Abstract
The mammalian cerebral cortex is a dense network composed of local, subcortical, and intercortical synaptic connections. As a result, mapping cell type-specific neuronal connectivity in the cerebral cortex in vivo has long been a challenge for neurobiologists. In particular, the development of excitatory and inhibitory interneuron presynaptic input has been hard to capture. We set out to analyze the development of this connectivity in the first postnatal month using a murine model. First, we surveyed the connectivity of one of the earliest populations of neurons in the brain, the Cajal-Retzius (CR) cells in the neocortex, which are known to be critical for cortical layer formation and are hypothesized to be important in the establishment of early cortical networks. We found that CR cells receive inputs from deeper-layer excitatory neurons and inhibitory interneurons in the first postnatal week. We also found that both excitatory pyramidal neurons and inhibitory interneurons received broad inputs in the first postnatal week, including inputs from CR cells. Expanding our analysis into the more mature brain, we assessed the inputs onto inhibitory interneurons and excitatory projection neurons, labeling neuronal progenitors with Cre drivers to study discrete populations of neurons in older cortex, and found that excitatory cortical and subcortical inputs are refined by the fourth week of development, whereas local inhibitory inputs increase during this postnatal period. Cell type-specific circuit mapping is specific, reliable, and effective, and can be used on molecularly defined subtypes to determine connectivity in the cortex.
SIGNIFICANCE STATEMENT Mapping cortical connectivity in the developing mammalian brain has been an intractable problem, in part because it has not been possible to analyze connectivity with cell subtype precision. Our study systematically targets the presynaptic connections of discrete neuronal subtypes in both the mature and developing cerebral cortex. We analyzed the connections that Cajal-Retzius cells make and receive, and found that these cells receive inputs from deeper-layer excitatory neurons and inhibitory interneurons in the first postnatal week. We assessed the inputs onto inhibitory interneurons and excitatory projection neurons, the major two types of neurons in the cortex, and found that excitatory inputs are refined by the fourth week of development, whereas local inhibitory inputs increase during this postnatal period.
Keywords: Cajal-Retzius cells, cerebral cortex, circuit development, interneurons, neuronal connectivity, viral circuit tracing
Introduction
Neuronal diversity is generated during embryonic cortical development, and neuronal connectivity is formed during postnatal development (McConnell, 1995; Price et al., 1995; Guillemot, 2005; Corbin et al., 2008). This connectivity is refined with the onset of neuronal activity and sensory inputs (Katz and Shatz, 1996; Wong and Ghosh, 2002; Fox and Wong, 2005); however, the refinement of inputs at the circuit level has not been thoroughly elucidated. Neuronal subtypes are generally defined by a combination of morphology, immunohistochemical properties, and connectivity (Ascoli et al., 2008). Excitatory neurons are largely pyramidal-shaped projection neurons that make connections extending long distances, often to subcortical or contralateral cortical regions, as well as local connections (Sur and Rubenstein, 2005; Molyneaux et al., 2007). Inhibitory interneurons have inputs onto both excitatory and inhibitory neurons within their local cortical area (Fino and Yuste, 2011; Packer and Yuste, 2011; Gittis and Kreitzer, 2012; Pfeffer et al., 2013; Sun et al., 2014). The Cajal-Retzius (CR) cells are horizontal in shape and sit above the pyramidal neurons, mixing with inhibitory interneurons in layer I. While their role in cortical layer formation in embryonic development has been well characterized (Haas et al., 2002; Soriano and Del Río, 2005; Meyer, 2010), their connectivity during postnatal cortical development is unexplored.
Most work on presynaptic inputs has involved dual-cell recording or analyses of neurotransmitter receptor subtypes to determine the types of inputs that neurons receive (Lin and Bergles, 2004; Lin et al., 2005; Nikolenko et al., 2007; Fino and Yuste, 2011; Bell et al., 2013). It has not been possible with these approaches to determine the cytoarchitecture of presynaptic inputs onto different neuronal subclasses during early postnatal development. Furthermore, little has been done to systematically link the embryonic origins of neuronal subtypes with their adult circuitry. We set out to address these questions, combining genetic targeting of embryonic neuronal subtypes with monosynaptic circuit tracing to analyze cortical presynaptic connectivity in the first postnatal month.
In this study, we addressed two main issues. Can monosynaptic circuit tracing be used to analyze the dense connectivity of the cerebral cortex, particularly the presynaptic connections of discrete neuronal subclasses in development? And, can we determine the developmental maturation of inputs onto discrete neuronal subtypes in early postnatal development? We show that this approach is a powerful means of examining CR cell circuitry in early postnatal development. We also examine the mature connectivity in excitatory projection neurons and inhibitory interneuron subclasses, and reveal that these inputs change during synaptogenesis, between 1 and 4 weeks of age. We find that excitatory neurons receive numerous contralateral and subcortical connections from both excitatory and inhibitory neurons, while inputs onto inhibitory interneurons are largely local. Finally, we map the inputs of distinct cortical layers by analyzing connectivity in layer V corticofugal neurons. We also use this approach to generate connectivity maps for specific cell subtypes and targeted brain regions; this a powerful tool to not only map the dynamics of presynaptic connectivity in development, but to create whole-brain approaches to determine changes in connectivity in mouse mutants.
Materials and Methods
Animal use.
ROSA-Ai14, ROSA-tTA (tetracycline transactivator protein; The Jackson Laboratory), Emx1-Cre (Chan et al., 2001; Gorski et al., 2002), Dlx5/6-Cre (Wang et al., 2010), Fzd10-CreERT2 (Zhao et al., 2006; Gu et al., 2009), Shh-Cre (Harfe et al., 2004), and pTRE-Bi-G-TVA (avian tumor virus receptor A; Rowland et al., 2013) mice of either sex used in these studies were maintained according to the protocols approved by the Institutional Care and Use Committee at University of California, San Francisco. For postnatal animals, the day of birth was considered to be postnatal day 0 (P0). The genotyping of animals were performed as described previously (Gorski et al., 2002). Timed pregnant dams were generated for the Fzd10-CreERT2 experiments; the plug date was treated as embryonic day 0.5 (E.0.5); at least three brains was analyzed for each age. Tamoxifen was delivered via gavage to pregnant dams at E.12.5 at a dose of 5 mg/40 g, using a solution of 20 mg/ml tamoxifen dissolved in corn oil.
Production of avian envelope protein-pseudotyped, glycoprotein-deficient rabies virus.
SADΔG GFP un-pseudotyped virus was produced, ultracentrifuged, and titered manually, as described by Wickersham et al. (2010). Avian envelope protein (EnvA)-pseudotyped SADΔG GFP virus was then produced from un-pseudotyped stocks, ultracentrifuged, and tittered, as in the study by Wickersham et al. (2010), with the exception that ultracentrifuged SADΔG GFP virus was used instead of supernatant to infect BHK-EnvA cells at a multiplicity of infection of 2, and cells were washed in PBS three times after 24 and 48 h, and replaced with fresh media; then the supernatant was ultracentrifuged, as in the study by Wickersham et al. (2010). SADΔG GFP (EnvA; hereafter referred to as RabV-GFP) virus was titered on TVA-800 cells. HEK293T cells were used as a control for infection; both TVA-800 and HEK cells were FACS sorted to determine a viral titer of 1 × 109; no green cells were detected by FACS in the HEK cell-titering wells.
Viral injection procedures.
For all viral tracing experiments, Emx1-Cre, Dlx5/6-Cre, Shh-Cre, or Fzd10-CreERT2 mice were crossed with ROSA-tTA, ROSA-Ai14, or pTRE-Bi-G-TVA mice. For the adult viral tracing experiments, P21–P25 mice were injected with RabV-GFP and killed 1 week later. For the developmental experiments, P7, P14, and P21 mice were injected with RabV-GFP and killed 1 week later. Mice were anesthetized with isoflurane inhalation and injected with 300 nl of RabV-GFP virus at bregma at coordinates x = 3.1/y = −0.7/z = 1.85. Stereotaxic procedures were followed as described by Lasek and Azouaou (2010). Mice were allowed to recover and were killed 1 week after viral injection, except where noted. For experiments performed in the first postnatal week in the Cajal-Retzius cell circuit tracing experiments, mice were anesthetized with ice or with Nembutal, as appropriate, and injected as above, with the exception that virus was delivered using a glass capillary before P5.
Tissue preparation and histology.
All animals were perfused first with cold 1× PBS followed by 4% PFA. Dissected brains were postfixed for 2 h and then cryoprotected by sucrose immersion; and P7 brains were embedded in Tissue Tek Optimum Cutting Temperature, frozen, and stored at −80°C. Serial coronal sections of embedded tissue were cut at 30 μm thickness using a cryostat and mounted directly onto slice. P28 brains in sucrose 30% in PBS were frozen and cut in coronal sections using a sliding microtome at 30 μm, and then mounted on the slides serially.
Immunohistochemistry.
Cryostat- or microtome-mounted sections were air dried and rinsed 3× in PBS plus 0.3%Triton before blocking for 1 h in 10% normal lamb serum diluted in PBS with 0.3% Triton to prevent nonspecific binding. Primary antibodies were diluted in 10% serum diluted in PBS with 0.3% Triton; sections were incubated in primary antibody overnight at 4°C. The primary antibodies used were as follows: rat anti-somatostatin (SST; 1:200; Millipore); mouse anti-parvalbumin (1:5000; Millipore); mouse anti-reelin (1:1000; Millipore); rabbit anti-calretinin (1:1000; Millipore); rat anti-Ctip2 (1:500; Abcam); and rabbit anti-calbindin (1:2000; Swant). To detect primary antibodies, we used Alexa Fluor-conjugated secondary antibodies (rat, rabbit, and mouse, 1:500; Invitrogen) in the same blocking buffer for 2 h at room temperature and counterstained with DAPI for 0.5 h, and then were washed with PBS and coverslipped with gel mount (Sigma-Aldrich).
Microscopy.
Fluorescent photographs were taken using Zeiss LSM 510 and 710 confocal microscopes. For confocal image analysis, each fluorophore was scanned sequentially, and z-stacks of the images obtained were collapsed onto a single projection image or presented as individual optical sections. Images for reconstruction analyses were taken on a Zeiss Axioscan Z.1 microscope; N = 4 brains for each Cre line were mounted in serial sections and were scanned sequentially for image analysis. Figures were prepared using ImageJ and Adobe Illustrator; brightness and contrast adjustments were applied equally across all images.
Image processing.
Images were stitched and exported from Zen Software (Zeiss) to .tiff format and processed in several stages. Custom Python scripts applying NumPy (www.numpy.org) and OpenCV (www.opencv.org) libraries were used to define regions of interest (ROIs) representing each brain, centering and aligning each ROI, and removing extraneous intensity. All steps were automated, and several quality control checkpoints helped to flag images that required rescanning or correction. Briefly, fluorescence from nuclear staining was used as a reference to identify areas of intensity associated with the brain region, and coordinates surrounding these regions were collected as contours. To obtain the optimal number of contours representing the complete brain region and excluding portions of adjacent slices, algorithms integrating slice depth, region area, and similarity scoring were applied to each image. Contour coordinates were used to define a mask for the ROI in each image and to discard all other regions of intensity. Each image was centered and aligned by fitting an ellipse to the contour coordinates bounding the ROI. The mask and transformation coordinates are collected and applied to each channel.
Reconstructions and animations.
Aligned images were further thresholded automatically by stretching the histogram and keeping the cluster of pixels with the highest intensity. When compared with manually thresholded images, this approach worked well for Emx1-Cre and Shh-Cre experiments but did not generalize to the Dlx5/6-Cre set of images. To define a universal coordinate system for the 170–210 z-slice images representing each brain, and all varying in size, maximum width, and height, were obtained for each experiment; and each slice was centered within a new, larger background. Thresholded, binary, red and green images for each slice were merged, falsely colored, and used to produce the animations in this article.
Thresholded images were also used to obtain the pixel coordinates for each channel. The pixel coordinates were imported into Igor Pro (WaveMetrics) and plotted in 3D with the help of the Gizmo Tool. Off-angle representations of coronal, sagittal, and horizontal views were selected for clarity. The views are shown with the following Euler angles: off-coronal (A, 10°; B, 10°; C, −90°), off-sagittal (A, −80°; B, 30°; C, −90°), and off-transverse (A, −55°; B, 81°; C, 34°). Scatter plots are shown with the z direction representing the stack of images (slices) and the x and y directions representing the columns and rows, respectively, of each image.
Results
Labeling diverse neuronal subtypes and their presynaptic connections with genetically targeted RabV-GFP circuit tracing
We combined existing neuronal progenitor-specific Cre alleles with a floxed allele for the tTA, an additional floxed allele for a fluorescent reporter (Tomato), and a tet-dependent bicistronic allele containing the viral proteins necessary for infection and spread of, respectively, the avian TVA and the rabies glycoprotein (GRab). When these mice are injected with deletion mutant EnvA-pseudotyped rabies virus, only the cells of interest, genetically marked with Tomato, are infected, and their presynaptic partner cells are labeled, resulting in green GFP+ presynaptic cells and their yellow (GFP+ and Tomato+) postsynaptic partners (Fig. 1).
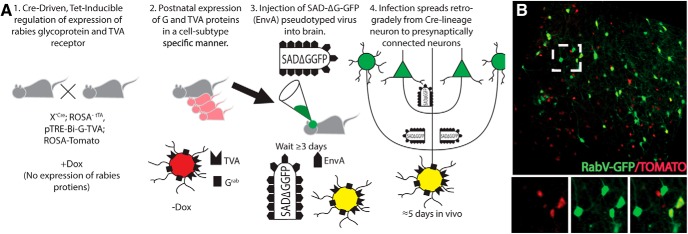
Figure 1.
Strategy to target cell subtype-specific presynaptic connections using a deletion mutation rabies virus. A, 1, The rabies glycoprotein GRab and avian sarcoma leukosis virus (ASLV) avian TVA proteins are required for viral spread in targeted neurons using a cell subtype-specific driver. Cre recombines the ROSA-tTA reporter; tTA is necessary for the removal of inhibition by TRE (tetracycline response element), which prevents expression in the presence of doxycycline. 2, Doxycycline removal activates G and TVA protein expression; ROSA-driven Tomato marks these cells. 3, Targeted infection with RabV GFP pseudotyped virus in neurons expressing both G and TVA proteins, labeled with ROSA-Tomato. 4, GFP+ virus spreads upstream to neurons presynaptically connected to virus-infected G- and TVA-expressing neurons. B, Example of labeling; yellow cells are initially infected G/TVA/Tomato+ neurons; green cells are presynaptically connected neurons. Inset illustrates a pyramidal connected cell in green.
We first determined the efficacy of this approach (i.e., that viral gene expression was expressed in a doxycycline-dependent fashion). Using Emx1-Cre to target pyramidal neurons, we show that doxycycline repressed expression of the pTRE-driven TVA protein necessary for the infection of RabV-GFP, and that, after removal of doxycycline, the TVA protein was re-expressed at levels similar to those of untreated animals (Fig. 2A,B).
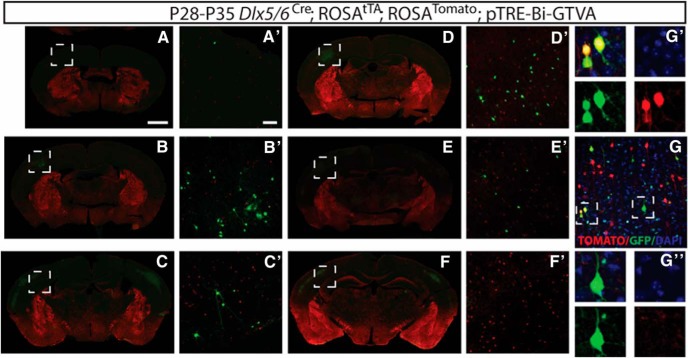
Figure 2.
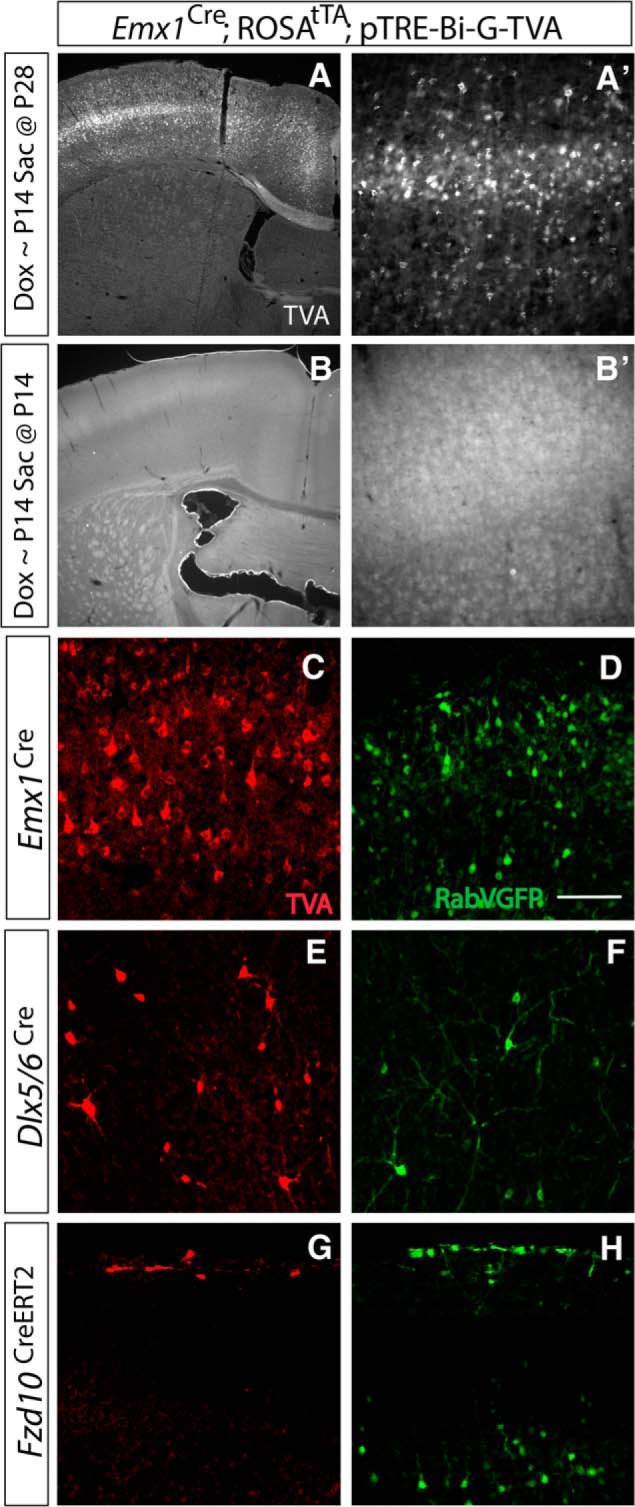
Genetically targeted monosynaptic circuit tracing allows cell subtype-specific labeling of presynaptic inputs. A, A′, Magnification (4×) of somatosensory region (A), shown at higher power at 20× (A′), illustrating TVA protein expression in the absence of doxycycline. Doxycycline feed was administered until P14 with analysis at P28. Removal of doxycycline allowed for TVA protein expression. B, B′, Magnification (4×) of somatosensory region, shown at higher power at 20× (B′), illustrating no TVA protein expression in the presence of doxycycline. Doxycycline feed administered until P14 and analyzed at P14. The presence of doxycycline prevented TVA expression. C, E, G, Cre-driven recombination of TVA receptor protein (Red) in Emx1 lineage excitatory pyramidal cells, Dlx5/6 lineage inhibitory interneurons, and Fzd10 lineage Cajal-Retzius cells. D, F, H, Specific infection using RabV-GFP (Green) to label excitatory pyramidal neurons, inhibitory interneurons, and Cajal-Retzius cells reveals unique patterns of presynaptic connectivity in each cell type. Emx1 lineage and Dlx5/6 lineages were injected at P28 and analyzed at P35. Fzd10 lineage injected at P4 and analyzed at P7. Scale bar, 100 μm.
We then examined whether this approach reliably recombined only in our cell subtypes of interest, as cellular specificity is required for our analyses of connectivity. In three different cell subtypes, we found that our cell-specific monosynaptic tracing labeled the targeted cell population and their presynaptic connections exclusively (Fig. 2). We show using an antibody for TVA that this approach reliably recombines in cells of interest, but not in cells outside of the pyramidal lineage (Emx1-Cre), inhibitory interneuron lineage (Dlx5/6-Cre), or CR cell lineage (Fzd10-CreERT2; Fig. 2C,F,H). Injection of virus into the cortex in each of these neuronal models resulted in unique patterns of labeled connected neurons (Fig. 2E,G,I).
We next wanted to confirm that this approach was effective for targeting cell subtype-specific connections through development and into adulthood. Again using Emx1-Cre, we targeted pyramidal neurons in the first postnatal week, and showed numerous initially infected pyramidal neurons, as well as connected subcortical neurons in thalamus (data not shown). We targeted pyramidal neurons late at P60 as well, and had connectivity patterns similar to our mature P28 data (data not shown; see Fig. 4), indicating that the targeting of embryonically defined progenitors and their mature circuits in adulthood is also possible with this approach.
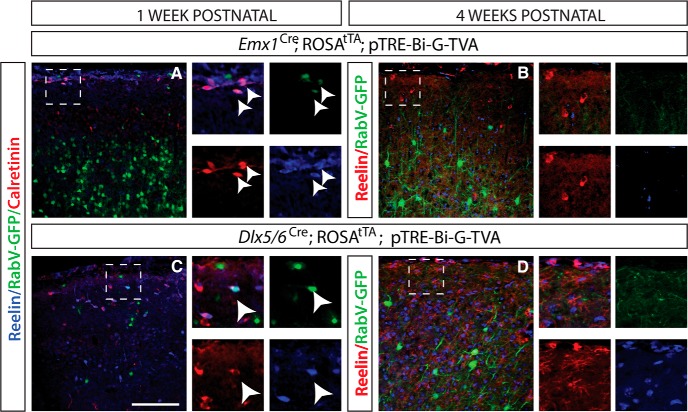
Figure 4.
Inhibitory interneurons and pyramidal neurons receive presynaptic inputs from Cajal-Retzius cells. A, Emx1 infection of excitatory pyramidal neurons at P3 and analysis at P7. Cajal-Retzius cells labeled with calretinin (red) and reelin (blue) are labeled with RabV-GFP virus (arrowheads), indicating that they are presynaptically connected to Emx1 lineage pyramidal neurons. B, Emx1 infections of excitatory pyramidal neurons at P21 and analysis at P28. No Cajal-Retzius cells labeled with reelin (red) are labeled with RabV-GFP virus, indicating the lack of presynaptic connections due to the absence of CR cells. C, Dlx5/6 infection of inhibitory interneurons at P3 and analysis at P7. Cajal-Retzius cells, labeled with calretinin (red) and reelin (blue), are labeled with RabV-GFP virus (green; arrowhead), indicating that they are presynaptically connected to Dlx5/6 lineage interneurons. D, Dlx5/6 infection of inhibitory interneurons at P21 and analysis at P28, showed no Cajal-Retzius cells labeled with reelin (red) and are labeled with RabV-GFP virus (green), indicating the lack presynaptic connections due to absence of CR cells. DAPI is in blue in B and D. Scale bar, 100 μm.
CR cell circuits in early postnatal cerebral cortex
To gain insight into the formation of the earliest cortical circuits in postnatal development, we began an examination of the CR cells and their presynaptic and postsynaptic connections. CR cells secrete reelin, an extracellular matrix protein critical for migration and lamination in embryonic development (Soriano and Del Río, 2005). In mice, the majority of CR cells in the cortex are eliminated during the second week of life, after cortical circuitry has been established (Del Río et al., 1996). In contrast, humans retain large numbers of CR cells (Meyer, 2010). These cells may be important in maintaining normal brain architecture, since their loss is associated with cytological defects found in patients with temporal lobe epilepsy (Haas et al., 2002). CR cells are most prevalent in the first postnatal week in mice, after which their numbers decline. During this time, CR cells make a dense horizontal network of axonal outputs within layer I, and receive both GABAergic and non-GABAergic input, as analyzed via electron microscopy and electrophysiology (Beaulieu et al., 1994; Hestrin and Armstrong, 1996). In the hippocampus, they make connections onto excitatory pyramidal cells, determined by patch-clamp recordings, suggesting that these cells are part of an early cortical network (Kirmse et al., 2007). To understand the role of these cells in the development of cortical microcircuits, we explored the inputs onto CR cells, as well as the outputs of CR cells, by determining whether deeper-layer neurons received inputs from CR cells.
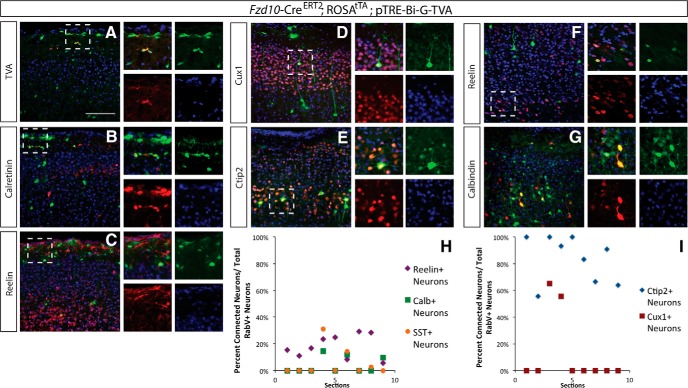
We used this approach to determine CR cell circuitry in the first postnatal week, analyzing presynaptic inputs onto CR cells. We confirmed that we were able to target these neurons using the Frizzled10-CreERT2 line, showing that TVA was only expressed in these cells, and that RabV-GFP+ cells colocalized with calretinin and reelin (Fig. 3A–C). We found that these neurons receive local connections within layer I from inhibitory interneurons, as labeled by RabV-GFP+ calretinin+ (Fig. 3C). We also found that they receive longer-distance inputs from deep-layer neurons in the cortex, particularly layer V Ctip2+ pyramidal neurons (although a small subset of upper layer Ctip2+ neurons were also labeled; Fig. 3D,E). Interestingly, we also found that a number of reelin+ neurons in layer VI made synapses onto the CR cells, indicating that inputs onto these neurons span the length of the developing cortex (Fig. 3F). When we analyzed the distribution of inputs onto CR cells, we found that deep-layer reelin+ and Ctip2+ neurons were the most prevalent subtypes to make inputs onto CR cells (Fig. 3H,I).
Figure 3.
Cajal-Retzius cells receive presynaptic inputs from Ctip2+ excitatory neurons as well as deeper layer reelin+ neurons early in postnatal development. Viral infection into marginal zone at P3, analysis at P7. A, TVA expression (red) from Fzd10-CreER recombination of G and TVA (tamoxifen at E12.5) is specific to the Cajal-Retzius cell lineage (green). B, C, Cajal-Retzius cells labeled by CR (red) and reelin (red) are infected with RabV-GFP virus (green). D, E, Few Cux1+ (red) top-layer neurons show monosynaptic connection, but many Ctip2+ (red) lower-layer pyramidal neurons are connected to the Cajal-Retzius cells (green) early in postnatal development. F, G, Reelin+ interneurons (red) make presynaptic connections onto Cajal-Retzius Cells (green) in the marginal zone; calbindin+ interneurons (red) in the cortex also make connections onto Cajal-Retzius cells (green) near the ventricular surface. DAPI is in blue in A–G. H, Quantification of the pyramidal neuron inputs onto Cajal-Retzius cells. Connected neuronal subtypes were analyzed for each antigen separately and were calculated as follows: percentage of Ctip2+/RabV+ neurons in layers IV–V over the total number of RabV cells in layers IV—V; the percentage of Cux1+/RabV+ neurons in layer II/III over the total number of RabV cells in layer II/III. I, Quantification of the inhibitory interneuron inputs onto Cajal-Retzius cells. Connected neuronal subtypes were analyzed for each antigen separately and were calculated as follows: percentage of reelin+/RabV+ neurons in the layer over the total number of RabV cells; the percentage of SST+/RabV+ neurons over the total number of RabV cells; and the percentage of Calb+, RabV+ neurons over the total number of RabV+ cells. N = 3 brains, 3 slides per brain, per antigen. Scale bar, 100 μm.
We wanted to know whether CR cells make reciprocal connections onto neurons deeper in the cortex, so we injected virus into P4 Emx1-Cre and Dlx5/6 Cre mice to label excitatory neuron or inhibitory neuron inputs, respectively, and killed them at P7. We then stained for reelin and calretinin to label presynaptic CR cells. Significantly, a small number of CR cells was labeled by RabV-GFP+ using this approach (Fig. 4A,C). The prevalence of presynaptic Cajal-Retzius cells was similar in both excitatory and inhibitory targeting experiments. We also examined these inputs at P28, given that CR cells are largely absent from the cerebral cortex by this time, and found that at this age there were no longer connections between either excitatory or inhibitory neurons and CR cells (Fig. 4B,D). These data indicate that CR cells in layer I also make reciprocal connections onto excitatory and inhibitory neurons located in deeper layers of the cortex during the first postnatal week, but these connections are not present by the fourth postnatal week.
Analysis of presynaptic inputs during the first postnatal month
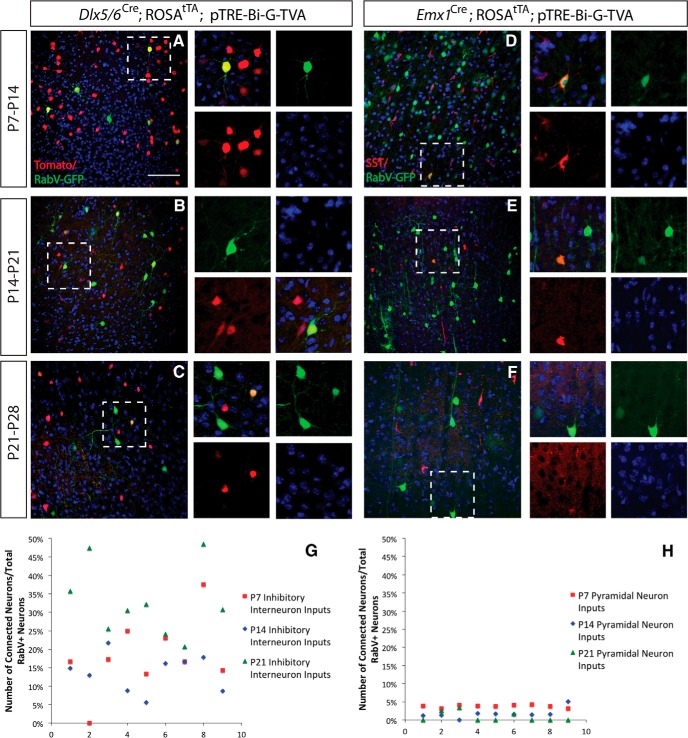
Given our results in CR cell circuits, we wanted to know whether there were large differences in the numbers of inputs onto excitatory and inhibitory neurons during the first month of postnatal development, when synaptogenesis is at its peak. We examined excitatory pyramidal neuron connections onto inhibitory interneurons at the following three stages: P7, P14, and P21 (with analysis 1 week later). We compared inputs from SST+ inhibitory interneurons onto pyramidal neurons at each of these ages, as these interneurons express somatostatin at embryonic stages, making it possible to examine these inputs across all three ages. We found that at P7, 3.78% (SE = 0.13) of the pyramidal neuron inputs were from SST+ interneurons; at P14, 1.75% (SE = 0.45) of the pyramidal neuron inputs were from SST+ interneurons; and at P21, 0.84% (SE = 0.44) of the pyramidal neuron inputs were from SST+ interneurons. A one-way ANOVA indicated that there was a significant difference between these inputs at these three postnatal stages (p = 0.00003). A post hoc Tukey test indicated that there was a significant difference between the numbers of inputs found at P7, compared with P14 (p < 0.01) and at P7 compared with P21 (p < 0.01), although there was not a significant difference between inputs at P14 and P21 (Fig. 5D–F,H). We also compared inhibitory inputs onto excitatory pyramidal neurons at the same postnatal ages, and found the opposite effect: there was an increase in the number of excitatory inputs onto inhibitory neurons during postnatal maturation. We found that at P7, 18.2% (SE = 3.37) of the inhibitory interneuron inputs were from excitatory pyramidal neurons; at P14, 13.69% (SE = 1.72) of the inhibitory interneuron inputs were from excitatory pyramidal neurons; and at P21, 32.82% (SE = 3.23) of the inhibitory interneuron inputs were from excitatory pyramidal neurons. A one-way ANOVA indicated that there was a significant difference among these inputs at these three postnatal stages (p = 0.0002). A post hoc Tukey test indicated that there was a significant difference between the numbers of inputs found at P7, compared with P21 (p < 0.01), and at P14 compared with P21 (p < 0.01), although there was not a significant difference between inputs at P7 and P14 (Fig. 5A–C,G).
Figure 5.
Inhibitory interneuron and pyramidal neuron inputs are developmentally regulated in the first postnatal month. A, Dlx5/6-Cre infection of inhibitory interneurons at P7 and analysis at P14. Excitatory interneurons labeled with Tomato (red) are labeled with RabV-GFP virus, indicating excitatory input onto inhibitory interneurons. B, Dlx5/6-Cre infection of inhibitory interneurons at P14 and analysis at P21. C, Dlx5/6-Cre infection of inhibitory interneurons at P21 and analysis at P28. D, Emx1-Cre infection of excitatory pyramidal neurons at P7 and analysis at P14. Inhibitory interneurons labeled with somatostatin (red) are labeled with RabV-GFP virus, indicating inhibitory input onto excitatory neurons. E, Emx1-Cre infection of excitatory pyramidal neurons at P14 and analysis at P21. Inhibitory interneurons labeled somatostatin (red) and labeled with RabV-GFP. F, Emx1-Cre infection of excitatory pyramidal neurons at P21 and analysis at P28. G, H, Quantification of the number of inputs relative to total labeled neurons (RabV-labeled presynaptic input neurons vs total RabV-labeled presynaptic and postsynaptic neurons) to Dlx5/6 lineage interneurons and Emx1 lineage pyramidal neurons and at P7, P14, and P21. DAPI is in blue in all panels. Scale bar, 100 μm.
Analysis of presynaptic inputs onto cortical interneurons and pyramidal neurons in mature brain
We turned to analyses of larger populations of cortical neurons in mature mice, examining excitatory pyramidal neurons and inhibitory interneurons in somatosensory cortex. We examined the inputs of excitatory neurons onto inhibitory neurons by targeting the interneurons themselves in one set of experiments (Fig. 6), and then we examined the inputs of inhibitory interneurons onto excitatory neurons by targeting excitatory pyramidal neurons in another set of injections (Fig. 7). We wanted to determine whether we could use this approach for both single-cell resolution analyses of the cell subtypes and cellular proteins expressed by presynaptically connected neurons, as above, but also create whole-brain analyses of circuits in the cerebral cortex.
Figure 6.
Presynaptic connectivity mapping shows sparse excitatory inputs onto interneurons in the somatosensory cortex. RabV-GFP infection of primary somatosensory cortex between P21 and P25, and analysis 1 week later. A, Section of bregma at −0.58 mm showing the initial site of injection in barrel field of the primary somatosensory cortex. A′, Magnified site with infected presynaptic excitatory neurons (green) and Dlx5/6 lineage neurons Tomato+ (red). B, Section of bregma at −0.82 mm showing the initial site of injection in barrel field of the primary somatosensory cortex. B′, Magnified site with infected presynaptic excitatory neurons (green) and Dlx5/6 lineage neurons Tomato+ (red). C, Section of bregma at −1.34 mm showing the initial site of injection in barrel field of the primary somatosensory cortex. C′, Magnified site with infected presynaptic excitatory neurons (green) and Dlx5/6 lineage neurons Tomato+ (red). D, Section of bregma at −1.46 mm showing the initial site of injection in barrel field of the primary somatosensory cortex. D′, Magnified site with infected presynaptic excitatory neurons (green) and Dlx5/6 lineage neurons Tomato+ (red). E, Section of bregma at −1.70 mm showing the initial site of injection in barrel field of the primary somatosensory cortex. E′, Magnified site with infected excitatory neurons (green) and Dlx5/6 lineage neurons Tomato+ (red). F, Section of bregma at −2.06 mm showing initial site of injection in the primary somatosensory cortex. F′, Magnified site with infected excitatory neurons (green) and Dlx lineage neurons Tomato+ (red). G, Excitatory neurons RabV-GFP (green) connected presynaptically to local Dlx lineage inhibitory cells, Tomato+ (red), and DAPI (blue). Scale bars: A, 1 mm; A′, 100 μm.
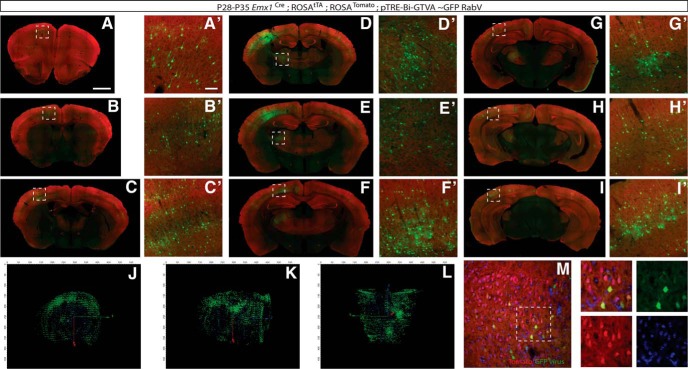
Figure 7.
Presynaptic connectivity mapping of excitatory neurons shows multiple types of interneuron inputs in the somatosensory cortex. RabV-GFP infection of Emx1 lineage cells in the cortex at P21 and analysis at P28. Connected cells are located throughout the cortex. A, Section of bregma at 1.98 mm. A′, Magnification of secondary motor cortex showing infection of connected cells. B, Section of bregma at 0.50 mm. B′, Magnification of second motor cortex showing infected connected cells. C, Section of bregma at −0.82 mm. C′, Magnification of dysgranular region and shoulder region of the primary somatosensory cortex showing infected connected cells. D, Section of bregma at −1.06 mm. D′, Magnification of the ventral thalamus showing infected connected cells. E, Section of bregma at −1.34 mm. E′, Magnification of ventral thalamus depicting infected connected cells. F, Section of bregma at −1.70 mm. F′, Magnification of primary somatosensory cortex showing infected connected cells. G, Section of bregma at −2.46 mm. G′, Magnification of primary and secondary visual cortex showing infected connected cell. H, Section of bregma −2.80 mm. H′, Magnification of primary visual cortex showing infected connected cell. I, Section of bregma at −328 mm. I′, Magnification of primary visual cortex and lateral secondary visual cortex showing infected connected cells. J–L, 3D representation of thresholded GFP pixels (green), red- and green-colabeled pixels within connected cells (yellow), and cell nuclei (blue), providing a visual reference. For clarity, views were chosen slightly off angle from coronal, sagittal, and horizontal views. M, Higher-magnification optical sections illustrate initially infected (yellow) and connected cells (green only), and DAPI (blue). Scale bars: A, 1 mm; A′, 100 μm.
Analysis of mature inhibitory neurons and their presynaptic inputs
We analyzed the excitatory presynaptic inputs onto inhibitory interneurons in the somatosensory cortex, injecting it with virus at P23 (±2 d), and analyzing it 1 week later to determine the distribution of excitatory and inhibitory interneurons onto Dlx5/6 lineage RabV-targeted cells. In this experiment, pyramidal neurons were defined by morphology and by the absence of Tomato expression (Fig. 6). RabV-GFP+ (green only) pyramidal neurons were present predominantly locally, near the injection site in the somatosensory cortex. Unlike the inputs onto pyramidal neurons that have wide regions of collateralization and cortical–cortical connections across hemispheres, we found the excitatory inputs onto inhibitory interneurons were sparse and localized within the same brain region. Furthermore, few inhibitory interneuron inputs were found outside of the injection region, consistent with previous work that has shown very local inputs of inhibitory neurons (Xu and Callaway, 2009; Kinnischtzke et al., 2014), as displayed in single sections from a reconstructed Dlx-5/6 brain (Fig. 6A–F). Excitatory inputs onto inhibitory neurons were found locally as well and were confirmed with confocal imaging of optical sections (Fig. 6I). While this has been shown previously with electrophysiological slice recordings (Fino and Yuste, 2011; Packer and Yuste, 2011), this is, to our knowledge, the first cell subtype circuit-labeling approach to capture this phenomenon within the cytoarchitecture of the whole brain.
Analysis of mature excitatory neurons and their presynaptic inputs
We then analyzed the presynaptic inputs onto excitatory neurons in the somatosensory cortex. We used the Emx1-Cre driver to target pyramidal neurons in the somatosensory cortex, injecting it with virus (±2 d), and analyzing it 1 week later (Fig. 7). Consistent with previously studied inputs, pyramidal neurons from somatosensory cortex received inputs from motor cortex, prefrontal cortex, piriform cortex, basolateral amygdala, and several thalamic nuclei (Fig. 7A–I). We also found reciprocal cortical projections, particularly in somatosensory and motor cortex, also consistent with known pyramidal inputs (Dantzker and Callaway, 2000; Hooks et al., 2013; Lee et al., 2013; Fig. 7C–I). The labeled inputs were much more extensive than those found in the Dlx5/6 mice, and so we took a different approach to analyze these connections. In addition to representative single sections illustrating the span of labeled neurons (Fig. 7A–I), we also reconstructed the entire brain, sectioning from olfactory bulb through the entire cortex, and aligned the sections in order from anterior to posterior. We then imaged these sections on an Axioscan microscope, and again aligned the sections anterior to posterior to recreate the whole brain at a single-cell resolution. These data are illustrated in single panels of sections from the Emx1-infected brains scanned on the Axioscan (Fig. 7A–I), as well as a 3D reconstruction of a single brain, indicating connected and infected cells (Fig. 7J,K,L; details of alignment and reconstruction as well as image processing are discussed in Materials and Methods). The movie of the entire reconstructed brain (Movie 1) indicates region of initially infected and connected neurons. This is, to our knowledge, one of the most comprehensive approaches for imaging neuronal circuits at both a single-cell and whole-brain level. Confocal optical sections of high-resolution images from the same brain illustrate initially infected and connected neurons (Fig. 7M). This pattern of infection was reliable across several brains, indicating that this would also be a useful tool for analyzing defects in pyramidal neuron inputs in these mice. We found that this approach offers a reliable method to determine populations of inputs to target cells of interest.
Reconstructed rendering of serial sections of the entire forebrain, illustrating the pattern of infection and neural connectivity. Initially infected cells are yellow, connected cells are green, and Emx1 lineage cells are red. The movie begins with most anterior sections and contains serial images of one single brain.
Examination of layer-specific cortical connectivity
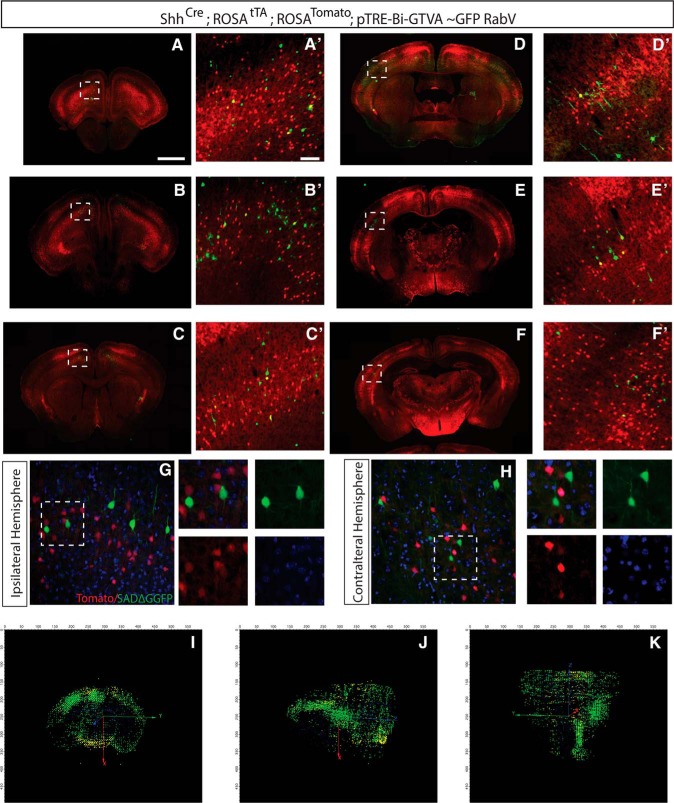
While analyses of large populations of pyramidal cell inputs are useful for broad examinations of inputs, we wanted to determine whether this approach could be used to examine inputs to specific layers of pyramidal neurons. Our viral targeting did not allow layer-specific analyses, and so we used a genetic approach to gain layer-specific targeting. We used a Sonic Hedgehog promoter to drive Cre expression, allowing us to target layer V projection neurons and label their presynaptic connections, injecting into motor cortex and examining the presynaptic connectivity patterns of these neurons. Layer V Shh+ neurons received connections locally within motor cortex, as well as long-distance projections into somatosensory cortex, prefrontal cortex, and contralateral motor cortex (Fig. 8A–H). Subcortical ipsilateral inputs from amygdala and thalamus were also labeled (Fig. 8E,F). 3D reconstructed whole-brain images confirm this pattern of connectivity (Fig. 8I–K). While these patterns are similar to the patterns of inputs seen in the Emx1 pan-pyramidal neuron analyses, we found fewer contralateral and subcortical projections, and the densest input from cingulate and somatosensory cortical areas onto corticofugal neurons; we show this in our reconstruction of the whole brain (Movie 2).
Figure 8.
Labeling of presynaptic input onto Sonic Hedgehog-expressing neurons in the motor cortex. Using Shh-Cre, ROSA-Tomato, ROSA-tTA, and pTRE-Bi-G-TVA mice injected with RabV-GFP virus at P21, layer V Shh+ cortical pyramidal neurons were targeted. Contralateral and subcortical inputs were complementary to injections in Emx1 somatosensory cortex, with many connections to ipsilateral somatosensory cortex and contralateral motor cortex. A, Section of bregma at 2.58 mm. A′, Magnification of motor cortex, yellow initially infected cells, green connected cells, and red Tomato+ cell. B, Section of bregma at 2.22 mm. B′, Magnification of secondary motor cortex, yellow initially infected cells, green connected cells, and red Tomato+ cell. C, Section of bregma at 1.10 mm. C′, Magnification of part of primary and secondary motor cortex, yellow initially infected cells, green connected cells, and red Tomato+ cell. D, Section of bregma at −0.22 mm. D′, Magnification of primary somatosensory cortex, yellow initially infected cells, green connected cells, and red Tomato+ cell. E, Section of bregma at −0.94 mm. E′, Magnification of primary somatosensory cortex, yellow initially infected cells, green connected cells, and red Tomato+ cell. F, Section of bregma at −1.58 mm. F, Magnification of primary somatosensory cortex, yellow initially infected cells, green connected cells, and red Tomato+ cell. G, Ipsilateral somatosensory cortex with RabV-GFP+ connected cells (green only). H, Contralateral motor cortex with RABV-GFP+ connected cells green only. G, H, DAPI staining of cell nuclei (blue). I–K, Reconstruction of pixels representing 3D distribution of connected cells (green) and initially infected cells (colabeled, yellow). Coronal, sagittal, and horizontal views of reconstructed brains shown slightly off angle for clarity. Scale bars: A, 1 mm; A′, 100 μm.
Reconstructed rendering of serial sections of the entire forebrain, illustrating the pattern of infection and neural connectivity. Initially infected cells are yellow, connected cells are green, and Shh lineage cells are red. The movie begins with most anterior sections and contains serial images of one single brain.
In summary, we examined cortical microcircuitry in early postnatal development and in mature brain, examining the Cajal-Retzius cells and their inputs and outputs, as well as the inputs and outputs of excitatory pyramidal neurons and inhibitory interneurons in the cerebral cortex. We examined the development of inhibitory presynaptic input onto excitatory neurons, as well as excitatory input onto inhibitory neurons, and found that these connections mature differently early in postnatal development. Finally, we show that corticofugal pyramidal neurons in motor cortex receive inputs from prefrontal and somatosensory cortex, as well as subcortical inputs from amygdala and thalamus. These findings are summarized in Figure 9.
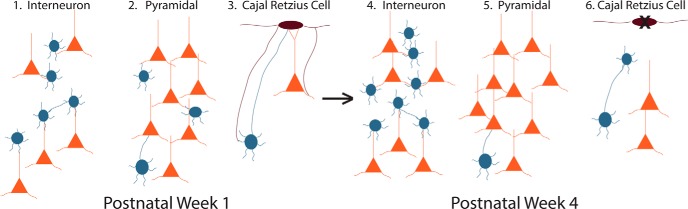
Figure 9.
Summary of findings. We compared the circuit connections at week 1 vs week 4 between inhibitory interneurons (blue), excitatory pyramidal cells (orange), and Cajal-Retzius cells (purple). During week 1, we observe (1) high inhibitory interneuron input onto excitatory pyramidal neurons; (2) high excitatory pyramidal cell input onto inhibitory interneurons; and (3) layer I Cajal-Retzius cells receive input from Ctip2+ pyramidal neurons and inhibitory interneurons as well as input onto both. During postnatal week 4, we observe (4) decreased inhibitory interneuron input onto excitatory pyramidal neurons; (5) increased excitatory input onto inhibitory interneurons in mature mice brains; and (6) no presynaptic connection of Cajal-Retzius cells to inhibitory interneurons or excitatory pyramidal cells.
Discussion
This is the first study to systematically target the presynaptic connectivity of discrete neuronal subtypes in the cerebral cortex in both developing and mature cerebral cortex. Mapping cortical connectivity in the developing mammalian brain has been an intractable problem, in part because it has not been possible to analyze connectivity with cell subtype precision. We have created a methodology for reconstructing these circuits in the entire brain, allowing analyses at a single-cell level as well as whole-brain analyses of connectivity. We illustrate that this approach is a powerful tool to analyze cell subtype-specific circuits in vivo, during development, and throughout the life span. Our approach allows precise genetic control of the starting population of cells for tracing, and we show here that it reliably labels those connections in excitatory pyramidal neurons, in corticofugal pyramidal neurons, in inhibitory interneurons, and in Cajal-Retzius cells.
CR cells have long been an enigma in developmental neuroscience, critical for their expression of reelin and for the regulation of layer formation and migration in embryonic development, they are ultimately lost in the murine brain in the second postnatal week (although they persist longer in the human neocortex; Deguchi et al., 2003; Frotscher et al., 2003). Their role in postnatal development has been largely unexplored; it has been shown in the hippocampus that these cells fire action potentials and receive synapses from inhibitory neurogliaform cells, and GABAergic input from oriens lacunosum-moleculare cells that serves as excitatory input onto CR cells (Quattrocolo and Maccaferri, 2013). Their inputs in the cerebral cortex have been less explored. We show here that Cajal-Retzius cells receive connections from excitatory pyramidal neurons in layer V as well as inhibitory reelin+ interneurons in the subplate. We also find that Cajal-Retzius cells make reciprocal connections onto excitatory and inhibitory interneurons in the first postnatal week. The significance of these connections, present just before the peak of synaptogenesis, may be important for establishing the initial network activity of the cortex: there may be an interesting functional role for CR cells postnatally in refining cortical networks, perhaps even as neurons reach their final destination in the cortical layers (analogous to the situation in the hippocampus; see from a decade ago Soriano and Del Río). Future research analyzing the electrical properties of these inputs would provide additional insight into their function. While we see loss of CR cell connections by P28, it is difficult to determine whether this is due to the loss of CR cells or loss of connectivity, as most CR cells are lost in the cortex by P28.
We find that cortical inhibitory interneurons have largely local connections labeled using our approach: only local excitatory pyramidal neurons are labeled with this approach, and only a small number of pyramidal neurons makes inputs onto interneurons from the contralateral hemisphere. We find that the number of excitatory inputs onto inhibitory interneurons increases during the first month of postnatal development, and this is consistent with the slow electrophysiological maturation of inhibitory neurons, compared with excitatory neurons, in the cortex (Chu and Anderson, 2015). The limitations of our approach make it difficult to determine whether the inhibitory labeling that we observe are local homotypic connections or are primarily coinfected neurons, and so we have largely limited our discussion to the excitatory inputs of inhibitory neurons.
In contrast, the inhibitory inputs onto excitatory neurons appear to undergo refinement during the first month of postnatal life: we find fewer inhibitory inputs onto excitatory neurons at P21, compared with P7, which is consistent with the peak of synaptogenesis and pruning in excitatory neurons of the cortex (Colón-Ramos, 2009). In comparison, we find that pyramidal neuron inputs from other pyramidal neurons are extensive, spanning both hemispheres and coming from many subcortical areas. The density of the subcortical and cortical–cortical connections is apparent even when we examine connections from a single cortical layer: corticofugal neurons from the Sonic Hedgehog lineage receive numerous dense inputs from contralateral cortex, including the thalamus, amygdala, and hippocampus. Whole-brain reconstructions of connectivity maps of unique neuronal subtypes, which we have included here in two analyses of cortical pyramidal neurons, may be a powerful means to examine neuronal circuitry across the entire brain in future studies.
Analysis of the presynaptic connections of discrete neuronal subtypes is a powerful tool for studying the plasticity of circuits in development, changes in connectivity after perturbation by gain or loss of function experiments, and analysis of circuit-level defects in animal models of human neurological and psychiatric disease. Importantly, because this approach targets particular populations of cells, it would also be a useful tool to modulate activity using optogenetics to drive activity in a given neuronal population or its presynaptic partners. This approach lends itself to analysis in development and in the mature brain, making it possible to answer developmental questions about circuit formation and function using the same methodology. While large numbers of cells are targeted with this approach, the reliability of connected input regions is consistent across animals, validating this approach as a tool for analyses of circuits in gain-of-function and loss-of-function experiments.
It is important to note that in the Emx1 lineage-tracing experiments we find it difficult to analyze cortical–cortical connections due to the sheer number of labeled cells using this approach; regardless, the reliability of the infected and connected cells labeled across animals makes it possible to analyze differences in inputs onto pyramidal cells in a region-specific fashion in mouse mutants, and during development. Additionally, our approach does not allow us to validate that the spread of virus occurs only via synaptic connections early in postnatal development. While adult studies have confirmed that rabies virus is only able to spread via synaptic contacts in adult brains, we have not tested here whether the virus is able to spread by other means at early postnatal ages, and so it is theoretically possible that not all labeled connections are synaptic. However, there is no evidence supporting the spread of the virus using any other mechanism, and our data are consistent with previous studies using single-cell electrophysiological analyses (Wickersham et al., 2007).
Several recent studies have examined mature circuitry in the basal ganglia, olfactory bulb, dorsal root ganglion, and hippocampus (Miyamichi et al., 2011; Wall et al., 2013; Sun et al., 2014). Previous retrograde tracing approaches used a two-virus delivery system, with a “helper virus” to deliver complementary proteins necessary for infection and spread, and a second deletion mutant rabies virus to target initially infected and connected cells with the fluorescent proteins necessary for analysis (Wall et al., 2010). Early approaches lacked cell subtype specificity. Modified approaches with floxed adeno-associated virus (AAV) helper viruses are not useful for developmental studies where the long timelines required for AAV-driven expression of viral proteins make early developmental analyses impossible (Miyamichi et al., 2011).
This research builds upon existing monosynaptic circuit tracing techniques developed by the Weible et al. (2010). The primary tools available required a dual-virus system with an AAV helper virus to place the TVA and G-proteins necessary for infection and spread, respectively, into cells before infection with pseudotyped G-deleted rabies virus. The long AAV viral infection rate, combined with the necessity of two injections, has made it impossible to analyze early connectivity. Furthermore, the recombination of G and TVA embryonically, as we have done here, allows for the targeting of populations of neurons that are defined by progenitor pools and by gene expression early in embryonic development, offering a powerful approach to analyze related progenitor populations in the developing brain. A recent floxed GFP-G-TVA mouse, while a simpler approach, has proven to be difficult to use in longitudinal experiments, as the floxed allele is apparently not able to maintain the expression of G and TVA well into adulthood (Li et al., 2013). We show here that our approach, by combining Cre- and Tet-driven genetics, is able to efficiently label presynaptic connections at stages throughout development, and does so in a tightly controlled fashion, with only the neuronal subtypes of interest able to express the genes necessary for infection and spread. This is an important tool to be able to analyze circuits in development and in disease, and will be beneficial to design gain-of-function and loss-of-function experiments at the level of cell type-specific circuits in vivo. Furthermore, our tracing experiments here illustrate both macroanalyses and microanalyses of circuitry in vivo, using a whole-brain approach while retaining single-cell resolution. We are able to reconstruct the entire brain with both connected and initially infected cells, and have also shown that we can use this to analyze the specific neuronal subtypes that are presynaptically connected to our cells of interest. These datasets contain both single-cell resolution and whole-brain resolution, making analyses of cellular as well as circuit-level defects possible. These tools will not only be powerful for analyzing large circuit-level defects in mouse mutants, but the cellular resolution will allow us to finally begin to untangle the complex wiring diagrams made by the billions of neurons in mammalian cerebral cortex, a task originally thought to be possible only in invertebrates.
Footnotes
We thank Trung Huynh for help with genotyping; Dr. Andrew Levitt for sharing the TVA antibody; Dr. Clifford Kentros for sharing the pTRE-Bi-G-TVA mice; Dr. Ian Wickersham and Dr. Ed Callaway for sharing the SADdeltaGmcherry and SADdeltaGGFP viruses, as well as BHK-EnvA and BHK-19G cells; Dr. Ian Wickersham for help with troubleshooting viral amplification; Dr. Luis Fuentealba for sharing TVA-800 cells for tittering; Dr. Dorit Ron for use of the stereotaxic rig; Dr. Vincent Warnault for help with optimizing viral injections; and Konstantin Sudakov for help with developing the tools for image alignment.
References
- Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsáki G, Cauli B, Defelipe J, Fairén A, Feldmeyer D, Fishell G, Fregnac Y, Freund TF, Gardner D, Gardner EP, Goldberg JH, Helmstaedter M, Hestrin S, Karube F, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C, Campistron G, Crevier C. Quantitative aspects of the GABA circuitry in the primary visual cortex of the adult rat. J Comp Neurol. 1994;339:559–572. doi: 10.1002/cne.903390407. [DOI] [PubMed] [Google Scholar]
- Bell LA, Bell KA, McQuiston AR. Synaptic muscarinic response types in hippocampal CA1 interneurons depend on different levels of presynaptic activity and different muscarinic receptor subtypes. Neuropharmacology. 2013;73:160–173. doi: 10.1016/j.neuropharm.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Godinho LN, Thomaidou D, Tan SS, Gulisano M, Parnavelas JG. Emx1 is a marker for pyramidal neurons of the cerebral cortex. Cereb Cortex. 2001;11:1191–1198. doi: 10.1093/cercor/11.12.1191. [DOI] [PubMed] [Google Scholar]
- Chu J, Anderson SA. Development of cortical interneurons. Neuropsychopharmacology. 2015;40:16–23. doi: 10.1038/npp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Ramos DA. Synapse formation in developing neural circuits. Curr Top Dev Biol. 2009;87:53–79. doi: 10.1016/S0070-2153(09)01202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E, Haydar TF. Regulation of neural progenitor cell development in the nervous system. J Neurochem. 2008;106:2272–2287. doi: 10.1111/j.1471-4159.2008.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzker JL, Callaway EM. Laminar sources of synaptic input to cortical inhibitory interneurons and pyramidal neurons. Nat Neurosci. 2000;3:701–707. doi: 10.1038/76656. [DOI] [PubMed] [Google Scholar]
- Deguchi K, Inoue K, Avila WE, Lopez-Terrada D, Antalffy BA, Quattrocchi CC, Sheldon M, Mikoshiba K, D'Arcangelo G, Armstrong DL. Reelin and disabled-1 expression in developing and mature human cortical neurons. J Neuropathol Exp Neurol. 2003;62:676–684. doi: 10.1093/jnen/62.6.676. [DOI] [PubMed] [Google Scholar]
- Del Río JA, Heimrich B, Supèr H, Borrell V, Frotscher M, Soriano E. Differential survival of Cajal-Retzius cells in organotypic cultures of hippocampus and neocortex. J Neurosci. 1996;16:6896–6907. doi: 10.1523/JNEUROSCI.16-21-06896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fino E, Yuste R. Dense inhibitory connectivity in neocortex. Neuron. 2011;69:1188–1203. doi: 10.1016/j.neuron.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K, Wong RO. A comparison of experience-dependent plasticity in the visual and somatosensory systems. Neuron. 2005;48:465–477. doi: 10.1016/j.neuron.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Frotscher M, Haas CA, Förster E. Reelin controls granule cell migration in the dentate gyrus by acting on the radial glial scaffold. Cereb Cortex. 2003;13:634–640. doi: 10.1093/cercor/13.6.634. [DOI] [PubMed] [Google Scholar]
- Gittis AH, Kreitzer AC. Striatal microcircuitry and movement disorders. Trends Neurosci. 2012;35:557–564. doi: 10.1016/j.tins.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Yan Y, Li H, He D, Pleasure SJ, Zhao C. Characterization of the Frizzled10-CreERT2 transgenic mouse: an inducible Cre line for the study of Cajal-Retzius cell development. Genesis. 2009;47:210–216. doi: 10.1002/dvg.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr Opin Cell Biol. 2005;17:639–647. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Haas CA, Dudeck O, Kirsch M, Huszka C, Kann G, Pollak S, Zentner J, Frotscher M. Role for reelin in the development of granule cell dispersion in temporal lobe epilepsy. J Neurosci. 2002;22:5797–5802. doi: 10.1523/JNEUROSCI.22-14-05797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hestrin S, Armstrong WE. Morphology and physiology of cortical neurons in layer I. J Neurosci. 1996;16:5290–5300. doi: 10.1523/JNEUROSCI.16-17-05290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks BM, Mao T, Gutnisky DA, Yamawaki N, Svoboda K, Shepherd GM. Organization of cortical and thalamic input to pyramidal neurons in mouse motor cortex. J Neurosci. 2013;33:748–760. doi: 10.1523/JNEUROSCI.4338-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kinnischtzke AK, Simons DJ, Fanselow EE. Motor cortex broadly engages excitatory and inhibitory neurons in somatosensory barrel cortex. Cereb Cortex. 2014;24:2237–2248. doi: 10.1093/cercor/bht085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmse K, Dvorzhak A, Henneberger C, Grantyn R, Kirischuk S. Cajal Retzius cells in the mouse neocortex receive two types of pre- and postsynaptically distinct GABAergic inputs. J Physiol. 2007;585:881–895. doi: 10.1113/jphysiol.2007.145003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, Azouaou N. Virus-delivered RNA interference in mouse brain to study addiction-related behaviors. Methods Mol Biol. 2010;602:283–298. doi: 10.1007/978-1-60761-058-8_17. [DOI] [PubMed] [Google Scholar]
- Lee S, Kruglikov I, Huang ZJ, Fishell G, Rudy B. A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nat Neurosci. 2013;16:1662–1670. doi: 10.1038/nn.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Stam FJ, Aimone JB, Goulding M, Callaway EM, Gage FH. Molecular layer perforant path-associated cells contribute to feed-forward inhibition in the adult dentate gyrus. Proc Natl Acad Sci U S A. 2013;110:9106–9111. doi: 10.1073/pnas.1306912110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Bergles DE. Synaptic signaling between neurons and glia. Glia. 2004;47:290–298. doi: 10.1002/glia.20060. [DOI] [PubMed] [Google Scholar]
- Lin SC, Huck JH, Roberts JD, Macklin WB, Somogyi P, Bergles DE. Climbing fiber innervation of NG2-expressing glia in the mammalian cerebellum. Neuron. 2005;46:773–785. doi: 10.1016/j.neuron.2005.04.025. [DOI] [PubMed] [Google Scholar]
- McConnell SK. Strategies for the generation of neuronal diversity in the developing central nervous system. J Neurosci. 1995;15:6987–6998. doi: 10.1523/JNEUROSCI.15-11-06987.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G. Building a human cortex: the evolutionary differentiation of Cajal-Retzius cells and the cortical hem. J Anat. 2010;217:334–343. doi: 10.1111/j.1469-7580.2010.01266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, Callaway EM, Horowitz MA, Luo L. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Nikolenko V, Poskanzer KE, Yuste R. Two-photon photostimulation and imaging of neural circuits. Nat Methods. 2007;4:943–950. doi: 10.1038/nmeth1105. [DOI] [PubMed] [Google Scholar]
- Packer AM, Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J Neurosci. 2011;31:13260–13271. doi: 10.1523/JNEUROSCI.3131-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat Neurosci. 2013;16:1068–1076. doi: 10.1038/nn.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Williams BP, Götz M. The generation of cellular diversity in the cerebral cortex. Ciba Found Symp. 1995;193:71–84. doi: 10.1002/9780470514795.ch4. [DOI] [PubMed] [Google Scholar]
- Quattrocolo G, Maccaferri G. Novel GABAergic circuits mediating excitation/inhibition of Cajal-Retzius cells in the developing hippocampus. J Neurosci. 2013;33:5486–5498. doi: 10.1523/JNEUROSCI.5680-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland DC, Weible AP, Wickersham IR, Wu H, Mayford M, Witter MP, Kentros CG. Transgenically targeted rabies virus demonstrates a major monosynaptic projection from hippocampal area CA2 to medial entorhinal layer II neurons. J Neurosci. 2013;33:14889–14898. doi: 10.1523/JNEUROSCI.1046-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano E, Del Río JA. The cells of Cajal-Retzius: still a mystery one century after. Neuron. 2005;46:389–394. doi: 10.1016/j.neuron.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nguyen AQ, Nguyen JP, Le L, Saur D, Choi J, Callaway EM, Xu X. Cell-type-specific circuit connectivity of hippocampal CA1 revealed through cre-dependent rabies tracing. Cell Rep. 2014;7:269–280. doi: 10.1016/j.celrep.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci U S A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall N, De La Parra M, Callaway EM, Kreitzer AC. Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron. 2013;79:347–360. doi: 10.1016/j.neuron.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dye CA, Sohal V, Long JE, Estrada RC, Roztocil T, Lufkin T, Deisseroth K, Baraban SC, Rubenstein JL. Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J Neurosci. 2010;30:5334–5345. doi: 10.1523/JNEUROSCI.5963-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weible AP, Schwarcz L, Wickersham IR, Deblander L, Wu H, Callaway EM, Seung HS, Kentros CG. Transgenic targeting of recombinant rabies virus reveals monosynaptic connectivity of specific neurons. J Neurosci. 2010;30:16509–16513. doi: 10.1523/JNEUROSCI.2442-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Sullivan HA, Seung HS. Production of glycoprotein-deleted rabies viruses for monosynpatic tracing and high-level expression in neurons. Nat Protocols. 2010;3:595–606. doi: 10.1038/nprot.2009.248. [DOI] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Xu X, Callaway EM. Laminar specificity of functional input to distinct types of inhibitory cortical neurons. J Neurosci. 2009;29:70–85. doi: 10.1523/JNEUROSCI.4104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Guan W, Pleasure SJ. A transgenic marker mouse line labels Cajal-Retzius cells from the cortical hem and thalamocortical axons. Brain Res. 2006;1077:48–53. doi: 10.1016/j.brainres.2006.01.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reconstructed rendering of serial sections of the entire forebrain, illustrating the pattern of infection and neural connectivity. Initially infected cells are yellow, connected cells are green, and Emx1 lineage cells are red. The movie begins with most anterior sections and contains serial images of one single brain.
Reconstructed rendering of serial sections of the entire forebrain, illustrating the pattern of infection and neural connectivity. Initially infected cells are yellow, connected cells are green, and Shh lineage cells are red. The movie begins with most anterior sections and contains serial images of one single brain.