Abstract
The Hippo pathway regulates tissue growth and cell fate. In colon cancer, Hippo pathway deregulation promotes cellular quiescence and resistance to 5-Fluorouracil. In this study 14 polymorphisms in 8 genes involved in the Hippo pathway (MST1, MST2, LATS1, LATS2, YAP, TAZ, FAT4 and RASSF1A) were evaluated as recurrence predictors in 194 patients with stages II/III colon cancer treated with 5-Fu-based adjuvant chemotherapy. Patients with a RASSF1A rs2236947 AA genotype had higher 3-year recurrence rate than patients with CA/CC genotypes (56% vs 33%, HR: 1.87; p=0.017). Patients with TAZ rs3811715 CT or TT genotypes had lower 3-year recurrence rate than patients with a CC genotype (28% vs 40%; HR: 0.66; p=0.07). In left-sided tumors, this association was stronger (HR: 0.29; p=0.011) and a similar trend was found in an independent Japanese cohort. These promising results reveal polymorphisms in the Hippo pathway as biomarkers for stage II and III colon cancer.
Keywords: polymorphisms, Hippo pathway, colon cancer, recurrence, biomarkers
1. Introduction
Tumor recurrence following resection of stage II and III colon cancer occurs in approximately 25–40% of the patients 1. Adjuvant chemotherapy with 5-Fluorouracil (5-Fu) reduces the risk of recurrence 2 and, the addition of oxaliplatin to 5-Fu can further decrease this risk in stage III colon cancer patients 3. However, in current practice the majority of patients does not benefit from adjuvant chemotherapy and will relapse despite treatment. The underlying mechanisms of tumor recurrence after curative treatment are not fully understood. Several processes have been proposed to influence tumor relapse and promote chemotherapy resistance such as the presence of cancer stem cells (CSCs) or the epithelial mesemchymal transition process 4, 5. Disruption of the Salvador-Warts-Hippo pathway, commonly known as the Hippo pathway, is the newest contributor to these recurrence mechanisms. The Hippo pathway is a highly evolutionary conserved pathway, whose main physiological function is to control tissue growth and hence organ size 6, 7. The core signaling consists of several kinases, STE20-like kinase 1 and 2 (MST1 and MST2), large tumor suppressor 1 and 2 (LATS1 and LATS2) and the adaptor proteins MOB kinase activator 1A and 1B (MOB1A and MOB1B). Together, these proteins facilitate the phosphorylation of homologous oncoproteins Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ). Phosphorylation of YAP/TAZ leads to their accumulation in the cytoplasm and stimulates their proteosomal degradation 8. Inactivation of this cascade results in YAP/TAZ nuclear translocation. In the nucleus, YAP/TAZ exert their function by activating transcription factors such as SMAD1–3 and TEAD1–4 that induce the transcription of multiple target genes. Among others, these target genes include, Axin2, Birc5, Myc, Ctgf, and β2-integrin, which are involved in stem cell maintenance, epithelial mesenchymal transition (EMT), metastasis development and regulation of microRNA biogenesis 9–11. The upstream regulation of the Hippo pathway still remains poorly understood however, several upstream branches have been described 12. One of them is the Ras-association domain 1 (RASSF1). RASSF1a is a putative tumor suppressor gene that is methylated in several tumor types including colorectal cancer 13. RASSF1a can activate Hippo signaling by protein-protein interaction by binding MST2 through its SARAH (Sav/Rassf/Hpo) domain 14.
In colon cancer the Hippo effectors YAP/TAZ have been reported to contribute to 5-Fluorouracil (5-Fu) resistance by inducing cellular quiescence 15 and, their expression has been correlated with the patients’ prognosis 16–18. Furthermore, Hippo signaling is interconnected with several other pathways that are well-established major role players in the development and progression of colorectal cancer. Wnt/β-catenin pathway crosstalks with Hippo signaling through a mechanism scarcely understood. β-catenin interacts with TAZ/YAP favoring their translocation to the nucleus, thus increasing the transcription of the Hippo targeted genes 19. Other colon cancer-associated pathways that engage in regulatory crosstalk with the Hippo signaling include among others transforming growth factor β (TGF-β), Hedgehog, and Notch pathways 20.
Based on the importance of Hippo signaling in processes possibly implicated in colon cancer recurrence, this work was designed to evaluate the potential role as prognostic biomarkers of single nucleotide polymorphisms (SNPs) in genes involved in the Hippo pathway, in patients with resected stages II and III colon cancer.
2. Material and Methods
2.1 Eligible patients
A total of 194 patients with high-risk stage II and III colon cancer were included. Patients with stage II were classified as high risk if they presented at least one the following characteristics: poorly differentiated tumor, lymph node sampling <12, lymphatic or perineural invasion, obstruction or perforation as tumor presentation and pT4. All patients received adjuvant chemotherapy based on 5-Fu at the Norris Comprehensive Cancer Center/University of Southern California (NCC/USC) or the Los Angeles County/USC-Medical in Los Angeles, California, USA. Data were collected retrospectively from clinical records. The USC Review Board approved this study. All the participating patients signed informed consent for tissue and blood collection and analysis. A second exploratory cohort comprised of 350 Japanese patients with stage III colorectal cancer patients mostly treated with adjuvant chemotherapy based on 5-Fu in the Cancer Institute Hospital in Tokyo, Japan. Clinical data were collected retrospectively and the study was approved by the Institute’s Ethical Committee. Table 1 shows in detail the patients’ basal characteristics. This study was performed following the REMARK recommendations for the reporting of biomarkers 21.
Table 1.
Baseline characteristics and treatment of USC and Japanese cohorts
| USC* (n=194) | Japanese (n=350) | P value† | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Age, years | |||||
| <55 | 70 | 36.1 | 70 | 20.0 | |
| 55–64 | 69 | 35.6 | 101 | 28.9 | <0.001 |
| ≥65 | 55 | 28.4 | 179 | 51.1 | |
| Sex | |||||
| Female | 88 | 45.4 | 175 | 50.0 | 0.30 |
| Male | 106 | 54.6 | 175 | 50.0 | |
| T | |||||
| T1 | 2 | 1.0 | 0 | ||
| T2 | 13 | 6.7 | 5 | 1.4 | |
| T3 | 156 | 80.4 | 232 | 66.3 | <0.001 |
| T4 | 19 | 9.8 | 113 | 32.3 | |
| Tx | 4 | 2.1 | |||
| N | |||||
| Negative | 85 | 43.8 | 0 | ||
| N1 | 59 | 30.4 | 229 | 65.4 | |
| N2 | 50 | 25.8 | 121 | 34.6 | <0.001 |
| Grade | |||||
| Well | 10 | 5.2 | 75 | 21.4 | |
| Moderate | 119 | 61.3 | 236 | 67.4 | |
| Poor/undifferentiated | 48 | 24.7 | 24 | 6.9 | <0.001 |
| Missing | 17 | 8.8 | 15 | 4.3 | |
| Stage | |||||
| II | 85 | 43.8 | 0 | ||
| III | 109 | 56.2 | 229 | 65.4 | <0.001 |
| IIIC | 121 | 34.6 | |||
| N of resected lymph nodes | |||||
| ≤12 | 61 | 31.4 | 47 | 13.5 | |
| >12 | 115 | 59.3 | 302 | 86.5 | <0.001 |
| Missing | 18 | 9.3 | 1 | ||
| Tumor side | |||||
| Right | 95 | 49.0 | 110 | 31.4 | |
| Left | 92 | 47.4 | 238 | 68.0 | |
| Left and right | 2 | 1.0 | 2 | 0.6 | <0.001 |
| Missing | 5 | 2.6 | |||
| Adjuvant treatment | |||||
| Fluoropyrimidines | 129 | 66.5 | 206 | 58.9 | |
| 5-FU/LV/Oxaliplatin | 48 | 24.7 | 68 | 19.4 | |
| 5-FU/LV/Irinotecan | 17 | 8.8 | 0 | ||
| None | 76 | 21.7 | |||
| Ethnicity | |||||
| Asian | 27 | 13.9 | 350 | 100.0 | |
| African American | 13 | 6.7 | NA | ||
| Caucasian | 108 | 55.7 | |||
| Hispanic | 46 | 23.7 | |||
40 patients were excluded from the original cohort due to depletion of DNA specimen.
Based on χ2 test and excluded patients with missing characteristics.
2.2 Genetic studies
We studied 14 SNPs in 8 genes involved in the Hippo pathway: MST1, MST2, LATS1, LATS2, YAP1, TAZ, FAT4 and RASSF1a. The polymorphisms were selected based on the following predefined criteria: more than 10% minor allele frequency (MAF); previously reported associations in literature resources (PubMed, dbSNP, Ensembl and Genecards) and potential functionality based on genomic location and/or in silico analysis (F-SNP and SNPinfo NIH database). The characteristics of the selected SNPs are shown in Table 2. DNA was extracted from peripheral blood using the QIAmp-kit (Qiagen, Valencia, CA, USA). All samples were genotyped using PCR-based direct sequencing. To ensure the accuracy of the genotypes, 5% of the samples were re-sequenced showing a concordance of >99%. The researcher performing the genotyping of samples was blinded to the clinical data set.
Table 2.
Primary information on the analyzed polymorphisms
| GENE | SNP | LOCATION | SNP FUNCTION/ASSOCIATION | F-SNP SCORE |
|---|---|---|---|---|
| MST1 | rs17420378 | Exon 11 | Missense Val312Met |
0.533 |
| rs6073629 | 3′UTR | Transcriptional regulation | 0.5 | |
| MST2 | rs10955176 | 3′UTR | NA | NFI |
| LATS1 | rs12174349 | 5′UTR | NA | NFI |
| LATS2 | rs558614 | Exon 4 | Missense Ala324Aval |
0.156 |
| LATS2 | rs9552315 | 3′UTR | Transcriptional regulation | 0.5 |
| YAP | rs8504 | 3′UTR | NA | NFI |
| rs1820453 | Upstream | Survival in NSCLC27 | NFI | |
| TAZ | rs3811715 | Intron | Splice donor | 0.242 |
| rs6783790 | Intron | Splice donor | 0.389 | |
| FAT4 | rs1014867 | Exon 17 | Missense Pro4972Ser Esophageal cancer risk26 |
0.59 |
| rs1039808 | Exon 1 | Missense Ala807Val Esophageal cancer risk26 |
NFI | |
| RASSF1 | rs2073498 | Exon 3 | Missense Ala133Ser Breats cancer risk28 |
0.5 |
| rs2236947 | Intron | Transcriptional regulation | 0.268 |
Abbreviations: MST1: STE20-like kinase 1; MST2: STE20-like kinase 2; LATS1: large tumor suppressor 1; LATS2: large tumor suppressor 2; YAP1: yes associated protein; TAZ: transcriptional co-activator with PDZ-binding motif; FAT4: atypical cadherin 4; RASSF1: Ras-association domain 1; NA: not analyzed; NFI: no functional information; NSCLC: non small cell lung cancer.
2.3 Statistical analysis
The endpoint of this study was time to recurrence (TTR) that was defined as a period from the date of diagnosis to the date of first documented tumor recurrence. TTR was censored at the time of last follow-up or death if patients remained recurrence free. With samples from 194 patients available (79 events) for genotyping the selected SNPs, this study had 80% power to detect a hazard ratio of 1.89–2.13 in a dominant model with a minor frequency of 0.1–0.4 and 2.10–3.23 in a recessive model with a minor frequency of 0.25–0.45 using a two-sided log-rank test at a significance level of 0.05.
Deviations from the Hardy-Weinberg equilibrium were tested using χ2 test. The association between the allelic distribution of the SNPs and their potential association with the baselines characteristics was examined using χ2 or Fisher’s exact test. The true inheritance mode of the analyzed polymorphisms is unknown, therefore a co-dominant, dominant or recessive model was assumed wherever appropriate. The association of the SNPs and time to recurrence was analyzed using- Kaplan Meier curves and log-rank test. In the multivariable Cox regression analysis, the model was adjusted by stage, type of adjuvant chemotherapy and stratified by race.
No correction for multiple testing was performed.
Recursive partitioning (RP) analysis was conducted to explore patterns of recurrence by SNPs of Hippo pathway.
All calculations were performed using SAS statistical package version 9.4 and R package version 3.1.0. All tests were 2-sided at a significance level of 0.05.
3. Results
The median follow up of the USC cohort was 4.4 years (range 0.4–16.8 years) and the 3-year recurrence rate was 36% (± 4% standard error, SE). The median overall survival for this cohort has not yet been reached.
The median follow up of the Japanese cohort was 5 years (rage 0.3–8.6) and the 3-year recurrence rate was 29% (± 2% SE). The median overall survival of this series has not been reached.
Genotypes were achieved in at least 90% of the analyzed samples for each polymorphism. In failed cases, genotyping was not successful due to low quality of DNA or limited DNA quantity. All the analyzed SNPs but one (rs9552315) were within the probability limits of Hardy-Weinberg equilibrium. There were significant differences in some polymorphisms in the allele frequencies across races in the USC cohort (Supplementary table 1).
3.1 Genetic determinants and outcome
Located in the Rassf1a gene, the rs2236947 polymorphism was associated with the 3-year recurrence probability: patients homozygous for the variant A allele had a 56% (±10% SE) 3-year recurrence probability compared to 33% (±4%) for patients with a CC or CA genotype (HR: 1.87; 95% CI, 1.10–3.17; p=0.017). In multivariable analysis this association remained significant (1HR: 1.78; 95% CI, 1.03–3.06; p=0.039).
In the TAZ gene, the variant allele of the rs3811715 polymorphism was associated with a lower 3-year recurrence rate. Patients with a CT or TT genotype had a 28% (±5% SE) 3-year recurrence probability compared to 40% (±5% SE) for patients with a homozygous wild type CC genotype, although this association did not reach statistical significance (HR: 0.66; 95% CI, 0.41–1.05; p=0.077).
No association was found in the overall population in the Japanese cohort for these two SNPs. The MAF for these polymorphisms in the Japanese cohort was 25% for both SNPs. In the Asian population included in the USC cohort, the MAF for these SNPs were 27% and 34% for Rassf1a rs2236947 and TAZ rs3811715, respectively.
3.2 Subgroup analysis by gender and tumor location
Differences were detected for the association of the analyzed SNPs and the 3-year recurrence probability based on gender and tumor location.
Based on tumor location, TAZ rs3811715 correlated strongly with the 3-year recurrence probability in patients with left-sided tumors. The genotype frequencies in this subgroup were CC=55, CT=28 and TT=6. Patients harboring a CT or TT genotype had a 10% (± 5% SE) 3-year recurrence probability whereas patients harboring a CC genotype had 48% (± 7% SE) (HR: 0.25; 95% CI, 0.10–0.60; p=0.001). Patients with a TT genotype (n=6) had no recurrence. This association remained significant after adjusting for the relevant clinical parameters (HR: 0.29; 95% CI, 0.11–0.78; p= 0.011).
In the Japanese exploratory cohort in patients bearing left-side tumors, patients carrying a TAZ rs3811715 TT genotype (TT=14, CT=78, CC=129) had 7% (± 7% SE) 3-year recurrence rate compared 27% (± 3% SE) for patients with at least a C genotype (HR: 0.21; 95% CI, 0.03–1.54; p= 0.091). Additionally, in left-sided tumors a polymorphism located in MST1, rs17420378, was associated with the recurrence probability. Patients with a GA or AA genotypes had a higher recurrence probability than patients with a GG genotype (HR: 2.31; 95% CI, 1.21–4.43; p=0.009). However, in multivariate analysis this association was not maintained (HR: 2.01; 95% CI, 0.98–4.10; p=0.057). This polymorphism was not tested in the Japanese cohort, as the reported MAF is <10%.
Based on gender, the association of TAZ rs3811715 with the 3-year recurrence rate was stronger in the female population (HR: 0.46; 95% CI, 0.22–0.96; p=0.031), although this association did not retain significance in the multivariable analysis (p=0.06) (Table 4).
Table 4.
Subgroup analyses of Hippo SNPs and time to recurrence in patients with stage II or III colon cancer at USC
| N | 3-year recurrence probability ± SE* | HR(95%CI)† Univariate | P† value | HR(95%CI)† Multivariate | P† value | |
|---|---|---|---|---|---|---|
| LEFT-SIDED COLON CANCER | ||||||
| FAT4rs1014867 | 0.55 | 0.37 | ||||
| C/C | 81 | 0.35±0.06 | 1(reference) | 1(reference) | ||
| C/T | 10 | 0.44±0.17 | 1.33(0.52,3.42) | 1.57(0.58,4.28) | ||
| FAT4rs1039808 | 0.93 | 0.73 | ||||
| C/C | 36 | 0.42±0.09 | 1(reference) | 1(reference) | ||
| C/T | 35 | 0.32±0.08 | 0.87(0.42,1.79) | 0.77(0.35,1.65) | ||
| T/T | 18 | 0.27±0.12 | 0.95(0.39,2.30) | 1.07(0.41,2.81) | ||
| LATS1rs12174349 | 0.32 | 0.68 | ||||
| G/G | 21 | 0.44±0.11 | 1(reference) | 1(reference) | ||
| G/A | 44 | 0.31±0.07 | 0.57(0.27,1.20) | 0.70(0.31,1.57) | ||
| A/A | 24 | 0.41±0.11 | 0.73(0.32,1.67) | 0.87(0.36,2.10) | ||
| LATS2rs558614 | 0.49 | 0.78 | ||||
| A/A | 40 | 0.41±0.08 | 1(reference) | 1(reference) | ||
| A/G | 38 | 0.33±0.08 | 0.82(0.42,1.61) | 0.87(0.43,1.77) | ||
| G/G | 12 | 0.32±0.15 | 0.49(0.15,1.66) | 0.64(0.18,2.33) | ||
| LATS2rs9552315 | 0.83 | 0.55 | ||||
| C/C | 56 | 0.41±0.07 | 1(reference) | 1(reference) | ||
| C/T | 26 | 0.21±0.08 | 0.82(0.38,1.76) | 0.92(0.40,2.11) | ||
| T/T | 6 | 0.50±0.25 | 0.75(0.18,3.16) | 2.50(0.41,15.36) | ||
| MST1rs6073629 | 0.25 | 0.35 | ||||
| G/G | 69 | 0.38±0.06 | 1(reference) | 1(reference) | ||
| G/A or A/A | 19 | 0.28±0.11 | 0.62(0.27,1.43) | 0.67(0.29,1.56) | ||
| MST1rs17420378 | 0.009 | 0.057 | ||||
| G/G | 54 | 0.29±0.07 | 1(reference) | 1(reference) | ||
| G/A or A/A | 33 | 0.47,0.09 | 2.31(1.21,4.43) | 2.01(0.98,4.10) | ||
| MST2rs10955176 | 0.62 | 0.71 | ||||
| C/C | 24 | 0.44±0.11 | 1(reference) | 1(reference) | ||
| C/T | 49 | 0.35±0.07 | 0.83(0.41,1.69) | 0.86(0.41,1.79) | ||
| T/T | 16 | 0.30±0.13 | 0.60(0.21,1.70) | 0.63(0.22,1.87) | ||
| RASSF1rs2073498 | 0.93 | 0.95 | ||||
| C/C | 72 | 0.38±0.06 | 1(reference) | 1(reference) | ||
| C/A or A/A | 19 | 0.29±0.11 | 0.97(0.45,2.05) | 1.02(0.46,2.29) | ||
| RASSF1rs2236947 | 0.26 | 0.92 | ||||
| C/C or C/A | 76 | 0.34±0.06 | 1(reference) | 1(reference) | ||
| A/A | 13 | 0.54±0.15 | 1.59(0.70,3.63) | 1.04(0.43,2.57) | ||
| TAZrs3811715 | 0.001 | 0.011 | ||||
| C/C | 55 | 0.48±0.07 | 1(reference) | 1(reference) | ||
| C/T or T/T | 34 | 0.10±0.05 | 0.25(0.10,0.60) | 0.29(0.11,0.76) | ||
| TAZrs6783790 | 0.26 | 0.16 | ||||
| C/C | 35 | 0.39±0.09 | 1(reference) | 1(reference) | ||
| C/T | 38 | 0.40±0.09 | 0.75(0.38,1.49) | 0.53(0.25,1.10) | ||
| T/T | 14 | 0.22±0.11 | 0.42(0.14,1.25) | 0.43(0.13,1.40) | ||
| YAP1rs8504 | 0.95 | 0.95 | ||||
| G/G | 38 | 0.37±0.08 | 1(reference) | 1(reference) | ||
| G/A | 40 | 0.34±0.08 | 0.90(0.45,1.79) | 1.10(0.52,2.34) | ||
| A/A | 9 | 0.31±0.19 | 0.90(0.26,3.08) | 1.18(0.32,4.34) | ||
| YAP1rs1820453 | 0.65 | 0.77 | ||||
| A/A | 30 | 0.32±0.10 | 1(reference) | 1(reference) | ||
| A/C | 42 | 0.37±0.08 | 1.10(0.52,2.34) | 0.96(0.43,2.13) | ||
| C/C | 14 | 0.36±0.13 | 1.50(0.60,3.72) | 1.32(0.49,3.57) | ||
| N | 3-year recurrence probability ± SE* | HR(95%CI)† Univariate | P† value | HR(95%CI)† Multivariate | P† value | |
|---|---|---|---|---|---|---|
| FEMALE POPULATION | ||||||
| FAT4rs1014867 | 0.66 | 0.67 | ||||
| C/C | 82 | 0.35±0.06 | 1(reference) | 1(reference) | ||
| C/T | 4 | 0.25±0.22 | 0.64(0.09,4.72) | 0.64(0.09,4.85) | ||
| FAT4rs1039808 | 0.30 | 0.08 | ||||
| C/C | 46 | 0.33±0.07 | 1(reference) | 1(reference) | ||
| C/T | 32 | 0.35±0.09 | 1.03(0.48,2.18) | 0.98(0.44,2.21) | ||
| T/T | 10 | 0.42±0.16 | 2.09(0.77,5.71) | 4.13(1.13,15.12) | ||
| LATS1rs12174349 | 0.49 | 0.50 | ||||
| G/G | 21 | 0.30±0.10 | 1(reference) | 1(reference) | ||
| G/A | 35 | 0.33±0.08 | 0.81(0.33,1.99) | 0.68(0.27,1.71) | ||
| A/A | 29 | 0.42±0.11 | 1.31(0.54,3.17) | 1.10(0.43,2.85) | ||
| LATS2rs558614 | 0.49 | 0.74 | ||||
| A/A | 38 | 0.39±0.08 | 1(reference) | 1(reference) | ||
| A/G | 40 | 0.35±0.08 | 0.84(0.42,1.70) | 0.99(0.43,2.26) | ||
| G/G | 10 | 0.21±0.13 | 0.43(0.10,1.85) | 0.55(0.11,2.64) | ||
| LATS2rs9552315 | 0.33 | 0.24 | ||||
| C/C | 58 | 0.42±0.07 | 1(reference) | 1(reference) | ||
| C/T | 26 | 0.19±0.09 | 0.52(0.21,1.26) | 0.47(0.18,1.24) | ||
| T/T | 2 | 0.50±0.35 | 0.97(0.13,7.14) | 1.74(0.19,15.73) | ||
| MST1rs6073629 | 0.44 | 0.12 | ||||
| G/G | 60 | 0.38±0.07 | 1(reference) | 1(reference) | ||
| G/A or A/A | 26 | 0.28±0.09 | 0.73(0.33,1.64) | 0.50(0.21,1.19) | ||
| MST1rs17420378 | 0.047 | 0.14 | ||||
| G/G | 49 | 0.31±0.07 | 1(reference) | 1(reference) | ||
| G/A or A/A | 36 | 0.40,0.09 | 1.98(0.99,3.96) | 1.78(0.83,3.84) | ||
| MST2rs10955176 | 0.34 | 0.67 | ||||
| C/C | 20 | 0.36±0.11 | 1(reference) | 1(reference) | ||
| C/T | 49 | 0.40±0.08 | 1.04(0.47,2.27) | 1.18(0.51,2.71) | ||
| T/T | 18 | 0.24±0.11 | 0.48(0.15,1.56) | 0.73(0.21,2.53) | ||
| RASSF1rs2073498 | 0.96 | 0.79 | ||||
| C/C | 71 | 0.33±0.06 | 1(reference) | 1(reference) | ||
| C/A or A/A | 13 | 0.41±0.14 | 1.03(0.39,2.68) | 1.15(0.41,3.22) | ||
| RASSF1rs2236947 | 0.17 | 0.26 | ||||
| C/C or C/A | 73 | 0.32±0.06 | 1(reference) | 1(reference) | ||
| A/A | 13 | 0.58±0.14 | 1.77(0.76,4.11) | 1.66(0.69,4.00) | ||
| TAZrs3811715 | 0.031 | 0.060 | ||||
| C/C | 48 | 0.43±0.08 | 1(reference) | 1(reference) | ||
| C/T or T/T | 39 | 0.23±0.07 | 0.46(0.22,0.96) | 0.47(0.21,1.03) | ||
| TAZrs6783790 | 0.85 | 0.75 | ||||
| C/C | 30 | 0.32±0.09 | 1(reference) | 1(reference) | ||
| C/T | 42 | 0.40±0.08 | 0.91(0.44,1.90) | 1.21(0.56,2.61) | ||
| T/T | 13 | 0.23±0.12 | 0.72(0.23,2.22) | 1.67(0.41,6.84) | ||
| YAP1rs8504 | 0.94 | 0.79 | ||||
| G/G | 36 | 0.33±0.08 | 1(reference) | 1(reference) | ||
| G/A | 35 | 0.38±0.09 | 0.88(0.41,1.87) | 0.77(0.35,1.71) | ||
| A/A | 16 | 0.29±0.12 | 0.99(0.38,2.58) | 1.00(0.37,2.72) | ||
| YAP1rs1820453 | 0.64 | 0.52 | ||||
| A/A | 27 | 0.30±0.09 | 1(reference) | 1(reference) | ||
| A/C | 46 | 0.33±0.08 | 0.98(0.44,2.20) | 0.90(0.38,2.12) | ||
| C/C | 8 | 0.38±0.17 | 1.56(0.53,4.64) | 1.68(0.54,5.23) | ||
Greenwood SE.
Estimates were not reached.
Based on log-rank test in the univariable analysis and based on Wald test within multivariatble Cox proportional hazards model adjusting for stage and type of adjuvant therapy and stratified by race.
In the dominant model.
3.3 Recursive partitioning analysis
Recursive partitioning analysis was applied to construct a decision tree as a model to classify patients according to their 3-year recurrence risk (Figure 1). In the overall population, four terminal nodes arose showing significantly different 3-year recurrence probabilities ranging from 12 % (± 6% SE) for patients in node 1 to 56% (± 9% SE) for patients allocated in node 4. The initial split was due to Rassf1a rs2236947 indicating that this SNP was the main contributor to the variation in the recurrence probability rate, followed by TAZ rs3811715 and FAT4 rs1039808 (Figure 1).
Figure 1.
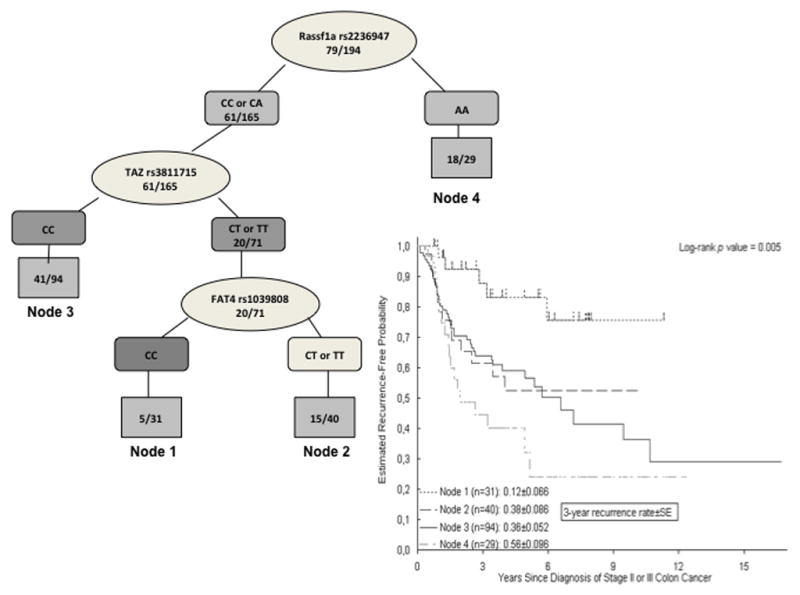
Recursive partitioning analysis and estimated recurrence-free probability for all patients
Recursive partitioning analysis also confirmed the influence of tumor location and revealed different patterns for patients bearing left or right-sided tumors. For patients with right colon carcinomas, Rassf1a rs2236947 remained the most important polymorphism to predict recurrence probability followed by YAP rs8504 and LATS rs9552315, whereas for patients with left-sided tumors TAZ rs3811715 was responsible for the tree’s initial split (Figure 2).
Figure 2.
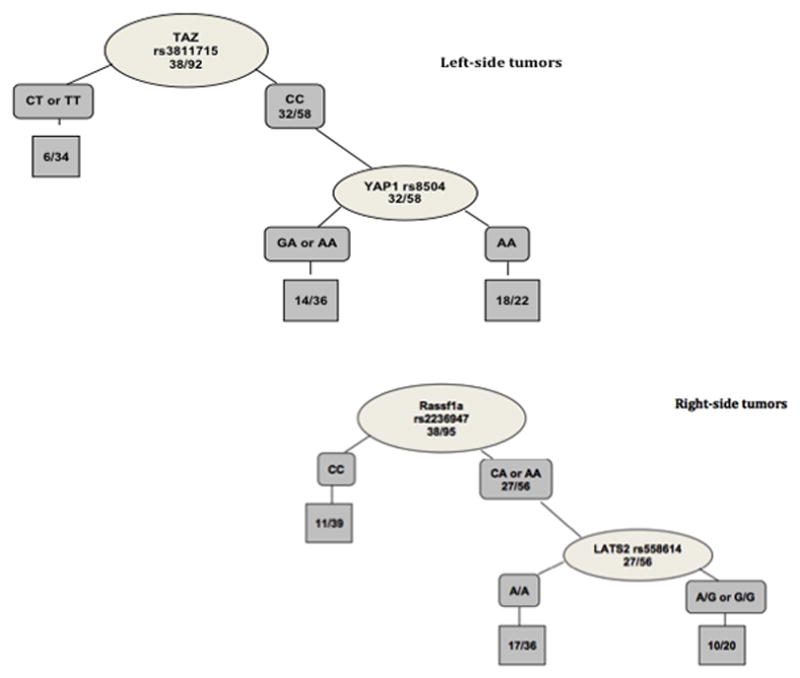
Recursive partitioning analyses based on tumor location
4. Discussion
The present study identifies polymorphisms within genes involved in the Hippo pathway as predictors of recurrence in patients with high-risk stage II and stage III colon cancer treated with adjuvant 5-Fu based chemotherapy. Moreover, our data suggest that the value of these polymorphisms as biomarkers for localized colon cancer is influenced by tumor location and gender.
The Hippo signaling pathway has gained notoriety over the past few years. Despite of this increasing interest, to our knowledge, polymorphisms located in genes involved in this pathway had never been evaluated as biomarkers for colon cancer. As an emerging cascade involved in cancer, in Hippo signaling neither the upstream regulators nor the downstream effectors are fully understood. One of the upstream regulators is Rassf1a, a tumor suppressor that is frequently methylated in colon cancer and that can activate Hippo signaling by binding to MST and ultimately promote apoptosis through p53. In this work, the Rassf1a rs2236947 polymorphism, correlated with the recurrence probability in this cohort of patients. Although no functionality is known for this SNP, in silico analysis revealed that this SNP could affect transcriptional regulation 22.
At the center of the Hippo signaling cascade, the highly homologous YAP and TAZ are the main effectors of the pathway. When phosphorylated YAP/TAZ remain in the cytoplasm, Hippo signaling acts as a tumor suppressor pathway. In the cytoplasm YAP/TAZ interact with β-catenin, which can lead to inhibition of Wnt signaling. Moreover, YAP/TAZ form cytoplasmic complexes with junctional proteins like Scribble or α-catenin maintaining cell polarity. Disruption of the pathway leads to increased YAP/TAZ translocation into the nucleus, which promotes tissue growth, cell viability and stem cell maintenance by regulation of different transcription factors 12, 23. Even more, loss of cell polarity due to lack of TAZ regulation has been implicated in the epithelial-mesenchymal-transition (EMT) 24. In this work, the TAZ rs3811715 polymorphism correlated with the recurrence probability. This SNP is located intronically and prediction tools revealed that affects a splicing site leading to a frameshift coding change 25. In our work, patients with at least a variant allele at this locus had lower recurrence probability than patients with a homozygous wild type genotype, suggesting that the variant allele could reduce TAZ’s nuclear ability to promote cell proliferation, survival and EMT. The presence of a variant allele for TAZ rs3811715 and the correlation with a lower recurrence probability was stronger in patients bearing left-side tumors. Increasing data have underlined the fact that right and left side tumors are different entities 26. Particularly in the adjuvant setting, these molecular differences might influence, in part, the response and the benefit from 5-Fu-based adjuvant treatment 27. Interestingly, Hippo signaling has been implicated in resistance to 5-Fu in CRC cell lines as YAP overexpression has been shown to lead to cellular quiescence and chemoresistance 15. However, the potential differences in the Hippo signaling activity depending on the tumor location have not been studied. In an exploratory analysis performed in an independent Japanese cohort, a similar trend was found for TAZ rs3811715 in patients bearing a left-sided tumor. However, this association was found in a different genetic model, and did not reach statistical significance. Many reasons could account for this fact such as the differences in minor allele frequencies between the two cohorts. The American cohort comprises of different races including Caucasian, African-American, Hispanic as well as Asian and MAFs among these groups differ greatly. We also believe that the clear differences in the baseline characteristics of the patients in these two cohorts have clearly influenced these results. These differences include the percentage of stages II and III (the Japanese cohort is comprised of only stage III patients), the number of resected lymph nodes or the tumor location as it shown in table 1. Surprisingly, despite of being all stage III patients, the Japanese cohort had a lower recurrence rate than the American cohort (36% vs 29%). This fact could be explained by the higher rate of optimal lymphadenectomy in the Japanese cohort.
Overall, this work represents the first approach to the evaluation of polymorphisms within genes involved in the Hippo pathway as prognostic factors. This hypothesis generating study lacks correction for multiple testing and a more similar validation cohort, therefore these results should be interpreted with caution. Nonetheless, the critical implications of the Hippo signaling in several recurrence mechanisms like stem cell maintenance, EMT and resistance to 5-Fu, make this pathway a highly interesting target for colon cancer treatment. Therefore, further genetic studies are warranted.
Supplementary Material
Table 3.
Hippo SNPs and time to recurrence in patients with stage II or III colon cancer at USC
| N | 3-year recurrence probability ± SE* | HR(95%CI)† Univariate | P† value | HR(95%CI)† Multivariate | P† value | |
|---|---|---|---|---|---|---|
| FAT4rs1014867 | 0.19 | 0.17 | ||||
| C/C | 175 | 0.34±0.04 | 1(reference) | 1(reference) | ||
| C/T | 15 | 0.51±0.14 | 1.62(0.77,3.37) | 1.70(0.80,3.65) | ||
| FAT4rs1039808 | 0.53 | 0.40 | ||||
| C/C | 79 | 0.36±0.06 | 1(reference) | 1(reference) | ||
| C/T | 78 | 0.36±0.06 | 1.17(0.72,1.91) | 1.10(0.65,1.86) | ||
| T/T | 32 | 0.37±0.10 | 1.42(0.75,2.67) | 1.56(0.81,3.01) | ||
| LATS1rs12174349 | 0.92 | 1.00 | ||||
| G/G | 53 | 0.37±0.07 | 1(reference) | 1(reference) | ||
| G/A | 82 | 0.38±0.06 | 0.90(0.53,1.52) | 0.98(0.57,1.69) | ||
| A/A | 50 | 0.33±0.07 | 0.94(0.52,1.70) | 0.98(0.52,1.84) | ||
| LATS2rs558614 | 0.69 | 0.70 | ||||
| A/A | 98 | 0.36±0.05 | 1(reference) | 1(reference) | ||
| A/G | 72 | 0.38±0.06 | 1.08(0.67,1.72) | 1.14(0.68,1.91) | ||
| G/G | 20 | 0.34±0.11 | 0.73(0.31,1.73) | 0.79(0.32,1.93) | ||
| LATS2rs9552315 | 0.82 | 0.98 | ||||
| C/C | 128 | 0.39±0.05 | 1(reference) | 1(reference) | ||
| C/T | 46 | 0.30±0.07 | 0.92(0.53,1.59) | 1.04(0.58,1.87) | ||
| T/T | 10 | 0.42±0.19 | 0.71(0.22,2.27) | 1.10(0.31,3.92) | ||
| MST1rs6073629 | 0.37 | 0.40 | ||||
| G/G | 141 | 0.38±0.04 | 1(reference) | 1(reference) | ||
| G/A‡ | 45 | 0.29±0.07 | 0.78(0.45,1.34) | 0.79(0.46,1.36) | ||
| A/A‡ | 2 | |||||
| MST1rs17420378 | 0.20 | 0.56 | ||||
| G/G | 111 | 0.33±0.05 | 1(reference) | 1(reference) | ||
| G/A | 65 | 0.45±0.07 | 1.48(0.93,2.34) | 1.31(0.80,2.15) | ||
| A/A | 10 | 0.21±0.13 | 0.86(0.31,2.39) | 1.12(0.38,3.28) | ||
| MST2rs10955176 | 0.53 | 0.69 | ||||
| C/C | 50 | 0.33±0.07 | 1(reference) | 1(reference) | ||
| C/T | 106 | 0.39±0.05 | 1.17(0.70,1.96) | 1.16(0.68,1.98) | ||
| T/T | 35 | 0.32±0.08 | 0.83(0.41,1.68) | 0.90(0.43,1.86) | ||
| RASSF1rs2073498 | 0.95 | 0.95 | ||||
| C/C | 154 | 0.36±0.04 | 1(reference) | 1(reference) | ||
| C/A‡ | 34 | 0.32±0.09 | 1.02(0.58,1.79) | 0.98(0.55,1.75) | ||
| A/A‡ | 1 | |||||
| RASSF1rs2236947 | 0.017 | 0.039 | ||||
| C/C or C/A | 161 | 0.33±0.04 | 1(reference) | 1(reference) | ||
| A/A | 29 | 0.56±0.10 | 1.87(1.10,3.17) | 1.78(1.03,3.06) | ||
| TAZrs3811715 | 0.077 | 0.12 | ||||
| C/C | 108 | 0.40±0.05 | 1(reference) | 1(reference) | ||
| C/T or T/T | 83 | 0.28±0.05 | 0.66(0.41,1.05) | 0.67(0.41,1.10) | ||
| TAZrs6783790 | 0.27 | 0.33 | ||||
| C/C | 76 | 0.36±0.06 | 1(reference) | 1(reference) | ||
| C/T | 83 | 0.41±0.06 | 0.93(0.58,1.48) | 0.81(0.49,1.32) | ||
| T/T | 28 | 0.25±0.09 | 0.52(0.23,1.16) | 0.54(0.23,1.27) | ||
| YAP1rs8504 | 0.99 | 0.98 | ||||
| G/G | 82 | 0.36±0.06 | 1(reference) | 1(reference) | ||
| G/A | 76 | 0.37±0.06 | 0.97(0.60,1.58) | 0.98(0.59,1.63) | ||
| A/A | 29 | 0.34±0.10 | 1.00(0.51,1.98) | 0.93(0.45,1.88) | ||
| YAP1rs1820453 | 0.59 | 0.48 | ||||
| A/A | 62 | 0.39±0.07 | 1(reference) | 1(reference) | ||
| A/C | 93 | 0.35±0.05 | 0.77(0.47,1.27) | 0.75(0.45,1.25) | ||
| C/C | 26 | 0.29±0.09 | 0.84(0.43,1.66) | 1.00(0.50,2.02) |
Greenwood SE.
Estimates were not reached.
Based on log-rank test in the univariable analysis and based on Wald test within multivariatble Cox proportional hazards model adjusting for stage and type of adjuvant therapy and stratified by race.
In the dominant model.
Acknowledgments
The project described was supported in part by award number P30CA014089 from the National Cancer Institute.
Ana Sebio is a recipient of a Juan Rodés contract from the Instituto de Salud Carlos III (JR14/00006).
Stefan Stremitzer is a recipient of the Erwin Schrödinger Fellowship Grant from the Austrian Science Fund.
Footnotes
Conflicts of interest: none
‘Supplementary information is available at The Pharmacogenomics Journal’s website’
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Bibliography
- 1.Edge CC, SBBDRC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. 7. New York: Springer; 2010. [Google Scholar]
- 2.Andre T, Quinaux E, Louvet C, Colin P, Gamelin E, Bouche O, et al. Phase III study comparing a semimonthly with a monthly regimen of fluorouracil and leucovorin as adjuvant treatment for stage II and III colon cancer patients: final results of GERCOR C96. 1. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25(24):3732–3738. doi: 10.1200/JCO.2007.12.2234. [DOI] [PubMed] [Google Scholar]
- 3.Andre T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(19):3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 4.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 5.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 6.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(4):1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(4):1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138(1):9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7(3):182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 11.Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, et al. Hippo Signaling Regulates Microprocessor and Links Cell-Density-Dependent miRNA Biogenesis to Cancer. Cell. 2014;156(5):893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 13.van Engeland M, Roemen GM, Brink M, Pachen MM, Weijenberg MP, de Bruine AP, et al. K-ras mutations and RASSF1A promoter methylation in colorectal cancer. Oncogene. 2002;21(23):3792–3795. doi: 10.1038/sj.onc.1205466. [DOI] [PubMed] [Google Scholar]
- 14.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796(2):114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Touil Y, Igoudjil W, Corvaisier M, Dessein AF, Vandomme J, Monte D, et al. Colon cancer cells escape 5FU chemotherapy-induced cell death by entering stemness and quiescence associated with the c-Yes/YAP axis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013 doi: 10.1158/1078-0432.CCR-13-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuen HF, McCrudden CM, Huang YH, Tham JM, Zhang X, Zeng Q, et al. TAZ expression as a prognostic indicator in colorectal cancer. PloS one. 2013;8(1):e54211. doi: 10.1371/journal.pone.0054211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Xie C, Li Q, Xu K, Wang E. Clinical and prognostic significance of Yes-associated protein in colorectal cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34(4):2169–2174. doi: 10.1007/s13277-013-0751-x. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Shi S, Guo Z, Zhang X, Han S, Yang A, et al. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PloS one. 2013;8(6):e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493(7430):106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irvine KD. Integration of intercellular signaling through the Hippo pathway. Seminars in cell & developmental biology. 2012;23(7):812–817. doi: 10.1016/j.semcdb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK) European journal of cancer. 2005;41(12):1690–1696. doi: 10.1016/j.ejca.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 22.http://compbio.cs.queensu.ca/F-SNP/.
- 23.Chan SW, Lim CJ, Chen L, Chong YF, Huang C, Song H, et al. The Hippo pathway in biological control and cancer development. J Cell Physiol. 2011;226(4):928–939. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- 24.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 25.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36(Database issue):D820–824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer KM, Hummon AB, Buechler S. Right-side and left-side colon cancer follow different pathways to relapse. Mol Carcinog. 2012;51(5):411–421. doi: 10.1002/mc.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. European journal of cancer. 2009;45(10):1890–1896. doi: 10.1016/j.ejca.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 28.Du J, Ji J, Gao Y, Xu L, Xu J, Zhu C, et al. Nonsynonymous polymorphisms in FAT4 gene are associated with the risk of esophageal cancer in an Eastern Chinese population. International journal of cancer Journal international du cancer. 2013;133(2):357–361. doi: 10.1002/ijc.28033. [DOI] [PubMed] [Google Scholar]
- 29.Wu C, Xu B, Yuan P, Miao X, Liu Y, Guan Y, et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer research. 2010;70(23):9721–9729. doi: 10.1158/0008-5472.CAN-10-1493. [DOI] [PubMed] [Google Scholar]
- 30.Donninger H, Barnoud T, Nelson N, Kassler S, Clark J, Cummins TD, et al. RASSF1A and the rs2073498 Cancer Associated SNP. Frontiers in oncology. 2011;1:54. doi: 10.3389/fonc.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


