Abstract
Tibetan is a valuable Himalayan sheep breed classified as endangered. Knowledge of the level and distribution of genetic diversity in Tibetan sheep is important for designing conservation strategies for their sustainable survival and to preserve their evolutionary potential. Thus, for the first time, genetic variability in the Tibetan population was accessed with twenty five inter-simple sequence repeat markers. All the microsatellites were polymorphic and a total of 148 alleles were detected across these loci. The observed number of alleles across all the loci was more than the effective number of alleles and ranged from 3 (BM6506) to 11 (BM6526) with 5.920 ± 0.387 mean number of alleles per locus. The average observed heterozygosity was less than the expected heterozygosity. The observed and expected heterozygosity values ranged from 0.150 (BM1314) to 0.9 (OarCP20) with an overall mean of 0.473 ± 0.044 and from 0.329 (BM8125) to 0.885 (BM6526) with an overall mean 0.672 ± 0.030, respectively. The lower heterozygosity pointed towards diminished genetic diversity in the population. Thirteen microsatellite loci exhibited significant (P < 0.05) departures from the Hardy–Weinberg proportions in the population. The estimate of heterozygote deficiency varied from − 0.443 (OarCP20) to 0.668 (OarFCB128) with a mean positive value of 0.302 ± 0.057. A normal ‘L’ shaped distribution of mode-shift test and non-significant heterozygote excess on the basis of different models suggested absence of recent bottleneck in the existing Tibetan population. In view of the declining population of Tibetan sheep (less than 250) in the breeding tract, need of the hour is immediate scientific management of the population so as to increase the population hand in hand with retaining the founder alleles to the maximum possible extent.
Keywords: Bottleneck, Genetic diversity, Heterozygote deficiency, Microsatellite markers, Tibetan sheep
Highlights
-
•
First report on genetic variability of Tibetan, an endangered Indian sheep breed
-
•
Tibetan sheep possess sufficient allele diversity but high heterozygote deficiency
-
•
Population has not suffered major bottleneck despite decline in numbers
-
•
Genetic diversity estimates give chance for conservation of Tibetan breed
1. Introduction
Sheep biodiversity in India is characterized by high degree of endemism as variations in agro climatic conditions have led to the development of more than 40 breeds (Acharya, 1982). This vast ovine biodiversity is being eroded rapidly with more than 50% of sheep breeds currently under threat (Bhatia and Arora, 2005). Sheep provides employment and income to the socially and economically disadvantageous sections of the society being reared by the landless laborers and marginal farmers. Indigenous breeds must be considered as important reservoirs of non-exploited resources due to the presence of potentially unrecognized beneficial genetic variation. Moreover, autochthones breeds are the cultural properties due to their role in the agriculture tenures and in the social life of rural populations. Tibetan sheep is one such important breed of Indian North temperate Himalayan region. The sheep migrated to India with the Tibetan traders who used them as beasts of burden for transporting various merchandise besides wool (Government of Bengal, 1864). Independence of India in 1947 led to political and economic changes with concomitant cessation of their migration in the country.
Tibetan animals are of medium size, mostly white with black or brown face and brown and white spots on the body. The fleece is relatively fine and is among the best obtained from native ovine breeds of Indian subcontinent (Banerjee, 2009). The animals are unique being adapted to the harsh temperate climate and difficult terrains of Himalayas and survive even in the open housing system. Tibetan is regarded as one of the most rustic sheep breeds which thrives in conditions of extreme harshness and deprivation while providing meat and down for the people. They are the only source of livelihood in the area since agriculture is not suitable due to the geo-climatic characteristics. However, population of Tibetan sheep has been decreasing drastically in recent past (Banerjee, 2009) due to lack of regulated market, transport linkage, shrinkage of pasture land, increased inbreeding and occurrence of diseases. Population has gone down from 30,000 (Acharya, 1982) to less than 250 in Sikkim (Livestock Census, 2012, Kumar, 2015). Therefore, urgent specific management and conservation measures are required not only for the restoration of depleted natural population due to endangered status but also for its biological and economical relevance and ecological importance. Knowledge of genetic variability is very important for establishing any conservation and management program. The use of highly variable molecular genetic markers, such as microsatellites, is one of the most powerful means for studying genetic diversity because of their high degree of polymorphism, abundance, random distribution across the genome, co-dominant inheritance and neutrality with respect to selection (Barcaccia et al., 2013, Putman and Carbone, 2014). These are being used to estimate the diversity of autochthonous sheep breeds all over the world (Ghazyl et al., 2013, Ceccobelli et al., 2015) including India (Bhatia and Arora, 2005, Sharma et al., 2010). Unfortunately, no study has been undertaken so far to investigate the genetic characteristics of Tibetan sheep using molecular biology techniques. This is critical as urgent conservation efforts are required to conserve and utilize the Tibetan sheep genetic resource.
Hence, the aim of the present study was firstly to estimate the genetic intra-breed variability of Tibetan sheep using 25 microsatellite markers and secondly to detect population bottleneck, if any.
2. Materials and methods
2.1. Sample collection and polymerase chain reaction
The breeding tract of Tibetan sheep is now confined to the dry alpine zone, North District of Sikkim state in India (Fig. 1). Population is confined into six flocks only which are located in the Phalung valley (4697 m, latitude 27° 56′ longitude 88° 35′). Blood samples were acquired from twenty Tibetan sheep (about 10% of existing population) from the breeding region (Fig. 1) following the guidelines of MoDAD (Measurement of Domestic Animal Diversity) program (FAO, 2004). The native sheep were evaluated for their phenotypic breed characteristics as per the breed descriptor and individuals from each flock were sampled. Owners were questioned in detail to minimize the sampling of closely related individuals. Blood samples (5–6 ml) were collected in vacutainer containing Ethylene diamine tetra acetic acid (0.5 mM, pH 8.0). Genomic DNA was extracted from whole blood using phenol-chloroform protocol (Sambrook et al., 1989).
Fig. 1.
Distribution of Tibetan sheep in Sikkim (India).
2.2. Microsatellite genotyping
A panel consisting of 25 microsatellite markers was selected for the diversity analysis of Tibetan sheep population. These were chosen from literature related with sheep diversity studies aiming to analyze highly polymorphic markers spread across the genome. These markers also adhere to the guidelines of International Society for Animal Genetics and FAO (http://dad.fao.org/en/refer/library/guidelin/marker.pdf). Detailed information on primers is presented in Table 1. Forward primer of each marker was 5′ labeled with fluorescent dye, i.e. FAM, NED, PET and VIC. PCR amplification was performed in a reaction volume of 25 μl on i-cycler. Reaction mixture consisted of 50–100 ng of genomic DNA, 200 μM of each dNTP, 50 pM of each primer and 0.5 units of Taq DNA polymerase. The amplification was carried out using a Touchdown program for all microsatellite loci, which consists of initial denaturation of 95 °C for 1 min; 3 cycles of 95 °C for 45 s and 60 °C for 1 min, 3 cycle of 95 °C for 45 s and 57 °C for 1 min; 3 cycles of 95 °C for 45 s and 54 °C for 1 min, 3 cycles of 95 °C for 45 s and 51 °C for 1 min and 20 cycles of 95 °C for 45 s and 48 °C for 1 min. The PCR amplification was confirmed by electrophoresing the products in 1.8% agarose gel followed by staining with ethidium bromide (0.5 mg/ml). PCR products were multiplexed (Table 1) and genotyping was carried out on an automated ABI-3100 DNA sequencer (Applied Biosystems, USA) using LIZ 500 as the internal size standard (Applied Biosystems, USA). Allele sizing was done using GeneMapper™ software v 3.7. Stutter related scoring error, often seen in dinucleotide repeats, was absent and alleles could be scored unambiguously.
Table 1.
Sequence and characteristics of the 25 primers used for diversity estimation of Tibetan sheep.
| Microsatellite locus | Primer sequence (5′ → 3′) | Set | Chromosome location | Dye | Allele size range (bp) |
|---|---|---|---|---|---|
| BM0757 | F — tgg aaa caa tgt aaa cct ggg R — ttg agc cac caa gga acc |
1 | 9 | FAM | 182–198 |
| BM8125 | F — ctc tat ctg tgg aaa agg tgg g R — ggg ggt tag act tca aca tac g |
1 | 17 | FAM | 111–115 |
| OarCP49 | F — cag aca cgg ctt agc aac taa acg c R — gtg ggg atg aat att cct tca taa gg |
1 | 17 | NED | 80–112 |
| BM0827 | F — ggg ctg gtc gta tgc tga g R — gtt gga ctt gct gaa gtg acc |
1 | 3 | NED | 202–220 |
| OarHH47 | F — ttt att gac aaa ctc tct tcc taa ctc cac c R — gta gtt att taa aaa aat atc ata cct ctt aag g |
1 | 18 | VIC | 124–136 |
| CSSM47 | F — tct ctg tct cta tca cta tat ggc R — ctg ggc acc tga aac tat cat cat |
2 | 2 | VIC | 124–150 |
| MAF214 | F — aat gca gga gat ctg agg cag gga cg R — ggg tga tct tag gga ggt ttt gga gg |
2 | 16 | PET | 187–273 |
| OarCP20 | F — gat ccc ctg gag gag gaa acg g R — ggc att tca tgg ctt tag cag g |
2 | 21 | PET | 65–79 |
| OarHH41 | F — tcc aca ggc tta aat cta tat agc aac c R — cca gct aaa gat aaa aga tga tgt ggg ag |
2 | 10 | NED | 120–130 |
| OarVH72 | F — ctc tag agg atc tgg aat gca aag ctc R — ggc ctc tca agg ggc aag agc agg |
2 | 25 | FAM | 123–127 |
| INRA63 | F — gac cac aaa ggg att tgc aca agc R — aaa cca cag aaa tgc ttg gaa g |
3 | 14 | FAM | 169–193 |
| OarAE129 | F — aat cca gtg tgt gaa aga cta atc cag R — gta gat caa gat ata gaa tat ttt tca aca cc |
3 | 5 | NED | 147–159 |
| OarCP34 | F — gct gaa caa tgt gat atg ttc agg R — ggg aca ata ctg tct tag atg ctg c |
3 | 3 | FAM | 102–122 |
| OarFCB128 | F — cag ctg agc aac taa gac ata cat gcg R — att aaa gca tct tct ctt tat ttc ctc gc |
3 | 2 | PET | 91–121 |
| OarHH35 | F — aat tgc att cag tat ctt taa cat ctg gc R — atg aaa ata taa aga gaa tga acc aca cgg |
4 | 4 | NED | 117–133 |
| OarHH64 | F — cgt tcc ctc act atg gaa agt tat ata tgc R — cac tct att gta aga att tga atg aga gc |
4 | 4 | PET | 118–136 |
| OarJMP029 | F — gta tac acg tgg aca ccg ctt tgt ac R — gaa gtg gca aga ttc aga ggg gaa g |
4 | 24 | FAM | 98–144 |
| OarJMP08 | F — cgg gat gat ctt ctg tcc aaa tat gc R — cat ttg ctt tgg ctt cag aac cag ag |
4 | 6 | VIC | 115–131 |
| BM1314 | F— ttc ctc ctc ttc tct cca aac R — atc tca aac gcc agt gtg g |
5 | 22 | NED | 157–173 |
| BM6506 | F — gca cgt ggt aaa gag atg gc R — agc aac ttg agc atg gca c |
5 | 1 | FAM | 187–201 |
| CSRD247 | F — gga ctt gcc aga act ctg caa t R — cac tgt ggt ttg tat tag tca gg |
5 | 14 | NED | 215–239 |
| CSSM31 | F — cca agt tta gta ctt gta agt aga R — gac tct cta gca ctt tat ctg tgt |
5 | 9 | FAM | 144–166 |
| OarFCB48 | F — gag tta gta caa gga tga caa gag gca c R — gac tct aga gga tcg caa aga acc ag |
5 | 17 | PET | 144–164 |
| HSC | F — ctg cca atg cag aga cac aag a R — gtc tgt ctc ctg tct tgt cat c |
6 | X | FAM | 267–293 |
| BM6526 | F — cat gcc aaa caa tat cca gc R — tga agg tag aga gca agc agc |
6 | 26 | VIC | 142–170 |
2.3. Data analysis
Basic genetic parameters including allele frequencies, observed (No) and effective number of alleles (Ne), observed (Ho) and expected heterozygosity (He) and heterozygote deficit (FIS) in the whole population were calculated by analyzing the genetic data with GenAlEx 6.2 software (Peakall and Smouse, 2008). Tests of Hardy–Weinberg equilibrium and Ewens–Watterson Neutrality were applied using POPGENE 1.31 version (Yeh et al., 1999). Bottleneck events in the population were tested by three methods. The first method consisted of three excess heterozygosity tests developed by Cornuet and Luikart (1996): (i) Sign test, (ii) Standardized differences test, and (iii) a Wilcoxon sign-rank test. The probability distribution was established using 1000 simulations under three models–Infinite allele model (IAM), stepwise mutation model (SMM) and two-phase model of mutation (TPM). The second method was the graphical representation of the mode-shift indicator originally proposed by Luikart et al. (1998). Loss of rare alleles in bottlenecked populations is detected when one or more of the common allele classes have a higher number of alleles than the rare allele class (Luikart et al., 1998). These two methods were applied using Bottleneck v1.2.02 (http://www.ensam.inra.fr/URLB).
3. Results and discussion
Maintenance of genetic diversity is the major objective in conservation programs, so that population can face environmental challenges in the future and can respond to long term selection, either natural or artificial for traits of economic and cultural interest. Thus in this study, we characterized genetics of the Tibetan sheep based on 25 microsatellite loci in order to generate information for their conservation. Of course, it is better to obtain as many individual data as possible to understand the current status of the population in terms of genetic diversity. However, in case of Tibetan sheep, only 215 animals are left (Kumar, 2015) which are reared in quite inaccessible region, the higher reaches of the North District of Sikkim state. Thus genomic data on twenty animals may be considered sufficient to evaluate genetic diversity of highly endangered sheep breed.
3.1. Genetic variability of microsatellite loci
All the markers were polymorphic and a total of 148 alleles were detected across the 25 loci. An exact test for genotypic linkage disequilibrium yielded no significant P values across the population, and therefore independent assortment of all the loci was assumed. Reasonable amount of polymorphism in Tibetan sheep breed was evident from the allele frequency data (available on request) with the mean number of alleles (MNA) of 5.920 ± 0.387 (Table 2). BM6526 showed the highest number of observed alleles per locus (11) while BM8125, OarVH72 and BM6506 showed the lowest (3). Expected number of alleles varied from 1.490 (BM8125) to 8.667 (BM6526) with the mean of 3.70 (Table 2). Thus selection of microsatellites with a range of polymorphism reduced the risk of overestimating genetic variability, which might occur with the selective use of highly polymorphic loci. A microsatellite preferably should have at least 4 alleles to be useful for the evaluation of genetic diversity as per the standard selection of microsatellites loci (Barker, 1994). However, 3 alleles per locus have also been used to evaluate genetic diversity (Li et al., 2010). Therefore all the 25 microsatellites were retained for further analysis.
Table 2.
Observed and effective number of alleles, information index, observed and expected heterozygosity, FIS and average estimates of polymorphic microsatellite loci in Tibetan sheep.
| Locus | Na | Ne | I | Ho | He | uHe | FIS | PIC | χ2 value |
|---|---|---|---|---|---|---|---|---|---|
| BM0757 | 4 | 2.926 | 1.215 | 0.625 | 0.658 | 0.679 | 0.050 | 0.609 | 8.907 |
| BM8125 | 3 | 1.490 | 0.609 | 0.167 | 0.329 | 0.338 | 0.493 | 0.301 | 19.583a |
| BM0827 | 6 | 2.586 | 1.249 | 0.400 | 0.613 | 0.634 | 0.348 | 0.570 | 22.960 |
| CSSM47 | 4 | 1.800 | 0.855 | 0.222 | 0.444 | 0.471 | 0.500 | 0.411 | 21.130a |
| MAF214 | 6 | 4.800 | 1.651 | 0.250 | 0.792 | 0.826 | 0.684 | 0.760 | 36.078a |
| OarCP49 | 8 | 3.587 | 1.621 | 0.750 | 0.721 | 0.740 | − 0.040 | 0.695 | 35.075 |
| OarCP20 | 5 | 2.658 | 1.198 | 0.900 | 0.624 | 0.640 | − 0.443 | 0.576 | 13.388 |
| OarHH41 | 5 | 2.855 | 1.237 | 0.500 | 0.650 | 0.668 | 0.230 | 0.598 | 7.036 |
| OarHH47 | 6 | 4.781 | 1.662 | 0.421 | 0.791 | 0.812 | 0.468 | 0.762 | 45.735a |
| OarVH72 | 3 | 2.067 | 0.785 | 0.250 | 0.516 | 0.529 | 0.516 | 0.406 | 7.418 |
| BM6526 | 11 | 8.667 | 2.269 | 0.462 | 0.885 | 0.920 | 0.478 | 0.863 | 79.387a |
| INRA63 | 7 | 5.635 | 1.800 | 0.667 | 0.823 | 0.846 | 0.189 | 0.800 | 27.088 |
| OarAE129 | 4 | 2.941 | 1.221 | 0.600 | 0.660 | 0.733 | 0.091 | 0.610 | 6.800 |
| OarCP34 | 7 | 5.505 | 1.829 | 0.563 | 0.818 | 0.845 | 0.313 | 0.800 | 31.813 |
| OarFCB128 | 7 | 4.966 | 1.736 | 0.250 | 0.799 | 0.833 | 0.687 | 0.772 | 40.245a |
| HSC | 5 | 3.375 | 1.353 | 0.722 | 0.704 | 0.724 | − 0.026 | 0.654 | 28.439a |
| OarHH35 | 6 | 3.879 | 1.575 | 0.250 | 0.742 | 0.792 | 0.663 | 0.715 | 31.020a |
| OarHH64 | 6 | 4.346 | 1.578 | 0.412 | 0.770 | 0.793 | 0.465 | 0.732 | 42.689a |
| OarJMP029 | 9 | 5.333 | 1.871 | 0.750 | 0.813 | 0.833 | 0.077 | 0.788 | 59.949a |
| OarJMP08 | 7 | 4.042 | 1.607 | 0.412 | 0.753 | 0.775 | 0.453 | 0.716 | 33.819a |
| BM1314 | 5 | 1.681 | 0.797 | 0.150 | 0.405 | 0.415 | 0.630 | 0.368 | 20.497a |
| BM6506 | 3 | 1.629 | 0.644 | 0.250 | 0.386 | 0.396 | 0.353 | 0.329 | 5.788 |
| CSRD247 | 6 | 2.676 | 1.192 | 0.650 | 0.626 | 0.642 | − 0.038 | 0.557 | 28.553a |
| CSSM31 | 8 | 5.348 | 1.818 | 0.842 | 0.813 | 0.835 | − 0.036 | 0.787 | 20.559 |
| OarFCB48 | 7 | 2.935 | 1.383 | 0.368 | 0.659 | 0.677 | 0.441 | 0.620 | 27.125 |
| Mean | 5.920 | 3.700 | 1.390 | 0.473 | 0.672 | 0.696 | 0.302 | 0.632 | |
| SE | 0.387 | 0.334 | 0.085 | 0.044 | 0.030 | 0.032 | 0.057 | 0.032 |
Na = No. of different alleles.
Ne = No. of effective alleles = 1/(Sum pi^2).
I = Shannon's Information Index = − 1 ∗ Sum (pi ∗ Ln (pi)).
Ho = observed heterozygosity = no. of Hets/N.
He = expected heterozygosity = 1 − Sum pi^2.
uHe = unbiased expected heterozygosity = (2 N/(2 N − 1)) ∗ He.
FIS = Fixation Index = (He − Ho)/He = 1 − (Ho/He).
Pi is the frequency of the ith allele for the population & Sum pi^2 is the sum of the squared population allele frequencies.
PIC = polymorphism information content.
Significant deviation from Hardy–Weinberg equilibrium (P < 0.05).
Allelic diversity (mean number of observed alleles per locus) higher than that observed in Tibetan sheep has been reported in some of the Indian sheep breeds (Arora et al., 2008, Kumarasamy et al., 2009, Radha et al., 2011, Sharma et al., 2010). However, MNA in Tibetan breed is higher than that observed in several other Indian sheep breeds viz. 4.94 in Madras Red sheep (Prema et al., 2008), 5.0 in Nilagiri (Girish et al., 2007), 5.04 in Muzaffarnagari (Arora and Bhatia, 2004), 5.3 in Kheri (Bhatia and Arora, 2008), 5.6 in Shahabadi (Pandey et al., 2009), 5.7 in Magra (Arora and Bhatia, 2006) and 5.88 in Vembur (Pramod et al., 2009).Thus it can be concluded that sufficient genetic variability exists in Tibetan sheep despite its decreasing population trend.
The polymorphic information content (PIC) of a marker reveals its usefulness in diversity analysis of a breed. Following the criteria of Botstein et al. (1980), 84% of the investigated markers were observed to be highly informative (PIC > 0.5), 12% as reasonably informative (0.25 < PIC < 0.5) and only 4% were slightly informative (PIC < 0.25) across Tibetan breed (Table 2). An average value of 0.63 for the PIC once again indicated abundant genetic diversity in this population. The higher PIC further indicated the utility of these markers for population assignment (MacHugh et al., 1997) as well as genome mapping (Kayang et al., 2002) studies in addition to genetic diversity analysis.
3.2. Heterozygosity and Hardy–Weinberg equilibrium
Genetic variability is also measured as the amount of actual or potential heterozygosity, which is presented in Table 2. Expected heterozygosity was higher than the observed heterozygosity at all the loci. The observed and expected heterozygosity values ranged from 0.150 (BM1314) to 0.842 (CSSM31) and from 0.329 (BM8125) to 0.885 (BM6526) with an overall mean of 0.473 ± 0.044 and 0.672 ± 0.030, respectively. Genetic variation of similar magnitude (0.46–0.55) has also been reported in the endangered Namaqua Afrikaner sheep indigenous to South Africa (Qwabe et al., 2015). Whereas, higher heterozygosity has been reported in the two endangered Spanish breeds, Churra tensina (0.659) and Churra lebrijana (0.674) despite their small population size (Calvo et al., 2011). Most of the Indian sheep breeds with sufficient population exhibit higher heterozygosity (Ho) than the Tibetan sheep (Arora et al., 2011). Tibetan belongs to the sheep breeds of North temperate region of India. It has the least diversity estimates among the breeds of North temperate region also (Table 3). Similarly, much higher heterozygosity has been reported in several exotic sheep viz. Balearic sheep breeds (0.62, Pons and Landi, 2015), 12 sheep breeds of Croatia, Bosnia and Herzegovina (0.643–0.743, Salamon et al., 2014) and Turkish sheep breeds (0.66, Yilmaz and Sezenler, 2015). However, in assessing diversity estimates from different studies, it should be mentioned that the values are not directly comparable, as different microsatellite sets have been used by different workers. These values have only suggestive indication of diversity in the population.
Table 3.
Genetic diversity indices across sheep breeds of Northern temperate region of India.
| Breed | No. of alleles |
Heterozygosity |
Reference | ||
|---|---|---|---|---|---|
| Observed | Effective | Observed | Expected | ||
| Rampur Bushair | 6.000 | 3.471 | 0.515 | 0.675 | Pandey et al. (2008) |
| Gurej | 6.280 | 3.340 | 0.490 | 0.660 | Gupta et al. (2007) |
| Karnah | 6.830 | 3.960 | 0.530 | 0.680 | Gupta and Gannai (2007) |
| Changthangi | 8.760 | 4.539 | 0.691 | 0.716 | Sharma et al. (2010) |
| Tibetan | 5.920 | 3.700 | 0.473 | 0.672 | Present study |
Observed heterozygosity was lower than the expected showing a departure from Hardy–Weinberg equilibrium (HWE) and possibility of inbreeding. Significant deviation from HWE was observed in 13 out of 25 loci at P < 0.05 (Table 2). Departures from HWE of the similar magnitude were also reported in the endangered Spanish sheep breeds (Calvo et al., 2011), Italian (Ceccobelli et al., 2015), Turkish (Yilmaz and Sezenler, 2015) and in various Indian sheep breeds (Radha et al., 2011). Various factors can contribute towards excess of homozygotes. First, the locus is under selection. Second, ‘null alleles’ may be present which are leading to a false observation of excess homozygotes. Third, inbreeding may be common in the population. Fourth, the presence of population substructure may lead to Wahlunds' effect. (Nei, 1987, Peter et al., 2007). Distinguishing among these is generally difficult (Christiansen et al., 1974). The likelihood of each of these explanations must be assessed from additional data, such as demographic information, i.e. population distribution. Null alleles are most unlikely to be segregating at all the loci. Similarly possible Wahlunds' effects may not account significantly to the observed heterozygote deficit, as all the six flocks were at the same location. Ewens–Watterson Test for Neutrality revealed that all the microsatellite except OarHH47 and INRA63 (Table S1, observed F values lie outside of the upper and lower limits of 95% confidence region of the expected F values) were neutral. Since 92% loci were neutral, selection as a cause of the decrease in observed heterozygosity was ruled out. Thus the difference between the observed and expected heterozygosity can be attributed to the non-random mating among the individuals of the population. This was also reflected in the positive FIS value (0.302 ± 0.057) which ranged from − 0.443 to 0.687. Similar to our assumptions, the high FIS value (0.143) in the Spanish mouflon was considered as the major cause of the Hardy–Weinberg disequilibrium in that population (Calvo et al., 2011).
Higher heterozygote deficiency is generally observed in Indian sheep breeds; Magra (16%, Arora and Bhatia, 2006); Shahabadi (21.5%, Pandey et al., 2009); Rampur Bushair (22.7%, Pandey et al., 2008), and sheep breeds of Rajasthan-Nali (28.4%) and Chokla (28.6%) (Sodhi et al., 2006). However, Tibetan sheep (30.2%) exhibited the highest heterozygote deficiency among the Indian breeds, investigated so far. Authors agree that the major factor contributing towards the observed heterozygote deficiency in Tibetan sheep may be inbreeding due to extremely small population (< 250) and ignorance of sheep owners about the scientific management. Under field condition, only natural mating is being practiced where the dominant male generally excludes subordinate males, and presumably sires most of the offspring. The general breeding practice in the region was to castrate most of the males. This lead to reduced effective population size/or mating between relatives and consequent genetic drift. The consanguinity produced by mating between relatives can be one of the principal causes for loss of heterozygotes, but has to be evaluated with caution, as inbreeding with significant deficit of heterozygote affect all or most of the loci in a similar way. Since eighty percent of loci depicted heterozygote deficit (Table 2), therefore we might consider consanguinity as the foremost cause of heterozygote deficiency. Salamon et al. (2014) have also concluded that breeding practice lead to the higher FIS value in the study involving 12 eastern Adriatic and western Dinaric native sheep breeds. Such a high level of inbreeding is risky as it could lead to genetic diseases and moreover can adversely affect animal fitness.
3.3. Genetic bottleneck analysis
Bottleneck influences the distribution of genetic variation within and among populations. Population of Tibetan sheep has gone down drastically, which indicates the possibility of demographic bottleneck. In recently bottlenecked populations, the majority of loci will exhibit an excess of heterozygotes, exceeding the heterozygosity expected in a population at mutation drift equilibrium. To estimate the excess of such heterozygosity Sign, Standardized differences and Wilcoxon sign rank tests were utilized. The actual mutation model of evolution followed by our microsatellites is not known, thus all the three models (IAM, TPM and SMM) were selected for running the Bottleneck program. The values of average heterozygosity (He) and their probabilities (H > He) in the Sign test, under three models of microsatellite evolution were calculated and used to measure the expected number of loci with heterozygosity excess (Table 4). The expected numbers of loci with heterozygosity excess were 14.77, 14.94 and 14.62 in IAM, TPM and SMM with probabilities of 0.13211, 0.10276 and 0.39403 respectively, meaning that the null hypothesis was accepted when using the Sign test. These results indicate that, due to mutation-drift equilibrium, the Tibetan population has not undergone a recent genetic bottleneck. The standardized difference test provided the T2 statistics equal to 2.252, 0.269 and − 2.667 for the IAM, TPM and SMM models, respectively. The probability values were less than 0.05 for IAM and SMM, thus hypothesis of mutation-drift equilibrium was accepted under TPM only. Using the Wilcoxon rank test (a non-parametric test) the probability values were 0.00441 (IAM), 0.28009 (TPM) and 0.91775 (SMM) under these three models, indicating that the null hypothesis is accepted under TPM and SMM and thus the population under study has not undergone a recent bottleneck. It has been considered that the most useful markers for bottleneck detection are those evolving under IAM, and they provide guidelines for selecting sample sizes of individuals and loci (Cornuet and Luikart, 1996, Di Rienzo et al., 1994, Spencer et al., 2000); meanwhile, the TPM is thought to more closely simulate microsatellite mutation (Estoup and Cornuet, 2000). Unlike the SMM, which predicts all mutations corresponding to the increment or decrement of a single base-pair repeat, the TPM predicts the occurrence of an occasional multiple base-pair repeat (Di Rienzo et al., 1994). The strict SMM is obviously the most conservative model for testing for a significant heterozygosity excess caused by bottlenecks, because in some conditions it can produce a heterozygosity deficiency, and due to the heterozygosity excess it is always lower than other mutation models. Thus we have considered results from all the three tests together and it is clear that serious demographic bottlenecks have most probably not occurred in this breed.
Table 4.
Population bottleneck analysis under three microsatellite evolution models.
| Test/model | I.A.M. | T.P.M. | S.M.M. | |
|---|---|---|---|---|
| Sign test (number of loci with heterozygosity excess) | Exp | 14.77 | 14.94 | 14.62 |
| Obs | 18 | 15 | 11 | |
| P-value | 0.13211 | 0.10276 | 0.39403 | |
| Standardized differences test | T2 value | 2.252 | 0.269 | − 2.667 |
| P-value | 0.01216a | 0.39403 | 0.00382a | |
| Wilcoxon rank test (one tail for heterozygosity excess) | P-value | 0.00441a | 0.28009 | 0.91775 |
Bottleneck (rejection of null hypothesis of mutation drift equilibrium).
The Mode-shift indicator test was also utilized as a second method to detect potential bottlenecks, as the non-bottleneck populations that are near mutation-drift equilibrium are expected to have a large proportion of alleles with low frequency. This test discriminates many bottlenecked populations from stable populations (Luikart, 1997, Luikart and Cornuet, 1997). A graphical representation utilizing allelic class and proportion of alleles showed a normal ‘L’ shaped distribution (Fig. 2). The L shaped curve indicated the abundance of low frequency (< 0.10) alleles. This finding suggested the absence of any detectably large, recent genetic bottleneck (last 40–80 generations) in declining population, where the probability of low frequency allele's loss was very high. Taken together all the results indicate the absence of bottleneck events in the recent past history of this breed.
Fig. 2.
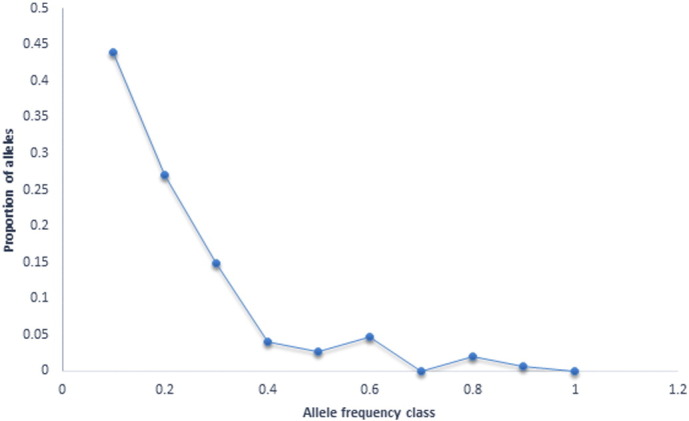
Graphic representation of proportion of alleles and their distribution in Tibetan sheep.
Shrinkage of pasture, inbreeding, absence of scientific animal husbandry practices and management in very harsh climate have resulted in diminished profits for the Tibetan sheep herders. As a result, sheep rearing has failed to attract younger generations. Additionally, lack of proper policy is leading to rapid decline of Tibetan sheep population which are less than two hundred and fifty at present. Therefore, timely intervention is required to prevent extinction of this valuable breed of sheep, which helps in sustaining the livelihood of highlanders and the fragile agro-ecosystem. The information reported here is important for sheep breeders for the establishment of conservation strategies.
4. Conclusions
The present work for the first time generated the knowledge of existing genetic diversity in the endangered Tibetan sheep based on microsatellite analysis. It is a unique gene pool with good adaptation to the extremely cold and low oxygen conditions. Genetic parameters show a high value of inbreeding and therefore this breed should be monitored due to very low number of individuals that compose it. Still there is scope for reviving the breed because of high allele diversity which suggests that unique alleles present in this breed may not have been lost and the absence of bottleneck.
The following are the supplementary data related to this article.
The Ewens–Watterson test for neutrality.
Acknowledgments
This work was financially supported by the Network Project on Animal Genetic Resources (ICAR).
References
- Acharya R.M. FAO Animal Production and Health Paper 30. FAO of United Nations; Rome, Italy: 1982. Sheep and goat breed of India. [Google Scholar]
- Arora R., Bhatia S. Genetic structure of muzzafarnagri sheep based on microsatellite analysis. Small Rumin. Res. 2004;54:227–230. [Google Scholar]
- Arora R., Bhatia S. Genetic diversity of Magra sheep from India using microsatellite analysis. Asian Australas. J. Anim. Sci. 2006;19:938–942. [Google Scholar]
- Arora R., Bhatia S. Genetic variability in Jalauni sheep of India inferred from microsatellite data. Livest. Res. Rural. Dev. 2008;20 (Article no. 4 retrieved from www.cipav.org.co/lrrrd20/1/ aror20004.htm) [Google Scholar]
- Arora R., Bhatia S. Diversity analysis of sheep breeds from Southern peninsular and Eastern regions of India. Trop. Anim. Health Prod. 2011;43:401–408. doi: 10.1007/s11250-010-9706-z. [DOI] [PubMed] [Google Scholar]
- Banerjee S. Shift from transhumance and subtle livelihood patterns of the bhotia community and its impact on Tibetan sheep population in Sikkim (India) World Appl. Sci. J. 2009;7:1540–1546. [Google Scholar]
- Barcaccia G., Felicetti M. Molecular analysis of genetic diversity, population structure and inbreeding level of the Italian Lipizzan horse. Livest. Sci. 2013;151:124–133. [Google Scholar]
- Barker J.S.F. Proceedings of the 5th World Congress on Genetics Applied to Livestock Production, Guelph and Ontario, Canada. Vol. 21. 1994. A global protocol for determining genetic distances among domestic livestock breeds; pp. 501–508. [Google Scholar]
- Bhatia S., Arora R. Biodiversity and conservation of Indian sheep genetic resources — an overveiw. Asian Australas. J. Anim. Sci. 2005;18:1387–1402. [Google Scholar]
- Bhatia S., Arora R. Genetic diversity in Kheri — a pastoralists developed Indian sheep using microsatellite markers. Indian J. Biotechnol. 2008;7:108–112. [Google Scholar]
- Botstein D., White R.L. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Calvo J.H., Alvarez-Rodriguez J. Genetic diversity in the Churra tensina and Churra lebrijana endangered Spanish sheep breeds and relationship with other Churra group breeds and spanish mouflon. Small Rumin. Res. 2011;95:34–39. [Google Scholar]
- Ceccobelli S., Karsli T. Genetic diversity of cornigliese sheep breed using STR markers. Small Rumin. Res. 2015;123:62–69. [Google Scholar]
- Christiansen F.B., Frydenberg O. Genetics of Zoarces populations VI. Further evidence, based on age group samples, of a heterozygote deficit ESTIII polymorphism. Hereditas. 1974;77:225–236. [PubMed] [Google Scholar]
- Cornuet J.M., Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo A., Peterson A.C. Mutational processes of simple-sequence repeat loci in human populations. Proc. Natl. Acad. Sci. U. S. A. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estoup A., Cornuet J.M. Microsatellite evolution: inferences from population data. In: Goldstein D.B., Schlotterer C., editors. Microsatellites: Evolution and Applications. Oxford University Press; Oxford: 2000. pp. 49–65. [Google Scholar]
- FAO Secondary guidelines for development of national farm animal genetic resources management plans for global management of cattle genetic resources using reference microsatellites global projects for the maintenance of domestic animal genetic diversity (MoDAD) 2004. http://www.fao.org/dad-is/
- Ghazyl A., Mokhtar S. Genetic diversity and distances of three Egyptian local sheep breeds using microsatellite markers. Zoology. 2013;3:1–9. [Google Scholar]
- Girish H., Sivaselvam S.N. Molecular characterisation of Nilagiri sheep (Ovis aries) of south India based on microsatellites. Asian Australas. J. Anim. Sci. 2007;20:633–637. [Google Scholar]
- Government of Bengal. Proceedings of Bengal Government General. July 1864. In: Wake H.C., editor. Superintendent of Darjeeling, to Undersecretary to Govt. of Bengal. Vol. 277. 1864. pp. 47–57. [Google Scholar]
- Gupta N., Gannai T.A.S. National Bureau of Animal Genetic Resources; Karnal, India: 2007. Sheep Genetic Resources of India. Karanah — A Finest Wool Breed, Monograph 50. [Google Scholar]
- Gupta S.C., Gannai T.A.S. National Bureau of Animal Genetic Resources; Karnal, India: 2007. Sheep Genetic Resources of India. Gurez — An Endangered Breed, Monograph 49. [Google Scholar]
- Kayang B.B., Inoue-Murayama M. Microsatellite loci in Japanese quail and cross-species amplification in chicken and guinea fowl. Genet. Sel. Evol. 2002;34:233–253. doi: 10.1186/1297-9686-34-2-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B. Proceedings of 13th Annual Review Meet of Network Project on Animal Genetic Resources (ICAR), Karnal, India. Vol. 4. 2015. Characterization of Tibetan sheep. [Google Scholar]
- Kumarasamy P., Prema S. Molecular characterization of Coimbatore breed of sheep (Ovis aries) in South India. IUP J. Genet. & Evol. 2009;2:56–65. [Google Scholar]
- Li H.F., Song W.T. Genetic diversity and population structure of 10 Chinese indigenous egg-type duck breeds assessed by microsatellite polymorphism. J. Genet. 2010;89:65–72. doi: 10.1007/s12041-010-0012-3. [DOI] [PubMed] [Google Scholar]
- Livestock Census . Ministry of Agriculture, Government of India; Krishi Bhavan, New Delhi: 2012. BAHS—Basic Animal Husbandry Statistics: Department of Animal Husbandry, Dairying & Fisheries. [Google Scholar]
- Luikart G. University of Montana; Missoula, USA: 1997. Usefulness of Molecular Markers for Detecting Population Bottlenecks and Monitoring Genetic Change. (Ph. D. thesis) [DOI] [PubMed] [Google Scholar]
- Luikart G., Cornuet J.M. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv. Biol. 1997;12:228–237. [Google Scholar]
- Luikart G.L., Allendrof F.W. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 1998;89:238–247. doi: 10.1093/jhered/89.3.238. [DOI] [PubMed] [Google Scholar]
- MacHugh D.E., Shriver M.D. Microsatellite DNA variation and the evolution domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus) Genetics. 1997;146:1071–1086. doi: 10.1093/genetics/146.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York USA: 1987. Molecular Evolutionary Genetics. [Google Scholar]
- Pandey A.K., Sharma R. Genetic variability in Rampur–Bushair sheep breed using microsatellite marker. Indian J. Anim. Sci. 2008;78:623–626. [Google Scholar]
- Pandey A.K., Sharma R. Variation of 18 STR loci in Shahabadi sheep of India. Russ. J. Genet. 2009;45:1–7. [PubMed] [Google Scholar]
- Peakall R., Smouse P.E. A heterogeneity test for fine-scale genetic structure. Mol. Ecol. Notes. 2008;17:3389–3400. doi: 10.1111/j.1365-294x.2008.03839.x. [DOI] [PubMed] [Google Scholar]
- Peter C., Bruford M. Genetic diversity and subdivision of 57 European and Middle–Eastern sheep breeds. Int'l. Soc. Anim. Genet. 2007;38:37–44. doi: 10.1111/j.1365-2052.2007.01561.x. [DOI] [PubMed] [Google Scholar]
- Pons A.L., Landi V. The biodiversity and genetic structure of balearic sheep breeds. J. Anim. Breed. Genet. 2015;132:268–276. doi: 10.1111/jbg.12129. [DOI] [PubMed] [Google Scholar]
- Pramod S., Kumarasamy P. Molecular characterization of Vembur sheep (Ovis aries) of South India based on microsatellites. Indian J. Sci. Technol. 2009;2:55–58. [Google Scholar]
- Prema S., Sivaselvam S.N. A note on genetic analysis in Madras Red sheep (Ovis aries) of India using microsatellite markers. Livest. Res. Rural. Dev. 2008;20:181–185. [Google Scholar]
- Putman A.I., Carbone I. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol. Evol. 2014;4:4399–4428. doi: 10.1002/ece3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qwabe S.O., Marle-Köster E.V. Genetic diversity and population structure of the endangered Namaqua Afrikaner sheep. J. Anim. Breed. Genet. 2015 doi: 10.1007/s11250-012-0250-x. [DOI] [PubMed] [Google Scholar]
- Radha P., Sivaselvam S.N. Genetic diversity and bottleneck analysis of Kilakarsal sheep by microsatellite markers. Indian J. Biotechnol. 2011;10:52–55. [Google Scholar]
- Salamon D., Gutierrez-Gil B. Genetic diversity and differentiation of 12 eastern Adriatic and western Dinaric native sheep breeds using microsatellites. Animal. 2014;8:200–207. doi: 10.1017/S1751731113002243. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F. Cold Spring Harbour Lab. Press; Cold Spring Harbour, NY: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Sharma R., Pandey A.K. Microsatellite based diversity estimation of Changthangi — a high altitude sheep breed. Indian J. Anim. Sci. 2010;80:436–440. [Google Scholar]
- Sodhi M., Mukesh M. Characterizing Nalli and Chokla sheep differentiation with microsatellite markers. Small Rumin. Res. 2006;65:185–192. [Google Scholar]
- Spencer C.C., Neigel J.E. Experimental evaluation of the usefulness of microsatellite DNA for detecting demographic bottlenecks. Mol. Ecol. 2000;9:1517–1528. doi: 10.1046/j.1365-294x.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- Yeh F.C., Yang R.C. University of Alberta; Edmonton: 1999. POPGENE Version 1.31: Microsoft Window-based Free Software for Population Genetic Analysis. [Google Scholar]
- Yilmaz O., Sezenler T. Genetic relationships among four Turkish sheep breeds using microsatellites. Turk. J. Vet. Anim. Sci. 2015;39:576–582. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Ewens–Watterson test for neutrality.