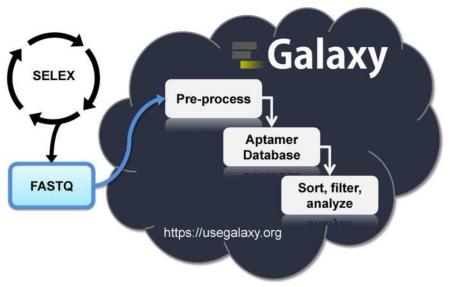
Graphical Abstract
Keywords: Aptamer, SELEX, bioinformatics, Galaxy, high-throughput sequencing (HTS), next-generation sequencing (NGS)
INTRODUCTION
Aptamers are small (20-100 nucleotide) structured DNA or RNA oligonucleotides that interact with a target molecule with a high degree of specificity and affinity. Aptamers can be generated with affinities similar to those of monoclonal antibodies and have been referred to as “chemical antibodies” or “nucleic acid antibodies” [1]. Several aptamers are currently undergoing various stages of clinical trials [2], and one aptamer, Macugen, has been FDA approved for the treatment of age-related macular degeneration [2].
Aptamers with affinity and selectivity for a target can be developed utilizing the SELEX process [3, 4], Systematic Evolution of Ligands by EXponential enrichment. The SELEX process begins with a DNA or RNA oligonucleotide library that contains 5′ and 3′ constant regions flanking a variable region. The length of the variable region, which typically ranges from 20 to 60 nucleotides, dictates the complexity of the starting aptamer library, yielding 1012 to 1036 different aptamer sequences, respectively. The SELEX process removes non-specific aptamers from the library and enriches for aptamers highly specific for the target molecule using a combination of negative and positive selection pressures. The SELEX process has been adapted to include a wide-range of conditions. For all SELEX processes, the sequence information of the enriched aptamers must be determined. Originally, the precise sequences of aptamers from the final round of selection was obtained using sub-cloning techniques followed by sequencing each clone one at a time. High-throughput sequencing (HTS) fundamentally changed the aptamer field by enabling the sequencing of hundreds of millions of reads from multiple rounds of a selection [5-8]. With aptamer HTS data came the need for more sophisticated aptamer bioinformatics methods [9]. Bioinformatics analysis of these HTS data is necessary to narrow down the hundreds of millions of sequences to a select few candidates that can be feasibly evaluated experimentally [5].
Aptamer bioinformatics begins with pre-processing the aptamer HTS data to remove adapter/barcode/constant region sequences and sequences with mismatches within the constant region. Pre-processing also consists of setting a variable region length cutoff and counting the number of identical reads [5, 10, 11]. The next step of aptamer bioinformatics is a first pass course filtering to separate non-selected sequences from selected sequences. This first pass filtering can be accomplished by a variety of methods: 1) calculating fold enrichment of an aptamer sequence between selection rounds [5, 10-12]; 2) comparing the round representation of each sequence to a non-selected round (e.g., round 0) [5]; 3) comparing the number of identical reads of a sequence (read count) to a non-selected round (e.g., round 0) [5]; and 4) isolating aptamer sequences with shared homology [13]. Frequently, a subsequent second pass in-depth analysis is performed to identify sequence families [5, 7, 12, 13], structure families [5], motifs[14, 15] and potential beneficial mutations [16]. Several bioinformatics tools have been generated to analyze aptamer HTS data [9, 10, 13, 16]. Unfortunately, many of these tools are either not easily accessible, have extensive requirements prior to use or require significant computational/coding expertise. These requirements exclude most molecular biologists with expertise in aptamer selections from conducting their own data analysis. To meet this critical need we have adapted the web-based Galaxy Project [17-19] tools for analyzing HTS genome, exome and transcriptome datasets for the analysis of HTS aptamer data. The Galaxy Project’s public webserver, termed the “Main instance,” is a freely available collection of bioinformatics tools that are powerful, flexible, dynamic, and most importantly easy to use with minimal computational expertise/knowledge. All that is needed to start using Galaxy is a web browser, a freely available account with Galaxy and aptamer HTS data. Herein, we describe methods where we have adapted the tools within Galaxy to pre-process and conduct a first-pass course analysis of aptamer HTS data.
EQUIPMENT
1. Computer
A computer is necessary to store and upload data files. These files will include HTS data (e.g. FASTQ) and text files with output data. Any computer capable of running a current web browser along with an internet connection will be sufficient.
2. Web browser
All major web browsers (e.g. Chrome, Firefox, Internet Explorer, Safari, Opera) are supported by Galaxy.
3. Galaxy Project Main webserver account
This article is based on a registered account of the web-based Main instance of Galaxy (https://usegalaxy.org/). The registration process (https://usegalaxy.org/user/create?use_panels=True) requires a valid email address and an approval with the Galaxy Web Portal Service Agreement (https://usegalaxy.org/static/terms.html). Each user is allowed one registered account, which includes 250GB server space for data and permits running a maximum of six concurrent jobs. The Galaxy Wiki includes additional information about Galaxy Main user accounts (https://wiki.galaxyproject.org/Learn/UserAccounts) and a video walk-through (https://vimeo.com/75925027) of the Galaxy Main registration is available online.
METHODS
1. Getting Started
1.1. Galaxy webserver workspace
The Main instance of the Galaxy webserver workspace includes three panels. The left-most panel contains all of the tools used for data analysis and is referred to as “Tools”. These tools are reference throughout this article (e.g. NGS: QC and manipulation\Collapse). The right-most panel keeps track of data files and work history of the data analysis and will be referred to as “History”. Selecting a data file within the History will expand the data files options. These options include view data, edit attributes, deleting and download. The central panel is a dynamic space and will contain the menu options for selected tools from Tools or a sample of data from files selected within the History.
1.2. Getting help
Galaxy includes extensive documentation through the Galaxy Wiki (https://wiki.galaxyproject.org/). When a tool under Tools is selected, the middle panel of Galaxy will include instructions and examples of data input and output.
1.3. Moving files
The analysis of HTS aptamer data using Galaxy will require uploading data files to the Galaxy web server. These files may include compressed HTS raw data files and text files for using the Barcode Splitter tool. The Galaxy webserver has multiple routes for uploading data files. Small files, such as the barcode text file, can be uploaded using The Get Data\Upload File > Choose Local File option. However, large files, such as the raw HTS data files, will need to be uploaded using FTP and may take several hours (10+) depending on network connection speeds and file sizes. More information on uploading data files by FTP is available on the Galaxy Wiki (https://wiki.galaxyproject.org/FTPUpload) and a video walk-through describing all of the methods for uploading data to the Galaxy web server can be found at (https://vimeo.com/75938324). Downloading data files from the Galaxy web server can be accomplished by selecting the download option of a file within the work history.
1.4. Running multiple jobs
Each tool running is referred to as a “job” and Galaxy allows registered users to run up to six jobs concurrently. Each “job” may take several minutes to several hours to complete depending on server load and the complexity of the “job”. Tools within Galaxy can be stacked sequentially in the History and several jobs may be executed prior to the previous tool completing a task. The History panel color codes each “job” to reflect status; green = completed, yellow = currently working; grey = queued; red = failed.
1.5. Edit attributes
Data files in the History are numbered sequentially and are referenced with the tool from the previous job. This information can be edited by expanding the data file in the History and selecting the “Edit attributes” pencil icon.
1.6. HTS aptamer data format
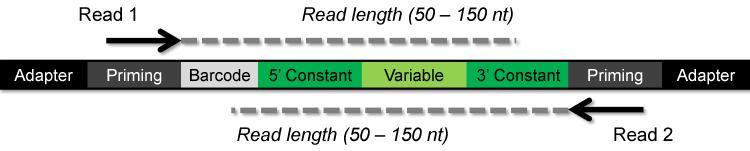
The characteristics of the HTS aptamer data (e.g. FASTQ) should be understood prior to bioinformatics analysis. To date, Illumina sequencers are the most common HTS platform for acquired HTS aptamer data. Three criteria should be noted from DNA amplicon molecule (Figure 1) used to attain HTS aptamer data:
Figure 1. Schematic of DNA Illumina amplicon.
The amplicon includes an adapter sequence, priming regions, a barcode and the aptamer sequence with two constant regions flanking a variable region. Single end read sequence (Read 1) will begin with the barcode and may include only a partial sequence of the 3′ constant region if the read length does not extend through the 3′ constant region. Paired end read (Read1 and Read 2) will include sequence information from both ends of the amplicon. Pre-processing HTS aptamer data involves isolating the variable region sequence and eliminating reads with errors.
1.6.1. Read length:
Read length refers to the number of nucleotides sequenced and is a fixed number ranging from 50 to 150 nucleotides. The read length is important with aptamer HTS data in determining how far the sequence data extends through the variable region into the 3′ constant region. Longer read lengths will likely extend through the entirety of the 3′ constant region into the Illumina priming/adapter sequence.
1.6.2. Single-end read or paired-end read
Single-end read describes HTS data attained from one end of the amplicon DNA molecule (Read 1). Single-end reads may start analysis with barcode splitting (Step 2.2). Single-end reads with 100 nucleotide read lengths will be sufficient for most aptamer libraries. Paired-end read refers to sequence attained from both ends of the same amplicon DNA molecule (Read 1 and Read 2). A paired-end read will include the single-end read data (Read 1) along with reads from the opposing end of the amplicon DNA molecule (Read 2). Paired-end reads may be used to extend the total sequence space to read lengths greater than attainable by Illumina sequencers or as additional error proofing. Currently the Galaxy webserver does not have tools to merge or error proof paired end reads with overlapping regions of sequence information.
1.6.3. Barcode index
The DNA amplicon library sequenced can be multiplexed to contain multiple rounds of selection. Each multiplexed round of selection is labeled by a unique six to eight nucleotide sequence identifier called a barcode. Barcodes may be indexed by Illumina, which will be barcode split during sequencing, or non-Illumina indexed barcodes, which will require barcode splitting (Step 2.2).
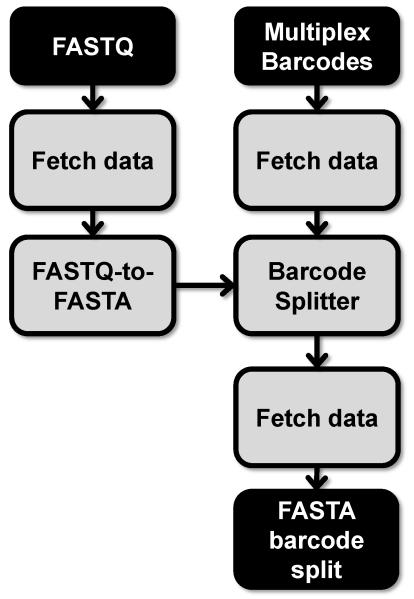
2. FASTQ barcode split (Figure 2)
Figure 2. Method for FASTQ barcode splitting (Section 2).
Files are noted in black and tools in grey. Multiplex Barcodes file refers to a barcode file containing the sequence identifiers for HTS data that has been multiplexed.
2.1. Upload raw HTS data
This article assumes the HTS data is from an Illumina sequencing run. Follow the Galaxy wiki guide to upload the raw FASTQ Illumina HTS data file by FTP (https://wiki.galaxyproject.org/FTPUpload). The Galaxy web server will decompress these files, if necessary.
2.2. Discard low quality reads
Inherent to Illumina HTS data are low quality reads, which should be removed from the data set. Read quality is assessed at each base call within the read by a Phred quality score (Q-score). The Q-score (Q) is a measure of the probability (Q = -10 log10 P where P = probability) that the base call is correct. Base calls with higher Q-score (e.g. Q-score 30) have a higher probability of being correct (e.g. Q = 30 has a 99.9% probability of being correct). Reads may be filtered by Q-score using a threshold for the minimum accepted Q-score along with a percentage of the base calls within a read that must meet the minimum accepted Q-score. Galaxy provides a tool to filter reads by Q-score, NGS: QC and manipulation\Filter by quality. The NGS: QC and manipulation\FASTQ Groomer tool may be necessary to first convert the FASTQ data file to a format that the NGS: QC and manipulation\Filter by quality tool recognizes. The NGS: QC and manipulation\Filter by quality default settings require 90% of the base calls for a given read to have a Q-score of 20 (99% probability of being correct). Aptamer HTS data may use less stringent thresholds (e.g. Q-score =20; Percent threshold = 50%) due to other error eliminating procedures, such as filtering sequences for intact constant region sequence, and that aptamer bioinformatics does not focus on identifying exceptionally rare events.
2.3. Convert FASTQ to FASTA
The FASTQ file should be converted to FASTA using the Convert Formats/FASTQ to FASTA tool.
2.4. Sort barcoded data
If the Illumina run was multiplexed using non-Illumina indexed barcodes the data needs to be sorted using the Barcode Splitter tool. Create a text file that contains the round identification and barcodes separated by a tab. Each round identification with the corresponding barcode should be placed on a new line. If the barcode precedes the 5′ constant region sequence this step may be combined with next step, 3.1 Filter data for intact 5′ constant region, by adding the 5′ constant region sequence to each barcode sequence. For additional help, follow the example detailed under the Barcode Splitter tool options. Upload this text file using Get Data\Upload File > Choose local file. Under Tools select NGS: QC and manipulation\Barcode Splitter. Under “Barcodes to use” select the text file containing the barcode information and under “Library to split” select the FASTA file of the HTS aptamer data. Select the appropriate “Barcodes found at” option, change the “Number of allowed mismatches” to 0 and “Number of allowed barcodes nucleotide deletions” to 0. Execute the Barcode Splitter tool. Upon completion the central panel of Galaxy will contain a table of barcode sorted data. These files should be downloaded and saved to a computer and then reuploaded to Galaxy using the Get Data\Upload File tool. Confirm that the uploaded files are labeled with the appropriate selection round.
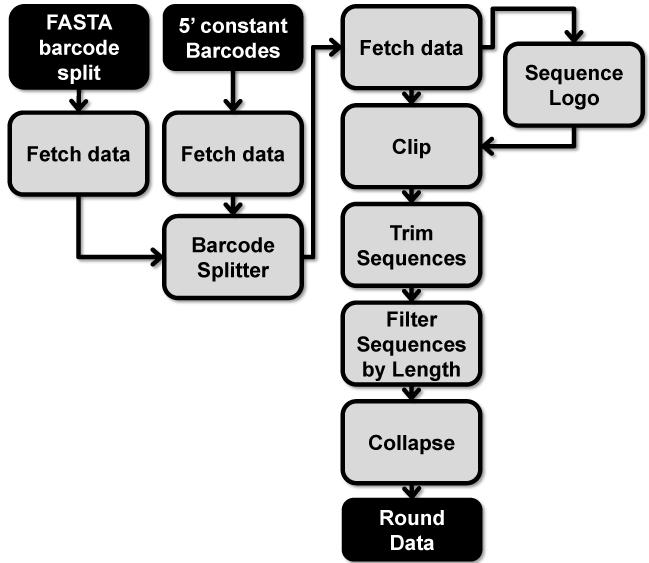
3. Pre-processing aptamer HTS data (Figure 3)
Figure 3. Method for pre-processing aptamer HTS data (Section 3).
Files are noted in black and too ls in grey. 5′ constant Barcodes file refers to the barcode file containing the 5′ constant region to verify an intact 5′ constant region sequence within each aptamer sequence.
3.1. Filter data for intact 5′ constant region
The Barcode Splitter tool may be used to filter selection round data for intact 5′ constant region. This may have already been accomplished in step 2.4 if the 5′ constant region sequence follows the barcode sequence. If not, then create a text file in the same format as previously described in step 2.4 using the 5′ constant sequence rather than the barcode. Follow the same steps as described in step 2.3 for using the NGS: QC and manipulation\Barcode Splitter tool and for downloading the sorted data back into Galaxy. These data will include aptamers that only have a full intact 5′ constant region. Download, save, and re-upload these data files as done in step 2.4 using Get Data\Upload File. Repeat for each round of selection.
3.2. Filter data for intact 3′ constant region, remove sequence with unknown bases (N)
Select the NGS: QC and manipulation\Clip tool. Under “Library to clip” select the round data to be processed. Set the “Minimum sequence length” to “1”. Select “Enter custom sequence” under “Source” and then enter the 3′ constant region sequence in the “Enter custom clipping sequence” box.
If the read length of the Illumina sequencing run does not cover the entire 3′ constant region, a partial match may be used with the clip tool. To determine an optimal 3′ constant region partial match select the Motif Tools\Sequence Logo tool. Select the data file under “Fasta File” and execute the tool. The output graph will provide a consensus sequence with the 5′ constant region and 3′ constant regions exhibiting significant homology. The consensus sequence of the 3′ constant region will give the optimal partial match. Be aware that too short of a sequence may cause unintended clipping of variable region sequence.
Leave the “enter non-zero value to keep the adapter sequence and x bases that follow it” at 0. Select “Yes” for “Discard sequences with unknown (N) bases”. Under “Output options” select “Output only clipped sequences (i.e. sequences which contained the adapter)” option. Execute the Clip tool and repeat for each round of selection.
3.3. Remove the barcode sequence, 5′ constant region sequence
The 5′ constant region and barcode sequence can be removed using NGS: QC and manipulation\Trim sequences tool. Select the appropriate file from the “Library to Clip” dropdown menu. Enter the starting position of the variable region as the “First base to keep”. For example, a 6 nucleotide barcoded aptamer library with a 15 nucleotide 5′ constant region (23 nucleotides total) would have a variable region starting position of 24. Set the “Last base to keep” to “999” to retain all of the different lengths of variable region sequence. Filtering the sequence length of the variable region will be handled in step 3.4. Execute the Trim tool and repeat for each round of selection.
3.4. Filter variable region sequences by length
The remaining variable region sequences may include sequences that are too short or too long. Select the FASTA manipulation\Filter sequences by length tool and highlight the appropriate data file under “Fasta file.” Enter the desired “Minimal length” and the desired “Maximum length” of the variable regions. A useful range of aptamer variable region lengths to retain would be within ±20% of the variable region length (e.g. For a 40 nucleotide variable region aptamer library: Minimum = 32 nucleotides; Maximum = 48 nucleotides). Execute the Filter sequences by length tool and repeat for each round of selection.
3.5. Collapse duplicate variable region sequences
The Collapse tool reduces identical sequences into a single sequence with a count corresponding to the number of duplicates reads (read count). Select NGS: QC and manipulation\Collapse tool. Select the data to be collapsed from the “Library to collapse” dropdown menu. Execute the Collapse tool and repeat for each round of selection.
4. Assessing selection progression from aptamer HTS data
4.1. Determine unique reads and total reads
The number of total reads and unique reads can be obtained from the output of the Collapse tool. Select and expand the Collapse output file within History. The expanded Collapse output file will contain a summary of the number of unique sequences and the total number of reads those sequences represent (e.g. “Output: 100,000 sequences (representing 10,000,000 reads)”). These data may be copied into a data analysis program such as Microsoft Excel.
4.2. Calculate sequence enrichment
Sequence complexity is the relative amount of unique sequences as compared to total reads within each round of selection (% Complexity = Unique/Total) [5]. The percent sequence enrichment can be determined by taking the complement of sequence complexity (% Enrichment = 1-[Unique/Total]). Lower selection rounds (e.g. round 0) will have sequence enrichment near 0% (sequence complexity near 100%) and higher selection rounds will exhibit significant increases in sequence enrichment (lower sequence complexity). Sequence enrichment plotted against the round of selection can then be analyzed to assess the progression of an aptamer selection.
5. Compile all HTS data across all selection rounds into a non-redundant aptamer database (Figure 4)
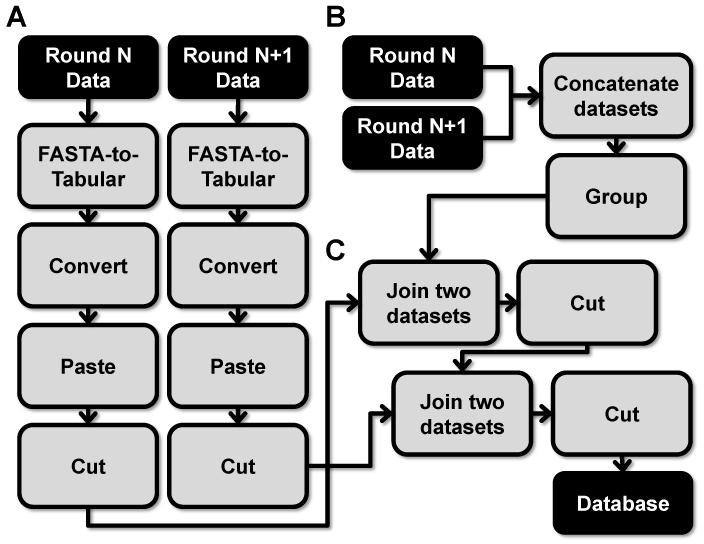
Figure 4. Methods to compile all sequence data across all selection rounds (N, N=1 ...) into a non-redundant database (Section 5).
A) Format round data. B) Create key file. C) Generate non-redundant database.
5.1. Tracking each sequence across all rounds of selection
Compiling a comprehensive database of aptamer HTS data allows for additional analysis described in section 6. The database tracks both the representation (round representation) and distribution (number of reads) of each unique sequence across all selection rounds. The creation of this database requires three steps. First, the formatting of each selection round data (step 5.2). Second, the creation of a key file that contains every unique sequence across all of the aptamer HTS data (step 5.3). Third, the key file is used to merge the data from each round of selection to create a non-redundant database of all unique sequences across all selection rounds (step 5.4).
5.2. Formatting data
Formatting data to create the non-redundant aptamer database requires two steps. Conversion of the FASTA formatted data to tabular format and a reordering of the tabular data.
5.2.1. Convert data from FASTA to tabular
To generate the non-redundant database the key data file and all of the unique sequence data from each selection round needs to be first converted from FASTA format to tabular format. Select the Convert Formats\FASTA-to-Tabular tool and select the appropriate data file from “Convert these sequences.” Set the “How many columns to divide title string into?” to 1 and the “How many title characters to keep?” to 0. Execute the FASTA-to-Tabular tool and repeat for each round of selection.
5.2.2. Restructure the Tabular data
To generate the non-redundant database the structure of the tabular data needs to be re-ordered with sequence in the first column and number of reads in the second column. This re-ordering uses three Galaxy tools sequentially, the Convert tool, the Paste tool and the Cut tool. Select the Text Manipulation\Convert tool and under “Convert all” select from the dropdown options “Dashes.” Under “in Dataset select the appropriate data file. Execute the Convert tool. The columns need to be duplicated within the tabular data file. Select the Text Manipulation\Paste tool and select the same data file under both the “Paste” menu and the “and” menu. Set the “Delimit by” to “Tabs.” Execute the Paste tool. The tabular data can be re-ordered by cutting the data in the order desired. Select the Text Manipulation\Cut tool and enter “c3,c5” under “Cut columns.” Set the “Delimited by” to “Tab” and the “From” to the appropriate data file. Execute the Cut tool. The resulting tabular data should contain sequence within the first column and the number of reads in the second column (or round representation for the key file). Rename the final data file appropriately using the “Edit attribute” option (see section 1.5). These tabular data files will be used in step 5.5 to create the non-redundant database.
5.3. Create a key file and obtain round representation data
The creation of the key file requires appending all of the round data into a single table and grouping these data using column containing the sequence information. Select the Text Manipulation\Concatenate datasets tool. Under “Concatenate Dataset” add each selection round starting with the earliest round data file first. Execute the Concatenate datasets tool. Select the Join, Subtract and Group/Group tool. Under “Select data” select the concatenated data file. Under “Operation” expand the “Insert Option”. Under “Type” select the “Count” option and select “Column: 1” under “On Column.” Execute the Group tool.
5.4. Generate a non-redundant aptamer database
Creating the non-redundant aptamer database requires iteratively joining the data from each selection round. This process starts with the key file and the lowest round of selection (e.g. round 0) using the sequence as the common field for the joining of the two databases. Each subsequent selection rounds will then be joined to the growing non-redundant aptamer database.
5.4.1. Joining two datasets
Select the Join, Subtract and Group\Join two Datasets tool. Under “Join” select the key data file and under “using Column” select “Column: 1”. For the “with” field select the earliest selection round tabular data file and select “Column: 1” under the “and column” options. Select “Yes” for “Keep lines of the first input that do not join with second input” and select “Yes” for “Keep lines of first input that are incomplete.” Select “Yes” for “Fill empty columns”, which will expand revealing additional options. Select “Yes” for “Only fill unjoined rows”, then select “Single fill value” under “Fill Columns by” and enter “0” for “Fill value.” Execute the Join tool.
5.4.2. Remove extra sequence column
The resulting file from the Join tool will include an extra sequence data within the second to last column. Following each Join tool job this extra column of sequence data should be removed. Select the Text Manipulation\Cut tool and enter “c1,c2,cN...,skip,cNlast” under “Cut columns”. The c1 column contains the variable sequence, the c2 column contains the round representation of the unique variable region sequence and the “cN” columns contain the read count of the variable region sequence from each round, the “skip” represents the second to last column containing the extra sequence data and the “cNlast” represents the last column containing the newly joined read count data of the variable region sequence (e.g. “c1,c2,c3,c4,c6” where “c5” was the extra column of sequence data). Set the “Delimited by” to “Tab” and the “From” to the appropriate data file. Execute the Cut tool. Confirm that the appropriate column of extra sequence data was removed. Repeat these steps with each selection round data using the previous output file.
5.4.3. Final aptamer non-redundant database
The final non-redundant aptamer database should contain multiple columns of data with column 1 containing sequence data, column 2 containing round representation data, and all subsequent columns containing the number of reads from each round.
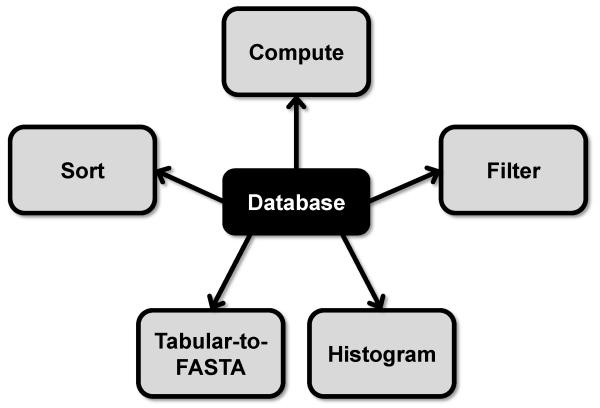
6. Analyzing and manipulating data from non-redundant database
6.1. Simple computation
Galaxy may be used to perform other simple calculations including addition, multiplication and division. For example, Galaxy may be used to sum the total number of reads across all selection rounds or Galaxy may be used to calculate the fold change in read number between two selection rounds. Select the Text Manipulation\Compute tool. Under “Add expression” enter the simple calculation desired, select the non-redundant database under “as a new column to” and select “YES” or “NO” under “Round result?”. Execute the Compute tool.
6.2. Variable region length histogram
Select the Convert Formats\Tabular-to-FASTA tool and highlight the non-redundant database file under “Tab-delimited file.” Select any desired columns as the “Title column(s)” and “Column: 1” under the “Sequence column.” Execute the Tabular-to-FASTA tool. Select FASTA manipulation\Compute sequence length tool and highlight the previous Tabular-to-FASTA output file. Enter “0” for “How many title characters to keep?”. Select the Text Manipulation\Cut tool and enter “c2” under “Cut columns”, “Tab” under “Delimited by” and the appropriate data file under “From”. Execute the Cut tool. These data can be downloaded via the History “Download data” diskette icon and analyzed by another program such as R Studio or within Galaxy. To analyze and plot these data within Galaxy, select the Graph//Display Data\Histogram tool and the data to be analyzed under “Dataset”. Select “Column: 1” under “Numerical column for x axis”, enter “0” for “Number of breaks (bars)” and fill in the remaining fields as desired. Execute the Histogram tool. The histogram plot is outputted as a pdf file that can be downloaded from the History by selecting the “Download” diskette icon or viewed in Galaxy by selecting the “View data” eye icon.
6.3. Round representation histogram
Round representation may be used to identify selected sequences over background non-selected sequences. Selected sequences exhibit persistence throughout a selection. A selected sequence will be observed in multiple rounds of selection whereas non-selected sequences will not be observed in multiple rounds of selection. Round representation of all sequences from each selection round can be compared to the round representation of sequences found within round 0 as a non-selected control. From these data a threshold of round representation may be established to use for filtering the non-redundant database (step 6.6).
Select the Text Manipulation\Cut tool and highlight the non-redundant database under “From” and under “Delimited by” select “Tab”. Under “Cut columns” enter the column with the round representation data and the column with the read data of the selection round to be analyzed separated by a comma (e.g. “c2,c6). Execute the Cut tool. Select the Filter and Sort\Filter tool and select the Cut data under “Filter”. Enter under “With following condition” the expression “c2>0”. This logic statement will remove from the data file any sequences not found within the round being analyzed. Select the Graph//Display Data\Histogram tool and highlight the filtered data under “Dataset”. For the “Numerical column for x axis” field select “Column: 1” and enter “0” for “Number of breaks (bars)”. Execute the histogram tool and repeat with each selection round. Alternatively, the filtered data may be downloaded and analyzed using other programs such as R studio [20], which give significantly more flexibility in data analysis and data plotting.
6.4. Read count histogram
The number of reads for each sequence within a selection round may be used as a cutoff for separating selected sequences from non-selected sequences. To establish this cutoff the read number of sequences within selected rounds may be compared to the read number of round 0 as a non-selected control. Select the Text Manipulation\Cut tool and highlight the non-redundant database under “From” and under “Delimited by” select “Tab”. Under “Cut columns” enter the column with the reads for the round to be analyzed (e.g. “c6”). Execute the cut tool. Select the Graph/Display Data\Histogram tool and highlight the cut data under “Dataset”. For the “Numerical column for x axis” field select “Column: 1” and enter “0” for “Number of breaks (bars)”. Execute the histogram tool and repeat with each selection round. As with round representation, other programs like R Studio [20] give more flexibility in data analysis and data plotting.
6.5. Normalizing reads
The total reads within each round will likely vary. To compare the changes in reads of a unique sequence across multiple selection rounds these data should be normalized using the total reads (obtained in step 4.1) from the round. Select the Text Manipulation\Compute tool. Under “Add expression” enter the round column number divided by the total reads for that selection round attained in step 4.1 then multiplied by a 1,000,000 (e.g. “(c3/3384957)*1000000). Select the non-redundant database under “as a new column to” and select “NO” under “Round result?”. Execute the Compute tool and repeat for each selection round.
6.6. Filtering and sorting data
Galaxy is capable of using simple expressions to filter and sort data within the non-redundant database. This may be used to filter these data based upon a threshold identified during the round representation (section 6.4) and read count (section 6.5) analysis. Both the Filter tool and Sort tool use Regular Expression, which is documented best under the Filter and Sort\Sort tool.
6.6.1. Filter data
To filter data, select the Filter and Sort\Filter tool and select the non-redundant database under “Filter”. Enter the expression to be used as a filter (e.g. for round representation of at least 3 enter “c2>=3”) under “With the following condition” and enter “0” under “number of header lines to skip”. The expressions understood by Galaxy are described under “Syntax” within the Filter tool. Execute the Filter tool. Under the History, the filtered file will indicate the number of sequences that fulfilled the criteria and these data may be downloaded using the “Download” eye icon.
6.6.2. Sort data
To sort data, select the Filter and Sort\Sort tool and highlight the non-redundant database under “Sort Dataset”. Select the appropriate column under “on column”, the type of sort under “with flavor” and the order under “everything in”. Multiple sort criteria may be used by selecting and expanding the “Insert Column selection” under “Column selection”. Execute the Sort tool.
7. Exporting data
7.1. Exporting data into into an excel spreadsheet
The table within Galaxy may be transferred into an Excel spreadsheet. However, Excel has a limitation on the number of lines supported (Excel 97/200/2002/2003 support 65,536 lines; Excel 2007/2010/2013 1,048,576 lines). From the History, download the tabular data file. In Excel, select “Data” and find the “Get External Data” option “From Text”. Highlight “All Files” and select the Galaxy tabular data file. Excel will open a dialogue box that will guide importing the data from Galaxy, which is tab delimited.
7.2. Adding constant region sequences back
The 5′ and 3′ constant region sequences can be added back using the Text Manipulation\Add column tool. Enter the constant region to be added under the “Add this value”, highlight the non-redundant database under “to Dataset”, select “NO” for “Iterate?” and execute the tool. Use the Text Manipulation\Paste tool and Text Manipulation\Cut tool to reorder the sequence data appropriately (section 5.4). To create the full-length aptamer sequence, select the Text Manipulation\Merge Columns tool and select the re-ordered sequence data file under “Select data”. Under “Merge Column” highlight the 5′ constant region column and the variable region column under “with column”. Add the 3′ constant region by selecting and expanding the “Insert Columns” option and enter the 3′constant region column under “Add column”. Execute the Merge Columns tool.
7.3. As a FASTA formatted file
The non-redundant database may be converted back into FASTA format using the Convert Formats\Tabular-to-FASTA tool. Select the non-redundant database under “Tab-delimited file” and “Column: 1” under “Sequence column”. The title column may include any data from any columns, including sequence data. Data from each column will be separated by an underscore “_” in the title. Execute the Tabular-to-FASTA tool.
7.4. Sharing data
A History, and all associated data, within Galaxy may be made available publically or shared with other registered Galaxy users. To share a History, select “History Options” gear icon within the History panel and select “Saved Histories” within the dropdown menu options. This should produce a list of all saved Histories within the central panel. Select the drop down menu next to the name of the History to be shared and select “Share or Publish”. Several options are available to share the History, select and appropriate option.
7.5. Automation
Galaxy includes the ability to automate jobs through “Workflows”, which involves a visual scripting language (https://wiki.galaxyproject.org/Learn/AdvancedWorkflow). Workflows may be accessed by selecting “Workflow” from the top menu options. Workflows shared by other Galaxy users may be searched by selecting “Shared Data” from the top menu options and selecting “Published Workflows.” Search terms such as “aptamer” and “SELEX” may be used to find workflows relevant for aptamer HTS data.
8. What next?
8.1. Other available aptamer bioinformatics tools
Aptamer bioinformatics is a growing field with a variety of tools available for data analysis. These tools include motif searches [7, 14, 15], aptamer clustering [5, 10, 13] and other methods of in-depth analysis [9].
8.2. Citing Galaxy
Work published that uses the Galaxy web server should follow the recommendation within the Galaxy Wiki (https://wiki.galaxyproject.org/CitingGalaxy) and cite the following three publications:
Goecks, J, Nekrutenko, A, Taylor, J and The Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010 Aug 25;11(8):R86.
Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. "Galaxy: a web-based genome analysis tool for experimentalists". Current Protocols in Molecular Biology. 2010 Jan; Chapter 19:Unit 19.10.1-21.
Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. "Galaxy: a platform for interactive large-scale genome analysis." Genome Research. 2005 Oct; 15(10):1451-5.
DISCUSSION
The methods described are designed to enable aptamer HTS bioinformatics analysis using a platform that was originally designed to analyze HTS genomic data. Using the Galaxy tools, users can perform extensive pre-processing of HTS aptamer data, including removing adapter/barcode/constant region sequences, removing sequences with mismatches within the constant region, setting a variable region length cutoff, and counting the number of duplicate reads. In addition, users can determine total reads and unique reads of each selection round. This information is used to calculate sequence enrichment and determine the progression of the aptamer selection. All aptamer selection data can be compiled into a single database where the round representation and number of reads for each sequence is tracked across all rounds of selection. This compiled aptamer selection database can then be easily analyzed, sorted and filtered.
The application of Galaxy public web-based tools for aptamer bioinformatics has several key benefits. First, Galaxy tools are “ready to use” and do not require any set up, such as compiling code or installing/updating programs. Moreover, Galaxy tools are available as a graphical interface rather than command line, further enhancing usability. The Galaxy Project is supported by multiple different entities and is thus well maintained, with extensive wiki guides and a strong user base. The web-based nature of Galaxy makes it a truly platform-independent working environment allows for easy sharing of data and workflows and does not constrain the user to a single computer. For example, Galaxy tools can be executed from a mobile device, where a user can analyze gigabytes of data representing millions of reads using only a web browser.
Since the application of Galaxy tools for aptamer HTS data analysis is an emerging method, there are limitations. For example, there are no tools in Galaxy that were created specifically for aptamer bioinformatics, such as paired end read merge with overlapping regions of sequence data. The current Galaxy tool for merging paired end reads (NGS: QC and manipulation/FASTQ joiner) will join paired end reads, but does not take into account regions of overlapping sequence information. There are also limited workflows available for aptamer bioinformatics. Finally, the capacity for in-depth analysis (e.g., motif searching and clustering) is limited as compared to other stand-alone aptamer bioinformatics programs [10, 13, 15]. However, it is important to note that Galaxy has the potential to overcome all of these limitations as more aptamer scientists begin to utilize this resource to analyze aptamer HTS data.
Figure 5.
Database analysis, sorting, filtering and exporting (Section 6).
HIGHLIGHTS.
Analyzing aptamer high-throughput sequencing (HTS) data using the Galaxy platform.
Pre-process aptamer HTS data to isolate the variable region sequence information.
Compile multiple rounds of aptamer selection data into a single database.
Use the aptamer database for additional bioinformatics analyses.
Histogram analyses, sorting and filtering to identify key aptamer sequences.
ACKNOWLEDGMENTS
This work was supported by the American Heart Association (11POST7620018, 13POST17070101, and 14SDG18850071 to WHT), the National Institutes of Health (R01CA138503 and R21DE019953 to PHG), Mary Kay Foundation (9033-12 and 001-09 to PHG), Elsa U Pardee Foundation (E2766 to PHG), and the Roy J Carver Charitable Trust (RJCCT 01-224 to PHG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ABBREVIATIONS
- SELEX
Systematic Evolution of Ligands by EXponential enrichment
- HTS
high-throughput sequencing
Footnotes
DISCLOSURES None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Mol Ther Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sundaram P, Kurniawan H, Byrne ME, Wower J. Eur J Pharm Sci. 2013;48:259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- [3].Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- [4].Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- [5].Thiel WH, Bair T, Peek AS, Liu X, Dassie J, Stockdale KR, Behlke MA, Miller FJ, Jr., Giangrande PH. PLoS ONE. 2012;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Thiel WH, Bair T, Wyatt Thiel K, Dassie JP, Rockey WM, Howell CA, Liu XY, Dupuy AJ, Huang L, Owczarzy R, Behlke MA, McNamara JO, Giangrande PH. Nucleic Acid Ther. 2011;21:253–263. doi: 10.1089/nat.2011.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, II, Giangrande PH. Nucl Acids Res. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zimmermann B, Gesell T, Chen D, Lorenz C, Schroeder R. PLoS ONE. 2010;5:e9169. doi: 10.1371/journal.pone.0009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Blind M, Blank M. Mol Ther Nucleic Acids. 2015;4:e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Alam KK, Chang JL, Burke DH. Mol Ther Nucleic Acids. 2015;4:e230. doi: 10.1038/mtna.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cho M, Xiao Y, Nie J, Stewart R, Csordas AT, Oh SS, Thomson JA, Soh HT. Proc Natl Acad Sci U S A. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schutze T, Wilhelm B, Greiner N, Braun H, Peter F, Morl M, Erdmann VA, Lehrach H, Konthur Z, Menger M, Arndt PF, Glokler J. PLoS ONE. 2011;6:e29604. doi: 10.1371/journal.pone.0029604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hoinka J, Berezhnoy A, Sauna ZE, Gilboa E, Przytycka TM. In: Research in Computational Molecular Biology. Sharan R, editor. Springer International Publishing; Pittsburgh PA: 2014. pp. 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hoinka J, Zotenko E, Friedman A, Sauna ZE, Przytycka TM. Bioinformatics. 2012;28:i215–i223. doi: 10.1093/bioinformatics/bts210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jiang P, Meyer S, Hou Z, Propson NE, Soh HT, Thomson JA, Stewart R. Bioinformatics. 2014;30:2665–2667. doi: 10.1093/bioinformatics/btu348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hoinka J, Berezhnoy A, Dao P, Sauna ZE, Gilboa E, Przytycka TM. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Goecks J, Nekrutenko A, Taylor J. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19. Unit 19 10 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Racine JS. Journal of Applied Econometrics. 2012;27:167–172. [Google Scholar]