Abstract
Protein arginine methyltransferases (PRMTs) mediate the methylation of a number of protein substrates of arginine residues and serve critical functions in many cellular responses, including cancer development, progression, and aggressiveness, T-lymphocyte activation, and hepatic gluconeogenesis. There are nine members of the PRMT family, which are divided into 4 types (types I–IV). Although most PRMTs do not require posttranslational modification (PTM) to be activated, fine-tuning modifications, such as interactions between cofactor proteins, subcellular compartmentalization, and regulation of RNA, via micro-RNAs, seem to be required. Inflammation is an essential defense reaction of the body to eliminate harmful stimuli, including damaged cells, irritants, or pathogens. However, chronic inflammation can eventually cause several types of diseases, including some cancers, atherosclerosis, rheumatoid arthritis, and periodontitis. Therefore, inflammation responses should be well modulated. In this review, we briefly discuss the role of PRMTs in the control of inflammation. More specifically, we review the roles of four PRMTs (CARM1, PRMT1, PRMT5, and PRMT6) in modulating inflammation responses, particularly in terms of modulating the transcriptional factors or cofactors related to inflammation. Based on the regulatory roles known so far, we propose that PRMTs should be considered one of the target molecule groups that modulate inflammatory responses.
1. Introduction
Inflammation, the body's physiological protective response to infection by pathogens, is an important component of innate immunity. Inflammation can be categorized as either acute or chronic. Recognition of pathogen-specific molecules, such as lipopolysaccharides, by pattern recognition receptors (PRRs) triggers acute inflammation; among PRRs, toll-like receptors (TLRs) have been intensively studied [1]. In response to stimulation of TLRs by an appropriate pathogen, many molecular events, including activation of nuclear factor- (NF-) κB and activator protein- (AP-) 1 signal pathways, are instigated and, consequently, transcription of genes that code proinflammatory enzymes, such as inducible nitric oxide (NO) synthase (iNOS) and cyclooxygenase- (COX-) 2, is increased [2, 3]. iNOS-induced NO and COX-2-derived prostaglandin E2 (PGE2) act as key mediators of active inflammation, affecting essentially all stages of the inflammation process [4, 5]. Because acute inflammation, a generic response that attempts to remove the initial cause of infection, requires constant stimulation to be maintained, it begins to attenuate as stimuli decline [6, 7]. A serious complication comes up during long-lasting inflammation condition, known as chronic inflammation. Chronic inflammation can cause inflammatory-related or autoimmune diseases, including rheumatoid arthritis, Alzheimer's disease, systemic lupus erythematosus, asthma, atherosclerosis, cancer, and ischemic heart disease [8, 9].
Biologically, methylation is a reaction that adds a methyl group to substrates, including DNA, RNA, and proteins, via various methyltransferases. DNA methylation mainly occurs at cytosine-phosphate-guanine (CpG) sites, where a cytosine follows a guanine in the DNA sequence. The cytosine in CpG sites is methylated by DNA methyltransferases to form 5-methylcytosine. In mammals, 70 to 80% of CpG cytosines exist as methylated form, and CpG methylation is a key reaction in epigenetics because it acts as a switch that turns a gene on or off. Additionally, protein methylation, a posttranslational modification, is a reaction that covalently adds a methyl group to specific amino acid residues; these reactions can be divided into two main categories: N-methylation and O-methylation (carboxymethylation). The methylation of lysine, arginine, histidine, alanine, proline, glutamine, phenylalanine, asparagine, and methionine is a type of N-methylation, while O-methylation involves the methylation of glutamic acid and aspartic acid. The creation of these methylated amino acids is catalyzed by methyltransferases that utilize S-adenosyl-L-methionine (AdoMet) as the methyl donor. Three types of AdoMet-dependent methyltransferases are defined, based on their structural features. The largest class (class I) has a seven-strand twisted β-sheet structure [10]. Class II encompasses SET lysine methyltransferases [11]. Finally, class III contains membrane-associated enzymes [12].
Arginine methylation is catalyzed by the protein arginine-N-methyltransferase (PRMT) family and is observed in both cytoplasmic and nuclear proteins. Methylation of arginine residues is involved in many cellular responses, including RNA splicing, signaling transduction, DNA damage repair, and protein-protein interactions. The functions of arginine-methylated proteins are listed in Table 1. Arginine is methylated in three different ways: monomethylated arginine (MMA), symmetrically dimethylated arginine (sDMA), and asymmetrically dimethylated arginine (aDMA). MMA is considered an intermediate form of DMA. Each type of methylarginine is produced by one of nine PRMTs (Figure 1).
Table 1.
The biological function of methylated arginine.
| Postulated or proven function | Proteins involved |
|---|---|
| mRNA splicing | Motor neuron proteins Small nuclear ribonucleoprotein |
| Signal transduction | Interferon receptor |
| Cellular proliferation | Transcription factor |
| Chromatin remodeling | Histones |
| Transcriptional coactivator | Nuclear receptor, p53 |
| Protein-protein interaction | Inter- and intramolecules |
| Translocation | hnRNP |
| Myelogenesis | Myelin basic protein |
Figure 1.
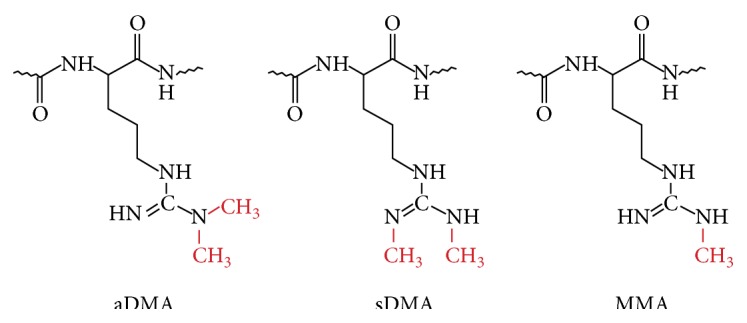
Conformation of three different methylarginines.
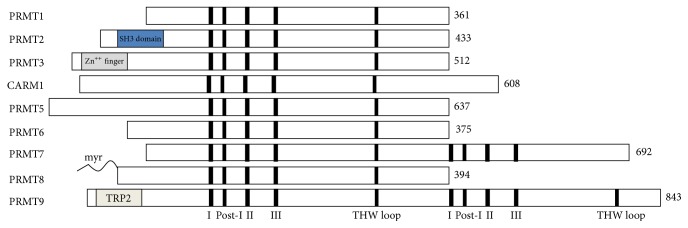
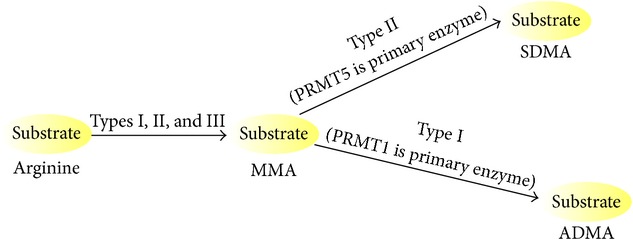
PRMT family members are a class I enzyme, having a set of four signature motifs (I, post-I, II, and III) and a conserved THW loop [13]. Motif I, post-I, and the THW loop are important to the formation of the AdoMet-binding pocket (Figure 2) [14]. Glycine- and arginine-rich patches (GAR motifs) in substrates are mainly methylated by PRMTs, but coactivator-associated arginine methyltransferase 1 (CARM1) is an exception. PRMTs are divided into 4 types (types I–IV) of enzymes (Figure 2). Type I arginine methyltransferase, the most common type of PRMT, induces asymmetric dimethylation, adding two methyl groups to the terminal nitrogen atoms (ω-NG,NG-dimethylarginine). Six enzymes are categorized as type I PRMTs: PRMT1, PRMT2, PRMT3, PRMT4, PRMT6, and PRMT8 [15]. Among them, PRMT1 is the predominant type I enzyme. Type II PRMTs add one methyl group to the terminal nitrogen atoms (ω-NG,NG-dimethylarginine) by catalyzing the symmetric dimethylation of arginine side chains. PRMT5 is associated with this type, whereas PRMT7 is thought to belong to type II, although this is still controversial [16]. Recently, PRMT9 turned out to belong to the type II enzyme that methylates RNA splicing factor SF3B2 [17, 18]. Type III enzymes generate monomethyl arginine as their final product, even though monomethylated arginine at terminal nitrogen atoms (ω-NG-methylarginine) is an essential intermediate of both types I and II PRMT reactions [19]. Type IV methyltransferases catalyze monomethylation of the internal nitrogen atom (N-methylarginine), which is found only in fungi (Table 2) [20].
Figure 2.
Structure of the PRMT family.
Table 2.
Classification of the PRMT family.
| PRMTs | Family | Methylarginine formation by PRMTs |
|---|---|---|
| PRMT1 | Type I |
|
| PRMT2 | Type I | |
| PRMT3 | Type I | |
| PRMT4 (CARM1) | Type I | |
| PRMT5 | Type II | |
| PRMT6 | Type I | |
| PRMT7 | Type II (controversial) and type III | |
| PRMT8 | Type I | |
| PRMT9 | Type II |
Most PRMTs are active when they are expressed as purified recombinant proteins, which indicates that PRMTs do not require additional processing or PTMs to maintain their activity. However, there are mechanisms for fine-tuning PRMT activity, such as PTMs, interacting with regulatory proteins, subcellular compartmentalization, and control of RNA levels by micro-RNAs. Regulation of PRMTs is explained in Table 3.
Table 3.
Regulatory mechanisms of PRMTs.
(a) PTMs: PRMT activities are modulated by PTMs
| PRMTs | Types of PTM | Enzymes involved in PTM | Biological role of PTM | Reference |
|---|---|---|---|---|
| CARM1 | Phosphorylation at S217 | Unidentified kinase | Activating by regulation to bind with AdoMet | [21, 22] |
| Glycosylation | N-acetylglucosamine transferase (OGT) | Activating by controlling the phosphorylation of CARM1 | [23, 24] | |
| Auto-arginine methylation | Unidentified | Unclear | [25] | |
|
| ||||
| PRMT5 | Amino-terminal phosphorylation | Janus kinase 2 (JAK2) | Inactivating via blocking the interaction between PRMT5 and methylosome protein 50 (MEP50) | [26] |
|
| ||||
| PRMT6 and PRMT8 | Auto-arginine methylation | Unidentified | Unclear | [27, 28] |
(b) Regulatory partner: interaction with regulatory proteins can control the activity of PRMTs
| PRMTs | Regulatory partner | Biological role | Reference |
|---|---|---|---|
| PRMT5 | MEP50 | Required for PRMT5 activation | [29] |
| SWI/SNF chromatin complex | Elevates the activity of MEP50-PRMT5 toward histone substrates | [30] | |
| Exon junction complex component and RNA-binding protein Y14 | Enhances MEP50-PRMT5 activity toward Sm proteins | [31] | |
|
| |||
| PRMT1 | Orphan nuclear receptor TR3 (NR4A1) | Inhibits PRMT1 enzyme activity | [32] |
| BTG1-binding chromatin assembly factor 1 (CAF1) | Negatively regulates PRMT1 activity | [33] | |
| BTG1 | Increases PRMT1 activity | [34] | |
| PRMT2 | Stimulates PRMT1 activation | [35] | |
|
| |||
| PRMT3 | Tumor suppressor DAL1 | Blocks PRMT3 methyltransferase ability | [36] |
|
| |||
| PRMT7 | CCCTC-binding factor like (CTCFL) | Increases PRMT7 activity | [37] |
(c) miRNA regulation
| PRMTs | Type of miRNA | Reference |
|---|---|---|
| PRMT5 | miR-19a, miR-25, miR-32, miR-92, miR-92b, and miR-96 | [38] |
2. The Role of PRMTs in Inflammatory Responses
According to a number of recent reports, four PRMTs are currently thought to be correlated with inflammatory responses: CARM1, PRMT1, PRMT5, and PRMT6.
2.1. CARM1 (PRMT4)
CARM1, also known as PRMT4 (protein arginine N-methyltransferase 4), regulates many proteins involved in DNA packing, transcription regulation, pre-mRNA splicing, and mRNA stability by inducing methylation of the guanidine nitrogen of arginine residues of substrates. There are two classes of CARM1 substrates: chromatin remodeling proteins (histone H3 and p300/CBP), which are included in class 1 substrates, and proteins possessing RNA-binding activity, such as PABP1, TARPP, HuR, HuD, and splicing factors [46]. CARM1 acts as a secondary coactivator and is associated with the p160 family (SRC-1, GRIP1, and AIB) of transcriptional coactivators, which are involved in gene activation by steroid hormone receptors [47]. CARM1 also associates with CBP/p300 transcriptional coactivators that activate steroid hormone receptors and C/EBP-mediated gene expression; it functions as a coactivator or corepressor of CBP/P300 molecules. As a coactivator, CARM1 is recruited to nuclear receptors by p160 coactivator, which is activated by hormone. Chromatin remodeling occurs through histone acetylation and methylation proximate to the hormone response element (HRE); as a consequence, transcription is stimulated. In contrast, CARM1 acts as a corepressor of cyclic AMP-induced signaling when accompanied by CBP/p300. CARM1-methylated CBP was found to inhibit transcriptional activity of CREB by blocking the interaction between the KIX domain (the CREB and MYB interaction domain in CBP) and the kinase-inducible domain (KID) of CREB [48].
2.1.1. The Role of CARM1 in Inflammation through the Regulation of NF-κB
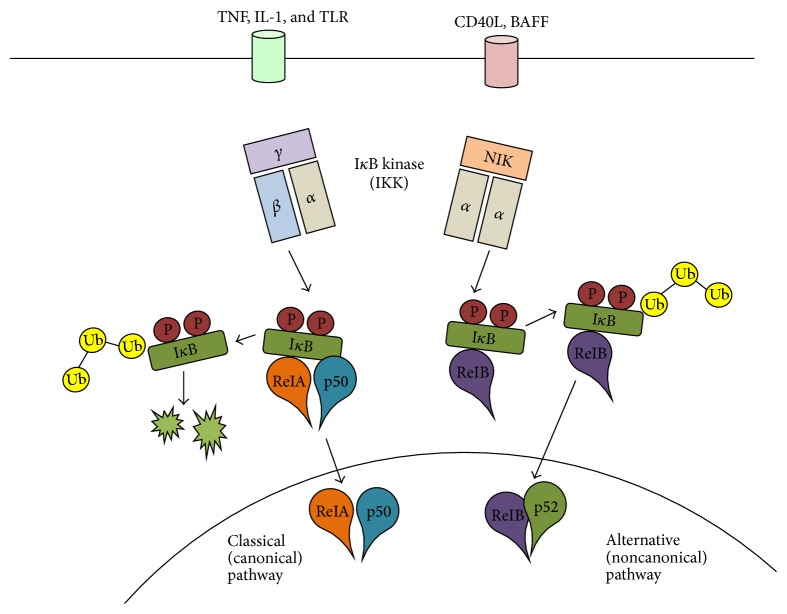
Nuclear factor- (NF-) κB is one of the most important transcriptional factors because it regulates the transcription of many proteins involved in inflammatory diseases, autoimmune diseases, septic shock, viral infection, and immune development. Indeed, elevated NF-κB activity has been detected in the airways of asthmatic patients [49]. Increased expression of NF-κB has been noted in Crohn's disease and ulcerative colitis patients [50]. Additionally, upregulated NF-κB was observed in the synovial fluid of patients with rheumatoid arthritis [51]. The transcription factor, NF-κB, is composed of homo- or heterodimers of subunits (members of the Rel family), including p50, p52 (p49), p65 (Rel A), c-Rel, and Rel B. All these proteins contain a highly conserved domain known as the Rel homology domain (RHD), which consists of about 300 amino acids and plays a role in homologous or heterologous dimerization, DNA binding, and nuclear translocation. Among the various NF-κB subunit combinations, the p65/p50 and c-Rel/p50 heterodimers are the most commonly described forms.
There are two pathways leading to NF-κB activation: the classic (canonical) pathway and the alternative (noncanonical) pathway (Figure 3) [52]. The pathways are defined based on different requests for IKK subunits that regulate NF-κB activation at an upstream stage. The IKK complexes are composed of three subunits, including IKKα (IKK1) and IKKβ (IKK2), which are kinase subunits, and a regulatory subunit IKKγ (NEMO). In the canonical pathway, IKKβ and IKKγ, but not IKKα, regulate degradation of IκB through phosphorylation of IκB and, consequently, free NF-κB is translocated into the nucleus [53]. The alternative pathway, however, requires only IKKα, in which p100, a precursor of p52, is phosphorylated and matured by IKKα [54]. The stimuli of each pathway are also different. The major triggers for the canonical pathway are proinflammatory cytokines and microbial products, such as tumor necrosis factor- (TNF-) α, IL-1, and lipopolysaccharide (LPS), resulting in activation of complexes comprised of Rel A or c-Rel [55], whereas the alternative pathway is activated by the TNF family, leading to activation of Rel B/p52 complexes [56].
Figure 3.
Two pathways leading to NF-κB activation.
There are coactivators of NF-κB, CREB-binding protein (CBP), and its homolog, p300, including the p300/CBP-associated factor (P/CAF) and members of the steroid receptor coactivator (p160/SRC) family. These coactivators directly interact with NF-κB subunits, such as p50 and p60 [57]. Interestingly, CARM1 is closely associated with the transcriptional coactivator, CBP/p300, in p53-mediated and nuclear receptor- (NR-) mediated transcriptional regulation. This implies that CARM1 would be a coactivator of NF-κB, because NF-κB utilizes a similar set of coactivator proteins with p53 and NR [58]. Indeed, CARM1 was found to be a novel transcriptional coactivator of NF-κB [59]. The NF-κB-dependent genes, such as ICAM-1, G-CSF, MCP-1, IP-10, and MIP-2, were impaired in Carm1(−/−) fibroblasts that underwent TNF-α and LPS stimulation. TNF-α- and LPS-induced NF-κB reporter gene activities were also lower in Carm1(−/−) cells compared to controls. However, IκB degradation and p65/Rel A translocation were not affected by the absence of CARM1. Instead, it seems that CARM1 regulates NF-κB-mediated gene expression through complex formations with p65 and p300. Consequently, CARM1 acts as a primary coactivator by enhancing NF-κB recruitment to cognate sites and by controlling transcription in a gene-specific manner [59].
However, CARM1's functional role in NF-κB-dependent gene expression remains unclear and even controversial. According to the recovery experiments using complemented carm1−/− mouse embryonic fibroblast cells by retroviral transduction, either with wild-type CARM1 or with an enzymatic inactive E267Q mutant of CARM1, CARM1 enzymatic activity was not essential for NF-κB-dependent gene expression which was stimulated by TNF-α or PMA. Additionally, CARM1 is not needed for recruitment of Rel A/p65 to chromatin, indicating that CARM1 contributes to the stabilization of complex proteins. These observations generate two hypotheses: (1) CARM1 might recruit Brg1, an enzymatic ATPase subunit of the SWI/SNF complex, to promoters of specific genes, because CARM1 interacts with Brg1 [60]; (2) a third interaction partner, whose enzymatic activity is independent of CARM1, might also be recruited by CARM1. Therefore, a more rigorous investigation of CARM1's role in transcriptional regulation is required to understand its exact role in inflammatory responses.
2.2. PRMT1
PRMT1 is the most common form of PRMT and is expressed in most tissues, constituting up to 85% of all PRMT activity in cultured RAT1 cells and in mouse liver tissue under experimental conditions [61]. PRMT1 is broadly thought to be the main enzyme on histone H4 for monomethylation and asymmetric dimethylation of Arg-3, which are required for transcriptional activation by nuclear hormone receptors [62]. Nonhistone proteins have also been reported to act as substrates of PRMT1. Through the methylation of PIAS1, PRMT1 can control STAT1 transcriptional activity in the late phase of interferon-γ (IFN-γ) signaling [63]. PRMT1 acts as an activator of estrogen receptor- (ER-) mediated transactivation through TAF15 methylation [64]. Also, PRMT1 methylates FOXO1 and increases its transcriptional activity by retaining it in the nucleus [65].
2.2.1. The Role of PRMT1 in Antigen-Induced Pulmonary Inflammation (AIPI) in Rats
An antigen-induced pulmonary inflammation (AIPI) model is the in vivo rat asthma model that shares many pathological features with human asthma. Interestingly, there are remarkable differences in the gene expression of PRMTs in rats with AIPI comparing to normal rats [66]. In particular, the expression of PRMT1 was significantly higher in the AIPI model, implying putative involvement of PRMT1 in asthma.
During pulmonary inflammation, eosinophils, the most critical immune cells in asthmatic conditions, are recruited into the lungs through a process mediated by eotaxins. Interleukin- (IL-) 4 boosts eosinophilic inflammation by producing eotaxin-1 [67]. PRMT1 has been shown to be associated with the mechanisms underlying the recruitment of eosinophils into airways by IL-4 [68]. The upregulation of PRMT1 was induced by Th2 cytokine IL-4 in the AIPI model. According to a transcription factor search program, IL-4 seems to increase PRMT1 expression through activation of STATs, CREB, NF-κB, and GATA3, which are all involved in the promoter region of PRMT1. AMI, a PRMT1 inhibitor, suppressed eotaxin-1 production and eosinophil infiltration in the AIPI model, implying that PRMT1, when activated by IL-4, functions as a pulmonary inflammation regulator via the modulation of eotaxin-1 release [68]. It has also been reported that PRMT1 can methylate the STAT family, which is responsible for IL-4 expression [69]. Therefore, it has been suggested that IL-4 induces overexpression of PRMT1, leading to increased transcription of eotaxin-1 by elevated STAT signaling. Additionally, transforming growth factor- (TGF-) β-induced PRMT1 also contributes to pulmonary inflammation in chronic AIPI. TGF-β is produced by IL-4 stimulated epithelial cell, and subsequently proliferation of fibroblast and PRMT1 expression are elevated. Then, increased PRMT1 is regarded to regulate pulmonary inflammation through inducing COX-2 expression [70].
2.2.2. Regulation of CITED2 by PRMT1
CBP/p300-interacting transactivator 2 (CITED2) is a coactivator of the p300/CBP-mediated transcript complex. It also acts as a transcriptional corepressor of HIF-dependent genes [71]. It serves multiple functions in several biological processes; for example, in knockout analyses, CITED2 was observed to play important roles in mouse embryo development [72–74]. Moreover, CITED2 is required for maintenance of adult hemopoietic stem cell functions [75]. A recent study on CITED2 function in immunity and inflammation led to the observation that CITED2 is induced by LPS and acts as a novel repressor of NF-κB by preventing p65 from binding to p300 [76].
Interestingly, PRTM1 and CARM1 regulate CITED2 expression under IL-4 stimulation conditions. According to Uta-Maria's work, PRMT1 and CARM1 were observed to recruit CITED2 gene promoter sites when stimulated with fetal calf serum (FCS)/IL-4 [77]. Additionally, both PRMTs interact with STAT5 in an IL-4-dependent manner [77]. The data indicate that PRMT1 and CARM1 cooperatively increase the expression of CITED2 through STAT5-dependent transcriptional activation when induced by IL-4 signaling. Interaction of the two PRMTs with STAT5 and their recruitment to the promoter region are also enhanced by IL-4 stimulation [77]. These findings imply that CARM1 and PRMT1 might participate in immune responses by regulating CITED2 transcription.
2.2.3. Modulation of NF-κB by PRMT1
PRMT1 is considered an inflammation regulator because of its NF-κB regulation capacity. PRMT1 controls NF-κB-dependent gene expression in collaboration with other coactivators. Hassa et al. found that, under TNF-α stimulation, PRMT1 forms a nuclear complex with p65 and poly[ADP-ribose] polymerase 1 (PARP1), and PRTM1 is recruited to p65-containing complexes that are associated with promoters [78]. Moreover, PRMT1 was required for PARP1 and p300-dependent NF-κB gene transcription, based on luciferase reporter gene assays, suggesting that PRMT1 synergistically coactivates NF-κB-dependent transcription in cooperation with PARP1 and p300.
How these coactivators (PRMT1, PARP1, and p300) cross-talk as part of their associated activity is still unclear. One possible explanation is that histone acetylation by CBP/p300 might be succeeded by PRMT1-mediated methylation of Arg-3 on histone H4, because it has been revealed that Arg-3 methylation on H4 by PRMT1 is essential to maintain “active” chromatin modification [79]. Another possibility is that PRMT1 may methylate other NF-κB transcriptional coactivators; for example, CARM1 catalyzes p300/CBP methylation, which is linked to the alteration of transcription states [48]. Similar to CARM1, PRMT1 might directly methylate promoter-associated coactivators, such as PARP1 or p160 family members. This explanation is consistent with Stallcup et al.'s findings regarding the functional regulation of nonhistone proteins by PRMT1 [80]. Further studies are needed to fully understand how PRMT1 regulates NF-κB-dependent gene expression through its collaboration with other cofactors.
2.3. PRMT6
Protein arginine methyltransferase 6 (PRMT6) mainly catalyzes asymmetric dimethylation of histone H3 Arg-2 (H3R2me2a). Histone methylation by this enzyme has been confirmed in vivo [81]. Thus, alteration of PRMT6 expression could affect universal gene expression. In addition to methylation on histone proteins, PRMT6 has been observed to control gene expression by direct interaction with transcription factors, including NF-κB and G-protein pathway suppressor 2 (GPS2). Because those two molecules are directly involved in inflammatory responses, it is possible that PRMT6 also plays a role in inflammation responses.
2.3.1. Regulation of NF-κB via PRMT6
In total, 4 PRMTs (PRMT1, CARM1, PRMT5, and PRMT6) are known to be regulators of NF-κB, despite having different regulatory mechanisms. The role of PRMT6 in NF-κB was first described by Di Lorenzo et al., using a gain-of-function mouse model [82]. This group established a transgenic mouse model that overexpressed PRMT6 and explored the role of PRMT6 in inflammatory responses in these mice. They observed that PRMT6 elevated IL-6 expression levels by regulating NF-κB. Detailed experiments led to the conclusion that PRMT6 directly binds to the NF-κB subunit, Rel A, allowing shuttling of Rel A into the nucleus. Consistent with this, colocalization of Rel A and PRMT6 at NF-κB binding promoters was observed to be elevated and, subsequently, NF-κB target gene expression, including IL-6, was elevated when stimulated by TNF-α.
However, the role of arginine methylation by PRMT6 during activation of NF-κB-related gene expression has not been thoroughly explored. According to in vitro methylation assay results, PRMT6-mediated Rel A methylation was not directly observed, indicating that NF-κB activation is indirectly regulated by methylation of NF-κB coactivators, such as p160/steroid receptor coactivator (SRC) proteins. Indeed, PRMT6 interacts with activation domain 2 (AD2) of SRC-1, increasing the possibility that p160/SRC could be methylated by PRMT6 [83].
2.3.2. Regulation of G-Protein Pathway Suppressor 2 (GPS2) by PRMT6
GPS2 is a multifunctional protein belonging to a transcriptional cofactor. GPS2 serves a function in G-protein mitogen-activated protein kinase (MAPK) signaling pathways, appearing to have a negative effect on RAS-, MAPK-, and JAK-mediated signaling cascades. Because these enzymes are major signaling regulators associated with inflammatory responses, the modulation of GPS2 by PRMT6 implies involvement of PRMT6 in controlling inflammation [84].
Recently, Huang et al. observed that PRMT6 modulates GPS2 by arginine methylation at Arg-323 and Arg-312 [85]. Of these, Arg-323 methylation was found to be an essential reaction that prevented proteasomal degradation of GPS2, resulting in its increased stability. Huang et al.'s findings indicate that methylation of Arg-323 is needed for recognition by transducer beta-like protein 1 (TBL1), which prevents degradation of polyubiquitinated GPS2. TBL1 subsequently binds to the ubiquitinated GPS2 by recognizing the methylated Arg-323, which was catalyzed by PRMT6. Consequently, proteasomal degradation is diminished. Although the direct relationship between inflammation and PRMT6 and GPS2 is still unclear, accumulated reports indicate a strong likelihood that PRMT6 is an inflammation regulator.
2.4. PRMT5
Protein arginine methyltransferase 5 (PRMT5) catalyzes the production of a symmetrically dimethylated (SDMA) guanidine group and belongs to a type II PRMT. Similar to other PRMTs, PRMT5 also controls gene expression, mainly by modulating histone methylation. PRMT5 represses gene transcription by inducing dimethylation of Arg-8 on histone H3 (H3R8) and Arg-3 on histone H4 (H4R3) [86]. Contrary to this, PRMT5 can also upregulate gene transcription under particular conditions [87].
2.4.1. The Role of PRMT5 in HOXA9-Mediated Endothelial Cell (EC) Inflammation
EC inflammatory responses are mediated by proinflammatory endothelial-leukocyte adhesion molecules (ELAM), such as E-selectin and vascular cell adhesion molecule 1 (VCAM-1) [88]. When ECs receive inflammation signals, transcription factors that induce adhesion molecules are activated [89]; a key transcriptional factor involved in this reaction is HOXA9 [90]. HOXA9 belongs to the homeobox family, and its posttranslational modifications, including phosphorylation and ubiquitination, play a critical role in hematopoietic differentiation [91]. Interestingly, HOXA9 is methylated at Arg-140 by PRMT5 and this reaction is activated by TNF-α. Methylation of HOXA9 plays a critical role in the upregulation of ELAM (E-selectin and VCAM-1), indicating that PRMT5 could play an important role in EC inflammation [92]. In contrast, PRMT5 has an inhibitory role in the induction of E-selectin by mediating histone H4R3, which leads to gene silencing [93]. Therefore, it seems that PRMT5 could contribute to EC inflammation as an on-off switch.
2.4.2. The Function of PRMT5 in Immune Responses through NF-κB Regulation
There are several reports indicating that PRMT5 regulates NF-κB activity. At first, PRMT5 was reported to be a NF-κB regulator during TRAIL-induced apoptosis [94]. According to Hiroshi et al., PRMT5 binds to the TRAIL receptor and, consequently, TRAIL-induced apoptosis is activated via IKK activation and IκB degradation. Because TRAIL may stimulate inflammatory cytokine expression, such as CCL20, and because NF-κB is a key regulator of inflammation in immune cells, PRMT5 involvement in inflammatory responses was also explored. In practice, PRMT5 appears to be associated with DR4-dependent immune regulation by controlling the NF-κB pathway [95]. DR4 binds to TRAIL, leading to recruitment of RIP1 and TRAF2 in DISC, as well as activation of NF-κB. In these reactions, PRMT5 acts as a competitor of TRAIL to bind to DR4, resulting in suppression of NF-κB activation and CCL20 expression.
Additionally, PRMT5 was directly revealed to regulate NF-κB activity by inducing methylation of the p65 subunit. Wei et al. demonstrated that PRMT5 methylates Arg-30 (R30) residues of the p65 subunit and regulates NF-κB-dependent gene expression, such as interleukin-1α (IL-1α) and TNF receptor-associated factor 1 (TRAF1). According to microarray analyses, about 85% of NF-κB-dependent gene expression seems to be required for R30 and p65 methylation [96]. With that, it has also been reported that R35, as well as R30, is methylated by PRMT5. Methylated R30 and R35 at p65 participate in elevating p65 and in transcription of a subset of TNF-α-induced proinflammatory genes, especially CXCL10, in endothelial cells [97].
3. Conclusions
Based on our review, there is evidence for a correlation between PRMTs and inflammatory responses. In particular, transcription regulation by NF-κB, a key molecule of inflammation, appears to be a main function of PRMTs in the regulation of inflammation system. However, studies linking PRMTs to inflammation are in a very nascent stage, and the current evidence is circumstantial. Therefore, an introduction of key model systems is needed in order to understand the biological role of PRMTs in inflammation. Mouse knockout models could be useful in this process (Table 4). In fact, mouse models can be a powerful tool for investigating the in vivo inflammation-regulatory roles of PRMTs and understanding their molecular mechanisms. The relevance of the association between PRMTs and inflammatory diseases can also be estimated using in vivo inflammatory knockout models, such as hepatitis, gastritis, colitis, peritonitis, and dermatitis. Additionally, phenomenological access is likely to be required to understand the connection between PRMTs and inflammatory disease. For that, a good approach would be to examine the expression pattern of PRMT activity in immune cells obtained from in vivo inflammatory disease models or chronic inflammatory disease patients. Moreover, testing the inhibitory efficacy of PRMT inhibitors against inflammatory diseases will contribute to the development of a new anti-inflammatory drug.
Table 4.
PRMTs of knockout mice.
| PRMT | Knockout mouse phenotype | Reference |
|---|---|---|
| Prmt1 | Embryonic lethality | [39] |
| Loss of PRMT1 in mouse embryonic fibroblasts (MEFs) leads to spontaneous DNA damage, cell cycle progression delay, checkpoint defects, aneuploidy, and polyploidy | [40] | |
|
| ||
| Carm1 | Neonatal lethality | [41] |
| Mutant embryos have defects in many systems, including adipose tissue, hematopoietic system, immune system (T-cell differentiation), and the respiratory system | [42] | |
|
| ||
| Prmt5 | Early embryonic lethality (dies by E6.5) | [43] |
| PRMT5 is required for NPC homeostasis | [44] | |
|
| ||
| Prmt6 | No lethality | [45] |
| MEFs from PRMT6(−/−) mice show growth defects and undergo cellular senescence | ||
Lastly, the possibility of functional involvement of arginine demethylase in arginine methylation cannot be excluded. Many studies have indicated the existence of arginine demethylases, even though arginine methylation is a stable modification [98]. For example, H3R17me2A exhibits cyclic expression with 20-minute fluctuation intervals [99], and PRMT1-induced transient methylation of ERα with estrogen treatment reaches peak levels within 5 minutes and then disappears within 10 minutes [100]. In practice, Jumonji C domain-containing protein 6 (JMJD6), also known as a lysine hydroxylase, was identified as a first putative arginine demethylase [101]. Therefore, recent reports on PRMTs and their counterpart arginine demethylase propose that these enzymes are a functionally important unit in the regulation of inflammatory responses. Further verification of involvement of these enzymes in each inflammatory disease will be of considerable interest.
Acknowledgment
This work was supported by research grant from the National Cancer Center, Korea (NCC-1610600).
Disclaimer
The authors alone are responsible for the content and writing of the paper.
Conflict of Interests
The authors have no conflict of interests to report.
Authors' Contribution
Ji Hye Kim and Byong Chul Yoo contributed equally to this work.
References
- 1.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence T. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harbor Perspectives in Biology. 2009;1(6) doi: 10.1101/cshperspect.a001651.a001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moon S.-J., Jeong J.-H., Jhun J. Y., et al. Ursodeoxycholic acid ameliorates pain severity and cartilage degeneration in monosodium iodoacetate-induced osteoarthritis in rats. Immune Network. 2014;14(1):45–53. doi: 10.4110/in.2014.14.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guzik T. J., Korbut R., Adamek-Guzik T. Nitric oxide and superoxide in inflammation and immune regulation. Journal of Physiology and Pharmacology. 2003;54(4):469–487. [PubMed] [Google Scholar]
- 5.Hwang Y.-J., Song J., Kim H.-R., Hwang K.-A. Oleanolic acid regulates NF-κB signaling by suppressing MafK expression in RAW 264.7 cells. BMB Reports. 2014;47(9):524–529. doi: 10.5483/bmbrep.2014.47.9.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotran S., Kumar C., Collins T., Robbins W. Pathologic Basis of Disease. Philadelphia, Pa, USA: Saunders; 1999. [Google Scholar]
- 7.Kim K. H., Kim T. S., Lee J. G., et al. Characterization of proinflammatory responses and innate signaling activation in macrophages infected with Mycobacterium scrofulaceum . Immune Network. 2014;14(6):307–320. doi: 10.4110/in.2014.14.6.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McInnes I. B., Schett G. The pathogenesis of rheumatoid arthritis. The New England Journal of Medicine. 2011;365(23):2205–2219. doi: 10.1056/nejmra1004965. [DOI] [PubMed] [Google Scholar]
- 9.Lim S., Park S. Role of vascular smooth muscle cell in the inflammation of atherosclerosis. BMB Reports. 2014;47(1):1–7. doi: 10.5483/BMBRep.2014.47.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng X., Blumenthal R. S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. Singapore: World Scientific; 1999. [Google Scholar]
- 11.Yeates T. O. Structures of SET domain proteins: protein lysine methyltransferases make their mark. Cell. 2002;111(1):5–7. doi: 10.1016/s0092-8674(02)01010-3. [DOI] [PubMed] [Google Scholar]
- 12.Romano J. D., Michaelis S. Topological and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Molecular Biology of the Cell. 2001;12(7):1957–1971. doi: 10.1091/mbc.12.7.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz J. E., Dlakić M., Clarke S. Automated identification of putative methyltransferases from genomic open reading frames. Molecular & Cellular Proteomics. 2003;2(8):525–540. doi: 10.1074/mcp.M300037-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X., Zhou L., Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. The EMBO Journal. 2000;19(14):3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bedford M. T. Arginine methylation at a glance. Journal of Cell Science. 2007;120(24):4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.-H., Cook J. R., Yang Z.-H., et al. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. The Journal of Biological Chemistry. 2005;280(5):3656–3664. doi: 10.1074/jbc.m405295200. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y., Hadjikyriacou A., Xia Z., et al. PRMT9 is a Type II methyltransferase that methylates the splicing factor SAP145. Nature Communications. 2015;6:p. 6428. doi: 10.1038/ncomms7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadjikyriacou A., Yang Y., Espejo A., Bedford M. T., Clarke S. G. Unique features of human protein arginine methyltransferase 9 (PRMT9) and its substrate RNA splicing factor SF3B2. The Journal of Biological Chemistry. 2015;290(27):16723–16743. doi: 10.1074/jbc.m115.659433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zurita-Lopez C. I., Sandberg T., Kelly R., Clarke S. G. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. Journal of Biological Chemistry. 2012;287(11):7859–7870. doi: 10.1074/jbc.m111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niewmierzycka A., Clarke S. S-adenosylmethionine-dependent methylation in Saccharomyces cerevisiae: identification of a novel protein arginine methyltransferase. The Journal of Biological Chemistry. 1999;274(2):814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 21.Feng Q., He B., Jung S.-Y., et al. Biochemical control of CARM1 enzymatic activity by phosphorylation. The Journal of Biological Chemistry. 2009;284(52):36167–36174. doi: 10.1074/jbc.M109.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashimoto K., Kuhn P., Desai D., Cheng X., Xu W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(30):12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung W. D., Sakabe K., Housley M. P., Dias W. B., Hart G. W. O-linked β-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. The Journal of Biological Chemistry. 2008;283(49):33935–33941. doi: 10.1074/jbc.m806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakabe K., Hart G. W. O-GlcNAc transferase regulates mitotic chromatin dynamics. Journal of Biological Chemistry. 2010;285(45):34460–34468. doi: 10.1074/jbc.m110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn P., Chumanov R., Wang Y., Ge Y., Burgess R. R., Xu W. Automethylation of CARM1 allows coupling of transcription and mRNA splicing. Nucleic Acids Research. 2011;39(7):2717–2726. doi: 10.1093/nar/gkq1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F., Zhao X., Perna F., et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19(2):283–294. doi: 10.1016/j.ccr.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankel A., Yadav N., Lee J., Branscombe T. L., Clarke S., Bedford M. T. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. The Journal of Biological Chemistry. 2002;277(5):3537–3543. doi: 10.1074/jbc.m108786200. [DOI] [PubMed] [Google Scholar]
- 28.Sayegh J., Webb K., Cheng D., Bedford M. T., Clarke S. G. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. Journal of Biological Chemistry. 2007;282(50):36444–36453. doi: 10.1074/jbc.m704650200. [DOI] [PubMed] [Google Scholar]
- 29.Friesen W. J., Wyce A., Paushkin S., et al. A novel WD repeat protein component of the methylosome binds Sm proteins. The Journal of Biological Chemistry. 2002;277(10):8243–8247. doi: 10.1074/jbc.m109984200. [DOI] [PubMed] [Google Scholar]
- 30.Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Molecular and Cellular Biology. 2004;24(21):9630–9645. doi: 10.1128/mcb.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuang T.-W., Peng P.-J., Tarn W.-Y. The exon junction complex component Y14 modulates the activity of the methylosome in biogenesis of spliceosomal small nuclear ribonucleoproteins. The Journal of Biological Chemistry. 2011;286(11):8722–8728. doi: 10.1074/jbc.m110.190587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei N.-Z., Zhang X.-Y., Chen H.-Z., et al. A feedback regulatory loop between methyltransferase PRMT1 and orphan receptor TR3. Nucleic Acids Research. 2009;37(3):832–848. doi: 10.1093/nar/gkn941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robin-Lespinasse Y., Sentis S., Kolytcheff C., Rostan M.-C., Corbo L., Le Romancer M. hCAF1, a new regulator of PRMT1-dependent arginine methylation. Journal of Cell Science. 2007;120(4):638–647. doi: 10.1242/jcs.03357. [DOI] [PubMed] [Google Scholar]
- 34.Lin W.-J., Gary J. D., Yang M. C., Clarke S., Herschman H. R. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. The Journal of Biological Chemistry. 1996;271(25):15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 35.Pak M. L., Lakowski T. M., Thomas D., Vhuiyan M. I., Hüsecken K., Frankel A. A protein arginine N-methyltransferase 1 (PRMT1) and 2 heteromeric interaction increases PRMT1 enzymatic activity. Biochemistry. 2011;50(38):8226–8240. doi: 10.1021/bi200644c. [DOI] [PubMed] [Google Scholar]
- 36.Singh V., Branscombe Miranda T., Jiang W., et al. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene. 2004;23(47):7761–7771. doi: 10.1038/sj.onc.1208057. [DOI] [PubMed] [Google Scholar]
- 37.Jelinic P., Stehle J.-C., Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biology. 2006;4(11, article e355) doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Pal S., Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Molecular and Cellular Biology. 2008;28(20):6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlak M. R., Scherer C. A., Chen J., Roshon M. J., Ruley H. E. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Molecular and Cellular Biology. 2000;20(13):4859–4869. doi: 10.1128/mcb.20.13.4859-4869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Z., Chen T., Hébert J., Li E., Richard S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Molecular and Cellular Biology. 2009;29(11):2982–2996. doi: 10.1128/MCB.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D., Lee J., Cheng D., et al. Enzymatic activity is required for the in vivo functions of CARM1. The Journal of Biological Chemistry. 2010;285(2):1147–1152. doi: 10.1074/jbc.m109.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yadav N., Lee J., Kim J., et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(11):6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tee W.-W., Pardo M., Theunissen T. W., et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes & Development. 2010;24(24):2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bezzi M., Teo S. X., Muller J., et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes & Development. 2013;27(17):1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neault M., Mallette F. A., Vogel G., Michaud-Levesque J., Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Research. 2012;40(19):9513–9521. doi: 10.1093/nar/gks764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng D., Côté J., Shaaban S., Bedford M. T. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Molecular Cell. 2007;25(1):71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 47.Chen D., Ma M., Hong H., et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284(5423):2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 48.Xu W., Chen H., Du K., et al. A transcriptional switch mediated by cofactor methylation. Science. 2001;294(5551):2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 49.Hart L. A., Krishnan V. L., Adcock I. M., Barnes P. J., Chung K. F. Activation and localization of transcription factor, nuclear factor-κB, in asthma. American Journal of Respiratory and Critical Care Medicine. 1998;158(5):1585–1592. doi: 10.1164/ajrccm.158.5.9706116. [DOI] [PubMed] [Google Scholar]
- 50.Neurath M. F., Becker C., Barbulescu K. Role of NF-κB in immune and inflammatory responses in the gut. Gut. 1998;43(6):856–860. doi: 10.1136/gut.43.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feldmann M., Brennan F. M., Maini R. N. Role of cytokines in rheumatoid arthritis. Annual Review of Immunology. 1996;14(1):397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 52.Karin M. The beginning of the end: IκB kinase (IKK) and NF-κB activation. The Journal of Biological Chemistry. 1999;274(39):27339–27342. doi: 10.1074/jbc.274.39.27339. [DOI] [PubMed] [Google Scholar]
- 53.Zandi E., Rothwarf D. M., Delhase M., Hayakawa M., Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for Iκb phosphorylation and NF-κB activation. Cell. 1997;91(2):243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 54.Senftleben U., Cao Y., Xiao G., et al. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science. 2001;293(5534):1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 55.Karin M., Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annual Review of Immunology. 2000;18(1):621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 56.Bonizzi G., Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends in Immunology. 2004;25(6):280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 57.Gerritsen M. E., Williams A. J., Neish A. S., Moore S., Shi Y., Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(7):2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.An W., Kim J., Roeder R. G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117(6):735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Covic M., Hassa P. O., Saccani S., et al. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-κB-dependent gene expression. The EMBO Journal. 2005;24(1):85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee Y.-H., Coonrod S. A., Kraus W. L., Jelinek M. A., Stallcup M. R. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang J., Frankel A., Cook R. J., et al. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. The Journal of Biological Chemistry. 2000;275(11):7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 62.Wang H., Huang Z.-Q., Xia L., et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293(5531):853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 63.Weber S., Maaß F., Schuemann M., Krause E., Suske G., Bauer U.-M. PRMT1-mediated arginine methylation of PIAS1 regulates STAT1 signaling. Genes & Development. 2009;23(1):118–132. doi: 10.1101/gad.489409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Jobert L., Argentini M., Tora L. PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Experimental Cell Research. 2009;315(7):1273–1286. doi: 10.1016/j.yexcr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 65.Yamagata K., Daitoku H., Takahashi Y., et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Molecular Cell. 2008;32(2):221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 66.Sun Q.-Z., Jiao F.-F., Yang X.-D., et al. Expression of protein arginine N-methyltransferases in E3 rat models of acute asthma. Journal of Southern Medical University. 2010;30(4):716–719. [PubMed] [Google Scholar]
- 67.Banwell M. E., Tolley N. S., Williams T. J., Mitchell T. J. Regulation of human eotaxin-3/CCL26 expression: modulation by cytokines and glucocorticoids. Cytokine. 2002;17(6):317–323. doi: 10.1006/cyto.2002.1021. [DOI] [PubMed] [Google Scholar]
- 68.Sun Q., Yang X., Zhong B., et al. Upregulated protein arginine methyltransferase 1 by IL-4 increases eotaxin-1 expression in airway epithelial cells and participates in antigen-induced pulmonary inflammation in rats. Journal of Immunology. 2012;188(7):3506–3512. doi: 10.4049/jimmunol.1102635. [DOI] [PubMed] [Google Scholar]
- 69.Igarashi H., Kuwahara K., Yoshida M., et al. GANP suppresses the arginine methyltransferase PRMT5 regulating IL-4-mediated STAT6-signaling to IgE production in B cells. Molecular Immunology. 2009;46(6):1031–1041. doi: 10.1016/j.molimm.2008.08.272. [DOI] [PubMed] [Google Scholar]
- 70.Sun Q., Liu L., Roth M., et al. PRMT1 upregulated by epithelial proinflammatory cytokines participates in COX2 expression in fibroblasts and chronic antigen-induced pulmonary inflammation. The Journal of Immunology. 2015;195(1):298–306. doi: 10.4049/jimmunol.1402465. [DOI] [PubMed] [Google Scholar]
- 71.Bhattacharya S., Michels C. L., Leung M.-K., Arany Z. P., Kung A. L., Livingston D. M. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes & Development. 1999;13(1):64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bamforth S. D., Bragança J., Eloranta J. J., et al. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nature Genetics. 2001;29(4):469–474. doi: 10.1038/ng768. [DOI] [PubMed] [Google Scholar]
- 73.Weninger W. J., Floro K. L., Bennett M. B., et al. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development. 2005;132(6):1337–1348. doi: 10.1242/dev.01696. [DOI] [PubMed] [Google Scholar]
- 74.Yin Z., Haynie J., Yang X., et al. The essential role of Cited2, a negative regulator for HIF-1α, in heart development and neurulation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10488–10493. doi: 10.1073/pnas.162371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kranc K. R., Schepers H., Rodrigues N. P., et al. Cited2 is an essential regulator of adult hematopoietic stem cells. Cell Stem Cell. 2009;5(6):659–665. doi: 10.1016/j.stem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lou X., Sun S., Chen W., et al. Negative feedback regulation of NF-κB action by CITED2 in the nucleus. The Journal of Immunology. 2011;186(1):539–548. doi: 10.4049/jimmunol.1001650. [DOI] [PubMed] [Google Scholar]
- 77.Kleinschmidt M. A., Streubel G., Samans B., Krause M., Bauer U.-M. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Research. 2008;36(10):3202–3213. doi: 10.1093/nar/gkn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hassa P. O., Covic M., Bedford M. T., Hottiger M. O. Protein arginine methyltransferase 1 coactivates NF-κB-dependent gene expression synergistically with CARM1 and PARP1. Journal of Molecular Biology. 2008;377(3):668–678. doi: 10.1016/j.jmb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 79.Strahl B. D., Briggs S. D., Brame C. J., et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Current Biology. 2001;11(12):996–1000. doi: 10.1016/S0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 80.Lee D. Y., Ianculescu I., Purcell D., Zhang X., Cheng X., Stallcup M. R. Surface-scanning mutational analysis of protein arginine methyltransferase 1: roles of specific amino acids in methyltransferase substrate specificity, oligomerization, and coactivator function. Molecular Endocrinology. 2007;21(6):1381–1393. doi: 10.1210/me.2006-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guccione E., Bassi C., Casadio F., et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449(7164):933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 82.Di Lorenzo A., Yang Y., Macaluso M., Bedford M. T. A gain-of-function mouse model identifies PRMT6 as a NF-κB coactivator. Nucleic Acids Research. 2014;42(13):8297–8309. doi: 10.1093/nar/gku530.gku530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harrison M. J., Tang Y. H., Dowhan D. H. Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucleic Acids Research. 2010;38(7):2201–2216. doi: 10.1093/nar/gkp1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frazier W. J., Xue J., Luce W. A., Liu Y. MAPK signaling drives inflammation in LPS-stimulated cardiomyocytes: the route of crosstalk to G-protein-coupled receptors. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0050071.e50071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang J., Cardamone M. D., Johnson H. E., et al. Exchange factor TBL1 and arginine methyltransferase PRMT6 cooperate in protecting G protein pathway suppressor 2 (GPS2) from proteasomal degradation. Journal of Biological Chemistry. 2015;290(31):19044–19054. doi: 10.1074/jbc.m115.637660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lacroix M., El Messaoudi S., Rodier G., Le Cam A., Sardet C., Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Reports. 2008;9(5):452–458. doi: 10.1038/embor.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dacwag C. S., Ohkawa Y., Pal S., Sif S., Imbalzano A. N. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Molecular and Cellular Biology. 2007;27(1):384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bevilacqua M. P., Stengelin S., Gimbrone M. A., Jr., Seed B. Endothelial leukocyte adhesion molecule 1: an inducible receptor for neutrophils related to complement regulatory proteins and lectins. Science. 1989;243(4895):1160–1165. doi: 10.1126/science.2466335. [DOI] [PubMed] [Google Scholar]
- 89.Edelstein L. C., Pan A., Collins T. Chromatin modification and the endothelial-specific activation of the E-selectin gene. The Journal of Biological Chemistry. 2005;280(12):11192–11202. doi: 10.1074/jbc.m412997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bandyopadhyay S., Ashraf M. Z., Daher P., Howe P. H., DiCorleto P. E. HOXA9 participates in the transcriptional activation of E-selectin in endothelial cells. Molecular and Cellular Biology. 2007;27(12):4207–4216. doi: 10.1128/MCB.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vijapurkar U., Fischbach N., Shen W., et al. Protein kinase C-mediated phosphorylation of the leukemia-associated HOXA9 protein impairs its DNA binding ability and induces myeloid differentiation. Molecular and Cellular Biology. 2004;24(9):3827–3837. doi: 10.1128/mcb.24.9.3827-3837.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bandyopadhyay S., Harris D. P., Adams G. N., et al. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Molecular and Cellular Biology. 2012;32(7):1202–1213. doi: 10.1128/MCB.05977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao Q., Rank G., Tan Y. T., et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature Structural & Molecular Biology. 2009;16(3):304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tanaka H., Hoshikawa Y., Oh-hara T., et al. PRMT5, a novel TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis via nuclear factor-κB activation. Molecular Cancer Research. 2009;7(4):557–569. doi: 10.1158/1541-7786.mcr-08-0197. [DOI] [PubMed] [Google Scholar]
- 95.Wang D., Liu D., Gao J., et al. PRMT5 suppresses DR4-mediated CCL20 release via NF-κB pathway. Chinese Science Bulletin. 2012;57(33):4351–4355. doi: 10.1007/s11434-012-5511-6. [DOI] [Google Scholar]
- 96.Wei H., Wang B., Miyagi M., et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(33):13516–13521. doi: 10.1073/pnas.1311784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harris D. P., Bandyopadhyay S., Maxwell T. J., Willard B., DiCorleto P. E. Tumor necrosis factor (TNF)-α induction of CXCL10 in endothelial cells requires protein arginine methyltransferase 5 (PRMT5)-mediated nuclear factor (NF)-κB p65 Methylation. The Journal of Biological Chemistry. 2014;289(22):15328–15339. doi: 10.1074/jbc.m114.547349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang Y., Bedford M. T. Protein arginine methyltransferases and cancer. Nature Reviews Cancer. 2013;13(1):37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 99.Métivier R., Penot G., Hübner M. R., et al. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115(6):751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 100.Le Romancer M., Treilleux I., Leconte N., et al. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Molecular Cell. 2008;31(2):212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 101.Chang B., Chen Y., Zhao Y., Bruick R. K. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]