: U 67/68 Forward- TTCCGGTATATGACCTTCGTAAGC; U67/68 Reverse- GATGTCTCACCTCCAAATCTTTAGAAAT; U67/68 Probe B [6FAM]CATTATATATCGAATCTGACGCTACCTTCCG[TAMRA]; U67/68 probe A [6VIC]ACATTATATGTCGAACTTGACACTA CCTTCCG[TAMRA]. These assays have high specificity for HHV-6 [18,19], and the use of EBV-induced LCLs should not affect the ability to quantitate HHV-6 in extracted DNA.
Inherited ciHHV-6 was identified using a novel droplet digital PCR (ddPCR) method to test samples for HHV-6 DNA and human ribonuclease P DNA (RPP30, a reference gene for cell count; two RPP30/cell) [14]. Briefly, droplet digital PCR (ddPCR) was performed on the Bio-Rad QX100/200 system as previously described [14]. Data were analyzed with QuantaSoft analysis software (Version 1.7.4), and quantification of target molecules was presented as copies per μl of PCR reaction. Inherited ciHHV-6 was presumed in samples with a ratio of HHV-6 DNA copies to cells of 1 ± 0.07. This assay has been validated against FISH-confirmed inherited ciHHV-6 cell lines [14].
3.3. In vitro proof of concept
To test the feasibility of group testing for identification of patients with inherited ciHHV-6, we first demonstrated the ability of our HHV-6 qPCR assay to detect a cell line with inherited ciHHV-6 in large pools of negative samples. We spiked DNA from a cell line without HHV-6 infection with DNA from a FISH-confirmed inherited ciHHV-6 positive cell line (Hector-2 cell line, Bioworld Consulting Laboratories, MD [7]) at varying concentrations. This demonstrated that our assay decisively detected high levels of HHV-6 DNA even in dilutions of greater than 1:100 (Table 1).
Table 1.
A cell line with inherited ciHHV-6 can easily be detected in large sample pools.
| Inherited ciHHV-6 cell line (ηg DNA) | Genomic background (ηg DNA) | Pool ratio | HHV-6 DNA (copies/ηg)a |
|---|---|---|---|
| 75 | 1400 | 1:19 | 1.09 × 105 |
| 50 | 1400 | 1:28 | 2.68 × 104 |
| 25 | 1400 | 1:56 | 2.81 × 104 |
| 12.5 | 1400 | 1:112 | 1.40 × 104 |
Results of HHV-6 quantitative PCR assay.
3.4. Screening algorithm for inherited ciHHV-6
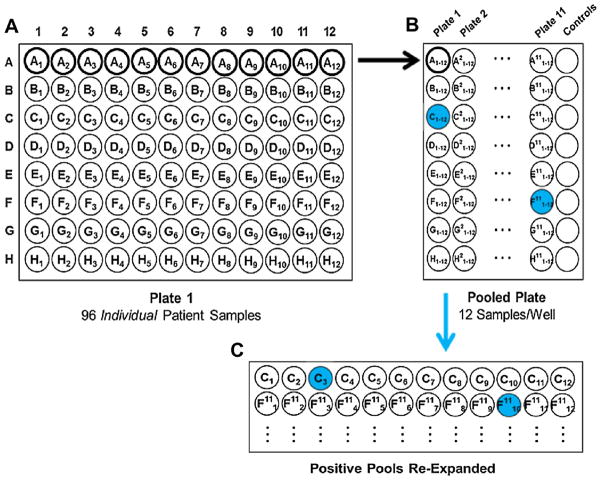
Based on the estimated 1% prevalence of inherited ciHHV-6, group testing is most efficient using pools of 11 or 12 samples, which allows for up to 80% less testing (Table 2). We created pools of 12 samples to facilitate separation of positive pools into individual samples using 8 × 12-well PCR plates (Fig. 1). Two μL from each of 12 samples were used to create 24 μL pools with a manual multichannel pipette, and 10 μL from each pool was used for HHV-6 DNA testing by qPCR. The expected minimum HHV-6 DNA copy number from a pool with at least one inherited ciHHV-6 sample is >103, and pools were categorized as high positive or low positive based on this threshold. Every individual sample from a high positive pool was screened with qPCR. Low positive pools were not further tested (aside from a subset used for confirmatory testing), as they were unlikely to contain a sample with inherited ciHHV-6.
Table 2.
Pooling strategy to screen for inherited ciHHV-6 reduces testing by up to 80%.
| Assuming 1% prevalence of inherited ciHHV-6 | Assuming 2% prevalence of inherited ciHHV-6 | ||||
|---|---|---|---|---|---|
|
| |||||
| Pool size | Expected number of tests for 2496 samples | Reduction in number of tests | Pool size | Expected number of tests for 2496 samples | Reduction in number of tests |
| 1 | 2496 | 0% | 1 | 2496 | 0% |
| 6 | 562 | 77% | 6a | 701 | 72% |
| 12a | 492 | 80% | 12 | 745 | 70% |
| 24 | 639 | 76% | 24 | 1063 | 57% |
Optimal pool size based on expected prevalence.
Fig. 1.
Specimen pooling and screening schematic. (a) Plate 1 is an 8 × 12 plate with individual DNA samples. Each row was combined into 1 well in the (b) pooled plate, which was screened for HHV-6 with qPCR (high positive pools are shaded). (c) Individual samples from high positive pools were screened for HHV-6 by qPCR, and inherited ciHHV-6 was confirmed in high positive individual samples (shaded) using ddPCR.
3.5. Sensitivity, positive predictive value and negative predictive value
To establish the sensitivity of our approach for identifying pools containing an inherited ciHHV-6 sample, or p(high qPCR | inherited ciHHV-6), 11 randomly selected samples with confirmed inherited ciHHV-6 were spiked into pools of 11 samples without inherited ciHHV-6. These positive samples had 1.8 × 104–1.1 × 105 copies/ml HHV-6 DNA, representative of the dynamic range of viral loads observed for all other samples identified as inherited ciHHV-6. Each pool was screened for HHV-6 using qPCR, repeated five times, for a total of 55 tests. Pools without inherited ciHHV-6 were randomly included, and all testing was performed in a blinded fashion. To establish the positive predictive value of our approach, or p(inherited ciHHV-6 | high qPCR), all high positive individual samples were tested for inherited ciHHV-6 using our novel ddPCR assay [14]. To establish the negative predictive value of our approach, or p(not inherited ciHHV-6 | negative or low positive qPCR), 50 low positive individual samples were tested with ddPCR to ensure that they did not harbor inherited ciHHV-6. Sample sizes were calculated using the Clopper–Pearson method to generate results with no more than a 5–7% margin of error for the 95% confidence interval.
3.6. Study approval
The study was approved by the Institutional Review Board at Fred Hutchinson Cancer Research Center, and written informed consent was obtained from study participants or their legally authorized representatives.
4. Results
4.1. Screening algorithm and virologic results
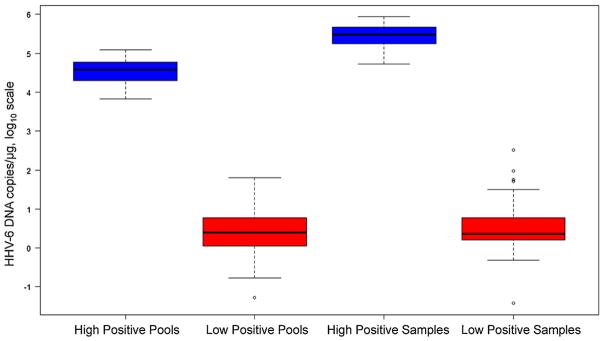
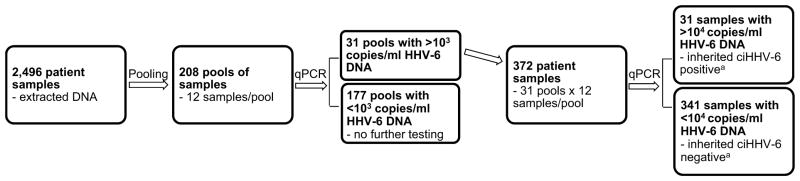
We extracted DNA from beta LCLs or PBMCs of 1248 donor–recipient HCT pairs (2496 samples). The samples were 85% LCLs and 15% PBMCs. All samples were obtained prior to HCT. Using the pooling algorithm outlined in Fig. 1, 208 pools (12 samples/pool) were aliquoted onto 8 × 12 well PCR plates such that all individual samples from an 8 × 12 plate were contained in one column on the pooled plate. Screening for HHV-6 DNA identified 31 high positive pools (i.e. >103 copies/μg) with median viral load 38,110 copies/μg (range, 6630–121,554) (Fig. 2). Individual samples from high positive pools were subsequently aliquoted onto PCR plates and screened by qPCR. One high positive individual sample was identified in each high positive pool with median viral load 306,270 copies/μg (range, 53,079–874,363). There were also 68 pools and 57 individual samples with low positive HHV-6 DNA detection (i.e. <103 copies/μg). Half of the low positive individual samples were PBMCs. Fig. 3 demonstrates a flow chart of the screening algorithm.
Fig. 2.
HHV-6 DNA detection in pooled and individual samples. This box and whisker plot demonstrates values of high versus low positive HHV-6 DNA detection in pooled and individual samples. A threshold of HHV-6 DNA >103 copies/μg clearly distinguishes high positive pools and individual samples from low positive pools and individual samples. This correlates with detection of pools and samples with and without inherited ciHHV-6, respectively.
Fig. 3.
Flow chart of the inherited ciHHV-6 screening algorithm. 31 individuals with inherited ciHHV-6 were identified in a cohort of 2496 patients using only 580 qPCR tests (208 pools + 372 individual samples) through the application of specimen pooling. aAll 31 samples were confirmed to have inherited ciHHV-6 by ddPCR; a subset of negative samples were also tested by ddPCR and confirmed to be without inherited ciHHV-6.
All 31 high positive individual samples were confirmed to have inherited ciHHV-6 by ddPCR demonstrating a ratio within 1% of 1 HHV-6 copy per cell (Table 3). The prevalence of inherited ciHHV-6 was 1.5% (19/1248) in HCT recipients and 0.96% (12/1248) in donors. Inherited ciHHV-6 was identified in similar proportions of beta LCL (28/2,128; 1.3%) and PBMC (3/368; 0.8%) samples. HHV-6B and HHV-6A accounted for 65% (n = 20) and 35% (n = 11) of cases, respectively, and were equally distributed among donors and recipients.
Table 3.
Inherited chromosomally integrated HHV-6 screening results.
| Number | |
|---|---|
| Samples (HCT donor–recipient pairs) | 2496 (1248) |
| Pools | 208 |
| Total quantitative PCR tests | 580 |
| Reduction in testing | 77% |
| Prevalence of inherited ciHHV-6 (n = 31) | |
| - Recipients (n = 19/1248) | 1.5% |
| - Donors (n = 12/1248) | 0.96% |
| Species distribution of inherited ciHHV-6 | |
| - HHV-6A (n = 11/31) | 35% |
| - HHV-6B (n = 20/31) | 65% |
This algorithm only required 580 HHV-6 qPCR assays to screen 2496 patient samples for inherited ciHHV-6, a 77% reduction compared to testing each sample individually. This reduction in testing will lead to a commensurate decrease in laboratory costs and personnel time. Furthermore, it obviates the need for less widely available and more labor intensive testing such as ddPCR and FISH when screening large populations.
4.2. Sensitivity, positive predictive value and negative predictive value
Among 55 pools containing a known inherited ciHHV-6 sample, all had high positive HHV-6 DNA detection, indicating ≥93% sensitivity (with 95% confidence) for identifying true positive pools (data not shown). The positive predictive value was estimated to be ≥88% (with 95% confidence) based on the finding that all high positive samples identified in this cohort (n = 31) were confirmed to have inherited ciHHV-6. Among 50 individual samples with low positive HHV-6 DNA detection, none had ddPCR results consistent with inherited ciHHV-6, indicating a negative predictive value ≥93% (with 95% confidence; data not shown). In practice, the predictive values are likely to approach 100% given the performance characteristics of the HHV-6 qPCR assay and study design.
5. Discussion
We demonstrated that specimen pooling coupled with sensitive qPCR efficiently identifies patients with inherited ciHHV-6 with high accuracy and precision. Using a novel application of the Dorfman group testing scheme [16], we identified 31 patients with inherited ciHHV-6 in a large cohort of 2496 individuals with a 77% reduction in the number of qPCR tests required to screen all samples individually.
The accumulating but limited evidence for disease associations with inherited ciHHV-6 underscores the importance of large-scale research of this condition [6–12]. The only large study performed to date required vast resources to test 20,000 patient samples for inherited ciHHV-6 and reported an association with angina pectoris [15]. This landmark paper implicates inherited ciHHV-6 as a possible risk factor for coronary artery disease, a condition of significant public health importance. This and another study [8] demonstrate evidence for chromosomal instability and shortening due to telomeric deletions in chromosomes with integrated HHV-6, which may have important downstream effects. The biologic plausibility for pathology due to inherited ciHHV-6 can also be inferred from the effect of Marek’s disease virus infection in fowl, an alpha-herpesvirus that similarly integrates into telomeres with resultant T-cell lymphomas and high mortality [20]. Indeed, an expert has recently called for caution in the use of stem cells or organs from donors with inherited ciHHV-6 [11]. Further study of affected individuals in specific patient populations, such as HCT recipients, may provide critical insights into unique immunologic features of these patients and the effect of telomeric chromosomal disruption.
We are confident that our approach accurately identified patients with and without inherited ciHHV-6 because (1) cellular samples from patients with inherited ciHHV-6 have high viral loads, and (2) high level HHV-6 detection in healthy donors and pre-HCT patients is unlikely, particularly with a HHV-6 DNA to cell ratio of 1 [17]. A recent study testing pre and post-HCT blood samples from 16 of these patients also provided evidence of inherited ciHHV-6 in the recipient or their donor [19]. Additionally, the prevalence of inherited ciHHV-6 in healthy individuals and cancer patients identified in this cohort closely matches that reported in other studies [4,15]. Importantly, our strategy confirmed detection of >103 HHV-6 DNA copies/μg as a clear threshold distinguishing between pools and samples containing inherited ciHHV-6 and those without (Fig. 2). Low positive HHV-6 DNA detection in some samples may be due to low grade reactivation or identification of patients with latently infected cells generated through natural infection, as lymphocytes are a site of HHV-6 latency [21]. Thus, low positives pools/samples do not need further testing, and one does not require advanced techniques such as FISH or ddPCR to confirm inherited ciHHV-6 [4,14], expanding the generalizability of this approach. Despite the large range of qPCR values for samples identified to have inherited ciHHV-6, each sample only had 1 copy of HHV-6 per cell as confirmed by ddPCR; this was likely due to differences in the cellularity of samples.
Strengths of this study include the use of a large patient cohort with cellular samples obtained at a time point during which HHV-6 reactivation is rare, minimizing confounding due to HHV-6 detection from non-inherited ciHHV-6 sources. Additionally, the use of both LCL and PBMC samples underscores the utility of this testing strategy with a variety of cellular specimens. Limitations of this algorithm include the need for cellular samples, which may not be as readily available. Manual aliquoting also increased the risk for contamination and/or error, although we found no evidence that this occurred. Alternative pooling strategies like the square array could be utilized to further improve efficiency [22], but the incremental gain in test reduction did not warrant the increased labor and margin for error based on our laboratory setup.
In conclusion, we demonstrate an efficient method for identifying inherited ciHHV-6 in patient samples through application of modern molecular diagnostic techniques to a group testing model. This approach provides a novel strategy for the study of inherited ciHHV-6 in large retrospective and prospective cohorts, as well as for real-time clinical utilization. Given the biologically plausible association of inherited ciHHV-6 with important diseases, larger studies will be critical to establish the clinical significance of this condition.
Acknowledgments
Funding
This work was supported by a pilot grant from the HHV-6 Foundation (J.A.H.), a New Investigator award from the American Society for Blood and Marrow Transplantation (J.A.H.), and by the National Institutes of Health [1K23AI119133 to J.A.H., CA18029 to M.B., and HL093294 to M.B.].
We would like to thank the Research Cell Bank at Fred Hutchinson Cancer Research Center for providing DNA from beta LCL and PBMC samples. We would also like to acknowledge Jon Guan and Michelle Cho for performing the laboratory work, as well as Zachary Stednick for help with figures.
Abbreviations
- HHV-6
human herpesvirus 6
- ciHHV-6
chromosomally integrated HHV-6
- HCT
hematopoietic cell transplantation
- qPCR
quantitative polymerase chain reaction
- ddPCR
droplet digital PCR
- HLA
human leukocyte antigen
- LCL
lymphoblastoid cell line
- PBMC
peripheral blood mononuclear cell
- PCR
polymerase chain reaction assay
Footnotes
Presented as a poster at the 15th International CMV/Beta Herpesvirus Workshop, Brisbane, Australia 2015.
Author contributions
J.A.H., M.B., R.H.-S., A.M., K.R.J., D.M.Z. and M.-L.H. contributed to the study design and analysis. M.-L.H. oversaw all laboratory work. The paper was drafted by J.A.H. with input from all other authors. All authors reviewed and approved the final version.
Conflict of interest
J.A.H. and D.M.Z. received research support from Chimerix Inc. M.B. has served as a consultant and has received research support from Chimerix Inc. and Genentech/Roche in addition to consulting for Clinigen. All authors have read the journal’s authorship agreement and policy on disclosure of potential conflicts of interest.
References
- 1.Zerr DM, Meier AS, Selke SS, Frenkel LM, Huang ML, Wald A, et al. A population-based study of primary human herpesvirus 6 infection. N Engl J Med. 2005;352:768–776. doi: 10.1056/NEJMoa042207. [DOI] [PubMed] [Google Scholar]
- 2.Hill JA, Zerr DM. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol. 2014;9:53–60. doi: 10.1016/j.coviro.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufer BB, Flamand L. Chromosomally integrated HHV-6: impact on virus, cell and organismal biology. Curr Opin Virol. 2014;9C:111–118. doi: 10.1016/j.coviro.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Pellett P, Ablashi D. Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol. 2012:144–155. doi: 10.1002/rmv.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tweedy J, Spyrou MA, Hubacek P, Kuhl U, Lassner D, Gompels UA. Analyses of germline, chromosomally integrated human herpesvirus 6A and B genomes indicate emergent infection and new inflammatory mediators. J Gen Virol. 2015;96:370–389. doi: 10.1099/vir.0.068536-0. [DOI] [PubMed] [Google Scholar]
- 6.Lee SO, Brown aR, Razonable RR. Clinical significance of pretransplant chromosomally integrated human herpesvirus-6 in liver transplant recipients. Transplantation. 2011;92:224–229. doi: 10.1097/TP.0b013e318222444a. [DOI] [PubMed] [Google Scholar]
- 7.Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A. 2010;107:5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, Hidalgo-Bravo A, Zhang E, Cotton VE, Mendez-Bermudez A, Wig G, et al. Human telomeres that carry an integrated copy of human herpesvirus 6 are often short and unstable, facilitating release of the viral genome from the chromosome. Nucleic Acids Res. 2014;42:315–327. doi: 10.1093/nar/gkt840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kühl U, Lassner D, Wallaschek N, Gross UM, Krueger GRF, Seeberg B, et al. Chromosomally integrated human herpesvirus 6 in heart failure: prevalence and treatment. Eur J Heart Fail. 2015;17:9–19. doi: 10.1002/ejhf.194. [DOI] [PubMed] [Google Scholar]
- 10.Endo A, Watanabe K, Ohye T, Suzuki K, Matsubara T, Shimizu N, et al. Molecular and virological evidence of viral activation from chromosomally integrated human herpesvirus 6A in a patient with X-linked severe combined immunodeficiency. Clin Infect Dis. 2014;59:545–548. doi: 10.1093/cid/ciu323. [DOI] [PubMed] [Google Scholar]
- 11.Flamand L. Pathogenesis from the reactivation of chromosomally integrated human herpesvirus type 6: facts rather than fiction. Clin Infect Dis. 2014;59:549–551. doi: 10.1093/cid/ciu326. [DOI] [PubMed] [Google Scholar]
- 12.Hill JA, Sedlak RH, Zerr DM, Huang ML, Yeung C, Myerson D, et al. Prevalence of chromosomally integrated human herpesvirus 6 in patients with human herpesvirus 6-central nervous system dysfunction. Biol Blood Marrow Transpl. 2015;21:371–373. doi: 10.1016/j.bbmt.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill JA, Sedlak RH, Jerome KR. Past, present, and future perspectives on the diagnosis of Roseolovirus infections. Curr Opin Virol. 2014;9:84–90. doi: 10.1016/j.coviro.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sedlak RH, Cook L, Huang ML, Magaret A, Zerr DM, Boeckh M, et al. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin Chem. 2014;60:765–772. doi: 10.1373/clinchem.2013.217240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravel A, Dubuc I, Morissette G, Sedlak RH, Jerome KR, Flamand L. Inherited chromosomally integrated human herpesvirus 6 as a predisposing risk factor for the development of angina pectoris. Proc Natl Acad Sci U S A. 2015;112:8058–8063. doi: 10.1073/pnas.1502741112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorfman R. The Detection of defective members of large populations. Ann Math Stat. 1943;14:436–440. [Google Scholar]
- 17.Razonable RR, Fanning C, Brown RA, Espy MJ, Rivero A, Wilson J, et al. Selective reactivation of human herpesvirus 6 variant a occurs in critically ill immunocompetent hosts. J Infect Dis. 2002;185:110–113. doi: 10.1086/324772. [DOI] [PubMed] [Google Scholar]
- 18.Zerr DM, Gupta D, Huang ML, Carter R, Corey L. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2002;34:309–317. doi: 10.1086/338044. [DOI] [PubMed] [Google Scholar]
- 19.Hall Sedlak R, Hill JA, Nguyen T, Cho M, Levin G, Cook L, et al. Detection of HHV-6B reactivation in hematopoietic cell transplant recipients with inherited chromosomally integrated HHV-6A by droplet digital PCR. J Clin Microbiol. 2016 doi: 10.1128/JCM.03275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. Marek’s disease virus: from miasma to model. Nat Rev Microbiol. 2006;4:283–294. doi: 10.1038/nrmicro1382. [DOI] [PubMed] [Google Scholar]
- 21.Géraudie B, Charrier M, Bonnafous P, Heurté D, Desmonet M, Bartoletti MA, et al. Quantitation of human herpesvirus-6A, -6B and -7 DNAs in whole blood, mononuclear and polymorphonuclear cell fractions from healthy blood donors. J Clin Virol. 2012;53:151–155. doi: 10.1016/j.jcv.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 22.Hudgens MG, Kim HY. Optimal configuration of a square array group testing algorithm. Commun Stat Theory Methods. 2011;40:436–448. doi: 10.1080/03610920903391303. [DOI] [PMC free article] [PubMed] [Google Scholar]