Abstract
Adipose tissue engineering is a diverse area of research where the developed tissues can be used to study normal adipose tissue functions, create disease models in vitro, and replace soft tissue defects in vivo. Increasing attention has been focused on the highly specialized metabolic pathways that regulate energy storage and release in adipose tissues which affect local and systemic outcomes. Non-invasive, dynamic measurement systems are useful to track these metabolic pathways in the same tissue model over time to evaluate long term cell growth, differentiation, and development within tissue engineering constructs. This approach reduces costs and time in comparison to more traditional destructive methods such as biochemical and immunochemistry assays and proteomics assessments. Towards this goal, this review will focus on important metabolic functions of adipose tissues and strategies to evaluate them with noninvasive in vitro methods. Current non-invasive methods, such as measuring key metabolic markers and endogenous contrast imaging will be explored.
Keywords: tissue engineering, non-destructive characterization, optical imaging
Introduction
Adipose tissues not only act as thermal insulators, provide cushion for internal organs, and serve as a reservoir for fat; they also have highly specialized metabolic pathways that regulate energy storage and release affecting local and systemic functions.32,57 For tissue engineering applications, proper metabolic regulation within adipose tissues is essential to promote and to monitor differentiation and functionality in vitro in order to allow extrapolations to in vivo scenarios.
Adipose tissue engineering is a diverse area of research where the developed tissues can be used to study normal adipose tissue function, create disease models in vitro,38 and replace soft tissue defects from disease, trauma, or injury in vivo.13 While advances in our understanding of adipose tissue metabolism are significant, new experimental approaches are required to provide insights into mechanisms of energy balance and adipokine (cytokines released by adipocytes) regulation and action.32 In particular, many diseases, including obesity, dyslipidemia, type II diabetes, thyroid hormone disorders, and different cancers (breast, colon, etc.), disturb normal metabolic regulation in adipose tissues.28,40,50
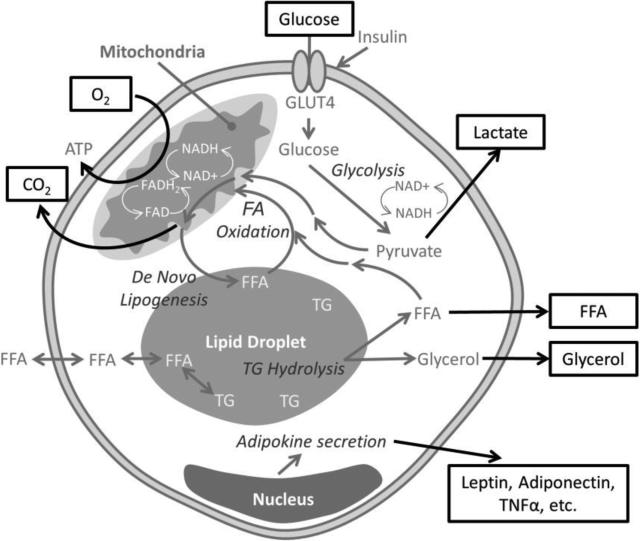
Techniques such as the polymerase chain reaction, immunohistochemistry, flow cytometry, mass spectroscopy, and electron microscopy are all destructive endpoints for cell or tissue analysis that require sacrificing samples to acquire the desired information. Consumption of reagents to maintain the large sample sizes required for these endpoints is not practical due to logistics and cost, especially when cultures are extended for long periods of time. Long term in vitro adipose tissue models will likely be required to improve physiological relevance1 since differentiation in vitro can take multiple weeks,25 and disease mechanisms that affect the adipose tissues develop in vivo over weeks to months.29 Additionally, specimen variability can affect the endpoint readouts of destructive assays that do not enable baseline readings and repeated measurements of the same tissue in culture. By focusing on non-invasive, dynamic measurement systems, cell growth and development can be quantified in the same tissue construct over time,12,46 minimizing sample reagents, increasing statistical power, and allowing for long term studies. Towards this goal, this review will focus on important metabolic functions of adipose tissues (Figure 1) and strategies to evaluate them with non-invasive methods (Table 1).
Figure 1.
Schematic representation of an adipocyte's metabolism, where factors that can be evaluated non-invasively are outlined. In response to insulin stimulation, adipocytes translocate glucose transporter type 4 (GLUT4) to the plasma membrane, resulting in glucose uptake. Therefore, extracellular changes in glucose levels can be monitored. Once glucose is taken up in the cell, it is metabolized to the final end products of: lipids, lactate (released from the cell) or carbon dioxide (CO2, released from the cell). Triglycerides (TG), are stored in lipid droplets (which can be visualized by microscopes) as an energy resource, from two routes: synthesis of fatty acids from non-lipid substrates (de novo lipogenesis) or uptake of free fatty acids. In times of fasting, triglycerides are hydrolyzed (lipolysis) into glycerol and free fatty acids (FFA). Fatty acids (FA) can be secreted, oxidized in the mitochondria resulting in CO2 as an end-product, or enter the nucleus. Glycerol is secreted. Throughout all of these metabolic cycles, different adipokines are expressed, resulting in secretion of proteins that can be monitored.
Table 1.
Summary of non-destructive in vitro assays for studying metabolism in adipose tissues
| Metabolic Activity | Technique | Reference |
|---|---|---|
| Lipolysis | Colorimetric/Fluorometric quantification of glycerol, FFA | (22, 23) |
| Fatty Acid Oxidation | Quantification of radio-labelled CO2 from uptake of [14C] palmitic acid | (24, 25) |
| Glucose consumption | Quantification of glucose/lactate levels | (26, 27) |
| Thermogenesis | Measurement of oxygen consumption | (25, 30) |
| Adipokine secretion | ELISA or protein array of adipokines | (23) |
Abbreviations: FFA = free fatty acids, ELISA = enzyme-linked immunosorbent assay
Different metabolically active human adipose tissues
In humans, there are many metabolically distinct adipose tissue depots throughout the body. White adipose tissue is found in visceral, subcutaneous, and intramuscular regions. Visceral white adipose tissue wraps around inner organs and is divided into different regions accordingly: omental (begins near the stomach and spleen and extends into the ventral abdomen), mesenteric (intestine), retroperitoneal (kidney), gonadal (in females it surrounds the uterus/ovaries and in males it surrounds the epididymis/testis), and pericardial (heart).10 Subcutaneous white adipose tissue is situated beneath the skin predominately in the abdomen and gluteofemoral region (measured by hip, thigh, and leg circumference)10 Finally, white adipose tissue can be found between and within the skeletal muscles of the body and increases in abundance with age.34
Brown adipose tissue is located in the supraclavicular and subscapular region, and is the smallest mass of adipose tissue in the body.15 Unlike white adipose tissues which store energy, brown adipose tissue dissipates energy by thermogenesis. In addition, brown adipose tissues have a higher uptake (and shorter half-life) of lipids compared to other adipose tissues.33 They also have a different morphology in which brown adipocytes contain multilocular lipid droplets and a large quantity of mitochondria, while white adipocytes have a large unilocular lipid droplet and a smaller quantity of mitochondria.
Finally, there are specialized adipose tissue depots, mainly the mammary gland and bone marrow, which are functionally distinct from white and brown adipose tissues. Mammary gland adipose tissue is involved in milk production and regulates epithelial cell growth, whereas bone marrow is involved in hematopoiesis and osteogenesis and regulates skeletal metabolism.48
Non-destructive evaluation of triglyceride storage and release
Triglycerides are stored in lipid droplets of adipocytes as an energy resource from two routes: de novo lipogenesis (synthesis of fatty acids from non-lipid substrates) or uptake of free fatty acids. White and mammary adipose tissues are capable of de novo lipogenesis; however, uptake of free fatty acids is the main source of triglyceride storage.11,32,52 While triglyceride storage cannot be quantified directly without cell lysis, the levels can be monitored nondestructively using different imaging techniques (see section on tracking cells non-invasively over time with optical imaging).
Lipolysis breaks down lipid stores by the hydrolysis of triglycerides into free fatty acids and glycerol. This process is important to monitor, as it plays a large role in energy balance. Free fatty acids can either be transported to the mitochondria for β-oxidation (providing cellular ATP), enter the nucleus (acting as ligands for nuclear hormone receptors and regulating gene transcription), or released from the cells (where it fatty acids are used as fuel and signaling molecules for tissues).26 While free fatty acids can be reutilized by adipocytes, glycerol cannot and is a more accurate measurement of lipolysis.59 Both fatty acid and glycerol assay kits are commercially available and rely on spectrophotometric measurements. These assays make it possible to monitor lipolysis out to extended time points without putting additional stress on tissue constructs or cells,2,8 since fatty acids and glycerol can be collected in culture media.
Byproduct monitoring of fatty acid oxidation
Fatty acids oxidation allows for the production of ATP to fuel metabolic activity.16 Fatty acid oxidation can be measured non-invasively by adding a low concentration of [14C] palmitic acid to the culture media for a brief period, and measuring the production of radio-labeled CO 62.2 In comparing the oxidation rates observed in human brown and white adipose tissues, brown adipose tissues are able to use fatty acids as an energy source more efficiently. This method has been used to quantify fatty acid oxidation in human white and brown progenitors.62 Given this assays non-destructive nature, it may be used for repeated metabolic analysis of the same tissue construct.
Tracking glucose metabolism non-invasively
In response to insulin stimulation, adipocytes translocate glucose transporter type 4 (GLUT4) to the plasma membrane, resulting in increased glucose uptake. Glucose transport can be evaluated in a number of destructive ways using radioactive glucose analogs (e.g., 2-deoxy- glucose, 3-O-methyl glucose, [3-3H]-glucose) and evaluating the cell lysate.5 However, for nondestructive purposes, indirect measurement of glucose metabolism can be determined by tracking glucose levels in the supernatant over time to indicate glucose consumption.39 Once glucose is taken up by the adipocytes, it is metabolized to either lipids (~20% de novo lipogenesis), lactate (~70%) or carbon dioxide (~10%) end products.35 Therefore, another indirect measurement of glucose metabolism involves measuring lactate released into the media, which can be measured with spectrophotometric kits5
Non-destructive evaluation of thermogenesis
In brown adipose tissue, thermogenesis is characterized by high levels of cellular catabolism and thus oxygen consumption.27 Therefore, assessing the metabolic capabilities of in vitro adipose cultures is important for brown adipose tissue engineering,56 and if replicated, could provide a way to combat obesity.55 One technique used to monitor this thermogenic process in vitro requires the use of a commercially available probe that exhibits fluorescence upon oxygen depletion. In order to ensure the availability of a finite amount of oxygen, the cells are covered by a non-toxic mineral oil,56 making the addition of chemicals or compounds and repeated measures on the same samples more difficult.49 Commercially available devices such as extracellular flux analyzers, have also been used to evaluate mitochondrial respiration and are label free and non-destructive21,49 and have been used to determine thermogenesis in human brown adipose progenitors.62
Adipokine secretion for non-destructive monitoring of differentiation and function
As an endocrine gland, adipose tissue secretes pleiotropic adipokines that regulate appetite, insulin sensitivity, angiogenesis, blood pressure, and immune responses.36 Non-destructive analysis techniques used on adipose tissues and their isolated cell types often rely on the collection of supernatant which contains adipokines for monitoring adipogenic differentiation 58. These proteins also provide important insight into obesity and other diseases involving white adipose tissue. Leptin and adiponectin are two commonly studied proteins that play an important role in metabolism.4,6,36 Leptin negatively regulates body weight and food intake through receptors in the brain, and is increased in obesity.4 Adiponectin is secreted by adipocytes into the circulation and acts predominantly on the skeletal muscle and liver (causing an increase in fatty acid oxidation and reduction in liver glucose synthesis), and is reduced with insulin resistance and obesity.4 Other adipokines, like tumor necrosis factor alpha, produce inflammation in adipose tissues. Because these proteins are more prevalent in obese individuals, chronic inflammation has been linked to obesity. This observation indicates that inflammatory adipokines may play a role in diseases associated with obesity, like type 2 diabetes, metabolic complications, and atherosclerosis.30 Adipokines can be detected in the supernatant using enzyme-linked immunosorbent assays. This method has been used to quantify leptin release at extended time points up to 6 months.8,60 Additionally, cytokine protein arrays can be used with a variety of proteins as a more general, less quantitative screening of protein secretion.7
Adipokine secretion can be monitored to track adipogenic differentiation and to evaluate the accuracy of long-term in vitro models in replicating obesity and its associated complications. While analysis of extracellular concentrations of proteins provides an overall view of the metabolic function of the tissue, it does not provide a spatial map of development within the tissue. To assess variability within tissue constructs, optical imaging can be utilized to quantify the morphological and biochemical heterogeneity of tissues.
Tracking cells non-invasively over time with optical imaging
Non-destructive, optical imaging modalities are ideally suited for tracking adipose tissue engineered constructs over time (Table 2). They concurrently provide information on the scaffold, matrix organization (collagen and elastin) and cell morphology and functionality.12 For further background, the basis for various analytical imaging methods are reviewed elsewhere.22,23 By taking advantage of the inherent optical properties of cells and tissues, optical imaging of adipose tissue does not necessarily require additional cellular tagging to assess cell function and tissue organization. However, when necessary, non-toxic tags can supplement label-free imaging to provide additional information.
Table 2.
Non-destructive imaging techniques for adipose tissue engineered models
| Technique | Label | Measurement | Reference |
|---|---|---|---|
| TPEF, FLIM | None | NADH and FAD (endogenous contrast) | (11, 43, 53) |
| THG, SRS, CARS | None | Lipid droplet morphology and content (endogenous contrast) | (12, 47-50) |
| Ultrasound and photoacoustic imaging | Gold Nanomarkers | Location of tagged cells | (54, 55) |
| MRI | None | Distinguishes fat versus water based tissues due to differences in relaxation times | (56, 57) |
Abbreviations: TPEF = Two-Photon Excitation Fluorescence, THG = Third Harmonic Generation, MRI = Magnetic resonance imaging
For imaging human derived stem cells in 3D scaffolds, the use of two-proton excited fluorescence (TPEF) has been shown to be effective in assessing naturally fluorescent endogenous markers.12,46,47,61 Natural markers include nicotinamide adenine dinucleotide (phosphate) (NAD(P)H), flavin adenine dinucleotide (FAD), keratins, lipofuscin, retinol, and porphyrins.12,43,63 When specifically considering mesenchymal stem cells, an optical redox ratio of FAD/(FAD+NADH) autofluorescence intensity provides information on the stage of adipogenic differentiation (Figure 2).46 Since TPEF has the ability to provide optical tissue sections at depths up to 500 microns, individual cells can be tracked over a period of time within the 3D environment. TPEF-derived optical redox ratios have been used to track human adipose derived stem cell (hASC) proliferation and adipogenic differentiation in silk protein scaffolds for over six months.46 This approach has been supplemented by a perfusion bioreactor system to analyze in vitro adipose tissue structure and function under flow as well. 61 Third harmonic generation (THG), coherent anti-stokes Raman scattering (CARS), and stimulated Raman scattering (SRS) are additional non-linear optical methods that can be used to characterize lipid droplet dynamics and provide context for TPEF-based metabolic outcomes.12,20,42,44,53 THG in particular has been paired with redox ratio outcomes to evaluate hASC differentiation during 3D in vitro development.12 Furthermore, the use of fluorescent lifetime imaging (FLIM) can be used in conjunction with TPEF microscopy to help discriminate naturally fluorescent endogenous markers and provide additional context to redox ratio based outcomes.18,22 A variety of visual markers of differentiation can also be evaluated with microscopy techniques, such as CARS, THG, or phase contrast imaging. For instance cell volume, lipid volume per cell, lipid volume normalized to cell volume, lipid droplet count per cell, and average droplet size per cell can all be quantified.12 In addition, mitochondria can be quantified and visualized in live cell cultures noninvasively,47,51 which is particularly relevant for brown adipose tissues which have a greater volume of mitochondria.
Figure 2.
A sample redox ratio image of differentiating adipocytes at 4 weeks. The differentiating cells have a lower redox ratio compared to the stem cells, which have high redox ratios. On a heat map the blue represents a low redox ratio while red represents a high redox ratio. The scale bar is 50 μm.
Both SRS and CARS can be used to identify droplets via C-H bond vibrations, and also may offer insights into more subtle differences in the chemical composition of droplets 20,37,42,44,53 . One of the benefits of SRS is that it lacks a non-resonant background signal often found in CARS37,45,53, but there are more technical challenges to implement a SRS system, and consequently it is not available through commercial vendors. The practical implementation of CARS is a little more straightforward, and it has recently become commercially available. THG does not have the molecular specificity of SRS and CARS; since a signal is produced at any interface with distinct optical properties 12,19,54 (e.g. the edge of a lipid droplet). The implementation of THG requires only a single pulsed laser, resulting in a simpler, more cost effective approach however.
Biocompatible, non-toxic labeling of hASCs has been implemented to supplement label-free imaging.12,24,47 Non-toxic gold nanorods were used to label and image hASCs using spectroscopic photoacoustic imaging.41 In addition, 20 nm gold nanospheres were used to nontoxically tag hASCs and were imaged via ultrasound and photoacoustic methods and tracked in vitro for over two weeks.14 The use of ultrasound in conjunction with photoacoustic imaging has also been used to collect data on the quantitative factors related to hASC growth and development when implanted in vivo. 14,41 Ultrasound and spectroscopic photoacoustic imaging were used to examine injured tissue recovery,41 and to track the progress of stem cell therapies in the subcutaneous adipose tissue.41
Additionally, imaging using fast spin echo based magnetic resonance can be used to quantify and distinguish different types of adipose tissues in vivo9 and has been shown to be an effective imaging technique in vitro. 3 Other in vivo imaging modalities, such as CT-volume data, which is used to evaluate body fat,31 could be important for assessing tissue engineering constructs that have been implanted in vivo for regenerative applications. In the future, the use of in vivo techniques to assess in vitro cultures will be an important step towards correlating clinical outcomes with in vitro tissue models.
While imaging has many advantages, there are some challenges that should be considered. For instance, some scaffolds auto-fluoresce, which can interfere with the collection and isolation of signals from other fluorophores. Silk scaffolds, in particular, can be excited and emit fluorescence due to tyrosines, tryptophan, and cross-links.24 To address these issues, algorithms and techniques such as linear discriminant analysis can be implemented to differentiate between cells and silk fluorescence.12,46,61 Phototoxicity can also be a problem of extended imaging, and can be countered by changing media supplements and decreasing exposure times.17 Finally, the penetration depth of high-resolution optical imaging approaches (TPEF, CARS, SRS, THG) is typically limited to 100-200 microns in lipid rich tissues because of the highly scattering nature of lipid droplets. However, this depth is usually sufficient for studying most 3D in vitro adipose tissue models.
Conclusions
As the need for in vitro adipose tissue engineering increases, further advances in nondestructive analyses will be required to minimize sample sizes and evaluate the same tissue construct throughout culture time. Current non-destructive methods of evaluating differentiation and functionality, such as optical imaging, are important areas of research that allow multiple time points to be taken on the same tissue sample. This non-destructive approach enables tracking of tissue features relative to baseline measurements, which helps account for variability that may exist among tissue samples. Moving forward, it will be essential to focus on non-invasive methods for analyzing long-term changes in architecture and metabolism in adipose tissue.
Acknowledgments
The authors would like to thank the National Institutes of Health (Tissue Engineering Resource Center P41 EB002520, R01EB007542 and K99EB017723) and AFIRM (W81XWH-08-2-0032) for supporting our tissue engineering studies.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Abbott RD, Kaplan DL. Strategies for improving the physiological relevance of human engineered tissues. Trends in biotechnology. 2015;33:401–407. doi: 10.1016/j.tibtech.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbott RD, Raja WK, Wang RY, Stinson JA, Glettig DL, Burke KA, Kaplan DL. Long term perfusion system supporting adipogenesis. Methods. 2015 doi: 10.1016/j.ymeth.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed N, Ahmad NM, Fessi H, Elaissari A. In vitro MRI of biodegradable hybrid (iron oxide/polycaprolactone) magnetic nanoparticles prepared via modified double emulsion evaporation mechanism. Colloids and Surfaces B: Biointerfaces. 2015;130:264–271. doi: 10.1016/j.colsurfb.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Andrade-Oliveira V, Camara NO, Moraes-Vieira PM. Adipokines as drug targets in diabetes and underlying disturbances. Journal of diabetes research. 2015;2015:681612. doi: 10.1155/2015/681612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arner P. Techniques for the measurement of white adipose tissue metabolism: a practical guide. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 1995;19:435–442. [PubMed] [Google Scholar]
- 6.Baboota RK, Singh DP, Sarma SM, Kaur J, Sandhir R, Boparai RK, Kondepudi KK, Bishnoi M. Capsaicin induces “brite” phenotype in differentiating 3T3-L1 preadipocytes. PLoS One. 2014;9:e103093. doi: 10.1371/journal.pone.0103093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer D, Mazzio E, Soliman KF, Taka E, Oriaku E, Womble T, Darling-Reed S. Diallyl disulfide inhibits TNFalpha-induced CCL2 release by MDA-MB-231 cells. Anticancer research. 2014;34:2763–2770. [PMC free article] [PubMed] [Google Scholar]
- 8.Bellas E, Marra KG, Kaplan DL. Sustainable three-dimensional tissue model of human adipose tissue. Tissue engineering. Part C, Methods. 2013;19:745–754. doi: 10.1089/ten.tec.2012.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhanu Prakash KN, Gopalan V, Lee SS. Quantification of abdominal fat depots in rats and mice during obesity and weight loss interventions. PLoS ONE. 9:2014. doi: 10.1371/journal.pone.0108979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjorndal B, Burri L, Staalesen V, Skorve J, Berge RK. Different adipose depots: their role in the development of metabolic syndrome and mitochondrial response to hypolipidemic agents. Journal of obesity. 2011;2011:490650. doi: 10.1155/2011/490650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjorntorp P, Sjostrom L. Carbohydrate storage in man: speculations and some quantitative considerations. Metabolism: clinical and experimental. 1978;27:1853–1865. doi: 10.1016/s0026-0495(78)80004-3. [DOI] [PubMed] [Google Scholar]
- 12.Chang T, Zimmerley MS, Quinn KP, Lamarre-Jouenne I, Kaplan DL, Beaurepaire E, Georgakoudi I. Non-invasive monitoring of cell metabolism and lipid production in 3D engineered human adipose tissues using label-free multiphoton microscopy. Biomaterials. 2013;34:8607–8616. doi: 10.1016/j.biomaterials.2013.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JH, Gimble JM, Lee K, Marra KG, Rubin JP, Yoo JJ, Vunjak-Novakovic G, Kaplan DL. Adipose tissue engineering for soft tissue regeneration. Tissue engineering. Part B, Reviews. 2010;16:413–426. doi: 10.1089/ten.teb.2009.0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung E, Nam SY, Ricles LM, Emelianov SY, Suggs LJ. Evaluation of gold nanotracers to track adipose-derived stem cells in a PEGylated fibrin gel for dermal tissue engineering applications. International Journal of Nanomedicine. 2013;8:325–336. doi: 10.2147/IJN.S36711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cinti S. The role of brown adipose tissue in human obesity. Nutrition, metabolism, and cardiovascular diseases : NMCD. 2006;16:569–574. doi: 10.1016/j.numecd.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Coppack SW, Frayn KN, Humphreys SM, Whyte PL, Hockaday TD. Arteriovenous differences across human adipose and forearm tissues after overnight fast. Metabolism. 1990;39:384–390. doi: 10.1016/0026-0495(90)90253-9. [DOI] [PubMed] [Google Scholar]
- 17.Danmark S, Gladnikoff M, Frisk T, Zelenina M, Mustafa K, Russom A, Finne-Wistrand A. Development of a novel microfluidic device for long-term in situ monitoring of live cells in 3-dimensional matrices. Biomedical microdevices. 2012;14:885–893. doi: 10.1007/s10544-012-9668-1. [DOI] [PubMed] [Google Scholar]
- 18.Datta R, Alfonso-Garcia A, Cinco R, Gratton E. Fluorescence lifetime imaging of endogenous biomarker of oxidative stress. Scientific reports. 2015;5:9848. doi: 10.1038/srep09848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debarre D, Supatto W, Pena AM, Fabre A, Tordjmann T, Combettes L, Schanne-Klein MC, Beaurepaire E. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nature methods. 2006;3:47–53. doi: 10.1038/nmeth813. [DOI] [PubMed] [Google Scholar]
- 20.Evans CL, Xie XS. Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annual review of analytical chemistry. 2008;1:883–909. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- 21.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug discovery today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Georgakoudi I, Quinn KP. Optical imaging using endogenous contrast to assess metabolic state. Annual review of biomedical engineering. 2012;14:351–367. doi: 10.1146/annurev-bioeng-071811-150108. [DOI] [PubMed] [Google Scholar]
- 23.Georgakoudi I, Rice WL, Hronik-Tupaj M, Kaplan DL. Optical spectroscopy and imaging for the noninvasive evaluation of engineered tissues. Tissue engineering. Part B, Reviews. 2008;14:321–340. doi: 10.1089/ten.teb.2008.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgakoudi I, Tsai I, Greiner C, Wong C, Defelice J, Kaplan D. Intrinsic fluorescence changes associated with the conformational state of silk fibroin in biomaterial matrices. Optics Express. 2007;15:1043–1053. doi: 10.1364/oe.15.001043. [DOI] [PubMed] [Google Scholar]
- 25.Gerlach JC, Lin YC, Brayfield CA, Minteer DM, Li H, Rubin JP, Marra KG. Adipogenesis of human adipose-derived stem cells within three-dimensional hollow fiber-based bioreactors. Tissue engineering. Part C, Methods. 2012;18:54–61. doi: 10.1089/ten.tec.2011.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y, Cordes KR, Farese RV, Walther TC. Lipid droplets at a glance. Journal of cell science. 2009;122:749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Himms-Hagen J. Brown adipose tissue thermogenesis: interdisciplinary studies. FASEB J. 1990;4:2890–2898. [PubMed] [Google Scholar]
- 28.Iyengar NM, Hudis CA, Dannenberg AJ. Obesity and inflammation: new insights into breast cancer development and progression. American Society of Clinical Oncology educational book / ASCO. American Society of Clinical Oncology. Meeting. 2013:46–51. doi: 10.1200/EdBook_AM.2013.33.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawasaki N, Asada R, Saito A, Kanemoto S, Imaizumi K. Obesity-induced endoplasmic reticulum stress causes chronic inflammation in adipose tissue. Scientific reports. 2012;2:799. doi: 10.1038/srep00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim TK, Park KS. Inhibitory effects of harpagoside on TNF-alpha-induced pro-inflammatory adipokine expression through PPAR-gamma activation in 3T3-L1 adipocytes. Cytokine. 2015 doi: 10.1016/j.cyto.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Kim YJ, Lee SH, Kim TY, Park JY, Choi SH, Kim KG. Body fat assessment method using CT images with separation mask algorithm. Journal of digital imaging. 2013;26:155–162. doi: 10.1007/s10278-012-9488-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lafontan M. Advances in adipose tissue metabolism. International journal of obesity. 2008;32(Suppl 7):S39–51. doi: 10.1038/ijo.2008.237. [DOI] [PubMed] [Google Scholar]
- 33.Li M, Yang S, Bjorntorp P. Metabolism of different adipose tissues in vivo in the rat. Obesity research. 1993;1:459–468. doi: 10.1002/j.1550-8528.1993.tb00028.x. [DOI] [PubMed] [Google Scholar]
- 34.Marcus RL, Addison O, Kidde JP, Dibble LE, Lastayo PC. Skeletal muscle fat infiltration: impact of age, inactivity, and exercise. The journal of nutrition, health & aging. 2010;14:362–366. doi: 10.1007/s12603-010-0081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marin P, Rebuffe-Scrive M, Smith U, Bjorntorp P. Glucose uptake in human adipose tissue. Metabolism: clinical and experimental. 1987;36:1154–1160. doi: 10.1016/0026-0495(87)90242-3. [DOI] [PubMed] [Google Scholar]
- 36.Maury E, Brichard SM. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Molecular and cellular endocrinology. 2010;314:1–16. doi: 10.1016/j.mce.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 37.Min W, Freudiger CW, Lu S, Xie XS. Coherent nonlinear optical imaging: beyond fluorescence microscopy. Annual review of physical chemistry. 2011;62:507–530. doi: 10.1146/annurev.physchem.012809.103512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minteer DM, Gerlach JC, Marra KG. Bioreactors Addressing Diabetes Mellitus. Journal of diabetes science and technology. 2014;8:1227–1232. doi: 10.1177/1932296814548215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minteer DM, Young MT, Lin YC, Over PJ, Rubin JP, Gerlach JC, Marra KG. Analysis of type II diabetes mellitus adipose-derived stem cells for tissue engineering applications. Journal of tissue engineering. 2015;6:2041731415579215. doi: 10.1177/2041731415579215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittendorfer B. Origins of metabolic complications in obesity: adipose tissue and free fatty acid trafficking. Current opinion in clinical nutrition and metabolic care. 2011;14:535–541. doi: 10.1097/MCO.0b013e32834ad8b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nam SY, Chung E, Suggs LJ, Emelianov SY. Combined Ultrasound and Photoacoustic Imaging to Noninvasively Assess Burn Injury and Selectively Monitor a Regenerative Tissue-Engineered Construct. Tissue Engineering - Part C: Methods. 2015;21:557–566. doi: 10.1089/ten.tec.2014.0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nan X, Cheng JX, Xie XS. Vibrational imaging of lipid droplets in live fibroblast cells with coherent anti-Stokes Raman scattering microscopy. Journal of lipid research. 2003;44:2202–2208. doi: 10.1194/jlr.D300022-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Palero JA, de Bruijn HS, van der Ploeg van den Heuvel A, Sterenborg HJ, Gerritsen HC. Spectrally resolved multiphoton imaging of in vivo and excised mouse skin tissues. Biophysical journal. 2007;93:992–1007. doi: 10.1529/biophysj.106.099457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pezacki JP, Blake JA, Danielson DC, Kennedy DC, Lyn RK, Singaravelu R. Chemical contrast for imaging living systems: molecular vibrations drive CARS microscopy. Nature chemical biology. 2011;7:137–145. doi: 10.1038/nchembio.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popov KI, Pegoraro AF, Stolow A, Ramunno L. Image formation in CARS and SRS: effect of an inhomogeneous nonresonant background medium. Optics letters. 2012;37:473–475. doi: 10.1364/OL.37.000473. [DOI] [PubMed] [Google Scholar]
- 46.Quinn KP, Bellas E, Fourligas N, Lee K, Kaplan DL, Georgakoudi I. Characterization of metabolic changes associated with the functional development of 3D engineered tissues by noninvasive, dynamic measurement of individual cell redox ratios. Biomaterials. 2012;33:5341–5348. doi: 10.1016/j.biomaterials.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinn KP, Sridharan GV, Hayden RS, Kaplan DL, Lee K, Georgiakoudi I. Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Scientific Reports. 2013:3. doi: 10.1038/srep03432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. G, Kahn CR. Adipose Tissue Biology. Springer Science & Business Media; 2011. pp. 93–95. [Google Scholar]
- 49.Salabei JK, Gibb AA, Hill BG. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nature protocols. 2014;9:421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santini F, Marzullo P, Rotondi M, Ceccarini G, Pagano L, Ippolito S, Chiovato L, Biondi B. Mechanisms in endocrinology: the crosstalk between thyroid gland and adipose tissue: signal integration in health and disease. European journal of endocrinology / European Federation of Endocrine Societies. 2014;171:R137–152. doi: 10.1530/EJE-14-0067. [DOI] [PubMed] [Google Scholar]
- 51.Sikder S, Reyes JM, Moon CS, Suwan-apichon O, Elisseeff JH, Chuck RS. Noninvasive mitochondrial imaging in live cell culture. Photochemistry and photobiology. 2005;81:1569–1571. doi: 10.1562/2005-06-18-RC-580. [DOI] [PubMed] [Google Scholar]
- 52.Sjostrom L. Fatty acid synthesis de novo in adipose tissue from obese subjects on a hypercaloric high-carbohydrate diet. Scandinavian journal of clinical and laboratory investigation. 1973;32:339–349. doi: 10.3109/00365517309084357. [DOI] [PubMed] [Google Scholar]
- 53.Slipchenko MN, Le TT, Chen H, Cheng JX. High-speed vibrational imaging and spectral analysis of lipid bodies by compound Raman microscopy. The journal of physical chemistry. B. 2009;113:7681–7686. doi: 10.1021/jp902231y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Squier J, Muller M, Brakenhoff G, Wilson KR. Third harmonic generation microscopy. Optics express. 1998;3:315–324. doi: 10.1364/oe.3.000315. [DOI] [PubMed] [Google Scholar]
- 55.Stanford KI, Middelbeek RJ, Townsend KL, An D, Nygaard EB, Hitchcox KM, Markan KR, Nakano K, Hirshman MF, Tseng YH, Goodyear LJ. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest. 2013;123:215–223. doi: 10.1172/JCI62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Than A, He HL, Chua SH, Xu D, Sun L, Leow MK, Chen P. Apelin Enhances Brown Adipogenesis and Browning of White Adipocytes. J Biol Chem. 2015;290:14679–14691. doi: 10.1074/jbc.M115.643817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trayhurn P, Beattie JH. Physiological role of adipose tissue: white adipose tissue as an endocrine and secretory organ. The Proceedings of the Nutrition Society. 2001;60:329–339. doi: 10.1079/pns200194. [DOI] [PubMed] [Google Scholar]
- 58.Trayhurn P, Wood IS. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br J Nutr. 2004;92:347–355. doi: 10.1079/bjn20041213. [DOI] [PubMed] [Google Scholar]
- 59.Vaughan M, Steinberg D. Effect of Hormones on Lipolysis and Esterification of Free Fatty Acids during Incubation of Adipose Tissue in Vitro. J Lipid Res. 1963;4:193–199. [PubMed] [Google Scholar]
- 60.Vermette M, Trottier V, Menard V, Saint-Pierre L, Roy A, Fradette J. Production of a new tissue-engineered adipose substitute from human adipose-derived stromal cells. Biomaterials. 2007;28:2850–2860. doi: 10.1016/j.biomaterials.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 61.Ward A, Quinn KP, Bellas E, Georgakoudi I, Kaplan DL. Noninvasive Metabolic Imaging of Engineered 3D Human Adipose Tissue in a Perfusion Bioreactor. PLoS ONE. 8:2013. doi: 10.1371/journal.pone.0055696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue R, Lynes MD, Dreyfuss JM, Shamsi F, Schulz TJ, Zhang H, Huang TL, Townsend KL, Li Y, Takahashi H, Weiner LS, White AP, Lynes MS, Rubin LL, Goodyear LJ, Cypess AM, Tseng YH. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat Med. 2015;21:760–768. doi: 10.1038/nm.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nature biotechnology. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]