Abstract
The goal of untargeted lipidomics is to have high throughput, yet comprehensive and unambiguous identification and quantification of lipids. Novel stationary phases in LC separation and new mass spectrometric instruments capable of high mass resolving power and faster scanning rate are essential to achieving this goal. In this work, 4 reversed phase LC columns coupled with a high field quadrupole orbitrap mass spectrometer (Q Exactive HF) were thoroughly compared using complex lipid standard mixture and rat plasma and liver samples. A good separation of all lipids was achieved in 24 min of gradient. The columns compared include C30 and C18 functionalization on either core–shell or totally porous silica particles, with size ranging from 1.7 to 2.6 μm. Accucore C30 column showed the narrowest peaks and highest theoretical plate number, and excellent peak capacity and retention time reproducibility (<1% standard deviation). As a result, it resulted in 430 lipid species identified from rat plasma and rat liver samples with highest confidence. The high resolution offered by the up-front RPLC allowed discrimination of cis/trans isomeric lipid species, and the high field orbitrap mass spectrometer afforded the clear distinction of isobaric lipid species in full scan MS and the unambiguous assignment of sn-positional isomers for lysophospholipids in MS/MS. Taken together, the high efficiency LC separation and high mass resolving MS analysis are very promising tools for untargeted lipidomics analysis.
Keywords: Lipidomics, Reverse-phase liquid chromatography, Accucore C30 column, Q Exactive HF, Mass spectrometry, Untargeted analysis, LipidSearch
1. Introduction
Lipids are small molecules of great importance due to their functions in energy storage, biological membrane structure and signal transduction [1–3]. Dysregulations of lipids have been related with many diseases and the interest for their study has grown in the past decade. In this respect, Lysophosphatidic acid (LPA) has been linked with cancer, since it has been found to stimulate cell proliferation, migration and survival by acting on its cognate G-protein-coupled receptors [2]. Insulin resistance and type 2 diabetes have been associated with a clustering of interrelated plasma lipid and lipoprotein abnormalities, which include reduced HDL cholesterol, a predominance of small dense LDL particles, elevated triglyceride levels, and increased risk of atherosclerosis and coronary heart disease [4]. In addition, dysregulated lipid metabolism has been involved in neurological disorders [5]. For example, Alzheimer’s disease has been linked with aberrant cholesterol metabolism [6], while the abnormal glycolipid metabolism was shown to be associated with Parkinson’s disease [7].
Different methods have been developed for global lipidomic analysis [8]. Lipids, particularly phospholipids and glycerolipids, generate lipid-class characteristic fragment ions and neutral losses when the molecular ions are fragmented under low energy collision induced dissociation in a tandem mass spectrometer. Based on this, various direct infusion (shotgun) tandem mass spectrometric (MS) methods have been developed for quickly profiling lipid species [9–11]. However, the omission of chromatographic separation of lipid samples in these methods make them incapable of differentiating isobaric and isomeric lipids; in addition, ion suppression could also be an issue if lipid class based fractionation is not performed prior to mass spectrometric analysis [8]. To overcome these limitations, new developments in this technique have been made recently, including novel multiplexed approach for extraction of lipids, implementation of multiple informative dimensions for MS interrogation and the development of new bioinformatics approaches for enhanced identification and quantitation [11].
Chromatographic methods have also been used widely for separation of lipids. In this case, thin layer chromatography, solid phase extraction and normal phase liquid chromatography (LC) have been used for purification and fractionation of different lipid classes, while gas chromatography (GC) and reversed phase LC for analysis of the different lipid molecular species. In particular, reversed phase LC separates lipid molecules based on the interaction between the hydrophobic stationary phase and the hydrophobicity of fatty acyls (including the number and position of unsaturation), as well as the polarity of lipid head group [12]. Among the various stationary phases, C18 has been the mostly used packing material, with particle sizes ranging from 5 to sub-2 μm for enhanced resolving power. To overcome the ultrahigh backpressure resulted from porous sub-2 μm particles, core–shell particles has gained popularity in recent years for their reduced resistance to mass transfer and high particle uniformity [13,14]. For example, Witting et al. [15] compared sub-2 μm core–shell columns and sub-2 μm porous columns for in depth lipidomic study of Caenorhabditis elegans, and demonstrated that the Cortecs C18—a core–shell column, showed superior performance in case of chromatographic peak characteristics (plate number, number of detected lipid features) in comparison with the others. In addition, C18 particles with low level surface charge has also been used to enhance the separation and increase the loading capacity [16]. For the separation of cis and trans phospholipid isomers, Bird et al. [12] observed that the column incorporating charged surface produced sharper peaks when compared with a fused-core or a fully porous core non-charged C18 columns.
Although C30 stationary phase is much less used in untargeted profiling of lipidome, its potential has been demonstrated in separation of phospholipids [9]. The aim of the present work is to compare different reversed phase columns, including core–shell and porous particles, with C18 and C30 stationary phases, for the most reproducible, high resolving power, and comprehensive lipidomic analysis. The evaluation of column performance was based on both complex lipid standard mixture and complex biological samples, i.e. rat liver and plasma lipid extracts. The optimized LC method, when combined with the newer generation, high resolution, fast scanning mass spectrometer (Q Exactive HF) has demonstrated to be powerful in global lipidomic analysis.
2. Experimental
2.1. Chemicals and columns
Ammonium formate, isopropanol (IPA) and water (Optima® LC–MS grade) were obtained from Fisher Scientific (Fair Lawn, NJ). Acetonitrile (ACN) and formic acid were of LC/MS quality and acquired from Fluka (Germany). Chloroform (HPLC grade) and methanol (LC/MS grade) were provided by Merck (Germany) and EMD Chemicals (Gibbstown, NJ) respectively. Columns used (Table 1) were supplied by Phenomenex (USA), Waters (Ireland) and Thermo Scientific (USA).
Table 1.
Characteristics of the chromatographic columns used in this study.
| Column | Supplier | Phase type | Dimensions (length × ID) (mm) | Pore size (Å) | Particle size (μm) | Pressure (bar) |
|---|---|---|---|---|---|---|
| Kinetex C18 | Phenomenex | Core–shell | 100 × 2.1 | 100 | 1.7 | 400–650 |
| HSS T3C18 | Waters | High strength silica porous particles | 100 × 2.1 | 100 | 1.8 | 380–570 |
| Cortecs C18 | Waters | Core–shell | 100 × 2.1 | 90 | 1.6 | 470–790 |
| Accucore C30 | Thermo | Core–shell | 150 × 2.1 | 150 | 2.6 | 300–500 |
The pressure ranges listed indicate the operating pressures of the LC system at the beginning and end of the analysis.
Phospholipids standards were acquired from Avanti Polar Lipids (Birmingham, AL), while acylglycerols, fatty acids, cholesterol and cholesterol esters were obtained from Nu-Chek-Prep, Inc. (Elysian, MN). Cardiolipin from bovine heart was supplied by Sigma–Aldrich (St. Louis, MO). Standard lipid solutions were prepared by dissolution in ACN/IPA/water (65:30:5, v/v/v) in a concentration of 0.025 μg/μL.
Pierce LTQ Velos ESI Positive Ion Calibration Solution and Pierce LTQ Velos ESI Negative Ion Calibration Solution were provided by Thermo Fisher Scientific (Waltham, MA).
Rat plasma and liver were obtained from BioreclamationIVT (Baltimore, MD), both of them came from the same animal.
2.2. Lipid extraction from rat plasma and liver samples
Prior to lipid extraction, the liver was homogenized by using an Omni TH homogenizer (Omni International, Warrenton, VA). Lipids were extracted in duplicate from 50 μL aliquot of plasma and 0.05 g of liver according to the Folch method [17] under cold conditions (−20 °C) using chloroform/methanol (2:1, v/v) in a 5:1 and 20:1 ratios over the sample volume, respectively. The mixtures were vortexed for 10 s and allowed to stand on ice for 10 min. After being mixed again by vortexing, they were centrifuged at 9279 × g for 10 min. For plasma samples, 200 μL of the organic layers were collected, while for the liver samples, all the organic solvents were collected and filtered through filter paper (Fisher Scientific). Then, the extracts were evaporated to dryness under vacuum. Prior to LC–MS analysis, the lipid extracts were reconstituted in 100 μL of chloroform for plasma samples while in the case of liver samples, the residue was re-dissolved at a final concentration of 0.05 μg/μL. The whole procedure was carried out using glass vials to avoid contaminations from container.
2.3. Chromatographic and mass spectrometric conditions
A Vanquish UHPLC system (ThermoFisher Scientific) with a binary pump and an autosampler was used for this study. The separation was performed using different columns. The column oven temperature suitable for all columns was 40 °C. A generic binary gradient elution was carried out using different ratios of eluents A (ACN:water, 60:40,v/v) and B (IPA:ACN, 90:10, v/v), both containing 10 mM ammonium formate and 0.1% formic acid [18]. A better separation for all the columns studied was achieved using the following gradient: −3 −0 min isocratic elution with 30% B for the equilibration of the column; 0–5 min, 30–43% B; 5–5.1 min, 43–50%; 5.1–14 min, 50–70% B; 14.1–21 min, 70–99% B; 21–24 min, 99% B; 24–24.1 min, 99–30%; 24.1–28 min, 30% B for column washing and equilibration. The separation time for the analytes was 24 min and the total analysis time including column re-equilibration was 31 min. The flow rate was set to 350 μL/min, the temperature of the sample tray was set to 15 °C and the injection volume was 5 μL.
The comparison of the columns was completed using the lipid standard mixture on a TSQ Quantiva triple quadrupole mass spectrometer (ThermoFisher Scientific) operated in full scan mode. All MS experiments were performed in positive and negative ion modes using a Heated Electro Spray Ionization (HESI) source. Tune parameters were optimized using PC(18:1(9Z)/18:1(9Z)), Cer(d18:1/18:1(9Z)), PG(18:1(9Z)/18:1(9Z)) in both negative and positive ion modes, and TG(16:0/16:0/16:0), DG(18:1(9Z)/18:1(Z)/0:0) and 18:1 choresteryl ester in positive ion mode. Due to the high flow rate used, a sweep cone was used for best performance and protection of the system. The spray voltage for positive ion mode was 3 kV, while 2 kV was used for negative ion mode. The flow rates of sheath, auxiliary and sweep gases were 20, 7 and 1 (arbitrary units), respectively, for both ionization modes. Ion transfer tube and vaporizer temperatures were at 350 and 400 °C, respectively. Scan range was 100–1900 m/z, while the scan rate was 1000 amu/s. The resolution was set at 0.7 (FWHM).
For the untargeted lipidomic analysis, a Q Exactive HF (QEHF) hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific, USA) was employed. All MS experiments were performed in positive and negative ion modes using a HESI source. Tune parameters were optimized using PC(18:1(9Z)/18:1(9Z)), Cer(d18:1/18:1(9Z)), PG(18:1(9Z)/18:1(9Z)) in both negative and positive ion modes, and TG(16:0/16:0/16:0), DG(18:1(9Z)/18:1(9Z)/0:0) and 18:1 cholesteryl ester in positive ion mode. A sweep cone was also used in this system. The flow rates of sheath gas and sweep gas for both polarities were adjusted to 20 and 1 (arbitrary units), while the auxiliary gas rate was 5 for positive and 7 for negative. For both ionization modes, the spray voltage, the capillary temperature and the heater temperature were maintained at 3 kV, 350 °C and 400 °C, respectively. The S-Lens RF level was set at 50. The Orbitrap mass analyzer was operated at a resolving power of 120,000 in full-scan mode (scan range: 114–1700 m/z; automatic gain control target: 1e6) and of 30,000 in the Top20 data-dependent MS2 mode (HCD fragmentation with stepped normalized collision energy: 25 and 30 in positive ion mode, and 20, 24 and 28 in negative ion mode; Injection time: 100 ms; Isolation window: 1m/z; automatic gain control target: 1e5) with dynamic exclusion setting of 15.0 s.
The QEHF was externally calibrated within a mass accuracy of 1 ppm every week using the Thermo Scientific Pierce LTQ Velos ESI Negative Ion Calibration Solution (mixture containing sodium dodecyl sulfate, sodium taurocholate and Ultramark 1621 in an acetonitrile–methanol–acetic solution) and the Thermo Scientific Pierce LTQ Velos ESI Positive Ion Calibration Solution (containing a mixture of caffeine, tetra peptide MRFA, Ultramark 1621, and N-butylamine in an acetonitrile–methanol–acetic solution).
2.4. Data processing
All MS data were acquired and processed using the software package Xcalibur 3.0. LipidSearch software version 4.1 (Mitsui Knowledge Industry, Tokyo, Japan) was used for lipid molecular species identification and quantification in complex biological samples. Key processing parameters were: target database: QExactive; precursor tolerance: 5 ppm; product tolerance: 5 ppm; product ion threshold: 5%; m-score threshold: 1; Quan m/z tolerance: ±5 ppm; Quan RT (retention time) range: ±0.5 min; use of main isomer filter and ID quality filters A, B, C and D; Adduct ions: +H and +NH4 for positive ion mode, and −H, +HCOO and −2H for negative ion mode. The lipid classes selected for the search were: LPC (lysophosphatidylcholine), PC (phosphatidylcholine), LysoPE (lysophosphatidylethanolamine), PE (phosphatidylethanolamine), LysoPS (lysophosphatidylserine), PS (phosphatidylserine), LysoPG (lysophosphatidylglycerol), PG (phosphatidylglycerol), LysoPI (lysophosphatidylinositol), PI (phosphatidylinositol), LysoPA (lysophosphatidic acid), PA (phosphatidic acid), SM (sphingomyelin), MG (monoacylglycerol), DG (diacylglycerol), TG (triacylglycerol), CL (cardiolipin), So (sphingosine), Cer (ceramides), Che (cholesterol ester). The same lipid annotations within ±0.1 min were merged into the aligned results. These parameters were first optimized using lipid standards before being applied to untargeted lipidomic analysis. Positions of the fatty acyls in lipid species identified from real samples were also manually confirmed according to the well-recognized rules established by tandem mass spectrometry [19,20], where the fatty acyls were either separated by “/” when the sn-position can be confirmed, or by “,” when it is unambiguous for the assignment. Free fatty acids have not been considered in the identification because the main fragment ion resulted from CO2 loss requires NCE > 50. Consequently, LipidSearch will not be able to identify free fatty acids using productions if [M−H-44]− is not in the product ion spectrum.
3. Results and discussion
3.1. Optimization of chromatographic conditions
By using a complex mixture of lipid standards that represents different classes of lipids with long and short chain fatty acyls (Table 2), we carefully optimized the chromatographic conditions to achieve the best separation for the maximum number of lipid species. Four commercially available reversed phase columns with either core–shell or porous particles from three manufacturers were evaluated (Table 1). The chosen gradient allowed a better separation between lipid standards (Table 2) for all the columns studied. In addition, the first 3 min of column equilibration prior to sample injection allowed the same column conditions to be applied for all runs, and the 4 min equilibration time applied at the end of the gradient resulted in minimal sample carryover as confirmed in subsequent analysis of blank samples.
Table 2.
Lipid standards used in the optimization of the LC–MS method conditions.
| Lipid name | Supplier | Catalog number |
Formula | Formula mass (isotopic) | Expected m/z | Ion type |
|---|---|---|---|---|---|---|
| 12:0 Cholesteryl ester | Nu-Chek | CH-800 | C39H68O2 | 568.5219 | 586.5562 | [M+NH4]+ |
| 18:1 Cholesteryl ester | Nu-Chek | CH-828 | C45H78O2 | 650.6002 | 668.6345 | [M+NH4]+ |
| 20:0 Cholesteryl ester | Nu-Chek | CH-819 | C47H84O2 | 680.6472 | 698.6815 | [M+NH4]+ |
| Cholesterol | Nu-Chek | CH-800 | C27H46O | 386.3549 | 404.3892 | [M+NH4]+ |
| PC(18:1(9Z)/16:0) | Avanti | 850475C | C42H82NO8P | 759.5778 | 760.5856 | [M+H]+ |
| PC(18:1(9Z)/18:1(9Z)) | Avanti | 850375C | C44H84NO8P | 785.5934 | 786.6012 | [M+H]+ |
| PC(18:2(9Z,12Z)/18:2(9Z,12Z)) | Avanti | 850385C | C44H80NO8P | 781.5621 | 782.57 | [M+H]+ |
| PC(22:1(13Z)/22:1(13Z)) | Avanti | 850398C | C52H100NO8P | 897.7186 | 898.7264 | [M+H]+ |
| PC(12:0/12:0) | Avanti | 850335C | C32H64NO8P | 621.4769 | 622.4448 | [M+H]+ |
| LysoPC(14:0/0:0) | Avanti | 855575C | C22H46NO7P | 467.3011 | 468.309 | [M+H]+ |
| LysoPC(22:0/0:0) | Avanti | 855779C | C30H62NO7P | 579.4264 | 580.4342 | [M+H]+ |
| PC(P-18:0/18:1(9Z)) | Avanti | 852467C | C44H86NO7P | 771.6142 | 772.622 | [M+H]+ |
| PC(P- 18:0/20:4(5Z,8Z,11Z,14Z) |
Avanti | 852469C | C46H84NO7P | 793.5985 | 794.6063 | [M+H]+ |
| PE(12:0/12:0) | Avanti | 850702P | C29H58NO8P | 579.39 | 580.3978 | [M+H]+ |
| PE(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/ 22:6(4Z,7Z,10Z,13Z,16Z,19Z)) |
Avanti | 850797C | C49H74NO8P | 835.5152 | 836.523 | [M+H]+ |
| PE(18:1(9Z)/18:1(9Z)) | Avanti | 850725C | C41H78NO8P | 743.5465 | 744.5543 | [M+H]+ |
| LysoPE(18:1(9Z)/0:0) | Avanti | 846725C | C23H46NO7P | 479.3012 | 480.309 | [M+H]+ |
| PE(P-18:0/18:1(9Z)) | Avanti | 852758C | C41H80NO7P | 729.567 | 730.5748 | [M+H]+ |
| PS(14:0/14:0) | Avanti | 840033P | C34H66NO10P | 679.4424 | 680.4502 | [M+H]+ |
| PS(18:1(9Z)/18:1(9Z)) | Avanti | 840035C | C42H78NO10P | 787.5363 | 788.5441 | [M+H]+ |
| LysoPS(18:1(9Z)/0:0) | Avanti | 858143C | C24H46NO9P | 523.291 | 524.2988 | [M+H]+ |
| PG(18:1(9Z)/18:1(9Z)) | Avanti | 840475C | C42H79O10P | 774.5411 | 775.5489 | [M+H]+ |
| PG(14:0/14:0) | Avanti | 840445P | C34H67O10P | 666.4472 | 667.455 | [M+H]+ |
| PG(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/ 22:6(4Z,7Z,10Z,13Z,16Z,19Z)) |
Avanti | 840492C | C50H75O10P | 866.5098 | 867.5176 | [M+H]+ |
| LysoPG(14:0/0:0) | Avanti | 858120P | C20H41O9P | 456.2488 | 457.2566 | [M+H]+ |
| LysoPG(18:1(9Z):0:0) | Avanti | 858125C | C24H47O9P | 510.2958 | 511.3036 | [M+H]+ |
| PI(16:0/18:1(9Z)) | Avanti | 850142P | C43H81O13P | 836.5415 | 837.5493 | [M+H]+ |
| LysoPI(16:0/0:0) | Avanti | 850102P | C25H49O12P | 572.2962 | 573.304 | [M+H]+ |
| LysoPI(20:4(5Z,8Z,11Z,14Z)/0:0) | Avanti | 850105P | C29H49O12P | 620.2961 | 621.304 | [M+H]+ |
| PA(18:1(9Z)/18:1(9Z)) | Avanti | 840875C | C39H73O8P | 700.5043 | 669.4965 | [M−H]− |
| Cer(d18:1/18:1(9Z)) | Avanti | 860519C | C36H69NO3 | 563.5277 | 564.5356 | [M+H]+ |
| Cer(d18:1/12:0) | Avanti | 860512P | C30H59NO3 | 481.4495 | 482.4573 | [M+H]+ |
| FA(12:0) | Nu-Chek | N-12-A | C12H24O2 | 200.1776 | 199.1698 | [M−H]− |
| FA(18:1(9Z)) | Nu-Chek | U-46-A | C18H34O2 | 282.2559 | 281.2481 | [M−H]− |
| FA(26:0) | Sigma–Aldrich | H0388 | C26H52O2 | 396.3967 | 395.3889 | [M−H]− |
| MG(12:1(11Z)/0:0/0:0) | Nu-Chek | M-199 | C15H28O4 | 272.1987 | 290.2331 | [M+NH4]+ |
| MG(18:1(9Z)/0:0/0:0) | Sigma–Aldrich | M7765 | C21H40O4 | 356.2927 | 374.327 | [M+NH4]+ |
| MG(22:1/0:0/0:0) | Nu-Chek | M-304 | C25H48O4 | 412.3552 | 430.3896 | [M+NH4]+ |
| DG(12:1(11Z)/12:1(11Z)/0:0) | Nu-Chek | D-196 | C27H48O5 | 452.3502 | 470.3845 | [M+NH4]+ |
| DG(14:0/14:0/0:0) | Nu-Chek | D-141 | C31H60O5 | 512.4441 | 530.4784 | [M+NH4]+ |
| DG(18:1(9Z)/18:1(9Z)/0:0) | Nu-Chek | D-236 | C39H72O5 | 620.5379 | 638.5723 | [M+NH4]+ |
| DG(22:1(13Z)/22:1(13Z)/0:0) | Nu-Chek | D-301 | C47H88O5 | 732.6631 | 750.6975 | [M+NH4]+ |
| TG(12:1(11Z)/12:1(11Z)/12:1(11Z)) | Nu-Chek | T-195 | C39H68O6 | 632.5016 | 650.5359 | [M+NH4]+ |
| TG(14:0/14:0/14:0) | Nu-Chek | T-140 | C45H86O6 | 722.6424 | 740.6768 | [M+NH4]+ |
| TG(16:0/16:0/16:0) | Nu-Chek | T-150 | C51H98O6 | 806.7363 | 824.7706 | [M+NH4]+ |
| TG(22:1(13Z)/22:1(13Z)/22:1(13Z)) | Nu-Chek | T-300 | C69H128O6 | 1052.971 | 1071.005 | [M+NH4]+ |
| CL(18:2)4 | Sigma–Aldrich | C1649 | C81H142O17 P2 | 1448.972 | 1447.964 | [M−H]− |
| CL(18:2)3 (18:1) | C81H144O17P2 | 1450.988 | 1449.980 | [M−H]− | ||
| CL(18:2)2 (18:1)2 | C81H146O17P2 | 1453.004 | 1451.996 | [M−H]− | ||
| SM(d18:1/12:0) | Avanti | 860583P | C35H71N2O6P | 646.5049 | 647.5127 | [M+H]+ |
| SM(d18:1/24:1(15Z)) | Avanti | 860593P | C47H93N2O6P | 812.6771 | 813.6849 | [M+H]+ |
Due to the high proportion of IPA in mobile phase B, and its high viscosity, we evaluated whether back pressure would pose a problem for these columns with small internal diameter and small size particles. Different flow rates were tested at 200, 300, 350, 400 and 500 μL/min. The highest flow presented problems of overpressure in all the columns tested during the incremental increase of mobile phase B, except for the Accucore C30 column. The same problem was observed at 400 μL/min when using Kinetex C18 and Cortecs C18 columns. Raising column temperature to 40 °C from room temperature helped reducing back pressure, however the temperature was not further increased because two columns, Cortecs and HSST3 can only accept operating temperature up to 45 °C. Peak tailing was observed when the flow rate was decreased to 200 μL/min. Finally, 350 μL/min was selected for all the columns to yield narrower peaks and best resolution. The operating pressures of the LC system at the beginning and end of the analysis for all evaluated columns are shown in Table 1.
Under the optimized gradient and chromatographic conditions, lysoglycerophospholipids and fatty acids (more polar lipids) eluted at the beginning under a higher concentration of mobile phase A, while higher ratios of mobile phase B at the end of the gradient resulted in elution of the most hydrophobic and larger lipids, such as cholesterol esters and triacylglycerols. The glycerophospholipids, sphingolipids, monoacylglycerols and diacylglycerols were eluted in the middle of the gradient.
We also tested three other columns, i.e. CSH C18 (100 × 2.1 mm, 1.7 μm, Waters), Cortecs C18+ (100 × 2.1 mm, 1.6 μm, Waters) and Accucore Vanquish C18 (100 × 2.1 mm, 1.5 μm, ThermoFisher Scientific). However, the results obtained with these columns are not presented here due to their overall underperformance, in particular in the region of the chromatogram where glycerophospholipid species elute.
3.2. Optimization of MS conditions
A TSQ Quantiva triple quadrupole mass spectrometer operated in full scan mode was coupled to the Vanquish UHPLC system for comparison of the columns using the lipid standard mixture listed in Section 2.3. Different tune parameters were optimized for the best operating conditions. The optimal vaporizer tempera-ture observed was 400–450©C, but no significant differences wereobserved between them and 400©C was chosen. At a vaporizer tem-perature of 500©C, the signals of TG species started to decrease, anddisappeared at 550©C due to thermal instability. Different sprayvoltages influenced the base line of total ion chromatogram. Underboth ionization modes, higher spray voltages were observed toimprove S/N and decrease the low-frequency oscillations of thebaseline that was caused by spray instability. A spray voltage of3 kV was chosen for positive and 2 kV for negative ion mode, andno significant improvements were observed when higher voltageswere used. The optimum flow rates of sheath, auxiliary and sweepgases were 20, 7 and 1 (arbitrary units), respectively, for both ion-ization modes. No changes in the signal were observed at highervalues of sweep gas. However, the signal had a significant reduc-tion when the values of sheath and auxiliary gases were higher than 40 and 12, respectively.
The optimized ionization conditions in the Quantiva MS were applied as the starting settings to optimize the electrospray ionization parameters of the QEHF system, and only slight modifications were deemed necessary as the electrospray interfaces on these two instruments were very similar. A mass resolution of 120 K at m/z 200 was chosen for effective differentiation of isobaric mass peaks, which could be as small as 0.03 mass units (see Fig. S1) [21,22]. The 60 K setting is the minimum mass resolution required for clear identification of both [M+H]+ ions (m/z 478.32 and 478.29 of the two lysophospholipid species, LysoPC(16:1p/0:0) and LysoPE (18:2)), which overlapped partially chromatographically at ~2.2 min. For unambiguous lipid molecular species identification and quantification, high mass resolution is essential to differentiate lipid species between co-eluting isobaric molecular ions in the complex biological samples.
The optimized ionization conditions in the Quantiva MS were applied as the starting settings to optimize the electrospray ionization parameters of the QEHF system, and only slight modifications were deemed necessary as the electrospray interfaces on these two instruments were very similar. A mass resolution of 120 K at m/z 200 was chosen for effective differentiation of isobaric mass peaks, which could be as small as 0.03 mass units (see Fig. S1) [21,22]. The 60 K setting is the minimum mass resolution required for clear identification of both [M+H]+ ions (m/z 478.32 and 478.29 of the two lysophospholipid species, LysoPC(16:1p/0:0) and LysoPE (18:2)), which overlapped partially chromatographically at ~2.2 min. For unambiguous lipid molecular species identification and quantification, high mass resolution is essential to differentiate lipid species between co-eluting isobaric molecular ions in the complex biological samples.
MS/MS data were acquired using a data dependent top-20 method with a mass resolution setting of 30 K. Even under this high mass resolution setting in the MS/MS scans, the cycle time of 0.04 min still allowed on average 9 MS survey scans performed on a typical chromatographic peak (mean of 0.39 min at the base, see Tables S1–S4), which makes accurate MS-level peak integration and therefore lipid quantification possible. An isolation width of 1 m/z provided a more pure precursor ion, which allowed a higher quality MS/MS spectrum for accurate identification of individual lipid species based on product ions. This was achieved as a result of the segmented quadrupole in the QEHF, which allows a narrow precursor ion isolation width with minimum transmission losses, besides of the very fast isolation and fragmentation capabilities [23].
It is known that fragmentation efficiency of lipid ions is dependent upon the collision energy applied during collision induced dissociation, and optimal collision energy varies with lipid classes and fatty acyl compositions of lipids. To this end, stepped collision energy has been used in both ionization modes. By combining fragmentation at low and high energy to give a broader range of fragment ions, all the lipid classes can be effectively fragmented under the same MS/MS experimental settings, which circumvents the need for multiple LC–MS/MS runs each with a different collision energy setting optimized for each lipid class.
3.3. Column comparison using lipid standards
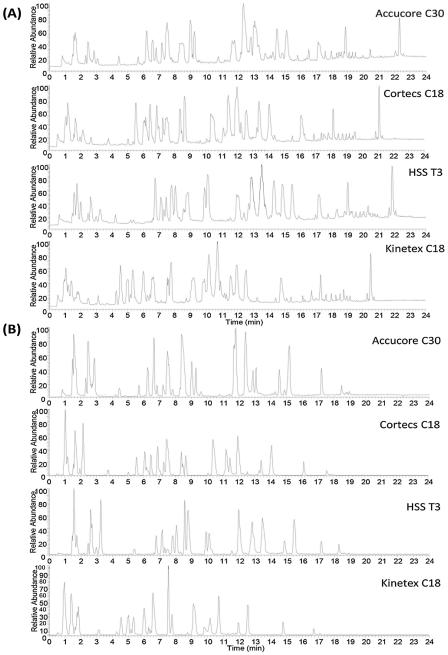
Initial evaluation of the selected columns was conducted using a mix of different lipid standards (Table 2), under identical gradient conditions in positive and negative ion modes (Fig. 1). All columns presented comparable time of analysis for the standard mixture used, peak elution started ~1 min after sample injection and the last lipid standard eluted between 17.00–18.91 min in negative ion mode and between 20.48–22.33 min in positive ion mode. Kinetex C18 column showed the lowest retentivity for the standard lipids.
Fig. 1.
Total ion chromatograms of lipid standard mixture obtained under positive (A) and negative (B) ion modes. Identity and retention time of each peak are listed in Tables S1–S4.
Retention time, peak area, height and width were extracted for each standard lipid (Tables S1–S4), and their means and standard deviations (SD) were used as the basis for evaluating the performance of each column. Remarkably, Accucore C30, Cortecs C18 and HSS T3 columns presented excellent retention time reproducibility from several injections of the lipid standard mixture, with no change in retention time in most of the cases, with the standard deviation always <1%. The only exception is the Kinetex C18 column, in positive ion mode one peak at 0.91 min had retention time SD of 3.30%, which is likely due to the co-elution of three lysophospholipids, i.e. LysoPG(14:0/0:0) + LysoPI(20:4/0:0) + LysoPC(14:0/0:0) at the beginning of the gradient (Table S4). Witting et al. [15] compared the Cortecs C18 and Kinetex C18 columns, and their results also showed that the SD of retention time were higher when using Kinetex C18. However, it looks like a better time reproducibility was obtained in the present work than that of Witting et al. [15], since most of retention time differences were 0.00 min.
In terms of the peak widths, Accucore C30 yielded the narrowest peaks from the four columns investigated with a range of 0.58–0.10 min in positive ion mode, followed by HSS T3 (0.66–0.16 min), Cortecs C18 (0.63–0.18 min) and Kinetex C18 (0.75–0.16 min). However, in negative ion mode, Kinetex C18 had the narrowest peaks with a range of 0.69–0.11 min, followed by Cortecs C18 (0.83–0.13 min), Accucore C30 (0.93–0.16 min) and HSS T3 (0.88–0.23 min), but the average peak widths on Accucore C30 and Kinetex C18 were the same (0.39 min) and higher on Cortecs C18 (0.41 min) and HSS T3 (0.44 min).
Theoretical plate numbers for the different lipid standards are also reported in Tables S1–S4 to measure the separation efficiency of the different columns. The number of theoretical plates (N) was calculated using the following formula: N = 16(tR/w)2 , where tR is the peak retention time and w is the peak width. In general, it was observed that the number of theoretical plates increased with the fatty acid chain length for the same class of lipids, for example, a plate number of 9589 was obtained for the peak of PE(12:0/12:0) using the Accucore C30 column in negative ion mode, while it was 29383 for PE(18:1(9Z)/18:1(9Z)) species. In positive ion mode, Accucore C30 presented the highest number of plates for the lysophospholipids and PE species, followed by the HSS T3 column. The number of plates for SM and Cer species was comparable for Accucore C30, Cortecs C18 and HSS T3. The corresponding peaks of those species on Kinetex C18 were broader, showed a lower retention time and as a consequence, a smaller plate number than the other three columns. For PC species, all columns performed equally well. HSS T3 and Kinetex C18 yielded two separate peaks for PS(14:0/14:0) and PG(14:0/14:0) species, but only one peak for PS(18:1(9Z)/18:1(9Z)) and PG(18:1(9Z)/18:1(9Z)). The Cortecs C18 and Accucore C30 columns also presented one peak for the above PS and PG species containing the same fatty acids and had smaller plate number than the other two columns. Interestingly, HSS T3 showed the highest number of theoretical plates for DG and TG species, except for TG 22:1. Regarding to the results obtained in negative ion mode, Accucore C30 performed better than the other columns, showing a higher plate number for PC, PE, plasmalogen, Cer, PI, PG, PS and SM species. Although PI can ionize in either negative or positive ion mode, negative ion mode is used more frequently because of the carboxylate anions of the fatty acids and unique fragments produced by the polar head group [24]. All columns performed equally for fatty acids and for lysophospholipids, except that the highest number of plates for LysoPC(22:0/0:0) was obtained from Accucore C30, and HSS T3 presented the highest number for LysoPG(14:0/0:0) and LysoPI(20:4/0:0). In the case of CL species, no clear trend was observed. Overall, using the lipid standard mixture, Accucore C30 showed higher performance than the other columns in negative ion mode, but in positive ion mode, Accucore C30 and HSS T3 were comparable in performance depending on the type of lipid studied.
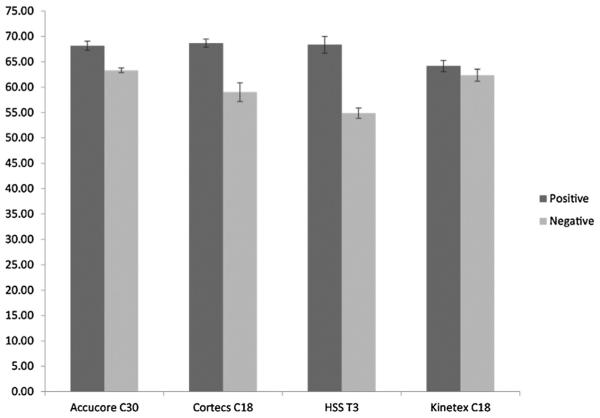
Peak capacity is a good measurement of separation efficiency and can be calculated from the peak widths w according to the method of Neue [25]. Fig. 2 shows a bar plot of the peak capacity for all columns, bar height representing the mean of five injections and error bars showing ±standard deviation. It can be observed that the peak capacity was higher in positive ion mode than that in negative ion mode. Besides, Accucore C30, Cortecs C18 and HSS T3 column presented similar peak capacity (~68) in positive ion mode and all higher than Kinetex C18. However, in negative ion mode, the highest peak capacities were obtained with Accucore C30 (peak capacity = 63.3) and Kinetex C18 (peak capacity = 62.4).
Fig. 2.
Bar plot showing the peak capacity obtained from the lipid standard mixture (see Tables 1, S1–S4) for every column under positive and negative ion modes. Bar heights represent the mean of five injections and error bars show ± standard deviation.
The ionization efficiency of different lipid species within the same polar lipid class is mainly dependent on their identically charged head group, while their differential acyl chains including the length and unsaturation only slightly affect the ionization efficiency under certain conditions [26]. The physicochemical properties of lipid molecules will affect their binding strength to the different stationary phases used in each column. With relatively polar mobile phase at the initial stage of the gradient, this may induce solubility problems in a species-dependent manner that leads to differential ionization efficiency. In addition, the applied gradient can also introduce alterations in ionization efficiency and cause ionization instability during elution [26]. This variation could explain some of the differences observed between the different columns used, since the same gradient was used.
Previously, Gao et al. showed that HSS T3 column has better performance when compared with Jupiter C18 and Waters C8 columns using lipid standards [27]. On the other hand, Damen et al. [28] demonstrated the superior separation of different isomers using the CSH (charged surface hybrid) C18 column in comparison with the HSS T3. Despite its bigger particle size (2.6 μm), Accucore C30 column showed higher performance than the other three columns with sub 2 μm particles. This very likely is due to the stronger interaction between the fatty acids moieties and the C30 stationary phase, while the influence of the fatty acyl chains is greater than that of the polar head group. Although a good separation of lipids can be achieved in reversed-phase chromatography, co-elution of lipids belonging to different classes in reversed-phase separations is quite commonly observed due to the fact that the mechanism of action of lipids is based on their lipophilicity, which is governed by the carbon chain length and the number of double bonds [29]. As an alternative, hydrophilic interaction liquid chromatography (HILIC) can provide a simple and effective means to separate lipids by class, offering a complementary separation technique to the reversed-phase methods [30]. Because of the orthogonality in separation mechanisms between HILIC and RPLC, oftentimes they are coupled for 2D lipid analysis. However, HILIC typically lacks the separating power achievable on RPLC, therefore it is used mostly in the first dimension for fractionation of lipids into classes, while RPLC in the second dimension for further separation of lipids in the same class into molecular species. Although the 2D method is comprehensive in coverage of lipids, it will increase substantially the time for analysis, which is not feasible when hundreds of samples need to be analyzed in a lipidomics study. Recently, one dimension HILIC–ESI–MS method has been used for analysis of phospholipids, good separation of lysophopholipids was achieved, while co-elution of other different lipid classes, like PS and PE, was also observed [31]. Although its performance has greatly improved over other HILIC-based lipid separations, its capability in complete separation of phospholipid classes is less desirable compared with a normal phase column, where no co-elution was achieved using a Chromolith–HPLC–ESI–MS [32]. Another disadvantage of using HILIC is that most of the methods in the literature are not applicable for the quantification of nonpolar lipid classes due to their elution in the void volume [33].
Very limited use of C30 as stationary phase has been reported. For the study of phospholipids from rat liver and PE mixture from bovine brain, the Deverosil C30 (5 μm, 100 mm × 0.3 mm i.d.) column has been applied to separate the minor molecular species [9]. For the profiling and separation of triacylglycerols in palm and canola oils [34], a C30 column showed good separation for several triacylglycerol regioisomers. In addition, a C30 column (YMC Carotenoid 5 μm, 250 × 4.6 mm id) was reported for carotenoid analysis [35].
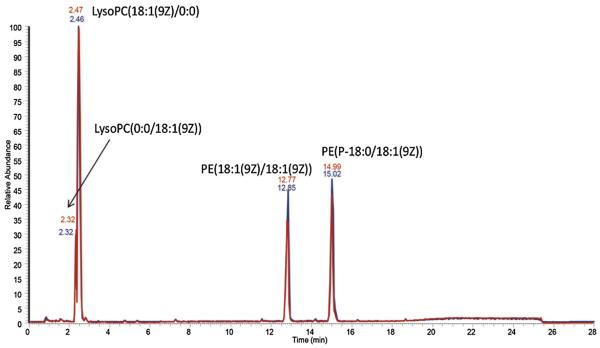
Due to its high performance, Accucore C30 column is also being evaluated for the long-term stability in terms of variations in retention time and peak width. A mixture composed of four standard lipids was used as the quality control sample, and it has been injected periodically onto the column amidst a large batch of biological samples. Fig. 3 showed that the chromatogram obtained for this sample after 250 injections is almost identical to the original chromatogram, proving that this column has the required stability in large scale lipidomics studies.
Fig. 3.
Overlaid chromatograms of lipid standards after 250 injections (red line) using Accucore C30 column in negative ion mode. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Profiling of lipids in rat liver and rat plasma lipid extracts
To further evaluate the performance of different columns in analysis of complex biological samples, we analyzed the lipids extracted from rat liver and rat plasma (Figs. S2 and S3, respectively). Four injections of total lipid extracts from each sample type were performed in every column in both positive and negative ion modes.
Various software have been developed to identify and quantify lipids in global lipidomics analysis, such as LipidSearch [22], LipID [36] and Progenesis QI [37]. The LC–MS/MS data acquired from the rat liver and plasma samples were searched using Lipid-Search, which has a database containing more than 1.5 million lipid ions and their predicted fragment ions. For the mobile phases used in our separation, [M+H]+ and [M+NH4]+ adduct ions were being considered for the precursor ions in positive ion mode, and [M−H]− and [M+HCOO]− for negative ion mode, respectively. Examples of MS/MS spectrum of different lipids from real samples (rat liver and rat plasma) are shown in Figs. S4–S20, which were obtained using HCD fragmentation in either positive or negative and ion modes.
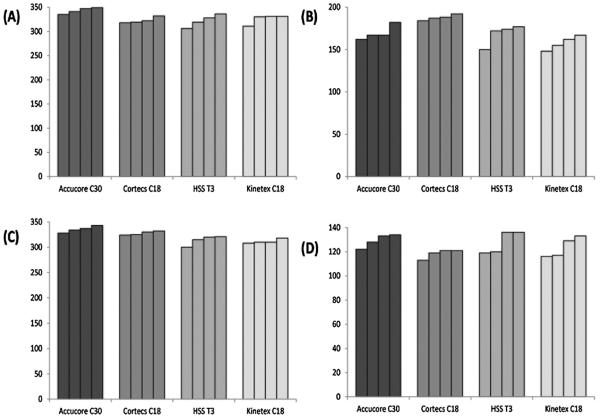
The total number of lipids identified from each replicate of rat liver lipid extract sample is listed in the bar graph of Fig. 4A and B. Accucore C30 column yielded the most number of identifications in positive ion mode. However, Cortecs C18 showed the highest number of lipids identified in negative ion mode, followed by Accucore C30. Similarly, the results obtained for lipid extract samples from rat plasma are shown in Fig. 4C and D for the positive and negative ion mode, respectively. Again, the most lipid species identified were using the Accucore C30 column in positive ion mode. In negative ion mode, on average Accucore C30 still yielded the highest number of identifications, while the lowest was found using Cortecs C18.
Fig. 4.
Number of lipid compounds identified in rat liver (A: positive ion mode, B: negative ion mode) and rat plasma (C: positive ion mode, D: negative ion mode) by LC–MS/MS.
The lipid species identified from the positive and negative ion modes can be aligned using LipidSearch to combine search results, so the total number of lipids identified from each sample under both ionization modes can be obtained without duplicated compound identifications. Table 3 summarizes the total number of lipids identified in rat liver and rat plasma extracts using all the columns, including the ones mentioned in Section 3.1. The highest number of lipids identified in both type of samples was achieved using Accucore C30, followed by Cortecs C18, HSS T3 and Kinetex C18. In comparison, much lower number of identifications was obtained with the other three columns, Cortecs UPLC C18+, Accucore Vanquish C18 and Acquity UPLC CSH C18. The surface charged columns (Cortecs C18+ and CSH) showed the lowest number of identifications. Although having the largest particle size in all of the columns tested, Accucore C30 outperformed all other columns in the number of lipid molecules identified from complex biological samples, which could be contributed from the better separation resulted from stronger interaction between the fatty acyls and the longer alkyl chains of the stationary phase.
Table 3.
Number of lipids identified using LipidSearch from the data acquired by LC–MS/MS, lipids from positive and negative ion modes were combined after alignment of the four replicates.
| Column | Rat liver | Rat plasma |
|---|---|---|
| Accucore C30 | 432 | 424 |
| Kinetex C18 | 398 | 373 |
| HSS T3C18 | 419 | 390 |
| Cortecs C18 | 424 | 386 |
| Cortecs C18+ | 346 | 322 |
| Accucore Vanquish C18 | 366 | 323 |
| CSH C18 | 350 | 348 |
All of the lipids identified in rat plasma and rat liver extract are presented in the Supplemental material (Tables S5 and S6). Different molecular species of Cer, DG, LysoPC, LysoPE, PC, PE, PI, PS and TG were found in both types of samples. However, sphingosine (So) species were only present in rat plasma samples, while CL, LysoPI, LysoPS, LysoPG, PA, PG and SM species were found just in rat liver samples. Besides PE, PI and PA species ionized much more efficiently in negative ion mode, CL and PG species were obtained just in negative ion mode and DG and TG in positive ion mode. Therefore, combination of both ionization modes is necessary for a comprehensive characterization of lipid species in a complex lipidomic sample.
3.5. Lipid isomer separation
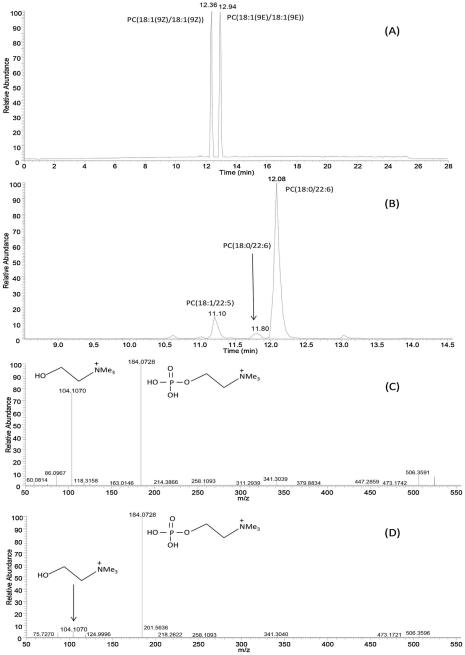
It is of great interest to separate cis/trans isomers in the chromatographic analysis, for example humans only synthetize cis fatty acids endogenously but synthetic trans fatty acids are also present in the human body due to the dietary intake, and they play an important role in several metabolic diseases [28]. To this end, the ability of differentiating between cis/trans isomers was evaluated on the Accucore C30 column. The separation of cis and trans isomeric PC species, such as PC(18:1(9Z)/18:1(9Z)) and PC(18:1(9E)/18:1(9E)), can be easily achieved, as shown in Fig. 5A. Similarly, very good separation was obtained for the PE(18:1(9Z)/18:1(9Z)) and PE(18:1(9E)/18:1(9E)), and for the PG(18:1(9Z)/18:1(9Z)) and PG(18:1(9E)/18:1(9E)) isomers, with a difference in the retention time of 0.6 min between the two isomers. Cis isomers appeared always at a lower retention time than the trans isomers, because the cis fatty acyl moieties experience a weaker interaction with the alkyl chains of the stationary phase due to the U shaped geometry of the cis isomers. For the lipid molecular species identified in rat liver or rat plasma extracts, we noticed that lipid species eluted at different retention times could be annotated as the same lipid molecule by LipidSearch, this observation is likely due to the position of the double bonds or to the cis/trans configurations. An example is shown in Fig. 5B, three PC 40:6 species (m/z of 833.59) were observed from the rat liver lipid extract, and the latter two peaks were both annotated as PC (18:0/22:6); however, the exact molecular configurations of these two lipids cannot be determined due to lack of standards and many possible conformations for the 6 C=C double bonds.
Fig. 5.
(A) Total ion chromatogram of lipid standards PC (18:1(9Z)/18:1(9Z)) and PC (18:1(9E)/18:1(9E)) in positive ion mode. (B) Extracted ion chromatogram of m/z 833.59 obtained in the positive ion mode for [M+H]+ of PC (40:6) from rat liver lipid extract. Product-ion spectrum of the [M+H]+ ion of LysoPC (18:0/0:0) (C) and LysoPC (0:0/18:0)
With respect to positional isomers, we found that PC(16:0/18:1(9Z)) and PC(18:1(9Z)/16:0) could not be chromatographically separated on this column, nor could the positional isomers of diacylglycerols. However, these positional isomers could be differentiated using MS/MS. Although the same fragmentation ions are expected from these sn-positional isomers [38], they have different intensities, and the intensity ratio between certain ions can help to distinguish them. For example, the intensity ratio of fragment ions m/z 184.1 to 104.1 can be used to distinguish between the sn-1 and sn-2 isomers of LysoPCs, with the sn-2 isomer has higher ratio than the sn-1 isomer [38]. As demonstrated in Fig. 5(C) and (D), similar pattern was observed for the MS/MS spectra for LysoPC(18:0/0:0) and LysoPC(0:0/18:0) from rat plasma extracts. The intensity ratio difference between these two ions was used by us to assign the sn-positions for the LysoPC isomers, since LipidSearch program does not assign the sn-position to these isomers. Levels of specific positional isomers of LysoPC has been reported as useful clinical diagnostic markers to reveal the pathophysiological changes [38]. Therefore, annotation of the exact sn-position for the lysophospholipids is a very important step in the accurate quantification of their levels. In this respect, the methods that we adopted in this study enable the most accurate annotation of lipid molecular species, as detailed in Tables S5 and S6, which could be adapted for other studies for unambiguous characterization of sn-positional isomers of lysophospholipids. Previously, offline 2D combination of HILIC fractionation in the first dimension followed by the reversed phase LC analysis of collected fractions in the second dimension allowed a comprehensive characterization of lipids in complex biological samples [30], and the separation of sn1- and sn2-lysophospholipid regioisomers was achieved in HILIC and the higher relative abundance of fatty acyl ions formed by the cleavage of fatty acid from sn2-position in negative ion ESI mass spectra enabled the differentiation of lysophospholipid regioisomers. Similar results to those obtained previously can be achieved with the present method while only using one dimensional reversed phase analysis. Most recently, one dimension HILIC–ESI–MS coupled with HCD–MS/MS was used for the identification of regioisomeric species of LPCs [31]. However, the fragmentation patterns reported by those authors are different from the ones obtained in the present work; for example, neither the m/z 104.11 for the sn-2 LPC nor the [M+H-(P-Chol)]+ ion for the sn-1 LPC were obtained in their study. This may be due to different collision energies used in these two studies, i.e. NCE 20 was used in their study compared with NCE 25 or 30 used by the present method in positive ion mode.
4. Conclusion
In this work, four different reversed phase LC columns were compared extensively, with three containing core–shell particles and one porous particles, and three having C18 and one C30 stationary phase. Using a complex lipid standard mixture for column performance evaluation, the narrowest peaks and highest theoretical plate number were typically obtained on the Accucore C30 column, while the Accucore C30, Cortecs C18 and HSS T3 column showed similar peak capacity in positive ion mode, and the C30 and Kinetex C18 had highest peak capacity in negative ion mode. Further evaluation was performed on complex biological samples of rat plasma and rat liver, and we found that again the highest number of lipids were identified using Accucore C30. In addition, the LC–MS/MS method that we developed allowed differentiation of isobaric lipid ions using ultra high mass resolving power, cis/trans lipid isomers based on retention time, and the co-eluting snpositional isomers based on relative fragment ion intensity ratios using tandem mass spectrometry. The fast scan rates of QEHF provided sufficient scans across the chromatographic peak even under very high mass resolution settings for both MS and MS/MS scans, which make accurate quantification and unambiguous identification readily achievable. This reversed phase LC–MS/MS method is very promising for high throughput lipidomic analysis.
Supplementary Material
Acknowledgement
The authors gratefully acknowledge the National Institutes of Health (GM104678) for support of this work.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.chroma.2016.02.054.
References
- [1].Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 2003;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- [2].Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- [3].Yamada T, Uchikata T, Sakamoto S, Yokoi Y, Fukusaki E, Bamba T. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J. Chromatogr. A. 2013;1292:211–218. doi: 10.1016/j.chroma.2013.01.078. [DOI] [PubMed] [Google Scholar]
- [4].Krauss RM. Lipids and lipoproteins in patients with type 2 diabetes. Diabetes Care. 2004;27:1496–1504. doi: 10.2337/diacare.27.6.1496. [DOI] [PubMed] [Google Scholar]
- [5].Wenk MR. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- [6].Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease, Proc. Natl. Acad. Sci. U. S. A. 2004;101:2070–2075. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lwin A, Orvisky E, Goker-Alpan O, LaMarca ME, Sidransky E. Glucocerebrosidase mutations in subjects with parkinsonism. Mol. Genet. Metab. 2004;81:70–73. doi: 10.1016/j.ymgme.2003.11.004. [DOI] [PubMed] [Google Scholar]
- [8].Cajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography–mass spectrometry, Trends Anal. Chem. 2014;61:192–206. doi: 10.1016/j.trac.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Houjou T, Yamatani K, Imagawa M, Shimizu T, Taguchi R. A shotgun tandem mass spectrometric analysis of phospholipids with normal-phase and/or reverse-phase liquid chromatography/electrospray ionization mass spectrometry, Rapid Commun. Mass Spectrom. 2005;19:654–666. doi: 10.1002/rcm.1836. [DOI] [PubMed] [Google Scholar]
- [10].Yang K, Zhao Z, Gross RW, Han X. Shotgun lipidomics identifies a paired rule for the presence of isomeric ether phospholipid molecular species. PLoS One. 2007;2:e1368. doi: 10.1371/journal.pone.0001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Han X. Multi-dimensional mass spectrometry-based shotgun lipidomics and the altered lipids at the mild cognitive impairment stage of Alzheimer’s disease. Biochim. Biophys. Acta. 2010;1801:774–783. doi: 10.1016/j.bbalip.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bird SS, Marur VR, Stavrovskaya IG, Kristal BS. Separation of cis-trans phospholipid isomers using reversed phase LC with high resolution MS detection. Anal. Chem. 2012;84:5509–5517. doi: 10.1021/ac300953j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chester TL. Recent developments in high-performance liquid chromatography stationary phases. Anal. Chem. 2013;85:579–589. doi: 10.1021/ac303180y. [DOI] [PubMed] [Google Scholar]
- [14].Omamogho JO, Glennon JD. Comparison between the efficiencies of sub-2 mum C18 particles packed in narrow bore columns. Anal. Chem. 2011;83:1547–1556. doi: 10.1021/ac102139a. [DOI] [PubMed] [Google Scholar]
- [15].Witting M, Maier TV, Garvis S, Schmitt-Kopplin P. Optimizing a ultrahigh pressure liquid chromatography-time of flight-mass spectrometry approach using a novel sub-2mum core–shell particle for in depth lipidomic profiling of Caenorhabditis elegans. J. Chromatogr. A. 2014;1359:91–99. doi: 10.1016/j.chroma.2014.07.021. [DOI] [PubMed] [Google Scholar]
- [16].Gallart-Ayala H, Courant F, Severe S, Antignac JP, Morio F, Abadie J, Le Bizec B. Versatile lipid profiling by liquid chromatography-high resolution mass spectrometry using all ion fragmentation and polarity switching. Preliminary application for serum samples phenotyping related to canine mammary cancer. Anal. Chim. Acta. 2013;796:75–83. doi: 10.1016/j.aca.2013.08.006. [DOI] [PubMed] [Google Scholar]
- [17].Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- [18].Hu C, van Dommelen J, van der Heijden R, Spijksma G, Reijmers TH, Wang M, Slee E, Lu X, Xu G, van der Greef J, Hankemeier T. RPLC-ion-trap-FTMS method for lipid profiling of plasma: method validation and application to p53 mutant mouse model. J. Proteome Res. 2008;7:4982–4991. doi: 10.1021/pr800373m. [DOI] [PubMed] [Google Scholar]
- [19].Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- [20].Nakanishi H, Iida Y, Shimizu T, Taguchi R. Separation and quantification of sn-1 and sn-2 fatty acid positional isomers in phosphatidylcholine by RPLC-ESIMS/MS. J. Biochem. 2010;147:245–256. doi: 10.1093/jb/mvp171. [DOI] [PubMed] [Google Scholar]
- [21].Ishida M, Yamazaki T, Houjou T, Imagawa M, Harada A, Inoue K, Taguchi R. High-resolution analysis by nano-electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry for the identification of molecular species of phospholipids and their oxidized metabolites. Rapid Commun. Mass Spectrom. 2004;18:2486–2494. doi: 10.1002/rcm.1650. [DOI] [PubMed] [Google Scholar]
- [22].Taguchi R, Ishikawa M. Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine LipidSearch. J. Chromatogr. A. 2010;1217:4229–4239. doi: 10.1016/j.chroma.2010.04.034. [DOI] [PubMed] [Google Scholar]
- [23].Scheltema RA, Hauschild JP, Lange O, Hornburg D, Denisov E, Damoc E, Kuehn A, Makarov A, Mann M. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol. Cell. Proteomics. 2014;13:3698–3708. doi: 10.1074/mcp.M114.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hsu FF, Turk J. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4 5-bisphosphate by electrospray ionization tandem mass spectrometry: a mechanistic study. J. Am. Soc. Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- [25].Neue UD. Theory of peak capacity in gradient elution. J. Chromatogr. A. 2005;1079:153–161. doi: 10.1016/j.chroma.2005.03.008. [DOI] [PubMed] [Google Scholar]
- [26].Yang K, Han X. Accurate quantification of lipid species by electrospray ionization mass spectrometry—meet a key challenge in lipidomics. Metabolites. 2011;1:21–40. doi: 10.3390/metabo1010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gao X, Zhang Q, Meng D, Isaac G, Zhao R, Fillmore TL, Chu RK, Zhou J, Tang K, Hu Z, Moore RJ, Smith RD, Katze MG, Metz TO. A reversed-phase capillary ultra-performance liquid chromatography–mass spectrometry (UPLC–MS) method for comprehensive top–down/bottom–up lipid profiling. Anal. Bioanal. Chem. 2012;402:2923–2933. doi: 10.1007/s00216-012-5773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Damen CW, Isaac G, Langridge J, Hankemeier T, Vreeken RJ. Enhanced lipid isomer separation in human plasma using reversed-phase UPLC with ion-mobility/high-resolution MS detection. J. Lipid Res. 2014;55:1772–1783. doi: 10.1194/jlr.D047795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Perona JS, Ruiz-Gutierrez V. Simultaneous determination of molecular species of monoacylglycerols, diacylglycerols and triacylglycerols in human very-low-density lipoproteins by reversed-phase liquid chromatography. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003;785:89–99. doi: 10.1016/s1570-0232(02)00861-9. [DOI] [PubMed] [Google Scholar]
- [30].Lisa M, Cifkova E, Holcapek M. Lipidomic profiling of biological tissues using off-line two-dimensional high-performance liquid chromatography–mass spectrometry. J. Chromatogr. A. 2011;1218:5146–5156. doi: 10.1016/j.chroma.2011.05.081. [DOI] [PubMed] [Google Scholar]
- [31].Granafei S, Losito I, Palmisano F, Cataldi TR. Identification of isobaric lyso-phosphatidylcholines in lipid extracts of gilthead sea bream (Sparus aurata) fillets by hydrophilic interaction liquid chromatography coupled to high-resolution Fourier-transform mass spectrometry. Anal. Bioanal. Chem. 2015;407:6391–6404. doi: 10.1007/s00216-015-8671-9. [DOI] [PubMed] [Google Scholar]
- [32].Narvaez-Rivas M, Gallardo E, Rios JJ, Leon-Camacho M. A new high-performance liquid chromatographic method with evaporative light scattering detector for the analysis of phospholipids. Application to Iberian pig subcutaneous fat. J. Chromatogr. A. 2011;1218:3453–3458. doi: 10.1016/j.chroma.2011.03.067. [DOI] [PubMed] [Google Scholar]
- [33].Cifkova E, Holcapek M, Lisa M, Vrana D, Melichar B, Student V. Lipidomic differentiation between human kidney tumors and surrounding normal tissues using HILIC-HPLC/ESI-MS and multivariate data analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015;1000:14–21. doi: 10.1016/j.jchromb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- [34].Lee JW, Nagai T, Gotoh N, Fukusaki E, Bamba T. Profiling of regioisomeric triacylglycerols in edible oils by supercritical fluid chromatography/tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014;966:193–199. doi: 10.1016/j.jchromb.2014.01.040. [DOI] [PubMed] [Google Scholar]
- [35].Garcia-de Blas E, Mateo R, Vinuela J, Alonso-Alvarez C. Identification of carotenoid pigments and their fatty acid esters in an avian integument combining HPLC-DAD and LC–MS analyses. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2011;879:341–348. doi: 10.1016/j.jchromb.2010.12.019. [DOI] [PubMed] [Google Scholar]
- [36].Hubner G, Crone C, Lindner B. lipID—a software tool for automated assignment of lipids in mass spectra. J. Mass Spectrom. 2009;44:1676–1683. doi: 10.1002/jms.1673. [DOI] [PubMed] [Google Scholar]
- [37].Holcapek M, Cervena B, Cifkova E, Lisa M, Chagovets V, Vostalova J, Bancirova M, Galuszka J, Hill M. Lipidomic analysis of plasma, erythrocytes and lipoprotein fractions of cardiovascular disease patients using UHPLC/MS MALDI-MS and multivariate data analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015;990:52–63. doi: 10.1016/j.jchromb.2015.03.010. [DOI] [PubMed] [Google Scholar]
- [38].Dong J, Cai X, Zhao L, Xue X, Zou L, Zhang X, Liang X. Lysophosphatidylcholine profiling of plasma: discrimination of isomers and discovery of lung cancer biomarkers. Metabolomics. 2010;6:478–488. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.