Abstract
Several endpoints associated with cellular responses to DNA damage as well as overt cytotoxicity were multiplexed into a miniaturized, “add and read” type flow cytometric assay. Reagents included a detergent to liberate nuclei, propidium iodide and RNase to serve as a pan-DNA dye, fluorescent antibodies against γH2AX, phospho-histone H3, and p53, and fluorescent microspheres for absolute nuclei counts. The assay was applied to TK6 cells and 67 diverse reference chemicals that served as a training set. Exposure was for 24 hrs in 96 well plates, and unless precipitation or foreknowledge about cytotoxicity suggested otherwise, the highest concentration was 1 mM. At 4 and 24 hrs aliquots were removed and added to microtiter plates containing the reagent mix. Following a brief incubation period robotic sampling facilitated walk-away data acquisition. Univariate analyses identified biomarkers and time points that were valuable for classifying agents into one of three groups: clastogenic, aneugenic, or non-genotoxic. These mode of action predictions were optimized with a forward-stepping process that considered Wald test p-values, receiver operator characteristic curves, and pseudo R2 values, among others. A particularly high performing multinomial logistic regression model was comprised of four factors: 4hr γH2AX and phospho-histone H3 values, and 24 hr p53 and polyploidy values. For the training set chemicals, the four-factor model resulted in 94% concordance with our a priori classifications. Cross validation occurred via a leave-one-out approach, and in this case 91% concordance was observed. A test set of 17 chemicals that were not used to construct the model were evaluated, some of which utilized a short-term treatment in the presence of a metabolic activation system, and in 16 cases mode of action was correctly predicted. These initial results are encouraging as they suggest a machine learning strategy can be used to rapidly and reliably predict new chemicals’ genotoxic mode of action based on data from an efficient and highly scalable multiplexed assay.
Keywords: γH2AX, phospho-histone H3, p53, flow cytometry, mode of action
Introduction
New mammalian cell genotoxicity assays continue to be developed for a variety of reasons, especially as a means to address current practices’ relatively low specificity, lack of information relating to mode of action (MoA), and modest throughput capacity. While some efforts have focused on improving the performance of conventional tests [Fowler et al., 2012; Bryce et al., 2008; Wood et al., 2010], others have incorporated newer/non-traditional endpoints that are more amenable to automated analysis platforms and/or provide greater amounts of MoA information [Huang et al., 2006; Westerink et al., 2009; Ridpath et al., 2011; Walmsley and Tate, 2012; Hendriks et al., 2012; Nagel et al., 2014; Li et al., 2015].
γH2AX is a biomarker of DNA double strand breaks that appears to be applicable to in vitro genotoxicity testing strategies [Audebert et al., 2010; Smart et al., 2011; Garcia-Canton et al., 2013; Nikolova et al., 2014]. As several previous reports have suggested, when γH2AX readings are coupled with mitotic cell (phospho-histone H3-positive event) readings, additional information is gained that is useful for discriminating between clastogenic and aneugenic activities [Bryce et al., 2014; Khoury et al., 2015; Cheung et al., 2015].
The work reported herein takes this multi-endpoint approach a step further by adding biomarkers that were predicted to increase the robustness of clastogenic, aneugenic, and non-genotoxicant signatures, and by multiplexing each of the endpoints into one simple assay. More specifically, we describe a comprehensive strategy with the following characteristics: i) use of p53 competent human cells, ii) homogeneous assay format, iii) automated data acquisition, iv) compatible with microtiter wells, v) multiplexed biomarkers that provide genotoxic MoA information, and vi) accompanied by algorithms that provide probability scores for categorization of clastogen, aneugen, or non-genotoxicant group membership. We consider this last point to be important, as the high dimensionality of the data relative to standard in vitro genotoxicity assays requires new approaches for efficiently synthesizing results into information on which decisions can be made. The generation of MoA predictions with associated probability scores was accomplished by multinomial logistic regression algorithms and is described in detail herein.
This work was performed with TK6 cells, and except for those chemicals requiring metabolic activation exposure was continuous for 24 hrs, with flow cytometric analysis occurring at both early (4 hr) and late (24 hr) time points. Once endpoints and time points that were effective at discriminating among different MoA were identified and a provisional multi-factorial model was developed, it underwent leave-one-out cross-validation. Subsequently, the computation model was used to predict genotoxic MoA on an external test set of chemicals, including several pro-genotoxicants that require the presence of an exogenous activation system to exert appreciable DNA damage. Based on the promising results described herein, it appears that several multiplexed biomarkers and a companion machine learning tool have the potential to endow mammalian cell genotoxicity assays with greater throughput, higher specificity, and important MoA information.
Materials and Methods
Chemicals
The identity of the 67 training set and 17 test set chemicals is provided in Table I, along with our a priori predictions of genotoxic potential. Clastogens (31) and aneugens (14) were selected from the literature and represent a broad range of genotoxic activities that have generally been observed in multiple laboratories and in both in vitro as well as in vivo systems. The non-genotoxicants (39) were selected for their diverse range of activities/toxic mechanisms, and many are part of an ECVAM list of agents that should be negative in in vitro genotoxicity testing [Kirkland et al., 2008, 2015]. Collectively these chemicals represented reference agents that were useful for identifying the time points and biomarkers that can discriminate clastogens, aneugens, and non-genotoxicants, developing supervised machine learning algorithms, and for validating the strategy.
Table I.
Training and Test Set Chemicals with a priori Classification.
| Chemical | CAS No. | Set | a priori Clasification; notes | References |
|---|---|---|---|---|
| AMG 900 | 945595-80-2 | Training | Aneugen; pan-Aurora kinase inhibitor; top conc. 0.1 µM | Payton et al., 2010 |
| Carbendazim | 10605-21-7 | Training | Aneugen; mitotic spindle poison | Van Hummelen et al., 1995 |
| Colchicine | 64-86-8 | Training | Aneugen; mitotic spindle poison; top conc. 0.1 µM | Kirkland et al., 2015 |
| Diethylstilbestrol | 56-53-1 | Training | Aneugen; synthetic estrogen | Elhajouji et al., 1997 |
| Griseofulvin | 126-07-8 | Training | Aneugen; mitotic spindle poison | Oliver et al., 2006 |
| Mebendazole | 31431-39-7 | Training | Aneugen; mitotic spindle poison; top conc. 10 µM | Van Hummelen et al., 1995 |
| Nocodazole | 31430-18-9 | Training | Aneugen; top conc. 10 µM | Attia, 2013 |
| Noscapine | 128-62-1 | Training | Aneugen; mitotic spindle poison | Schuler et al., 1999 |
| Paclitaxel | 33069-62-4 | Training | Aneugen; mitotic spindle poison; top conc. 1 µM | Kirkland et al., 2015 |
| Vinblastine sulfate | 143-67-9 | Training | Aneugen; mitotic spindle poison; top conc. 0.01 µM | Kirkland et al., 2015 |
| 17β-Estradiol | 50-28-2 | Test | Aneugen; steroid hormone | Hernández et al., 2013 |
| Fisetin | 345909-34-4 | Test | Aneugen; Aurora B kinase inhibitor; clastogenic properties reported at high concentrations that are attributed to topoisomerase II inhibition | Olaharski et al., 2005; Gollapudi et al., 2014 |
| Flubendazole | 31430-15-6 | Test | Aneugen; inhibits tubulin polymerization | Tweats et al., 2015 |
| Vincristine sulfate | 2068-78-2 | Test | Aneugen; mitotic spindle poison | Kondo et al., 1992 |
| 4-Nitroquinoline 1-oxide | 56-57-5 | Training | Clastogen; likely several modes of action that may include ROS; top conc. 10 µM | Kirkland et al., 2015 |
| 5-Fluorouracil | 51-21-8 | Training | Clastogen; anti-metabolite | Kirkland et al., 2015 |
| Aphidicolin | 38966-21-1 | Training | Clastogen; DNA polymerase inhibitor; top conc. 10 µM | Glover et al., 1984 |
| Azathioprine | 446-86-6 | Training | Clastogen; prodrug of mercaptopurine, purine analog | Henderson et al., 1993 |
| Azidothymidine | 30516-87-1 | Training | Clastogen; nucleoside analog | Kirkland et al., 2015 |
| Camptothecin | 7689-03-4 | Training | Clastogen; topoisomerase I inhibitor; top conc. 0.1 µM | Attia et al., 2009 |
| Chlorambucil | 305-03-3 | Training | Clastogen; nitrogen mustard-type alkylator; top conc. 100 µM | Dertinger et al., 2012 |
| Cisplatin | 15663-27-1 | Training | Clastogen; atypical alkylator; prepared immediately before use | Kirkland et al., 2015 |
| Cytosine arabinoside | 147-94-4 | Training | Clastogen; anti-metabolite; top conc. 10 µM | Kirkland et al., 2015 |
| Ethyl methanesulfonate | 62-50-0 | Training | Clastogen; alkylator | Gocke et al., 2009 |
| Etoposide | 33419-42-0 | Training | Clastogen; topoisomerase II inhibitor; top conc. 10 µM | Kirkland et al., 2015 |
| Methyl methanesulfonate | 66-27-3 | Training | Clastogen; alkylator | Kirkland et al., 2015 |
| Hydralazine HCl | 304-20-1 | Training | Clastogen; prepared in RPMI medium | Martelli et al., 1995 |
| Hydrogen peroxide | 7722-84-1 | Training | Clastogen; ROS; prepared in RPMI medium immediately before use | Kimura et al., 2013 |
| Hydroxyurea | 127-07-1 | Training | Clastogen; anti-metabolite, ribonucleotide reductase inhibitor | Dertinger et al., 2012 |
| Melphalan | 142-82-3 | Training | Clastogen; nitrogen mustard-type alkylator | Dertinger et al., 2012 |
| Menadione | 58-27-5 | Training | Clastogen; ROS implicated | Cojocel et al., 2006 |
| Methotrexate | 59-05-2 | Training | Clastogen; anti-metabolite; top conc. 10 µM | Keshava et al., 1998 |
| Methyl methanesulfonate | 66-27-3 | Training | Clastogen; alkylator | Kirkland et al., 2015 |
| Mitomycin C | 50-07-7 | Training | Clastogen; DNA cross-linker; top conc. 10 µM | Kirkland et al., 2015 |
| N-Ethyl-N-nitrosourea | 759-73-9 | Training | Clastogen; alkylator | Kirkland et al., 2015 |
| Temozolomide | 85622-93-1 | Training | Clastogen; alkylator | Chinnasamy et al., 1997 |
| Thiotepa | 52-24-4 | Training | Clastogen; alkylator | Dertinger et al., 2012 |
| Topotecan | 123948-87-8 | Training | Clastogen; topoisomerase I inhibitor; top conc. 0.1 µM | Aydemir and Bilaloğlu, 2003 |
| 1,3-Propane sultone | 1120-71-4 | Test | Clastogen; alkylator | Dertinger et al., 2011 |
| 2-Acetylaminofluorene | 53-96-3 | Test | Clastogen with rat liver S9 | Kirkland et al., 2015 |
| 7,12-Dimethylbenzanthracene | 57-97-6 | Test | Clastogen with rat liver S9 | Kirkland et al., 2015 |
| Benzo[a]pyrene | 50-32-8 | Test | Clastogen with rat liver S9 | Kirkland et al., 2015 |
| Bleomycin sulfate | 9041-93-4 | Test | Clastogen; radiomimetic | |
| Cyclophosphamide monohydrate | 6055-19-2 | Test | Clastogen with rat liver S9 | Kirkland et al., 2015 |
| Glycidamide | 5694-00-8 | Test | Clastogen; major in vivo metabolite of acrylamide | Paulsson et al., 2003 |
| Lynparza™ (Olaparib) | 763113-22-0 | Test | Clastogen; PARP inhibitor | FDA approved label |
| Amitrole | 61-82-5 | Training | Non-genotoxicant | Kirkland et al., 2015 |
| Anthranilic acid | 118-92-3 | Training | Non-genotoxicant | Kirkland et al., 2015 |
| Brefeldin A | 20350-15-6 | Training | Non-genotoxicant; ER-golgi transporter inhibitor, ER stress-induced apoptosis; top conc. 10 µM | Moon et al., 2012 |
| Caffeine | 58-08-2 | Training | Non-genotoxicant; mitochondria-dependent apoptosis, ROS involvement likely | Lu et al., 2008 |
| Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) | 555-60-2 | Training | Non-genotoxicant; uncoupler of oxidative phosphorylation; top conc. 100 µM | de Graaf et al., 2004 |
| Clofibrate | 637-07-0 | Training | Non-genotoxicant; antilipidemic agent | IARC monograph |
| Cyclohexanone | 108-94-1 | Training | Non-genotoxicant; industrial chemical | Kirkland et al., 2008 |
| Cycloheximide | 66-81-9 | Training | Non-genotoxicant; protein synthesis inhibitor; top conc. 100 µM | Youngblom et al., 1989 |
| D-Mannitol | 69-65-8 | Training | Non-genotoxicant; polyol | Kirkland et al., 2015 |
| Dexamethasone | 50-02-2 | Training | Non-genotoxicant; glucocorticoid receptor agonist | Krishna et al., 1995 |
| Dextrose | 50-99-7 | Training | Non-genotoxicant; sugar | Lotz et al., 2009 |
| Di-(2-ethylhexyl)phthalate (DEHP) | 117-81-7 | Training | Non-genotoxicant; organic plasticizer | Kirkland et al., 2015 |
| Diethanolamine | 111-42-2 | Training | Non-genotoxicant; secondary amine | Kirkland et al., 2015 |
| Erythromycin | 114-07-8 | Training | Non-genotoxicant; antibiotic | Kirkland et al., 2015 |
| Pepcid® (Famotidine) | 76824-35-6 | Training | Non-genotoxicant; histamine H2 receptor antagonist | FDA approved label |
| Gleevec® (imatinib mesylate) | 152459-95-5 | Training | Non-genotoxicant; protein-tyrosine kinase inhibitor | FDA approved label |
| Hexachloroethane | 67-72-1 | Training | Non-genotoxicant; industrial chemical | Kirkland et al., 2015 |
| Lidoderm® (Lidocaine) | 137-58-6 | Training | Non-genotoxicant; amide local anesthetic | FDA approved label |
| Mevacor® (Lovastatin) | 75330-75-5 | Training | Non-genotoxicant; HMG-CoA reductase inhibitor | FDA approved label |
| Melamine | 108-78-1 | Training | Non-genotoxicant: industrial organic base | Kirkland et al., 2015 |
| Methyl carbamate | 598-55-0 | Training | Non-genotoxicant; industrial intermediate | Kirkland et al., 2015 |
| N-Butyl chloride | 109-69-3 | Training | Non-genotoxicant; fumigant | Kirkland et al., 2015 |
| Floxin® (Ofloxacin) | 82419-36-1 | Training | Non-genotoxicant; fluoroquinoline antibiotic; top conc. 500 µM due to solubility | FDA approved label |
| Paxil® (Paroxetine) | 61869-08-7 | Training | Non-genotoxicant; SSRI antidepressant | FDA approved label |
| Phenanthrene | 85-01-8 | Training | Non-genotoxicant; polycyclic aromatic hydrocarbon | Kirkland et al., 2008 |
| Phenformin HCl | 834-28-6 | Training | Non-genotoxicant; biguanide antidiabetic | Kirkland et al., 2015 |
| Progesterone | 57-83-0 | Training | Non-genotoxicant; steroid hormone | Kirkland et al., 2008 |
| Pyridine | 110-86-1 | Training | Non-genotoxicant; heterocyclic organic compound | Kirkland et al., 2015 |
| Sodium chloride | 7647-14-5 | Training | Non-genotoxicant; prepared in RPMI medium | Matsushima et al., 1999 |
| Sodium dodecyl sulfate | 151-21-3 | Training | Non-genotoxicant; ionic detergent | NTP website |
| Sucrose | 57-50-1 | Training | Non-genotoxicant | Diaz et al., 2007 |
| Tert-butyl alcohol | 75-65-0 | Training | Non-genotoxicant | Kirkland et al., 2015 |
| Thapsigargin | 67526-95-8 | Training | Non-genotoxicant; ER stress-induced apoptosis | Futami et al., 2005 |
| Tunicamycin | 11089-65-9 | Training | Non-genotoxicant; glycosylation inhibitor, ER stress-mediated apoptosis; top conc. 500 µM due to solubility | Han et al., 2008 |
| Alosetron HCl | 122852-42-0 | Test | Non-genotoxicant; 5-HT3 antagonist; top conc. 250 µM due to solubility | Kirkland et al., 2015 |
| D-Limonene | 5989-27-5 | Test | Non-genotoxicant; male rat kidney tumors due to α2μ-globulin nephropathy | Kirkland et al., 2015 |
| Nutlin-3 | 548472-68-0 | Test | Non-genotoxicant; inhibitor of Mdm2 and p53 interaction, stabilizes p53, | Secchiero et al., 2006 |
| Tolterodine L-tartrate | 124937-52-6 | Test | Non-genotoxicant; muscarinic receptor antagonist | Kirkland et al., 2015 |
| Zonisamide | 68291-97-4 | Test | Non-genotoxicant; sulfonamide anticonvulsant | Kirkland et al., 2015 |
DNA Damage Assay
Reagents necessary to prepare nuclei for flow cytometric analysis were part of a prototype kit, i.e., MultiFlow™ DNA Damage Kit—p53, γH2AX, Phospho-Histone H3 (Litron Laboratories, Rochester, NY). The proprietary working solution was used to simultaneously digest cytoplasmic membranes, stain chromatin with a fluorescent nucleic acid dye, and label several epitopes with fluorescent antibodies. With this kit, anti-γH2AX-Alexa Fluor® 647 is used as a DNA double strand break marker, anti-phospho-histone H3-PE serves as a mitotic cell marker, and anti-p53-FITC recognizes an N-terminal domain of p53. RNase and propidium iodide provide cell cycle and polyploidization information, and a known concentration of latex microspheres (“counting beads”; i.e., Sphero™ Multi-Fluorophore Particles, cat. no. FP-3057-2; Spherotech, Inc.,Lake Forest, IL) is used to calculate nuclei density.
Cell culture and treatments
TK6 cells were obtained from ATCC® (cat. no. CRL-8015), and grown in a humid atmosphere at 37°C with 5% CO2. For routine culturing, the cells were maintained at or below 1 × 106 cells/mL. The culture medium consisted of RPMI 1640 with 200 µg/mL sodium pyruvate, 200 µM L-glutamine, 50 units/mL penicillin, 50 µg/mL streptomycin, and 10% v/v heat-inactivated horse serum.
On the day of treatment, logarithmically growing cells were exposed to 20 concentrations of test chemical. Each concentration was evaluated in a single culture except for solvent control that was evaluated in 4 replicate cultures. The solvent was DMSO except in several instances noted in Table I. In all cases final solvent concentration was 1% v/v in culture. Top test chemical concentration was 1000 µM unless precipitation was observed in the 100× stock solution or upon addition to culture medium; in these cases the highest non-precipitating concentration was evaluated. Table I notes instances when less than 1000 µM was used as the top concentration based on foreknowledge about cytotoxicity. Regardless of top starting concentration, every successively lower concentration differed by a factor of square root 2, for example 1000, 707, 500 µM, etc.
Treatment of cells took place after they were adjusted to 2 × 105 cells/mL and 198 µL of this cell suspension was added to each well of round-bottom 96-well plates. Addition of test article (2 µL/well) was immediately followed by re-incubation in a humid atmosphere at 37°C with 5% CO2. As described below, 25 µL of cells were removed from each well for flow cytometric analysis at the 4 and 24 hr time points.
Four test set chemicals (benzo[a]pyrene, cyclophosphamide, 7,12-dimethylbenzanthracene, and 2-acetylaminofluorene) were studied in conjunction with rat liver S9 metabolic activation. In these cases treatment occurred in 96 well plates as above, however a commercially available metabolic activation system was used to provide a final S9 concentration of 2% (Mutazyme Lyophilized Rat Liver S9; Molecular Toxicology, Inc., Boone, NC). At the 4 hr time point cells were washed out of exposure medium by two successive centrifugation (340 × g, 5 min) and resuspension steps (200 µL/well). The cells were finally resuspended with 200 µL/well fresh growth medium. At this point 25 µL of washed cells were removed from each well for flow cytometric analysis and the remainder were returned to the incubator for an additional 20 hrs when cells were removed for a final analysis.
Flow cytometric analysis
At 4 and 24 hr time points cells were resuspended with pipetting and then 25 µL were removed from each well and added to a new 96-well plate containing 50 µL/well of pre-aliquoted working MultiFlow Kit reagent. Mixing occurred by pipetting the contents of each well several times. After a 10 min room temperature incubation period, flow cytometric analysis occurred using either a FACSCanto™ II flow cytometer equipped with a BD™ High Throughput Sampler or a Miltenyi Biotec MACSQuant® Analyzer 10 flow cytometer with integrated 96-well MiniSampler device. Both of these instruments provided automated data acquisition. The user-defined mixing and fluidics parameters for the FACSCanto were programmed as follows: 40 µL of sample were mixed 4 times at a mixing speed of 180 µL/sec, and then 20 µL were analyzed at a flow rate of 1 µL/sec until 10,000 total events were acquired or the entire 20 µL volume was exhausted, whichever came first. Between samples the High Throughput Sampler was programmed to wash the probe with 400 µL of sheath fluid. The user-defined mixing and fluidics parameters for the MACSQuant were as follows: 40 µL of sample were mixed, and then 20 µL were analyzed at a flow rate of 50 µL/min until the 20 µL volume was exhausted. The “screen mode” setting ensured that the probe was rinsed with 208.4 µL of sheath fluid between samples. Given these programs, each 4 hr sample tended to provide approximately 1,700 2n and greater nuclei for analysis, and each 24 hr sample tended to provide approximately 4,800 2n and greater nuclei.
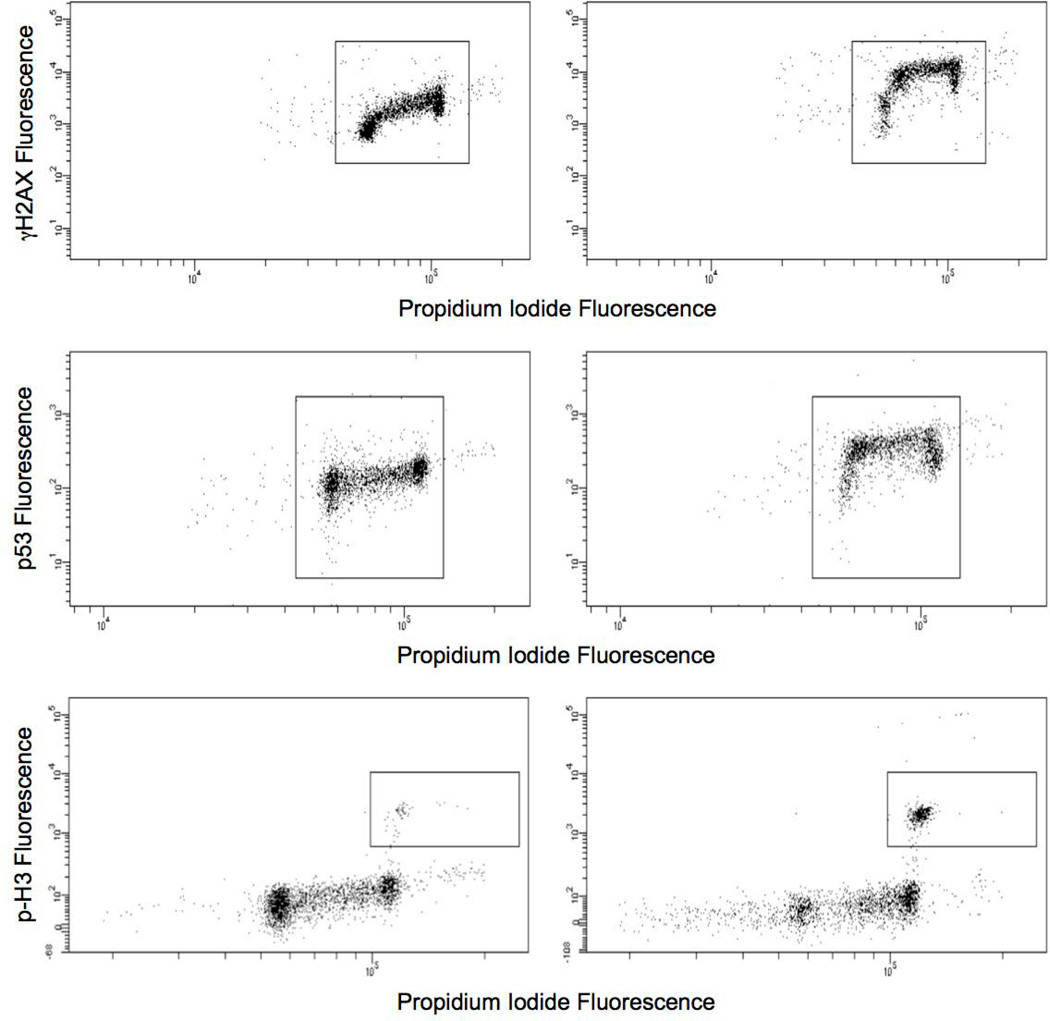
Two endpoints, γH2AX and p53, were based on median fluorescence intensity, and for all graphical representation and statistical analyses they were expressed as fold-change relative to a plate-specific solvent control arithmetic mean value. Gating logic required these events to exhibit propidium iodide-associated fluorescence corresponding to 2n – 4n DNA content (see Figure 1, top and middle panels). In order to limit the influence that mitotic and apoptotic cells might have on these measurements, phospho-histone H3 (p-H3) positive cells and highly fluorescent γH2AX-positive events were excluded from analysis [McManus and Hendzel, 2005; Huang et al., 2006; Rogakou et al., 2000].
Figure 1.
Top panel: TK6 nuclei with γH2AX-associated fluorescence plotted against propidium iodide fluorescence. These nuclei were prepared from cells treated with solvent (left) or mitomycin C (right). Increased γH2AX fluorescence is indicative of clastogenic (double strand-breaking) activity. Middle panel: TK6 nuclei with p53-associated fluorescence plotted against propidium iodide fluorescence. These nuclei were prepared from cells treated with solvent (left) or etoposide (right). Increased p53 fluorescence is indicative of genotoxicant-induced translocation of cytoplasmic p53 to the nucleus. Bottom panel: TK6 nuclei with phospho-histone H3-associated fluorescence plotted against propidium iodide fluorescence. These nuclei were prepared from cells treated with solvent (left) or griseofulvin (right). Phospho-histone H3-positive events are mitotic cells, and increased frequencies are a typical result of exposure to aneugenic agents.
p-H3 and polyploidy data were based on their frequency among other cells. The p-H3 measurements were the proportion of p-H3-positive events that exhibited propidium iodide-associated fluorescence of 4n and greater DNA content relative to the number of total events with 2n and greater DNA content (see Figure 1, bottom panel). Polyploidy was quantified as the proportion of 8n-positive events relative to the number of total events with 2n and greater DNA content. For all graphical representations and statistical analyses presented herein the p-H3 and polyploidy data were converted to fold-change relative to a plate-specific solvent control arithmetic mean value.
Latex microspheres were included in the working dye/antibody solution at a known concentration (between 8.1 and 9.5 × 104/mL) and this allowed these particles to serve as counting beads. Nuclei to counting bead ratios were calculated for each specimen, and this was used to determine absolute nuclei counts (those with 2n and greater DNA-associated propidium iodide fluorescence). These values were used to calculate 24 hr cytotoxicity values relative to plate-specific mean solvent control wells, and included relative nuclei counts (RNC), relative population doublings (RPD), and relative increased nuclei counts (RINC). The equations for calculating these values are provided in Lorge et al., 2008. As described in more detail below, for graphical representations and statistical analyses associated with this report, %cytotoxicity refers to a derivation of the RNC values, that is 100 – RNC at 24 hrs.
Electronic compensation was used to eliminate propidium iodide's fluorescence spillover into the PE channel. When viewing a PE versus propidium iodide bivariate plot without compensation, nuclei show increased PE and propidium iodide fluorescence as DNA content increases. With proper subtraction of the propidium iodide's fluorescence spillover, nuclei events show increased propidium iodide-associated fluorescence with increasing DNA content, but the associated increase in the PE channel is eliminated. This provides for optimal resolution of p-H3-positive events based on anti-p-H3-PE labeling. Further details about the flow cytometric analyses, including gating logic and other considerations, are provided in a Supplemental file.
Supervised machine learning
Multinomial logistic regression was used to construct and evaluate predictive models for categorizing chemicals into one of three MoA groups—clastogens, aneugens, or non-genotoxicants (JMP® software, v12.0.1, SAS Institute Inc., Cary, NC). While any number of machine learning tools may have been suitable, we focused our efforts on logistic regression based on several advantageous characteristics including: its widespread use in biological sciences; simplicity of execution; the fact that it provides probability scores with each group membership prediction; and the relatively few assumptions that are made about the nature of the data, for instance continuous data need not be normally distributed [Hosmer et al., 2013].
Initially univariate analyses performed with training set data were used to identify biomarkers and time points that may be valuable for classifying agents into one of the three groups. Statistics that were useful for this purpose included Wald test p-values, McFadden’s pseudo R2, receiver operator characteristic (ROC) curves, and percentage of correctly categorized chemicals. To evaluate whether multiple biomarker data would improve the model, an iterative forward-stepping approach was used. As covariates identified for inclusion were added to the multivariate model, the importance of each endpoint was assessed in a manner similar to the univariate analyses described above. A particularly high performing model consisted of four-factors: 4hr γH2AX, 4 hr p-H3, 24 hr p53, and 24 hr polyploidy fold-change data. Weighting the model on cytotoxicity (i.e., 100% – RNC at 24 hrs) resulted in further improvements to Wald p-values, McFadden’s pseudo R2, ROC values, and percentage of correctly categorized chemicals.
Note that we previously described a multinomial logistic regression strategy whereby each chemical provided data from only one concentration [Bryce et al., 2014]. In that report RNC data were used to select one concentration for each chemical that were to a first approximation equitoxic. That data reduction technique was not employed for the work described herein. Rather, for the current machine learning strategy data from every concentration were utilized. One exception, described in greater detail below, is that we excluded data associated with exposures that resulted in greater than 80% cytotoxicity.
A consequence of building a logistic regression model with data from multiple concentrations is that it calculates numerous group membership (MoA) probabilities. This explains why the graphs presented herein show a range of probability scores for each chemical. Since a prediction as to genotoxic MoA occurred at every concentration, this also explains the need to synthesize the model output (probability scores) into a final judgment as to MoA, and this is described in detail below.
Call criteria, definitions
A clastogenic MoA call required two successive concentrations to exhibit clastogen probability scores ≥ 90%. Aneugenic MoA required two successive concentrations to exhibit aneugen probability scores ≥ 90%. A non-genotoxicant MoA call was defined as no two successive concentrations exhibiting clastogenic or aneugenic probability scores ≥ 90%. An equivocal result was defined as only one concentration exhibiting a clastogenic or aneugenic probability score ≥ 90%.
Assay sensitivity was calculated by determining the percentage of clastogens and aneugens that were identified as such. Assay specificity was calculated by determining the percentage of non-genotoxicants that were identified as such. Assay concordance was calculated by determining the percentage of correct calls compared to total calls.
Cross-validation, external validation
The four-factor weighted model was evaluated using a cross-validation approach known as leave-one-out [Hosmer et al., 2013]. With this method, each one of the 67 training set chemicals was in turn removed from the model, and a prediction was made as to its genotoxic MoA. The model was further evaluated by testing an additional 17 chemicals that were not part of the training set.
Results and Discussion
Multiplexed endpoints
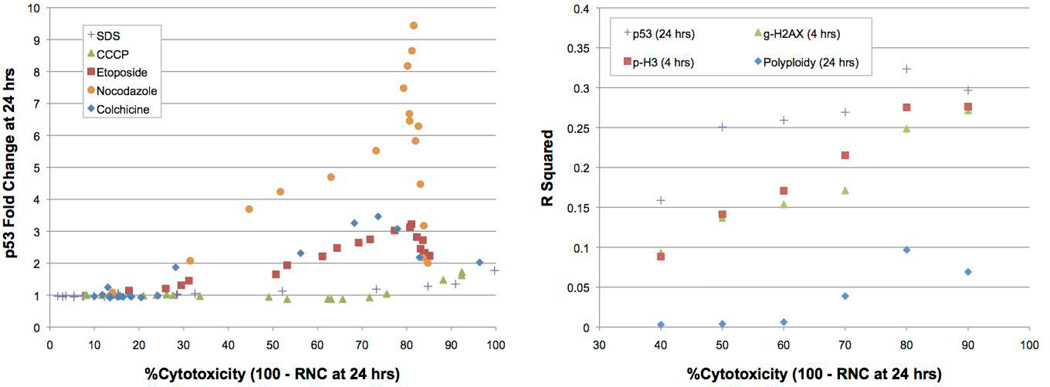
We used absolute nuclei counts to derive several related cytotoxicity measurements: RNC, RINC, and RPD. A graph that directly compares these measurements is provided as a Supplemental file. Furthermore, all flow cytometry data, including these statistics, are provided for each chemical at each concentration in an annotated Supplemental (Excel) file. For the purposes of this manuscript we chose to focus on the simplest of the cytotoxicity measurements, RNC, which unlike the others does not require an initial (time zero) reading. Figure 2 (left panel) depicts 24 hr p53 responses for several diverse chemicals plotted against 100% - RNC values. As was the case for γH2AX, p-H3, and polyploidy (data not shown), the p53 results indicate that maximal responses to clastogens as well as aneugens occurs in the context of some degree of cytotoxicity, whereas higher levels were observed to saturate or even reduce the effects. Additionally, by considering non-genotoxicants, it is also apparent that the specificity can be adversely affected once cytotoxicity becomes excessive. In other words, as with other mammalian cell genotoxicity test systems, it is perhaps not surprising that this multiplexed assay benefits from a cytotoxicity limit in order to maximize signal to noise ratios.
Figure 2.
Left panel: Fold-increases in p53 median channel are graphed against an index of cytotoxicity at the 24 hr time point (i.e., 100% - Relative Nuclei Count). Responses are graphed for several genotoxic agents, etoposide, nocodazole, and colchicine, as well as two cytotoxic non-genotoxicants, sodium dodecyl sulfate (SDS) and carbonyl cyanide m-chlorophenyl hydrazone (CCCP). From these data it is apparent that p53 responses can plateau or downturn after a certain level of cytotoxicity occurs, and non-genotoxicants begin to modestly affect the p53 biomarker at a certain level of cytotoxicity. Right panel: Univariate logistic regression analyses for each of several biomarkers were performed for 67 training set chemicals using variable cytotoxicity limits (40 to 90%). Resulting R2 values are graphed against each of the six cytotoxicity limits that were considered. These data indicate that 80% is useful for optimizing the assay to distinguish among three modes of action.
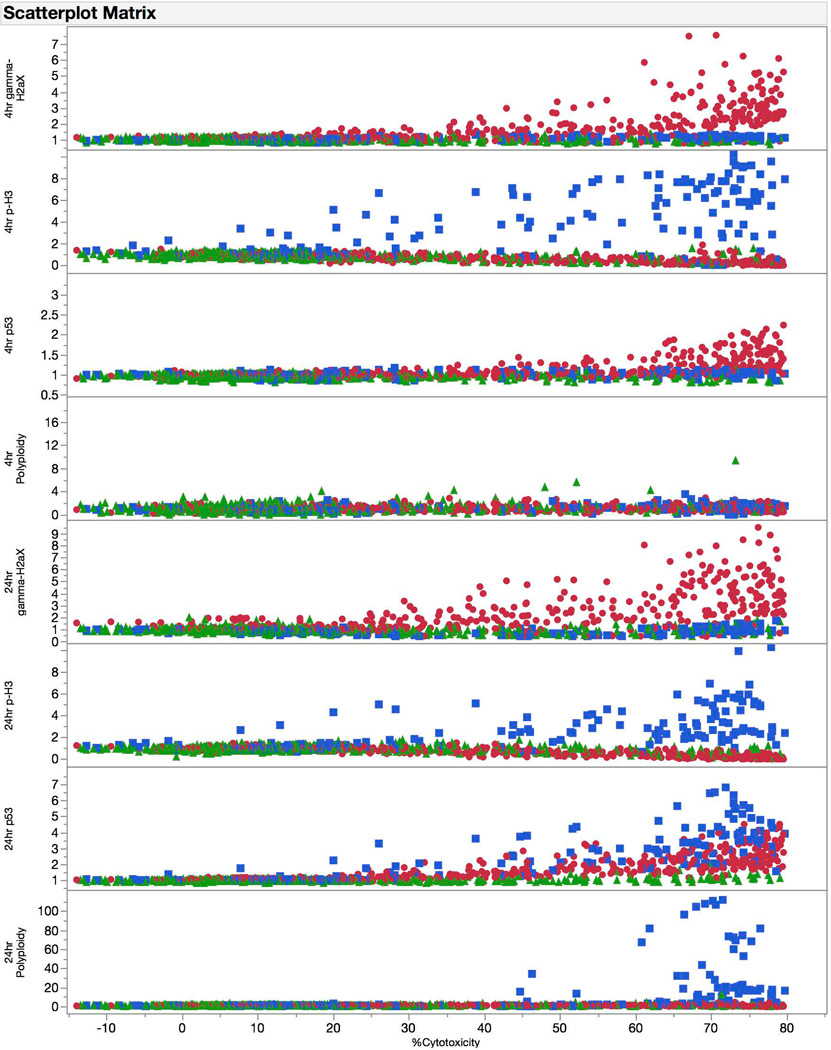
In an effort to optimize the cytotoxicty cutoff value, we performed univariate logistic regression analyses with all 67 training test chemicals (see Figure 2, right panel). For this exercise the cytotoxicity limit was varied from 40 to 90%. Each resulting R2 value, a measure of fit used to assess model accuracy, is graphed against the cytotoxicity limit. Collectively these data demonstrate that a cytotoxicity cutoff value of approximately 80% represents a good compromise across multiple endpoints. This is evident from another aggregate view of the data provided in Figure 3. Here, γH2AX, p-H3, p53, and polyploidy data for all 67 training set chemicals are graphed as a function of cytotoxicity. This graphical presentation of the data reinforces the premise that even for these disparate biomarkers and time points, it is possible to optimize signal to noise ratios by testing to one pre-defined cytotoxicity limit.
Figure 3.
Fold-increase responses for each biomarker at each of two time points are graphed against a measure of cytotoxicity (i.e., 100% - Relative Nuclei Count at 24 hrs). The graphs are aggregate data for 67 training set chemicals, and are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. These graphs provide insight into which biomarkers are effective at discriminating among chemical classes, and provide further evidence that testing up to cytotoxic concentrations, to a point, maximizes signal to noise ratios across these disparate endpoints.
When considering the appropriateness of the 80% cytotoxicity cutoff value several points should be kept in mind. First, as described above, it was empirically chosen to balance assay sensitivity with specificity—the same can be said of conventional mammalian cell genotoxicity assays. Second, the cytotoxicity measurements are made after 24 hrs of continuous exposure. Obviously biomarkers evaluated at early time points (e.g., 4 hrs) have not experienced this level of cytotoxicity. Finally, at this juncture we consider the 80% cutoff value provisional in nature. While it appears optimal based on results from the current data set, it is important to recognize that the analyses considered a single cell line, one treatment schedule, and only 67 chemicals.
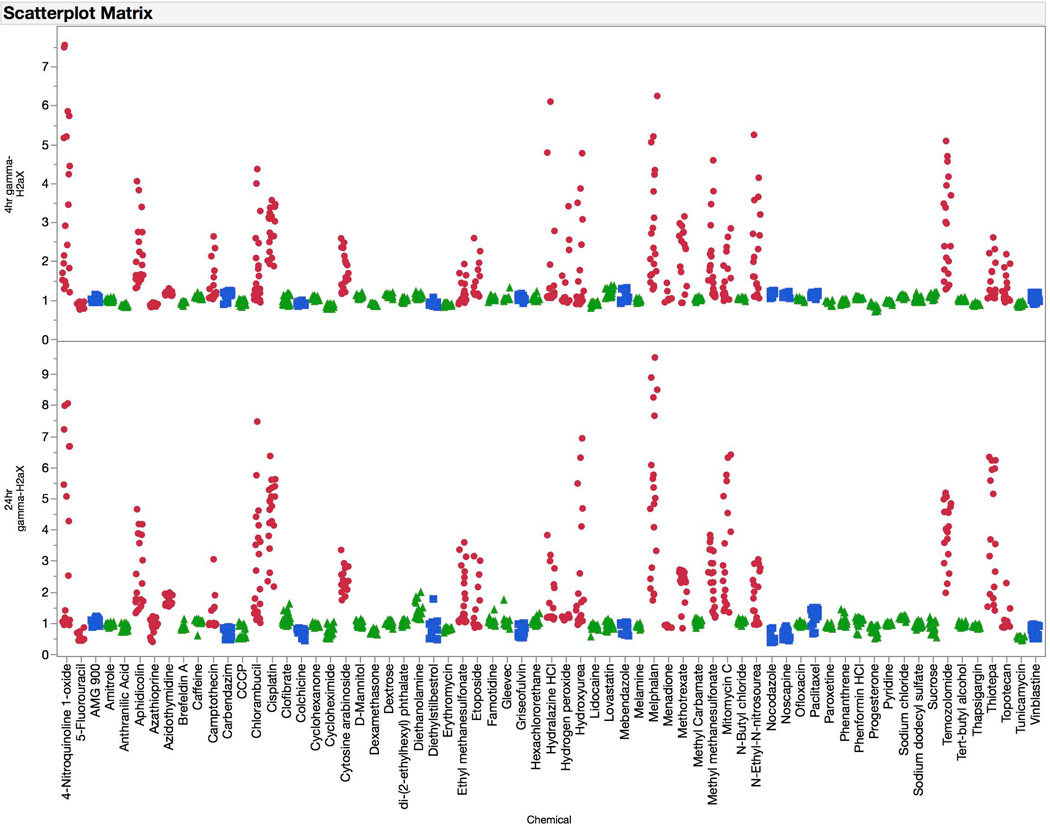
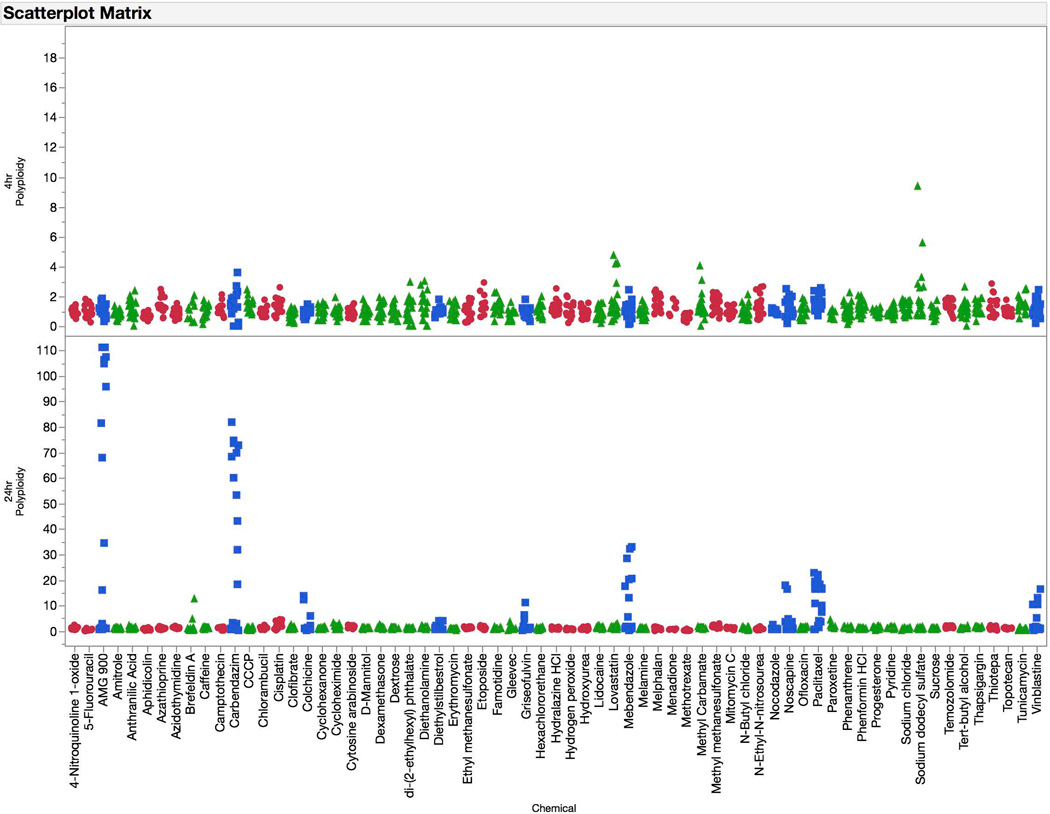
Training set chemicals’ 4 and 24 hr γH2AX, p-H3, p53, and polyploidy data (expressed as fold-change versus mean solvent control) are depicted in Figures 4 – 7. As shown in Figure 4, increased γH2AX-associated fluorescence was characteristic of clastogen exposure. In general the responses were quite similar at the 4 and 24 hr time points. However, there were some agents that exhibited markedly higher responses at 24 hrs. Mitomycin C is a good example of this. On the other hand, some advantages to the earlier time point were also apparent, for instance while H2O2 and menadione were responsive at this time there were no effects at 24 hrs. Additionally, while the non-genotoxicant diethanolamine showed no effect at 4 hrs, it caused modest increases in γH2AX-associated fluorescence at 24 hrs.
Figure 4.
Fold-increases in γH2AX median channel fluorescence are graphed for each of 67 training set chemicals for both the 4 hr (top panel) and 24 hr (bottom panel) time points. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of data points are plotted for each chemical, as each represents a different concentration. These data highlight the sensitivity of the γH2AX biomarker to clastogenic activity at both the 4 and 24 hr time points.
Figure 7.
Fold-increases in polyploid cells are graphed for each of 67 training set chemicals for both the 4 hr (top panel) and 24 hr (bottom panel) time points. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of data points are plotted for each chemical, as each represents a different concentration. These data highlight the sensitivity of polyploidization to aneugenic activity at the 24 hr time point.
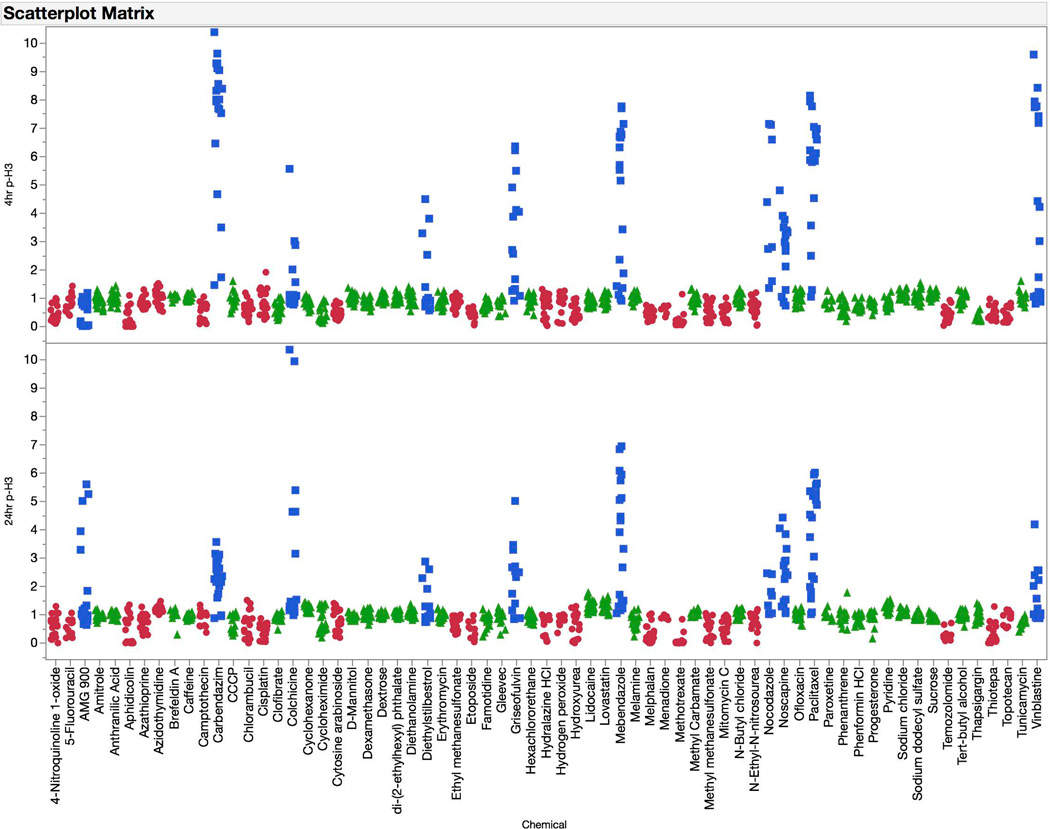
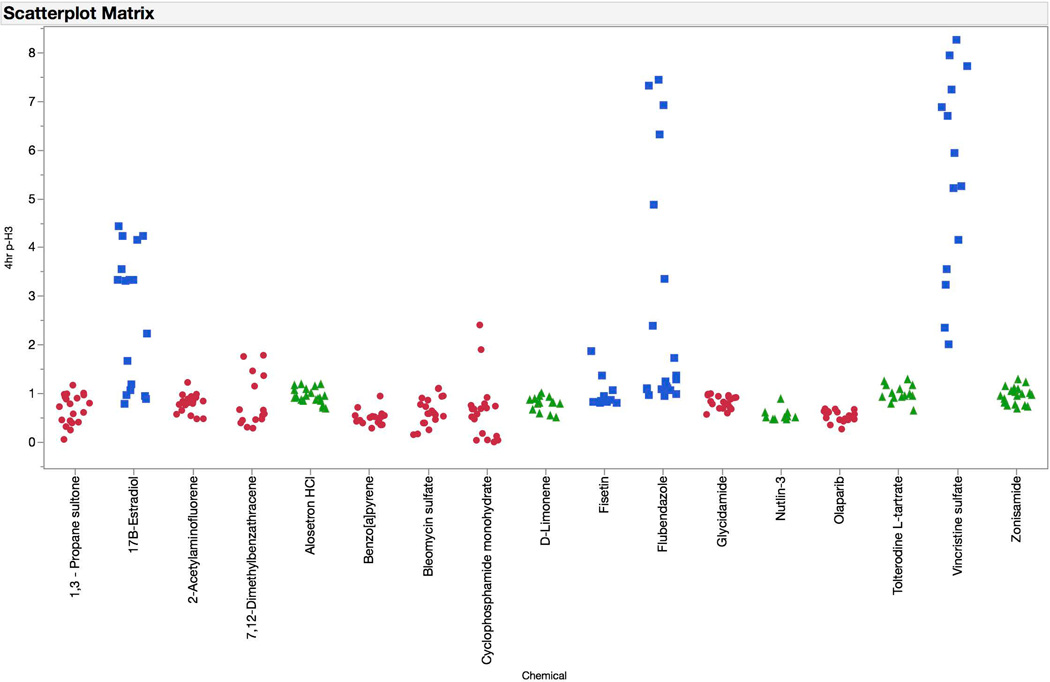
Increased proportions of p-H3-positive events were quite specific for aneugenic agents (see Figure 5). As with the γH2AX endpoint, the responses observed at 4 hrs were generally similar to those found at the later time point. However, in the case of p-H3 there was a greater tendency for 4 hr responses to be higher in magnitude—carbendazim and nocodazole are good examples of this phenomenon.
Figure 5.
Fold-increases in phospho-histone H3 (p-H3) positive events are graphed for each of 67 training set chemicals for both the 4 hr (top panel) and 24 hr (bottom panel) time points. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of data points are plotted for each chemical, as each represents a different concentration. These data highlight the sensitivity of the p-H3 biomarker to aneugenic activity at both the 4 and 24 hr time points.
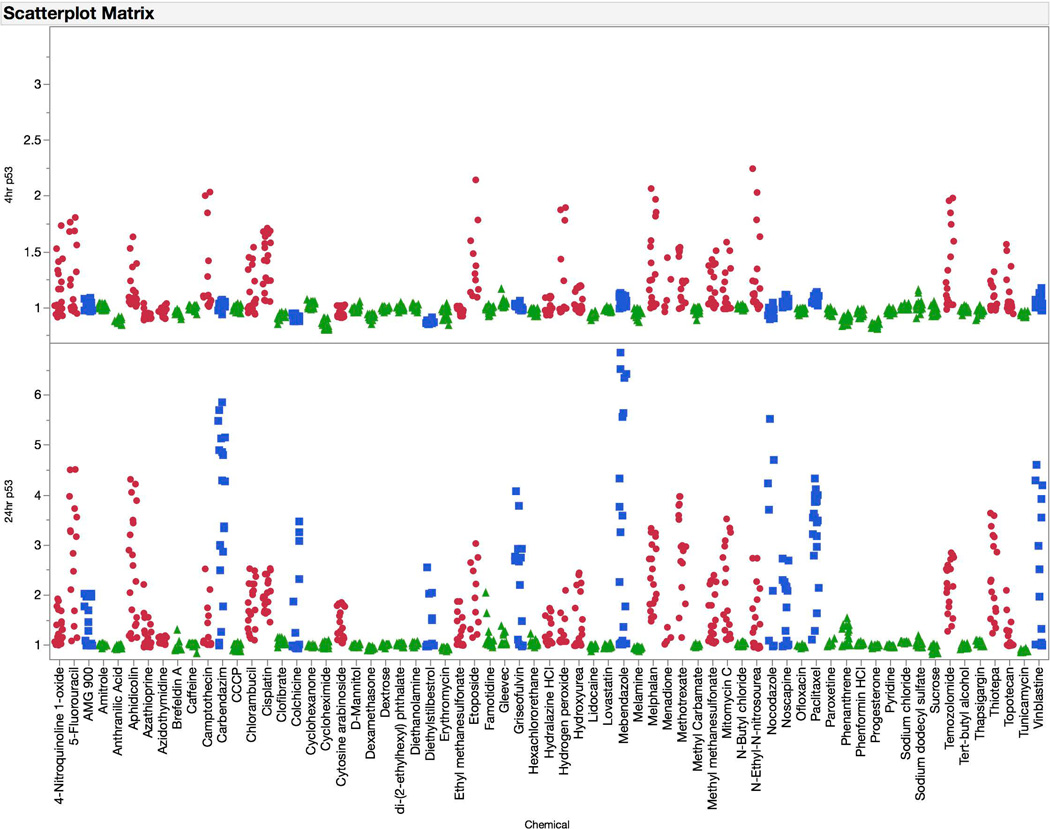
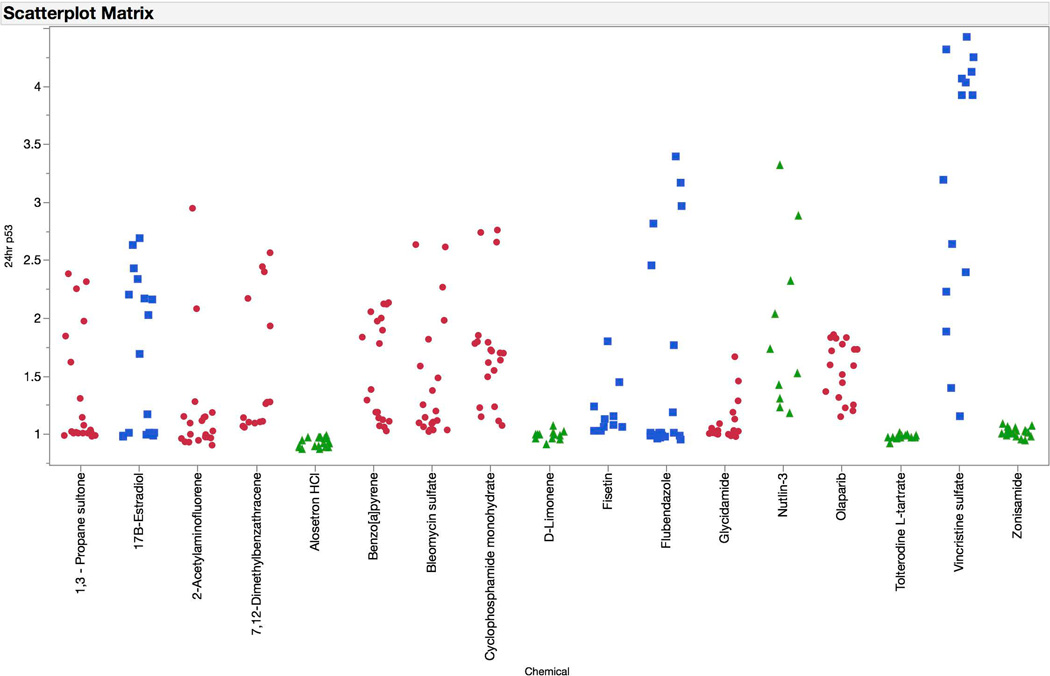
The p53 biomarker exhibited an interesting time-dependent response profile (see Figure 6). Clastogen exposures were associated with modest increases in p53-associated fluorescence at 4 hrs. At the later time point, these responses were markedly higher. The later reading was also required for aneugen-induced effects to manifest. At no time did non-genotoxicants cause appreciable effects to the p53 biomarker, even when the cytotoxicity cutoff value of 80% was reached.
Figure 6.
Fold-increases in p53 median channel fluorescence are graphed for each of 67 training set chemicals for both the 4 hr (top panel) and 24 hr (bottom panel) time points. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of data points are plotted for each chemical, as each represents a different concentration. These data highlight the sensitivity of the p53 biomarker to clastogenic activity at the 4 hr time point, and both clastogenic and aneugenic activity at the 24 hr time point.
Changes to the proportion of polyploid nuclei are shown in Figure 7. As expected, the early time point was insensitive to diverse chemical exposures. On the other hand the 24 hr time point was relatively specific for aneugenic agents, and at times very large magnitude effects were observed.
Model building
Multinomial logistic regression was used to construct predictive models for categorizing chemicals as clastogenic, aneugenic, or non-genotoxic. Univariate analyses conducted with training set data identified several promising explanatory factors: 4 and 24 hr γH2AX, 4 and 24 hr p-H3, 24 hr p53, and 24 hr polyploidy. We proceeded to build and evaluate multivariate models using these factors. While there were not great differences between models that incorporated 4 or 24 hr γH2AX and p-H3 data, we made a decision to utilize 4 hr γH2AX and p-H3 data. Given the similarity of the various models’ performance, this decision was largely made on theoretical grounds. That is, by focusing on early time points whenever possible, we hypothesized that the assay may be more resistant to effects that are not caused by direct DNA damage but rather by secondary consequences of overt toxicity. Additionally, while the magnitude of several chemicals’ γH2AX responses were moderately higher at 24 hrs, there were two examples of a 4 hr response that was entirely lost at the later time point.
The multivariate model that was built based on these and other considerations (e.g., Wald test p-values) consisted of four biomarkers: 4 hr γH2AX; 4 hr p-H3; 24 hr p53; and 24 hr polyploidy. Because the model utilized data from every concentration, and since each of the biomarkers generally exhibited greater signal to noise ratios as cytotoxicity increased (to a point, see Figure 2), we tested whether a weight function using %cytotoxicity values would improve the model. This was found to be the case. Table II quantitatively summarizes the benefits of using data from four biomarkers instead of any one, and of applying a weight function. Note that additional summary statistics for the four factor weighted model, for example parameter estimates and standard errors, are included as a Supplementary file. To our knowledge providing a logistic regression model with chemicals’ complete dose-response data and weighting on cytotoxicity is novel, and are features of this machine learning strategy that warrant further study.
Table II.
Model Building Statistics.
| Endpoint | Wald Test p-value | Pseudo R2 | ROC |
|---|---|---|---|
| γH2AX, 4 hr | < 0.0001 | 0.2486 | ≥ 0.6927 |
| p-H3, 4 hr | < 0.0001 | 0.2753 | ≥ 0.7579 |
| p53, 24 hr | < 0.0001 | 0.3235 | ≥ 0.7234 |
| Polyploidy, 24 hr | < 0.0001 | 0.0967 | ≥ 0.5370 |
| All Four Biomarkers | ≤ 0.0002 | 0.5569 | ≥ 0.9003 |
| All Four and Weighted on Cytotoxicity | < 0.0001 | 0.7444 | ≥ 0.9614 |
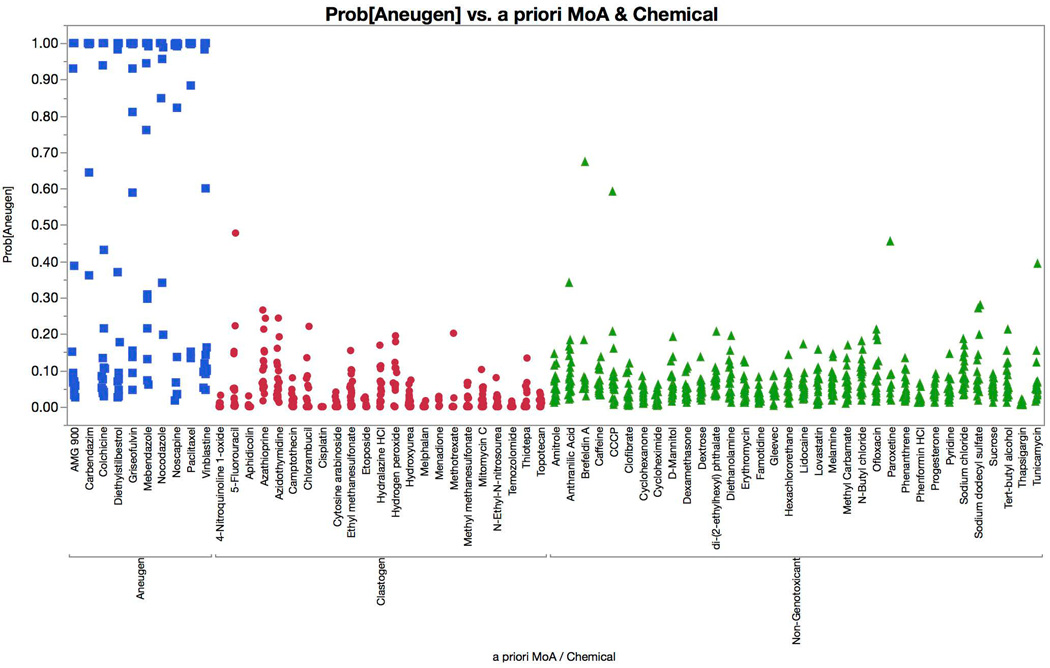
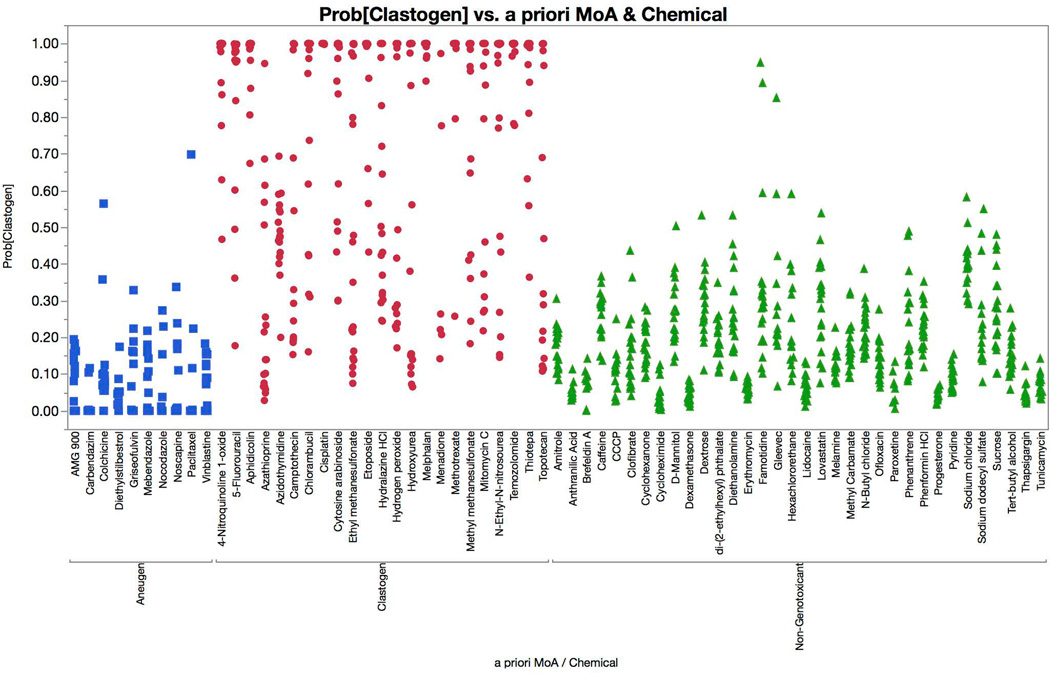
Figure 8 displays logistic regression model predictions, with probabilities of an aneugen call graphed for each of the 67 training set chemicals. According to our positive call criteria, each of the 10 aneugens were correctly identified, with at least two successive concentrations showing ≥ 90% probability values. The prototypical aneugen signature was increases in p-H3, p53, and polyploidization. AMG 900 was a notable exception—while it caused marked polyploidization and a modest p53 response, its known ability to interfere with histone H3 phosphorylation led to a pronounced decrease in p-H3-positive events [Payton et al., 2010]. None of the clastogens or non-genotoxicants were misclassified as an aneugen.
Figure 8.
Multinomial logistic regression probabilities for an aneugen classification are graphed for each of 67 training set chemicals. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of probabilities are plotted for each chemical, as each represents a different concentration. These data show that each of the aneugens were correctly classified by the four-factor model (two successive concentrations with probabilities in excess of 0.90, i.e., 90%), while none of the clastogens or non-genotoxicants were misclassified as aneugens.
Figure 9 shows clastogen probabilities for each of the training set chemicals. According to our positive call criteria, 20 of the 23 clastogens were correctly identified. The prototypical clastogen signature was increases in γH2AX and p53, typically with a modest reduction in p-H3. Azidothymidine was misclassified as a non-genotoxicant, while azathioprine and menadione were equivocal, as only one concentration exceeded the 90% probability threshold. Another notable clastogen was 5-fluorouracil. While its genotoxic MoA was correctly predicted, this was attributable to a remarkable p53 response (Figure 6), as it did not cause increased γH2AX-associated fluorescence as other clastogens did (Figure 4). None of the aneugens were misclassified as a clastogen, and only one of the 34 non-genotoxicants generated an equivocal call (famotidine).
Figure 9.
Multinomial logistic regression probabilities for a clastogen classification are graphed for each of 67 training set chemicals. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of probabilities are plotted for each chemical, as each represents a different concentration. These data show that 20 of 23 clastogens were correctly classified by the four-factor model (two successive concentrations with probabilities in excess of 0.90, i.e., 90%). None of the aneugens were misclassified as clastogens, and 1 of 34 non-genotoxicants met our definition of equivocal (famotidine).
If one considers equivocal calls as incorrect predictions, then the assay’s overall concordance with a priori MoA classifications was 94%, with 91% sensitivity and 97% specificity.
Validation
Cross-validation using the leave-one-out process was used to further evaluate the performance of the model. In this case AMG 900, azidothymidine, azathioprine, and famotidine were misclassified, while menadione and brefeldin A met our definition of an equivocal result. This analysis yielded an overall concordance value of 91%. This provided evidence that the four-factor model, weighted on cytotoxicity, was not overfitting the data, and it would likely be generalizable.
We proceeded to evaluate the model with 17 diverse chemicals that had not been studied previously—that is, with an external test set of chemicals. As described previously, four of the clastogens require rat liver S9 using a short-term treatment. It was not clear whether the model would be appropriate given the different experimental design, but this initial work was considered an important first step in understanding the robustness of the method for studying metabolically activated genotoxicants and more generally short-term treatment schedules.
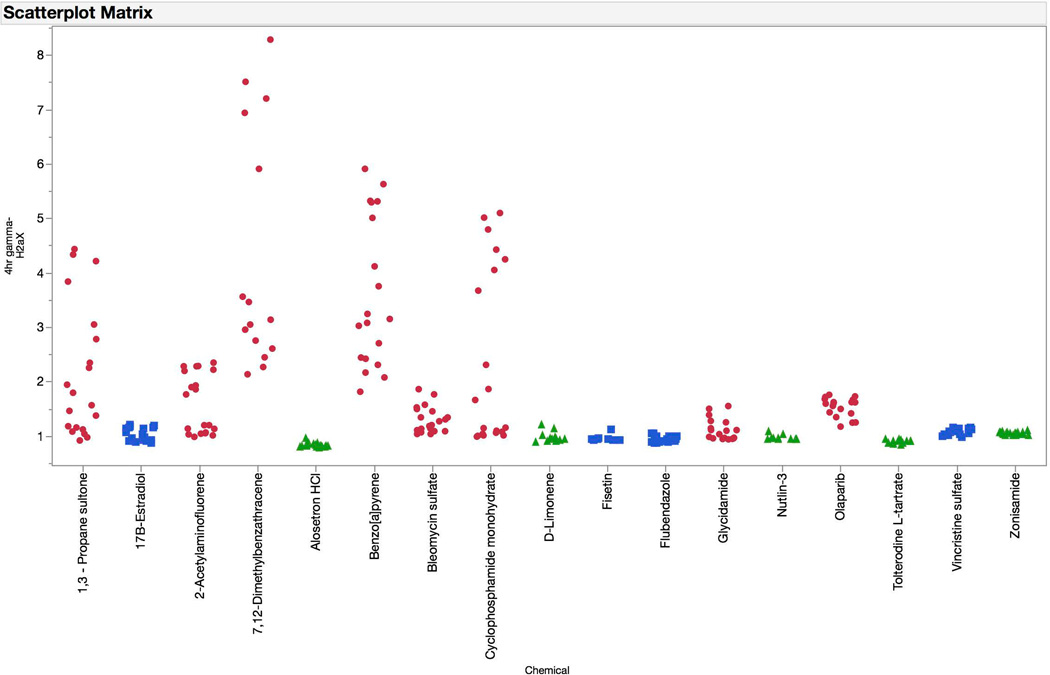
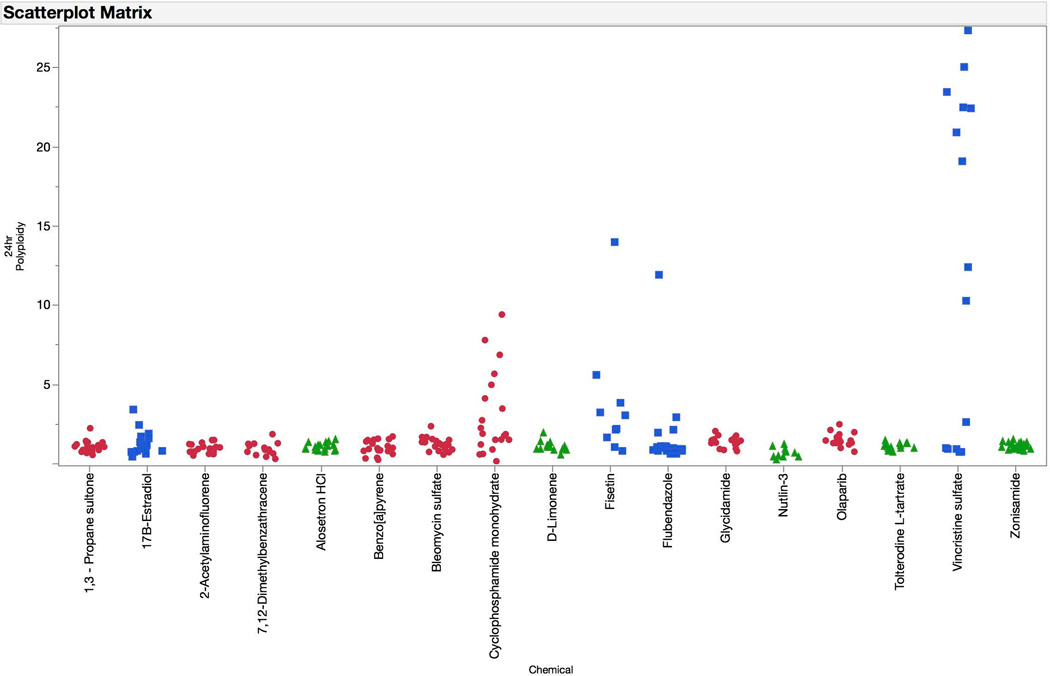
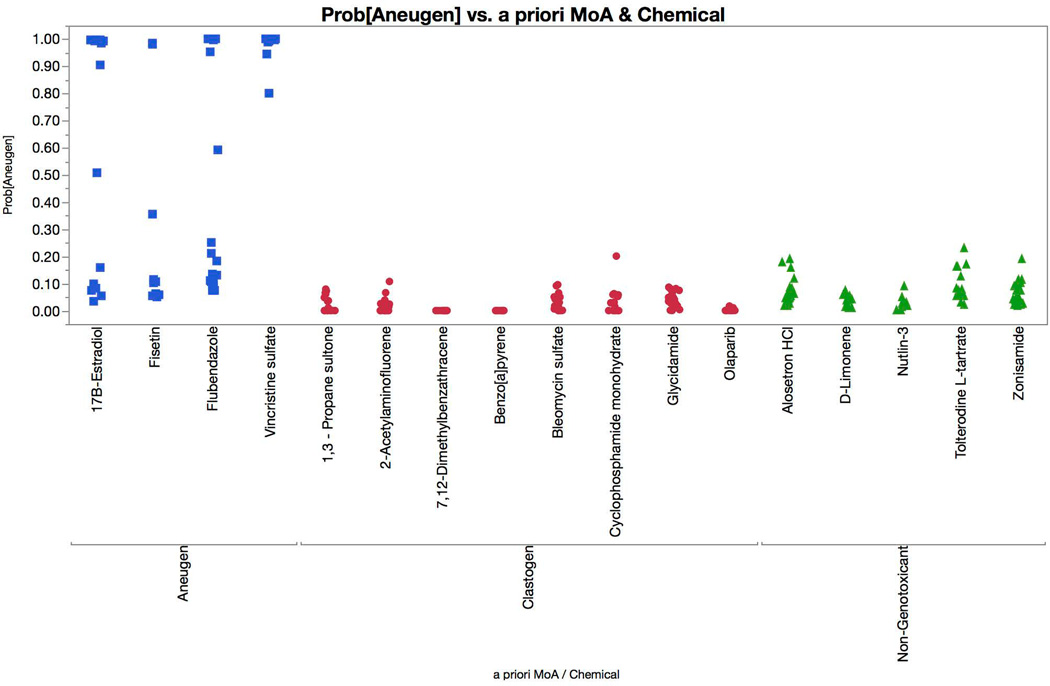
γH2AX, p-H3, p53, and polyploidization responses are graphed for the test set chemicals (Figures 10–13). The logistic regression probability results are shown in Figures 14 and 15. The test set chemicals were correctly classified in 16 of 17 cases, including all four of the agents that required S9 activation. Interestingly, for each of the four promutagens, γH2AX and p53 responses were observed to saturate before the 80% cytotoxicity limit was reached. This suggests that for treatments that involve S9 activation, and perhaps for short-term treatments in general, a lower cytotoxicity limit may be advantageous. This speculation deserves further investigation.
Figure 10.
Fold-increases in γH2AX median channel fluorescence are graphed for each of the test set chemicals for the 4 hr time point. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of data points are plotted for each chemical, as each represents a different concentration. These data show the relative double strand-breaking activity of these agents, with the clastogenic agents showing the greatest fold-increase values. Note that in the case of 2-acetylaminofluorene, 7,12-dimethylbenzanthracene, benzo[a]pyrene, and cyclophosphamide, TK6 cells were exposed to test article in the presence of a rat liver S9 activation system, and after 4 hrs were washed with fresh growth medium.
Figure 13.
Fold-increases in polyploidy cells are graphed for each of the test set chemicals for the 24 hr time point. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of data points are plotted for each chemical, as each represents a different concentration. These data show chemicals’ polyploidization responses, with aneugens showing the greatest effects. Note that in the case of 2-acetylaminofluorene, 7,12-dimethylbenzanthracene, benzo[a]pyrene, and cyclophosphamide, TK6 cells were exposed to test article in the presence of a rat liver S9 activation system, and after 4 hrs were washed with fresh growth medium.
Figure 14.
Multinomial logistic regression probabilities for an aneugen classification are graphed for each of the test set chemicals. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of probabilities are plotted for each chemical, as each represents a different concentration. These data show that each of the aneugens were correctly classified by the four-factor model (two successive concentrations with probabilities in excess of 0.90, i.e., 90%), while none of the clastogens or non-genotoxicants were misclassified as aneugens.
Figure 15.
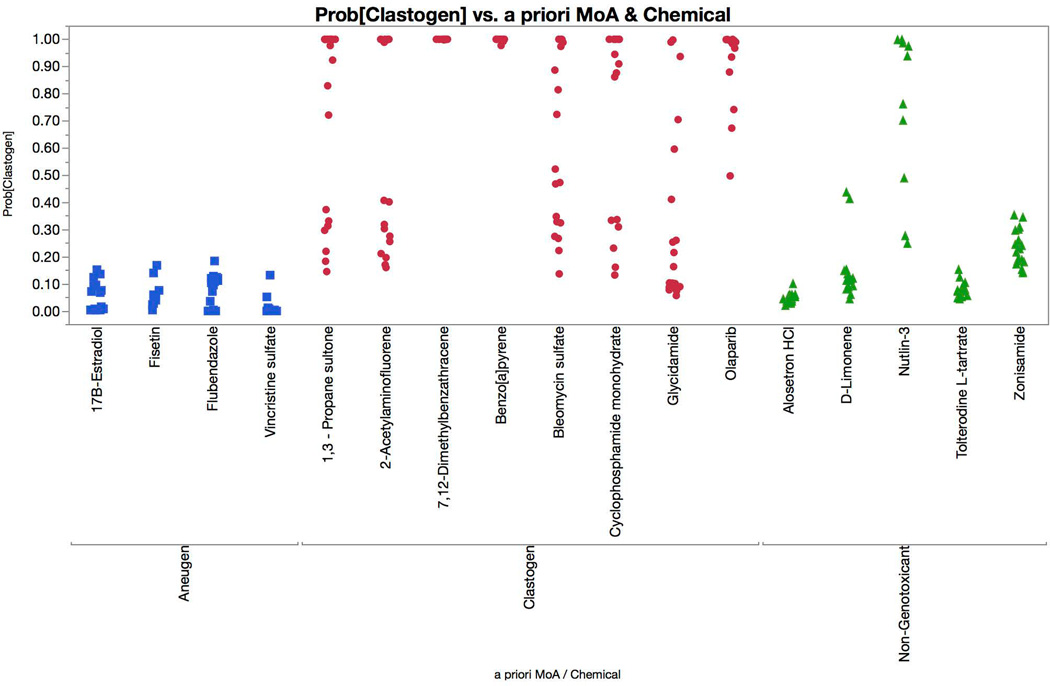
Multinomial logistic regression probabilities for a clastogen classification are graphed for each of the test set chemicals. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of probabilities are plotted for each chemical, as each represents a different concentration. These data show that each of the clastogens were correctly classified by the four-factor model (two successive concentrations with probabilities in excess of 0.90, i.e., 90%), while 4 of 4 aneugens and 4 of 5 non-genotoxicants were classified correctly.
Two other test set chemicals are noteworthy. Nutlin 3 was misclassified as a clastogen owing to the high p53 response that occurred, an expected result given its inhibitory action on p53 and mdm2 heterodimerization and the consequential translocation of p53 to the nucleus [Secchiero et al., 2006]. Fisetin also produced interesting results. The statistical model produced high aneugen probability scores that abruptly switched to high clastogen scores, the latter only occurring at concentrations that exceeded our cytotoxicity limit. It is not clear whether these were spurious results associated with overt toxicity, or whether they were related to observations made by Eastmond and colleagues who reported that fisetin is aneugenic at low concentrations, with clastogenic activity evident at higher concentrations [Olaharski et al., 2005; Gollapudi et al., 2014].
Conclusions
It was possible to devise an efficient add and read-type multiplexed assay that effectively reports on γH2AX, p-H3, p53, polyploidization, as well as cytotoxicity. Multinomial logistic regression was found to be one useful machine learning approach for evaluating the various endpoints and time points for their ability to differentiate between clastogens, aneugens, and non-genotoxicants. Aside from establishing the merit of this particular panel of endpoints, we also demonstrated that this computational platform was useful for predicting responses for compounds that are outside of the training set. This is evidenced by high concordance values for leave-one-out validation, as well as the predictivity observed for the 17 chemical test set. This approach represents a useful strategy for synthesizing results from an assay that is providing higher data dimensionality than is generated from conventional genotoxicity assays. Importantly, it is robust enough to accommodate deviations from prototypical signatures, for instance as was the case with the clastogen 5-fluorouracil. As our results demonstrate, such atypical responses can be categorized correctly, so long as the supervised training component includes sufficient numbers and types of chemicals.
Overall, the characteristics of this assay make it well suited for early screening programs, including high throughput environments that utilize automatic liquid handling instruments. The genotoxic MoA information that is provided would also be valuable for follow-up testing when other test system(s) suggest a chemical or series of chemicals may possess DNA damaging potential. Future work will be needed to extend these promising initial results in order to evaluate the compatibility of the biomarkers with other cell lines, additional chemicals and treatment/harvest schedules, and the transferability of the method across laboratories.
Supplementary Material
Figure 11.
Fold-increases in phospho-histone H3 (H3) positive events are graphed for each of the test set chemicals for the 4 hr time point. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of data points are plotted for each chemical, as each represents a different concentration. These data show mitotic delay/arrest responses, with aneugenic agents showing the greatest effects. Note that in the case of 2-acetylaminofluorene, 7,12-dimethylbenzanthracene, benzo[a]pyrene, and cyclophosphamide, TK6 cells were exposed to test article in the presence of a rat liver S9 activation system, and after 4 hrs were washed with fresh growth medium.
Figure 12.
Fold-increases in p53 median channel fluorescence are graphed for each of the test set chemicals for the 24 hr time point. Chemicals are coded according to genotoxic MoA: clastogens = red circles, aneugens = blue squares, and non-genotoxicants = green triangles. A series of data points are plotted for each chemical, as each represents a different concentration. These data show p53 nuclear translocation activity, with clastogens and aneugens generally showing the greatest fold-increase values. Note that in the case of 2-acetylaminofluorene, 7,12-dimethylbenzanthracene, benzo[a]pyrene, and cyclophosphamide, TK6 cells were exposed to test article in the presence of a rat liver S9 activation system, and after 4 hrs were washed with fresh growth medium.
Acknowledgments
This work benefited from genetic toxicology experts that contributed to the experimental design used herein, and made valuable suggestions about chemical selection as well as the graphical representation of the data. We are indebted to David Kirkland, Maik Schuler, Randy Spellman, Maria Engel, Jennifer Cheung, Andreas Zeller, Melanie Guérard, Valerie Naessens, Andreas Sutter, Sabrina Wilde, Marian Raschke, Marlies Nern, Elisabeth Lorge, Véronique Thybaud, Ulrike Hemmann, Michael Ruppert, Birgit Meyerhoefer, Pekka Heikkinen, and Marc Audebert.
This work was funded by a grant from the National Institute of Health/National Institute of Environmental Health Sciences (NIEHS; grant no. R44ES024039). The contents are solely the responsibility of the authors, and do not necessarily represent the official views of the NIEHS.
CONFLICT OF INTEREST STATEMENT
The authors are employed by Litron Laboratories. Litron has filed a patent covering the flow cytometry-based assay described in this manuscript and plans to sell a commercial kit based on these procedures.
Footnotes
AUTHOR CONTRIBUTIONS
All authors contributed to experimental design. DTB and SMB executed various aspects of the experiments. An outline of the paper was developed by SDD, and all authors contributed to the writing of the final version.
REFERENCES
- Attia SM, Aleisa AM, Bakheet SA, Al-Yahya AA, Al-Rejaie SS, Ashour AE, Al-Shabanah OA. Molecular cytogenetic evaluation of the mechanism of micronucleus formation induced by camptothecin, topotecan, and irinotecan. Environ Mol Mutagen. 2009;50:145–151. doi: 10.1002/em.20460. [DOI] [PubMed] [Google Scholar]
- Attia SM. Molecular cytogenetic evaluation of the mechanism of genotoxic potential of amsacrine and nocodazole in mouse bone marrow cells. J Appl Toxicol. 2013;33:426–433. doi: 10.1002/jat.1753. [DOI] [PubMed] [Google Scholar]
- Audebert M, Riu A, Jacques C, Hillenweck A, Jamin EL, Zalko D, Cravedi JP. Use of the gH2AX assay for assessing the genotoxicity of polycyclic aromatic hydrocarbons in human cell lines. Toxicol Lett. 2010;199:182–192. doi: 10.1016/j.toxlet.2010.08.022. [DOI] [PubMed] [Google Scholar]
- Aydemir N, Bilaloğlu R. Genotoxicity of two anticancer drugs, gemcitabine and topotecan, in mouse bone marrow in vivo. Mutat Res. 2003;537:43–51. doi: 10.1016/s1383-5718(03)00049-4. [DOI] [PubMed] [Google Scholar]
- Bryce SM, Avlasevich SL, Bemis JC, Lukamowicz M, Elhajouji A, Van Goethem F, De Boeck M, Beerens D, Aerts H, Van Gompel J, et al. Interlaboratory evaluation of a flow cytometric, high content in vitro micronucleus assay. Mutat Res. 2008;650:181–195. doi: 10.1016/j.mrgentox.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce SM, Bemis JC, Mereness JA, Spellman RA, Moss J, Dickinson D, Schuler MJ, Dertinger SD. Interpreting in vitro micronucleus positive results: simple biomarker matrix discriminates clastogens, aneugens, and misleading positive agents. Environ Mol Mutagen. 2014;55:542–555. doi: 10.1002/em.21868. [DOI] [PubMed] [Google Scholar]
- Cheung JR, Dickinson DA, Moss J, Schuler MJ, Spellman RA, Heard PL. Histone markers identify the mode of action for compounds positive in the TK6 micronucleus assay. Mutat Res. 2015;777:7–16. doi: 10.1016/j.mrgentox.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Chinnasamy N, Rafferty JA, Hickson I, Ashby J, Tinwell H, Margison GP, Dexter TM, Fairbairn LJ. O6-benzylguanine potentiates the in vivo toxicity and clastogenicity of temozolomide and BCNU in mouse bone marrow. Blood. 1997;89:1566–1573. [PubMed] [Google Scholar]
- Cojocel C, Novotny L, Vachalkova A. Mutagenic and carcinogenic potential of menadione. Neoplasma. 2006;53:316–323. [PubMed] [Google Scholar]
- de Graaf AO, van den Heuvel LP, Dijkman HB, de Abreu RA, Birkenkamp KU, de White T, van der Reijden BA, Smeitink JA, Jansen JH. Bcl-2 prevents loss of mitochondria in CCCP-induced apoptosis. Exp Cell Res. 2004;299:533–540. doi: 10.1016/j.yexcr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Phonethepswath S, Weller P, Avlasevich S, Torous DK, Mereness JA, Bryce SM, Bemis JC, Bell S, Portugal S, et al. Interlaboratory Pig-a gene mutation assay trial: Studies of 1,3-propane sultone with immunomagnetic enrichment of mutant erythrocytes. Environ Mol Mutagen. 2011;52:748–755. doi: 10.1002/em.20671. [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Phonethepswath S, Avlasevich SL, Torous DK, Mereness J, Bryce SM, Bemis JC, Bell S, Weller P, MacGregor JT. Efficient monitoring of in vivo Pig-a gene mutation and chromosomal damage: Summary of 7 published studies and results from 11 new reference compounds. Toxicol Sci. 2012;130:328–348. doi: 10.1093/toxsci/kfs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz D, Scott A, Carmichael P, Shi W, Costales C. Evaluation of an automated in vitro micronucleus assay in CHO-K1 cells. Mutat Res. 2007;630:1–13. doi: 10.1016/j.mrgentox.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Elhajouji A, Tibaldi F, Kirsch-Volders M. Indication for thresholds of chromosome non-disjunction versus chromosome lagging induced by spindle inhibitors in vitro in human lymphocytes. Mutagenesis. 1997;12:133–140. doi: 10.1093/mutage/12.3.133. [DOI] [PubMed] [Google Scholar]
- Raritan, NJ: Ortho-McNeil; 2008. Floxin [package insert] http://www.accessdata.fda.gov/drugsatfda_docs/label/2008/019735s059lbl.pdf. [Google Scholar]
- Fowler P, Smith K, Young J, Jeffrey L, Kirkland D, Pfuhler S, Carmichael P. Reduction of misleading (“false”) positive results in mammalian cell genotoxicity assays. I. Choice of cell type. Mutat Res. 2012;742:11–25. doi: 10.1016/j.mrgentox.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Futami T, Miyagishi M, Taira K. Identification of a network involved in thapsigargin-induced apoptosis using a library of small interfering RNA expression vectors. J Biol Chem. 2005;280:826–831. doi: 10.1074/jbc.M409948200. [DOI] [PubMed] [Google Scholar]
- Garcia-Canton C, Anadon A, Meredith C. Assessment of the in vitro γH2AX assay by high content screening as a novel genotoxicity test. Mutat Res. 2013;757:158–166. doi: 10.1016/j.mrgentox.2013.08.002. [DOI] [PubMed] [Google Scholar]
- East Hanover, NJ: Novartis; 2001. Gleevec [package insert] http://www.accessdata.fda.gov/drugsatfda_docs/label/2008/021588s024lbl.pdf. [Google Scholar]
- Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- Gocke E, Bürgin H, Müller L, Pfister T. Literature review on the genotoxicity, reproductive toxicity, and carcinogenicity of ethyl methanesulfonate. Toxicol Lett. 2009;190:254–265. doi: 10.1016/j.toxlet.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Gollapudi P, Hasegawa LS, Eastmond DA. A comparative study of the aneugenic and polyploidy-inducing effects of fisetin and two model Aurora kinase inhibitors. Mutat Res. 2014;767:37–43. doi: 10.1016/j.mrgentox.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Nam MK, Park HJ, Seong YM, Kang S, Rhim H. Tunicamycin-induced ER stress upregulates the expression of mitochondrial HtrA2 and promotes apoptosis through the cytosolic release of HtrA2. J Microbiol Biotechnol. 2008;18:1197–1202. [PubMed] [Google Scholar]
- Huang X, Halicka HD, Traganos F, Tanaka T, Kurose A, Darzynkiewicz Cytometric assessment of DNA damage in relation to cell cycle phase and apoptosis. Cell Prolif. 2006;38:223–243. doi: 10.1111/j.1365-2184.2005.00344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC monograph. Clofibrate. http://monographs.iarc.fr/ENG/Monographs/vol66/mono66-17.pdf. [Google Scholar]
- Henderson L, Fedyk J, Windebank S, Smith M. Induction of micronuclei in rat bone marrow and peripheral blood following acute and subchronic administrate of azathioprine. Mutat Res. 1993;291:79–85. doi: 10.1016/0165-1161(93)90019-v. [DOI] [PubMed] [Google Scholar]
- Hendriks G, Atallah M, Morolli B, Calléja F, Ras-Verloop N, Huijskens I, Raamsman M, van de Water B, Vrieling H. The ToxTracker assay: novel GFP reporter systems that provide mechanistic insight into the genotoxic properties of chemicals. Toxicol Sci. 2012;125:285–298. doi: 10.1093/toxsci/kfr281. [DOI] [PubMed] [Google Scholar]
- Hernández LG, van Benthem J, Johnson GE. A mode-of-action approach for the identification of genotoxic carcinogens. PLoS ONE. 2013;8(5):e64532. doi: 10.1371/journal.pone.0064532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmer DW, Jr, Lemeshow S, Sturdivant RX. Applied Logistic Regression. 3rd. New Jersey: John Wiley & Sons, Inc.; 2013. [Google Scholar]
- Keshava C, Keshava N, Whong WZ, Nath J, Ong TM. Inhibition of methotrexate-induced chromosomal damage by folinic acid in V79 cells. Mutat Res. 1998;397:221–228. doi: 10.1016/s0027-5107(97)00216-9. [DOI] [PubMed] [Google Scholar]
- Khoury L, Zalko D, Audebert M. Complementarity of phosphorylated histones H2AX and H3 quantification in different cell lines for genotoxicity screening. Arch Toxicol. 2015 doi: 10.1007/s00204-015-1599-1. In press. [DOI] [PubMed] [Google Scholar]
- Kimura A, Miyata A, Honma M. A combination of in vitro comet assay and micronucleus test using human lymphoblastoid TK6 cells. Mutagenesis. 2013;28:583–590. doi: 10.1093/mutage/get036. [DOI] [PubMed] [Google Scholar]
- Kirkland D, Kasper P, Müller L, Corvi R, Speit G. Recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests: a follow-up to an ECVAM workshop. Mutat Res. 2008;653:99–108. doi: 10.1016/j.mrgentox.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Kirkland D, Kasper P, Martus H-J, Müller L, van Benthem J, Madia F, Corvi R. Updated recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests. Mutat Res. 2015 doi: 10.1016/j.mrgentox.2015.10.006. In press. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Honda S, Nakajima M, Miyahana K, Hayashi M, Shinagawa Y, Sato S, Inoue K, Nito S, Ariyuki F. Micronucleus test with vincristine sulfate and colchicine in peripheral blood reticulocytes of mice using acridine orange supravital staining. Mutat Res. 1992;278:187–191. [PubMed] [Google Scholar]
- Krishna G, Urda G, Tefera W, Lalwani ND, Theiss J. Simultaneous evaluation of dexamethasone-induced apoptosis and micronuclei in rat primary spleen cell cultures. Mutat Res. 1995;332:1–8. doi: 10.1016/0027-5107(95)00075-3. [DOI] [PubMed] [Google Scholar]
- Li HH, Hyduke DR, Chen R, Heard P, Yauk CL, Aubrecht J, Fornace AJ., Jr Development of a toxicogenomics signature for genotoxicity using a dose-optimization and informatics strategy in human cells. Environ Mol Mutagen. 2015;56:505–519. doi: 10.1002/em.21941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chad’s Ford, PA: Endo Pharmaceuticals; 2004. Lidoderm [package insert] http://www.accessdata.fda.gov/drugsatfda_docs/label/2005/020612s007lbl.pdf. [Google Scholar]
- Lorge E, Hayashi M, Albertini S, Kirkland D. Comparison of different methods for an accurate assessment of cytotoxicity in the in vitro micronucleus test: I. Theoretical aspects. Mutat Res. 2010;655:1–3. doi: 10.1016/j.mrgentox.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Lotz AS, Havla JB, Richter E, Frölich K, Staudenmaier R, Hagen R, Kleinsasser NH. Cytotoxic and genotoxic effects of matrices for cartilage tissue engineering. Toxicol Lett. 2009;190:128–133. doi: 10.1016/j.toxlet.2009.06.880. [DOI] [PubMed] [Google Scholar]
- Lu P-Z, Lai C-Y, Chan W-H. Caffeine induces cell death via activation of apoptotic signal and inactivation of survival signal in human osteoblasts. Int J Mol Sci. 2008;9:698–718. doi: 10.3390/ijms9050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmington, DE: AstraZeneca Pharmaceuticals LP; 2014. Lynparza [package insert] http://www.accessdata.fda.gov/drugsatfda_docs/label/2014/206162lbl.pdf. [Google Scholar]
- Martelli A, Allavena A, Campart GB, Canonero R, Ghia M, Mattioli F, Mereto E, Robbiano L, Brambilla G. In vitro and in vivo testing of hydralazine genotoxicity. J Pharmacol Exp Ther. 1995;273:113–120. [PubMed] [Google Scholar]
- Matsushima T, Hayashi M, Matsuoka A, Ishidate M, Jr, Miura KF, Shimizu H, Suzuki Y, Morimoto K, Ogura H, Mure K, Koshi K, Sofuni T. Validation study of the in vitro micronucleus test in a Chinese hamster lung cell line (CHL/IU) Mutagenesis. 1999;14:569–580. doi: 10.1093/mutage/14.6.569. [DOI] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Molecular Biology of the Cell. 2005;16:5013–5025. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse Station, NJ: Merck & Co., Inc.; 2012. Mevacor [package insert] http://www.accessdata.fda.gov/drugsatfda_docs/label/2012/019643s085lbl.pdf. [Google Scholar]
- Moon JL, Kim SY, Shin SW, Park J-W. Regulation of brefeldin A-induced ER stress and apoptosis by mitochondrial NADP+ -dependent isocitrate dehydrogenase. Biochem Biophys Res Commun. 2012;417:760–764. doi: 10.1016/j.bbrc.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Nagel ZD, Margulies CM, Chaim IA, McRee SK, Mazzucato P, Ahmad A, Abo RP, Butty VL, Forget AL, Samson LD. Multiplexed DNA repair assays for multiple lesions and multiple doses via transcription inhibition and transcriptional mutagenesis. Proc Natl Acad Sci USA. 2014;111:E1823–E1832. doi: 10.1073/pnas.1401182111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Toxicology Program Report. Sodium Dodecyl Sulfate. http://ntp.niehs.nih.gov/testing/status/agents/ts-10604-g.html.
- Nikolova T, Dvorak M, Jung F, Adam I, Krämer E, Gerhold-Ay A, Kaina B. The γH2AX assay for genotoxic and nongenotoxic agents: Comparison of H2AX phosphorylation with cell death response. Toxicol Sci. 2014;140:103–117. doi: 10.1093/toxsci/kfu066. [DOI] [PubMed] [Google Scholar]
- Olaharski AJ, Mondrala ST, Eastmond DA. Chromosomal malsegregation and micronucleus induction in vitro by the DNA topoisomerase II inhibitor fisetin. Mutat Res. 2005;582:79–86. doi: 10.1016/j.mrgentox.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Oliver J, Meunier JR, Awogi T, Elhajouji A, Ouldelhkim MC, Bichet N, Thybaud V, Lorenzon G, Marzin D, Lorge E. SFTG international collaborative study on in vitro micronucleus test V. Using L5178Y cells. Mutat Res. 2006;607:125–152. doi: 10.1016/j.mrgentox.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Paulsson B, Kotova N, Grawé J, Henderson A, Granath F, Golding B, Törnqvist M. Induction of micronuclei in mouse and rat by glycidamide, genotoxic metabolite of acrylamide. Mutat Res. 2003;535:15–24. doi: 10.1016/s1383-5718(02)00281-4. [DOI] [PubMed] [Google Scholar]
- Research Triangle Park, NC: GlaxoSmithKline; 2011. Paxil [package insert] http://www.accessdata.fda.gov/drugsatfda_docs/label/2011/020031s058s066,020710s022s030lbl.pdf. [Google Scholar]
- Payton M, Bush TL, Chung G, Ziegler B, Eden P, McElroy P, Ross S, Cee VJ, Deak HL, Hodous BL, Nguyen HN, et al. Preclinical evaluation of AMG 900, a novel potent and highly selective pan-aurora kinase inhibitor with activity in taxane-resistant tumor cell lines. Cancer Res. 2010;70:9846–9854. doi: 10.1158/0008-5472.CAN-10-3001. [DOI] [PubMed] [Google Scholar]
- Whitehouse Station, NJ: Merck & Co, Inc.; 2011. Pepcid [package insert] http://www.accessdata.fda.gov/drugsatfda_docs/label/2011/019462s037lbl.pdf. [Google Scholar]
- Ridpath JR, Takeda S, Swenberg JA, Nakamura J. Convenient, multi-well plate-based DNA damage respons analysis using DT40 mutants in applicable to a high-throughput genotoxicity assay with characterization of modes of action. Environ Mol Mutagen. 2011;52:153–160. doi: 10.1002/em.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem. 2000;275:9390–9395. doi: 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- Schuler M, Muehlbauer P, Guzzie P, Eastmond DA. Noscapine hydrochloride disrupts the mitotic spindle in mammalian cells and induces aneuploidy as well as polyploidy in cultured human lymphocytes. Mutagenesis. 1999;14:51–56. doi: 10.1093/mutage/14.1.51. [DOI] [PubMed] [Google Scholar]
- Secchiero P, Barbarotto E, Tiribelli M, Zerbubati C, di Iasio MG, Gonelli A, Cavazzini F, Campioni D, Fanin R, Cuneo A, Zauli G. Functional integrity of the p53-mediated apoptotic pathway induced by the nongenotoxic agent nutlin-3 in B-cell chronic lymphocytic leukemia (B-CLL) Blood. 2006;107:4122–4129. doi: 10.1182/blood-2005-11-4465. [DOI] [PubMed] [Google Scholar]
- Smart DJ, Ahmedi KP, Harvey JS, Lynch AM. Genotoxicity screening via the γH2AX by flow assay. Mutat Res. 2011;715:25–31. doi: 10.1016/j.mrfmmm.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Tweats DJ, Johnson GE, Scandale I, Whitwell J, Evans DB. Genotoxicity of flubendazole and its metabolites in vitro and the impact of a new formulation on in vivo aneugenicity. Mutagenesis. 2015 doi: 10.1093/mutage/gev070. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hummelen P, Elhajouji A, Kirsch-Volders M. Clastogenic and aneugenic effects of three benzimidazole derivates in the in vitro micronucleus test using human lymphocytes. Mutagenesis. 1995;10:23–29. doi: 10.1093/mutage/10.1.23. [DOI] [PubMed] [Google Scholar]
- Walmsley RM, Tate M. The GADD45a-GFP GreenScreen HC assay. Methods Mol Biol. 2012;817:231–250. doi: 10.1007/978-1-61779-421-6_12. [DOI] [PubMed] [Google Scholar]
- Westerink WM, Stevenson JC, Lauwers A, Griffioen G, Horbach GJ, Schoonen WG. Evaluation of the Vitotox and RadarScreen assays for the rapid assessment of genotoxicity in the early research phase of drug development. Mutat Res. 2009;676:113–130. doi: 10.1016/j.mrgentox.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proc Natl Acad Sci USA. 2010;107:10008–10013. doi: 10.1073/pnas.1004056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblom JH, Wiencke JK, Wolff S. Inhibition of the adaptive response of human lymphocytes to very low doses of ionizing radiation by the protein synthesis inhibitor cycloheximide. Mutat Res. 1989;227:257–261. doi: 10.1016/0165-7992(89)90107-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.