Abstract
Pseudomonas aeruginosa causes acute and chronic infections in human. Its increasing resistance to antibiotics requires alternative treatments that are more effective than available strategies. Among the alternatives is the unconventional usage of conventional antibiotics, of which the macrolide antibiotic azithromycin (AZM) provides a paradigmatic example. AZM therapy is associated with a small but consistent improvement in respiratory function of cystic fibrosis patients suffering from chronic P. aeruginosa infection. Besides immunomodulating activities, AZM represses bacterial genes involved in virulence, quorum sensing, biofilm formation, and motility, all of which are due to stalling of ribosome and depletion of cellular tRNA pool. However, how P. aeruginosa responds to and counteracts the effects of AZM remain elusive. Here, we found that deficiency of PA3297, a gene encoding a DEAH-box helicase, intensified AZM-mediated bacterial killing, suppression of pyocyanin production and swarming motility, and hypersusceptibility to hydrogen peroxide. We demonstrated that expression of PA3297 is induced by the interaction between AZM and ribosome. Importantly, mutation of PA3297 resulted in elevated levels of unprocessed 23S-5S rRNA in the presence of AZM, which might lead to increased susceptibility to AZM-mediated effects. Our results revealed one of the bacterial responses in counteracting the detrimental effects of AZM.
Keywords: RNA helicase, antibiotic resistance, azithromycin, rRNA processing, Pseudomonas aeruginosa
Introduction
Pseudomonas aeruginosa is a versatile Gram-negative pathogenic bacterium, that can cause various infections in human (de Bentzmann and Plésiat, 2011; Campa et al., 2012). During infection, P. aeruginosa produces multiple virulence factors to facilitate colonization (Sadikot et al., 2005; Kipnis et al., 2006; Hauser, 2009; Liu et al., 2015). Meanwhile, its highly intrinsic antibiotic resistance and biofilm forming ability greatly hinder the eradication of this pathogen (Høiby et al., 2005). In patients suffering from cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD), P. aeruginosa caused chronic respiratory infections are responsible for most of the morbidity and mortality (Rabin et al., 2004; Rada and Leto, 2013). Intensive antibiotic treatment has been used to maintain the lung function and extend lifespan of the patients (Doring et al., 2000). However, the increasing antibiotic resistance has been compromising clinical efficacy of traditional antibiotics. Thus, alternatives or unconventional usage of the antibiotics are urgently needed (Breidenstein et al., 2011; Poole, 2011; Imperi et al., 2014).
The macrolide antibiotic azithromycin (AZM) provides a paradigmatic example of an unconventional antibacterial drug for P. aeruginosa treatment. Although, P. aeruginosa is highly resistance to macrolides owning to its low outer membrane permeability and the resistance-nodulation-cell division (RND) systems, AZM treatment benefits patients suffering from both intermittent and chronic P. aeruginosa infections (Saiman et al., 2003; Lister et al., 2009; Blasi et al., 2010; Steel et al., 2012; Aminov, 2013; Morita et al., 2013). AZM has been shown to have immunomodulatory activity, which attenuates the inflammatory response and promotes macrophage phagocytic activity (Legssyer et al., 2006; Steinkamp et al., 2008; Tsai et al., 2009). Furthermore, AZM exhibits bactericidal effect on stationary growth phase P. aeruginosa cells (Lovmar et al., 2004, 2009; Imamura et al., 2005; Köhler et al., 2007; Starosta et al., 2010; Gödeke et al., 2013). And sub-inhibitory concentrations of AZM suppress biofilm formation, motility, and production of multiple virulence factors, including proteases, pyocyanin, exotoxin A, phospholipase C (PLC), exopolysaccharides, and other quorum-sensing (QS) regulated genes in P. aeruginosa (Molinari et al., 1992, 1993; Tateda et al., 2001; Favre-Bonté et al., 2003; Gillis and Iglewski, 2004). The AZM-mediated killing of stationary-phase bacterial cells and reduced expression of QS-regulated virulence factors require interaction between AZM and ribosome (Köhler et al., 2007). AZM binds in the nascent peptide exit tunnel (NPET), resulting in ribosome stalling and depletion of the intracellular pools of aminoacyl-tRNAs (Lovmar et al., 2004, 2009; Köhler et al., 2007; Starosta et al., 2010; Gödeke et al., 2013). The effects of AZM on P. aeruginosa can be counteracted by over expression of ErmBP or a peptidyl-tRNA hydrolase, which blocks the interaction between AZM and ribosome by modifying the 23S rRNA or increases the intracellular aminoacyl-tRNA level, respectively (Köhler et al., 2007; Gödeke et al., 2013). However, how P. aeruginosa response to AZM treatment remains unclear. Understanding the mechanisms that P. aeruginosa uses to counteract AZM treatment may provide clues to enhance AZM-mediated virulence inhibitory and bacterial killing effects.
A large RNA helicase family named DExD/H box helicases are characterized by a conserved DExD/H box sequence (Cordin et al., 2006; Linder and Jankowsky, 2011), and play crucial roles in rRNA processing, translation initiation, and mRNA decay (Iost et al., 2013; Linder and Fuller-Pace, 2013). In addition, the DExD/H box helicases have been shown to participate in bacterial responses to various stresses, such as cold shock, pH, osmotic, and oxidative stresses (Owttrim, 2013). And several DEAD family RNA helicases, which belong to a specific subfamily of DExD/H box helicases, have been shown to regulate virulence factors in Escherichia coli, Borrelia burgdorferi, Staphylococcus aureus, Listeria monocytogenes, and P. aeruginosa (Koo et al., 2004; Salman-Dilgimen et al., 2011, 2013; Oun et al., 2013; Bareclev et al., 2014; Intile et al., 2015). The pleiotropic functions of DExD/H box family RNA helicases intrigued us to suspect that they might be involved in the bacterial response to AZM treatment. In this study, we found that deficiency in a DEAH box helicase, PA3297, renders P. aeruginosa more susceptible to the killing and virulence suppression by AZM. Our results suggest that the expression of PA3297 was up regulated in the presence of AZM, which might promote 23S rRNA maturation to counteract the inhibitory effect of AZM on protein elongation.
Materials and Methods
Strains and Plasmids
The bacterial strains and plasmids used in this study are listed in Table 1 (Simon et al., 1983; Hoang et al., 1998; Choi and Schweizer, 2006; Liberati et al., 2006). The E. coli strains DH5α, S17-1 and P. aeruginosa strains were routinely cultured in Luria-Bertani (LB) broth at 37°C. Antibiotics were used at the following concentrations: for E. coli, ampicillin 100 μg/ml, tetracycline 10 μg/ml, and gentamicin 10 μg/ml; for P. aeruginosa, carbenicillin 150 μg/ml, tetracycline 50 μg/ml, and gentamicin 50 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Descriptiona | Reference or origin |
|---|---|---|
| E. coli strains | ||
| DH5α | F-, φ80dlacZΔM15,Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk-,mk+), phoA, supE44, λ-, thi1, gyrA96, relA1 | TransGen |
| S17-1 | thi pro hsdR recA Tra+ | Simon et al., 1983 |
| P. aeruginosa strains | ||
| PA14 | Wild type Pseudomonas aeruginosa strain PA14 | Liberati et al., 2006 |
| PA0426::Tn | PA14 with a transposon inserted at PA0426 | Liberati et al., 2006 |
| PA0455::Tn | PA14 with a transposon inserted at PA0455 | Liberati et al., 2006 |
| PA2840::Tn | PA14 with a transposon inserted at PA2840 | Liberati et al., 2006 |
| PA3002::Tn | PA14 with a transposon inserted at PA3002 | Liberati et al., 2006 |
| PA3272::Tn | PA14 with a transposon inserted at PA3272 | Liberati et al., 2006 |
| PA3297::Tn | PA14 with a transposon inserted at PA3297 | Liberati et al., 2006 |
| PA3308::Tn | PA14 with a transposon inserted at PA3308 | Liberati et al., 2006 |
| PA3861::Tn | PA14 with a transposon inserted at PA3861 | Liberati et al., 2006 |
| PA3950::Tn | PA14 with a transposon inserted at PA3950 | Liberati et al., 2006 |
| ΔPA3297 | PA14 with PA3297 in frame deletion | This study |
| ΔPA3297/att7::PA3297 | PA14 ΔPA3297 with insertion of a single copy of PA3297 driven by its own promoter at attTn7 sites | This study |
| ΔPA3297/att7::PA3297 K101A | PA14 ΔPA3297 complemented with a single copy of PA3297 acquired a lysine101 mutation to alanine | This study |
| ΔPA3297/att7::PA3297 D192A | PA14 ΔPA3297 complemented with a single copy of PA3297 acquired an aspartate192 mutation to alanine | This study |
| ΔPA3297/att7::PA3297SAT224AAA | PA14 ΔPA3297 complemented with a single copy of PA3297 acquired a serine224 and threonine226 mutation to alanines | This study |
| Plasmids | ||
| pEX18Tc | Broad-host-range gene replacement vector; sacB TETr | Hoang et al., 1998 |
| pUC18t-mini-Tn7T-Gm | For gene insertion in chromosome; GENr | Choi and Schweizer, 2006 |
| pTNS3 | Helper plasmid | Choi and Schweizer, 2006 |
| pFLP2 | Source of Flp recombinase; sacB, AMPr/CARr | Hoang et al., 1998 |
| pTH1501 | pEX18Tc::ΔPA3297; TETr | This study |
| pTH1502 | PA3297 gene of PA14 on pUC18T-Mini-Tn7T-Gm with its own promoter; GENr | This study |
aAMPr, ampicillin resistant; TETr, tetracycline resistant; GENr, gentamicin resistance; CARr, carbenicillin resistant.
DNA Methods
DNA manipulations were performed according to standard protocols or following manufacturers’ instructions (Hoang et al., 1998; Zheng et al., 2004; Choi and Schweizer, 2006). The pEX18Tc::ΔPA3297 (pTH1501) was constructed by cloning the 1002-bp upstream and 964-bp downstream fragments of PA3297 coding region into the KpnI-HindIII sites of plasmid pEX18Tc. The fragments were amplified from the PA14 chromosome with primers PA14-UPA3297-FF, PA14-UPA3297-FR, PA14-DPA3297-FF, and PA14-DPA3297-FR (Table 2), respectively. Deletion of the PA3297 gene was confirmed by PCR with primers PA14-PA3297-FF and PA14-PA3297-FR (Table 2). For the complementation of PA3297, the PA3297 gene was amplified from the PA14 chromosome by PCR with the primers PA14-PA3297-FF and PA14-PA3297-FR (Table 2). The PCR product was ligated into the EcoRI- SacI sites of pUC18t-mini-Tn7T-Gm, resulting in pTH1502. The plasmid was introduced into the ΔPA3297 mutant by electroporation, along with the helper plasmid pTNS3 (Choi and Schweizer, 2006). Insertion of the PA3297 gene into the chromosome was confirmed by PCR with primers PTn7R and PglmS-down (Table 2; Choi and Schweizer, 2006). The site-directed mutagenesis was performed as previously described (Zheng et al., 2004). The mutation sites were chosen based on the conserved critical residues of other bacterial DExD-box proteins (Koo et al., 2004; Cordin et al., 2006). Briefly, PCR amplification was performed with pTH1502 as template and with primers listed in Table 2, for K101A, D192A, and SAT224-226AAA mutations, respectively. The PCR products were treated by DpnI for 3 h at 37°C and purified before transformation. The correctly mutated clones were identified by DNA sequencing.
Table 2.
Primers used in this study.
| Primersa | Nucleotide sequence (53)b |
|---|---|
| Cloning of upstream and downstream fragments for PA3297 deletion | |
| PA14-UPA3297-FF | GAAAGCGGTACCGAAGTAAGTCCGCCGTTGCC (Kpn I) |
| PA14-UPA3297-FR | CAGCTTTCTAGAGGTGCTGTCGTCGCTCTGGT (Xba I) |
| PA14-DPA3297-FF | TTGCAGTCTAGACGCTGGATGCTGGAGGAGTA (Xba I) |
| PA14-DPA3297-FR | CGCCGGAAGCTTCACCGAGCAGTGGCTGAAGAC (Hind III) |
| Cloning of gene PA3297 for complementation | |
| PA14-PA3297-FF | TGAAGAGAATTCGCCAGAAGTAAGTCCGCCGTTGCC (EcoR I) |
| PA14-PA3297-FR | CACCGGGAGCTCCGACCAGACCGACCTGTTCTTCACCAT (Sac I) |
| PTn7R | CACAGCATAACTGGACTGATTTC |
| PglmS-down | GCACATCGGCGACGTGCTCTC |
| Primers used for site-directed mutagenesis of PA3297 | |
| PA3297-K101A-FF | GCGAGACCGGCTCGGGC ACCACCCAG ACCACCCAG |
| PA3297-K101A-FR | T GCCCGAGCCGGTCTCGCCGGCGATC GCCCGAGCCGGTCTCGCCGGCGATC |
| PA3297-D192A-FF | TACGACACGCTGATCGTC GAAGCCCAC GAAGCCCAC |
| PA3297-D192A-FR | C GACGATCAGCGTGTCGTAGCGCTCCAG GACGATCAGCGTGTCGTAGCGCTCCAG |
| PA3297-SAT224-226AAA-FF | GCTGATCATCACC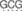 GCG GCG ATCGACCTGGAG ATCGACCTGGAG |
| PA3297-SAT224-226AAA-FR | T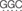 CGC CGC GGTGATGATCAGCTTCAGGTCC GGTGATGATCAGCTTCAGGTCC |
| RT-qPCR primers for gene expression measurements | |
| 23S-R-FF | AAAGATAACCGCTGAAAG |
| 23S-R-FR | CTATCAACGTCGTAGTCT |
| 5S-R-FF | CGAACTCAGAAGTGAAAC |
| 5S-R-FR | CTTGACGATGACCTACTC |
| 23S-5S-R-FF | GTACTAATTGCCCGTGAG |
| 23S-5S-R-FR | GTTCCAACGCTCTATGAT |
aThe direction of the primer is indicated at the end of the primer designation as follows: FF for forward and FR for reverse. bThe solid underlines are the sites of listed restriction enzymes; the dotted underlines are the sites for mutagenesis, with all the residues changed into alanine.
Assay for Pyocyanin Production
The pyocyanin concentration was determined as described previously (Essar et al., 1990). Briefly, 1 ml supernatant from each 24-h-old bacteria culture grown in the absence or presence of AZM was extracted with 0.5 ml of chloroform. Then, 0.4 ml solution from the lower organic phase was re-extracted into 0.3 ml of 0.2 N HCl to give a pink solution, whose absorbance was measured at 520 nm. Concentrations of pyocyanin (mg/ml) were calculated by multiplying the OD520 by 32.01 (Kurachi, 1958).
Antibiotic Susceptibility Assay
Minimum inhibitory concentrations (MICs) of P. aeruginosa to antimicrobial agents were determined by serial twofold broth dilution in LB medium, as described previously (Jo et al., 2003). MICs were recorded as the lowest concentration of antibiotic inhibiting visible growth after 24 h of incubation at 37°C.
Stationary-Phase Bacterial Cell Killing Assay
The killing assay was performed as described previously (Köhler et al., 2007). Briefly, bacteria were inoculated in LB medium and grown for 16 h at 37°C. The culture of each strain was diluted to an OD600 of 0.05 and cultured at 37°C. After reaching stationary phase (OD600, 3.0), indicated concentrations of AZM were added to the 2-ml aliquots of the cultures. Then the bacteria were cultured for 20–22 h at 37°C. The viable bacterial numbers were determined by serial dilution and plating on drug-free LB agar plates. The survival rate of each strain was calculated as live bacterial number in AZM treated sample divided by the bacterial number of the corresponding untreated sample.
Biofilm Tolerance to AZM
The biofilm resistance was measured as previously described with minor modifications (Bjerkan et al., 2009; Billings et al., 2013; Liao et al., 2013). Briefly, overnight bacterial cultures were diluted to an OD600 of 0.025. 150 μl of the bacteria were incubated in each well of a 96-well plate at 37°C without agitation (Brencic et al., 2009). After 24 h, the planktonic bacteria were discarded by aspiration. Then, the biofilms were treated with 150 μl LB medium containing indicated concentrations of AZM for 2 h. The medium in each well was replaced with fresh LB medium, and subjected to sonication at a frequency of 40 kHz, with a power output of 300 W, at 37°C for 5 min. The live bacteria were enumerated by serial dilution and plating.
Growth Assay
Overnight culture of each strain was diluted into fresh LB (150 μl) to an OD600 of 0.05 in each well of a 96-well plate without or with different concentrations of AZM. The plate was incubated at 37°C with constant agitation (Lau et al., 2012; Guénard et al., 2014). The bacterial growth was monitored by measuring the OD600 every 30 min for 12 h by a Varioskan Flash microplate reader (Thermo Electron Corporation).
H2O2 Susceptibility Assay
Overnight cultures of the P. aeruginosa strains were diluted to an OD600 of 0.05 and cultured at 37°C. When the OD600 reached 0.3 (about 1.5–2 h later), AZM (0.5 μg/ml) was added if needed. When the OD600 reached 2.0 (about 3 h later), bacteria from 500 μl culture were collected and washed twice with phosphate buffered saline (PBS). Then the bacteria were resuspended in PBS with or without 10% H2O2 and incubated for 15 min. The live bacterial numbers were determined by serial dilution and plating.
Motility Assay
The swarming motility was tested on modified M9 medium plates supplemented with 0.2% glucose, 1 mM MgSO4, and 0.05% glutamate as the nitrogen source. 0.5% agar was used for solidification. Two microliters of exponential growth phase P. aeruginosa was deposited on the plates, then incubated for 18 h at 37°C (Köhler et al., 2007).
Total RNA Isolation and Quantitative Real-Time PCR
Overnight cultures of P. aeruginosa strain PA14 and ΔPA3297 were diluted into fresh LB medium to an OD600 of 0.05. The bacteria were grown at 37°C with agitation (200 rpm). When the OD600 reached 0.3 (about 1.5–2 h later), AZM was added to reach indicated concentrations. Samples were harvested when the OD600 reached 2.0 (about 3 h later; Skindersoe et al., 2008). Total RNA was isolated with an RNeasy Minikit (Tiangen Biotech). The cDNA from each RNA sample was synthesized with reverse transcriptase and random primers (Takara). Real-time PCR was performed with SYBR premix Ex Taq (Roche). The conserved hypothetical protein coding gene PA1769 was used as an internal control (Son et al., 2007). The primers used in quantitative real-time PCR were listed in Table 2, with a designation of “RT-qPCR.”
Statistical Analysis
When indicated, Student’s t-test (two-tailed) was used to determine whether the deletion of PA3297 resulted in any significant differences compared to the wild-type cells treated with the same concentrations of AZM.
Results
Deficiency of PA3297 Intensifies the Effects of AZM on Pyocyanin Production
As pyocanin production is suppressed by AZM (Molinari et al., 1992, 1993; Tateda et al., 2001; Favre-Bonté et al., 2003; Gillis and Iglewski, 2004), which can be easily observed and quantified, we used this phenotype to test whether DExD/H box RNA helicases are involved in bacterial response to AZM treatment. There are 17 DExD/H box RNA helicases in the genome of P. aeruginosa strain PA14 (www.pseudomonas.com; Winsor et al., 2011). There are nine DExD/H box helicase mutants in the non-redundant PA14 transposon mutants library, however, the other eight DExD/H box helicase mutants are not available (Table 1; Liberati et al., 2006; Breidenstein et al., 2011). Thus, we examined pyocyanin production of the available mutants in the absence and presence of AZM at an OD600 of 2.0. A PA3297::Tn mutant displayed a significant decrease in pyocyanin production in the presence of AZM, whereas no difference was observed in the absence of AZM (Figure 1A). To confirm the role of PA3297, we generated an in frame deletion of PA3297 in PA14. The wild type PA14 and the ΔPA3297 mutant were grown in the absence or presence of 2, 5, or 10 μg/ml AZM. When the bacteria reached same density (OD600 of 2.0), we measured the pyocyanin levels. AZM inhibited the production of pyocyanin in the wild type strain in a dose dependent manner. However, the production of pyocyanin by the mutant was repressed more severely in the presence of AZM at all the tested concentrations (Figure 1B). Complementation with a PA3297 gene restored the production of pyocyanin (Figure 1B).
FIGURE 1.
Mutation of PA3297 intensified the AZM mediated inhibition of pyocyanin production. (A) Wild type PA14 and the mutants of DExD/H box helicases were grown to an OD600 of 2.0 in the absence or presence of 5 μg/ml AZM at 37°C and the pyocyanin concentrations were measured. (B) Bacteria of wild type PA14, ΔPA3297 mutant and the complementation strain were grown in the absence or presence of 2, 5, or 10 μg/ml AZM. When the OD600 reached 2.0, the pyocyanin concentrations were measured. The values are the means of three replicates and the error bars display the standard deviations. “ND” standards for “not detected.” ∗p < 0.05 compared to PA14 or the complemented strain by student’s t-test.
Increased Killing of the ΔPA3297 Mutant by AZM
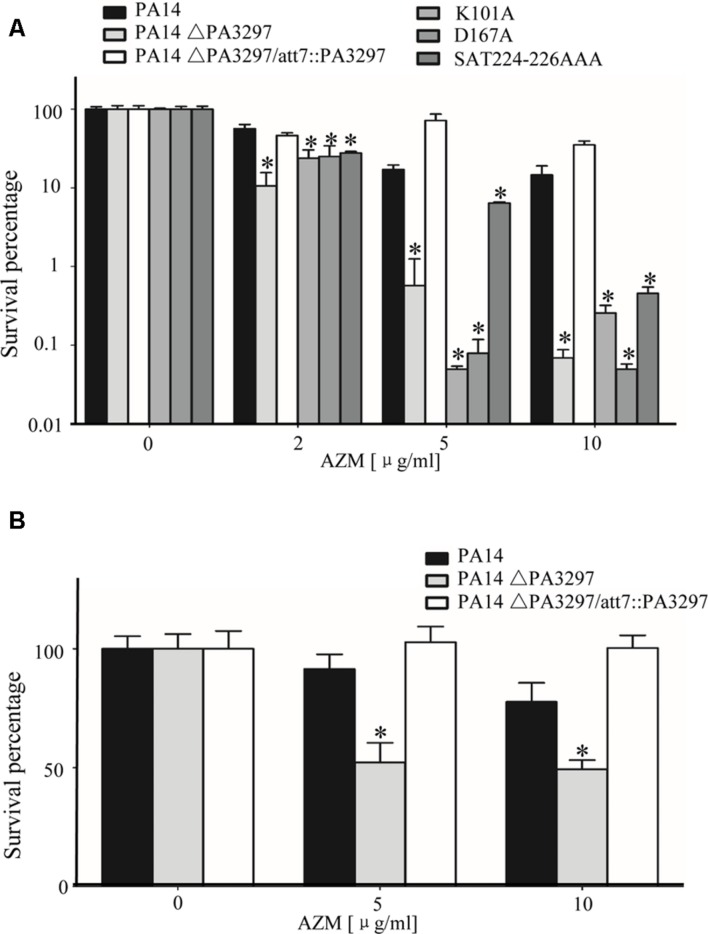
Stationary growth phase P. aeruginosa cells are susceptible to AZM (Lovmar et al., 2004, 2009; Imamura et al., 2005; Köhler et al., 2007; Starosta et al., 2010; Gödeke et al., 2013). To test whether mutation of PA3297 renders higher susceptibility, we performed the stationary-phase cells killing assay as previously described (Imamura et al., 2005; Köhler et al., 2007; Gödeke et al., 2013). Stationary-phase cells of PA14, the ΔPA3297 mutant and the complementation strain were subjected to treatment with AZM at the concentrations of 2, 5, and 10 μg/ml. As shown in Figure 2A, the AZM-mediated killing of the ΔPA3297 mutant was significantly increased at all of the AZM concentrations tested. The most significant difference was observed at the concentration of 10 μg/ml, where the survival rate of the mutant was approximately 1% of those of the wild type and complemented strains. In addition, the ΔPA3297 mutant was more susceptible to another macrolid antibiotic, erythromycin, with a fourfold lower MIC compared to the wild type strain PA14 (Table 3).
FIGURE 2.
Increased killing of the ΔPA3297 mutant by AZM. (A) AZM-mediated killing of wild type PA14, the ΔPA3297 mutant and the mutant complemented with wild type or the mutated PA3297. Stationary-phase bacteria were treated with 2, 5, or 10 μg/ml AZM for 20 h at 37°C. The numbers of live bacteria were determined by serial dilution and plating. (B) AZM-mediated killing of biofilm. 24-h-old biofilms formed by wild type PA14, the ΔPA3297 mutant and the complemented strain were treated with 5 μg/ml, 10 μg/ml AZM or not for 2 h. Bacteria in the biofilm were dissociated from the wells by gentle sonication. The viable bacteria were determined by serial dilution and plating. The averages and associated standard deviations from three replicates are shown. ∗p < 0.05 compared to PA14 or the complemented strain by student’s t-test.
Table 3.
Bacterial susceptibilities to macrolides and lincosamides.
| Strain | MIC (μg/ml)a | |||
|---|---|---|---|---|
| ERY | AZM | LIN | CLI | |
| PA14 | 300 | 400 | 12800 | 4800 |
| ΔPA3297 | 75 | 100 | 3200 | 2400 |
| ΔPA3297/att7::PA3297 | 300 | 400 | >12800 | 9600 |
| ΔPA3297/att7::PA3297 K101Ab | 75 | 100 | 6400 | 1600 |
| ΔPA3297/att7::PA3297 D192Ab | 75 | 100 | 3200 | 2400 |
| ΔPA3297/att7::PA3297 SAT224AAAb | 75 | 100 | 6400 | 2400 |
aERY, erythromycin; AZM, azithromycin; LIN, lincomycin; CLI, clindamycin.
bMutants with single or triple amino acid changes.
Next, we tested the susceptibility of the ΔPA3297 mutant to lincosamides, whose bactericidal mechanism is similar to macrolides (Tenson et al., 2003; Wilson, 2014). Indeed, mutation in PA3297 increased the bacterial susceptibility to lincomycin and clindamycin (Table 3). However, no increase of susceptibility was observed to other antibiotics, including ciprofloxacin, carbenicillin, meropenem, tetracycline, tobramycin, kanamycin, chloramphenicol, or polymyxin B (Supplementary Table S1). These results suggest that PA3297 plays an important role in the resistance against antibiotics targeting the peptide exit tunnel of ribosome.
PA3297 is a putative RNA helicase belonging to the DEAH-box family proteins, which are characterized by the presence of seven to nine conserved motifs (Tanner and Linder, 2001). In the E. coli RNA helicases DbpA and HrpA, the conserved residues GETGSGKT in motifI, DEAH in motifII, and SAT in motifIII have been shown to be required for interaction with and hydrolysis of NTP (Koo et al., 2004; Linder and Fuller-Pace, 2013). To determine whether these critical residues within motifs I, II, and III are important for PA3297 in the resistance to macrolides and lincosamides, we altered the residues by site-directed mutagenesis. Specifically, the K101 in motif I or the D167 in motif II was mutated to alanine. The S224 and T226 in motif III were both replaced with alanine. Each mutated PA3297 was transferred into the ΔPA3297 mutant and the susceptibility to antibiotics was tested. None of the mutated PA3297 was able to restore the survival rate of the mutant (Figure 2A). In addition, the mutated PA3297 was unable to restore the resistance of the ΔPA3297 mutant to macrolides, lincomycin, and clindamycin (Table 3). These results suggest that the RNA helicase function of PA3297 is required for its role in the resistance to macrolides and lincosamides.
Clinically, AZM has been used in the treatment of chronic P. aeruginosa infection (Saiman et al., 2003; Blasi et al., 2010), which is characterized by biofilm formation (Singh et al., 2000). The biofilm is notorious for high antibiotic tolerance, which severely hinders eradication of the bacteria (López et al., 2010; Breidenstein et al., 2011). We suspected that mutation of PA3297 might increase the killing efficacy of AZM on the biofilm. Indeed, the survival rates of the ΔPA3297 mutant in biofilm were lower than those of the wild type PA14 at various AZM concentrations, which were restored by complementation with a wild type PA3297 (Figure 2B).
Mutation of PA3297 Increases the Bacterial Susceptibility to Hydrogen Peroxide in the Presence of AZM
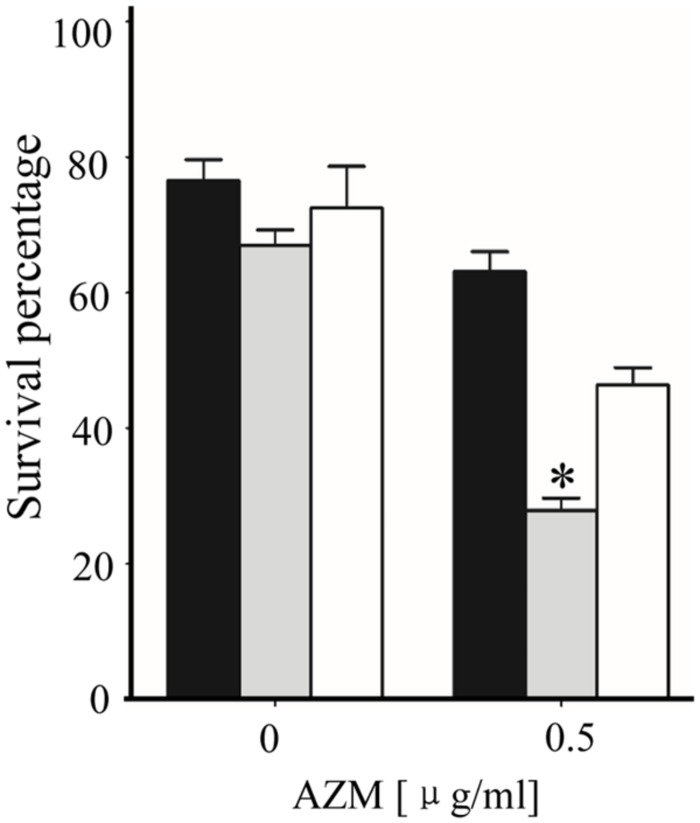
It has been shown that AZM treatment impairs the oxidative stress response in P. aeruginosa (Nalca et al., 2006), which intrigued us to test whether mutation of PA3297 leads to further impairment. The H2O2 susceptibility assay was performed in the presence of 0.5 μg/ml AZM, as the growth rates of wild type PA14 and the ΔPA3297 mutant were similar at this concentration of AZM (Supplementary Figure S1). The wild type PA14, ΔPA3297 mutant and the complemented strain were grown without or with AZM to an OD600 of 2.0, and then treated with 10% H2O2 for 15 min. In the presence of AZM, the ΔPA3297 mutant was more susceptible to H2O2 than the wild type and complemented strains, whereas no difference was observed in the absence of AZM (Figure 3). These results suggest that PA3297 is involved in the bacterial oxidative stress response in the presence of AZM.
FIGURE 3.
Hydrogen peroxide (H2O2) mediated killing of bacteria. PA14, the ΔPA3297 mutant and complemented strain were grown at 37°C in the absence or presence of 0.5 μg/ml AZM to an OD600 of 2.0. Bacteria were collected and washed with PBS. Then the bacteria were incubated in PBS with or without 10% H2O2 for 15 min. The live bacterial numbers were determined by serial dilution and plating. ∗p < 0.05 compared to PA14 or the complemented strain by student’s t-test.
Mutation of PA3297 Intensifies the Inhibitory Effect of AZM on Swarming Motility
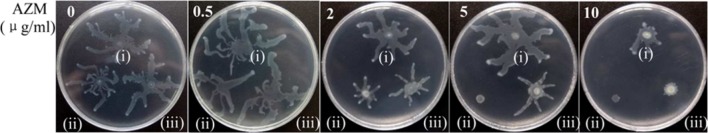
Besides oxidative stress response, AZM suppresses swarming motility (Tateda et al., 2001; Köhler et al., 2007; Gödeke et al., 2013). Same numbers of wild type PA14 and the ΔPA3297 mutant were inoculated on the plates containing various concentrations of AZM. As shown in Figure 4, 10 μg/ml AZM suppressed the swarming motility of wild type PA14 obviously, whereas the lower concentrations of AZM showed no inhibitory effect. However, starting from 2 μg/ml, AZM suppressed the swarming motility of the ΔPA3297 mutant in a dose dependent manner. Complementation with a PA3297 gene restored the swarming motility in the presence of AZM (Figure 4).
FIGURE 4.
Bacterial swarming motility. PA14 (i), the ΔPA3297 mutant (ii) and the complemented strain (iii) were inoculated on plates containing indicated concentrations of AZM.
Interaction between AZM and Ribosome Induces the Expression of PA3297
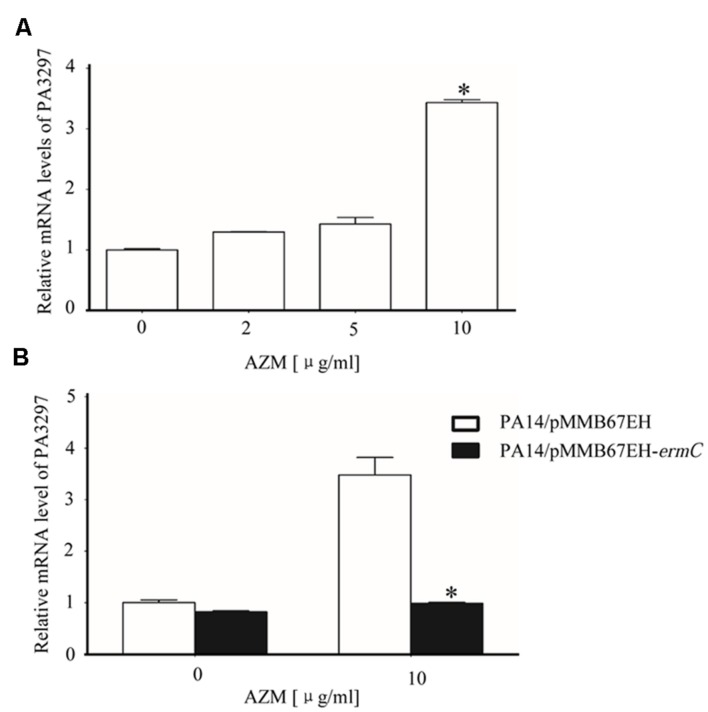
Our results so far suggested that PA3297 is involved in the bacterial response to AZM treatment. To test whether the expression of PA3297 is induced by AZM treatment, wild type PA14 was grown in the absence or presence of AZM at various concentrations and the relative RNA levels of PA3297 were determined by real time PCR. Indeed, the expression of PA3297 was induced by AZM (Figure 5A). However, overexpression of PA3297 in wild type PA14 did not further increase the bacterial tolerance to AZM (Table 4). We suspect that since the bacteria at exponential growth phase are highly resistant to macrolides and lincosamides, overexpression of PA3297 might not further increase the resistance significantly in the MIC test. Another possibility is that with the endogenous up regulation of PA3297, additional expression of PA3297 might be redundant.
FIGURE 5.
Expression of PA3297 in the presence of AZM. (A) Wild type PA14 was grown to an OD600 of 0.3 in LB medium. The bacteria were grown further in the absence or presence of 2, 5, or 10 μg/ml AZM. When the OD600 reached 2.0, total bacterial RNA was isolated and the mRNA levels of PA3297 were determined with real time PCR. ∗p < 0.05 compared to bacteria in the absence of presence of 2 or 5 μg/ml AZM by student’s t-test. (B) PA14 harboring pMMB67EH or the ErmC over expressing plasmid (pMMB67EH-ermC) was grown at 37°Cin the absence of AZM. When the OD600 reached 0.3, 1 mM IPTG was added to the medium. Meanwhile, no AZM or AZM at the final concentration of 10 μg/ml was added to the medium. At the OD600 of 2.0, total RNA was isolated and the mRNA levels of PA3297 were determined with real time PCR. The mRNA levels of lacI from the plasmid were used as internal control. ∗p < 0.05 compared to PA14/pMMB67EH by student’s t-test.
Table 4.
Bacterial susceptibilities to macrolides and lincosamides.
| Strain | MIC (μg/ml)a | |||
|---|---|---|---|---|
| ERY | AZM | LIN | CLI | |
| PA14/pMMB67EH | 600 | 200 | 12800 | 12800 |
| PA14/pMMB67-PA3297 | 600 | 200 | 12800 | 6400 |
| PA14/pMMB67-ermC | 2400 | 800 | >12800 | >12800 |
| ΔPA3297/pMMB67EH | 75 | 50 | 6400 | 3200 |
| ΔPA3297/pMMB67-PA3297 | 300 | 200 | 12800 | 3200 |
| ΔPA3297/pMMB67-ermC | 2400 | 400 | >12800 | >12800 |
aERY, erythromycin; AZM, azithromycin; LIN, lincomycin; CLI, clindamycin.
Köhler et al. (2007) previously demonstrated that ribosome is the only target of AZM in bacteria. To test whether the induction of PA3297 is caused by the interaction between AZM and ribosome, we performed the ribosomal protection assay by overexpressing ErmC, a 23S rRNA methylase that blocks the binding of macrolide antibiotics to the NPET (Köhler et al., 2007; Lawrence et al., 2008). In the presence of 10 μg/ml AZM, the growth speed of the ErmC overexpressing strain was similar with that of the wild type strain containing an empty vector. However, overexpression of ErmC abolished the induction of PA3297 by AZM (Figure 5B). In addition, antibiotics in the other categories, including ciprofloxacin, tobramycin, and carbenicillin did not affect the expression level of PA3297 (Supplementary Figure S2). These results suggest that the expression of PA3297 is regulated in response to AZM-mediated ribosome stalling.
Ribosome Protection Rescues the ΔPA3297 Mutant from AZM-Mediated Hyperlethality
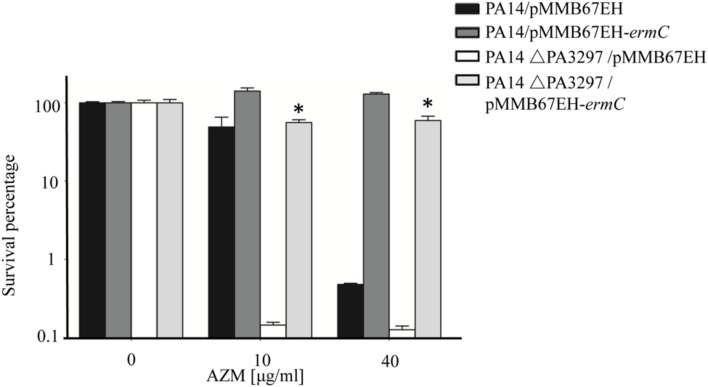
So far, we have demonstrated that the expression of PA3297 is induced by AZM and that mutation of PA3297 renders P. aeruginosa hypersusceptible to AZM. These results suggest that PA3297 might play a role in counteracting the detrimental effects caused by the interaction between AZM and ribosome (Köhler et al., 2007; Gödeke et al., 2013). Thus, ribosome protection should be able to increase the tolerance of the ΔPA3297 mutant to AZM. Indeed, overexpression of ErmC increased both the growth speed and the survival rate of the ΔPA3297 mutant when treated with 10 or 40 μg/ml AZM (Supplementary Figure S3, Figure 6). In addition, overexpression of ErmC increased the MICs of both wild type PA14 and the ΔPA3297 mutant to AZM, erythromycin, lincomycin and clindamycin (Table 4). Mutation of PA3297 did not increase the bacterial susceptibility to a variety of other antibiotics (Supplementary Table S1). These results suggest a specific role of PA3297 in responding to lincosamides and macrolides.
FIGURE 6.
Effects of Ribosome protection on the AZM mediated killing of the ΔPA3297 mutant. Wild type PA14 or the ΔPA3297 mutant harboring pMMB67EH or pMMB67EH-ermC was grown to an OD600 of 0.5 when 1 mM IPTG was added to the medium. When the OD600 reached 2.0, the bacteria were treated without or with 10 or 40 μg/ml AZM for 20 h at 37°C. The numbers of live bacteria were determined by serial dilution and plating. ∗p < 0.05 compared to the ΔPA3297 mutant harboring pMMB67EH by student’s t-test.
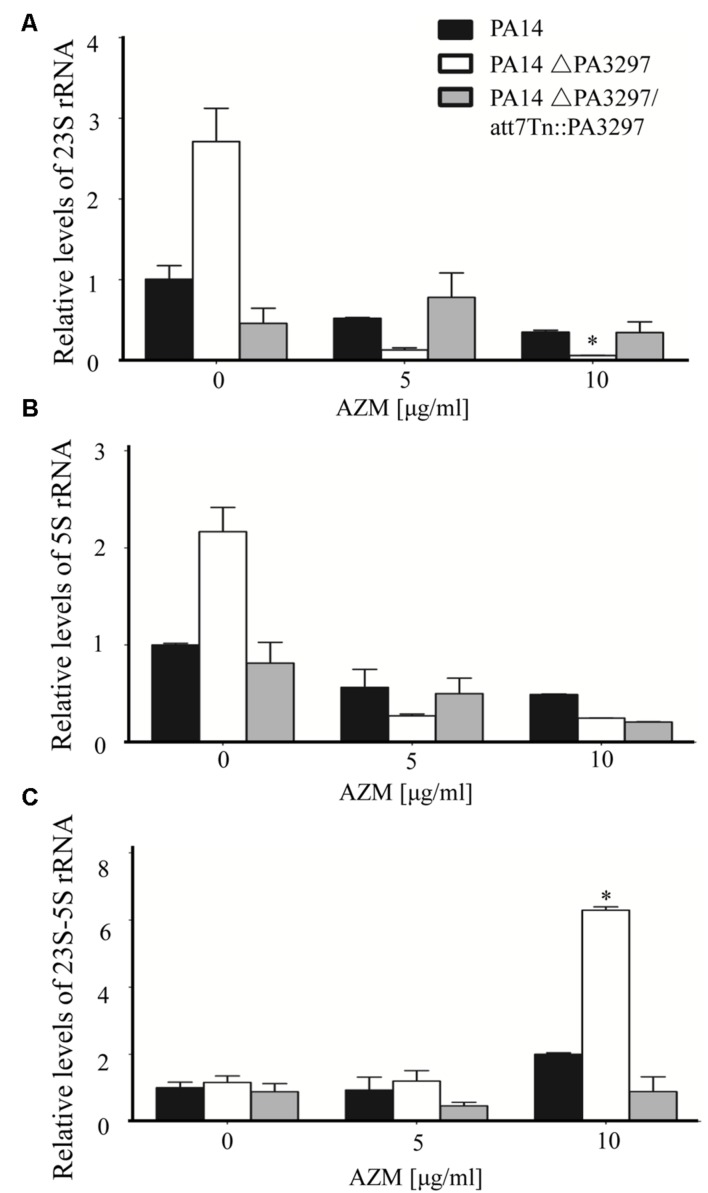
Deficiency in PA3297 Compromises rRNA Processing in the Presence of AZM
Studies in E. coli demonstrated that the DExD/H box play crucial roles in rRNA processing (Iost et al., 2013; Linder and Fuller-Pace, 2013). Therefore, we suspected that the up regulated PA3297 might participate in rRNA maturation, which facilitates ribosome biogenesis to compensate for AZM inactivated ribosome. The rRNA coding region in the chromosome of P. aeruginosa PA14 is shown in Supplementary Figure S4. To examine the processing of the rRNA transcript, we designed real-time PCR primers to analyze the total 23S and 5S rRNA levels as well as primers across the 23S and 5S rRNA coding region to analyze the level of unprocessed rRNA (Supplementary Figure S4). In wild type PA14, AZM at the concentrations of 5 and 10 μg/ml reduced the 23S and 5S rRNA levels (Figures 7A,B), and 10 μg/ml AZM slightly increased the unprocessed 23S-5S rRNA level (Figure 7C). In the absence of AZM, the total 23S and 5S rRNA levels were higher in the ΔPA3297 mutant than those in the wild type strain (Figures 7A,B), whereas the unprocessed 23S-5S levels were similar between the mutant and wild type strain. Interestingly, treatment with 10 μg/ml AZM resulted in a higher level of unprocessed 23S-5S rRNA in the ΔPA3297 mutant (Figure 7C), although its total 23S and 5S rRNA levels were lower than those in the wild type strain (Figures 7A,B).
FIGURE 7.
Effects of AZM on the processing of rRNA. PA14, the ΔPA3297 mutant and the complemented strain were grown at 37°C in the absence of AZM. When the OD600 reached 0.3, AZM (0, 5, and 10 μg/ml) was added. Total RNA was harvested when the OD600 reached 2.0. The levels of total 23S (A), 5S (B), and unprocessed 23S-5S rRNA (C) were determined by real time PCR. The mRNA levels of PA1769 were used as an internal control. ∗p < 0.05 compared to PA14 by student’s t-test.
Next, we calculated the percentages of unprocessed 23S-5S rRNA in wild type PA14 and the ΔPA3297 mutant with or without AZM treatment. Standard curves were generated to determine the amplification efficiencies of the primer pairs for the detection of total 23S, 5S and unprocessed 23S-5S rRNA levels in real time PCR (Supplementary Figure S5). Considering 5S rRNA is more prone to be lost during RNA purification, we calculated the ratio of unprocessed 23S-5S rRNA by dividing the levels of 23S-5S rRNA by those of 23S rRNA calibrated with the amplification efficiencies (Table 5). In the absence of AZM, the ratios of unprocessed 23S-5S rRNA were approximately 0.014 and 0.009% in wild type PA14 and the ΔPA3297 mutant, respectively. It seems that, without PA3297, the processing of 23S-5S rRNA is even more efficient. In the presence of 5 and 10 μg/ml AZM, the ratios of unprocessed 23S-5S rRNA rose to approximately 0.05 and 0.09% in wild type PA14, respectively. However, the ratios of unprocessed 23S-5S rRNA in the ΔPA3297 mutant were 0.33 and 1.61%, which were approximately 5- and 18-fold higher than those in PA14 under the same condition (Table 5).
Table 5.
The ratio of unprocessed 23S-5S rRNA in total 23S rRNA (%).
| Strain | AZM (μg/ml) | ||
|---|---|---|---|
| 0 | 5 | 10 | |
| PA14 | 0.014 | 0.051 | 0.089 |
| ΔPA3297 | 0.009 | 0.335 | 1.608 |
The unprocessed 23S-5S rRNA ratios were calculated according to the equation: Ratio = (1+e23S)Ct(23S)/(1+e23S-5S)Ct(23S-5S); e: Amplification efficiency. Ct: Cycles when fluorescence intensity reaches detection threshold.
Since, the growth speed of the ΔPA3297 mutant in the presence of 10 μg/ml AZM was similar as that of the wild type PA14 in the presence of 40 μg/ml AZM (Supplementary Figure S3), we compared the rRNAs levels of the two strains grown under the two conditions. The relative levels of 23S-5S rRNA, total 5S and 23S rRNA in the ΔPA3297 mutant were 150, 55, and 100% of those in the wild type PA14, respectively, indicating a similar rRNA processing status with the two different AZM concentrations. In combination, these results suggest that PA3297 might contribute to rRNA processing in response to AZM. And mutation of PA3297 might impair the biosynthesis of ribosome under AZM treatment, which renders the bacterium more susceptible to AZM.
Discussion
By binding to 23S rRNA in the 50S subunit of bacterial ribosome, AZM blocks polypeptide elongation and diminishes the intracellular pools of aminoacyl-tRNAs (Tenson et al., 2003; Gödeke et al., 2013; Wilson, 2014). And it has been demonstrated that AZM reduces the expression of gacA and the small RNAs rsmY and rsmZ, as well as quorum sensing genes (Kai et al., 2009; Pérez-Martínez and Haas, 2011). And the stationary phase killing by sub-MIC AZM was demonstrated to be correlated with increased outer membrane permeability (Imamura et al., 2005). Here, we found that mutation of PA3297 intensified the AZM mediated inhibitory effects on pyocyanin production and swarming motility of P. aeruginosa. In addition, the PA3297 mutant is more susceptible to oxidative stress in the presence of AZM. During infection, host generated reactive oxygen species (ROS) is an important bacterial killing mechanism. Therefore, inhibition of PA3297 together with the treatment with AZM, might render the bacteria more susceptible to host killing.
The MIC of the ΔPA3297 mutant is a quarter of that of the wild type strain. Consistently, the growth speed of the ΔPA3297 mutant in the presence of 10 μg/ml AZM was similar as that of the wild type strain in the presence of 40 μg/ml AZM (Supplementary Figure S3). However, in the stationary phase cell killing assay, the survival rate of the ΔPA3297 mutant treated with 10 μg/ml AZM was approximately 20% of that of the wild type strain treated with 40 μg/ml AZM (Figure 6). Therefore, PA3297 might play a more important role in the survival of stationary phase cells under AZM treatment.
Macrolides and lincosamides bind to the 50S of ribosome and block the NPET (Tenson et al., 2003). In our experiments, mutation in PA3297 did not alter the bacterial resistance to other antibiotics, including those binding to 30S or other parts of 50S ribosome. In addition, the expression of PA3297 was upregulated by AZM, which was abolished by ribosome protection. Therefore, PA3297 might specifically play a role in bacterial response to macrolides and lincosamides.
PA3297 is also named HrpA, both in PAO1 and PA14 (Winsor et al., 2016). According to the Profiles from GEO Expression Database at NCBI, the expression level of PA3297 was higher in biofilm than that in planktonic cells (Anderson et al., 2008). Isolates from CF lungs displayed higher expression levels of PA3297 levels than PAO1 (Son et al., 2007; Bielecki et al., 2013). And artificial medium that mimics CF lung sputum could increase the expression level of PA3297 slightly (Fung et al., 2010). In addition, increase of PA3297 expression was also observed in antibiotic-resistant small colony variants (Wei et al., 2011). However, the expression level of PA3297 showed a significant decrease in response to airway epithelia or low oxygen conditions (Alvarez-Ortega and Harwood, 2007; Chugani and Greenberg, 2007). These results indicate that the expression of PA3297 is regulated in response to various environmental stesses.
PA3297 is predicted to locate at the cytoplasmic membrane, with a molecular weight of 149.8 kDa (Winsor et al., 2016). It belongs to the DExD/H box helicase family. Members of this family have been found to play crucial roles in RNA metabolism and gene regulation (Linder and Jankowsky, 2011; Iost et al., 2013; Kaberdin and Bläsi, 2013; Linder and Fuller-Pace, 2013; Putnam and Jankowsky, 2013). According to NCBI protein blast, there are 81 homologous proteins with identities of 78% or more in other microorganisms. The homolog of PA3297 in Borrelia burgdorferi was found to be required for mouse infectivity and tick transmission and involved global gene regulation (Salman-Dilgimen et al., 2011; Owttrim, 2013). Another homolog in E. coli was found to be involved in fimbrial biogenesis (Koo et al., 2004). The identities they shared with PA3297 are 34 and 49%, respectively. Both of them possess the conserved residues in motif I, motif II, and motif III. Meanwhile, they are also involved in RNA processing. In Listeria monocytogenes, it has been reported that defect of a DExD-box RNA helicase, Lmo1722, reduced the maturation of 23S RNA (Bareclev et al., 2014) at low temperatures.
In E. coli, the primary transcript rRNA is cleaved by RNase III, yielding precursors of the 16S rRNA (17S rRNA), 23S rRNA, and 5S rRNA (9S rRNA; Shajani et al., 2011). Mutation of RNase III led to slower growth rate, reduction in cell viability and protein synthesis rates in the presence of AZM (Silvers and Champney, 2005). In E. coli, under certain stress conditions, DEAD box helicase may substitute for RhlB in the degradosome, such as CsdA under cold shock conditions (Prud’homme-Généreux et al., 2004). We found that mutation of PA3297 increased the percentage of unprocessed 23S-5S rRNA in the presence of AZM, which indicates that the cleaving function of RNase III might be impaired under the stress caused by AZM. Therefore, PA3297 might assist RNase III in rRNA processing in the presence of AZM. In addition, although the growth speed and rRNA processing in the ΔPA3297 mutant treated with 10 μg/ml AZM were similar to those in the wild type strain treated with 40 μg/ml AZM (Supplementary Figure S3), the survival rate of the ΔPA3297 mutant was lower than that of the wild type strain under those conditions. Thus, PA3297 might play other roles in counteracting the effects of AZM.
Overall, mutation of PA3297 renders P. aeruginosa more susceptible to AZM mediated inhibition on virulence factors and killing effect. Therefore, targeting the regulatory pathway or the function of PA3297 might further increase the beneficial effects of AZM in the treatment of chronic P. aeruginosa infections.
Author Contributions
Conceived and designed the experiments: WW, SJ, HT, ZC. Performed the experiments: HT, LZ, YW, RC, FZ, YJ. Analyzed the data: HT, LZ, SJ, WW. Wrote the paper: HT, SJ, WW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by National Science Foundation of China (31370168 to WW, and 31370167 to SJ); National Basic Research Program of China (973 Program, 2012CB518700 to SJ), Program of international S&T cooperation (2015DFG32500 to SJ) and Science and Technology Committee of Tianjin (13JCYBJC36700 to WW, 15JCZDJC33000 to SJ).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00317
References
- Alvarez-Ortega C., Harwood C. S. (2007). Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65 153–165. 10.1111/j.1365-2958.2007.05772.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov R. I. (2013). Biotic acts of antibiotics. Front. Microbiol. 4:241 10.3389/fmicb.2013.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G. G., Moreau-Marquis S., Stanton B. A., O’Toole G. A. (2008). In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect. Immun. 76 1423–1433. 10.1128/IAI.01373-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareclev C., Vaitkevicius K., Netterling S., Johansson J. (2014). DExD-box RNA-helicases in Listeria monocytogenes are important for growth, ribosomal maturation, rRNA processing and virulence factor expression. RNA Biol. 11 1458–1467. 10.1080/15476286.2014.996099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecki P., Komor U., Bielecka A., Müsken M., Puchałka J., Pletz M. W., et al. (2013). Ex vivo transcriptional profiling reveals a common set of genes important for the adaptation of Pseudomonas aeruginosa to chronically infected host sites. Environ. Microbiol. 15 570–587. 10.1111/1462-2920.12024 [DOI] [PubMed] [Google Scholar]
- Billings N., Millan M. R., Caldara M., Rusconi R., Tarasova Y., Stocker R., et al. (2013). The extracellular matrix component psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 9:e1003526 10.1371/journal.ppat.1003526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerkan G., Witso E., Bergh K. (2009). Sonication is superior to scraping for retrieval of bacteria in biofilm on titanium and steel surfaces in vitro. Acta Orthop. 80 245–250. 10.3109/17453670902947457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi F., Bonardi D., Aliberti S., Tarsia P., Confalonieri M., Amir O., et al. (2010). Long-term azithromycin use in patients with chronic obstructive pulmonary disease and tracheostomy. Pulm. Pharmacol. Ther. 23 200–207. 10.1016/j.pupt.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Breidenstein E. B., de la Fuente-Núñez C., Hancock R. E. (2011). Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19 419–426. 10.1016/j.tim.2011.04.005 [DOI] [PubMed] [Google Scholar]
- Brencic A., McFarland K. A., McManus H. R., Castang S., Mogno I., Dove S. L., et al. (2009). Th GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73 434–445. 10.1111/j.1365-2958.2009.06782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa M., Bendinelli M., Friedman H. (2012). Pseudomonas aeruginosa as an Opportunistic Pathogen. Berlin: Springer Science & Business Media. [Google Scholar]
- Choi K.-H., Schweizer H. P. (2006). Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1 153–161. 10.1038/nprot.2006.24 [DOI] [PubMed] [Google Scholar]
- Chugani S., Greenberg E. (2007). The influence of human respiratory epithelia on Pseudomonas aeruginosa gene expression. Microb. Pathog. 42 29–35. 10.1016/j.micpath.2006.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N. K., Linder P. (2006). The DEAD-box protein family of RNA helicases. Gene 367 17–37. 10.1016/j.gene.2005.10.019 [DOI] [PubMed] [Google Scholar]
- de Bentzmann S., Plésiat P. (2011). The Pseudomonas aeruginosa opportunistic pathogen and human infections. Environ. Microbiol. 13 1655–1665. 10.1111/j.1462-2920.2011.02469.x [DOI] [PubMed] [Google Scholar]
- Doring G., Conway S., Heijerman H., Hodson M., Hoiby N., Smyth A., et al. (2000). Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis: a European consensus. Eur. Respir. J. 16 749–767. 10.1034/j.1399-3003.2000.16d30.x [DOI] [PubMed] [Google Scholar]
- Essar D., Eberly L., Hadero A., Crawford I. (1990). Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172 884–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre-Bonté S., Köhler T., Van Delden C. (2003). Biofilm formation by Pseudomonas aeruginosa: role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 52 598–604. 10.1093/jac/dkg397 [DOI] [PubMed] [Google Scholar]
- Fung C., Naughton S., Turnbull L., Tingpej P., Rose B., Arthur J., et al. (2010). Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J. Med. Microbiol. 59 1089–1100. 10.1099/jmm.0.019984-0 [DOI] [PubMed] [Google Scholar]
- Gillis R. J., Iglewski B. H. (2004). Azithromycin retards Pseudomonas aeruginosa biofilm formation. J. Clin. Microbiol. 42 5842–5845. 10.1128/JCM.42.12.5842-5845.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gödeke J., Pustelny C., Häussler S. (2013). Recycling of peptidyl-tRNAs by peptidyl-tRNA hydrolase counteracts azithromycin-mediated effects on Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 571617–1624. 10.1128/AAC.02582-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénard S., Muller C., Monlezun L., Benas P., Broutin I., Jeannot K., et al. (2014). Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 58 221–228. 10.1128/AAC.01252-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7 654–665. 10.1038/nrmicro2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Høiby N., Frederiksen B., Pressler T. (2005). Eradication of early Pseudomonas aeruginosa infection. J. Cyst. Fibros. 4 49–54. 10.1016/j.jcf.2005.05.018 [DOI] [PubMed] [Google Scholar]
- Imamura Y., Higashiyama Y., Tomono K., Izumikawa K., Yanagihara K., Ohno H., et al. (2005). Azithromycin exhibits bactericidal effects on Pseudomonas aeruginosa through interaction with the outer membrane. Antimicrob. Agents Chemother. 49 1377–1380. 10.1128/AAC.49.4.1377-1380.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperi F., Leoni L., Visca P. (2014). Antivirulence activity of azithromycin in Pseudomonas aeruginosa. Front. Microbiol. 5:178 10.3389/fmicb.2014.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intile P. J., Balzer G. J., Wolfgang M. C., Yahr T. L. (2015). The RNA helicase DeaD stimulates ExsA translation To promote expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 197 2664–2674. 10.1128/jb.00231-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I., Bizebard T., Dreyfus M. (2013). Functions of DEAD-box proteins in bacteria: current knowledge and pending questions. BBA-Gene Regul. Mech. 1829 866–877. 10.1016/j.bbagrm.2013.01.012 [DOI] [PubMed] [Google Scholar]
- Jo J. T., Brinkman F. S., Hancock R. E. (2003). Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47 1101–1111. 10.1128/AAC.47.3.1101-1111.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdin V. R., Bläsi U. (2013). Bacterial helicases in post-transcriptional control. BBA-Gene Regul. Mech. 1829 878–883. 10.1016/j.bbagrm.2012.12.005 [DOI] [PubMed] [Google Scholar]
- Kai T., Tateda K., Kimura S., Ishii Y., Ito H., Yoshida H., et al. (2009). A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 22 483–486. 10.1016/j.pupt.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Kipnis E., Sawa T., Wiener-Kronish J. (2006). Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Maladies Infect. 36 78–91. 10.1016/j.medmal.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Köhler T., Dumas J.-L., Van Delden C. (2007). Ribosome protection prevents azithromycin-mediated quorum-sensing modulation and stationary-phase killing of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 51 4243–4248. 10.1128/AAC.00613-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo J. T., Choe J., Moseley S. L. (2004). HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 52 1813–1826. 10.1111/j.1365-2958.2004.04099.x [DOI] [PubMed] [Google Scholar]
- Kurachi M. (1958). Studies on the biosynthesis of pyocyanine. (II) : isolation and determination of pyocyanine. Bull. Inst. Chem. Res. Kyoto Univ. 36 174–187. [Google Scholar]
- Lau C. H.-F., Fraud S., Jones M., Peterson S. N., Poole K. (2012). Reduced expression of the rplU-rpmA ribosomal protein operon in mexXY-expressing pan-aminoglycoside-resistant mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56 5171–5179. 10.1128/AAC.00846-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M. G., Lindahl L., Zengel J. M. (2008). Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J. Bacteriol. 190 5862–5869. 10.1128/JB.00632-638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legssyer R., Huaux F., Lebacq J., Delos M., Marbaix E., Lebecque P., et al. (2006). Azithromycin reduces spontaneous and induced inflammation in DeltaF508 cystic fibrosis mice. Respir. Res. 7:134 10.1186/1465-9921-7-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Schurr M. J., Sauer K. (2013). The MerR-Like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J. Bacteriol. 195 3352–3363. 10.1128/jb.00318-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati N. T., Urbach J. M., Miyata S., Lee D. G., Drenkard E., Wu G., et al. (2006). An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U.S.A. 103 2833–2838. 10.1073/pnas.0511100103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Fuller-Pace F. V. (2013). Looking back on the birth of DEAD-box RNA helicases. BBA-Gene Regul. Mech. 1829 750–755. 10.1016/j.bbagrm.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Linder P., Jankowsky E. (2011). From unwinding to clamping—the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12 505–516. 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- Lister P. D., Wolter D. J., Hanson N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22 582–610. 10.1128/CMR.00040-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-C., Chan K.-G., Chang C.-Y. (2015). Modulation of host biology by Pseudomonas aeruginosa quorum sensing signal molecules: messengers or traitors. Front. Microbiol. 6:1226 10.3389/fmicb.2015.01226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López D., Vlamakis H., Kolter R. (2010). Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398 10.1101/cshperspect.a000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovmar M., Tenson T., Ehrenberg M. (2004). Kinetics of macrolide action: the josamycin and erythromycin cases. J. Biol. Chem. 279 53506–53515. 10.1074/jbc.M401625200 [DOI] [PubMed] [Google Scholar]
- Lovmar M., Vimberg V., Lukk E., Nilsson K., Tenson T., Ehrenberg M. (2009). Cis-acting resistance peptides reveal dual ribosome inhibitory action of the macrolide josamycin. Biochimie 91 989–995. 10.1016/j.biochi.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Molinari G., Guzman C., Pesce A., Schito G. (1993). Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. J. Antimicrob. Chemother. 31 681–688. 10.1093/jac/31.5.681 [DOI] [PubMed] [Google Scholar]
- Molinari G., Paglia P., Schito G. (1992). Inhibition of motility of Pseudomonas aeruginosa and Proteus mirabilis by subinhibitory concentrations of azithromycin. Eur. J. Clin. Microbiol. Infect. Dis. 11 469–471. 10.1007/BF01961867 [DOI] [PubMed] [Google Scholar]
- Morita Y., Tomida J., Kawamura Y. (2013). Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 4:422 10.3389/fmicb.2013.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalca Y., Jansch L., Bredenbruch F., Geffers R., Buer J., Hussler S. (2006). Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob. Agents Chemother. 50 1680–1688. 10.1128/aac.50.5.1680-1688.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oun S., Redder P., Didier J. P., Francois P., Corvaglia A. R., Buttazzoni E., et al. (2013). The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus. RNA Biol. 10 157–165. 10.4161/rna.22899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owttrim G. W. (2013). RNA helicases: diverse roles in prokaryotic response to abiotic stress. RNA Biol. 10 96–110. 10.4161/rna.22638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martínez I., Haas D. (2011). Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55 3399–3405. 10.1128/AAC.01801-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2011). Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2:65 10.3389/fmicb.2011.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme-Généreux A., Beran R. K., Iost I., Ramey C. S., Mackie G. A., Simons R. W. (2004). Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a cold shock degradosome. Mol. Microbiol. 54 1409–1421. 10.1111/j.1365-2958.2004.04360.x [DOI] [PubMed] [Google Scholar]
- Putnam A. A., Jankowsky E. (2013). DEAD-box helicases as integrators of RNA, nucleotide and protein binding. BBA-Gene Regul. Mech. 1829 884–893. 10.1016/j.bbagrm.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin H. R., Butler S. M., Wohl M. E. B., Geller D. E., Colin A. A., Schidlow D. V., et al. (2004). Pulmonary exacerbations in cystic fibrosis. Pediatr. Pulmonol. 37 400–406. 10.1002/ppul.20023 [DOI] [PubMed] [Google Scholar]
- Rada B., Leto T. L. (2013). Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 21 73–81. 10.1016/j.tim.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikot R. T., Blackwell T. S., Christman J. W., Prince A. S. (2005). Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171 1209–1223. 10.1164/rccm.200408-1044SO [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiman L., Marshall B. C., Mayer-Hamblett N., Burns J. L., Quittner A. L., Cibene D. A., et al. (2003). Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: a randomized controlled trial. JAMA 290 1749–1756. 10.1001/jama.290.13.1749 [DOI] [PubMed] [Google Scholar]
- Salman-Dilgimen A., Hardy P. O., Dresser A. R., Chaconas G. (2011). HrpA, a DEAH-Box RNA helicase, is involved in global gene regulation in the lyme disease spirochete. PLoS ONE 6: e22168 10.1371/journal.pone.0022168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman-Dilgimen A., Hardy P. O., Radolf J. D., Caimano M. J., Chaconas G. (2013). HrpA, an RNA helicase involved in RNA processing, is required for mouse infectivity and tick transmission of the lyme disease spirochete. PLoS Pathog. 9:e1003841 10.1371/journal.ppat.1003841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z., Sykes M. T., Williamson J. R. (2011). Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80 501–526. 10.1146/annurev-biochem-062608-160432 [DOI] [PubMed] [Google Scholar]
- Silvers J. A., Champney W. S. (2005). Accumulation and turnover of 23S ribosomal RNA in azithromycin-inhibited ribonuclease mutant strains of Escherichia coli. Arch. Microbiol. 184 66–77. 10.1007/s00203-005-0017-10 [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U., Puhler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat. Biotechnol. 1 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Singh P. K., Schaefer A. L., Parsek M. R., Moninger T. O., Welsh M. J., Greenberg E. P. (2000). Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407 762–764. 10.1038/35037627 [DOI] [PubMed] [Google Scholar]
- Skindersoe M. E., Alhede M., Phipps R., Yang L., Jensen P. O., Rasmussen T. B., et al. (2008). Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52 3648–3663. 10.1128/AAC.01230-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M. S., Matthews W. J., Jr., Kang Y., Nguyen D. T., Hoang T. T. (2007). In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75 5313–5324. 10.1128/IAI.01807-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta A. L., Karpenko V. V., Shishkina A. V., Mikolajka A., Sumbatyan N. V., Schluenzen F., et al. (2010). Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem. Biol. 17 504–514. 10.1016/j.chembiol.2010.04.008 [DOI] [PubMed] [Google Scholar]
- Steel H. C., Theron A. J., Cockeran R., Anderson R., Feldman C. (2012). Pathogen-and host-directed anti-inflammatory activities of macrolide antibiotics. Mediators Inflamm. 2012:584262 10.1155/2012/584262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkamp G., Schmitt-Grohe S., Döring G., Staab D., Pfründer D., Beck G., et al. (2008). Once-weekly azithromycin in cystic fibrosis with chronic Pseudomonas aeruginosa infection. Respir. Med. 102 1643–1653. 10.1016/j.rmed.2008.03.009 [DOI] [PubMed] [Google Scholar]
- Tanner N. K., Linder P. (2001). DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8251–262. 10.1016/S1097-2765(01)00329-X [DOI] [PubMed] [Google Scholar]
- Tateda K., Comte R., Pechere J.-C., Köhler T., Yamaguchi K., Van Delden C. (2001). Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45 1930–1933. 10.1128/AAC.45.6.1930-1933.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T., Lovmar M., Ehrenberg M. (2003). The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330 1005–1014. 10.1016/S0022-2836(03)00662-4 [DOI] [PubMed] [Google Scholar]
- Tsai W. C., Hershenson M. B., Zhou Y., Sajjan U. (2009). Azithromycin increases survival and reduces lung inflammation in cystic fibrosis mice. Inflamm. Res. 58 491–501. 10.1007/s00011-009-0015-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Tarighi S., Dötsch A., Häussler S., Müsken M., Wright V. J., et al. (2011). Phenotypic and genome-wide analysis of an antibiotic-resistant small colony variant (SCV) of Pseudomonas aeruginosa. PLoS ONE 6:e29276 10.1371/journal.pone.0029276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. N. (2014). Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12 35–48. 10.1038/nrmicro3155 [DOI] [PubMed] [Google Scholar]
- Winsor G. L., Griffiths E. J., Lo R., Dhillon B. K., Shay J. A., Brinkman F. S. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44 D646–D653. 10.1093/nar/gkv1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L., Lam D. K. W., Fleming L., Lo R., Whiteside M. D., Yu N. Y., et al. (2011). Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39 D596–D600. 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Baumann U., Reymond J.-L. (2004). An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115 10.1093/nar/gnh110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.