Abstract
Genetic susceptibility to type 1 diabetes (T1D) is well supported by epidemiologic evidence; however, disease risk cannot be entirely explained by established genetic variants identified so far. This study addresses the question of whether epigenetic modification of the inherited DNA sequence may contribute to T1D susceptibility. Using the Infinium HumanMethylation450 BeadChip array (450k), a total of seven long-term disease-discordant monozygotic (MZ) twin pairs and five pairs of HLA-identical, disease-discordant non-twin siblings (NTS) were examined for associations between DNA methylation (DNAm) and T1D. Strong evidence for global hypomethylation of CpG sites within promoter regions in MZ twins with TID compared to twins without T1D was observed. DNA methylation data were then grouped into three categories of CpG sites for further analysis, including those within: 1) the major histocompatibility complex (MHC) region, 2) non-MHC genes with reported T1D association through genome wide association studies (GWAS), and 3) the epigenome, or remainder of sites that did not include MHC and T1D associated genes. Initial results showed modest methylation differences between discordant MZ twins for the MHC region and T1D-associated CpG sites, BACH2, INS-IGF2, and CLEC16A (DNAm difference range: 2.2% – 5.0%). In the epigenome CpG set, the greatest methylation differences were observed in MAGI2, FANCC, and PCDHB16, (DNAm difference range: 6.9% – 16.1%). These findings were not observed in the HLA-identical NTS pairs. Targeted pyrosequencing of five candidate CpG loci identified using the 450k array in the original discordant MZ twins produced similar results using control DNA samples, indicating strong agreement between the two DNA methylation profiling platforms. However, findings for the top five candidate CpG loci were not replicated in six additional T1D-discordant MZ twin pairs. Our results indicate global DNA hypomethylation within gene promoter regions may contribute to T1D; however, findings do not support the involvement of large DNAm differences at single CpG sites alone in T1D.
Keywords: Type 1 diabetes, DNA methylation, Monozygotic Twins
1. Introduction
Type 1 diabetes (T1D) is an autoimmune disease characterized by the destruction of insulin-producing beta islet cells in the pancreas. The strongest genetic contribution to T1D comes from genes encoding the classical Human Leukocyte Antigens (HLA) in the Major Histocompatibility Complex (MHC) [1, 2]. Genome-wide association studies (GWAS) have identified and confirmed additional non-MHC loci with very modest effects [3, 4]. However, disease susceptibility is not explained by genetics alone; environmental factors, gene by environment interactions, and epigenetic influences are likely to play important roles in the etiology of T1D [5, 6]. Monozygotic (MZ) twin pairs, discordant for T1D, represent an ideal system to test susceptibility factors not attributable to genetic variation, especially epigenetic variation, since the genomes of the twins are identical. The ascertainment of disease-discordant MZ twin pairs for epigenetic studies is difficult; to date, only a few studies have utilized this design for autoimmune diseases, and the numbers of twins available for these studies range from 3 to 27 [7–10]. Most recently, a study by Stefan et al. identified 55 hyper- and 33 hypomethylated sites using 3 MZ twin pairs discordant for T1D [11].
Seven pairs of MZ twins, disease-discordant for at least eight years, were used for the current study. In addition, five pairs of HLA-identical, disease-discordant non-twin sibling (NTS) pairs were examined. Methylation of DNA (DNAm) at cytosine-guanine dinucleotides (CpG) within gene promoters is an established epigenetic mechanism of transcriptional down regulation [12]. Site-specific DNAm profiling data were generated for the disease-discordant MZ twins and HLA-identical disease-discordant NTS pairs using a comprehensive whole genome array of DNAm sites (CpGs). Hypo- and hypermethylation differences in promoter regions as proportions of total CpG sites characterized across the genome were investigated. DNAm was also assessed as a single global (mean) measure across all CpGs within three pre-selected CpG sets including: (1) sites within or neighboring MHC region genes, (2) within or neighboring T1D-associated genes (non-MHC, taken from results of the Type 1 Diabetes Genetics Consortium), and (3) sites within the remaining epigenome (excluding the MHC region and T1D-associated genes). Finally, individual site-specific DNAm differences were examined. Our study design utilized disease-discordant MZ twins and HLA-identical disease-discordant NTS pairs to test the hypothesis that peripheral blood DNAm is associated with the T1D disease phenotype.
2. Materials and Methods
2.1 Subjects
MZ twins were recruited from The Twin Family Study of Islet Cell Autoimmunity at the Barbara Davis Center (BDC); this study recruited twins locally at the BDC as well as nationally through TrialNet, previous Diabetes Prevention Trial (DPT–1), and Joslin study cohorts. Informed consent was obtained from the subjects or parents of each study subject. The Colorado Multiple Institutional Review Board (IRB) approved the study protocol. Seven long-term disease-discordant MZ twin pairs were selected initially for this study. An additional six twin pairs were selected for replication. All twin pairs had T1D discordance of at least 8 years and were of European descent. Five NTS pairs were recruited from Children’s Hospital and Research Center Oakland (CHRCO), with approval of the CHRCO IRB. Siblings were identical-by-descent for alleles in the Class I and II HLA genes; all individuals were of European descent. Demographic data for disease-discordant MZ twin pairs (TP1-TP7) and HLA-identical disease-discordant NTS pairs (SP1-SP5) are shown in Table 1. In the long-term disease-discordant twins, the age of onset for the affected twin ranged from 1 to 16; the length of disease discordance at sampling ranged from 8 to 20 years.
Table 1.
Demographic information on subjects used for DNA methylation study of T1D
| Pair ID | Sex (Affected/Unaffected) | Age at diagnosis | Age at sample draw (Affected) | Age at sample draw (Unaffected) | Years Discordant |
|---|---|---|---|---|---|
| TP1 | Female/Female | 10 | 30 | 30 | 20 |
| TP2 | Female/Female | 16 | 28 | 28 | 12 |
| TP3 | Female/Female | 5 | 21 | 21 | 16 |
| TP4 | Female/Female | 5 | 13 | 13 | 8 |
| TP5 | Male/Male | 8 | 16 | 16 | 8 |
| TP6 | Male/Male | 5 | 14 | 14 | 9 |
| TP7 | Female/Female | 1 | 9 | 9 | 8 |
| mean (SD) | 7.14 (4.8) | 18.7 (7.9) | 18.7 (7.9) | 11.6 (4.8) | |
|
| |||||
| SP1 | Female/Female | 2 | 4 | 7 | --- |
| SP2 | Male/Male | 10 | 11 | 12 | --- |
| SP3 | Female/Male | 9 | 9 | 16 | --- |
| SP4 | Male/Female | 13 | 14 | 2 | --- |
| SP5 | Male/Male | 19 | 27 | 25 | --- |
| mean (SD) | 10.6 (6.1) | 13.0 (8.6) | 12.4 (8.8) | --- | |
|
| |||||
| RP1 | Female/Female | 11 | 27 | 27 | 16 |
| RP2 | Male/Male | 2 | 20 | 20 | 13 |
| RP3 | Male/Male | 12 | 28 | 28 | 16 |
| RP4 | Female/Female | 11 | 22 | 22 | 10 |
| RP5 | Female/Female | 12 | 22 | 22 | 10 |
| RP6 | Male/Male | 7 | 17 | 17 | 10 |
| mean (SD) | 9.17 (4.0) | 22.7 (4.2) | 22.7 (4.2) | 12.5 (3.0) | |
Demographic data for disease-discordant MZ twin pairs (TP1-7), disease-discordant HLA-identical NTS pairs (SP1-5), and the additional set of twins used for replication of 450k findings (RP1-6).
2.2 Subjects and CpGs used for validation of 450k data
Jurkat control (cell line) DNA samples were used for validation by pyrosequencing of the top candidate CpGs loci, identified from analyses of 450k (defined below) data in the 7 discordant MZ twins. An additional independent set of 6 discordant MZ twin pairs was used for replication of 450k findings (RP1-RP6 in Table 1) by pyrosequencing. Based on available DNA and the performance of pyrosequencing primers, five CpGs were studied for validation and replication.
2.3 Genotyping and methylation assay QC
Peripheral blood DNA samples were bisulfite converted with the EZ DNA Methylation™ Kit (Zymo Research, Irvine, CA USA) and processed on the Infinium HumanMethylation450 (450k) BeadChip assay (Illumina, Inc., San Diego, CA USA) according to manufacturer protocols. The high quality and performance of the platform was supported by multiple quality control (QC) measures, including tests for proper bisulfite conversion, staining, and specificity of the internal controls, as determined by the Illumina GenomeStudio software. Average-beta values (proxy for DNAm level between 0, unmethylated, and 1, fully methylated) were normalized to internal controls and corrected by background subtraction. Non-autosomal CpGs and CpG probes with suboptimal detection (p<0.05 in at 80% of samples) were removed (n= 11,707). We additionally removed 13,961 CpGs from our analyses where single nucleotide polymorphisms (SNPs) were located within or near corresponding probes for DNAm detection on the BeadChip as recently described [13]; therefore, the potential for confounding due to genotype was minimized.
Several measures were conducted to ensure the quality and reliability of the 450k array. A total of 17 “within chip” and “between chip” duplicate sample comparisons showed a minimum r2 of 0.995, included sample duplicates and control DNA (Jurkat cell lines). No batch effects were detected, as indicated by “between chip” duplicate comparisons.
2.4 Pyrosequencing
Whole blood DNA (200ng) was bisulfite converted using the EpiTect Fast DNA Bisulfite Kit (cat no. 59824). During this process, unmethylated cytosine residues are converted into uracil, leaving the methylated cytosines unchanged. The converted single-stranded DNA was bound to the MinElute DNA spin column membrane, and then washed and desulphonated. Approximately 20ng of converted DNA was used in the PyroMark PCR kit (cat no. 978703) for amplification (see appendix A for amplification primers).
2.5 Statistical and descriptive analyses
In order to characterize genome-wide (global) DNAm patterns between paired subjects, we reported the ratio of hypo- to hypermethylated CpG sites in MZ twins and NTS pairs separately. We refer to CpGs as hypo- or hypermethylated if the mean of paired differences in beta-value between affected and unaffected subjects was negative or positive, respectively. Global comparisons were then stratified by CpG sites within promoter and non-promoter regions. CpG sites considered to be located in the promoter region of a gene were defined as anything 2500 bases upstream or 500 bases downstream of a transcription start site (according to the RefSeq Genes track in the UCSC Genome Browser). This method has previously been detailed by Whitaker et al. [14].
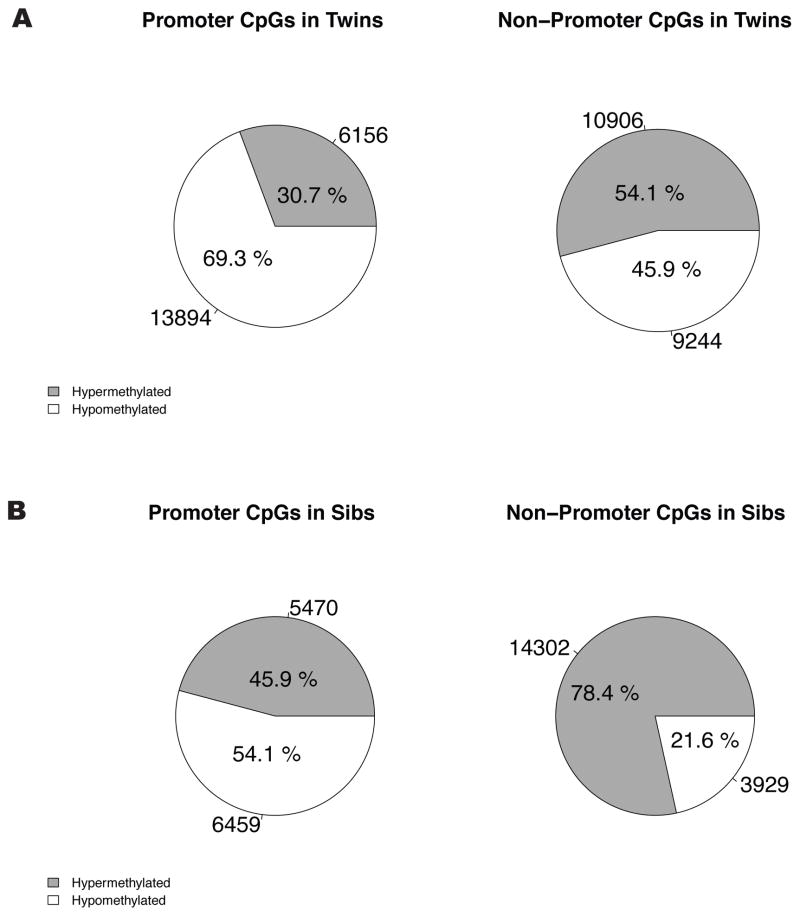
As a method of data reduction for global comparisons, we first employed site-specific statistical tests to limit our data to CpGs with the greatest likelihood of having true DNAm differences between paired subjects. A non-normal distribution of DNAm within subjects was observed (data not shown); thus, non-parametric testing was conducted using a paired, two-sample Wilcoxon signed-rank test. The alpha level for CpG removal from global comparisons was 0.1; this was chosen because no CpGs in the NTS pairs passed an alpha threshold of 0.05. However, we additionally employed CpG removal exclusively in discordant MZ twins at the alpha 0.05 level, for a more stringent evaluation. Due to limited power, Wilcoxon p-values were not corrected for multiple testing, and employed only as a method of data reduction and not for the purposes of identifying what can be considered “statistically significant.” The distribution of hypo- to hypermethylated sites was visualized using pie charts (see Figure 1); Comparisons were made between promoter and non-promoter sites within MZ twins and NTS pairs separately using Fisher’s exact tests.
Fig. 1.
Global comparisons of hypo- to hypermethylated CpGs across selected sites in discordant MZ twins and NTS pairs.
(A) The relative number of sites within promoter regions that were hypo- or hypermethylated and reached an alpha significance threshold of 0.1, based on a paired Wilcoxon signed-rank test. In affected twins, 69.3% of CpGs were hypomethylated out of a total, 20,760 sites. The proportion of hypomethylated sites in promoter region CpGs (left) was statistically different from non-promoter CpGs (right) (Fisher’s exact p-value <0.0001), where 46.0% of CpGs were hypomethylated in a total of 20,902 sites.
(B) HLA-identical NTS pairs had a much smaller difference in the ratio of hypo- to hypermethylated promoter region CpGs, with 53.8% hypomethylation out of 12,362 CpGs that reached the 0.1 alpha level. Among non-promoter CpGs, 21.6% were hypomethylated out of a total 18,963 CpGs. The ratio of hypo- to hypermethylated CpGs was also statistically different when comparing promoter region and non-promoter pies (Fisher’s exact p-value <0.0001).
450k data were then grouped into three distinct sets of CpG sites. Specifically, Set 1 was comprised of a subset of 8,682 CpGs, covering 245 genes in the MHC region. Set 2 included 629 CpGs from 33 non-MHC region genes with genetic variants reported to be T1D-associated in GWAS [3, 4, 15–19]. Set 3 (referred to in this report as “epigenome”) included the remainder of the 450,588 autosomal CpGs in the assay, excluding Set 1 and Set 2.
We examined differences among paired cases and controls (i.e., MZ twin pairs or NTS pairs) as a single global (mean) measure, across all sites in each of the three CpG sets, using a paired Wilcoxon signed-rank test. Additionally, individual CpGs were examined to find the largest mean DNAm differences, with top CpG sites ranked according to the magnitude of DNAm difference (not statistical difference). In order for DNAm differences to be associated with disease status, the direction of the DNAm difference (i.e., hypo- or hypermethylated in the affected twin) would be expected to show some consistency across all twin pairs. Therefore, only CpG sites that were consistent in direction in at least six of seven twin pairs are shown. CpGs with greatest DNAm differences identified in the discordant twins were then examined in NTS pairs.
Statistical computing was done using R (R v2.11.1 (2010–05-31)) and STATA (v11). Correlation values (r2) are given as Spearman’s coefficient. Data processing was performed with the help of the R-package, “IMA” [20]. Estimation of cell counts from 450k methylation data was performed using the R-package, “minfi” [21]. Identical statistical methods as described for 450k analyses were used in validation and replication analyses.
3. Results
3.1 Global comparisons of DNAm
Small differences in the proportion of all sites that were hypomethylated across non-promoter CpGs in the discordant MZ twins were observed; that is, the proportion of CpGs that were hypomethylated or hypermethylated was similar (Figure 1A). However, when analyses were restricted to CpGs within promoter regions, 69.3% of CpGs below the a priori 0.1 alpha threshold demonstrated hypomethylation in affected twins (total n=20,760 sites). The proportion of hypomethylated sites in promoter region CpGs was statistically different from non-promoter CpGs (Fisher’s exact p-value <0.0001; Figure 1A). Nearly identical patterns were observed when analyses were restricted to CpG sites at the alpha 0.05 level; within 7,298 promoter CpGs, 72.6% were hypomethylated and within 6,593 non-promoter CpGs, 47.4% were hypomethylated (Fisher’s exact p-value <0.0001).
A similar trend was observed in HLA-identical NTS pairs, although a smaller difference in the proportion of hypomethylated promoter region sites was present. Specifically, 53.8% of CpG sites demonstrated hypomethylation out of 12,362 CpGs that reached the 0.1 alpha level. A much smaller proportion of sites in the non-promoter CpGs were hypomethylated; a total of 21.6% hypomethylation out of 18,963 CpGs that reached the alpha 0.1 level (Figure 1B). The proportion of hypomethylated CpGs was also statistically different when comparing promoter region and non-promoter subsets (Fisher’s exact p-value <0.0001)
Mean DNAm for both the MHC region CpG set and non-MHC, T1D-associated gene CpG set differed significantly between twins in each pair (Wilcoxon p<0.05); the affected twin was hypomethylated compared to the unaffected twin. This difference was seen both for promoter (p<0.05) and non-promoter sites (p<0.05) (data not shown). No significant difference was observed for the set of remaining CpGs in twins or for any set in the NTS pairs (data not shown).
3.2 Site-specific comparisons of DNAm
Individual CpGs with the greatest DNAm differences for discordant MZ twins are shown in Table 2. For the MHC region, the ten CpGs with the greatest DNAm difference between affected and unaffected twins were in the following genes: ABT1 and TNXB, where hypomethylation was observed in the affected twin, and PRRT1, NUDT3, HSPA1B, RPP21, BTNL2, and PPP1R11, where hypermethylation was observed in the affected twin. The range in mean absolute average-beta difference for the MHC CpGs was 3.5% to 5.0% (Table 2).
Table 2.
CpGs with greatest DNAm differences and consistent direction in at least 6 of 7 disease-discordant MZ twin pairs
| TargetID | UCSC REFGENE Name* | DNAm direction in cases for at least 6 of 7twin pairs | Average- Beta Difference | Mean (cases) | Mean (controls) |
|---|---|---|---|---|---|
| MHC region genes | |||||
| cg18501647 | PRRT1 | Hyper | 0.050 | 0.652 | 0.601 |
| cg06526020 | NUDT3 | Hyper | 0.043 | 0.621 | 0.577 |
| cg00970279 | HSPA1B | Hypo | −0.040 | 0.231 | 0.271 |
| cg11502198 | ABT1 | Hypo | −0.037 | 0.125 | 0.162 |
| cg13064679 | TNXB | Hypo | −0.037 | 0.713 | 0.751 |
| cg10365886 | TNXB | Hypo | −0.037 | 0.520 | 0.557 |
| cg20856330 | RPP21 | Hyper | 0.036 | 0.252 | 0.215 |
| cg01337207 | TNXB | Hypo | −0.036 | 0.459 | 0.495 |
| cg15319032 | BTNL2 | Hyper | 0.036 | 0.826 | 0.790 |
| cg06083200 | PPP1R11 | Hyper | 0.035 | 0.469 | 0.434 |
| Non-MHC, T1D-associated genes | |||||
| cg08217526 | C14orf64 | Hyper | 0.042 | 0.486 | 0.444 |
| cg16413785 | C14orf64 | Hyper | 0.034 | 0.591 | 0.557 |
| cg09818385 | BACH2;BACH2 | Hypo | −0.028 | 0.647 | 0.675 |
| cg11717189 | INS-IGF2;IGF2 | Hyper | 0.028 | 0.437 | 0.410 |
| cg26316423 | IL2RA;IL2RA | Hypo | −0.027 | 0.676 | 0.703 |
| cg03217729 | CLEC16A | Hyper | 0.026 | 0.285 | 0.259 |
| cg17066529 | BACH2;BACH2 | Hypo | −0.024 | 0.837 | 0.861 |
| cg12929678 | CD226 | Hyper | 0.024 | 0.685 | 0.661 |
| cg23905216 | INS-IGF2;IGF2AS;IGF2 | Hyper | 0.024 | 0.255 | 0.231 |
| cg09058293 | SH2B3 | Hyper | 0.022 | 0.889 | 0.866 |
| Epigenome | |||||
| cg14464244 | MAGI2 | Hypo | −0.161 | 0.175 | 0.336 |
| cg13600149 | FANCC | Hyper | 0.093 | 0.186 | 0.093 |
| cg25340050 | PCDHB16 | Hyper | 0.090 | 0.582 | 0.492 |
| cg00758915 | PCDHGA4;PCDHGA6; | Hyper | 0.074 | 0.482 | 0.408 |
| cg12109260 | Non-Genic | Hyper | 0.074 | 0.861 | 0.787 |
| cg16579158 | PCDHGA2;PCDHGA1;PCDHG | Hyper | 0.072 | 0.585 | 0.513 |
| cg15116095 | PCDHA1;PCDHA1;PCDHA1 | Hyper | 0.071 | 0.665 | 0.594 |
| cg12813768 | SYCP1 | Hyper | 0.071 | 0.382 | 0.311 |
| cg14989243 | FILIP1 | Hyper | 0.070 | 0.548 | 0.478 |
| cg05982271 | CDKL2 | Hypo | −0.069 | 0.212 | 0.281 |
These findings were restricted to CpGs that showed consistent hypo or hypermethylation in at least 6 of 7 disease-discordant MZ twin pairs.
Genes with multiple gene names for the UCSC reference gene indicates that the probe interrogating the CpG contains transcripts within multiple genes.
For the set of CpGs in non-MHC, T1D-associated genes, those with the greatest DNAm difference included the following: BACH2 and IL2RA, where hypomethylation in the affected twin was observed, and C14orf64, INS-IGF2, CLEC16A, CD226, and SH2B3, where hypermethylation in the affected twin was observed. The range in mean absolute average-beta difference for sites in non-MHC, T1D-associated genes was 2.2% to 4.2% (see Table 2).
For the epigenome set, the greatest DNAm differences were observed in: MAGI2 and CDKL2, where hypomethylation was observed in the affected twin, and FANCC, PCDHB16, PCDHGA1, SYCP1, FILIP1, PCDHGA4, and a non-genic CpG, where hypermethylation was observed in the affected twin. The range in mean absolute average-beta difference for the epigenome sites was 6.9% to 16.1% (see Table 2).
NTS pair data were examined for the CpGs of interest from analysis of the disease-discordant MZ twin data described above. The NTS pair results were not consistent, in either magnitude or direction of DNAm, with those observed for MZ twins (Appendix A, Figure S1).
3.3 Variation in DNAm among cell types
For this study, only peripheral blood DNA was available for testing; however, DNAm can vary by cell type within an individual [22], and peripheral blood contains a diverse mixture of immune cells. To assess whether DNAm differences between MZ twins or NTS siblings in a pair could result from differences in proportions of cell types, we implemented a validated method, established by Houseman et. al, for cell mixture prediction of CD8+ and CD4+ T cells, NK cells, B cells, monocytes, and granulocytes among subjects, based on highly predictive CpGs present on the array [23]. Cell composition for each subject in the current study was predicted, adjusting for age and sex. The proportion of each cell type varied greatly in peripheral blood per individual (Appendix A, Table S1). However, correlation for the proportion of each cell type was high in the sample of disease-discordant MZ twins (mean r2=0.99; range: 0.96–0.99), suggesting that measurements of DNAm in twin pair should reflect a similar cell mixture in the T1D affected vs. unaffected twin, and, thus, observed DNAm differences between twins in a pair are likely to be accurate, rather than simply reflecting skewed cell proportions. Conversely, HLA-identical, disease-discordant NTS pairs had much lower correlation values (mean r2=0.88; range: 0.77–0.99).
3.4 Validation of 450k platform using pyrosequencing
We attempted to validate the 450k platform, and its ability to detect true DNAm values, by comparing 450k results with pyrosequenced results. We compared data derived for 19 control DNA samples from immortalized Jurkat cell lines, methylotyped on 6 separate 450k chips. Five CpGs were analyzed, from the following genes: C14orf64, BACH2, IL2RA, PCDHB16, and PRRT1 (Appendix A, Table S2 for additional details). We observed that pyrosequenced DNAm results for the five CpGs were highly correlated with 450k DNAm results in Jurkat controls. Figure S2 shows the discrepancies in DNAm beta-value between Jurkat replicates across two plates in the top five CpGs that were selected for pyrosequencing. We observed the differences are centered around zero, with a maximum absolute difference of roughly 0.05, indicating that DNAm average-beta values are accurate and not likely due to plate effects. A comparison of pyrosequenced DNAm levels in a Jurkat sample with 19 Jurkat controls that were methylotyped on the 450k array, yielded a minimum correlation of 0.98.
3.5 Replication of 450k results in additional set of MZ twins
Using available DNA from 6 independent MZ twin pairs that were not available for initial 450k analyses, we attempted to replicate CpG site-specific 450k findings by pyrosequencing. None of the top five CpGs selected for analysis approached statistical significance (minimum p-value = 0.44). Furthermore, none of these CpGs exhibited greater than a 0.051 magnitude of DNAm difference between co-twins, nor was the proportion of hypomethylation in the affected twins similar to original 450k findings. These results, therefore, verified that DNAm of the top five CpG sites selected from the original 450k findings were not significantly different (Table 3).
Table 3.
Top 5 CpGs pyrosequenced in an independent set of 6 disease-discordant MZ twin pairs
| TargetID | UCSC REFGENE Name | Fraction Hypermethylated in Affected Twin | Wilcoxon Signed-Rank P-value | Average- Beta Difference | Mean (cases) | Mean (controls) |
|---|---|---|---|---|---|---|
| MHC region genes | ||||||
| cg18501647 | PRRT1 | 66.7% | 0.438 | 0.027 | 0.792 | 0.766 |
| Non-MHC, T1D-associated Genes | ||||||
| cg16413785 | C14orf64 | 50.0% | 0.563 | 0.051 | 0.509 | 0.458 |
| cg09818385 | BACH2;BACH2 | 33.3% | 0.844 | 0.003 | 0.575 | 0.572 |
| cg26316423 | IL2RA;IL2RA | 83.3% | 0.219 | 0.044 | 0.756 | 0.713 |
| Epigenome | ||||||
| cg25340050 | PCDHB16 | 33.3% | 0.563 | −0.032 | 0.348 | 0.380 |
Average-Beta (DNAm) differences were calculated as affected-unaffected; a positive number indicates that a greater methylation percentage was observed in the affected individual.
4. Discussion
MZ twins are necessarily matched for age, sex, genome sequence, and, frequently, even environment. Disease-discordant MZ twins represent an ideal case-control sample; however, such twins are rare, and acquisition of large sample sets for disease studies is generally not feasible. Even so, much information can be determined from these sets, without the confounding of background genomic variation. In the current study, global DNAm profiles characterized across the genome showed clear differences in the relative proportions of hypo- to hypermethylated CpG sites within promoter region CpGs in disease discordant MZ twins compared to non-promoter CpGs. Specifically, the majority of genetic promoters were hypomethylated in affected twins, and thus, it is possible the corresponding gene products are over expressed in individuals with disease. A targeted approach was also employed to focus on potential DNAm differences in established T1D-associated genes, including genes within the MHC. If a candidate CpG methylation site were truly a disease risk factor, methylation differences at a given locus would be expected to be consistent in direction (i.e., the affected twin is consistently hypomethylated or hypermethylated compared to the unaffected twin). The results from 450k array data revealed DNAm differences at CpGs in several T1D-relevant genes that were consistent in direction for at least 6 of 7 disease-discordant MZ twins.
Results from analyses of HLA-identical non-twin sibling pairs were consistent with MZ twins; a greater proportion of hypomethylated sites within the promoter region CpGs and hypermethylated sites in non-promoter CpGs were observed. However, in contrast to MZ twin findings, only a slight majority of promoter region CpGs were hypomethylated in the sibling pairs (~54%), and a large majority of non-promoter CpGs were hypermethylated (~78%). More generally, the evidence suggests that affected twins were less methylated at promoter regions compared to affected non-identical siblings. One potential reason for the different results between MZ twins and HLA-identical non-twin siblings might be due to the age of subject at blood sample collection. MZ twins, on average, had samples drawn nearly 10 years after disease onset, while sibling pairs had samples drawn within three years. Therefore, differences in DNAm between subjects might be attributed to disease processes that are time dependent. Further, genetic backgrounds of T1D discordant HLA-identical non-twin siblings outside the MHC region will be more heterogeneous compared to the discordant MZ twins; that is, less sharing present in siblings, whereas the MZ twins are genetically identical. DNAm differences between the two groups of study subjects may also result from genetic differences.
Results from analyses of the original 7 MZ twin pairs showed some notable findings for individual loci. The top 10 sites from each of the three CpG sets were consistent for direction of DNAm difference between affected and unaffected twins; one CpG site showed an average DNAm difference of approximately 16% percent. The top three site-specific findings in the epigenome set (MAGI2:cg14464244, FANCC:cg13600149, and PCDHB16:cg25340050) had at least a 9% mean difference in methylation across twins in the current study. These gene candidates have also been highlighted in previous reports showing associations with non-T1D autoimmune disorders. For example, in a study cohort comprised of 681 subjects with Crohn’s disease (CD), 259 subjects with ulcerative colitis (UC) and 195 controls, significant associations were observed between disease status and genetic variation within MAGI2 introns [24]. The MAGI2 gene has been shown to inhibit cell proliferation of phosphatase and tensin homolog (PTEN) protein [25], which helps regulate cell growth and apoptosis, and also becomes stable after binding to MAGI2 [26]. Thus, there is a potential link between this gene and mechanisms involved in autoimmune reactivity.
The FANCC gene is a member of the fanconi anemia protein family, and to our knowledge, has no functional relevance to T1D. However, Cooke et al. observed differential methylation of CpG sites within FANCC when comparing DNA from rectal biopsies of CD and UC cases with healthy controls [27]. While little is understood about the mechanistic importance of FANCC with respect to autoimmune processes, it’s prior association with CD and UC indicates a need for more studies.
Lastly, Haas and colleagues showed a significantly higher expression of protocadherin beta 16 (PCDHB16) mRNA in MZ twins with rheumatoid arthritis compared to their healthy co-twin [28]. To date, little is known about the function of PCDHB16, but available results suggest it may have a role in broad autoimmune processes.
Our findings add to growing evidence that support a role for overlapping genetic contributions across different autoimmune disorders, including T1D. None of the top 10 single CpG sites in the discordant MZ twins were observed in HLA-identical NTS. These results are consistent with the genome-wide findings and differences in the two study groups described above. Although our top findings in discordant MZ twins could not be replicated using another small sample of subjects, more work is needed. GWAS results for autoimmune diseases, to date, have revealed a complex and heterogeneous genetic architecture with a large number of very modest individual effects underlying susceptibility. If DNAm influences are operating similarly, much larger samples will be needed for confirmation of any putative associations. Our top candidates in the current study do have support from studies of other autoimmune diseases, and therefore warrant further examination.
Using DNA from immortalized B-cell lines, Stefan et al. recently reported evidence for hypermethylation at CD226 in a sample of three discordant twin pairs [11]. Results from the current study are consistent with this finding (Table 2). MZ twins with T1D demonstrated evidence for hypermethylation of CD226:cg12929678, a site located within 200 base pairs of a transcription start site, compared to unaffected twins; albeit a very modest difference (2.4%). CD226 is known risk factor for T1D[29], and plays a mediatory role in cell activation and differentiation with respect to immune cells such as T-cell, monocytes, and natural killer cells [30]. CD226 is therefore an attractive candidate gene for further investigation in a larger study.
Other sites reported by Stefan et al. were not replicated in the current study. Additionally, our results did not overlap with the most differentially methylated candidate CpGs from another previous twin study conducted by Rakyan et al. [10]. Discrepancies in results from the two previous DNAm studies may be due to a number of factors. The prior studies utilized the earlier Illumina 27k DNA methylation BeadChip, which had low CpG density and several technical limitations; the current study incorporated data derived from the much more robust and comprehensive 450k platform. A comparison between the 27k and 450k chip CpG site density for MAGI2, FANCC, and PCDHB16, revealed tremendously different numbers. In the 27k chip, there are a total of 2 CpGs each for MAGI2 and FANCC and 1 CpG in PCDBH16, whereas there are 104, 25, and 19 CpGs respectively for each of these three genes in the 450k chip. Therefore, it is reasonable that more robust CpG coverage on the 450k chip might yield findings that could not have been observed using the 27k chip. Additionally, source DNA in Stefan et al. was derived from immortalized B-cells, while Rakyan et al. used DNA from peripheral blood monocytes [10, 11], similar to the current study. Though Rakyan et al. corrected for cell-composition, and comparisons in the current study showed little difference in cell-composition between co-twins, the possibility of residual confounding by cell-type cannot be discounted.
DNAm normalization techniques are performed in order to cancel out “noise” or variation in DNAm that might be attributable to the Illumina Chip or DNA processing plate. Both Stefan et al. and Rakyan et al. applied quantile-normalization, while the current study utilized Illumina’s standard protocol of background subtraction and internal control normalization. As normalization techniques have been shown to yield different DNAm findings [31], it is possible this had an effect on which top CpG-sites were discovered.
Sample size and the statistical tests performed are also likely to play a role in which top CpG sites are identified. Stefan et al. used a combination of T1D concordant (n=6 pairs) and discordant twin pairs (n=3 pairs), while Rakyan et al. used 15 discordant twin pairs [10, 11]; the current study utilized 7 discovery and 6 confirmatory, long-term discordant MZ twins. Regarding statistical testing, Stefan et al. performed a linear method in LIMMA regression with adjustment for multiple testing. Rakyan et al. performed non-parametric Wilcoxon signed-rank tests, without adjustment, similar to the current study. The current study, however, utilized both a candidate gene approach and a priori data reduction techniques.
A key strength of the current study is the testing of 450k results using quantitative pyrosequencing in an independent set of MZ twins. The reliability of the 450k platform in identifying DNAm levels was validated through the use of control DNA (Jurkat cell lines). Results showed strong correlation between pyrosequenced DNAm values and 450k average-beta values. These results demonstrate the strength of the 450k platform for accurate DNAm detection, and have important implications for future T1D studies that utilize this same platform. It should be possible to combine datasets if they become available for increased power.
Only peripheral blood DNA from a relatively small sample of MZ twins was available for the current study. Results show that DNAm patterns for MZ twins are not due to differences in cell type composition of whole blood, given the strong correlation of cell-types observed between identical twins. However, proportions of cell subsets can vary among closely related individuals, including the HLA-identical NTS pairs. Temporality could not be established in the current study, due to the fact that DNA was extracted from stored peripheral blood samples that had been collected after disease onset. Therefore, we cannot determine whether methylation differences are causal or a result from disease pathogenesis. Finally, failure to validate individual 450k CpG site findings by pyrosequencing indicates a larger sample of MZ twins is needed for future studies for increased power.
5. Conclusion
Characterization of global methylation differences showed the majority of promoter region CpGs in MZ twins with T1D are significantly hypomethylated compared to unaffected twins, in contrast with the rest of the genome. This interesting finding was not observed in NTS pairs and needs to be pursued further. Results suggest the contribution of DNA methylation differences to the pathophysiology of T1D are many and modest, similar to GWAS to date, and may require close matching of cases and controls; i.e., the use of MZ twins is beneficial for significant CpGs to be revealed.
Despite promising results from the initial 450k analysis and validation of the platform as a whole using control DNA, none of our top five CpG site-specific findings for T1D were replicated by pyrosequencing in an independent set of disease discordant MZ twins. Importantly, our findings inform design of future investigations and do not support involvement of large DNAm effects in T1D. Discordant MZ twin studies represent a unique approach for epigenetic studies of complex diseases, especially given the differences observed from HLA matched, discordant siblings. While such studies will not reveal missing heritability, observed DNAm differences in disease-discordant MZ twin pairs can help elucidate potential causal mechanisms underlying complex diseases [32].
Supplementary Material
Seven sets of T1D discordant MZ twins used to discover differentially methylated CpG sites within or near established T1D genes.
Strong evidence for overall global hypomethylation of gene promoter CpG sites in MZ twin with TID compared to twin without T1D was observed.
T1D discordant HLA-identical sibs show greater differences in patterns of DNA methylation compared to T1D discordant MZ twins.
Illumina’s 450k array results validated by pyrosequencing of control DNA samples.
Results based on analysis of T1D discordant MZ twin replication dataset suggest large DNA methylation differences at single CpG sites are not associated with T1D.
Acknowledgments
This work was supported in part by grants from the Juvenile Diabetes Foundation [11-2002-696], the National Institutes of Health [DK32083 to the late Dr. George Eisenbarth, DK61722 to J.A.N.], the Autoimmunity Prevention Center [AI50964], and the American Diabetes Association [1-04-RA-23, to P.R.F.]. Support for patient recruitment at Children’ s Hospital and Research Center Oakland was provided by National Institutes of Health Clinical and Translational Science Award grant UL1 RR024131. We thank Julie Roessig of Children’s Hospital Oakland Research Institute (CHORI) for assistance with identification of the Children’s Hospital, Oakland (CHO) NTS pairs.
Abbreviations
- T1D
type 1 diabetes
- DNAm
DNA methylation
- MZ
Monozygotic
- NTS
Non-twin sibling
- HLA
Human Leukocyte Antigen
- MHC
Major Histocompatibility Complex
- 450k
Illumina Human Methylation 450 Bead chip array
- CpG
DNA methylation site
- GWAS
Genome wide association study
Footnotes
Conflict of Interest
No conflicts of interest are reported by any author.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–92. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noble JA, Valdes AM, Varney MD, Carlson JA, Moonsamy P, Fear AL, et al. HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes. 2010;59:2972–9. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper JD, Howson JM, Smyth D, Walker NM, Stevens H, Yang JH, et al. Confirmation of novel type 1 diabetes risk loci in families. Diabetologia. 2012;55:996–1000. doi: 10.1007/s00125-012-2450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akirav EM, Lebastchi J, Galvan EM, Henegariu O, Akirav M, Ablamunits V, et al. Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc Natl Acad Sci U S A. 2011;108:19018–23. doi: 10.1073/pnas.1111008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahlquist G. Environmental risk factors in human type 1 diabetes--an epidemiological perspective. Diabetes Metab Rev. 1995;11:37–46. doi: 10.1002/dmr.5610110104. [DOI] [PubMed] [Google Scholar]
- 7.Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–6. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gervin K, Vigeland MD, Mattingsdal M, Hammero M, Nygard H, Olsen AO, et al. DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: identification of epigenetically dysregulated genes. PLoS Genet. 2012;8:e1002454. doi: 10.1371/journal.pgen.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2009;20:170–9. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7:e1002300. doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefan M, Zhang W, Concepcion E, Yi Z, Tomer Y. DNA methylation profiles in type 1 diabetes twins point to strong epigenetic effects on etiology. Journal of autoimmunity. 2014;50:33–7. doi: 10.1016/j.jaut.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shenker N, Flanagan JM. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. Br J Cancer. 2012;106:248–53. doi: 10.1038/bjc.2011.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Touleimat N, Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–41. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 14.Whitaker JW, Shoemaker R, Boyle DL, Hillman J, Anderson D, Wang W, et al. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome medicine. 2013;5:40. doi: 10.1186/gm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delepine M, Pociot F, Habita C, Hashimoto L, Froguel P, Rotter J, et al. Evidence of a non-MHC susceptibility locus in type I diabetes linked to HLA on chromosome 6. Am J Hum Genet. 1997;60:174–87. [PMC free article] [PubMed] [Google Scholar]
- 16.Fendler W, Klich I, Cieslik-Heinrich A, Wyka K, Szadkowska A, Mlynarski W. Increased risk of type 1 diabetes in Polish children - association with INS-IGF2 5'VNTR and lack of association with HLA haplotype. Endokrynol Pol. 2011;62:436–42. [PubMed] [Google Scholar]
- 17.Ludvigsson J, Faresjo M, Hjorth M, Axelsson S, Cheramy M, Pihl M, et al. GAD treatment and insulin secretion in recent-onset type 1 diabetes. N Engl J Med. 2008;359:1909–20. doi: 10.1056/NEJMoa0804328. [DOI] [PubMed] [Google Scholar]
- 18.Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, et al. Genetics of type 1 diabetes: what's next? Diabetes. 2010;59:1561–71. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy MV, Wang H, Liu S, Bode B, Reed JC, Steed RD, et al. Association between type 1 diabetes and GWAS SNPs in the southeast US Caucasian population. Genes Immun. 2011;12:208–12. doi: 10.1038/gene.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Yan L, Hu Q, Sucheston LE, Higgins MJ, Ambrosone CB, et al. IMA: an R package for high-throughput analysis of Illumina's 450K Infinium methylation data. Bioinformatics. 2012;28:729–30. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aryee KDHaM. R package. minfi: Analyze Illumina's 450k methylation arrays. p. [Google Scholar]
- 22.Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome research. 2011;21:1074–86. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGovern DP, Taylor KD, Landers C, Derkowski C, Dutridge D, Dubinsky M, et al. MAGI2 genetic variation and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:75–83. doi: 10.1002/ibd.20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Li Z, Guo L, Wang L, Zhang L, Cai X, et al. MAGI-2 Inhibits cell migration and proliferation via PTEN in human hepatocarcinoma cells. Arch Biochem Biophys. 2007;467:1–9. doi: 10.1016/j.abb.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 26.Valiente M, Andres-Pons A, Gomar B, Torres J, Gil A, Tapparel C, et al. Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinases. J Biol Chem. 2005;280:28936–43. doi: 10.1074/jbc.M504761200. [DOI] [PubMed] [Google Scholar]
- 27.Cooke J, Zhang H, Greger L, Silva AL, Massey D, Dawson C, et al. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:2128–37. doi: 10.1002/ibd.22942. [DOI] [PubMed] [Google Scholar]
- 28.Haas CS, Creighton CJ, Pi X, Maine I, Koch AE, Haines GK, et al. Identification of genes modulated in rheumatoid arthritis using complementary DNA microarray analysis of lymphoblastoid B cell lines from disease-discordant monozygotic twins. Arthritis Rheum. 2006;54:2047–60. doi: 10.1002/art.21953. [DOI] [PubMed] [Google Scholar]
- 29.Douroudis K, Nemvalts V, Rajasalu T, Kisand K, Uibo R. The CD226 gene in susceptibility of type 1 diabetes. Tissue antigens. 2009;74:417–9. doi: 10.1111/j.1399-0039.2009.01320.x. [DOI] [PubMed] [Google Scholar]
- 30.Shibuya A, Campbell D, Hannum C, Yssel H, Franz–Bacon K, McClanahan T, et al. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 1996;4:573–81. doi: 10.1016/s1074-7613(00)70060-4. [DOI] [PubMed] [Google Scholar]
- 31.Yousefi P, Huen K, Schall RA, Decker A, Elboudwarej E, Quach H, et al. Considerations for normalization of DNA methylation data by Illumina 450K BeadChip assay in population studies. Epigenetics : official journal of the DNA Methylation Society. 2013;8:1141–52. doi: 10.4161/epi.26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–50. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.