Abstract
Objectives
Recently, several meta-analyses have reported an association between interleukin (IL) gene polymorphisms and the risk of Alzheimer's disease (AD). Several further papers discussing the relationship with the risk of AD have recently been published. The aim of this meta-analysis was to re-evaluate and update the associations between IL gene polymorphisms and the risk of AD.
Methods
The search sources were PubMed, Science Direct, Scopus, and Google Scholar up to July 2015, and the following search terms were used: “interleukin 1 or interleukin 6 or interleukin 10” and “variant or polymorphism or SNP” in combination with “Alzheimer's disease”. A meta-analysis using the pooled odds ratios and 95% confidence intervals was carried out to assess the associations between four polymorphisms of IL genes (− 889C > T in IL-1α, − 511C > T in IL-1β, − 174G > C in IL-6 and − 1082G > A in IL-10) and the risk of AD under the heterozygous, homozygous, dominant, and recessive models with fixed- or random-effects models.
Results
A total of 21,864 cases and 40,321 controls from 93 individual studies were included in this meta-analysis. Our results indicated that the − 889C > T polymorphism was strongly associated with the increased risk of AD. However, three polymorphisms were not associated with the risk of AD.
Conclusions
Similar to previous meta-analyses, our updated meta-analysis suggested that the − 889C > T polymorphism may be a factor in AD. However, the results of our meta-analysis of the − 174G > C polymorphism differed from those of previous meta-analyses. Consequently, we suggest that the − 174G > C polymorphism may not be a risk factor for AD.
Abbreviations: OR, odds ratio; CI, confidence interval; HWE, Hardy–Weinberg equilibrium; SNP, sing nucleotide polymorphism; AD, Alzheimer's disease; IL, Interleukin
Keywords: Interleukin, Cytokine, Polymorphism, Meta-analysis, Alzheimer's disease
Highlights
-
•
889 C > T polymorphism of IL-1α was significantly associated with increased risk of Alzheimer's disease
-
•
Three polymorphisms (− 511C > T in IL − 1β, − 174G > C in IL-6 and − 1082G > A in IL-10) were no associated with risk of Alzheimer's disease
-
•
The results of our meta-analyses for three polymorphisms (− 889C > T, − 511C > T and − 1082G > A) were similar to those previous meta-analyses. However, the results of the − 174G > C polymorphism were different.
1. Introduction
Dementia is an overall term for conditions characterized by a decline in memory, cognitive and other thinking skills that affect a person's abilities. The total number of people with dementia worldwide was estimated at 35.6 million in 2010, and is projected to be 65.7 million in 2030 and 115.4 million in 2050 (WHO, 2012). Among the several types of dementia, Alzheimer's disease (AD) is the most common. AD was first identified more than 100 years ago. However, its symptoms, causes and risk factors were only discovered in the last 30 years (Alzheimer's Association, 2014).
Several cytokines including interleukin 1 (IL-1), IL-6, tumor necrosis factor-α (TNF-α) and transforming growth factor-β (TGF-β) have been reported to be associated with AD (Wilson et al., 2002). Interleukins (ILs) are important components of the immune system, and a deficiency in them may lead to autoimmune disease or immune deficiency. Several studies have suggested that IL-1 is related to the pathogenesis of AD. Griffin et al. reported that IL-1 immunoreactivity was increased in AD compared with non-AD subjects (Griffin et al., 1989). Sheng et al. suggested that overexpression of IL-1 was associated with evolution of neuritic plaques from diffuse amyloid-β (Aβ) deposits in AD (Sheng et al., 1995). In addition, IL-1 promotes the amyloid precursor protein (APP) cleavage pathway (Buxbaum et al., 1992). Similarly, IL-6 has been reported to be involved in AD pathogenesis. Quintanilla et al. reported that IL-6 was associated with increased levels of hyperphosphorylated tau protein in neurons (Quintanilla et al., 2004). Furthermore, Braida et al. suggested that IL-6 deficiency was associated with learning and memory skills in mice (Braida et al., 2004). These findings suggested ILs to be important factors in AD pathogenesis.
Several epidemiological studies have investigated the association between genetic polymorphisms of IL genes and the risk of AD, including − 889C > T (rs1800587) in IL-1α, − 511C > T (rs16944) in IL-1β, − 174C > G (rs1800795) in IL-6 and − 1082G > A (rs1800896) in IL-10 (Bagli et al., 2000, Bhojak et al., 2000, Du et al., 2000, Grimaldi et al., 2000, Minster et al., 2000, Nicoll et al., 2000, Rebeck, 2000, Ki et al., 2001, Prince et al., 2001, Combarros et al., 2002, Fidani et al., 2002, Green et al., 2002, Hedley et al., 2002, Mattila et al., 2002, Pirskanen et al., 2002, Pola et al., 2002, Shibata et al., 2002, Clarimon et al., 2003, Depboylu et al., 2003, Faltraco et al., 2003, Kuo et al., 2003, Licastro et al., 2003, Lio et al., 2003, Ma et al., 2003, McCarron et al., 2003, Sciacca et al., 2003, Tsai et al., 2003, Arosio et al., 2004, Capurso et al., 2004, Depboylu et al., 2004, Hayes et al., 2004, Li et al., 2004, McCulley et al., 2004, Nishimura et al., 2004, Scassellati et al., 2004, Zhang et al., 2004, Koivisto et al., 2005, Ma et al., 2005, Seripa et al., 2005, Wang et al., 2005, Culpan et al., 2006, Ramos et al., 2006, Ravaglia et al., 2006, Zhou et al., 2006, Bagnoli et al., 2007, Wang et al., 2007, Combarros et al., 2008, Deniz-Naranjo et al., 2008, Paradowski et al., 2008, Dursun et al., 2009, Hu et al., 2009, Klimkowicz-Mrowiec et al., 2009, Serretti et al., 2009, Vural et al., 2009, Capurso et al., 2010, Combarros et al., 2010, Klimkowicz-Mrowiec et al., 2010, Ribizzi et al., 2010, Shawkatova et al., 2010, Cousin et al., 2011, Vendramini et al., 2011, Heun et al., 2012, Mansoori et al., 2012, Payao et al., 2012, Moraes et al., 2013, Rasmussen et al., 2013, Torres et al., 2013, Flex et al., 2014, Kang et al., 2014, Tian et al., 2015, Toral-Rios et al., 2015). However, these epidemiological studies have reported inconsistent results. In addition, several previous meta-analyses have assessed the associations between four polymorphisms of the IL genes and the risk of AD. However, several further papers regarding this relationship between IL gene polymorphisms and the risk of AD have been published recently. It is thus necessary to update the data regarding the association between IL gene polymorphisms and the risk of AD.
Therefore, we have re-evaluated and updated the associations between the polymorphisms of four IL genes and the risk of AD using published studies.
2. Materials and methods
2.1. Search strategy
Two clinical researchers independently searched and reviewed the literature. We conducted a meta-analysis of the published literature to analyze the associations between IL gene polymorphisms and Alzheimer's disease. The search sources were the PubMed, Science Direct, Scopus, and Google Scholar databases, the search was conducted up to July 2015, and the following search terms were used: “interleukin 1 or interleukin 6 or interleukin 10” and “variant or polymorphism or SNP” in combination with “Alzheimer's disease”. The reference lists in the published articles were reviewed to identify any studies missing from the database search. The workflow of the literature search is shown in Fig 1.
Fig. 1.
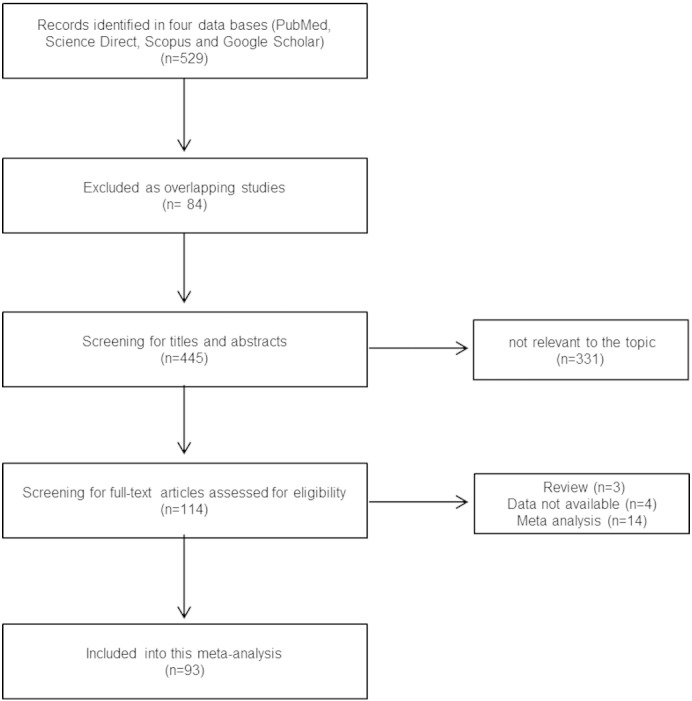
Flow chart of the selection of studies for inclusion in our meta-analysis.
2.2. Selection criteria
All articles reporting the genotype frequencies of the following IL gene single-nucleotide polymorphisms (SNPs) were included: − 889C > T, − 511C > T, − 174C > G and − 1082G > A. As the studies were heterogeneous in terms of the number of cases and controls, racial composition, and the polymorphisms analyzed, we used the following inclusion criteria: hospital-based or population-based case–control studies on the associations of IL gene polymorphisms with AD, genotype frequencies of each polymorphism provided for cases and controls, genotype distribution in the control group confirmed by Hardy–Weinberg equilibrium (HWE), and English-language articles only. If overlapping cases and controls between studies were identified, only the most-complete study was included in this meta-analysis.
2.3. Data extraction
Data extraction was performed by two reviewers. The following data were extracted from each study: last name of the first author, publication year, study region, participants' ethnicity, sample size, genotype distribution of the polymorphisms of four interleukin genes in both cases and controls, and p-values for the HWE of genotype distribution of controls (p value less than 0.05 of HWE was considered to indicate significance).
2.4. Statistical analysis
The chi-squared test was used to determine whether the distribution of genotypes in the control group was in agreement with HWE. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to assess the associations between four IL gene polymorphisms (− 889C > T, − 511C > T, − 174C > G and–1082G > A) and AD risk under the heterozygous, homozygous, dominant, and recessive models with fixed-effects (Mantel–Haenszel method) and random-effects models (Mantel–Haenszel method). Statistical heterogeneity between studies was evaluated using the I2 statistic. A random-effects model was used to calculate the pooled OR and 95% CI when I2 values > 50% were considered to indicate significant heterogeneity between studies. A fixed-effects model was used when I2 values < 50% were considered to indicate low heterogeneity between studies. We also performed subgroup analyses by ethnicity (Caucasian and Asian). The risk of small study bias, such as publication bias, was measured using funnel plots and further evaluated with Egger's linear regression test. It was assumed that large-sized studies would plot close to the mean in the absence of publication bias, whereas small-sized studies would be spread smoothly on both sides of the mean. All meta-statistical analyses were performed using the RevMan ver. 5.1 software (Cochrane Collaboration, Copenhagen, Denmark) and confirmed using the Comprehensive Meta-Analysis trial version. Two-sided p-values < 0.05 were considered to indicate significance.
3. Results
3.1. Characteristics of the included studies
A total of 529 papers published before July 2015 was identified in the search of the four databases. Of them, a total of 21,864 cases and 40,321 controls from 93 individual studies were included in our meta-analysis. A total of 8641 cases and 14,214 controls from 34 studies (42 subgroup studies) that reported on the association between the IL-1α gene polymorphism (− 889C > T) and risk of AD were included in the meta-analysis. A total of 3194 cases and 4621 controls from 18 studies (19 subgroup studies) that reported on the association between the IL-1β gene polymorphism (− 511C > T) and risk of AD were included in the meta-analysis. A total of 5755 cases and 12,456 controls from 24 studies (30 subgroup studies) of IL-6 gene polymorphism (− 174G > C) were included in the meta-analysis. Seventeen IL-10 gene polymorphism (− 1082G > A) studies (23 subgroup studies) involving 4274 cases and 9030 controls were included in the meta-analysis. Most of the studies were performed in Caucasian populations. However, several studies were conducted in Asian populations (nine subgroup studies in IL-1α, six subgroup studies in IL-1β, one subgroup study in IL-6, and one subgroup study in IL-10). The characteristics of the studies are summarized in Table 1.
Table 1.
Description of this meta-analysis of the association of four polymorphisms of IL genes with risk of Alzheimer's disease.
| IL-1α (− 889C > T) study (author/year) | Study region | Ethnicity | Criteria | Sample size (case/control) | Genotype distribution (case/control) |
HWE (p-value) | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | |||||||
| Clarimon et al. (2003) | Spain | Caucasian | NINCDS-ADRDA | 111/89 | 61/42 | 41/34 | 9/13 | 0.171 | Clarimon et al. (2003) |
| Combarros et al. (2002) | Spain | Caucasian | NINCDS-ADRDA | 298/306 | 161/195 | 119/104 | 18/7 | 0.108 | Combarros et al. (2002) |
| Combarros et al. (2010) (I) | Bonn | Caucasian | NINCDS-ADRDA-CERAD | 235/210 | 123/111 | 93/78 | 19/21 | 0.192 | Combarros et al. (2010)) |
| Combarros et al. (2010) (II) | Bristol | Caucasian | 198/56 | 87/24 | 8629 | 25/3 | 0.125 | ||
| Combarros et al. (2010) (III) | Nottingham | Caucasian | 83/96 | 36/46 | 38/38 | 9/12 | 0.353 | ||
| Combarros et al. (2010) (IV) | OPTIMA | Caucasian | 233/237 | 124/102 | 80/110 | 29/25 | 0.56 | ||
| Combarros et al. (2010) (V) | Oviedo | Caucasian | 187/109 | 95/52 | 77/50 | 15/7 | 0.269 | ||
| Combarros et al. (2010) (VI) | Rotterdam | Caucasian | 391/5110 | 185/2574 | 162/2111 | 44/425 | 0.789 | ||
| Combarros et al. (2010) (VII) | Santander | Caucasian | 302/374 | 162/220 | 114/127 | 26/27 | 0.15 | ||
| Cousin et al. (2011) | France | Caucasian | NINCDS-ADRDA | 129/190 | 60/90 | 61/85 | 8/15 | 0.409 | Cousin et al. (2011) |
| Deniz-Naranzo et al. (2008) | Spain | Caucasian | NINCDS-ADRDA | 282/312 | 138/168 | 118/121 | 26/23 | 0.85 | Deniz-Naranjo et al. (2008) |
| Du et al. (2000) | Germany | Caucasian | NINCDS-ADRDA | 259/191 | 141/126 | 97/62 | 21/3 | 0.131 | Du et al. (2000) |
| Dursun et al. (2009) | Turkey | Caucasian | DSM-IV | 104/103 | 60/45 | 41/52 | 3/6 | 0.07 | Dursun et al. (2009) |
| Fidani et al. (2002) | USA | Caucasian | NINCDS-ADRDA | 142/119 | 73/59 | 59/49 | 10/11 | 0.858 | Fidani et al. (2002) |
| Green et al. (2002) | UK/France | Caucasian | NINCDS-ADRDA-DSM-III-R | 294/503 | 134/221 | 126/217 | 34/65 | 0.309 | Green et al. (2002) |
| Grimaldi et al. (2000) | Italy | Caucasian | NINCDS-ADRDA | 318/335 | 140/142 | 125/163 | 53/30 | 0.08 | Grimaldi et al. (2000) |
| Hayes et al. (2004) | UK | Caucasian | CERAD | 68/503 | 30/221 | 31/220 | 7/62 | 0.528 | Hayes et al. (2004) |
| Hedley et al. (2002) | Australian | Caucasian | NINCDS-ADRDA | 221/351 | 98/153 | 94/168 | 29/30 | 0.087 | Hedley et al. (2002) |
| Hu et al. (2009) | China | Asian | NINCDS-ADRDA-DSM-III-R | 344/224 | 272/183 | 61/37 | 11/4 | 0.198 | Hu et al. (2009) |
| Ki et al. (2001) | Korean | Asian | NINCDS-ADRDA | 126/221 | 106/184 | 20/27 | 0/0 | 0.321 | Ki et al. (2001) |
| Kuo et al. (2003) | Taiwan | Asian | NINCDS-ADRDA | 125/93 | 104/72 | 20/21 | 1/0 | 0.22 | Kuo et al. (2003) |
| Li et al. (2004) | China | Asian | NINCDS-ADRDA-DSM-IV | 145/181 | 103/128 | 41/52 | 1/1 | 0.076 | Li et al. (2004) |
| Mattila et al. (2002) | Finland | Caucasian | NINCDS-ADRDA-CERAD | 110/73 | 42/33 | 39/29 | 29/11 | 0.281 | Mattila et al. (2002) |
| McCarron et al. (2003) | US/UK | Caucasian | CERAD | 232/167 | 103/82 | 99/74 | 30/11 | 0.291 | McCarron et al. (2003) |
| Minster et al. (2000) | USA | Caucasian | NINCDS-ADRDA-DSM-III-R | 297/204 | 139/102 | 126/86 | 32/16 | 0.717 | Minster et al. (2000) |
| Moraes et al. (2013) | Brazil | Caucasian | NINCDS-ADRDA | 120/412 | 64/209 | 45/168 | 11/35 | 0.88 | Moraes et al. (2013) |
| Nicoll et al. (2000) | US/UK | Caucasian | CERAD | 232/167 | 103/82 | 99/74 | 30/11 | 0.291 | Nicoll et al. (2000) |
| Nishmura et al. (2004) | Japan | Asian | NINCDS-ADRDA | 172/163 | 141/126 | 31/37 | 0/0 | 0.102 | Nishimura et al. (2004) |
| Pirskanen et al. (2002) | Finland | Caucasian | NINCDS-ADRDA | 237/513 | 123/248 | 91/209 | 23/56 | 0.235 | Pirskanen et al. (2002) |
| Prince et al. (2001) | Sweden | Caucasian | NINCDS-ADRDA | 198/175 | 89/93 | 89/65 | 20/17 | 0.264 | Prince et al. (2001) |
| Rebeck et al. (2000) | USA | Caucasian | CERAD | 247/187 | 119/97 | 103/74 | 25/16 | 0.725 | Rebeck (2000)) |
| Ribizzi et al. (2010) | Italy | Caucasian | NINCDS-ADRDA | 19/20 | 12/7 | 3/10 | 4/3 | 0.852 | Ribizzi et al. (2010) |
| Sciacca et al. (2003) | Italy | Caucasian | NINCDS-ADRDA | 353/482 | 165/229 | 153/219 | 35/34 | 0.057 | Sciacca et al. (2003) |
| Seripa et al. (2005) I) | Italy | Caucasian | NINCDS-ADRDA | 225/143 | 117/83 | 90/56 | 18/4 | 0.128 | Seripa et al. (2005) |
| Seripa et al. (2005) (II) | USA | Caucasian | NINCDS-ADRDA | 121/93 | 52/40 | 59/42 | 10/11 | 0.996 | |
| Serretti (2009) (I) | Greece | Caucasian | DSM-IV | 86/113 | 45/66 | 34/39 | 7/8 | 0.504 | Serretti et al. (2009) |
| Serretti (2009) (II) | Italy | Caucasian | NINCDS-ADRDA | 24/17 | 12/12 | 8/4 | 4/1 | 0.432 | |
| Tian et al. (2015) | China | Asian | NINCDS-ADRDA | 201/257 | 153/217 | 45/37 | 3/3 | 0.328 | Tian et al. (2015) |
| Tsai et al. (2003) | China | Asian | NINCDS-ADRDA | 234/170 | 212/147 | 21/22 | 1/1 | 0.858 | Tsai et al. (2003) |
| Vendramini et al. (2011) | Brazil | Caucasian | NINCDS-ADRDA | 199/241 | 96/136 | 84/91 | 19/14 | 0.811 | Vendramini et al. (2011) |
| Wang et al. (2007) | Taiwan | Asian | NINCDS-ADRDA | 219/209 | 182/174 | 37/33 | 0/2 | 0.756 | Wang et al. (2007) |
| Zhou et al. (2006) (abstract) | China | Asian | – | 520/505 | 369/407 | 134/92 | 17/6 | 0.756 | Zhou et al. (2006) |
| IL-1β (− 511C > T) study (author/year) | Study region | Ethnicity | Criteria | Sample size (case/control) | Genotype distribution (case/control) |
HWE (p-value) | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | |||||||
| Deniz-Naranzo et al. (2008) | Spain | Caucasian | NINCDS-ADRDA | 282/312 | 117/158 | 127/129 | 38/25 | 0.852 | Deniz-Naranjo et al. (2008) |
| Grimaldi et al. (2000) | Italy | Caucasian | NINCDS-ADRDA | 317/305 | 141/126 | 130/144 | 46/35 | 0.523 | Grimaldi et al. (2000) |
| Hayes et al. (2004) | UK | Caucasian | CERAD | 68/479 | 34/211 | 24/220 | 10/48 | 0.395 | Hayes et al. (2004) |
| Hedley et al. (2002) | Australian | Caucasian | NINCDS-ADRDA | 220/351 | 106/154 | 84/160 | 30/37 | 0.631 | Hedley et al. (2002) |
| Kang et al. (2014) | Korea | Asian | NINCDS-ADRDA-DSM-IV | 86/625 | 27/207 | 46/320 | 13/98 | 0.161 | Kang et al. (2014) |
| Klimkowicz-Mrowiec et al. (2009) | Poland | Caucasian | NINCDS-ADRDA | 331/219 | 152/118 | 147/85 | 32/16 | 0.897 | Klimkowicz-Mrowiec et al. (2009) |
| Li et al. (2004) | China | Asian | NINCDS-ADRDA-DSM-IV | 145/181 | 34/44 | 69/84 | 42/53 | 0.35 | Li et al. (2004) |
| Ma et al. (2003) | China | Asian | NINCDS-ADRDA | 90/100 | 26/22 | 26/33 | 38/45 | 0.002 | Ma et al. (2003) |
| Mattila et al. (2002) | Finland | Caucasian | NINCDS-ADRDA-CERAD | 92/52 | 35/25 | 47/25 | 10/2 | 0.159 | Mattila et al. 2002) |
| McCulley et al. (2004) | UK | Caucasian | NINCDS-ADRDA | 133/156 | 65/82 | 59/59 | 9/15 | 0.365 | McCulley et al. (2004) |
| Minster et al. (2000) | USA | Caucasian | NINCDS-DSM-III-R-ADRDA | 335/203 | 131/72 | 164/112 | 40/19 | 0.009 | Minster et al. (2000) |
| Nishmura et al. (2004) | Japan | Asian | NINCDS-ADRDA | 172/163 | 61/44 | 77/82 | 34/37 | 0.919 | Nishimura et al. (2004) |
| Payao et al. (2012) | Brazil | Caucasian | NINCDS-ADRDA | 188/263 | 38/48 | 107/132 | 43/83 | 0.722 | Payao et al. (2012) |
| Ravaglia et al. (2006) | Italy | Caucasian | NINCDS-ADRDA | 105/644 | 52/283 | 46/287 | 7/74 | 0.923 | Ravaglia et al. (2006) |
| Ribizzi et al. (2010) | Italy | Caucasian | NINCDS-ADRDA | 19/20 | 5/3 | 14/12 | 0/5 | 0.343 | Ribizzi et al. (2010) |
| Seripa et al. (2005) (I) | Italy | Caucasian | NINCDS-ADRDA | 225/143 | 103/54 | 97/70 | 25/19 | 0.62 | Seripa et al. (2005) |
| Seripa et al. (2005) (II) | USA | Caucasian | NINCDS-ADRDA | 121/93 | 50/38 | 60/40 | 11/15 | 0.419 | Wang et al. (2005) |
| Wang et al. (2005) | Taiwan | Asian | NINCDS-ADRDA | 46/103 | 17/27 | 13/52 | 16/24 | 0.915 | Wang et al. (2007) |
| Wang et al. (2007) | Taiwan | Asian | NINCDS-ADRDA-DSM-IV | 219/209 | 74/56 | 107/105 | 38/48 | 0.928 | Wang et al. (2005) |
| Il-6 (− 174g > c) study (author/year) | Study region | Ethnicity | Criteria | Sample size (case/control) | Genotype distribution (case/control) |
HWE (p-value) | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| GG | GC | CC | |||||||
| Arosio et al. (2004) | Italy | Caucasian | NINCDS-ADRDA-DSM-IV | 59/64 | 17/32 | 34/27 | 8/5 | 0.833 | Arosio et al. (2004) |
| Bagli et al. (2000) | Germany | Caucasian | NINCDS-ADRDA | 102/351 | 33/99 | 56/208 | 13/44 | < 0.001 | Bagli et al. (2000) |
| Bhojak et al. (2000) | USA | Caucasian | NINCDS-ADRDA | 464/337 | 178/126 | 221/155 | 65/56 | 0.478 | Bhojak et al. (2000) |
| Capurso et al. (2004) | Italy | Caucasian | NINCDS-ADRDA | 168/220 | 90/129 | 71/82 | 7/9 | 0.364 | Capurso et al. (2004) |
| Capurso et al. (2010) | Italy | Caucasian | NINCDS-ADRDA | 149/298 | 81/172 | 61/111 | 7/15 | 0.590 | Capurso et al. (2010) |
| Combarros et al. (2010) (i) | Bonn | Caucasian | NINCDS-ADRDA-CERAD | 241/224 | 81/77 | 123/95 | 37/52 | 0.035 | Combarros et al. (2010) |
| Combarros et al. (2010) (ii) | Bristol | Caucasian | 189/54 | 66/9 | 83/29 | 40/16 | 0.497 | ||
| Combarros et al. (2010) (iii) | Nottingham | Caucasian | 84/95 | 33/32 | 36/41 | 15/22 | 0.215 | ||
| Combarros et al. (2010) (iv) | OPTIMA | Caucasian | 243/240 | 88/65 | 106/141 | 49/34 | 0.002 | ||
| Combarros et al. (2010) (v) | Oviedo | Caucasian | 190/119 | 89/60 | 82/51 | 19/8 | 0.517 | ||
| Combarros et al. (2010) (vi) | Rotterdam | Caucasian | 391/5110 | 127/1824 | 191/2426 | 73/860 | 0.270 | ||
| Combarros et al. (2010) (vii) | Santander | Caucasian | 333/381 | 148/169 | 137/163 | 48/49 | 0.328 | ||
| Cousin et al. (2011) | France | Caucasian | NINCDS-ADRDA | 231/470 | 96/171 | 100/229 | 35/70 | 0.639 | Cousin et al. (2011) |
| Depboylu et al. (2004) | Germany | Caucasian | NINCDS-ADRDA | 113/108 | 33/26 | 65/64 | 15/18 | 0.046 | Depboylu et al. (2004) |
| Faltraco et al. (2003) | Germany | Caucasian | NINCDS-ADRDA | 101/133 | 44/43 | 47/70 | 10/20 | 0.326 | Faltraco et al. (2003) |
| Flex et al. (2014) | Italy | Caucasian | NINCDS-ADRDA | 533/713 | 216/160 | 241/337 | 76/216 | 0.192 | Flex et al. (2014) |
| Klimkowicz-mrowiec et al. (2010 | Poland | Caucasian | NINCDS-ADRDA | 361/200 | 119/66 | 185/91 | 57/43 | 0.271 | Klimkowicz-Mrowiec et al. (2010) |
| Koivisto et al. (2005) | Finland | Caucasian | – | 65/542 | 18/136 | 32/260 | 15/146 | 0.349 | Koivisto et al. (2005) |
| Licastro et al. (2003) | Italy | Caucasian | NINCDS-ADRDA-DSM-IV-R | 332/393 | 137/209 | 161/165 | 34/19 | 0.057 | Licastro et al. (2003) |
| Mansoori et al. (2012) | India | Caucasian | NINCDS-ADRDA | 80/120 | 55/88 | 24/29 | 1/3 | 0.743 | Mansoori et al. (2012) |
| Moraes et al. (2013) | Brazil | Caucasian | NINCDS-ADRDA-DSM-IV | 120/412 | 71/260 | 38/136 | 11/16 | 0.732 | Moraes et al. (2013) |
| Paradowski et al. (2008) | Poland | Caucasian | NINCDS-ADRDA | 51/36 | 11/12 | 31/16 | 9/8 | 0.549 | Paradowski et al. (2008) |
| Pola et al. (2002) | Italy | Caucasian | NINCDS-ADRDA | 124/134 | 56/29 | 51/58 | 17/47 | 0.170 | Pola et al. (2002) |
| Rasmussen et al. (2013) | Brazil | Caucasian | NINCDS-ADRDA-DSM-IV | 197/163 | 88/82 | 91/65 | 18/16 | 0.557 | Rasmussen et al. (2013) |
| Ravaglia et al. (2006) | Italy | Caucasian | NINCDS-ADRDA | 105/644 | 50/251 | 43/304 | 12/89 | 0.842 | Ravaglia et al. 2006) |
| Shawkatová et al. (2010) | Slovakia | Caucasian | NINCDS-ADRDA | 50/140 | 23/53 | 21/66 | 6/21 | 0.951 | Shawkatova et al. (2010) |
| Shibata et al. (2002) | Japan | Asian | NINCDS-ADRDA | 128/83 | 4/7 | 74/23 | 50/53 | 0.068 | Shibata et al. (2002) |
| Toral-rios et al. (2015) | Mexico | Caucasian | NINCDS-ADRDA | 94/100 | 5/3 | 23/15 | 66/82 | 0.040 | Toral-Rios et al. (2015) |
| Vural et al. (2009) | Turkey | Caucasian | NINCDS-ADRDA | 101/138 | 54/76 | 43/51 | 4/11 | 0.556 | Vural et al. (2009) |
| Zhang et al. (2004) | UK | Caucasian | NINCDS-ADRDA-DSM-III-R | 356/434 | 132/152 | 171/213 | 53/69 | 0.695 | Zhang et al. (2004) |
| IL-10 (− 1082G > A) study (author/year) | Study region | Ethnicity | Criteria | Sample size (case/control) | Genotype distribution (case/control) |
HWE (p-value) | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | |||||||
| Arosio et al. (2004) | Italy | Caucasian | NINCDS-ADRDA-DSM-IV | 63/63 | 4/14 | 28/29 | 31/20 | 0.573 | Arosio et al. (2004) |
| Bagnoli et al. (2007) | Italy | Caucasian | DSM-IV | 222/179 | 98/79 | 99/74 | 25/26 | 0.210 | Bagnoli et al. (2007) |
| Combarros et al. (2008) | Spain | Caucasian | NINCDS-ADRDA | 231/194 | 60/66 | 140/99 | 31/29 | 0.410 | Combarros et al. (2008) |
| Cousin et al. (2011) | France | Caucasian | NINCDS-ADRDA | 426/475 | 94/107 | 205/232 | 127/136 | 0.671 | Cousin et al. (2011) |
| Culpan et al. (2006) | Sweden | Caucasian | – | 160/92 | 41/24 | 79/50 | 40/18 | 0.380 | Culpan et al. (2006) |
| Depboylu et al. (2003) | Germany | Caucasian | NINCDS-ADRDA | 233/97 | 56/25 | 96/54 | 81/18 | 0.240 | Depboylu et al. (2003) |
| Heun et al. (2012) (I) | Bonn | Caucasian | NINCDS-ADRDA-CERAD | 245/216 | 54/45 | 118/109 | 73/62 | 0.819 | Heun et al. (2012) |
| Heun et al. (2012) (II) | Bristol | Caucasian | 162/52 | 45/12 | 72/25 | 45/15 | 0.799 | ||
| Heun et al. (2012) (III) | Nottingham | Caucasian | 67/76 | 21/22 | 28/29 | 18/25 | 0.040 | ||
| Heun et al. (2012) (IV) | OPTIMA | Caucasian | 237/241 | 72/58 | 112/123 | 53/60 | 0.747 | ||
| Heun et al. (2012) (V) | Oviedo | Caucasian | 186/110 | 24/25 | 97/61 | 65/24 | 0.252 | ||
| Heun et al. (2012) (VI) | Rotterdam | Caucasian | 391/5110 | 120/1339 | 190/2538 | 81/1233 | 0.656 | ||
| Heun et al. (2012) (VII) | Santander | Caucasian | 311/387 | 38/66 | 182/185 | 91/136 | 0.820 | ||
| Lio et al. (2003) | Italy | Caucasian | NINCDS-ADRDA | 132/213 | 32/86 | 91/118 | 9/9 | < 0.001 | Lio et al. (2003) |
| Ma et al. (2005) | China | Asian | NINCDS-ADRDA | 95/117 | 3/5 | 8/6 | 84/106 | < 0.001 | Ma et al. (2005) |
| Moraes et al. (2013) | Brazil | Caucasian | NINCDS-ADRDA | 120/412 | 15/35 | 68/189 | 37/188 | 0.192 | Moraes et al. (2013) |
| Ramos et al. (2006) | USA | Caucasian | NINCDS-ADRDA | 265/347 | 65/100 | 144/156 | 56/91 | 0.062 | Ramos et al. (2006) |
| Ribizzi et al. (2010) | Italy | Caucasian | NINCDS-ADRDA | 19/20 | 8/1 | 5/12 | 6/7 | 0.154 | Ribizzi et al. (2010) |
| Scassellati et al. (2004) | Italy | Caucasian | NINCDS-ADRDA | 215/153 | 35/26 | 109/64 | 71/63 | 0.168 | Scassellati et al. (2004) |
| Shawkatova et al. (2010) | Slovakia | Caucasian | NINCDS-ADRDA | 50/140 | 8/30 | 20/61 | 22/49 | 0.184 | Shawkatova et al. (2010) |
| Toral-Rios et al. (2015) | Mexico | Caucasian | NINCDS-ADRDA | 94/100 | 8/9 | 86/91 | 0/0 | < 0.001 | Toral-Rios et al. (2015) |
| Torres et al. (2013) | Brazil | Caucasian | NINCDS-ADRDA-CERAD | 249/98 | 25/12 | 103/40 | 121/46 | 0.476 | Torres et al. (2013) |
| Vural et al. (2009) | Turkey | Caucasian | NINCDS-ADRDA | 101/138 | 24/50 | 65/63 | 12/25 | 0.511 | Vural et al. (2009) |
Bonn, Ethics Review Board of the University of Bonn; Bristol, Frenchay Local Research Ethics committee Bristol; Nottingham, Nottingham Research Committee 2 (NHS); OPTIMA, Central Oxford Ethics Committee No 1656; Oviedo, Ethical Committee of the Hospital Central de Asturias; Rotterdam, Medical Ethical Committee of the Erasmus MC; Santander, Ethical Committee of the University Hospital “Marqués de Valdecilla”, Santander; NINCDS-ADRDA, National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's disease and Related Disorders Association; CERAD, The Consortium to Establish a Registry for Alzheimer's Disease; DSM, Diagnostic and Statistical Manual of Mental Disorder. *Zhou et al. data from abstract.
3.2. IL genes polymorphisms and risk of AD
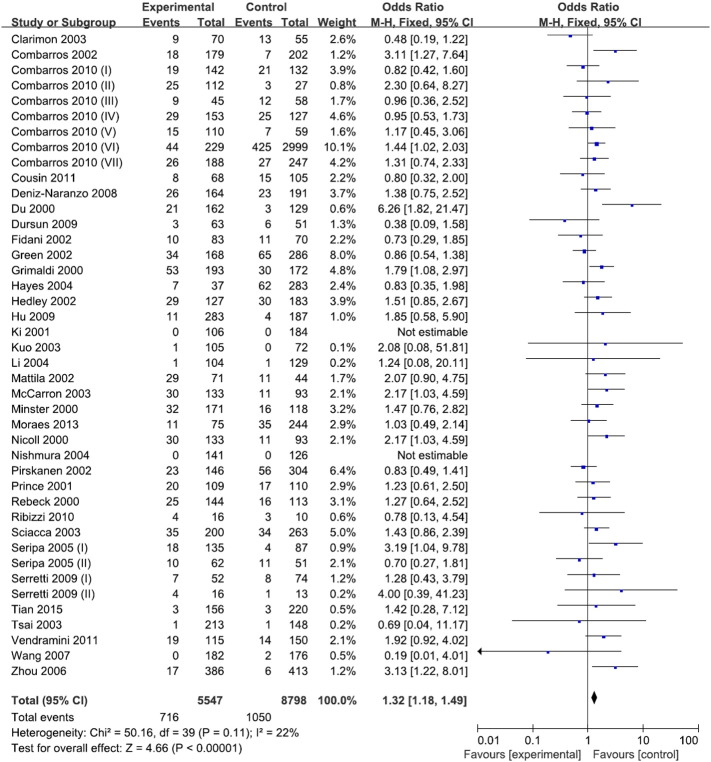
Forty-two subgroup studies involving 8641 cases and 14,214 controls identified an association between the − 889C > T polymorphism and risk of AD. The distributions of the genotypes in the control groups from all studies followed HWE. Our comprehensive meta-analysis indicated that the − 889C > T polymorphism was significantly associated with an increased risk of AD by three genetic models. The ORs of the homozygote (CC vs. TT), dominant (TT/CT vs. CC) and recessive (TT vs. CC/CT) models were 1.32, 1.09 and 1.32, respectively (95% CI: 1.18–1.49, 1.03–1.16 and 1.18–1.45, respectively) using a fixed-effects model (Fig. 2). However, heterozygote models (CC vs. TC) were not associated with risk of AD (OR: 1.05, 95% CI: 0.98–1.12). We also assessed the association between the − 889C > T polymorphism and risk of AD in Caucasian populations by excluding nine Asian studies (Ki et al., 2001, Kuo et al., 2003, Tsai et al., 2003, Li et al., 2004, Nishimura et al., 2004, Zhou et al., 2006, Wang et al., 2007, Hu et al., 2009, Tian et al., 2015). Data from the Caucasian studies showed that three genetic models (homozygote, dominant and recessive) were related to an increased risk of AD (OR: 1.30, 95% CI: 1.15–1.47; OR: 1.07, 95% CI: 1.00–1.15; OR: 1.30, 95% CI: 1.16–1.46, respectively). However, the heterozygote model was not related to risk of AD. Nineteen subgroup studies on the − 511C > T polymorphism of IL-1β included 3194 cases and 4621 controls. Of these, the distribution of genotypes in the control groups of two studies, Ma et al. 2003 and Minster et al. 2000, deviated from HWE (p < 0.05). Our meta-analysis with HWE revealed that the − 511C > T polymorphism was not associated with risk of AD (homozygote: OR = 0.95, 95% CI = 0.81–1.12 by fixed-effects model; heterozygote: OR = 0.94, 95% CI = 0.84–1.06 by fixed-effects model; dominant: OR = 0.95, 95% CI = 0.86–1.06 by fixed-effects model; recessive: OR = 0.98, 95% CI = 0.75–1.28 by random-effects model). Therefore, our meta-analysis suggested that the − 889C > T polymorphism was significantly associated with an increased risk of AD. However, the − 511C > T polymorphism was not related to risk of AD.
Fig. 2.
Forest plot for the association between the homozygote model (CC vs. TT) of the − 889C > T polymorphism of the IL-1α gene and risk of AD using a fixed-effects model.
Thirty subgroup studies on the − 174G > C polymorphism included 5755 cases and 12,456 controls. Of them, five studies deviated from HWE (p < 0.05) (Bagli et al., 2000, Depboylu et al., 2004, Combarros et al., 2010, Toral-Rios et al., 2015). The tendency of our meta-analysis indicated that the − 174G > C polymorphism was related to a decreased risk of AD. However, this polymorphism was statistically not associated with risk of AD (homozygote: OR = 0.85, 95% CI = 0.64–1.13; heterozygote: OR = 0.99, 95% CI = 0.85–1.15; dominant: OR = 0.95, 95% CI = 0.80–1.13; recessive: OR = 0.83, 95% CI = 0.67–1.03) by a random-effects model. Consequently, our results suggested that the − 174G > C polymorphism was not associated with risk of AD.
Twenty-three subgroup studies involving 4274 cases and 9030 controls identified an association between the − 1082G > A polymorphism and risk of AD. Two studies of the association between the − 1082G > A polymorphism and AD risk were conducted in Asian populations. Among previous studies, the results of four studies departed from HWE (p < 0.05) (Lio et al., 2003, Ma et al., 2003, Heun et al., 2012). Our meta-analysis results showed that the − 1082G > A polymorphism of IL-10 was not related to risk of AD. The ORs of four genetic models (homozygote, heterozygote, dominant and recessive) were 1.04, 1.12, 1.10 and 0.97, respectively, using a random-effects model (95% CIs: 0.85–1.28, 0.94–1.33, 0.93–1.29 and 0.83–1.14, respectively). The results of the meta-analysis are summarized in Table 2, Table 3.
Table 2.
The associations between four polymorphisms of IL genes and AD risk.
| SNP | Genetic models | Pooled OR (95% CI) |
Heterogeneity |
Publication bias |
|||
|---|---|---|---|---|---|---|---|
| Fixed effect model | Random effect model | I2 value | p-Value | p-Value | |||
| Overall | rs1800587 (IL-1α; − 889C > T) | Homozygote model (TT vs. CC) | 1.32 (1.18–1.49)⁎ | 1.31 (1.13–1.51) | 22% | 0.110 | 0.900 |
| Heterozygote model (CT vs. CC) | 1.05 (0.98–1.12) | 1.04 (0.97–1.13) | 24% | 0.080 | 0.174 | ||
| Dominant model (TT/CT vs. CC) | 1.09 (1.03–1.16)⁎ | 1.08 (1.00–1.17) | 31% | 0.030 | 0.164 | ||
| Recessive model (TT vs. CC/CT) | 1.32 (1.18–1.45)⁎ | 1.30 (1.14–1.49) | 18% | 0.160 | 0.897 | ||
| rs16944 (IL-1β; − 511C > T) | Homozygote model (TT vs. CC) | 0.95 (0.82–1.11) | 0.94 (0.77–1.16) | 37% | 0.050 | 0.381 | |
| Heterozygote model (CT vs. CC) | 0.93 (0.83–1.03) | 0.92 (0.81–1.04) | 24% | 0.160 | 0.323 | ||
| Dominant model (TT/CT vs. CC) | 0.94 (0.85–1.04) | 0.93 (0.82–1.05) | 27% | 0.140 | 0.223 | ||
| Recessive model (TT vs. CC/CT) | 0.97 (0.84–1.11) | 0.98 (0.77–1.25) | 61% | < 0.001 | 0.735 | ||
| rs1800795 (IL-6; − 174C > G) | Homozygote model (GG vs. CC) | 0.79 (0.71–0.88) | 0.83 (0.65–1.06) | 74% | < 0.001 | 0.579 | |
| Heterozygote model (GG vs. GC) | 0.95 (0.88–1.02) | 0.96 (0.84–1.10) | 62% | < 0.001 | 0.546 | ||
| Dominant model (CC/GC vs. GG) | 0.92 (0.85–0.99) | 0.92 (0.79–1.07) | 72% | < 0.001 | 0.831 | ||
| Recessive model (CC vs. GG/GC) | 0.80 (0.72–0.88) | 0.83 (0.68–1.005) | 68% | < 0.001 | 0.690 | ||
| rs1800896 (IL-10; − 1082G > A) | Homozygote model (AA vs. GG) | 0.99 (0.88–1.13) | 1.06 (0.87–1.29) | 49% | 0.005 | 0.146 | |
| Heterozygote model (GA vs. GG) | 1.11 (1.00–1.23) | 1.16 (0.98–1.37) | 50% | 0.004 | 0.517 | ||
| Dominant model (AA/GA vs. GG) | 1.08 (0.97–1.19) | 1.13 (0.96–1.33) | 51% | 0.002 | 0.331 | ||
| Recessive model (AA vs. GG/GA) | 0.93 (0.85–1.03) | 0.97 (0.83–1.13) | 49% | 0.005 | 0.177 | ||
| Caucasian | rs1800587 (IL-1α; − 889C > T) | Homozygote model (TT vs. CC) | 1.30 (1.15–1.47)⁎ | 1.28 (1.10–1.50) | 28% | 0.070 | 0.796 |
| Heterozygote model (CT vs. CC) | 1.03 (0.96–1.10) | 1.03 (0.95–1.11) | 12% | 0.280 | 0.435 | ||
| Dominant model (TT/CT vs. CC) | 1.07 (1.00–1.15)⁎ | 1.07 (0.99–1.16) | 21% | 0.150 | 0.490 | ||
| Recessive model (TT vs. CC/CT) | 1.30 (1.16–1.46)⁎ | 1.28 (1.11–1.48) | 26% | 0.090 | 0.780 | ||
| rs16944 (IL-1β; − 511C > T) | Homozygote model (TT vs. CC) | 1.04 (0.87–1.26) | 1.02 (0.77–1.35) | 47% | 0.030 | 0.438 | |
| Heterozygote model (CT vs. CC) | 0.96 (0.85–1.09) | 0.96 (0.83–1.11) | 25% | 0.190 | 0.378 | ||
| Dominant model (TT/CT vs. CC) | 0.98 (0.88–1.11) | 0.098 (0.85–1.13) | 32% | 0.130 | 0.284 | ||
| Recessive model (TT vs. CC/CT) | 1.02 (0.86–1.21) | 1.01 (0.73–1.40) | 66% | < 0.001 | 0.873 | ||
| rs1800795 (IL-6; − 174C > G) | Homozygote model (GG vs. CC) | 0.78 (0.70–0.88) | 0.82 (0.64–1.05) | 75% | < 0.001 | 0.521 | |
| Heterozygote model (GG vs. GC) | 0.94 (0.87–1.02) | 0.94 (0.83–1.08) | 60% | < 0.001 | 0.433 | ||
| Dominant model (CC/GC vs. GG) | 0.91 (0.85–0.98) | 0.91 (0.78–1.06) | 73% | < 0.001 | 0.652 | ||
| Recessive model (CC vs. GG/GC) | 0.82 (0.74–0.91) | 0.86 (0.71–1.04) | 66% | < 0.001 | 0.652 | ||
| rs1800896 (IL-10; − 1082G > A) | Homozygote model (AA vs. GG) | 0.99 (0.87–1.13) | 1.06 (0.87–1.29) | 51% | 0.004 | 0.229 | |
| Heterozygote model (GA vs. GG) | 1.11 (1.00–1.23) | 1.15 (0.97–1.36) | 52% | 0.003 | 0.628 | ||
| Dominant model (AA/GA vs. GG) | 1.11 (1.00–1.23) | 1.15 (0.97–1.36) | 52% | 0.003 | 0.334 | ||
| Recessive model (AA vs. GG/GA) | 0.93 (0.85–1.03) | 0.97 (0.83–1.14) | 52% | 0.003 | 0.172 | ||
Statistically significant (p < 0.05).
Table 3.
Associations between four polymorphisms of IL genes and AD risk in studies in Hardy–Weinberg equilibrium (HWE).
| SNP | Genetic models | Pooled OR (95% CI) |
Heterogeneity |
Publication bias |
Departed from the HWE | ||
|---|---|---|---|---|---|---|---|
| Fixed effect model | Random effect model | I2 value | P-value | P-value | |||
| rs1800587 (IL-1α; − 889C > T) | Homozygote model (TT vs. CC) | 1.32 (1.18–1.49)⁎ | 1.31 (1.13–1.51) | 22% | 0.110 | 0.900 | / |
| Heterozygote model (CT vs. CC) | 1.05 (0.98–1.12) | 1.04 (0.97–1.13) | 24% | 0.080 | 0.174 | ||
| Dominant model (TT/CT vs. CC) | 1.09 (1.03–1.16)⁎ | 1.08 (1.00–1.17) | 31% | 0.030 | 0.164 | ||
| Recessive model (TT vs. CC/CT) | 1.32 (1.18–1.45)⁎ | 1.30 (1.14–1.49) | 18% | 0.160 | 0.897 | ||
| rs16944 (IL-1β; − 511C > T) | Homozygote model (TT vs. CC) | 0.95 (0.81–1.12) | 0.94 (0.75–1.18) | 42% | 0.040 | 0.284 | Ma et al. (2003) and Minster et al. (2000) |
| Heterozygote model (CT vs. CC) | 0.94 (0.84–1.06) | 0.94 (0.82–1.08) | 29% | 0.130 | 0.924 | ||
| Dominant model (TT/CT vs. CC) | 0.95 (0.86–1.06) | 0.94 (0.82–1.08) | 32% | 0.100 | 0.528 | ||
| Recessive model (TT vs. CC/CT) | 0.96 (0.82–1.11) | 0.98 (0.75–1.28) | 63% | < 0.001 | 0.475 | ||
| rs1800795 (IL-6; − 174C > G) | Homozygote model (GG vs. CC) | 0.79 (0.70–0.88) | 0.85 (0.64–1.13) | 78% | < 0.001 | 0.670 | Bagli et al. (2000), Combarros et al. (2010) (I), Combarros et al. (2010) (IV), Depboylu et al. (2004) and Toral-Rios et al. (2015) |
| Heterozygote model (GG vs. GC) | 0.97 (0.89–1.05) | 0.99 (0.85–1.15) | 64% | < 0.001 | 0.953 | ||
| Dominant model (CC/GC vs. GG) | 0.93 (0.86–1.01) | 0.95 (0.80–1.13) | 76% | < 0.001 | 0.917 | ||
| Recessive model (CC vs. GG/GC) | 0.79 (0.71–0.88) | 0.83 (0.67–1.03) | 70% | < 0.001 | 0.616 | ||
| rs1800896 (IL-10; − 1082G > A) | Homozygote model (AA vs. GG) | 0.98 (0.86–1.12) | 1.04 (0.85–1.28) | 51% | 0.005 | 0.158 | Heun et al. (2012) (III), Lio et al. (2003), Ma et al. (2005) and Toral-Rios et al. (2015) |
| Heterozygote model (GA vs. GG) | 1.07 (0.96–1.20) | 1.12 (0.94–1.33) | 51% | 0.006 | 0.631 | ||
| Dominant model (AA/GA vs. GG) | 1.04 (0.94–1.16) | 1.10 (0.93–1.29) | 51% | 0.005 | 0.353 | ||
| Recessive model (AA vs. GG/GA) | 0.93 (0.84–1.03) | 0.97 (0.83–1.14) | 55% | 0.002 | 0.144 | ||
Combarros et al. (2010) (I), Bonn, Ethics Review Board of the University of Bonn; Combarros et al. (2010) (IV), OPTIMA, Central Oxford Ethics Committee No 1656; Heun et al. (2012), Nottingham, Nottingham Research Committee 2 (NHS).
− 889C > T polymorphism of IL-1α studies were not departed from HWE.
Statistically significant (p < 0.05).
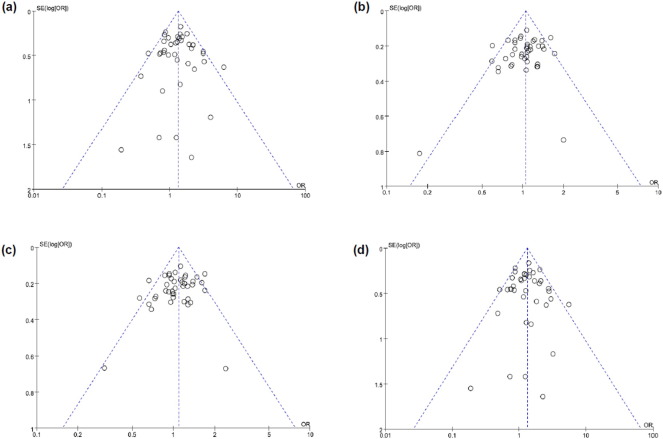
3.3. Publication bias
Publication bias is shown graphically with a funnel plot (Fig. 3). We confirmed publication bias using Egger's linear regression test, as the funnel plot shapes did not indicate distinct symmetry in all of the genetic models. We did not find any evidence of publication bias in most of the genetic models.
Fig. 3.
Funnel plot for the association between the − 889C > T polymorphism and Alzheimer's disease.
3.4. Heterogeneity and sensitivity
No significant heterogeneity was found among the studies of the − 889C > T polymorphism. However, significant heterogeneity was found in the recessive model for the − 511C > T polymorphism, all genetic models (homozygote, heterozygote, dominant and recessive) for the − 174C > G polymorphism and all genetic models for the − 1082G > A polymorphism. Therefore, we applied fixed-effects and random-effects models in the meta-analysis (Table 2, Table 3). We also performed a sensitivity test to assess the stability and reliability of the results by sequentially deleting each subgroup study from the meta-analysis. The sensitivity test results indicated that none of the subgroup studies altered the pooled OR, suggesting that our meta-analysis was stable and reliable.
4. Discussion
Our meta-analysis summarizes the evidence to date regarding the association between four polymorphisms (− 889C > T, − 511C > T, − 172G > C and − 1082G > A) and the risk of AD. The results indicate that − 889C > T was significantly associated with an increased risk of AD. However, three polymorphisms (− 511C > T, − 172G > C and − 1082G > A) were statistically not related to the risk of AD.
Over the past decades, many genetic studies and meta-analyses have been performed to investigate the relationship between IL gene polymorphisms and the risk of AD. The most recent meta-analyses of the association between the four IL gene polymorphisms (− 889C > T, − 511C > T, 174G > C and − 1082G > A) and the risk of AD were reported in 2012 and 2013 (Dai et al., 2012, Di Bona et al., 2012, Hua et al., 2012, Qi et al., 2012, Li et al., 2013, Yuan et al., 2013). A previous meta-analysis of − 889C > T polymorphism had included twenty-eight studies and a total 12,817 subjects (Li et al., 2013). They results indicated that − 889C > T polymorphism was significantly associated with increased risk of AD. Furthermore, Caucasian studies revealed that this polymorphism was associated with increased risk of AD. However, most of genetic models (dominant, recessive and T allele vs. C allele) showed that − 889C > T polymorphism was not associated with risk of AD in Asian. Similarly, our results showed that − 889C > T polymorphism was associated with increased risk of AD in overall and Caucasian subgroup studies. In − 511C > T polymorphisms, Yuan et al. reported that − 511C > T polymorphism was not associated with risk of AD. Furthermore, subgroup studies demonstrated that − 511C > T polymorphism was not related with AD in Europe, non-Europe, Caucasian and non-Caucasian. In addition, many genetic models showed that heterogeneity (Yuan et al., 2013). Similar to previous meta-analysis, our results indicated that − 511C > T polymorphism was not associated with risk of AD in overall and Caucasian subgroup studies. In 2012, Bona et al. suggested that GG vs. AG/AA model of − 1082G > A polymorphism was modestly associated with risk of AD (OR: 0.82, 95% CI: 0.65–1.02). In addition, results of meta-analysis showed that moderate degree of heterogeneity between studies (Di Bona et al., 2012). In contrast, our results suggested that − 1082G > A polymorphism was statistically not associated with risk of AD. However, degree of heterogeneity was similar to previous meta-analysis. As mentioned above, meta-analysis results of three polymorphisms (− 889C > T, − 511 C > T and − 1082G > A) were similar to previous meta-analysis. However, the results of the − 174G > C polymorphism were different. In 2012, Dai et al. reported an association between the − 174G > C polymorphism and the risk of AD in a meta-analysis including 3101 cases and 3860 controls. The overall analysis showed that the − 174G > C polymorphism was significantly associated with a decreased risk of AD using a recessive model (OR: 0.70, 95% CI: 0.54–0.90). In addition, the heterozygote model revealed that the − 174G > C polymorphism was strongly associated with a decreased risk of AD (OR: 0.83, 95% CI: 0.60–0.96) (Dai et al., 2012). Similarly, Qi et al.'s meta-analysis (4280 cases and 8788 controls) suggested that the recessive model (CC vs. GC/GG) was significantly associated with a decreased risk of AD (OR: 0.65, 95% CI: 0.52–0.82) (Qi et al., 2012). However, our meta-analysis (5755 cases and 12,456 controls) shows that all genetic models (homozygote, CC vs. GG; heterozygote, GC vs. GG; dominant CC/GC vs. GG; recessive models, CC vs. GC/GG) were significantly not associated with the risk of AD. The conflicting results between Qi et al. and our meta-analysis may be due to the included studies. Our meta-analysis contains an additional eight studies (Ravaglia et al., 2006, Combarros et al., 2010, Shawkatova et al., 2010, Cousin et al., 2011, Moraes et al., 2013, Rasmussen et al., 2013, Flex et al., 2014, Toral-Rios et al., 2015). In addition, we deleted four studies (Infante et al., 2004, Combarros et al., 2005, van Oijen et al., 2006, Fontalba et al., 2009). Three studies (Fontalba et al., 2009, Combarros et al., 2005 and Infante et al., 2004) provided deficient genotype data. Also, the genotype data presented by van Oijen et al.'s (2006) group may overlap with that of Combarros et al. 2010 (Rotterdam study). However, the Qi et al. meta-analysis included these four studies.
Three limitations of this meta-analysis should be mentioned. First, most of the cases and controls were Caucasians. Thus, the lack of studies involving Asian populations may limit the general application of our results. Second, the studies included in our meta-analysis were limited to published reports. Unpublished reports or those published in non-international journals could not be included in the analysis. These problems may have affected the stability of the meta-analysis data. Third, AD is a multifactorial disease. However, we did not consider gene–gene or gene–environmental interactions—such as age, smoking, alcohol status, and progression of AD—which may have influenced the associations between IL gene polymorphisms and AD risk. Nevertheless, this meta-analysis improves our understanding of the associations between four polymorphisms of IL genes and the risk of AD.
Many studies have reported the association between several gene polymorphisms and the risk of AD. Coon et al., suggested that ε2/ε4, ε3/ε4 and ε4/ε4 variant types of ApoE significantly increased the risk of AD (odds ratios: 3.49, 4.32 and 25.31, respectively) compared with ε3/ε3 (Coon et al., 2007). In addition, meta-analysis data suggested that ApoE e4/e4 type was significantly associated with the prevalence of AD. Interestingly, meta-analyses indicated that the highest estimates were in Northern Europe and the lowest estimates were in Asia (prevalence 14.1%, 95% CI: 12.2–16.0 in Northern Europe; prevalence: 7.70%, 95% CI: 5.84–9.55 in Asia) (Ward et al., 2012). In addition, it is known that mutations in the presenilin-1 (PSEN-1) and presenilin-2 (PSEN-2) genes are related to AD. Manotas-Rodriguez et al. reported that the PSEN-1 polymorphism (rs165932) was probably associated with the risk of AD in the European sub-group (fixed effect model. OR: 1.19, 95% CI: 1.02–1.37, p-value < 0.05) (Rodriguez-Manotas et al., 2007). In addition, a meta-analysis by Chen et al. suggested that the rs8383 polymorphism of PSEN-2 was associated with an increased risk of AD (C vs. T, OR: 1.16, 95% CI: 1.00–1.33, p-value: 0.043; CC vs. TT, OR: 1.37, 95% CI: 1.02–1.84, p-value: 0.037) (Chen et al., 2012). Furthermore, genome-wide association studies have provided several polymorphisms of candidate genes and loci for AD (Li et al., 2008, Harold et al., 2009). However, the associations between several polymorphisms of candidate genes and the risk of AD are still unclear. To better understand the genetic risk factors for AD, large scale studies are needed to validate the associations and further investigations should consider the effects of environmental factors and genetic interactions.
5. Conclusions
In summary, our updated meta-analysis of 93 studies showed that the results of − 889C > T polymorphism was statistically associated with the risk of AD. In contrast, three other polymorphisms were not associated with the risk of AD. In addition, our results of three polymorphisms (− 889C > T, − 511C > T and 1082G > A) were similar to those of previous meta-analyses. However, our results for the − 174G > C polymorphism differed from those of previous meta-analyses. Consequently, our results suggested that the − 889C > T polymorphism may be a potential risk factor in AD. However, the other three polymorphisms may not be a risk factor for AD.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Acknowledgment
The present research was conducted by the research fund of Dankook University in 2014.
References
- Alzheimer's A. 2014 Alzheimer's disease facts and figures. Alzheimers Dement. 2014;10:e47–e92. doi: 10.1016/j.jalz.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Arosio B., Trabattoni D., Galimberti L., Bucciarelli P., Fasano F., Calabresi C., Cazzullo C.L., Vergani C., Annoni G., Clerici M. Interleukin-10 and interleukin-6 gene polymorphisms as risk factors for Alzheimer's disease. Neurobiol. Aging. 2004;25:1009–1015. doi: 10.1016/j.neurobiolaging.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Bagli M., Papassotiropoulos A., Jessen F., Schmitz S., Rao M.L., Maier W., Heun R. Identical distribution of the alpha 2-macroglobulin pentanucleotide deletion in subjects with Alzheimer disease and controls in a German population. Am. J. Med. Genet. 2000;96:775–777. doi: 10.1002/1096-8628(20001204)96:6<775::aid-ajmg15>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Bagnoli S., Cellini E., Tedde A., Nacmias B., Piacentini S., Bessi V., Bracco L., Sorbi S. Association of IL10 promoter polymorphism in Italian Alzheimer's disease. Neurosci. Lett. 2007;418:262–265. doi: 10.1016/j.neulet.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Bhojak T.J., DeKosky S.T., Ganguli M., Kamboh M.I. Genetic polymorphisms in the cathespin D and interleukin-6 genes and the risk of Alzheimer's disease. Neurosci. Lett. 2000;288:21–24. doi: 10.1016/s0304-3940(00)01185-x. [DOI] [PubMed] [Google Scholar]
- Braida D., Sacerdote P., Panerai A.E., Bianchi M., Aloisi A.M., Iosue S., Sala M. Cognitive function in young and adult IL (interleukin)-6 deficient mice. Behav. Brain Res. 2004;153:423–429. doi: 10.1016/j.bbr.2003.12.018. [DOI] [PubMed] [Google Scholar]
- Buxbaum J.D., Oishi M., Chen H.I., Pinkas-Kramarski R., Jaffe E.A., Gandy S.E., Greengard P. Cholinergic agonists and interleukin 1 regulate processing and secretion of the Alzheimer beta/A4 amyloid protein precursor. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10075–10078. doi: 10.1073/pnas.89.21.10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurso C., Solfrizzi V., D'Introno A., Colacicco A.M., Capurso S.A., Capurso A., Panza F. Interleukin 6-174 G/C promoter gene polymorphism and sporadic Alzheimer's disease: geographic allele and genotype variations in Europe. Exp. Gerontol. 2004;39:1567–1573. doi: 10.1016/j.exger.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Capurso C., Solfrizzi V., Colacicco A.M., D'Introno A., Frisardi V., Imbimbo B.P., Lorusso M., Vendemiale G., Denitto M., Santamato A., Seripa D., Pilotto A., Fiore P., Capurso A., Panza F. Interleukin 6-174 G/C promoter and variable number of tandem repeats (VNTR) gene polymorphisms in sporadic Alzheimer's disease. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2010;34:177–182. doi: 10.1016/j.pnpbp.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Chen C., Zhou Z., Li M., Qu M., Ma Q., Zhong M., Zhang Y., Yu Z. Presenilin-2 polymorphisms and risk of sporadic AD: evidence from a meta-analysis. Gene. 2012;503:194–199. doi: 10.1016/j.gene.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Clarimon J., Bertranpetit J., Calafell F., Boada M., Tarraga L., Comas D. Joint analysis of candidate genes related to Alzheimer's disease in a Spanish population. Psychiatr. Genet. 2003;13:85–90. doi: 10.1097/01.ypg.0000056174.32550.7e. [DOI] [PubMed] [Google Scholar]
- Combarros O., Sanchez-Guerra M., Infante J., Llorca J., Berciano J. Gene dose-dependent association of interleukin-1 [− 889] allele 2 polymorphism with Alzheimer's disease. J. Neurol. 2002;249:1242–1245. doi: 10.1007/s00415-002-0819-9. [DOI] [PubMed] [Google Scholar]
- Combarros O., Infante J., Llorca J., Pena N., Fernandez-Viadero C., Berciano J. Interaction between interleukin-6 and intercellular adhesion molecule-1 genes and Alzheimer's disease risk. J. Neurol. 2005;252:485–487. doi: 10.1007/s00415-005-0658-6. [DOI] [PubMed] [Google Scholar]
- Combarros O., Sanchez-Juan P., Riancho J.A., Mateo I., Rodriguez-Rodriguez E., Infante J., Garcia-Gorostiaga I., Vazquez-Higuera J.L., Berciano J. Aromatase and interleukin-10 genetic variants interactively modulate Alzheimer's disease risk. J. Neural Transm. 2008;115:863–867. doi: 10.1007/s00702-008-0028-5. [DOI] [PubMed] [Google Scholar]
- Combarros O., Warden D.R., Hammond N., Cortina-Borja M., Belbin O., Lehmann M.G., Wilcock G.K., Brown K., Kehoe P.G., Barber R., Coto E., Alvarez V., Deloukas P., Gwilliam R., Heun R., Kolsch H., Mateo I., Oulhaj A., Arias-Vasquez A., Schuur M., Aulchenko Y.S., Ikram M.A., Breteler M.M., van Duijn C.M., Morgan K., Smith A.D., Lehmann D.J. The dopamine beta-hydroxylase -1021C/T polymorphism is associated with the risk of Alzheimer's disease in the Epistasis Project. BMC Med. Genet. 2010;11:162. doi: 10.1186/1471-2350-11-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon K.D., Myers A.J., Craig D.W., Webster J.A., Pearson J.V., Lince D.H., Zismann V.L., Beach T.G., Leung D., Bryden L., Halperin R.F., Marlowe L., Kaleem M., Walker D.G., Ravid R., Heward C.B., Rogers J., Papassotiropoulos A., Reiman E.M., Hardy J., Stephan D.A. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J. Clin. Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- Cousin E., Mace S., Rocher C., Dib C., Muzard G., Hannequin D., Pradier L., Deleuze J.F., Genin E., Brice A., Campion D. No replication of genetic association between candidate polymorphisms and Alzheimer's disease. Neurobiol. Aging. 2011;32:1443–1451. doi: 10.1016/j.neurobiolaging.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Culpan D., Prince J.A., Matthews S., Palmer L., Hughes A., Love S., Kehoe P.G., Wilcock G.K. Neither sequence variation in the IL-10 gene promoter nor presence of IL-10 protein in the cerebral cortex is associated with Alzheimer's disease. Neurosci. Lett. 2006;408:141–145. doi: 10.1016/j.neulet.2006.08.068. [DOI] [PubMed] [Google Scholar]
- Dai L., Liu D., Guo H., Wang Y., Bai Y. Association between polymorphism in the promoter region of interleukin 6 (− 174 G/C) and risk of Alzheimer's disease: a meta-analysis. J. Neurol. 2012;259:414–419. doi: 10.1007/s00415-011-6164-0. [DOI] [PubMed] [Google Scholar]
- Deniz-Naranjo M.C., Munoz-Fernandez C., Alemany-Rodriguez M.J., Perez-Vieitez M.C., Aladro-Benito Y., Irurita-Latasa J., Sanchez-Garcia F. Cytokine IL-1 beta but not IL-1 alpha promoter polymorphism is associated with Alzheimer disease in a population from the Canary Islands, Spain. Eur. J. Neurol. 2008;15:1080–1084. doi: 10.1111/j.1468-1331.2008.02252.x. [DOI] [PubMed] [Google Scholar]
- Depboylu C., Du Y., Muller U., Kurz A., Zimmer R., Riemenschneider M., Gasser T., Oertel W.H., Klockgether T., Dodel R.C. Lack of association of interleukin-10 promoter region polymorphisms with Alzheimer's disease. Neurosci. Lett. 2003;342:132–134. doi: 10.1016/s0304-3940(03)00231-3. [DOI] [PubMed] [Google Scholar]
- Depboylu C., Lohmuller F., Gocke P., Du Y., Zimmer R., Gasser T., Klockgether T., Dodel R.C. An interleukin-6 promoter variant is not associated with an increased risk for Alzheimer's disease. Dement. Geriatr. Cogn. Disord. 2004;17:170–173. doi: 10.1159/000076352. [DOI] [PubMed] [Google Scholar]
- Di Bona D., Rizzo C., Bonaventura G., Candore G., Caruso C. Association between interleukin-10 polymorphisms and Alzheimer's disease: a systematic review and meta-analysis. J. Alzheimers Dis. 2012;29:751–759. doi: 10.3233/JAD-2012-111838. [DOI] [PubMed] [Google Scholar]
- Du Y., Dodel R.C., Eastwood B.J., Bales K.R., Gao F., Lohmuller F., Muller U., Kurz A., Zimmer R., Evans R.M., Hake A., Gasser T., Oertel W.H., Griffin W.S., Paul S.M., Farlow M.R. Association of an interleukin 1 alpha polymorphism with Alzheimer's disease. Neurology. 2000;55:480–483. doi: 10.1212/wnl.55.4.480. [DOI] [PubMed] [Google Scholar]
- Dursun E., Gezen-Ak D., Ertan T., Bilgic B., Gurvit H., Emre M., Eker E., Engin F., Uysal O., Yilmazer S. Interleukin-1alpha-889C/T polymorphism in Turkish patients with late-onset Alzheimer's disease. Dement. Geriatr. Cogn. Disord. 2009;27:82–87. doi: 10.1159/000193627. [DOI] [PubMed] [Google Scholar]
- Faltraco F., Burger K., Zill P., Teipel S.J., Moller H.J., Hampel H., Bondy B., Ackenheil M. Interleukin-6-174 G/C promoter gene polymorphism C allele reduces Alzheimer's disease risk. J. Am. Geriatr. Soc. 2003;51:578–579. doi: 10.1046/j.1532-5415.2003.51177.x. [DOI] [PubMed] [Google Scholar]
- Fidani L., Goulas A., Mirtsou V., Petersen R.C., Tangalos E., Crook R., Hardy J. Interleukin-1 a polymorphism is not associated with late onset Alzheimer's disease. Neurosci. Lett. 2002;323:81–83. doi: 10.1016/s0304-3940(02)00114-3. [DOI] [PubMed] [Google Scholar]
- Flex A., Giovannini S., Biscetti F., Liperoti R., Spalletta G., Straface G., Landi F., Angelini F., Caltagirone C., Ghirlanda G., Bernabei R. Effect of proinflammatory gene polymorphisms on the risk of Alzheimer's disease. Neurodegener. Dis. 2014;13:230–236. doi: 10.1159/000353395. [DOI] [PubMed] [Google Scholar]
- Fontalba A., Gutierrez O., Llorca J., Mateo I., Vazquez-Higuera J.L., Berciano J., Fernandez-Luna J.L., Combarros O. Gene–gene interaction between CARD8 and interleukin-6 reduces Alzheimer's disease risk. J. Neurol. 2009;256:1184–1186. doi: 10.1007/s00415-009-5080-z. [DOI] [PubMed] [Google Scholar]
- Green E.K., Harris J.M., Lemmon H., Lambert J.C., Chartier-Harlin M.C., St Clair D., Mann D.M., Iwatsubo T., Lendon C.L. Are interleukin-1 gene polymorphisms risk factors or disease modifiers in AD? Neurology. 2002;58:1566–1568. doi: 10.1212/wnl.58.10.1566. [DOI] [PubMed] [Google Scholar]
- Griffin W.S., Stanley L.C., Ling C., White L., MacLeod V., Perrot L.J., White C.L., III, Araoz C. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7611–7615. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi L.M., Casadei V.M., Ferri C., Veglia F., Licastro F., Annoni G., Biunno I., De Bellis G., Sorbi S., Mariani C., Canal N., Griffin W.S., Franceschi M. Association of early-onset Alzheimer's disease with an interleukin-1alpha gene polymorphism. Ann. Neurol. 2000;47:361–365. [PubMed] [Google Scholar]
- Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., Jones N., Thomas C., Stretton A., Morgan A.R., Lovestone S., Powell J., Proitsi P., Lupton M.K., Brayne C., Rubinsztein D.C., Gill M., Lawlor B., Lynch A., Morgan K., Brown K.S., Passmore P.A., Craig D., McGuinness B., Todd S., Holmes C., Mann D., Smith A.D., Love S., Kehoe P.G., Hardy J., Mead S., Fox N., Rossor M., Collinge J., Maier W., Jessen F., Schurmann B., Heun R., van den Bussche H., Heuser I., Kornhuber J., Wiltfang J., Dichgans M., Frolich L., Hampel H., Hull M., Rujescu D., Goate A.M., Kauwe J.S., Cruchaga C., Nowotny P., Morris J.C., Mayo K., Sleegers K., Bettens K., Engelborghs S., De Deyn P.P., Van Broeckhoven C., Livingston G., Bass N.J., Gurling H., McQuillin A., Gwilliam R., Deloukas P., Al-Chalabi A., Shaw C.E., Tsolaki M., Singleton A.B., Guerreiro R., Muhleisen T.W., Nothen M.M., Moebus S., Jockel K.H., Klopp N., Wichmann H.E., Carrasquillo M.M., Pankratz V.S., Younkin S.G., Holmans P.A., O'Donovan M., Owen M.J., Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes A., Green E.K., Pritchard A., Harris J.M., Zhang Y., Lambert J.C., Chartier-Harlin M.C., Pickering-Brown S.M., Lendon C.L., Mann D.M. A polymorphic variation in the interleukin 1A gene increases brain microglial cell activity in Alzheimer's disease. J. Neurol. Neurosurg. Psychiatry. 2004;75:1475–1477. doi: 10.1136/jnnp.2003.030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley R., Hallmayer J., Groth D.M., Brooks W.S., Gandy S.E., Martins R.N. Association of interleukin-1 polymorphisms with Alzheimer's disease in Australia. Ann. Neurol. 2002;51:795–797. doi: 10.1002/ana.10196. [DOI] [PubMed] [Google Scholar]
- Heun R., Kolsch H., Ibrahim-Verbaas C.A., Combarros O., Aulchenko Y.S., Breteler M., Schuur M., van Duijn C.M., Hammond N., Belbin O., Cortina-Borja M., Wilcock G.K., Brown K., Barber R., Kehoe P.G., Coto E., Alvarez V., Lehmann M.G., Deloukas P., Mateo I., Morgan K., Warden D.R., Smith A.D., Lehmann D.J. Interactions between PPAR-alpha and inflammation-related cytokine genes on the development of Alzheimer's disease, observed by the Epistasis Project. Int. J. Mol. Epidemiol. Genet. 2012;3:39–47. [PMC free article] [PubMed] [Google Scholar]
- Hu J.L., Li G., Zhou D.X., Zou Y.X., Zhu Z.S., Xu R.X., Jiang X.D., Zeng Y.J. Genetic analysis of interleukin-1A C(− 889)T polymorphism with Alzheimer disease. Cell. Mol. Neurobiol. 2009;29:81–85. doi: 10.1007/s10571-008-9299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Zhao H., Kong Y., Lu X. Meta-analysis of the association between the interleukin-1A − 889C/T polymorphism and Alzheimer's disease. J. Neurosci. Res. 2012;90:1681–1692. doi: 10.1002/jnr.23062. [DOI] [PubMed] [Google Scholar]
- Infante J., Sanz C., Fernandez-Luna J.L., Llorca J., Berciano J., Combarros O. Gene–gene interaction between interleukin-6 and interleukin-10 reduces AD risk. Neurology. 2004;63:1135–1136. doi: 10.1212/01.wnl.0000138570.96291.a8. [DOI] [PubMed] [Google Scholar]
- Kang H.J., Kim J.M., Kim S.W., Shin I.S., Park S.W., Kim Y.H., Yoon J.S. Associations of cytokine genes with Alzheimer's disease and depression in an elderly Korean population. J. Neurol. Neurosurg. Psychiatry. 2014 doi: 10.1136/jnnp-2014-308469. [DOI] [PubMed] [Google Scholar]
- Ki C.S., Na D.L., Kim D.K., Kim H.J., Kim J.W. Lack of association of the interleukin-1alpha gene polymorphism with Alzheimer's disease in a Korean population. Ann. Neurol. 2001;49:817–818. doi: 10.1002/ana.1067. [DOI] [PubMed] [Google Scholar]
- Klimkowicz-Mrowiec A., Marona M., Wolkow P., Maruszak A., Styczynska M., Barcikowska M., Zekanowski C., Szczudlik A., Slowik A. Interleukin-1 gene − 511 CT polymorphism and the risk of Alzheimer's disease in a Polish population. Dement. Geriatr. Cogn. Disord. 2009;28:461–464. doi: 10.1159/000259460. [DOI] [PubMed] [Google Scholar]
- Klimkowicz-Mrowiec A., Wolkow P., Spisak K., Maruszak A., Styczynska M., Barcikowska M., Szczudlik A., Slowik A. Interleukin-6 gene (− 174 C/G) and apolipoprotein E gene polymorphisms and the risk of Alzheimer disease in a Polish population. Neurol. Neurochir. Pol. 2010;44:537–541. doi: 10.1016/s0028-3843(14)60149-3. [DOI] [PubMed] [Google Scholar]
- Koivisto A.M., Helisalmi S., Pihlajamaki J., Moilanen L., Kuusisto J., Laakso M., Hiltunen M., Keijo K., Hanninen T., Helkala E.L., Kervinen K., Kesaniemi Y.A., Soininen H. Interleukin-6 promoter polymorphism and late-onset Alzheimer's disease in the Finnish population. J. Neurogenet. 2005;19:155–161. doi: 10.1080/01677060600569721. [DOI] [PubMed] [Google Scholar]
- Kuo Y.M., Liao P.C., Lin C., Wu C.W., Huang H.M., Lin C.C., Chuo L.J. Lack of association between interleukin-1alpha polymorphism and Alzheimer disease or vascular dementia. Alzheimer Dis. Assoc. Disord. 2003;17:94–97. doi: 10.1097/00002093-200304000-00007. [DOI] [PubMed] [Google Scholar]
- Li X.Q., Zhang J.W., Zhang Z.X., Chen D., Qu Q.M. Interleukin-1 gene cluster polymorphisms and risk of Alzheimer's disease in Chinese Han population. J. Neural Transm. 2004;111:1183–1190. doi: 10.1007/s00702-004-0148-5. [DOI] [PubMed] [Google Scholar]
- Li H., Wetten S., Li L., St Jean P.L., Upmanyu R., Surh L., Hosford D., Barnes M.R., Briley J.D., Borrie M., Coletta N., Delisle R., Dhalla D., Ehm M.G., Feldman H.H., Fornazzari L., Gauthier S., Goodgame N., Guzman D., Hammond S., Hollingworth P., Hsiung G.Y., Johnson J., Kelly D.D., Keren R., Kertesz A., King K.S., Lovestone S., Loy-English I., Matthews P.M., Owen M.J., Plumpton M., Pryse-Phillips W., Prinjha R.K., Richardson J.C., Saunders A., Slater A.J., St George-Hyslop P.H., Stinnett S.W., Swartz J.E., Taylor R.L., Wherrett J., Williams J., Yarnall D.P., Gibson R.A., Irizarry M.C., Middleton L.T., Roses A.D. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch. Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- Li B.H., Zhang L.L., Yin Y.W., Pi Y., Guo L., Yang Q.W., Gao C.Y., Fang C.Q., Wang J.Z., Xiang J., Li J.C. Association between interleukin-1alpha C(− 889)T polymorphism and Alzheimer's disease: a meta-analysis including 12,817 subjects. J. Neural Transm. 2013;120:497–506. doi: 10.1007/s00702-012-0867-y. [DOI] [PubMed] [Google Scholar]
- Licastro F., Grimaldi L.M., Bonafe M., Martina C., Olivieri F., Cavallone L., Giovanietti S., Masliah E., Franceschi C. Interleukin-6 gene alleles affect the risk of Alzheimer's disease and levels of the cytokine in blood and brain. Neurobiol. Aging. 2003;24:921–926. doi: 10.1016/s0197-4580(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Lio D., Licastro F., Scola L., Chiappelli M., Grimaldi L.M., Crivello A., Colonna-Romano G., Candore G., Franceschi C., Caruso C. Interleukin-10 promoter polymorphism in sporadic Alzheimer's disease. Genes Immun. 2003;4:234–238. doi: 10.1038/sj.gene.6363964. [DOI] [PubMed] [Google Scholar]
- Ma S.L., Tang N.L., Lam L.C., Chiu H.F. Lack of association of the interleukin-1beta gene polymorphism with Alzheimer's disease in a Chinese population. Dement. Geriatr. Cogn. Disord. 2003;16:265–268. doi: 10.1159/000072811. [DOI] [PubMed] [Google Scholar]
- Ma S.L., Tang N.L., Lam L.C., Chiu H.F. The association between promoter polymorphism of the interleukin-10 gene and Alzheimer's disease. Neurobiol. Aging. 2005;26:1005–1010. doi: 10.1016/j.neurobiolaging.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Mansoori N., Tripathi M., Luthra K., Alam R., Lakshmy R., Sharma S., Arulselvi S., Parveen S., Mukhopadhyay A.K. MTHFR (677 and 1298) and IL-6-174 G/C genes in pathogenesis of Alzheimer's and vascular dementia and their epistatic interaction. Neurobiol. Aging. 2012;33(1003):e1–e8. doi: 10.1016/j.neurobiolaging.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Mattila K.M., Rinne J.O., Lehtimaki T., Roytta M., Ahonen J.P., Hurme M. Association of an interleukin 1B gene polymorphism (− 511) with Parkinson's disease in Finnish patients. J. Med. Genet. 2002;39:400–402. doi: 10.1136/jmg.39.6.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarron M.O., Stewart J., McCarron P., Love S., Vinters H.V., Ironside J.W., Mann D.M., Graham D.I., Nicoll J.A. Association between interleukin-1A polymorphism and cerebral amyloid angiopathy-related hemorrhage. Stroke. 2003;34:e193–e195. doi: 10.1161/01.STR.0000089294.85447.1E. [DOI] [PubMed] [Google Scholar]
- McCulley M.C., Day I.N., Holmes C. Association between interleukin 1-beta promoter (− 511) polymorphism and depressive symptoms in Alzheimer's disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;124B:50–53. doi: 10.1002/ajmg.b.20086. [DOI] [PubMed] [Google Scholar]
- Minster R.L., DeKosky S.T., Ganguli M., Belle S., Kamboh M.I. Genetic association studies of interleukin-1 (IL-1A and IL-1B) and interleukin-1 receptor antagonist genes and the risk of Alzheimer's disease. Ann. Neurol. 2000;48:817–819. [PubMed] [Google Scholar]
- Moraes C.F., Benedet A.L., Souza V.C., Lins T.C., Camargos E.F., Naves J.O., Brito C.J., Cordova C., Pereira R.W., Nobrega O.T. Cytokine gene polymorphisms and Alzheimer's disease in Brazil. Neuroimmunomodulation. 2013;20:239–246. doi: 10.1159/000350368. [DOI] [PubMed] [Google Scholar]
- Nicoll J.A., Mrak R.E., Graham D.I., Stewart J., Wilcock G., MacGowan S., Esiri M.M., Murray L.S., Dewar D., Love S., Moss T., Griffin W.S. Association of interleukin-1 gene polymorphisms with Alzheimer's disease. Ann. Neurol. 2000;47:365–368. [PMC free article] [PubMed] [Google Scholar]
- Nishimura M., Sakamoto T., Kaji R., Kawakami H. Influence of polymorphisms in the genes for cytokines and glutathione S-transferase omega on sporadic Alzheimer's disease. Neurosci. Lett. 2004;368:140–143. doi: 10.1016/j.neulet.2004.06.076. [DOI] [PubMed] [Google Scholar]
- Paradowski B., Celczynska D., Dobosz T., Noga L. Polymorphism 174 G/C of interleukin 6 gene in Alzheimer's disease–preliminary report. Neurol. Neurochir. Pol. 2008;42:312–315. [PubMed] [Google Scholar]
- Payao S.L., Goncalves G.M., de Labio R.W., Horiguchi L., Mizumoto I., Rasmussen L.T., de Souza Pinhel M.A., Silva Souza D.R., Bechara M.D., Chen E., Mazzotti D.R., Ferreira Bertolucci P.H., Cardoso Smith Mde A. Association of interleukin 1beta polymorphisms and haplotypes with Alzheimer's disease. J. Neuroimmunol. 2012;247:59–62. doi: 10.1016/j.jneuroim.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Pirskanen M., Hiltunen M., Mannermaa A., Iivonen S., Helisalmi S., Lehtovirta M., Koivisto A.M., Laakso M., Soininen H., Alafuzoff I. Interleukin 1 alpha gene polymorphism as a susceptibility factor in Alzheimer's disease and its influence on the extent of histopathological hallmark lesions of Alzheimer's disease. Dement. Geriatr. Cogn. Disord. 2002;14:123–127. doi: 10.1159/000063603. [DOI] [PubMed] [Google Scholar]
- Pola R., Flex A., Gaetani E., Lago A.D., Gerardino L., Pola P., Bernabei R. The − 174 G/C polymorphism of the interleukin-6 gene promoter is associated with Alzheimer's disease in an Italian population [corrected] Neuroreport. 2002;13:1645–1647. doi: 10.1097/00001756-200209160-00015. [DOI] [PubMed] [Google Scholar]
- Prince J.A., Feuk L., Sawyer S.L., Gottfries J., Ricksten A., Nagga K., Bogdanovic N., Blennow K., Brookes A.J. Lack of replication of association findings in complex disease: an analysis of 15 polymorphisms in prior candidate genes for sporadic Alzheimer's disease. Eur. J. Hum. Genet. 2001;9:437–444. doi: 10.1038/sj.ejhg.5200651. [DOI] [PubMed] [Google Scholar]
- Qi H.P., Qu Z.Y., Duan S.R., Wei S.Q., Wen S.R., Bi S. IL-6-174 G/C and − 572 C/G polymorphisms and risk of Alzheimer's disease. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla R.A., Orellana D.I., Gonzalez-Billault C., Maccioni R.B. Interleukin-6 induces Alzheimer-type phosphorylation of tau protein by deregulating the cdk5/p35 pathway. Exp. Cell Res. 2004;295:245–257. doi: 10.1016/j.yexcr.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Ramos E.M., Lin M.T., Larson E.B., Maezawa I., Tseng L.H., Edwards K.L., Schellenberg G.D., Hansen J.A., Kukull W.A., Jin L.W. Tumor necrosis factor alpha and interleukin 10 promoter region polymorphisms and risk of late-onset Alzheimer disease. Arch. Neurol. 2006;63:1165–1169. doi: 10.1001/archneur.63.8.1165. [DOI] [PubMed] [Google Scholar]
- Rasmussen L., Delabio R., Horiguchi L., Mizumoto I., Terazaki C.R., Mazzotti D., Bertolucci P.H., Pinhel M.A., Souza D., Krieger H., Kawamata C., Minett T., Smith M.C., Payao S.L. Association between interleukin 6 gene haplotype and Alzheimer's disease: a Brazilian case–control study. J. Alzheimers Dis. 2013;36:733–738. doi: 10.3233/JAD-122407. [DOI] [PubMed] [Google Scholar]
- Ravaglia G., Paola F., Maioli F., Martelli M., Montesi F., Bastagli L., Bianchin M., Chiappelli M., Tumini E., Bolondi L., Licastro F. Interleukin-1beta and interleukin-6 gene polymorphisms as risk factors for AD: a prospective study. Exp. Gerontol. 2006;41:85–92. doi: 10.1016/j.exger.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Rebeck G.W. Confirmation of the genetic association of interleukin-1A with early onset sporadic Alzheimer's disease. Neurosci. Lett. 2000;293:75–77. doi: 10.1016/s0304-3940(00)01487-7. [DOI] [PubMed] [Google Scholar]
- Ribizzi G., Fiordoro S., Barocci S., Ferrari E., Megna M. Cytokine polymorphisms and Alzheimer disease: possible associations. Neurol. Sci. 2010;31:321–325. doi: 10.1007/s10072-010-0221-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manotas M., Amorin-Diaz M., Canizares-Hernandez F., Ruiz-Espejo F., Martinez-Vidal S., Gonzalez-Sarmiento R., Martinez-Hernandez P., Cabezas-Herrera J. Association study and meta-analysis of Alzheimer's disease risk and presenilin-1 intronic polymorphism. Brain Res. 2007;1170:119–128. doi: 10.1016/j.brainres.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Scassellati C., Zanardini R., Squitti R., Bocchio-Chiavetto L., Bonvicini C., Binetti G., Zanetti O., Cassetta E., Gennarelli M. Promoter haplotypes of interleukin-10 gene and sporadic Alzheimer's disease. Neurosci. Lett. 2004;356:119–122. doi: 10.1016/j.neulet.2003.11.033. [DOI] [PubMed] [Google Scholar]
- Sciacca F.L., Ferri C., Licastro F., Veglia F., Biunno I., Gavazzi A., Calabrese E., Martinelli Boneschi F., Sorbi S., Mariani C., Franceschi M., Grimaldi L.M. Interleukin-1B polymorphism is associated with age at onset of Alzheimer's disease. Neurobiol. Aging. 2003;24:927–931. doi: 10.1016/s0197-4580(03)00011-3. [DOI] [PubMed] [Google Scholar]
- Seripa D., Matera M.G., Dal Forno G., Gravina C., Masullo C., Daniele A., Binetti G., Bonvicini C., Squitti R., Palermo M.T., Davis D.G., Antuono P., Wekstein D.R., Dobrina A., Gennarelli M., Fazio V.M. Genotypes and haplotypes in the IL-1 gene cluster: analysis of two genetically and diagnostically distinct groups of Alzheimer patients. Neurobiol. Aging. 2005;26:455–464. doi: 10.1016/j.neurobiolaging.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Serretti A., Olgiati P., Politis A., Malitas P., Albani D., Dusi S., Polito L., De Mauro S., Zisaki A., Piperi C., Liappas I., Stamouli E., Mailis A., Atti A.R., Morri M., Ujkaj M., Batelli S., Forloni G., Soldatos C.R., Papadimitriou G.N., De Ronchi D., Kalofoutis A. Lack of association between interleukin-1 alpha rs1800587 polymorphism and Alzheimer's disease in two independent European samples. J. Alzheimers Dis. 2009;16:181–187. doi: 10.3233/JAD-2009-0946. [DOI] [PubMed] [Google Scholar]
- Shawkatova I., Javor J., Parnicka Z., Vrazda L., Novak M., Buc M. No association between cytokine gene polymorphism and risk of Alzheimer's disease in Slovaks. Acta Neurobiol. Exp. (Wars) 2010;70:303–307. doi: 10.55782/ane-2010-1802. [DOI] [PubMed] [Google Scholar]
- Sheng J.G., Mrak R.E., Griffin W.S. Microglial interleukin-1 alpha expression in brain regions in Alzheimer's disease: correlation with neuritic plaque distribution. Neuropathol. Appl. Neurobiol. 1995;21:290–301. doi: 10.1111/j.1365-2990.1995.tb01063.x. [DOI] [PubMed] [Google Scholar]
- Shibata N., Ohnuma T., Takahashi T., Baba H., Ishizuka T., Ohtsuka M., Ueki A., Nagao M., Arai H. Effect of IL-6 polymorphism on risk of Alzheimer disease: genotype–phenotype association study in Japanese cases. Am. J. Med. Genet. 2002;114:436–439. doi: 10.1002/ajmg.10417. [DOI] [PubMed] [Google Scholar]
- Tian M., Deng Y.Y., Hou D.R., Li W., Feng X.L., Yu Z.L. Association of IL-1, IL-18, and IL-33 gene polymorphisms with late-onset Alzheimers disease in a Hunan Han Chinese population. Brain Res. 2015;1596:136–145. doi: 10.1016/j.brainres.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Toral-Rios D., Franco-Bocanegra D., Rosas-Carrasco O., Mena-Barranco F., Carvajal-Garcia R., Meraz-Rios M.A., Campos-Pena V. Evaluation of inflammation-related genes polymorphisms in Mexican with Alzheimer's disease: a pilot study. Front. Cell. Neurosci. 2015;9:148. doi: 10.3389/fncel.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres K.C., Araujo Pereira P., Lima G.S., Bozzi I.C., Rezende V.B., Bicalho M.A., Moraes E.N., Miranda D.M., Romano-Silva M.A. Increased frequency of T cells expressing IL-10 in Alzheimer disease but not in late-onset depression patients. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2013;47:40–45. doi: 10.1016/j.pnpbp.2013.07.021. [DOI] [PubMed] [Google Scholar]
- Tsai S.J., Liu H.C., Liu T.Y., Wang K.Y., Hong C.J. Lack of association between the interleukin-1alpha gene C(− 889)T polymorphism and Alzheimer's disease in a Chinese population. Neurosci. Lett. 2003;343:93–96. doi: 10.1016/s0304-3940(03)00333-1. [DOI] [PubMed] [Google Scholar]
- van Oijen M., Arp P.P., de Jong F.J., Hofman A., Koudstaal P.J., Uitterlinden A.G., Breteler M.M. Polymorphisms in the interleukin 6 and transforming growth factor beta1 gene and risk of dementia. The Rotterdam Study. Neurosci. Lett. 2006;402:113–117. doi: 10.1016/j.neulet.2006.03.050. [DOI] [PubMed] [Google Scholar]
- Vendramini A.A., de Labio R.W., Rasmussen L.T., Dos Reis N.M., Minett T., Bertolucci P.H., de Souza Pinhel M.A., Souza D.R., Mazzotti D.R., de Arruda Cardoso Smith M., Payao S.L. Interleukin-8-251T >, Interleukin-1alpha-889C > T and Apolipoprotein E polymorphisms in Alzheimer's disease. Genet. Mol. Biol. 2011;34:1–5. doi: 10.1590/S1415-47572010005000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vural P., Degirmencioglu S., Parildar-Karpuzoglu H., Dogru-Abbasoglu S., Hanagasi H.A., Karadag B., Gurvit H., Emre M., Uysal M. The combinations of TNFalpha-308 and IL-6 -174 or IL-10 -1082 genes polymorphisms suggest an association with susceptibility to sporadic late-onset Alzheimer's disease. Acta Neurol. Scand. 2009;120:396–401. doi: 10.1111/j.1600-0404.2009.01230.x. [DOI] [PubMed] [Google Scholar]
- Wang W.F., Liao Y.C., Wu S.L., Tsai F.J., Lee C.C., Hua C.S. Association of interleukin-I beta and receptor antagonist gene polymorphisms with late onset Alzheimer's disease in Taiwan Chinese. Eur. J. Neurol. 2005;12:609–613. doi: 10.1111/j.1468-1331.2005.01033.x. [DOI] [PubMed] [Google Scholar]
- Wang H.K., Hsu W.C., Fung H.C., Lin J.C., Hsu H.P., Wu Y.R., Hsu Y., Hu F.J., Lee-Chen G.J., Chen C.M. Interleukin-1alpha and -1beta promoter polymorphisms in taiwanese patients with dementia. Dement. Geriatr. Cogn. Disord. 2007;24:104–110. doi: 10.1159/000104829. [DOI] [PubMed] [Google Scholar]
- Ward A., Crean S., Mercaldi C.J., Collins J.M., Boyd D., Cook M.N., Arrighi H.M. Prevalence of apolipoprotein E4 genotype and homozygotes (APOE e4/4) among patients diagnosed with Alzheimer's disease: a systematic review and meta-analysis. Neuroepidemiology. 2012;38:1–17. doi: 10.1159/000334607. [DOI] [PubMed] [Google Scholar]
- WHO. Dementia; a Public Health Priority, 2012.
- Wilson C.J., Finch C.E., Cohen H.J. Cytokines and cognition–the case for a head-to-toe inflammatory paradigm. J. Am. Geriatr. Soc. 2002;50:2041–2056. doi: 10.1046/j.1532-5415.2002.50619.x. [DOI] [PubMed] [Google Scholar]
- Yuan H., Xia Q., Ge P., Wu S. Genetic polymorphism of interleukin 1beta − 511C/T and susceptibility to sporadic Alzheimer's disease: a meta-analysis. Mol. Biol. Rep. 2013;40:1827–1834. doi: 10.1007/s11033-012-2237-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Hayes A., Pritchard A., Thaker U., Haque M.S., Lemmon H., Harris J., Cumming A., Lambert J.C., Chartier-Harlin M.C., St Clair D., Iwatsubo T., Mann D.M., Lendon C.L. Interleukin-6 promoter polymorphism: risk and pathology of Alzheimer's disease. Neurosci. Lett. 2004;362:99–102. doi: 10.1016/j.neulet.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Zhou Y.T., Zhang Z.X., Zhang J.W., He X.M., Xu T. Association between interleukin-1 alpha-889 C/T polymorphism and Alzheimer's disease in Chinese Han population. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2006;28:186–190. [PubMed] [Google Scholar]