Abstract
Precision medicine in oncology requires not only identification of cancer-associated mutations, but also effective drugs for each cancer genotype, which is still a largely unsolved problem. One approach for the latter challenge has been large-scale testing of small molecules in genetically characterized cell lines. We hypothesized that compounds with high cell-line-selective lethality exhibited consistent results across such pharmacogenomic studies.
We analyzed the compound sensitivity data of 6,259 lethal compounds from the NCI-60 project. 2,565 cell-line-selective lethal compounds were identified and grouped into 18 clusters based on their GI50 profiles across the 60 cell lines, which were shown to represent distinct mechanisms of action. Further transcriptome analysis revealed a biomarker, NADPH abundance, for predicting sensitivity to ferroptosis-inducing compounds, which we experimentally validated. In summary, incorporating cell-line selectivity filters improves the predictive power of pharmacogenomic analyses and enables discovery of biomarkers that predict the sensitivity of cells to specific cell death inducers.
Keywords: Cancer pharmacology, pharmacogenomics, drug mechanisms of action, tumor biomarkers, ferroptosis, NADPH
Introduction
Cancers are often caused by somatic mutations in the genome. To understand the mechanisms of transformation, there have been efforts to collect inventories of DNA sequence variations relevant to transformation (Hudston et al., 2010; Weinstein et al., 2013). This information, in addition to providing insight into the tumorigenic process, has become the basis of the field of precision medicine (Barretina et al., 2012; Garnett et al., 2012; Basu et al., 2013; Schreiber et al., 2010; Eifert and Powers, 2012) .
Ideally, anticancer drugs should kill transformed cancer cells efficiently with minimal effects on non-transformed cells and tissues. However, except for a few successful cases of targeting cancers driven by a specific oncogene (e.g., imatinib targeting BCR-ABL in CML and vemurafenib targeting V600E BRAF in melanoma (Sala et al., 2008; Schindler et al., 2000)), two main obstacles prevent molecularly targeted precision therapies from being realized. First, mutated oncogenic proteins responsible for driving cancers are not necessarily viable candidates for targeted therapy, as these proteins may not be “druggable” or may not be needed for the ongoing survival and growth of cancers. Second, genes essential for cancers may also be important for the survival of normal cells (Luo et al., 2009). For these reasons, it has been challenging to extend the paradigm of directly targeting oncogenic drivers to diverse cancers beyond the known examples (Ginsburg and Kuderer, 2012).
A second approach in precision medicine is to identify biomarkers that predict sensitivity to targeted agents, even if these biomarkers are not themselves drivers of tumorigenesis. This approach is potentially more generalizable than directly targeting driver oncoproteins. To seek cancer biomarkers that predict sensitivity to therapeutic agents, large pharmacogenomic studies have been performed. The NCI-60 human tumor cell line anticancer drug screen (the NCI-60 project) started in the late 1980s, and is the first kind of such efforts. The NCI-60 project is a part of the Developmental Therapeutics Program, which aims for discovery and development of anticancer reagents. The NCI-60 project has been providing the research community with a service that tests lethal compounds in up to 60 different human cell lines from different tissue origins (Shoemaker, 2006). Other recent pharmacogenomic studies include the Cancer Cell Line Encyclopedia (CCLE) and the Cancer Genome Project (CGP) (Barretina et al., 2012; Garnett et al., 2012). In these studies, 138 and 24 compounds were tested for lethality in 727 and 1,036 cell lines, respectively. Not only somatic mutations, but also transcriptome and other molecular profiles were collected for each cell line in these studies. This potentially powerful approach seeks to correlate the sensitivity of cell lines to specific agents with the presence of specific biomarkers. However, recently, one controversial study suggested that these two projects, CCLE and CGP, generated inconsistent results when the same compounds and cell lines were tested (15 compounds tested in 471 cell lines); the implication of this study was that it may not currently be possible to perform compound sensitivity profiling on a large scale with consistent, reproducible results (Haibe-Kains et al., 2013).
We sought to test the hypothesis that the most selective compounds across cell lines would have the most consistent results, potentially addressing the reproducibility issue that has been recently discussed. We recognize that high-throughput technologies in modern biology are intrinsically noisy, but that they nonetheless provide valuable information. For example, transcriptome data, such as those obtained through microarrays or RNA-seq, is a widely accepted tool in biological research despite the noise that can be present; suitable data processing has overcome the limitations of noise using these technologies. We suspected that an appropriate technique for processing compound sensitivity data in large-scale studies might make the results more meaningful. The dispersion in relative compound sensitivity across cell lines, termed ‘cell-line selectivity’, was computed to see if it yielded more robust and consistent results across multiple studies. We then used this approach to correlate compound sensitivity and gene expression across NCI-60 cell lines and identified biomarkers that predict sensitivity of cell lines to ferroptosis, a recently reported form of regulated non-apoptotic cell death, for which predictive biomarkers have not been defined.
Results
Cell-line selectivity yields consistent results between the CCLE and CGP projects
It is often challenging to distinguish true signals from experimental noise. However, it is feasible to estimate noise in high-throughput experiments. In a single parameter estimation, one can argue that the larger sample distribution is more reliably estimated; a simple, but often appropriate, assumption is to model that an observation (x), its true signal (s) and experimental noise (n) have the relationship x = s+n, where the variables (x, s, n) are normally distributed with standard deviations of σ, σs, σn, respectively. Because signal and noise are independent of each other, the relationship between the variances holds true. When σ ≫ σn, we find that σ ≈ σs (≫ σn). On the other hand, when the signal is comparable to noise, or σ ≈ σn, care needs to be taken in estimating the signal (σs), as the noise (σn) is not insignificant.
In assessing relationships between two parameters, Pearson and Spearman correlations are popular measures because of their conceptual and computational succinctness. However, these correlations are scaled between −1 and 1, being independent of the sample dispersions (σ); therefore, we cannot assess the quality of the data associated with each variable from correlations.
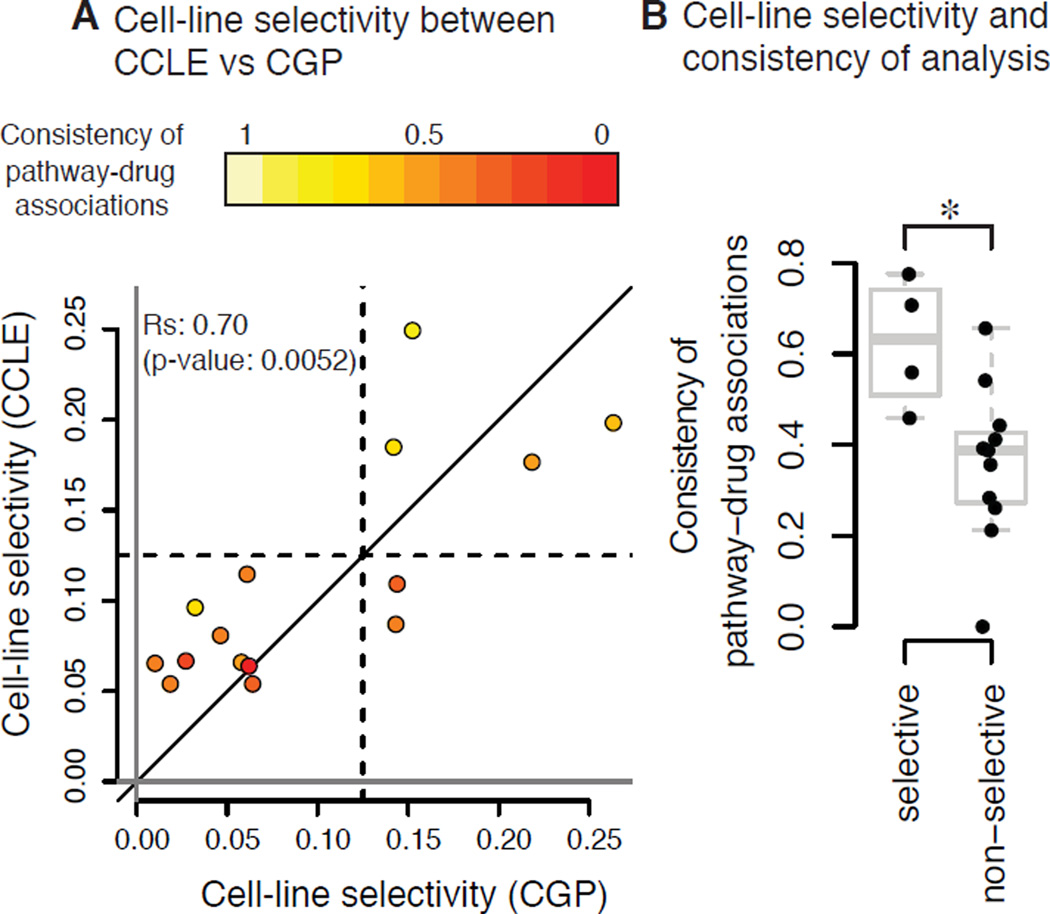
In transcriptome analysis, this is a common problem; one way to cope with this issue is to compute dispersion of expression of each gene across samples and remove genes from the data based on the magnitude of deviation of gene expression between samples before they are processed (Bourgon et al., 2010). Likewise, we hypothesized that filtering compounds based on the magnitude of the deviation in drug sensitivity across the samples would generate more robust results, which we termed cell-line selectivity. We thus computed cell-line selectivity of 15 compounds, as defined as the interquartile range (IQR) of the lethal compound across the cell line panel, for both CCLE and CGP data from the supplementary data of Haibe-Kains et al. (Haibe-Kains et al., 2013). This parameter was highly correlated between the two projects (the Spearman correlation coefficients were 0.70 and 0.67, with both p-values < 0.01 using two different measures of cell-line selectivity, ‘Log10 IC50 (IC50 hereafter)’and ‘area under dose-concentration curves (AUC)’, respectively) (Fig. 1A, Fig. S1A). Specifically, when AUC was used to compute cell-line selectivity of the 15 compounds, the parameter was similar between CCLE and CGP (i.e., the data were spread on both sides of the identity line); however, when IC50 was used, cell-line selectivity was overall different between the studies (i.e., the data were not on the identity line). Moreover, the cell-line selectivity of many compounds based on IC50 was close to zero in the CCLE data. It is likely that concentration ranges of these compounds did not cover the IC50 in many cell lines and thus, variations are not observed, while partial toxicity can still be measured using AUC. AUC is in such cases a better measure than IC50, as has been suggested previously (Haibe-Kains et al., 2013). Moreover, pathway-drug associations are also based on the drug sensitivity measures AUC and IC50 (see materials and methods); it is reasonable that cell-line selective compounds based on AUC are more consistent in pathway-drug associations than for non-selective compounds, while cell-line-selectivity based on IC50 (Fig. 1B, Fig. S1B) is not as powerful of a predictor of the pathway-drug associations because IC50 is a less accurate measure of toxicity than AUC. Overall, this analysis supports our notion that pharmacogenomic data are more robust and consistent when lethal compounds with high cell-line selectivity are analyzed, especially using AUC instead of IC50.
Figure 1. Cell-line-selective lethal compounds were more consistent between two large pharmacogenomic studies.
A. Cell-line selectivity of 15 lethal compounds based on AUC measure across the same 471 cell line panel in two studies, CCLE and CGP. Each point indicates one lethal compound. Colors indicate the consistency (the Spearman correlation coefficients) of pathway-compound associations between the two studies, given by Haibe-Kains et al. B. Consistency of pathway-drug associations of cell-line selective and non-selective compounds. Four lethal compounds showing larger cell-line selectivity in both studies in A were defined cell-line-selective lethal, and the others were defined non-selective. *: p-value = 0.0102 using Wilcoxon rank-sum test.
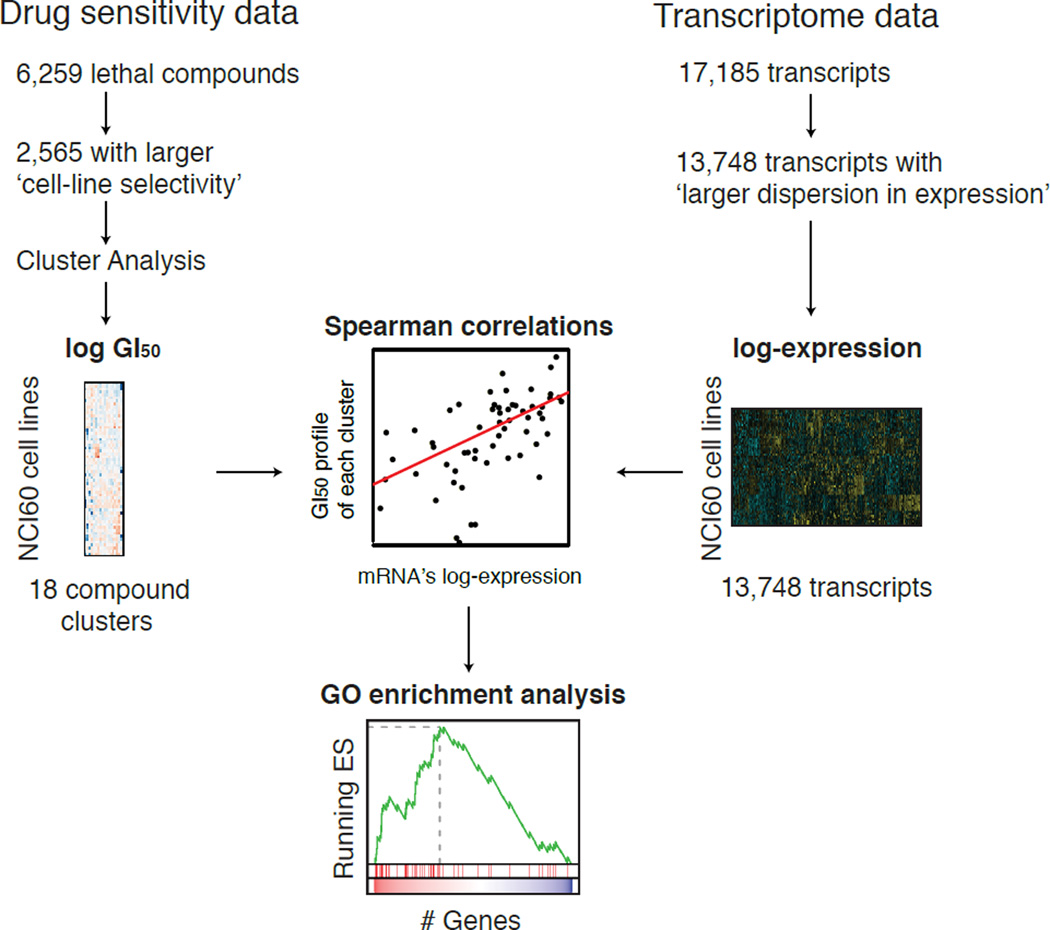
While AUC-derived cell-line-selective compounds gave more consistent results between CCLE and CGP, only 24 and 138 compounds, respectively, were analyzed in the two studies as of Haibe-Kains et al. published in 2013. We therefore focused on another pharmacogenomic study, the NCI-60 human cancer cell line screen (Shoemaker, 2006). Sensitivity profiles of 50,839 unique compounds from the NCI-60 screen (the NCI-60 data hereafter) are publicly available online (http://discover.nci.nih.gov/cellminer/loadDownload.do), 6,249 of which are without missing values. Although we prefer using AUC as a measure versus a single-concentration parameter, growth inhibitory (GI50) values were the only available measure of sensitivity in this dataset. Therefore, we analyzed the NCI-60 data using GI50 to test its suitability for the discovery of predictive biomarkers for lethal compounds (Fig. 2).
Figure 2. Scheme of the NCI-60 pharmacogenomic analysis.
(i) Drug sensitivity data were filtered using the ‘cell-line selectivity’ metric, followed by model-based clustering and consolidation of clustering into 18 clusters. (ii) 13,748 genes with larger dispersion in expression were selected based on IQR of their expression across the NCI-60 cell line panel. (iii) The Spearman correlation coefficient between drug sensitivity of each lethal compound cluster and the transcriptional profile of each gene across the NCI-60 panel was computed. (iv) The correlation coefficients between compound clusters and genes were used to compute GO enrichment analysis for each compound cluster.
Ferroptosis inducers are cell-line-selective lethal compounds in the NCI-60 data
Using the GI50-derived cell-line selectivity parameter, we focused on a set of lethal compounds inducing ferroptosis, a regulated, non-apoptotic form of oxidative cell death. Ferroptosis inducers were initially discovered to induce selective lethality in transformed cells over parental non-transformed cells (Yagoda et al., 2007). It is likely that ferroptosis occurs in certain pathological contexts, such as Huntington’s Disease, periventricular leukomalacia, and renal tubular cell death, whose death processes are suppressed by ferroptosis inhibitors such as ferrostatin-1 (Dixon et al., 2012; Linkermann et al., 2014a; Skouta et al., 2014). Thus, a better understanding of ferroptosis has significant therapeutic potentials. Ferroptosis is induced when the lipid repair enzyme glutathione peroxidase 4 (GPX4) is inhibited, either directly or indirectly (Dixon et al., 2012; Yang et al., 2014).
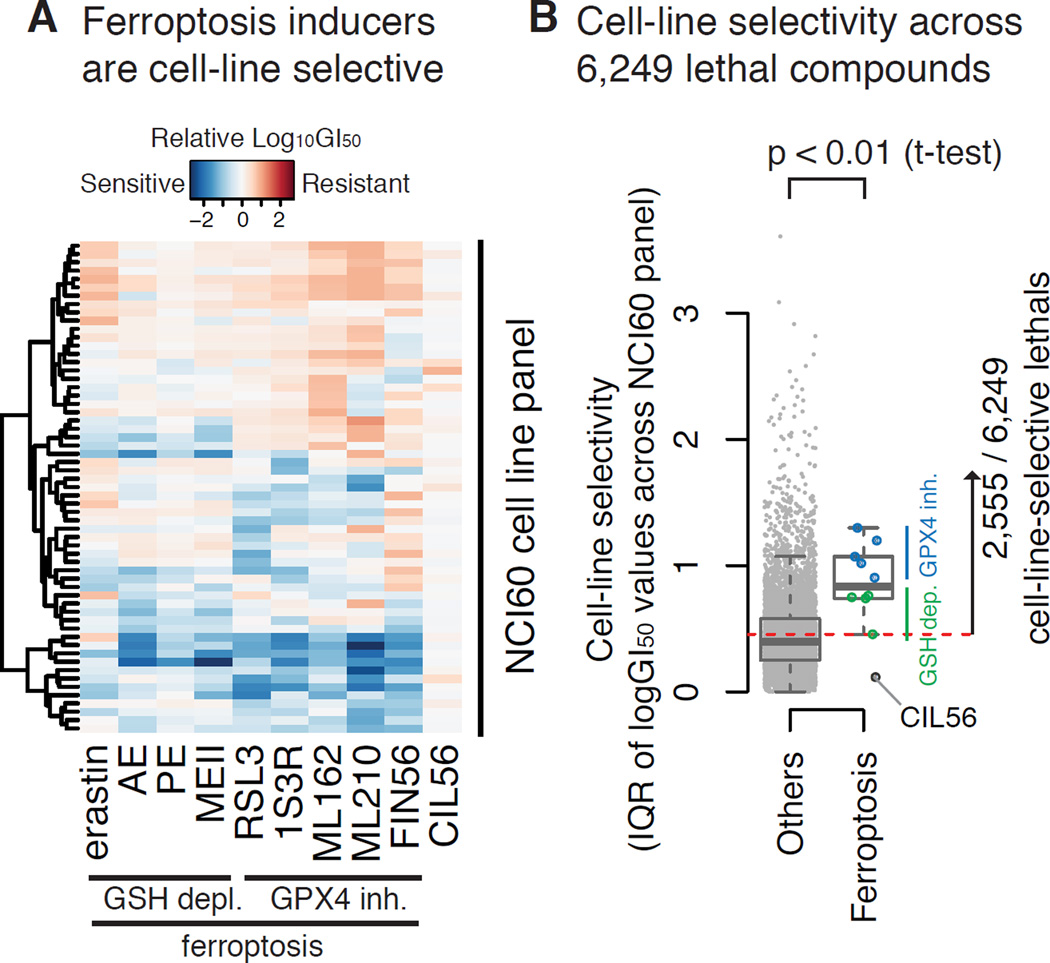
We sought to use cell-line-selectivity parameters to extract biomarkers that predict sensitivity to ferroptosis. We submitted ten ferroptosis inducers to the NCI-60 project to define their GI50 values across the NCI-60 cell lines (Fig. 3A). When their GI50 profiles (median-centered log10 GI50 values across the NCI-60 cell line panel) were analyzed, some cell lines were more resistant than others, and the difference in GI50 concentrations between the most sensitive and resistant cell lines was over 100-fold for all ferroptosis inducers except the dual-mechanism lethal compound CIL56. CIL56 induces necrotic cell death as well as ferroptosis. Overall, ferroptosis-specific lethal compounds were cell-line selective (Shimada et al., in revision). Between the two main mechanisms of ferroptosis, direct GPX4 inhibitors were more cell-line selective than compounds that induce glutathione (GSH) depletion (Fig. 3B), which ultimately leads to loss of GPX activity.
Figure 3. Selecting cell-line-selective lethal compounds from the NCI-60 drug sensitivity data.
A. GI50 profiles of 10 ferroptosis inducers. Log GI50 values were centered at the median across the NCI-60 cell lines. One of them, CIL56, is known to induce dual lethal mechanisms (ferroptosis + necrotic cell death). The others induce only ferroptosis. B. The cell-line selectivity of 6,249 lethal compounds from the public dataset in comparison with ferroptosis inducers. 2,555 compounds whose cell-line selectivity score was larger than at least one ferroptosis inducer were defined as cell-line selective lethal compounds (the threshold is shown as a dashed line).
We hypothesized that compounds with greater cell line selectivity would be more likely to yield predictive biomarkers. To test whether ferroptosis inducers were more cell-line selective than other lethal compounds, we computed cell-line selectivity for ferroptosis inducers and 6,249 other lethal compounds for which GI50 values were publicly available in the NCI-60 project. We found that ferroptosis inducers were more cell-line selective than other compounds (p-value < 0.01) (Fig. 3B): of the 6,249 compounds, we defined a subset of 2,555 lethal compounds that showed similar or larger cell-line-selective lethality than at least one of the ferroptosis inducers. 239 of the 2,555 publicly available compounds were known to induce death through specific mechanisms of action (MoA). Some, including topoisomerase I inhibitors, DNA synthesis inhibitors, antifolates (known to inhibit DNA synthesis), and serine/threonine kinase inhibitors, were more cell-line-selective than those compounds with other MoA, such as hormones (Ho) and apoptosis inducers (Apo) (Fig. S2).
Model-based clustering reveals that specific patterns of GI50 profiles correlate with mechanisms of action
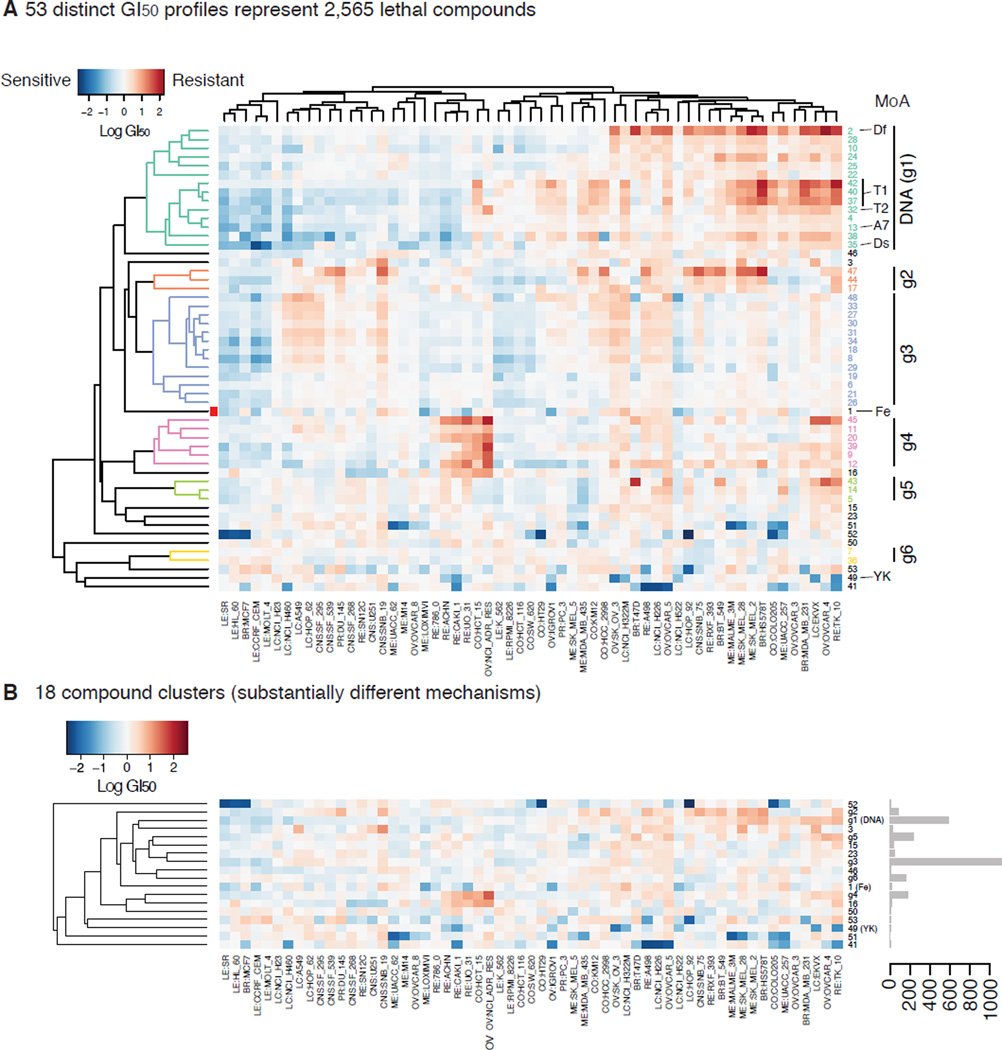
Most ferroptosis inducers were originally discovered in small molecule screening using engineered human fibroblasts (Smukste et al., 2006; Weïwer et al., 2012). Since most lethal compounds do not activate ferroptosis, we hypothesized that the GI50 profiles of the ferroptosis inducers would be distinct from other lethal compounds. We therefore applied cluster analysis of 2,565 compounds (10 ferroptosis inducers + 2,555 cell-line-selective lethal compounds) based on the similarity of their GI50 profiles. We used a ‘model-based clustering’ algorithm in the R statistical computing language (Fraley and Raftery, 2002). Without knowing the number of clusters or the mechanisms of action of compounds a priori, the optimal number of clusters was computed using the Bayesian Information Criterion. This clustering procedure assigned 2,565 compounds into 53 clusters (Fig. 4A). Seven of nine ferroptosis inducers were assigned to one cluster consisting of only 20 compounds (20/2,565 = 0.78%) (Table S1). This is consistent with the hypothesis that the GI50 profiles of ferroptosis inducers are similar to each other, but distinct from other cell-line-selective lethal compounds.
Figure 4. Clustering of cell-line-selective lethal compounds based on GI50 profiles A.
The GI50 profiles of 53 clusters computed by model-based clustering across 2,565 cell-line selective lethal compounds. The NCI-60 cell lines are represented in rows and 53 compound clusters are in columns. T1, T2: Topoisomerases 1 and 2 inhibitors, A7: A7, guanine alkylators at N7 position, Ds: DNA synthesis inhibitors, Df: antifolates (DNA/RNA synthesis inhibitors), Fe: ferroptosis inducers, YK: Tyr kinase inhibitors. Blue and red indicates a cell line is relatively sensitive or resistant to the compound cluster, respectively. Values are median-centered log10 GI50 values. B. 18 clusters resulting from merging clusters g1 through g6 into six clusters in A. Bar plot on the right indicates the number of compounds assigned to each cluster.
We then tested whether other compounds in the cluster containing ferroptosis inducers also induce ferroptosis or not. We found that one compound, NSC704397, was commercially available, and that it induces lethality in ferroptosis-susceptible HT-1080 fibrosarcoma cells, which was suppressed by 100 µM α-tocopherol (Fig. S3). We previously showed that lipophilic antioxidants such as α-tocopherol uniquely suppressed ferroptotic cell death among various oxidative cell death stimuli, strongly suggesting that NSC704397 induces ferroptosis (Dixon et al., 2012). Thus, this confirms that our cluster analysis is an effective approach to uncover mechanisms of action of lethal compounds systematically, and that the compound cluster we focused on represents ferroptosis inducers.
Topoisomerase I and II inhibitors (T1, T2), DNA alkylators at N-7 position of guanine (A7), DNA synthesis inhibitors (Ds), antifolates (Df), and tyrosine kinase inhibitors also exhibited significant enrichments in specific clusters (YK) (Fig. 4, Fig. S4). Other classes of compounds, such as microtubule inhibitors (Tu) and HSP90 inhibitors (P90), were also enriched within a single cluster. However, the number of compounds representing each class was too small compared to the total number of compounds of the cluster to make these latter results significant. Lethal compounds targeting DNA directly (Ds, Df, A7) or indirectly (T1, T2) share more similar GI50 profiles than other classes, such as ferroptosis or tyrosine kinase inhibitors, possibly reflecting their similarity in mechanism of action (Fig. 4). We therefore merged these 14 clusters into one DNA-targeting compound cluster. Other clusters sharing similar GI50 profiles to the same extent as these clusters based on the hierarchical clustering dendrogram (Fig. 4A left) were also merged, which resulted in 18 distinct clusters (Fig. 4B). We expect that these 18 clusters represent significantly distinct lethal mechanisms in sensitive cells.
Transcriptomic analysis revealed anticancer drugs were largely separated into two distinct groups
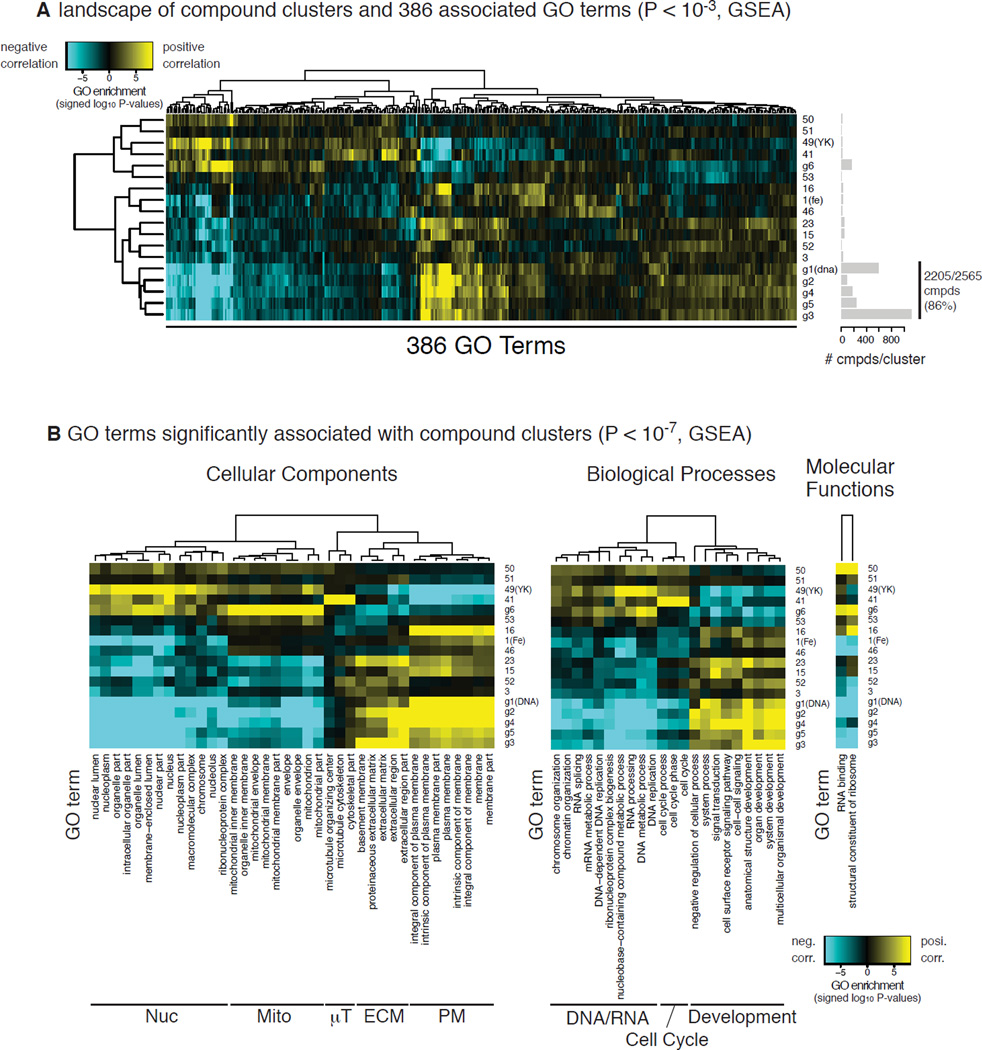
We then explored the transcriptome data of the NCI-60 cell lines to uncover biomarkers that could differentiate the cell-line selectivity of the 18 compound clusters. The transcriptome data were pruned so that 80% of the 17,185 genes across the NCI-60 panel were used. Pruning was based on IQR of their expression across the cell lines. We next computed the Spearman correlation coefficients between the GI50 profiles of each of the 18 compound clusters and each gene’s expression across the NCI-60 panel (Fig. 2). Here, positive and negative correlations were interpreted such that higher expression of the gene is associated with a resistance or sensitivity to the cluster compounds, respectively. We then performed gene set enrichment analysis and looked for gene ontology (GO) terms enriched in genes which were positively or negatively correlated with the GI50 profiles for each cluster (Subramanian et al., 2005). This analysis revealed 386 GO terms, whose activities were associated with the GI50 profiles of one or more compound clusters (p-value < 10−3, Gene Set Enrichment Analysis) (Fig. 5A).
Figure 5. GO enrichment analysis reveals GO terms associated with 18 compound clusters.
A. A heatmap showing 386 GO terms associated with the GI50 profiles of at least one of the 18 compound clusters (p-value < 10−3). Blue and yellow indicate negative and positive association, i.e., down- and up-regulation of genes in the GO term associated with resistance to the compound cluster, respectively. Bar plot on the right indicates the numbers of compounds assigned into each cluster. B. A subset of A, showing more significant association between GO terms and compound clusters (p-value < 10−7). The heatmap was further split based on categories of GO terms: BP (biological process), CC (cellular content), and MF (molecular function).
Hierarchical clustering of the GO term-compound associations split the 18 compound clusters roughly into two groups: the majority of compounds (~95%) including DNA-targeting compounds and ferroptosis inducers grouped together, while the remainder (~5%) including tyrosine kinase inhibitors (YK) belonged to the other. Notably, 2,205 compounds (86% of total) in the former group showed particularly similar GO term-compound associations within the larger group (Fig. 5A). This indicates that most of the 2,565 cell-line-selective lethal compounds in the NCI-60 project showed similar drug sensitivity profiles. Genes encoding extracellular matrix or plasma membrane proteins were positively and negatively correlated against the former and the latter groups, respectively, while genes encoding nucleic or mitochondrial proteins were negatively and positively correlated with them (Fig. 5B). These correlations may reflect upon the subcellular components that the compounds are ultimately targeting.
‘Specificity’ parameter predicts specific gene signatures as biomarkers for sensitivity to each cluster of compounds
The heatmap in Fig. 5B also revealed that some GO terms were exclusively associated with only one compound cluster. For example, genes encoding proteins in mitochondria (g6), microtubules (#41) and nuclei (#49) were positively correlated with only one compound cluster each. This inspired us to search for the GO terms that are exclusively associated with one of the 18 clusters. Because these GO terms are associated with only one compound cluster, they should represent biomarkers for sensitivity to the cluster’s mechanism of action. To identify such GO terms and the corresponding compound cluster, we not only computed the significance (p-value) of enrichment between all the GO terms and all the compound clusters, but also computed the ‘specificity’, or the difference between the first and second lowest p-values in log scale. This filter revealed that 38 GO terms were exclusively associated with 11 clusters (Table S2), indicating that the GO terms are potential biomarkers of sensitivity for these clusters.
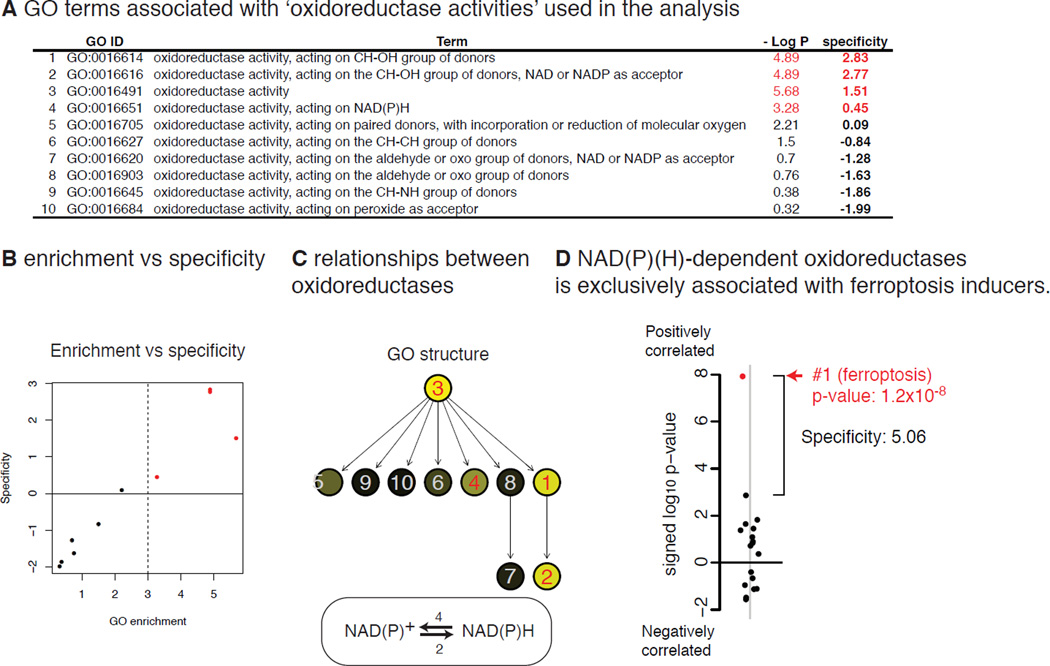
Activity of NAD(P)(H)-dependent oxidoreductases is associated with ferroptosis
We then focused on one cluster that contained ferroptosis inducers and their associated GO terms. The two most significant and specific GO terms associated with this cluster were ‘oxidoreductase activities’ (GO:0016614 and GO:0016616). There were ten GO terms related to oxidoreductase activity in the analysis (Fig. 6A, C), and five of them were most strongly associated with this cluster among the 18 clusters (i.e., specificity > 0) (Fig. 6B). Particularly, enzymes that reduce NAD+ or NADP+ (NAD(P)+ hereafter) (GO:0016616, GO:0016614) or oxidize NADH or NADPH (NAD(P)H hereafter) (GO:0016651), are included in this cluster; therefore, oxidoreductases using NAD(P)+ and NAD(P)H (NAD(P)(H) hereafter) appear to be genuine biomarkers of sensitivity to ferroptosis. When the two GO terms were merged into one term, representing oxidoredutases using NAD(P)(H), this new gene set became both more significant (p-value = 1.2 × 10−8) and more specific (specificity = 5.06) (Fig. 6D).
Figure 6. NAD(P)(H)-dependent oxidoreductases as biomarkers of the sensitivity to ferroptosis.
A. Ten GO terms associated with ‘oxidoreductases’. B. The significance in the GO enrichment (−log10 p-value) vs ‘specificity’ among the ten GO terms associated with oxidoreductases. c. Hierarchical relationship of the ten GO terms associated with oxidoreductases. D. The specificity of the NAD(P)(H)-dependent oxidoreductases (GO:0016616 + GO:0016651) against the 18 compound clusters.
Although the significance of this analysis appeared high, we needed to be cautious interpreting the analysis, as the large specificity of this gene set against the ferroptosis cluster may be due to overfitting the gene-drug correlations from the NCI-60 data specifically. We therefore performed similar analysis on an independent data set, the Cancer Therapeutic Response Portal (CTRP), which tested 255 compounds, 88 of which (including four ferroptosis inducers erastin, (1S,3R)-RSL3, ML162 and ML21) were cell-line-selective lethal in 115 cell lines from CCLE (Basu et al., 2013) (Fig. S5A). Although the transcriptional signature of NAD(P)(H)-dependent gene set was still strongly associated with sensitivity to the ferroptosis inducers, the association was not as significant or as unique to these compounds as in the NCI-60 analysis, possibly reflecting the noisy nature of the studies or that the collection of 88 compounds covers substantially distinct mechanisms of action from that of NCI-60 (Fig. S5B,C). Whatever the cause of the discrepancy between CTRP and NCI-60, it is important to note that the two independent analyses suggest that transcriptional signature of NAD(P)(H)-dependent oxidoreductases is strongly associated with sensitivity to ferroptosis, and therefore still a candidate biomarker for ferroptosis.
NAD(P)(H) is a biomarker for sensitivity to ferroptosis inducers across 12 cell lines
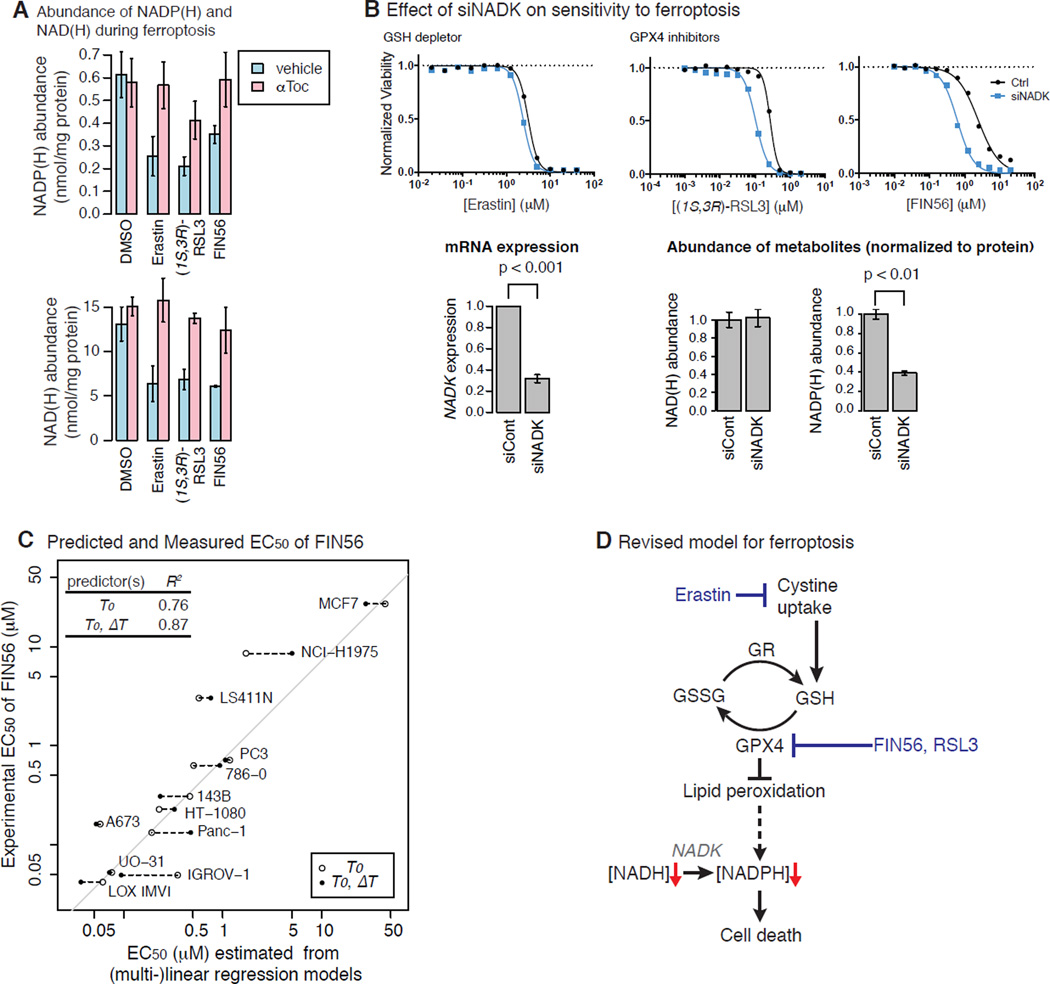
Based on this finding, we further hypothesized that not only is the expression of NAD(P)(H)-dependent genes, but also NAD(P)(H) level, correlated with sensitivity to ferroptosis. We found that three canonical ferroptosis inducers representing the three major mechanisms of inducing ferroptosis (erastin, (1S, 3R)-RSL3, and FIN56) decreased both NAD(H) and NADP(H) level (Fig. 7A). The decrease in NAD(P)(H) level following treatment with ferroptosis inducers was inhibited by the antioxidant α-tocopherol, suggesting that the loss of NAD(P)(H) during ferroptosis is a downstream consequence of lipid peroxidation upon GPX4 inhibition, thereby identifying for the first time an effect that is downstream of lipid ROS generation during ferroptosis.
Figure 7. Experimental validation of NADPH as Predictor of Sensitivity to Ferroptosis.
A. Total NAD(H) and NADP(H) levels upon treatment of ferroptosis inducers with or without α-tocopherol. B. The effect of NAD+ kinase (NADK) knockdown on the sensitivity to ferroptosis inducers (top), NADK messenger level (bottom left) and NAD(P)(H) level. C. The relationship between the sensitivity to FIN56 and the NADP(H) level across the 12 cell line panel. D. Revised model for ferroptosis.
NADPH is synthesized through phosphorylation of NAD+ by NAD+ kinase (NADK) (Pollak et al., 2007). NADPH is used by glutathione reductase (GR) to reduce oxidized glutathione (GSSG) to GSH (Deponte, 2013), further pointing to its potential relevance to ferroptosis. Therefore, we suspected that basal NADP(H) abundance might correlate with sensitivity to ferroptosis. Indeed, knockdown of NADK using siRNA, which decreased NADP(H) level without affecting NAD(H) level, sensitized HT-1080 cells to the three ferroptosis inducers, supporting this hypothesis (Fig. 7B). The extent to which siNADK enhanced the effects of the ferroptosis inducers was greater against GPX4 inhibitors ((1S,3R)-RSL3, FIN56) than against a GSH depletor (erastin). This corresponds to the fact that GPX4 inhibitors are more cell-line-selective lethal compounds than a GSH depletor (Fig. 3B).
NADP(H) is an essential metabolite for survival, and its depletion could be a critical consequence of ferroptotic cell death. Cells resistant to ferroptosis inducers may somehow overcome the depletion of NADP(H) upon ferroptosis induction. We hypothesized that there should be two ways for cells to achieve resistance; i) basal NADP(H) level in cells is high, and/or ii) cells are resistant to its depletion upon ferroptosis induction. To test these hypotheses, we expanded our analysis and measured the level of NADP(H) upon vehicle or FIN56 treatment for six hours in a panel of 12 cell lines from diverse tissues. Indeed, our analysis confirmed that resistant cell lines exhibited either (i) high basal level of NADP(H) and low [NADP+]/[NADPH] ratios, or (ii) resistance to depletion of NADP(H) upon FIN56 treatment (Fig. 7C, Fig. S6). The basal level of NADP(H) serves as a biomarker for sensitivity to FIN56-induced ferroptosis accounted for up to 70 % of variability of log EC50 values across 12 cell lines. Moreover, linear combination of basal NADP(H) level and the decrease in NADP(H) level can predict up to 90% of variability of the log10 EC50 values across the panel. It is worth noting that there is little to no correlation between the basal NADP(H) level and the extent of NADP(H) decrease upon FIN56 treatment across the 12 cell lines, indicating that cells can acquire resistance to FIN56 through these independent mechanisms.
In conclusion, analysis of compound sensitivity across the NCI-60 cell line panel using transcriptome data revealed that NADP(H) abundance is inversely correlated with sensitivity to ferroptosis inducers, validating the accuracy and usefulness of this approach to identify biomarkers for sensitivity to cell death induction.
Discussion
In this analysis, we computed cell-line selectivity in pharmacogenomic datasets. Compounds with low cell-line selectivity were removed from this analysis from the beginning, as this filter made the analysis more robust and consistent across independent studies. Among the cell-line-selective lethal compounds, we confirmed that compounds with the same mechanism of action showed similar drug sensitivity (GI50 profiles), indicating that the profiles likely reflect the compounds actual mechanism of action. We designed this analysis to make the result robust; first, using model-based clustering and further consolidating clusters of compounds with similar GI50 profiles, and thus similar mechanisms of action, and finally, computing the specificity metric revealed GO terms associated with individual compound clusters, helping us define biomarkers for sensitivity for compound clusters.
NADP(H) abundance was found to be a biomarker for sensitivity to the ferroptosis cluster; however, it may be too generous to conclude that the NADP(H) level is a exclusive predictor for ferroptosis-susceptibility. Although we discovered that another compound in the cluster, NSC704397, is a ferroptosis inducer, the cluster may possibly represent other cell death phenotypes, whose GI50 profiles are indistinguishable from ferroptosis inducers. The decrease of NAD(P)(H) is a known downstream consequence of some non-apoptotic regulated cell death, and for example, some associated with decreases in NAD+ downstream of cell death induction may fall into the same cluster (Linkermann et al., 2014b).
More compounds are being tested in CCLE (or equivalently, CTRP), CGP and similar efforts. For robust and consistent analysis of these studies, incorporation of data filters such as the ones described in this study will be beneficial for the discovery of biomarkers that predict sensitivity of lethal compounds.
Materials and Methods
CCLE and CGP Data Analysis
The comparison of CCLE and CGP were performed using the supplementary data of Haibe-Kains et al.(Haibe-Kains et al., 2013). In the original article, consistency of pathway-gene associations between the studies was assessed as follows: (i) first, the Spearman correlations between gene expression and the drug sensitivity (either AUC or IC50) to each lethal compound across 474 cell lines (which we termed consistency) were computed, (ii) these correlations were used to perform gene set enrichment analysis (GSEA) against GO terms. Significance of association between GO terms (pathways) and sensitivity to each drug was computed and the p-values were false discovery rate (FDR) adjusted, and (iii) for GO terms with only FDR adjusted p-values < 0.2 in at least one study, the Spearman correlations of normalized enrichment scores (NES) between the studies were computed for each lethal compound considered as ‘consistency of pathway-drug association’.
The cell-line selectivity across 474 cell lines for each lethal compound was then computed in each study. As in the original article, two measures, log10 IC50 values (IC50) and area under the dose-response curve (AUC), were used to compute the cell-line selectivity. Finally, relationship between consistency of pathway-drug association and the cell-line selectivity was evaluated for AUC (Fig. 1) and IC50 (Fig. S1), respectively.
NCI-60 Data Analysis
Symbols
We used the following symbols for the analysis of the NCI-60 data described below: a lethal compound (L), a cell line (C), a compound cluster (Γ), the ferroptosis cluster (F), the logGI50 value of L in C (gL,C), a vector of gL,C across the NCI-60 cell line panel (gL,*), the median value of gL,* among the cell lines (g̃L), the median subtracted logGI50 value of L in C (GL,C = gL,C – g̃L), and the GL,C of the NCI-60 cell lines, or ‘GI50 profile of L ‘ (GL,*). L can be a compound cluster T in GL,C; GI50 profile of a compound cluster (GΓ,C;GF,C for the ferroptosis cluster). For transcriptome analysis, the following symbols were used: a gene (T), the transcriptional expression of T in C (XT,C), the expression of T across the NCI-60 cell lines (XT,*), the Spearman correlation coefficient between XT,C and GΓ,C across the NCI-60 cell lines (ρT,Γ), a Gene Ontology term (GO), the significance of association between GO and Γ (pGO,Γ), and the specificity of GO against Γ(SGO,Γ).
Discovery of Cell Line-Selective Lethals from the NCI-60 data
Raw logGI50 data (gL,*) of 75,446 (50,839 unique) lethal compounds in NCI-60 cell lines were downloaded from CellMiner’s website (http://discover.nci.nih.gov/cellminer/loadDownload.do). For lethal compounds tested in more than one replicate, we computed the median of each cell line, so that there is a unique value, gL,C, for each L and each C. Of the 50,839 lethal compounds, we selected 6,249 that were tested in all 59 ell lines (breast cancer cell line MDA-MB-468 and melanoma cell line MDA-N were removed from this analysis because either pharmacology or transcriptome data were not available; the list of 59 cell lines used in the analysis is available at http://dtp.nci.nih.gov/docs/misc/common_files/cell_list.html). These gL,* (a matrix of 6,249 L by 59 C) have no missing values. Each L ‘s GI50 profile among NCI-60 cell lines was computed by GL,* = gL,* – g̃L because the relative cellular sensitivity to L among the NCI-60 panel was of our interest.
Regarding gL,* of ferroptosis inducers, we submitted ten lethal compounds to NCI: four GSH synthesis inhibitors (erastin, AE, PE, MEII); four covalent GPX4 inhibitors (racemic RSL3, (1S,3R)-RSL3, ML162, ML210); and two non-covalent GPX4 inhibitors (CIL56 and FIN56). gL,* of the ten ferroptosis inducers were acquired in biological duplicates at NCI. LogGI50 value of MEII in EKVX cell line (gMEII,EKVX) was estimated by averaging gL,EKVX of the other GSH depleting compounds (i.e., L=erastin, AE, and PE).
Next, we sought cell-line-selective lethal compounds. The parameter, cell-line selectivity score, was defined by the interquartile range (IQR; the difference between the upper and lower quartiles) of each GL,*. The cell-line selectivity scores of 6,249 compounds as well as ferroptosis inducers were computed. The cell-line selectivity score of ferroptosis inducers (excluding CIL56, which induces both ferroptosis and necrosis) was substantially larger than other compounds. 2,555 compounds whose scores were greater than that of at least one specific ferroptosis inducer were defined as cell line-selective lethal compounds.
Model-Based Clustering of Cell Line-Selective Lethals
Cluster analysis groups objects in a way where objects within a cluster are more similar than between clusters. Specifically, we chose to cluster 2,565 GL,* (10 ferroptosis inducers + 2,555 other cell-line-selective lethal compounds) using model-based clustering, which estimates the number of clusters as well as the membership of each object (Fraley and Raftery, 2002). The 2,565 compounds were grouped into 53 clusters.
The drug sensitivity data from the NCI-60 project came with mechanisms of action (MoA) of lethal compounds if known. We computed whether each MoA was overrepresented in any of the 53 clusters using Fisher’s exact test. A MoA is assigned to a compound cluster Γ when (i) Γ overrepresented MoA (FDR < 0.1) and (ii) Γ contained five or more compounds with the MoA. This analysis revealed that MoA targeting DNA were all relatively similar to each other. Therefore, we consolidated these adjacent clusters into one new cluster applying cutree function to the hierarchical clustering dendrogram of the 53 compound clusters. This ended up consolidating 53 clusters into 18 clusters.
A GI50 profile of a compound cluster Γ (GΓ,*) was represented by the median value of GL,* among L assigned to the same cluster Γ; we thus computed GΓ,* for both 53 and 18 clusters.
Transcriptional Analysis
Microarray data of NCI-60 cell lines (XT,C) using Affymetrix U133 Platform was available at https://wiki.nci.nih.gov/display/NCIDTPdata/Molecular+Target+Data. They are robust multi-array (RMA)-normalized microarray data of NCI-60 cell lines generated by Gene Logic, Inc. Based on their differential expression of XT,C (evaluated using IQR), 13,748 T (80% of the total genes on the chip) were chosen for further analyses. We first computed the Spearman correlation coefficients (ρT,Γ), i.e., ρT,Γ = corrSpearman(XT,C,GΓ,C)C between 13,748 T and 53 Γ.
We then performed Gene Ontology enrichment analysis using ρT,Γ computed above. GO were taken from Molecular Signatures Database (MSigDB) from the Broad Institute (http://www.broadinstitute.org), and GO containing ten T or more were examined in the Gene Set Enrichment Analysis (GSEA) framework (Subramanian et al., 2005). Significance of the enrichment was assessed using random sample permutation. We looked for GO terms satisfying two criteria, significance and specificity, defined as; (i) significance: GO’s enrichment against Γ should be pGO,Γ < 0.001 (or equivalently, -log10 pGO,Γ > 3). There were 386 GO’s satisfying the criteria against more than one Γ, and (ii) Specificity > 2. For each GO and each Γ, ‘signed log p-value’ was defined by slpGO,Γ = (−log10 pGO,Γ)*sign(eGO,Γ), where sign() is a sign function returning +1 or −1, depending on the sign of the enrichment score, eGO,Γ; e.g., GO is enriched in {T ρT,Γ > 0}, then slpGO,Γ > 0. Specificity ST,G is then defined by SGO,Γ = max((slpGO,Γ - slpGO,-Γ)*sign(eGO,Γ)), where slpGO,-Γis a vector of signed log p-values for GO and all the clusters except Γ; note that SGO,Γ is positive when and only when GO is most significantly associated with the compound cluster Γ, and it gets larger when GO’s association is more specific to Γ. In sum, we sought {(GO,Γ) -log10 pGO,Γ > 3,SGO,Γ > 2}, and expected such GO to be a specific and significant biomarker for the sensitivity to Γ.
Pathway-Drug Association in Cancer Therapeutic Response Portal (CTRP)
We analyzed drug sensitivity data of 354 lethal compounds in 242 cell lines from another project, CTRP, available in supplementary data of Basu et al. (Basu et al., 2013). Lethal compounds tested in CTRP included four ferroptosis inducers, all of which were tested in 115 cell lines, and 255 lethal compounds were also tested in the same 115 cell lines, which we analyzed in a similar way to our NCI-60 data analysis. We first computed cell-line selectivity of 255 compounds, and discovered 88 compounds were as cell-line selective as ferroptosis inducers. As we found the number of compounds were too few to seek compound clusters, as we did in the NCI-60 data analysis, we instead computed signed log p-values of the enrichment of NAD(P)(H)-dependent oxidoreductases (i.e., GO:0016616 + GO:0016651) against the drug sensitivity of the 88 compounds.
Cell Lines and Media
HT-1080 (fibrosarcoma), A-673 (Ewing’s sarcoma), MCF7 (breast), and PC-3 (prostate) cells were obtained from American Type Culture Collection. 143B cells (osteosarcoma) were obtained from Eric Schon (Columbia University). Renal cell carcinoma UO-31 and 786-0 cells were from National Cancer Institute. LOX-IMVI (melanoma), NCI-H1975 (lung), LS411N (large intestine), Panc-1 (pancreas), IGROV-1 (renal cell carcinoma) cells were from Columbia Genome Center. Four BJ cell lines were grown in DMEM High-Glucose media (Life Technologies), 20 % Medium 199 (Sigma), and 15 % heat-inactivated fetal bovine serum (FBS). HT-1080 cells were grown in DMEM High-Glucose media with 1 % non-essential amino acids (Life Technologies) and 10 % FBS. 143B, A-673, MCF7 and Panc-1 cells were grown in DMEM High-Glucose media with 1 % glutamine and 10% FBS. IGROV-1, PC-3, LOX-IMVI, UO-31, 786-0, NCI-H1975, and LC411N were grown in RPMI 1640 (Mediatech) with 10% FBS. All the cell lines were grown at 37 °C under 5% CO2.
Chemicals
Erastin and (1S, 3R)-RSL3 were synthesized as described previously (Yagoda et al., 2007; Yang et al., 2014). FIN56 was synthesized using the procedure reported previously (#WO2008140792A1) (Cholody et al., 2008). α-tocopherol was purchased from Sigma-Aldrich. NSC704397 was from Vitas-M labs.
Cell Viability Assay upon Lethal Compound Treatment
1000 cells/36 µl were seeded in each well in 384-well plates and incubated with 2-fold, 12-point dilution series of lethal compounds with different highest concentrations (erastin: 40 µM, (1S,3R)-RSL3: 4 µM, FIN56: 20 µM, NSC704397: 100 µM) at 5% CO2, 37 °C for 48 hours. 10 µl/well of 50% Alamar Blue (Life Technologies) prepared in 10% FBS in DMEM was added to cell culture, and further incubated at 37 °C for 6 hours. Fluorescence intensity (ex/em: 530/590) was measured with a Victor 3 plate reader (Perkin Elmer) and the viability was normalized. The viability was measured in biological quadruplicates. The representative dose-response curves, the mean and standard error of normalized viability from one replicate were plotted.
NADH and NADPH Quantification Assay
NADH and NADPH levels were analyzed using a fluorimetric NAD/NADH assay (Abcam) and Amplite fluorimetric NADP/NADPH ratio assay kit (AAT bioquest). NADH and NADPH measurements for Fig. 7A were performed simultaneously. 1 million cells were grown in 10 cm tissue culture dishes for 16 hrs. Cells were treated with either vehicle or a ferroptosis inducer for the indicated time (0.1% DMSO for 8 hrs, 5 µM FIN56 for 8 hrs, 10 µM erastin for 8 hrs, 0.5 µM (1S,3R)-RSL3 for 3 hrs), with or without 100 µM α-tocopherol supplementation. For the NADPH measurement for Fig. 7C and Fig. S6, 12 cell lines (HT-1080, 143B, A673, MCF7, Panc-1, IGROV-1, PC-3, LOX-IMVI, UO-31, 786-0, NCI-H1975, and LC411N) were seeded at 1 million cells per 10cm dish, incubated at 37°C for 16 hrs and treated with either vehicle (0.1% DMSO) or 5 µM FIN56 for 6 hrs. After treatment, cells were trypsinized, pelleted, and washed with PBS once. For NADH and NADPH simultaneous measurement, cells were aliquoted into two eppendorf tubes and flash frozen in an isopropanol bath containing dry ice and stored at-80°C overnight. On the analysis day, each cell pellet was lysed in 200 µl lysis buffer and processed with both kits following the manufacturers’ instructions with a slight modification; the fluorescence (ex/em: 530/590) was measured every minute for one hour upon enzyme-substrate mixture addition to cell extracts. The data from the time-course measurement were fitted using the equation derived from Michaelis-Menten kinetics: Yt =Y0 +(Ymax -Y0)*(1- exp(−K *(t -t0))), where Y and t were the fluorescence intensity and time, respectively. Y0, Yt, and Ymax were Y at time 0, t, and at the end point, respectively. The reaction and the measurement started at t=t0 (<0) and t=0, respectively. In this equation, K was confirmed to be proportional to the concentrations of NAD(P)H standards, and found to be a more accurate measurement than the fluorescence intensity at one time-point. The concentrations of these metabolites were normalized to the amount of protein. Measurements were done in biological triplicates (Fig. 7A) or duplicates (Fig. 7C and Fig. S6). The plots show the mean and its standard error.
Knockdown of NADK using siRNA
5 µL RNAiMAX and 20 pmol siRNA (control or NADK-targeting) added to 500 µL OptiMEM were incubated in an eppendorf tube for 15 minutes at room temperature. The mixture was then seeded to one well of a 6-well dish. The dish was incubated for 15 minutes at 37°C while HT-1080 cells were trypsinized, and the cells were seeded into three wells containing siRNA reagent at 120,000 HT-1080 cells in 1.5 mL culture media per well. The HT-1080 cells were incubated at 37 °C for 48 hrs and trypsinized again. For drug sensitivity assay, 1,000 HT-1080 cells were seeded, incubated with 14-point 2-fold dilution series of ferroptosis inducers for 48 hrs and their viability was measured. For qPCR, 150,000 HT-1080 cells were seeded, incubated for 12 hours and harvested. For NADH and NADPH assays, 500,000 to 1 million cells were seeded and incubated for 12 hours before harvest.
Statistical Analysis and Data Visualization
Dose-response curve plotting and EC50 computation were performed with Prism 5.0c. The rest of the statistics and plotting were performed using R language and the following R Packages and functions: wilcox.test (Wilcoxon rank sum test), fisher.test (Fisher’s exact test), heatmap.2 function in gplots package (plotting heatmap), mclust package (model-based clustering), rgraphviz package (a GO graph visualization), lm function (linear regressions), cor.test (significance of correlation), GSEA-P-R package (GSEA). P-values of enrichment scores in GSEA using random permutation was computed in Hotfoot HPC cluster at Columbia University.
Supplementary Material
Significance Statement.
In the era of precision medicine, mapping DNA sequence variants and transcriptional changes in individual patients’ cancers is feasible due to advances in sequencing technologies. There are large, collaborative efforts to assemble comprehensive molecular profiles of individual cancers. However, precision medicine requires not only identification of cancer-associated mutations, but also effective drugs for each cancer genotype, which is still a largely unsolved problem. One approach to address this problem has been to use large-scale testing of small molecules in genetically characterized cell lines. Although promising, this approach has not always led to reproducible results. We hypothesized that compounds that exhibited high cell-line-selective lethality would exhibit robust and consistent results across large pharmacogenomic studies and could be used to identify biomarkers predictive of sensitivity.
We analyzed the drug sensitivity data of 6,259 lethal compounds in the NCI-60 project and categorized lethal compounds as cell-line-selective or not based on interquartile ranges of log10 GI50 values across the NCI-60 cell line panel. We then clustered 2,565 cell-line-selective lethal compounds into 18 clusters. Correlating GI50 profiles of the 18 clusters and transcriptome data for the NCI-60 cell lines revealed biomarkers that may predict sensitivity of cell lines to specific lethal compound clusters. Notably, expression of NAD(P)(H)-dependent oxidoreductase genes was correlated with sensitivity to a cluster containing primarily ferroptosis-inducing compounds. This prediction was further experimentally validated: cellular NADP(H) abundance predicted sensitivity to ferroptosis inducers, and NADPH was depleted during ferroptosis.
Thus, analysis of cell-line selectivity improved the predictive power of pharmacogenomic analyses and enabled the discovery of a biomarker, NADP(H), that predicts the degree of sensitivity of cell lines to ferroptosis. The methodology described in this paper should be applicable to uncovering biomarkers for sensitivity of other cell-line-selective lethal compounds.
Highlights.
Cell-line-selective lethal compounds are consistent across pharmacogenomic studies
Compounds with distinct sensitivity profiles indicate distinct mechanisms of action
The GI50 profiles of ferroptosis inducers are cell-line-selective and unique
Basal NADP(H) level correlates with sensitivity to ferroptosis inducers
Acknowledgments
We acknowledge Yves Pommier and William Reinhold at National Cancer Institute for sharing the full names of drug mechanisms of action in the NCI-60 data. Brent R. Stockwell is an Early Career Scientist of the Howard Hughes Medical Institute, and this research was additionally funded by the National Institutes of Health (5R01CA097061, 5R01GM085081, R01CA161061), and New York Stem Cell Science (C026715).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.S. and B.R.S. conceived of the project, designed the experiments, analyzed the data and wrote the manuscript. K.S. performed all experiments and analyses except: M.H. participated in NADP(H) measurement in 12 cell lines (Fig. 7C, Fig. S6) and proofread the manuscript. N.C.P. tested the lethality of NSC704397 in HT-1080 cells (Fig. S3).
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, et al. An Interactive Resource to Identify Cancer Genetic and Lineage Dependencies Targeted by Small Molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc. Natl. Acad. Sci. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholody WM, Zang Y, Zuck K, Watthey JWH, Ohler Z, Strovel J, Ohler G, Chellappan S, Padia J. Derivatives of fluorene, anthracene, xanthene, dibenzosuberone and acridine and uses thereof. 2008 [Google Scholar]
- Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta BBA - Gen. Subj. 2013;1830:3217–3266. doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifert C, Powers RS. From cancer genomes to oncogenic drivers, tumour dependencies and therapeutic targets. Nat. Rev. Cancer. 2012;12:572–578. doi: 10.1038/nrc3299. [DOI] [PubMed] [Google Scholar]
- Fraley C, Raftery AE. Model-Based Clustering, Discriminant Analysis, and Density Estimation. J. Am. Stat. Assoc. 2002;97:611–631. [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg GS, Kuderer NM. Comparative Effectiveness Research, Genomics-Enabled Personalized Medicine, and Rapid Learning Health Care: A Common Bond. J. Clin. Oncol. 2012;30:4233–4242. doi: 10.1200/JCO.2012.42.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJWL, Quackenbush J. Inconsistency in large pharmacogenomic studies. Nature. 2013;504:389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudston TJ, Anderson W, Aretz A, Barker AD, Bell C, Bernabé RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkermann A, Skouta R, Himmerkus N, Mulay SR, Dewitz C, Zen FD, Prokai A, Zuchtriegel G, Krombach F, Welz P-S, et al. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. 2014a;111:16836–16841. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkermann A, Stockwell BR, Krautwald S, Anders H-J. Regulated cell death and inflammation: an auto-amplification loop causes organ failure. Nat. Rev. Immunol. 2014b;14:759–767. doi: 10.1038/nri3743. [DOI] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong K-K, Elledge SJ. A Genome-wide RNAi Screen Identifies Multiple Synthetic Lethal Interactions with the Ras Oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak N, Dölle C, Ziegler M. The power to reduce: pyridine nucleotides - small molecules with a multitude of functions. Biochem. J. 2007;402:205. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala E, Mologni L, Truffa S, Gaetano C, Bollag GE, Gambacorti-Passerini C. BRAF Silencing by Short Hairpin RNA or Chemical Blockade by PLX4032 Leads to Different Responses in Melanoma and Thyroid Carcinoma Cells. Mol. Cancer Res. 2008;6:751–759. doi: 10.1158/1541-7786.MCR-07-2001. [DOI] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural Mechanism for STI-571 Inhibition of Abelson Tyrosine Kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Shamji AF, Clemons PA, Hon C, Koehler AN, Munoz B, Palmer M, Stern AM, Wagner BK, Powers S, et al. Towards patient-based cancer therapeutics. Nat Biotech. 2010;28:904–906. doi: 10.1038/nbt0910-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Skouta R, Yang WS, Hayano M, Brown LM, Dixon SJ, Lee ED, Wolpaw AJ, Stockwell BR. Global Survey of Cell Death Mechanisms Reveals Metabolic Regulation of Ferroptosis. doi: 10.1038/nchembio.2079. (in revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Skouta R, Dixon SJ, Wang J, Dunn DE, Orman M, Shimada K, Rosenberg PA, Lo DC, Weinberg JM, Linkermann A, et al. Ferrostatins Inhibit Oxidative Lipid Damage and Cell Death in Diverse Disease Models. J. Am. Chem. Soc. 2014;136:4551–4556. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukste I, Bhalala O, Persico M, Stockwell BR. Using small molecules to overcome drug resistance induced by a viral oncogene. Cancer Cell. 2006;9:133–146. doi: 10.1016/j.ccr.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weïwer M, Bittker JA, Lewis TA, Shimada K, Yang WS, MacPherson L, Dandapani S, Palmer M, Stockwell BR, Schreiber SL, et al. Development of small-molecule probes that selectively kill cells induced to express mutant RAS. Bioorg. Med. Chem. Lett. 2012;22:1822–1826. doi: 10.1016/j.bmcl.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:865–869. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.