Abstract
We tested two pipelines developed for template-free protein structure prediction in the CASP11 experiment. First, the QUARK pipeline constructs structure models by reassembling fragments of continuously distributed lengths excised from unrelated proteins. Five free-modeling (FM) targets have the model successfully constructed by QUARK with a TM-score above 0.4, including the first model of T0837-D1, which has a TM-score=0.736 and RMSD=2.9 Å to the native. Detailed analysis showed that the success is partly attributed to the high-resolution contact map prediction derived from fragment-based distance-profiles, which are mainly located between regular secondary structure elements and loops/turns and help guide the orientation of secondary structure assembly. In the Zhang-Server pipeline, weakly scoring threading templates are re-ordered by the structural similarity to the ab initio folding models, which are then reassembled by I-TASSER based structure assembly simulations; 60% more domains with length up to 204 residues, compared to the QUARK pipeline, were successfully modeled by the I-TASSER pipeline with a TM-score above 0.4. The robustness of the I-TASSER pipeline can stem from the composite fragment-assembly simulations that combine structures from both ab initio folding and threading template refinements. Despite the promising cases, challenges still exist in long-range beta-strand folding, domain parsing, and the uncertainty of secondary structure prediction; the latter of which was found to affect nearly all aspects of FM structure predictions, from fragment identification, target classification, structure assembly, to final model selection. Significant efforts are needed to solve these problems before real progress on FM could be made.
Keywords: Protein structure prediction, CASP11, ab initio folding, QUARK, I-TASSER
INTRODUCTION
Ab initio folding (or free-modeling, FM) in protein structure prediction refers to the effort to construct 3D structure models without using homologous structures as templates. Although many efforts have been devoted to folding protein structures based on physics-based force fields, including molecular dynamics simulations that determine each atom’s position based on Newton’s laws of motion,1–3 the most efficient approach for FM structure prediction still exploits the knowledge and information derived from the Protein Data Bank (PDB) library and sequence databases. Information found useful for FM includes structural fragments excised from unrelated proteins in the PDB4–7 and residue-residue contact maps derived from co-evolution8,9 and machine learning.10,11
In a recent study, we found that the combination of QUARK-based ab initio folding6 with template-based I-TASSER structure assembly12 can help improve the quality and robustness of the final models for FM protein targets.13 In this approach, models built by QUARK are compared with threading templates by LOMETS;14 the templates are then re-ranked by their similarity to the QUARK models before they are subjected to the I-TASSER structure-assembly simulations. The idea behind the approach is that the ab initio models built from scratch are usually different from experimental structures in the PDB. A close match between the templates and QUARK models is usually an indication that the ab initio models (or the matched templates) adopt the correct fold.
The 11th CASP experiment contains an unprecedentedly high number of FM targets/domains that have no similar fold in the PDB and are of a moderate size modelable by FM (e.g. <200 residues); this created a unique opportunity to test the new pipeline of free modeling. Since the pipeline used in our human prediction is essentially the same as the server predictions, our report will mainly focus on the two server groups (‘QUARK’ and ‘Zhang-Server’) in which the structure predictions were generated fully automatically. In brief, while ‘QUARK’ is mainly based on the original QUARK programs,6,7 ‘Zhang-Server’ is built on the combination of QUARK and I-TASSER simulations that are further assisted by the restraints from distant-homology threading alignments14 and machine-learning based contact predictions.15,16
METHODS
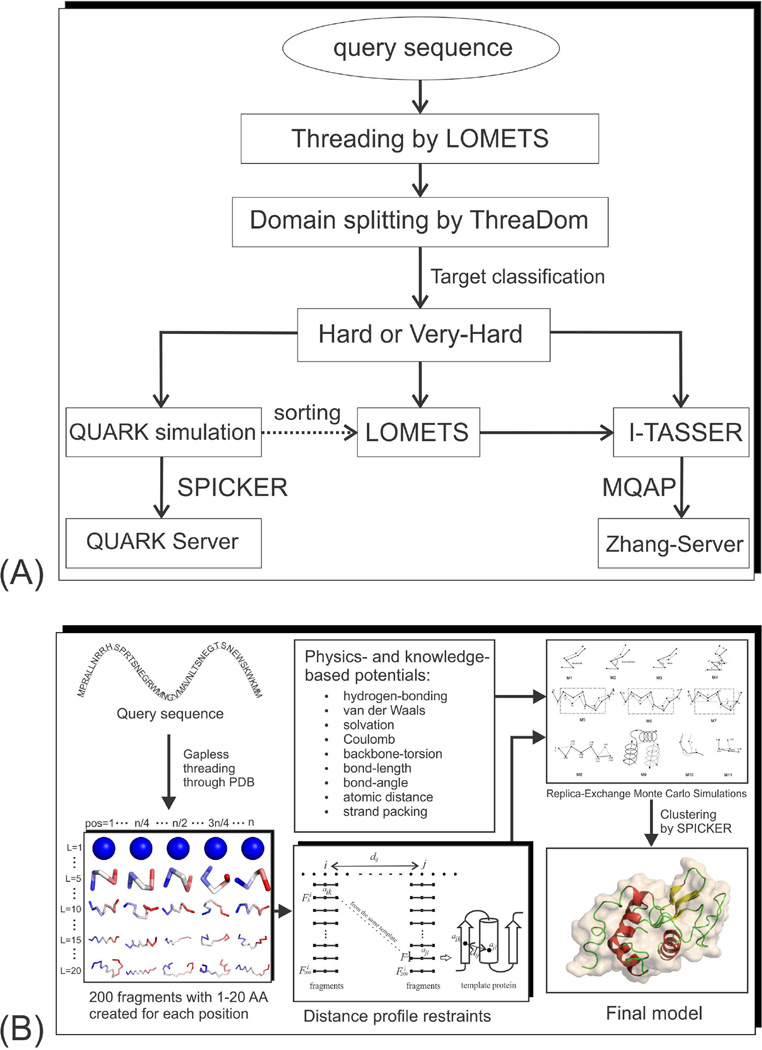
The pipelines for free modeling in ‘QUARK’ and ‘Zhang-Server’ in CASP11 are described in Figure 1A; both start from threading-based domain split and target classification, which are then followed by separated structure assembly simulations.
Figure 1.
The free-modeling pipelines in CASP11. (A) Flowchart for ‘QUARK’ and ‘Zhang-Server’ group predictions. (B) Flowchart of QUARK structure assembly simulations.
Domain split and target classification
Starting from the query sequence, LOMETS14 was used to generate threading alignments from the structures in the PDB. Subsequently, the target sequence is split into individual folding units (or domains) by ThreaDom17, which is built on the domain structure of the template proteins and the distribution of the gaps/insertions of the target-template alignments. Here the domain structure of template proteins is determined by the CATH database18 and the gap/insertion distributions are extracted from multiple LOMETS alignments. These two components are combined linearly with an equal weight into a domain conservation score, the profile distribution of which along the query sequence is used to determine the domain structure of the target.17
For proteins of multiple domains, a second round threading is conducted on the individual domains, where a difficulty score is assigned to each domain based on the normalized Z-score and the consensus of the top 20 threading alignments (Eq. 1 of Ref13). If a domain is assigned ‘Hard’ or ‘Very-Hard’, which means that both the significance and consensus scores of the top LOMETS threading alignments are low, a free-modeling process will be started for both QUARK and Zhang-Server groups (Figure 1A).
QUARK pipeline
The flowchart of QUARK is depicted in Figure 1B, which starts from the position-specific fragment structure generation. At each residue position, 4,000 (=200×20) structural fragments are generated, with continuous lengths ranging from 1 to 20 residues, based on gapless threading of the fragment sequence through a non-redundant set of 6,023 high-resolution PDB structures. The scoring function of the gapless threading consists of profile-profile, secondary structure, torsion angle and solvent accessibility matches.19 Two types of information are derived from the fragments to assist next step of structure folding simulations. First, a torsion angle (φ, ψ) distribution is collected from the 10-mer fragments at each residue position. Second, a residue-residue contact map is derived from the distance profiles between fragments. Here, a distance (dij) is recorded for each pair of fragments (at position i and j) that come from the same PDB structure although they were derived by independent gapless threading procedures. In general, the histogram of dij increases rapidly with the value of dij; this increase is trivial because it is mainly due to the increase of the geometrical phase space for larger distance dij as observed in random polymer chains.20 But if there is a peak appearing in the histogram at the position of dij <9 Å, which is non-trivial because it indicates that the histogram probability at a shorter distance is higher than that at the longer distance – against the general tendency of geometrical phase space in random chains, a contact between residue i and j will be predicted.7
In the next step, replica-exchange Monte Carlo (REMC) simulations are performed to assemble the fragments into full-length models under a composite physics- and knowledge-based potential, containing hydrogen bonding, van der Waals, solvation, Coulomb, backbone-torsion, bond-length and bond-angle, atomic distance, and strand pairing interactions. The conformational changes are driven by 11 local and global movements shown in the top-right panel of Figure 1B. While the first feature, the torsion-angle distribution as collected from the fragments, is used to constrain local torsion movement selection, the second feature, the contact map derived from the fragment distance profiles, is used as restraints to guide the simulations. The final models are selected by SPICKER21, which clusters all the decoys generated in the REMC simulations and ranks models by the size of the clusters.
Zhang-Server pipeline
Two versions of I-TASSER simulations were performed with the predicted models submitted by the ‘Zhang-Server’ group. The first I-TASSER simulation follows the standard I-TASSER pipeline,12 which reassembles fragments excised from the LOMETS threading alignments by a modified REMC simulation method.22 In the second version, the threading templates identified by LOMETS are sorted by their similarity to the QUARK models before being submitted to I-TASSER, where the similarity here is defined by the highest TM-score of the template to the top ten QUARK models. Because the LOMETS alignment scores are generally low and unreliable for the Hard and Very-Hard domains, the top templates re-ordered by QUARK models are usually completely different from the original LOMETS templates that are ordered by the Z-score of the threading alignments. Spatial restraints are collected from the new LOMETS templates and the QUARK models, which are then used by I-TASSER for structural assembly simulations. The structural decoys are submitted to SPICKER21 for structure clustering, and the models with the highest cluster density are further refined by fragment-guided molecular dynamic (FG-MD) simulations.23
The final models generated by both I-TASSER versions are scored by 5 Model Quality Assurance Programs (MQAPs) consisting of C-score,24 a structural consensus score that calculates the average TM-score of the target model to all other models,25 and three statistical potentials (RWplus,26 GOAP27 and DOPE28). An average rank is calculated for each model, defined as , where rij is the rank of the ith model by the jth MQAP program. The models with the lowest average rank are finally submitted in the ‘Zhang-Server’ group.
RESULTS AND DISCUSSION
There are 35 domains that were assigned by the CASP11 assessors as FM targets, meaning that these domains have no similar structure found in the PDB and their models must be constructed from scratch. These included 9 α-proteins, 5 β-proteins, 20 α β-proteins, and 1 protein (T0799-D2) with little secondary structure according to its experimental structure. The lengths of the 35 FM domains range from 45 to 345 residues with an average length equal to 143.5 residues.
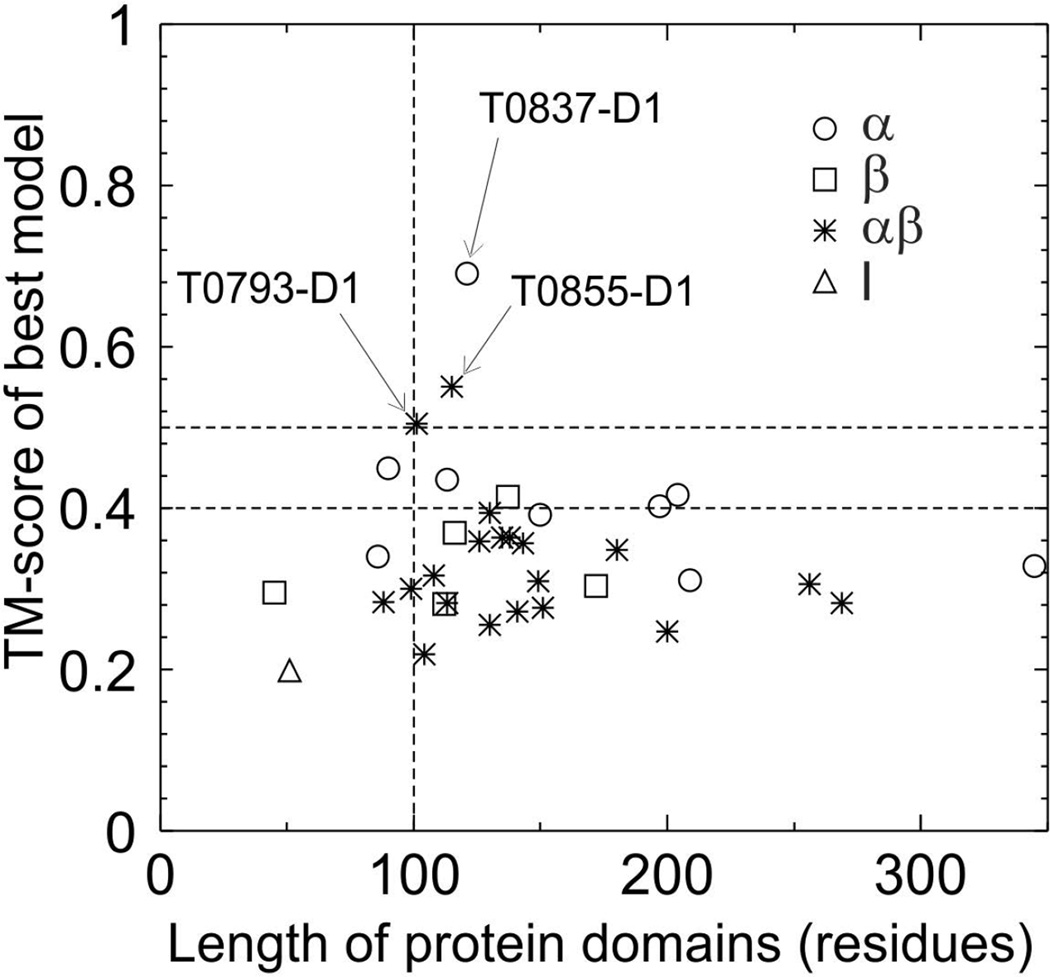
Figure 2 shows a summary of the TM-scores of the best models generated by Zhang-Server versus the size of the 35 FM targets (a similar plot for the best QUARK models is presented in Figure S1 in Supplemental Information). In general, proteins with a longer length should have a higher number of secondary structure elements, which render the structure conformations a more complicated topology. Due to the limits in the accuracy of the reduced force field and in the power of conformational search,29,30 it has been a significant challenge for the ab initio folding programs to construct such complicated structure models from scratch. In the previous CASP experiments, success on high-resolution FM targets was mainly limited to the domains with a size below 120 residues,31,32 with an exception in CASP10 where models on two FM targets (R0006-D1 with 169 residues and R0007-D1 with 161 residues) by an integrated QUARK and I-TASSER pipeline had the TM-score 0.622 and 0.620, respectively.13
Figure 2.
TM-score of the best models by Zhang-Server versus the length of the 35 FM protein domains. Dashed lines denote the TM-score cutoffs at 0.4 and 0.5 and the length cutoff of 100 to guide the eye. Circles, squares, stars and triangles denotes α-, β-proteins, αβ-proteins, and the protein with little secondary structure (l), respectively.
In this CASP, there are only three domains (T0837-D1, T0793-D2 and T0855-D1) by I-TASSER and QUARK that have a TM-score above 0.5, indicating correct fold.33 All the three domains have a size above 100 residues but below 150 residues. The best prediction was the first model built by QUARK for T0837-D1 that has a TM-score=0.736, which is higher than any models constructed by QUARK or I-TASSER in the former CASP experiments. However, there is no successfully modeled targets with length >150 AA, which demonstrates the challenge to fold big proteins by the current approaches. Nevertheless, there are two domains (T0831-D2 and T0836-D1), both being alpha-proteins, with a length near or above 200 residues that have a TM-score above 0.4, which indicates some promising progress towards folding larger size proteins at least for alpha-proteins.
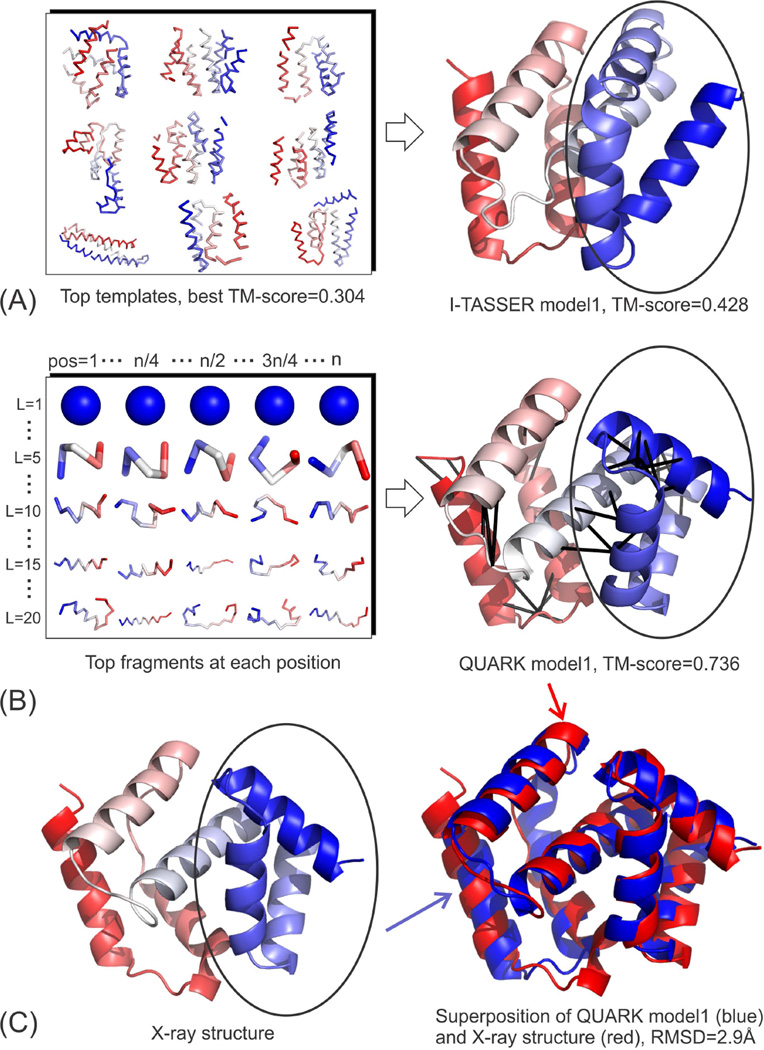
Case study on T0837-D1: QUARK successfully assembles the domain structure using local fragments and predicted contact maps
Figure 3 shows the modeling process and the first models predicted by I-TASSER and QUARK on T0837-D1. First, as a FM target, all the threading templates by LOMETS have an incorrect fold where the best template from 1kagA has only a low TM-score (0.304) (Figure 3A). I-TASSER reassembled the LOMETS fragments and generated a much improved first model that has a TM-score=0.428. This model has the C-terminal portion of the structure correctly constructed; but the N-terminal portion is a mirror image of the X-ray structure, as highlighted by the black circles in Figures 3A and 3C. This N-terminal structure is apparently resulted from the LOMETS alignments, as several top threading templates show a similar topology in the N-terminal domain (see the left panel of Figure 3A). The second I-TASSER model has the N-terminal domain constructed correctly, which resulted in a high TM-score=0.682 (image not shown). But this model has a much lower population in the I-TASSER simulation trajectories, which resulted in a low C-score. We anticipate that the success of this model in I-TASSER is mainly due to the restraints that were derived from the QUARK models that have a much higher TM-score than all the threading templates (see below). The consensus score by TM-score metric for this model is also low. Thus, although the statistical potentials give the model a high rank, the overall MQAP score is slightly lower than the first model, which resulted in an incorrect model selection in Zhang-Server group (Table 1). Nevertheless, we note that the post-CASP statistics on all domains showed that the average TM-score based on multiple MQAPs still outperforms either consensus or statistical scores; but for this specific target the statistical scores do better than the combined MQAP scores for model selection. Clearly, the selection of correct models remains as a significantly unsolved problem in protein structure prediction.
Figure 3.
Modeling procedure and the first model predicted for T0837-D1. (A) I-TASSER; (B) QUARK; (C) X-ray structure and the superposition of QUARK model1 and X-ray structure. The black lines on the QUARK model indicate the contacts predicted by the fragment-based distance profiles. Black circle highlights the N-terminal domain where I-TASSER model1 has the mirror image of native but I-TASSER model2 and QUARK model1 modeled it correctly.
Table I.
MQAP ranking results on the first two models generated by I-TASSER for T0837-D1. Data in parentheses are the relative ranks by different MQAP programs.
| Normalized MQAP score | I-TASSER model 1 | I-TASSER model 2 |
|---|---|---|
| TM-score to native | 0.428 | 0.682 |
| C-score (rank) | 3.580 (4) | 1.420 (99) |
| Consensus (rank) | 1.196 (1) | 1.071 (185) |
| Rwplus (rank) | −1.058 (24) | −1.069 (1) |
| GOAP (rank) | −1.198 (191) | −1.248 (2) |
| DOPE (rank) | −1.072 (27) | −1.093 (4) |
| Average rank | (49.0) | (59.2) |
Figure 3B shows the procedure of QUARK modeling on T0837-D1. As QUARK starts from small fragments from non-homologous proteins, the folding procedure is free from the influence of the LOMETS templates that include the incorrect secondary structure arrangement in the N-terminal domain. The QUARK trajectories were dominated by the model shown on the right panel of Figure 3B, which has a TM-score=0.736 and a RMSD=2.9 Å compared to the X-ray structure (Figure 3C). In addition to the QUARK force field that coordinates the overall folding simulations, two factors might have contributed to the success of QUARK on this target. First, the fragments identified by the profile-based gapless threading procedure are close to the native. In Table S1 in the Supplementary Information (SI), we present the average RMSD of the 10 best fragments at each position for each given fragment length. It shows that the average RMSD is below 2.7 Å with standard deviation below 0.5 Å for all sizes of fragments although the RMSD gradually increases with the fragment size as expected. This data indicates that there are at least 10 good fragments at every location that can be used to assemble the full-length models.
Second, the fragment-based distance profiles did a fine job in generating the residue-residue contact map predictions. Based on the distance profile distribution of fragments, QUARK collected 37 contacts for the query sequence, where 34 contacts appear in the sequence regions that have the X-ray structure solved (-- the other 3 contacts involve 7 residues in the C-terminal that have no X-ray structure). Table S2 displays all the 34 contacts with the predicted distances compared to the actual distance in the X-ray structure. 29 of the predicted contact distances are within 2 Å of the actual experimental distance, and the average distance error of all the contacts is 0.934 Å. The predicted contacts are also shown as black lines on the QUARK model at the right panel of Figure 3B. Although most of the QUARK contacts are short-range contacts (|i−j|<12 residues), there are still four long-range contacts (|i−j|>24) between the second and fourth helices that help constrain the packing of the N-terminal domain. Noticeably, almost all of the short- and medium-range contacts are located in the loop/turn regions (between the secondary structure elements and the loops/turns), which is important in constraining the relative orientations of the helix pairs and therefore help to guide the simulations toward the correct global topology of the target.
To obtain a general view of the contact predictions, we further counted the distant-profile based contact predictions for all 35 FM domains. The predictions generated overall 5,948 contacts for the 35 domains, where 4,013 have both involved residues with coordinates on the solved structure. 1,803 out of the 4,013 contacts (45%) are short-range with 5≤|i−j|≤11, 516 (13%) are medium-range with 12≤|i−j|≤24, and 1,694 (42%) are long-range with 25≤|i−j|. Figure S2 displays the data of the distance of all the 4,013 residue pairs that are predicted in contact versus the actual distance of the corresponding residue pairs on the native structure. Because these residue pairs were assumed by QUARK as in contact, the predicted distances are all below 9 Å. However, the actual distances of the residue pairs have a broad distribution up to 30 Å due to the prediction error. Therefore, there is nearly no correlation between the predicted and actual distances (--this is part of the reason that QUARK uses them as contact rather than distance restraints). Nevertheless, the average (or median) difference between the predicted and actual distance is 3.49 Å (or 2.39 Å), which suggests that many of the predicted residues are still in or near the contact distance. As expected, the average and median distance errors (1.42 and 0.88 Å respectively) for the short-range residue pairs are much lower than that of the medium- (4.05 and 3.05 Å) and long-range (5.51 and 5.04 Å) residue pairs. Compared to the data from T0837-D1 in Table S2, the average distance error of the overall FM target set is considerably worse, which partly explains the significantly better quality of the final models of T0837-D1 than the other FM targets.
Here, we note that the distance-profile based contact predictions do not assign a confidence score for any of the contact predictions and the QUARK simulations adopt all the residue pairs as contacts if they have a peak with dij <9 Å in the histogram distribution; this is in contrast to the traditional machine-learning prediction methods that use the training probability as the confidence score of the contact predictions. However, we noticed a modest correlation between the error of the predicted distances and the relative height of the histogram peaks with a Pearson correlation coefficient −0.41; the latter may be used as the confidence score to guide the weight setting of the contact restraints in future applications.
Implementation of multiple simulation processes enhances the robustness of I-TASSER pipeline for ab initio structure assembly
Two versions of the I-TASSER simulations were implemented in the Zhang-Server group, i.e., the standard I-TASSER12 and the I-TASSER with threading templates re-ordered by their structural similarity to the ab initio QUARK models. In the latter case, the correctness of the QUARK models has a strong impact on the final results of the I-TASSER simulations.
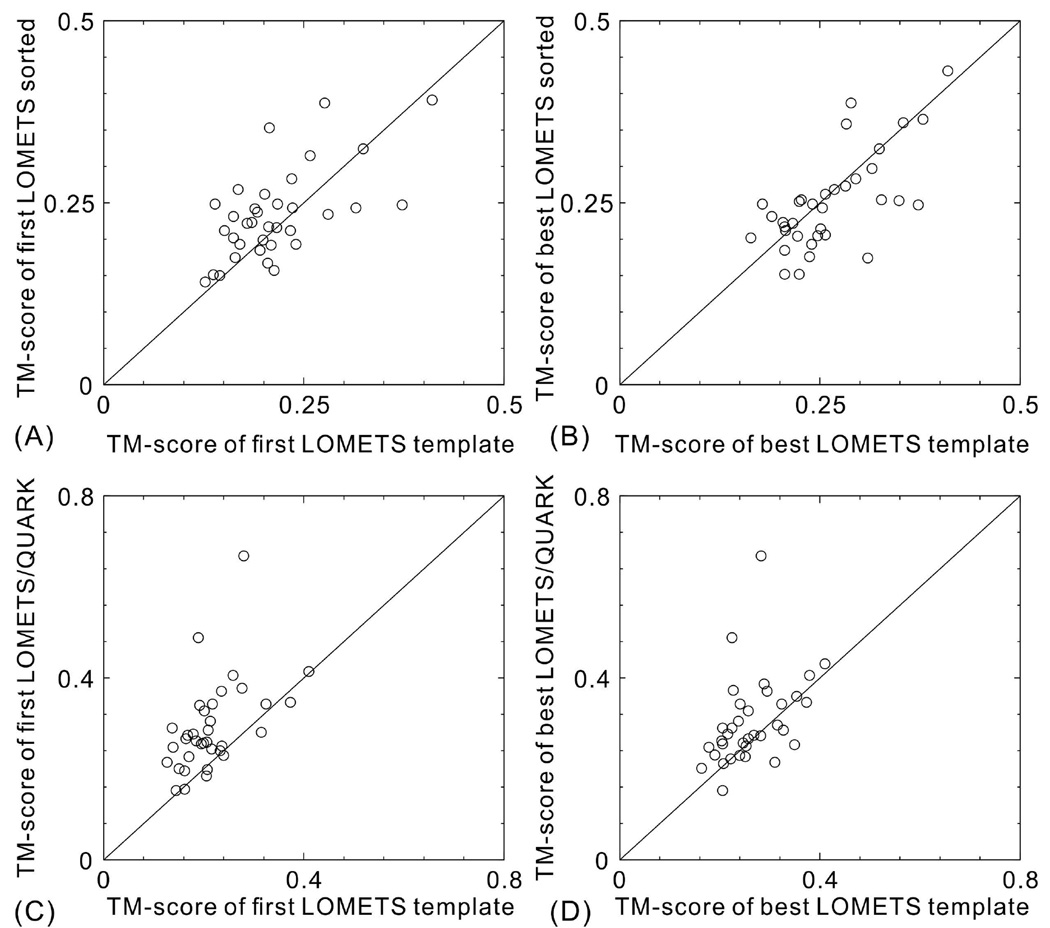
In Figure 4, we present a head-to-head comparison of the templates generated by the original LOMETS programs versus those re-ordered by the QUARK models. Since the domain prediction used in structure modeling was based on sequence, which can be different from the assessor’s domain definition that is based on X-ray structures, we first selected the domain models that cover the largest portion of the domains as defined by the assessors. The TM-score of the domain models is then normalized by the common length between the domain prediction and the domain assignment from the assessors. Therefore, the TM-score in Figure 4 may be different than the TM-score for the multi-domain proteins when the full-length X-ray structures are used; but the TM-scores are scaled equally for both templates with and without QUARK sorting. Meanwhile, for each target, around 200 LOMETS templates (i.e. top-20 templates collected from each of the 10 component threading programs) are used for sorting by the QUARK models. But only 20 templates are eventually used by I-TASSER to construct spatial restraints and initial conformation. Therefore, we only count in Figure 4 for the top-20 templates from each template set that are used by I-TASSER. Thus, the templates considered in Figure 4, with or without QUARK sort, not only have different order but also have different template identity.
Figure 4.
Comparison of threading templates with and without sorting using QUARK models. (A) TM-score of the first LOMETS templates; (B) TM-score of the best LOMETS templates; (C) TM-score of the first LOMETS templates including QUARK models; (D) TM-score of the best LOMETS templates including QUARK models.
Figures 4A and 4B show TM-score of the first and the best templates after QUARK-based sorting versus that from the original LOMETS templates. Apparently, the QUARK-based sorting considerably improved the quality of the first-rank templates, where the average TM-score increases from 0.215 to 0.243. However, the average TM-score of the best templates is slightly worse than the best of the original LOMETS templates (0.261 vs. 0.263), due to the fact that the structural variation of the top templates after sorting becomes lower than that of the original LOMETS templates. This result also highlights the rationale of running the standard I-TASSER version without using QUARK-based sorting, which should increase the robustness of I-TASSER pipeline.
Figures 4C and 4D show the TM-scores for the sorted templates plus the QUARK models, which are the input conformations that I-TASSER started from. The TM-score of the conformations is now significantly higher than that of the LOMETS templates for both the first and the best LOMETS templates, i.e., the average TM-score of the first and the best LOMETS templates increases from 0.215 and 0.263 to 0.291 and 0.304, respectively, when including QUARK models in the QUARK-based sorting templates. However, we note that there are two targets (T0793-D1 and T0837-D1) that have made the major contribution to the TM-score increase. If we remove these two targets, the average TM-score of the first and best LOMETS/QUARK models will be reduced to 0.274 and 0.287, while the TM-score increase on the first model is still quite significant (0.215 vs. 0.274) but that of the best models becomes comparable to the original LOMETS templates (0.263 vs. 0.287).
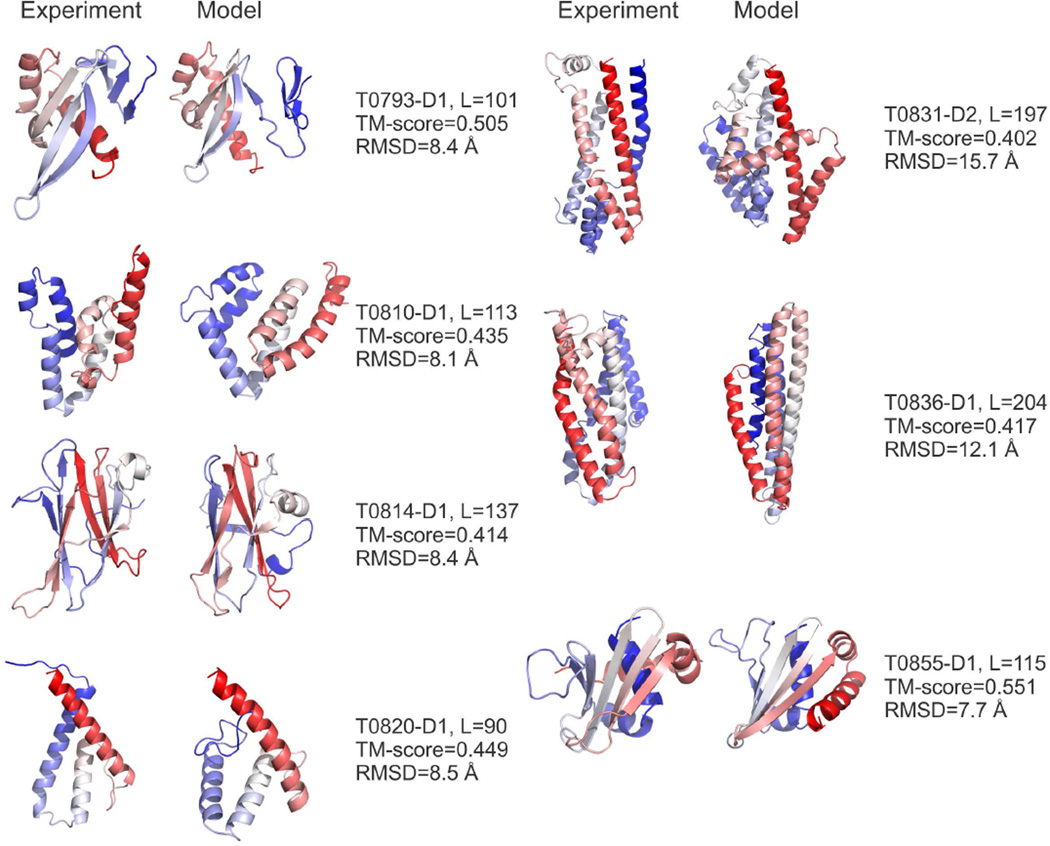
As a result, the I-TASSER predictions used in Zhang-Server showed an improved modeling quality in FM as compared to the QUARK pipeline, although QUARK outperforms Zhang-Server for a few specific cases (such as T0837-D1 discussed in the above section). Overall, there are eight cases where Zhang-Server models have a TM-score above 0.4, while QUARK has only five cases with the best TM-score above 0.4 (see Figure S1). All the five cases that QUARK successfully modeled are included in the eight cases where Zhang-Server succeeds. In addition to T0837-D1 shown in Figure 3, the structural models for the rest of the seven FM cases predicted by Zhang-Server are presented in Figure 5.
Figure 5.
The targets for which Zhang-Server generated models with a TM-score above 0.4, excluding T0837-D1 that is shown in Figure 3. Blue to red runs from the N- to C-terminals. The left cartoon of each panel shows the experimental structure and the right cartoon shows the predicted model. L is the length of the sequence having structure solved by X-ray, which is usually shorter than the length of the target sequence.
These results indicate that the composite I-TASSER pipelines are more robust than QUARK for the free-modeling simulations. This robustness might be due to several factors. First, I-TASSER simulations have the ability to refine template structures, which often draw the starting conformations closer to the native by the use of spatial restraints from multiple template alignments that are assisted with a highly optimal knowledge-based force field.34 Second, in addition to using QUARK models and the templates sorted by QUARK, Zhang-Server performs an additional I-TASSER simulation that is based on the original LOMETS templates. This additional structure-assembly simulation helps model the cases where QUARK failed to generate correct models, e.g., T0810-D2, T0831-D2, and T0836-D1; these cases are also manifested in Figure 4 where the LOMETS templates could have a better quality than QUARK models, even for the FM targets when considering the best templates.
Impact of secondary structure prediction on FM
The secondary structure in QUARK and I-TASSER was predicted by a combination of PSIPRED35 and PSSpred,12 where the secondary structure identity (helix, strand or coil) of a given residue is decided by the sum of the confidence scores from the two programs on the three secondary structure types. The average Q3 accuracy of the combined predictor is 78.3% for the 35 FM targets, which is slightly higher than the individual predictors, i.e. 76.1% for PSIPRED and 77.5% for PSSpred; but the Q3 value is considerably lower than that for the TBM targets, which are all above 82%.
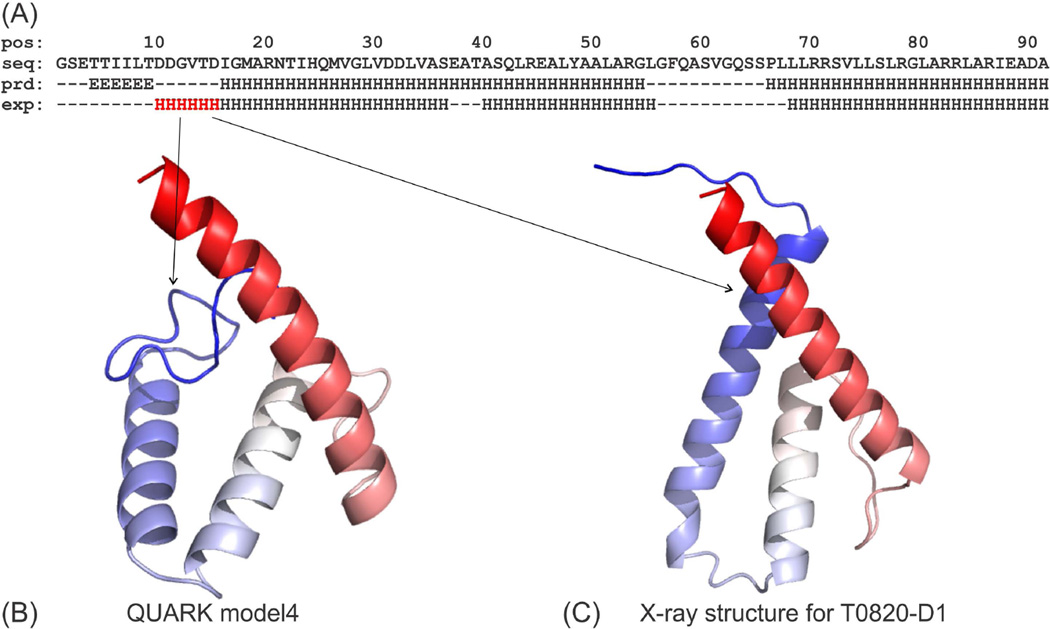
The relatively low accuracy of secondary structure prediction had an important impact on the results of the final FM models. One of the obvious examples is from T0820-D1 as shown in Figure 6. The X-ray structure of T0820-D1 has three helices; but the PSIPRED/PSSpred prediction generated two long helices with a beta-strand at the N-terminal (Figure 6A). In particular, the failure to predict alpha helix from D10-D15 in the SS prediction directly resulted in errors in the structure of the N-terminal. The predicted model has a random coil in the N-terminal instead of an alpha helix (Figure 6B).
Figure 6.
Structure prediction for T0820-D1. (A) Predicted secondary structure (‘prd’) compared to X-ray structure (‘exp’), where the α-helix region missed in secondary structure prediction is marked in red. ‘H’, ‘E’ and ‘-’ indicate helix, extended-strand and coil respectively. (B) QUARK model4. (C) X-ray structure for T0820-D1.
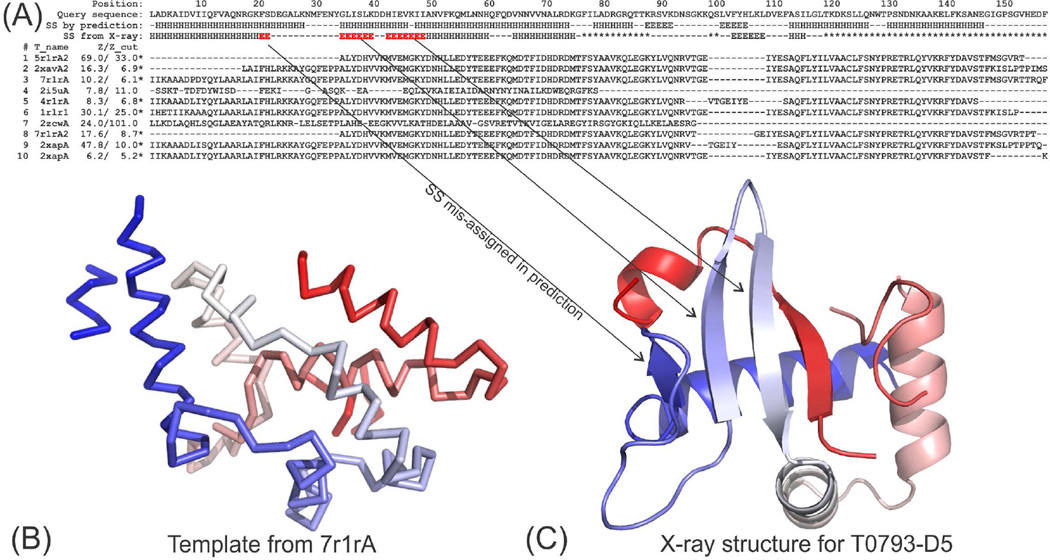
An example, for which secondary-structure errors resulted in an even larger impact in the final structure, is T0793-D5 (Figure 7). T0793 is a multiple-domain protein consisting of 595 residues and five domains. ThreaDom correctly predicted the domain boundary of T0793-D5. This is a medium-size domain of 156 residues, where the X-ray structure has only 103 solved residues (Figure 7C). The X-ray structure shows an alpha/beta fold but the secondary structure predicted by the PSIPRED/PSSpred programs shows mainly alpha helices in the regions that were solved by experiment. The secondary structure prediction resulted in a highly consensus template hit by many of the LOMETS threading programs from the ribonucleotide reductase R1 protein (7r1rA), which is an alpha-protein as shown in Figure 7B; the other top templates are from the same protein but with different PDB IDs (5r1rA, 2xavA, 4r1rA, 1r1rA, and 2xapA). This dominantly consensus and strong hit from LOMETS also misled the I-TASSER pipeline to define the domain as an Easy target, and thus no ab initio simulations were performed. The final models from Zhang-Server and QUARK thus had a completely incorrect model with a TM-score <0.22 for both cases.
Figure 7.
Structure prediction for T0793-D5. (A) Secondary structure prediction and LOMETS alignments. Z/Z_cut indicates the Z-score and the program-specific Z-score cutoffs that are used to define good or bad templates. β-strand regions missed in structure prediction are marked in red. Star ‘*” indicates the residues that are missed in the X-ray structure. (B) Consensus template 7r1rA that was identified by multiple LOMETS programs. (C) X-ray structure for T0793-D5.
We have re-run LOMETS on T0793-D5 using the correct secondary assignment taken from the X-ray structure after CASP11. Several LOMETS threading programs hit the correct template from 2ipqX, which has a TM-score=0.452. Further I-TASSER structure assembly simulations based on this template resulted in a model of TM-score=0.610 (data not shown). This target represents a typical case where secondary structure prediction errors impact almost all aspects of structure modeling, including template identification, target classification, and model refinement and selection.
SUMMARY
Two pipelines have been designed for free modeling (FM) in CASP11. In QUARK, structure models are assembled from small fragments with continuously distributed length of 1–20 residues using replica-exchange Monte Carlo simulations, guided by a composite physics- and knowledge-based force field.6 In Zhang-Server, the I-TASSER based structure assembly simulations12 start from both normal LOMETS templates and the templates that were re-ordered by their similarity to the QUARK models, where spatial restraints were collected from both template sets and used to guide the structure assembly simulations.
The modeling results showed that the QUARK simulations have the ability to build models with quality beyond the homologous modeling approach for FM domains – this improvement can be significant in some cases (e.g., T0837-D1). The success on these targets might be attributed to the quality of local fragment structures and the accuracy of the fragment-based contact-map prediction, in addition to the optimized composite force field. Although most of the fragment-derived contacts are short- to medium-range, the predicted contacts are mainly located between the secondary-structure elements and the turn/loop regions, which can help guide the orientation of secondary-structure element assembly and determination of the global fold.
Despite the fact that I-TASSER used the templates sorted by the QUARK models, the combination of both LOMETS threading alignments and the QUARK models shows that the I-TASSER pipeline can generate more robust modeling results than QUARK. Out of the 35 FM domains, the I-TASSER pipelines generated models with TM-score >0.4 for eight domains, whereas QUARK did so for five cases. This is partly due to the fact that I-TASSER contains both simulations using normal LOMETS templates as well as the templates including QUARK-sorted alignments. Meanwhile, the I-TASSER simulations have drawn the initial conformation closer to the native when combining multiple threading templates.
Despite the promising success on several domains, the data clearly showed that FM remains as a significantly unsolved problem, as highlighted by the fact that for the majority of the FM targets, the correct fold was not generated. Challenges include the difficulty in packing non-local beta-strand structures and the uncertainty of domain determination when no threading templates can be used to guide domain parsing, as shown in previous CASP experiments.13,31,36 In this CASP experiment, there are cases also showing that the low accuracy of secondary-structure predictions could have additional impacts on the critical stages of FM structure assembly, including template identification, target classification, and final model assembly and selection. Significant efforts are needed for solving these problems before real progress on FM could be made.
Supplementary Material
Acknowledgements
The project is supported in part by the National Institute of General Medical Sciences (R01GM083107 and R01GM084222).
References
- 1.Alder BJ, Wainwright TE. Phase transition for a hard sphere system. J Chem Phys. 1957;27:1208–1209. [Google Scholar]
- 2.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4(2):187–217. [Google Scholar]
- 3.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 4.Bowie JU, Eisenberg D. An evolutionary approach to folding small alpha-helical proteins that uses sequence information and an empirical guiding fitness function. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(10):4436–4440. doi: 10.1073/pnas.91.10.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268(1):209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 6.Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012;80(7):1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu D, Zhang Y. Toward optimal fragment generations for ab initio protein structure assembly. Proteins. 2013;81(2):229–239. doi: 10.1002/prot.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, et al. Protein 3D structure computed from evolutionary sequence variation. PloS one. 2011;6(12):e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(39):15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S, Szilagyi A, Zhang Y. Improving protein structure prediction using multiple sequence-based contact predictions. Structure. 2011;19(8):1182–1191. doi: 10.1016/j.str.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones DT, Buchan DW, Cozzetto D, Pontil M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28(2):184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Yan R, Roy A, Xu D, Poisson J, et al. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y. Interplay of I-TASSER and QUARK for template-based and ab initio protein structure prediction in CASP10. Proteins. 2014;82(Suppl 2):175–187. doi: 10.1002/prot.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucl Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun HP, Huang Y, Wang XF, Zhang Y, Shen HB. Improving accuracy of protein contact prediction using balanced network deconvolution. Proteins. 2015;83:485–496. doi: 10.1002/prot.24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Zhang Y. A comprehensive assessment of sequence-based and template-based methods for protein contact prediction. Bioinformatics. 2008;24(7):924–931. doi: 10.1093/bioinformatics/btn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Z, Xu D, Wang Y, Zhang Y. ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics. 2013;29(13):i247–i256. doi: 10.1093/bioinformatics/btt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, et al. CATH--a hierarchic classification of protein domain structures. Structure. 1997;5(8):1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Zhang Y. MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72(2):547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhou H, Ou-Yang Z. Stretching single-stranded DNA: interplay of electrostatic, base-pairing, and base-pair stacking interactions. Biophys J. 2001;81(2):1133–1143. doi: 10.1016/S0006-3495(01)75770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Skolnick J. SPICKER: A clustering approach to identify near-native protein folds. J Comput Chem. 2004;25(6):865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Kihara D, Skolnick J. Local energy landscape flattening: Parallel hyperbolic Monte Carlo sampling of protein folding. Proteins. 2002;48:192–201. doi: 10.1002/prot.10141. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19(12):1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Zhang Y. A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PloS one. 2010;5(10):e15386. doi: 10.1371/journal.pone.0015386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H, Skolnick J. GOAP: a generalized orientation-dependent, all-atom statistical potential for protein structure prediction. Biophys J. 2011;101(8):2043–2052. doi: 10.1016/j.bpj.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein science : a publication of the Protein Society. 2006;15(11):2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309(5742):1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18(3):342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinch L, Yong Shi S, Cong Q, Cheng H, Liao Y, et al. CASP9 assessment of free modeling target predictions. Proteins. 2011;79(Suppl 10):59–73. doi: 10.1002/prot.23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai CH, Bai H, Taylor TJ, Lee B. Assessment of template-free modeling in CASP10 and ROLL. Proteins. 2014;82(Suppl 2):57–83. doi: 10.1002/prot.24470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26(7):889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Kolinski A, Skolnick J. TOUCHSTONE II: A new approach to ab initio protein structure prediction. Biophys J. 2003;85:1145–1164. doi: 10.1016/S0006-3495(03)74551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins. 2009;77(S9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.